Abstract
Vascular endothelial cells (ECs) senescence plays a crucial role in vascular aging that promotes the initiation and progression of cardiovascular disease. The mutation of Grb10-interacting GYF protein 2 (GIGYF2) is strongly associated with the pathogenesis of aging-related diseases, whereas its role in regulating ECs senescence and dysfunction still remains elusive. In this study, we found aberrant hyperexpression of GIGYF2 in senescent human ECs and aortas of old mice. Silencing GIGYF2 in senescent ECs suppressed eNOS-uncoupling, senescence, and endothelial dysfunction. Conversely, in nonsenescent cells, overexpressing GIGYF2 promoted eNOS-uncoupling, cellular senescence, endothelial dysfunction, and activation of the mTORC1-SK61 pathway, which were ablated by rapamycin or antioxidant N-Acetyl-l-cysteine (NAC). Transcriptome analysis revealed that staufen double-stranded RNA binding protein 1 (STAU1) is remarkably downregulated in the GIGYF2-depleted ECs. STAU1 depletion significantly attenuated GIGYF2-induced cellular senescence, dysfunction, and inflammation in young ECs. Furthermore, we disclosed that GIGYF2 acting as an RNA binding protein (RBP) enhances STAU1 mRNA stability, and that the intron region of the late endosomal/lysosomal adaptor MAPK and mTOR activator 4 (LAMTOR4) could bind to STAU1 protein to upregulate LAMTOR4 expression. Immunofluorescence staining showed that GIGYF2 overexpression promoted the translocation of mTORC1 to lysosome. In the mice model, GIGYF2flox/flox Cdh-Cre+ mice protected aged mice from aging-associated vascular endothelium-dependent relaxation and arterial stiffness. Our work discloses that GIGYF2 serving as an RBP enhances the mRNA stability of STAU1 that upregulates LAMTOR4 expression through binding with its intron region, which activates the mTORC1-S6K1 signaling via recruitment of mTORC1 to the lysosomal membrane, ultimately leading to ECs senescence, dysfunction, and vascular aging. Disrupting the GIGYF2-STAU1-mTORC1 signaling cascade may represent a promising therapeutic approach against vascular aging and aging-related cardiovascular diseases.
Keywords: Vascular aging, Endothelial cell senescence, Endothelial dysfunction, eNOS uncoupling, Signaling
Graphical abstract

1. Introduction
Cardiovascular diseases (CVDs) are the leading cause of mortality and morbidity all over the world. Along with the changing lifestyles and aging population, the prevalence of CVDs tends to rise, imposing a heavy economic burden on society. Aging is the biological process of age-dependent progressive decline in intrinsic physiological functions [1,2]. Endothelial cell (ECs) senescence has been gaining recognition as a primary risk contributor to the pathogenesis of CVDs [3]. Vascular ECs are a single layer of flattened squamous epithelial cells located on the intravascular surface, which not only serve as a protective barrier between blood and the vessel wall/tissue, but also synthesize and release a variety of vasoactive substances, such as the vasodilator nitric oxide (NO) and the constrictor angiotensin II (Ang II), to moderate vascular function and maintain vascular homeostasis [4]. Senescent ECs can cause endothelial dysfunction as manifested by a range of molecular changes, e.g., increased reactive oxygen species (ROS) production, decreased NO generation, and enhanced intercellular cell adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1) secretion and pro-inflammatory cytokines, which are the common pathological triggers for the initiation and development of CVDs [5]. A substantial body of basic and clinical evidence has indicated that endothelial nitric oxide synthase (eNOS)-uncoupling in ECs results in decreased NO bioavailability and increased oxidative stress, thereby causing or exacerbating endothelial dysfunction and vascular aging [6].
The human Grb10-interacting GYF protein 2 (GIGYF2) gene, also termed TNRC15, is mapped in chromosome 2q36.1 with 72 exons encoding a protein of 1299 amino acids. GIGYF2 comprises a highly conserved GYF structural domain that interacts with the N-terminal end of the Grab10 bridging protein via the GYF region [7]. The functional studies reveal that GIGYF2 is an integral component of the translation repression complex and represses mRNA translation in a 4EHP-dependent and independent manner [8,9]. A multitude of current genome-wide screens and linkage analyses demonstrate that GIGYF2 is a candidate gene for aging-related diseases Parkinson's disease (PD) [[10], [11], [12]]. GIGYF2−/− mice display perinatal mortality, and Gigyf2+/- mice manifest an early-onset age-related neurodegenerative phenotype [13]. Mutants of the Gyf gene, whose product in Drosophila is homologous to GIGYF1 (Grb10-interacting GYF protein 1) and GIGYF2 in mammals, also exhibit an early-onset mortality rate and neurodegeneration-related mobility deficits [14]. Moreover, aberrant expression of GIGYF2 may contribute to the occurrence of aging-related diseases and diabetes-related cognitive impairment through a negatively regulated IGF1R signaling pathway [15]. These studies implicate that GIGYF2 may exert a unique function in other aging-related diseases. Whereas, the role of GIGYF2 in aging-related vascular diseases has been scarcely documented.
The mechanistic target of rapamycin complex 1 (mTORC1) functioning as a molecular hub and intracellular energy sensor plays a vital role in regulating various cellular processes [16]. The ribosome S6 protein kinase (S6K1), a serine/threonine kinase, is a classical downstream target of mTORC1 and can be phosphorylated and activated by mTORC1. Furthermore, S6K1 phosphorylates its substrate ribosome S6 protein, which promotes the initiation and processes of protein translation [17]. There is overwhelming evidence that the mTORC1-S6K1 pathway mediates a wide range of cellular functions including ECs senescence [18], and mammalian life span [19]. In Drosophila, Gyf (ortholog of GIGYF2) deficiency has been found to markedly suppress the phosphorylation of the mTORC1 substrate S6K1 [14]. On the basis of these, we are inspired to investigate the causal role of GIGYF2 in mTORC1 activation linking to the ECs senescence and vascular aging in mammals.
In the present study, we reveal a novel mechanism of GIGYF2 that leads to ECs senescence, dysfunction, and inflammation through the up-regulation of staufen double-stranded RNA binding protein 1 (STAU1), which activates the late endosomal/lysosomal adaptor MAPK and mTOR activator 4 (LAMTOR4)-mTORC1-S6K1 signaling axis via the recruitment of mTORC1 to the lysosomal membrane. This study yielded a promising therapeutic target for the treatment of vascular aging and aging-related CVDs.
2. Methods
2.1. Materials
All reagents used were from the following sources: rabbit antibodies against phospho-RPS6-Ser235/236 (#2317s), phospho-p70 S6 Kinase-Thr389 (#9234s), mTOR (#2983s), and PCNA (#PC10) were purchased from Cell Signaling (Danvers, USA); rabbit antibodies against GIGYF2 (24790-1-AP), STAU1 (14225-1-AP), and p70 S6K (14485-1-AP) were from Proteintech (Wuhan, China); mouse antibodies against HA (ab18181) and LAMP1 (ab253630) were from Abcam (Cambridge, UK); mouse antibody against β-actin (TA811000) was from Origene (Cambridge, UK); anti-mouse IgG-HRP (EK010) and anti-rabbit IgG-HRP (EK020) secondary antibodies were purchased from Xian Zhuangzhibio Biological Technology Co., Ltd (Xi'an, China); goat anti-rabbit IgG (H + L) secondary Antibody Alexa Fluor® 488 conjugate and goat anti-mouse IgG (H + L) secondary antibody Alexa Fluor® 594 conjugate were from Invitrogen/Thermo Fisher Scientific (Waltham, MA USA). N-Acetyl-l-cysteine (NAC, C8460), X-gal (X1010), and acetylcholine (Ach, 60-31-1) were purchased from Solarbio (Solarbio, Beijing, China); Rapamycin (53123-88-9) was from Aladin Reagent (Shanghai, China); phenylephrine (61-76-7), and sodium nitroprusside (SNP, 13755-38-9) were purchased from Macklin Inc. (Macklin, Shanghai, China); ROS Assay Kit (S0033S), diaminofluorescein-FM diacetate (DAF-FM DA) (S0019), and dihydroethidium (DHE, S0063) were purchased from Biyuntian Biotechnology Institute (Shanghai, China); Protein A/G agarose beads (Sc-2003) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA); The pmirGLO Dual-Luciferase expression vector was purchased from Promega (Madison, WI, USA). All cell culture media and materials were purchased from Gibco (Gibco, USA).
2.2. Cell culture and lentivirus transduction
Human umbilical vein endothelial cells (HUVEC) and senescent cells were prepared as previously described [18]. Briefly, 1×104 cells per cm2 were seeded in petri dishes coated with 1% gelatin and cultivated in RPMI-1640 complete medium supplemented with 5% fetal calf serum (FCS), 1% penicillin/streptomycin, and Endothelial Cell Growth Medium SupplementPack (C-39210, Promocell) at 37 °C in a 95% air and 5% CO2 atmosphere. Cells of passages 1-3 (P1 to P3) were defined as young cells. Young cells were further split in a ratio of 1-3 continuously over the course of several weeks until replicative senescence was observed as determined by SA-ꞵ-gal staining (P9 to P12). In this study, non-senescent endothelial cells are referenced as “young cells” [18]. The model of S6K1 driving endothelial cells senescence was performed as described by Rajapakse et al. [18]. Lentivirus was used to transduce cells for gene silencing or overexpression as described previously [18]. THP-1 cells were grown in RPMI-1640 containing 10% heat-inactivated fetal bovine serum (HIFBS) and 1% penicillin/streptomycin [20].
2.3. Generation of expression vectors
To generate the corresponding lentivirus, the pLKO.1-TRC plasmid with targeted shRNA sequences for knockdown or PLJMI plasmid with STAU1 gene sequences for overexpression and packaging plasmid (psPAX2) and envelope plasmid (pMD2.G) were co-transfected with HEK293T cells using Lipo6000™ Transfection Reagent. The transfected HEK293T cells were cultured for 24h and 48h, and the supernatant was collected and centrifuged (1000 rpm, 5 min) to obtain virus particles. Then, cells were infected with lentivirus and screened cells carrying the corresponding antibiotic resistance gene in a culture medium containing 2 μg/ml puromycin. The HA-GIGYF2 gene was amplified from template plasmid PX459-HA-GIGYF2 by polymerase chain reaction (PCR) using the following primer pairs, HA-HGIGYF2-F: 5′-CGCTAGCGCTACCGGCACCATGGCCTACCCATATGATG -3′ and the reverse primer: 5′-TCGAGGTCGAGAATTTCAGTAGTCATCCAACGTCTCGA -3’. The STAU1 gene was amplified from HUVECs genomic DNA via PCR, using the forward primer: 5′-CGCTAGCGCTACCGGCACCATGAAACTTGGAAAAAAACCA-3′ and the reverse primer: 5′-CCGAGGTCGAGAATTTCAGCACCTCCCACACACAG -3’.
The targeting sequences for hGIGYF2-shRNA, hSTAU1-shRNA and hLAMTOR4-shRNA are indicated in boldface below (only the sense strand is shown): pLKO.1-hGIGYF2-1-F: 5′-CCGGCACAGTACACTCCATTCAGTACTCGAGTACTGAATGGAGTGTACTGTGTTTTTG-3′ pLKO.1-hSTAU1-F: 5′-CCGGGCCTGCAGTTGAACGAGTAAACTCGAGTTTACTCGTTCAACTGCAGGCTTTTTG-3’; pLKO.1-hLAMTOR4-F: CCGGGTCTGTGGTCTTTGGAGAACACTCGAGTGTTCTCCAAAGACCACAGACTTTTTG.
2.4. Senescence-associated β-galactosidase (SA-β-gal) staining
SA-β-galactosidase staining was performed two days post-transduction as previously described [18]. Briefly, the cells were rinsed twice with phosphate-buffered saline (PBS), fixed in 4% of formaldehyde for 10-15 min at room temperature, and incubated with freshly prepared SA-β-gal staining solution (1 mg/ml X-gal, 40 mM citric acid, 150 mM sodium chloride, 2 mM magnesium chloride dissolved in phosphate buffer, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, pH 6.0) at 37 °C in a CO2-free atmosphere for overnight (12-16 h). The stained senescent cells were detected and quantified under a regular light microscope. Senescent cells were determined by the percentage of SA-β-gal-positive cells.
2.5. Immunoblotting
Proteins were separated by SDS-polyacrylamide gels (10%) and electrophoretically transferred to polyvinylidene difluoride membranes (BioRad Laboratories, Hertfordshire, UK). After blocking with 5% skimmed milk in tris buffered saline with tween 20 (TBST), the membranes were first incubated with the corresponding primary antibodies overnight at 4 °C. Then, the membranes were washed three times with phosphate-buffered saline with tween 20 (PBST), and corresponding anti-mouse or anti-rabbit HRP conjugated secondary antibodies were applied for 1.5 h at room temperature. Proteins bands were visualized by enhanced chemiluminescence (ECL) and quantified the level of proteins using the ImageJ software (National Institute of Health, Bethesda, MD, USA).
2.6. Quantitative real-time reverse transcription PCR (qRT-PCR) analysis
Total RNA from cells or aortas was extracted using the TRIzol extraction method (Solarbio, China) following the manufacturer's instructions. Total RNA (1 μg) was reversed transcribed into cDNA using the TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (Beijing Quanshijin Biotechnology Co., Ltd., Beijing, China) as described. The target genes were amplified using the PerfectStart Green qPCR SuperMix on the Bio-Rad CFX96 RealTime PCR system (Bio-Rad, US). The oligonucleotide primer sequences are listed in Table 1.
Table 1.
List of primers used for qRT-PCR.
| Target gene | Forward | Reverse |
|---|---|---|
| hGIGYF2 | ATCTTCCTCTGGACACCACG | GTCGCCGAAGAATTTCCTCC |
| hIL-6 | GGCACTGGCAGAAAACAACC | GCAAGTCTCCTCATTGAATCC |
| hMCP-1 | GATCTCAGTGCAGAGGCTCG | TGCTTGTCCAGGTGGTCCAT |
| hTNF-α | CCCAGGGACCTCTCTCTAATCA | GCTACAGGCTTGTCACTCGG |
| hVCAM-1 | CGAACCCAAACAAAGGCAGA | ACAGGATTTTCGGAGCAGGA |
| hICAM-1 | GATTCTGTGCCCACAGTAAGGC | TGGTCACAGAGCCACCTTCTTG |
| hGAPDH | TGCACCACCAACTGCTTAGC | GGCATGGACTGTGGTCATGAG |
| hNLRP3 | AAGGGCCATGGACTATTTCC | GACTCCACCCGATGACAGTT |
| hPPARG | AGCCTGCGAAAGCCTTTTGGTG | GGCTTCACATTCAGCAAACCTGG |
| hSTAU1 | TGCACTTAAACGGAACTTGCC | AATCGGATTGATCCCCTGGC |
| hEMP2 | ATTCTCTGCTGCATCGCCTT | CGTAGCTGCCTTCTCTGGTC |
| hTXNIP | AGATCAGGTCTAAGCAGCAGAACA | CCATATAGCAGGGAGGAGCTTC |
| hSERP1 | GAAGATGGTCGCCAAGCAAAG | TCTTCGGGGGCATTTCTCG |
| hUCP2 | CTACAGCCAGCGCCCAGTA | TCAGTACGCACCATGGTCAGA |
| hESR2 | ATGGAGTCTGGTCGTTGAAGG | TAACACTTCCGAAGTCGGCAGG |
| hDKK1 | TCCGAGGAAAATTGAGGAA | CCTTCTTGTCCTTGGTGTGA |
| hGBP2 | AATTAGGGGCCCAGTTGGAAG | AAGAGACGGTAACCTCCTGGT |
| hLAMTOR4 | AGTGCCATCTCTGAGCTGGTCA | CTGCCTCTTCACCACAAACACC |
| hp21 | AGGTGGACCTGGAGACTCTCAG | TCCTCTTGGAGAAGATCAGCCG |
| hp16INK4A | CTCGTGCTGATGCTACTGAGGA | GGTCGGCGCAGTTGGGCTCC |
| hSIRT1 | TAGACACGCTGGAACAGGTTGC | CTCCTCGTACAGCTTCACAGTC |
| hSIRT6 | TGGCAGTCTTCCAGTGTGGTGT | CGCTCTCAAAGGTGGTGTCGAA |
| mGIGYF2 | AGATGCAGCACCTCCAGTACCA | TCTTCAGAGCCTGGAACTGCTG |
| mSIRT1 | GGAGCAGATTAGTAAGCGGCTTG | GTTACTGCCACAGGAACTAGAGG |
| mSIRT6 | CAGTACGTCAGAGACACGGTTG | GTCCAGAATGGTGTCTCTCAGC |
| mp21 | TCGCTGTCTTGCACTCTGGTGT | CCAATCTGCGCTTGGAGTGATAG |
| mp16INK4A | TGTTGAGGCTAGAGAGGATCTTG | CGAATCTGCACCGTAGTTGAGC |
| mGAPDH | CATCACTGCCACCCAGAAGACTG | ATGCCAGTGAGCTTCCCGTTCAG |
| mSTAU1 | GCTCCTTACAGAACGAGGTCTTC | TCTCAGCAGCATTACGCTTGGC |
h: human; m: mouse.
2.7. Immunofluorescence staining
Cells grown plated onto coverslips were fixed with 4% paraformaldehyde for 15 min, permeabilized using 0.2% Triton X-100 in PBS for 5 min, and blocked with blocking buffer using 1 X PBS with 5% BSA and 0.3% Triton X-100 for 1 h. Primary antibodies were incubated in cells overnight at 4 °C, followed by Alexa Fluor-labeled secondary antibodies for 1 h at room temperature in the dark. All images were acquired through Leica TCS SP5 confocal laser microscope at the same exposure and magnification.
2.8. ROS and NO analysis
ROS and NO production in cultured endothelial cells were examined by staining cells with ROS and NO assay kits in accordance with the manufacturer's instructions, respectively. Briefly, the cells were washed twice with PBS and then incubated with an appropriate volume of diluted DCFH-DA (10 μM) in serum-free culture medium and DAF-FM DA (5 μM) in dilution supplied with the kit at 37 °C for 20 min in the dark. PBS was used to wash the cells three times to sufficiently remove the unbound DCFH-DA and DAF-FM DA, followed by observation by fluorescence microscopy (Olympus, Tokyo, Japan). Meanwhile, for quantitative detection of ROS and NO, the cells were cultured in 96-well plates, and respectively incubated with DHE (5 μM) diluted in PBS and DAF-FM DA (5 μM) diluted in the dilution solution for 20 min at 37 °C. Then, cells were washed with PBS to adequately remove unbound dyes, and quantified ROS and NO levels by reading the absorbance at excitation/emission wavelengths of 535/610 nm and 495/515 nm from a hybrid multi-mode microplate reader (Synergy H1, BioTek) [21].
2.9. Monocyte adhesion to endothelial cells
Monocyte labeling and adhesion assays were employed as previously described [20]. The human monocyte line THP-1 was pre-cultured in RPMI-1640 containing 10% heat-inactivated fetal bovine serum (HIFBS), then labeled with 5 μM CFDA-SE in PBS for 8 min at 37 °C. The labeled THP-1 cells were washed twice with PBS, suspended in RPMI 1640 medium containing 10% FBS. The experimental HUVECs were pretreated with low-serum RPMI-1640 medium containing 0.2% FBS for 12 h, and then incubated with CFDA-SE labeled THP-1 cells at 37 °C for 1 h. Non-adherent THP-1 cells were gently washed twice with PBS, and adherent THP-1 cells were fixed with 3.7% paraformaldehyde. The signals were observed and captured by fluorescence microscopy, and the number of adherent THP-1 cells was quantified by Image J software.
2.10. mRNA stability assay
To measure mRNA decay rates, cells were treated with Actinomycin D (10 μg/ml) post transfection, and harvested in TRIzol at 0, 2, 4, and 6 h after treatment. Subsequently, qRT-PCR assays were carried out to measure the percentage of remaining mRNA of the target gene. The mRNA levels were normalized to GAPDH.
2.11. RNA immunoprecipitation
Cells were washed briefly in ice-cold PBS and lysed on ice for 30 min with lysis buffer (40 mM HEPES pH 7.5, 120 mM NaCl, 1 mM EDTA, 10 mM pyrophosphate, 10 mM glycerophosphate, 0.3% CHAPS) supplemented with protease inhibitors (GLPBIO). Next, the cells were scraped, and centrifugated at 10,000×g for 15 min at 4 °C to remove debris, and the supernatant was transferred to a new 1.5 ml microcentrifuge tube. 1% of the supernatant sample was mixed with 490 μl TRIzol for the RNA input control. Lysates were pre-cleared with protein A/G agarose beads (Santa Cruz) and incubated with primary antibodies for 2 h at 4 °C followed by an incubation with 20 μL A/G beads overnight at 4 °C with gentle rotation. The immunoprecipitates were collected by centrifugation at 2500 rpm for 5 min at 4 °C and washed 3-4 times with washing buffer (50 mM HEPES pH 7.5, 40 mM NaCl, 2 mM EDTA). Subsequently, the beads containing the immunoprecipitated samples were resuspended in 500 μl of TRIzol, and subjected to qRT-PCR analysis.
2.12. Luciferase activity assay
Luciferase activity assay was performed using the Dual-Luciferase Assay Kit (Promega, USA) according to manufacturers’ manuals. The wild-type and mutant LAMTOR 4 DNA fragments (Table 2) were cloned into the pmirGLO Dual-Luciferase Vector (Promega, USA) between the Xba I and Sac I restriction sites, and co-transfected with the pLJM1-STAU1 plasmid into 70% confluent HEK-293T cells using Lipo6000™ transfection reagent. 48 h post transfection, the cells were lysed using 1X passive buffer, and centrifuged (12,000 rpm) at 4 °C for 10 min. Subsequently, the supernatant was collected and the luciferase activity was measured on a BioTek microplate reader.
Table 2.
List of DNA fragments of wild-type and mutant sequences.
| Position | Target | DNA fragments |
|---|---|---|
| chr7:100151697-100151717,chr7 | hSTAU1-LAMTOR4-WT-F1: | 5′-GTTTTTTTGTTTGTTTGTTTTTTTTTAAGATAGAGT-3′ |
| hSTAU1-LAMTOR4-WT-R1: | 5′-CTAG ACTCTATCTTAAAAAAAAACAAACAAACAAAAAAACAGCT-3′ | |
| hSTAU1-LAMTOR4-MUT-F1: | 5′-GTTTTTTTGTTTGTTTGTTTTTCAATTCGATAGAGT-3′ | |
| hSTAU1-LAMTOR4-MUT-R1: | 5′-CTAG ACTCTATCGAATTGAAAAACAAACAAACAAAAAAACAGCT-3′ | |
| chr7:100152017-100152037,chr7 | hSTAU1-LAMTOR4-WT-F2: | 5′-AGTTATTTTAAAGGAGGAATCAGTATATCTTGCTAA-3′ |
| hSTAU1-LAMTOR4-WT-R2: | 5′-CTAGTTAGCAAGATATACTGATTCCTCCTTTAAAATAACTAGCT-3′ | |
| hSTAU1-LAMTOR4-MUT-F2: | 5′-AGTTATTTTAAAGCATTCCTCAGTATATCTTGCTAA-3′ | |
| hSTAU1-LAMTOR4-MUT-R2: | 5′-CTAGTTAGCAAGATATACTGAGGAATGCTTTAAAATAACTAGCT-3′ | |
| chr7: 100152437-100152457,chr7 | hSTAU1-LAMTOR4-WT-F3: | 5′-AAAAAAGAAGTGGATTGGGAAGGAGGTGGATAAGGA-3′ |
| hSTAU1-LAMTOR4-WT-R3: | 5′-CTAGTCCTTATCCACCTCCTTCCCAATCCACTTCTTTTTTAGCT-3′ | |
| hSTAU1-LAMTOR4-MUT-F3: | 5′-AAAAAAGAAGTGGACAATCCAGGAGGTGGATAAGGA-3′ | |
| hSTAU1-LAMTOR4-MUT-R3: | 5′-CTAGTCCTTATCCACCTCCTGGATTGTCCACTTCTTTTTTAGCT-3′ |
The bases in bold are the predicted binding sites of STAU1 protein and LAMTOR4 RNA.
2.13. Animals
GIGYF2 conditional knockout mice (GIGYF2flox/flox Cdh-Cre+ mice) on the background of C57/BL6 were generated in endothelial cells using the Cre-loxP system [22]. Briefly, female GIGYF2flox/flox mice were first crossed with male mice carrying the Cdh5-Cre allele to generate GIGYF2flox/+; Cdh5-Cre mice, which were then mated again with GIGYF2flox/flox mice to obtain GIGYF2flox/flox; Cdh5-Cre mice. Genotyping was performed by PCR. Genomic DNA from mouse tails was extracted and PCR was performed using the KAPA mouse genotyping kit (Kapa Biosystems, Wilmington, USA) according to the manufacturer's instructions. PCR primers used were: 5′-AATTAAAGGCAGACCTAGCAGGTGGAGG-3′ (forward) and 5′-TAGCACCGC AGACTGCTATCTATTCCTG (reverse) for loxP sequence, 5′-GCGGTCTGGCAGTAAAAACTATC-3′ (forward) and 5′-GTGAAACAGCATTGCTGTCACTT-3′ (reverse) for Cdh5-Cre. All mice had ad libitum access to food and water and were maintained within a temperature (22 °C∼25 °C), humidity (50%), and light (12 h light/dark cycle) controlled room. At least 7 male mice in each group of young WT-mice (2 to 3 months old), young GIGYF2flox/flox Cdh-Cre+ mice (2 to 3 months old), old WT-mice (20 to 24 months old) and old GIGYF2flox/flox Cdh-Cre+ mice (20 to 24 months old) were anesthetized with 2% isoflurane, and humanely euthanized by cervical dislocation. The thoracic aorta of the mice was isolated and carefully cleaned of perivascular fat, and subjected to en face staining, vascular tone determination or directly snap frozen in liquid nitrogen and stored at -80 °C until further immunoblot analysis. All animal experiments were conducted under the approval of local authorities (NWU-AWC-20220202 M) and carried out in accordance with the guidelines from Directive 2010/63/EU.
2.14. En face confocal detection of ROS and NO in mouse aortas
The production of ROS in mouse aorta was assessed by DCFH-DA and DHE staining, and NO generation was evaluated by DAF-FM DA staining. Briefly, mice aortas cleaned of perivascular fat tissue were cut into rings 3 mm in length, and equilibrated in pre-warmed Krebs buffer at 37 °C for 30 min continuously aerated with 95% O2 and 5% CO2. After equilibration, the aortas were incubated with DCFH-DA (10 μM), DHE (5 μM) or DAF-FM DA (5 μM) reagents for 30 min, respectively, protected from light. The aorta was then washed three times with Krebs buffer, fixed with 4% paraformaldehyde for 30 min, and counterstained with DAPI (300 nmol/L) for 3 min. After washing with Krebs buffer, the aortas were carefully cut longitudinally and mounted En face (endothelial layer side down) on a slide with a drop of Vectashield mounting medium, and then covered with a coverslip for endothelial layer imaging. Fluorescence was analyzed with a Leica DM6000 confocal microscope within a few hours of preparation, with a Z-shaped scan of each sample, and images were collected after the signal was observed at the top of the sample (endothelial layer on the lumen boundary). Images stained with DCFH, DHE, DAF-FM and DAPI were quantified using Image J.
2.15. Aortic ring vasodilatory response
Endothelium-dependent and independent relaxations were performed as previously described [23]. Briefly, the aortas were isolated, immediately placed in ice-cold Krebs buffer, and followed by careful removal of surrounding adipose tissue. The aortas were cut into 3-4 mm rings and mounted in a Multi-Myograph System (620 M, DMT, Denmark). The vascular ring was pre-constricted with KCl (60 mM) or phenylephrine (1 μM), and after the constriction reached a plateau, cumulative doses of acetylcholine (ACh, 10-10-10-5 M) or sodium nitroprusside (SNP, 10-10-10-5 M) were added to evaluate vasodilation response. Vasodilation was expressed as percent relaxation.
2.16. Pulse wave velocity (PWV) assessment
Noninvasive aortic PWV in mice was measured using the Vevo 3100 ultrasound imaging platform (FUJIFILM VisualSonics) as previously described [24]. Briefly, mice were anesthetized for approximately 1-3 min in an anesthesia induction chamber filled with 2% isoflurane. Anesthesia was maintained via a dedicated nasal cone (1.5% isoflurane), and the mice were coated with conductive paste on the four limbs in a supine position secured on a heating plate (∼37 °C) to maintain body temperature. Remove abdominal hair of mice with depilatory cream and coat it with acoustic coupling gel. Velocity signals from the transverse aortic arch and abdominal aorta were obtained using a 10-MHz pulsed Doppler probe (Indus Instruments, Webster, TX) and collected using a Doppler signal processing workstation. Aortic PWV was calculated by dividing the separation distance between the two measurement sites by the transit time of the flow wave.
2.17. Measurement of blood pressure (BP)
Systolic blood pressure (SBP) and diastolic blood pressure (DBP) were monitored by the CODA® non-invasive BP monitoring system (Kent Scientific) using a volume-pressure recording (VPR) [25,26]. The mice were handled gently and well-trained to minimize handling stress for VPR tail-cuff measurement. No indication of stress was observed during BP testing. Blood pressures were measured at least 20 times per mouse at each time point until a steady state was reached, and the final value was calculated by the average of 10 repeats.
2.18. Statistical analysis
Results data are presented as mean ± SEM. In all experiments shown, the “n” represents the number of individual animals used or the number of individual experiments when conducted with cultured cells. Statistical significance between the data of two groups and more than two groups was analyzed by student's paired t-test or Analysis of variance (ANOVA) with multiple-comparison Bonferroni correction test. Statistical analysis was conducted using GraphPad Prism 9 software (GraphPad Software Inc.). The asterisks represent a significant difference between the data (*p < 0.05, **p < 0.01, ***p < 0.001).
3. Results
3.1. Senescent endothelial cells exhibit endothelial dysfunction and elevated expression of GIGYF2
ECs senescence is strongly associated with endothelial dysfunction and vascular aging [20]. As compared to non-senescent HUVECs (i.e., young cells), the number of SA-β-gal positively stained cells and mRNA expression levels of cyclin-dependent Kinase (CDK) inhibitors (p21, p16) were significantly elevated in senescent HUVECs (Figs. S1A–B), while the anti-aging gene markers SIRT1 and SIRT6 expression were significantly decreased (Fig. S1B). Meanwhile, senescent HUVECs displayed endothelial dysfunction phenotypes, including increased ROS (DCFH-DA staining and quantitative DHE signal), reduced NO (DAF-FM DA staining and quantitative DAF-FM DA signal) production (Figs. S1C–D), enhanced expression of pro-inflammatory cytokines (IL-6, TNF-α, and MCP-1), adhesion molecules (ICAM-1 and VCAM-1) (Fig. S1E) and elevated adhesion of THP-1 monocytes on endothelial cells (Fig. S1F). PCNA staining confirmed the decreased proliferation capability in senescent ECs (Fig. S1G). Strikingly, senescent HUVECs showed higher expression of GIGYF2 protein and mRNA in comparison with young cells (Figs. S1H–I), indicating its aberrant expression may be an event that contributes to HUVECs senescence.
3.2. GIGYF2 depletion in senescent ECs ameliorated endothelial senescence, dysfunction, and inflammation
Although GIGYF2 expression is significantly up-regulated in senescent ECs, whether this event is implicated with ECs senescence and dysfunction remains mysterious. For this, GIGYF2 was depleted in senescent HUVECs by lentivirus-mediated shRNA targeting GIGYF2, as validated by immunoblotting and qRT-PCR, respectively (Fig. 1A and B). In GIGYF2-depleted senescent HUVECs, we observed that GIGYF2 depletion significantly reduced the number of SA-β-gal positive cells (Fig. 1C), p21 and p16 mRNA expression (Fig. 1D) and ROS production (Fig. 1E and F), and conversely enhanced SIRT1 and SIRT6 mRNA expression (Fig. 1D) and NO production (Fig. 1E and F). In addition, GIGYF2 knockdown in senescent HUVECs reduced the mRNA levels of cytokines IL-6, TNF-α and MCP-1 (Fig. 1G) and the mRNA levels of adhesion molecules ICAM-1 and VCAM-1 (Fig. 1G), and suppressed the adhesion of THP-1 monocytes on endothelial cells (Fig. 1H). Furthermore, PCNA immunostaining showed that the decreased proliferation was also prevented by silencing GIGYF2 in senescent HUVECs (Fig. 1I), demonstrating a causal role of GIGYF2 in ECs senescence, endothelial dysfunction as well as inflammation.
Fig. 1.
GIGYF2 depletion in senescent ECs ameliorates endothelial senescence, dysfunction, and inflammation. Senescent HUVECs were transduced with pLKO.1- control (Con) or pLKO.1-GIGYF2 shRNA lentivirus. (A) Immunoblotting analysis of GIGYF2 silencing efficiency in senescent HUVECs. Bar graphs on the right represent the quantification of the relative level of GIGYF2 protein level normalized to β‐actin. (B) qRT-PCR analysis of GIGYF2 silencing efficiency in senescent HUVECs. (C) SA-β-gal staining. Quantifications of SA-β-gal positive cells are shown on the right. (D) qRT-PCR analysis of the mRNA expression of p21, p16, SIRT1 and SIRT6. (E) DCFH-DA staining for detection of ROS and DAF-FM DA staining for detection of NO. Quantifications of DCFH-DA and DAF-FM DA signals are shown right the images. (F) Quantification of DHE and DAF-FM DA fluorescence intensity by a hybrid multi-mode microplate reader. (G) qRT-PCR analysis of the expression of pro-inflammatory cytokines IL-6, TNF-α, MCP-1, and adhesion molecules VCAM-1 and ICAM-1 level. (H) CFDA-SE fluorescence labeled THP-1 monocyte adhesion to endothelial cells. The bar graphs on the right represent quantifications of the adhered monocytes. (I) Representative images of PCNA staining. Quantitation of the PCNA-positive staining is shown on the right. Statistical differences were examined by unpaired Student's t-test. n = 3–5. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 between the indicated groups. Scale bar = 0.1 mm.
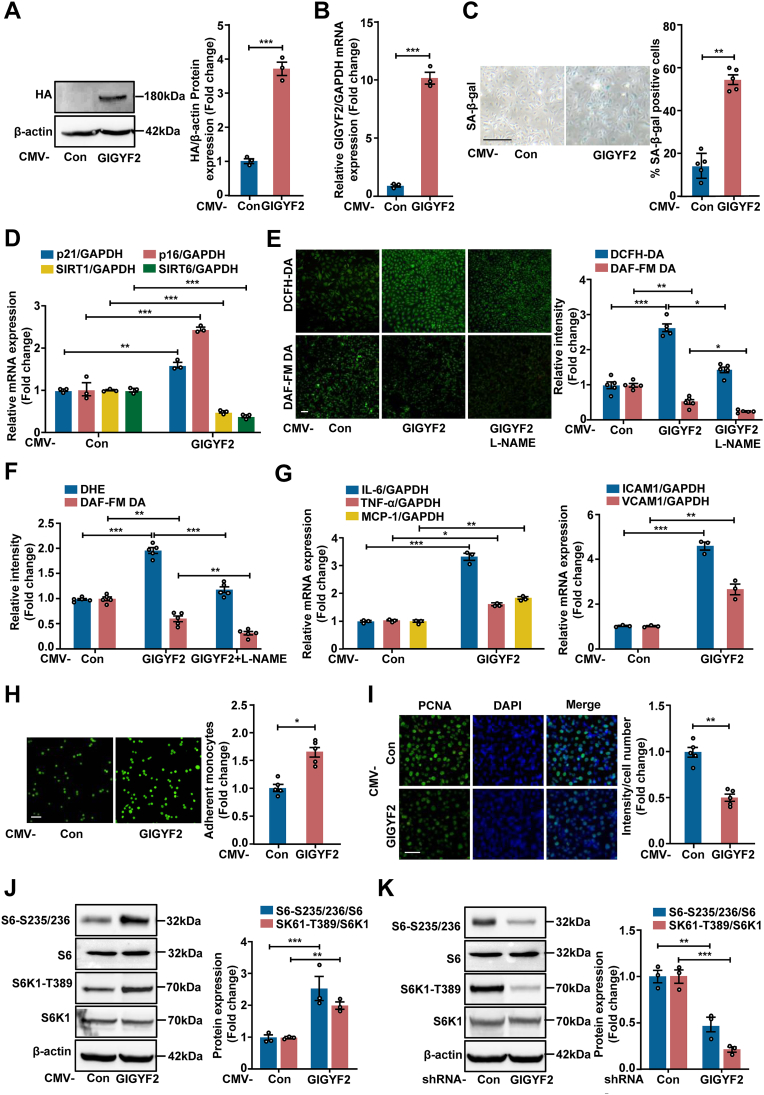
3.3. GIGYF2 overexpression in young ECs promotes cellular senescence, endothelial dysfunction, and mTORC1 activation
To further confirm the role of GIGYF2 in promoting vascular endothelial cell senescence and endothelial dysfunction, we overexpressed GIGYF2 in young ECs. As verified by immunoblotting and qRT-PCR (Fig. 2A and B), GIGYF2 overexpression leads to a significant increase in the number of SA-β-gal positive cells (Fig. 2C), and mRNA expression of p21 and p16 (Fig. 2D), and decrease in SIRT1 and SIRT6 mRNA expression (Fig. 2D), as well as eNOS-uncoupling, i.e., reduced NO level and increased intracellular ROS production (Fig. 2E and F). Following treatment with the eNOS inhibitor L-NAME (1 mM, 1 h), GIGYF2-induced eNOS-uncoupling could be abolished (Fig. 2E and F). The mRNA expression levels of IL-6, TNF-α, MCP-1, ICAM-1 and VCAM-1 (Fig. 2G) and monocyte adhesion to endothelial cells (Fig. 2H) were also remarkably enhanced, while cell proliferation capability is significantly reduced (Fig. 2I) in response to GIGYF2 overexpression in young endothelial cells. These results provide evidence for a definite role of GIGYF2 in promoting ECs senescence, dysfunction, and inflammation. A large body of contributions has identified mTORC1 signaling intimately linked to cellular senescence mediates organismal aging [27], which prompts us to investigate whether GIGYF2 mediates ECs senescence through the activation of the mTORC1-S6K1 signaling axis. Indeed, overexpressing GIGYF2 in the young HUVECs significantly enhances the phosphorylation expression levels of mTORC1 downstream substrates S6K1 (S6K1-T389) and S6 proteins (S6-S235/236) as confirmed by immunoblotting (Fig. 2J). Conversely, in senescent HUVECs, silencing GIGYF2 remarkably reduces the expression levels of S6K1-T389 and S6-S235/236 (Fig. 2K). In addition, similar results were also observed in HEK 293T cell line (Figs. S2A–B). Together, these results prompted us to presume that the GIGYF2 modulating mTORC1-S6K1 signaling axis may govern ECs senescence, dysfunction, and inflammation.
Fig. 2.
GIGYF2 overexpression promotes cellular senescence and activates mTORC1 in young ECs. Young HUVECs were transduced with pLJM1-EJMP as control (Con) or pLJM1-HA-GIGYF2 to overexpress GIGYF2 gene. (A) Immunoblotting analysis of GIGYF2 overexpression in young HUVECs. Bar graphs on the right show quantifications of GIGYF2/β-actin protein level. (B) qRT-PCR analysis of GIGYF2 overexpression in young HUVECs. (C) SA-β-gal staining. Quantifications of SA-β-gal positive cells are shown on the right. (D) qRT-PCR analysis of the mRNA expression of p21, p16, SIRT1 and SIRT6. (E) Cells with GIGYF2 overexpression were pre-treated with eNOS inhibitor l-NAME (1 mmol/L, 1 h), and DCFH-DA staining for detection of ROS and DAF-FM DA staining for detection of NO. Quantifications of DCFH-DA and DAF-FM DA signals are shown right the images. (F) Quantification of DHE and DAF-FM DA fluorescence intensity by a hybrid multi-mode microplate reader. (G) qRT-PCR analysis of the mRNA expression of cytokines IL-6, TNF-α, MCP-1, and adhesion molecules VCAM-1 and ICAM-1. (H) CFDA-SE fluorescence labeled THP-1 monocyte adhesion to endothelial cells. Bar graphs show quantifications of the adhered monocytes. (I) Representative images of PCNA staining. Quantitation results of the PCNA-positive staining are shown on the right. (J) Immunoblotting analysis of S6-S235/236, S6, S6K1-T389 and S6K1 expression levels. The bar chart shows the quantifications of signals on the right. (K) Senescent HUVECs were transduced with pLKO.1-control (Con) or pLKO.1-GIGYF2 shRNA, and immunoblotting analysis of S6-S235/236, S6, S6K1-T389 and S6K1 expression levels. The bar chart shows the quantifications of signals on the right. Statistical differences were examined by unpaired Student's t-test. n = 3–5. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 between the indicated groups. Scale bar = 0.1 mm.
3.4. Rapamycin ameliorates cell senescence evoked by GIGYF2 in young ECs
To validate the above presumption, we examined the effects of mTORC1 inhibitor rapamycin (RAPA) treatment on GIGYF2-induced ECs senescence, dysfunction as well as inflammation in young endothelial cells. As confirmed by immunoblotting, RAPA significantly represses GIGYF2-induced mTORC1-S6K1 activation (Fig. 3A). Meanwhile, we observed that the cellular senescence evoked by GIGYF2 in young cells was blockaded by RAPA treatment, which was envisioned by the reduced SA-β-gal positive cells (Fig. 3B) and p21 and p16 mRNA expression, and enhanced SIRT1 and SIRT6 mRNA expression (Fig. 3C). Furthermore, eNOS-uncoupling and pro-inflammatory responses caused by GIGYF2 overexpression, i.e., the elevated intracellular ROS generation, decreased NO production (Fig. 3D and E), enhanced mRNA expression levels of IL-6, TNF-α, MCP-1, VCAM-1 and ICAM-1 (Fig. 3F), and increased THP1 monocyte adhesion to endothelial cells (Fig. 3G) were all prevented by RAPA. GIGYF2 overexpression resulted in proliferation arrest, which was also ablated by RAPA (Fig. 3H). Accordingly, these results demonstrate that GIGYF2 promotes eNOS-uncoupling, cellular senescence, dysfunction and inflammation via triggering the mTORC1-S6K1 signaling pathway.
Fig. 3.
Rapamycin reverses cell senescence evoked by GIGYF2 overexpression in young ECs. Young HUVECs were transduced with pLJM1-EJMP as control (Con) or pLJM1-HA-GIGYF2 to overexpress GIGYF2. One group of GIGYF2-overexpressed HUVECs was treated with rapamycin (RAPA, 20 ng/ml) for 2 h. (A) immunoblotting analysis of S6-S235/236, S6, S6K1-T389 and S6K1 expression levels. (B) SA-β-gal staining. Bar graphs represent quantifications of SA-β-gal-positive cells. (C) qRT-PCR analysis of the mRNA expression of p21, p16, SIRT1 and SIRT6. (D) DCFH-DA staining for the detection of ROS and DAF-FM DA staining for the detection of NO. Quantifications of DCFH-DA and DAF-FM DA signals are shown right the images. (E) Quantification of DHE and DAF-FM DA fluorescence intensity by a hybrid multi-mode microplate reader. (F) qRT-PCR analysis for cytokines IL-6, TNF-α, MCP-1, and adhesion molecules VCAM-1 and ICAM-1 mRNA levels. (G) CFDA-SE fluorescence labeled THP-1 monocyte adhesion to endothelial cells. Bar graphs show quantifications of the adhered monocytes. (H) Representative images of PCNA staining. Quantitation results of the PCNA-positive staining are shown on the right. Statistical differences were examined by one-way ANOVA. Data are presented as mean ± SEM. n = 3–5. *p < 0.05, **p < 0.01, ***p < 0.001 between the indicated groups. Scale bar = 0.1 mm.
3.5. Inhibition of eNOS-uncoupling by NAC prevented GIGYF2-induced cells senescence and dysfunction
eNOS-uncoupling has been extensively reported to induce endothelial dysfunction and vascular aging [28]. Next, we are interested to explore if recoupling eNOS function is able to prevent GIGYF2-induced endothelial cell senescence and dysfunction. For this purpose, ROS scavenger NAC as an antioxidant was employed to reverse GIGYF2-induced eNOS-uncoupling as verified by the reduction of ROS generation and enhanced bioavailability of NO (Figs. S3A–B). Meanwhile, in GIGYF2-overexpressed young HUVECs, we observed that NAC significantly reduced the positive cell number of SA-β-gal staining (Fig. S3C), the mRNA expression levels of p21, p16, IL-6, MCP-1, TNF-α, ICAM-1 and VCAM-1 (Figs. S3D–E), and the monocyte adhesion to endothelial cells (Fig. S3F) in comparison with GIGYF2 per se group. Furthermore, reduced SIRT1 and SIRT6 mRNA expression levels and PCNA-positive staining upon GIGYF2 overexpression were also blocked by NAC (Fig. S3D and Fig. S3G). These results reveal that re-coupling eNOS protects against GIGYF2-indued ECs senescence and endothelial dysfunction.
3.6. STAU1 modulates GIGYF2-induced mTORC1 activation
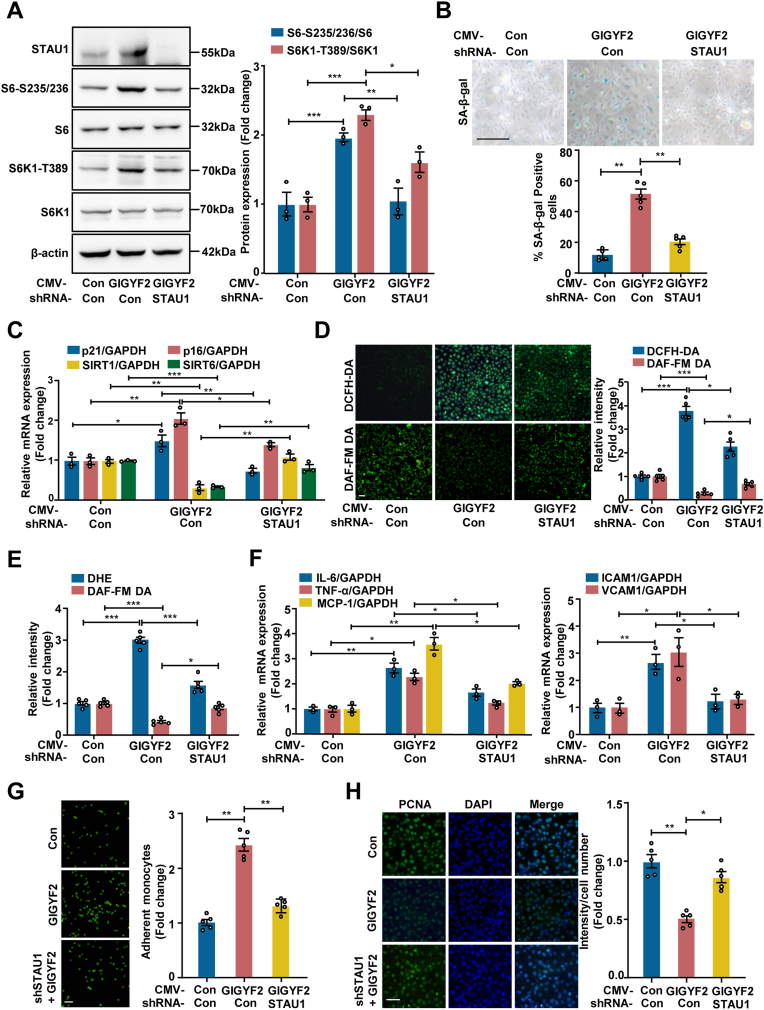
To gain further insight into the underlying mechanism by which GIGYF2 activates the mTORC1-S6K1 signaling axis to promote eNOS-uncoupling, endothelial cell senescence as well as senescence-related dysfunction, we performed transcriptome sequencing on two groups of wild-type and GIGYF2−/− HUVECs. As shown in Fig. S4A, the volcano map characterized the overall distribution of downregulated- and upregulated-differentially expressed genes (DEGs). The heat maps showed the top 29 upregulated- and downregulated- DGEs (Fig. S4B-C), respectively. To validate the RNA-seq results, we selected the top ten DEGs (EMP2, SERP1, NLRP3, GBP, UCP2, STAU1, PPARG, ESR2, DKK1 and TNXIP) with significant differences for qRT-PCR validation. It is worth noting that nine DEGs apart from PPARG appear to be consistent with the transcriptome sequencing results, of which, STAU1 showed the most pronounced alteration (Fig. S4D). Therefore, we imply that GIGYF2 induces mTORC1-S6K1 signaling activation via the up-regulation of STAU1, thereby modulating ECs senescence and endothelial dysfunction.
Next, to verify our hypothesis, we carried out lentivirus-mediated STAU1 depletion in young HUVECs with GIGYF2 overexpression. As shown in Fig. 4A, the increase in S6K1-T389 and S6-S235/236 induced by GIGYF2 was prevented by STAU1 knockdown. In parallel, overexpression of GIGYF2 promoting endothelial cells senescence and senescence-induced dysfunction and inflammation, i.e., increased number of positively stained cells for SA-β-gal (Fig. 4B), elevated p21 and p16 mRNA expression and intracellular ROS generation (Fig. 4C-E), decreased SIRT1 and SIRT6 mRNA expression and NO production (Fig. 4C-E), enhanced mRNA expression levels of IL-6, TNF-α, MCP-1, ICAM-1 and VCAM-1 (Fig. 4F), elevated monocyte adhesion to endothelial cells (Fig. 4G), and reduced of PCNA-positive staining (Fig. 4H) in young HUVECs, was also attenuated by silencing STAU1. Notably, as compared to young HUVECs, we observed that both STAU1 mRNA and protein expression were up-regulated in senescent HUVECs (Figs. S5A–B). In addition, silencing GIGYF2 markedly reduced the S6K1-T389 and S6-S235/236 expression levels in senescent HUVECs, which could be restored by STAU1 overexpression (Fig. S6A). Furthermore, overexpressing STAU1 ablated the protective effects that silencing GIGYF2 attenuated HUVECs senescence (Figs. S6B–C), eNOS-uncoupling (Figs. S6D–E), the mRNA expression levels of IL-6, TNF-α, MCP-1, VCAM-1 and ICAM-1 (Fig. S6F) and monocyte adhesion to endothelial cells (Fig. S6G), and enhanced cell proliferation capability evaluated by PCNA positive staining (Fig. S6H). Collectively, these data provide firm evidence to support our above-mentioned hypothesis that the GIGYF2-STAU1-mTORC1 signaling axis contributes to endothelial senescence, dysfunction, and inflammation.
Fig. 4.
Silencing STAU1 inhibits GIGYF2-induced mTORC1-S6K1 activation along with reduced cell senescence and improved endothelial function. Young HUVECs with GIGYF2 overexpression were transduced with pLJM1-EJMP as control (con) or pLJM1-HA-GIGYF2 alone or pLJM1-HA-GIGYF2 plus pLKO.1-STAU1 shRNA. (A) Immunoblotting analysis of STAU1, S6-S235/236, S6, S6K1-T389 and S6K1 protein expression. The bar graphs on the right show quantifications of protein levels. (B) SA-β-gal staining. Bar graphs represent quantifications of SA-β-gal-positive cells shown below. (C) qRT-PCR analysis of the mRNA expression of p21, p16, SIRT1 and SIRT6. (D) DCFH-DA staining for the detection of ROS and DAF-FM DA staining for the detection of NO. Quantifications of DCFH-DA and DAF-FM DA signals are shown right. (E) Quantification of DHE and DAF-FM DA fluorescence intensity by a hybrid multi-mode microplate reader. (F) qRT-PCR analysis of the expression of pro-inflammatory cytokines IL-6, TNF-α, MCP-1, and adhesion molecules VCAM-1 and ICAM-1. (G) CFDA-SE fluorescence labeled THP-1 monocyte adhesion to endothelial cells. Bar graphs show quantifications of the adhered monocytes. (H) Representative images of PCNA staining. Quantitation results of the PCNA-positive staining are shown on the right. Statistical differences were examined by one-way ANOVA. n = 3–5. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 between the indicated groups. Scale bar = 0.1 mm.
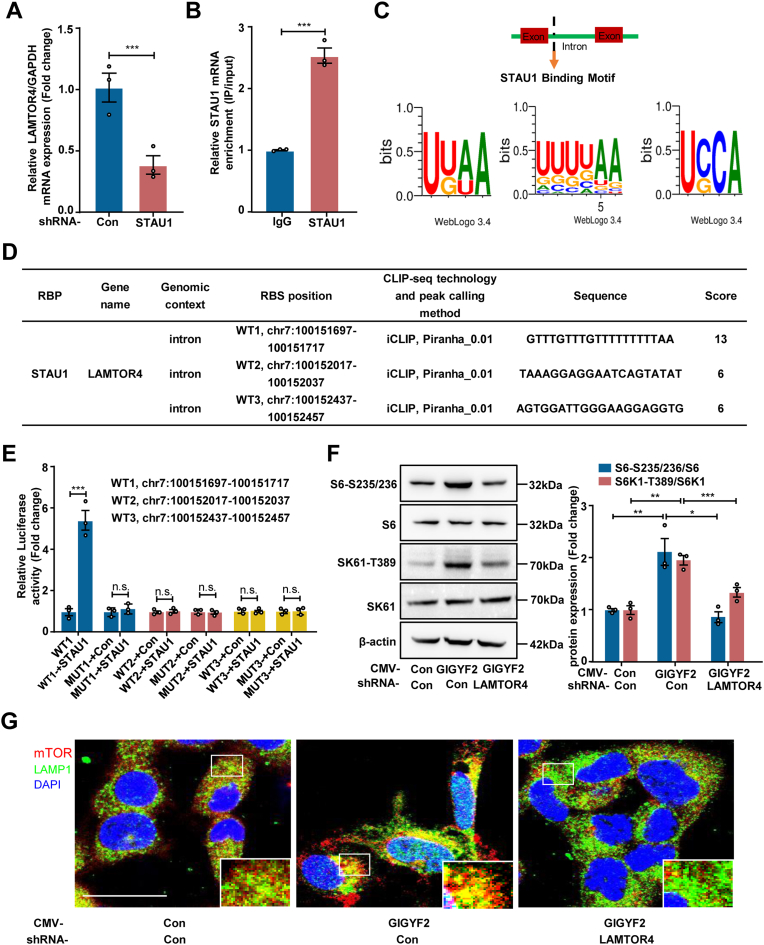
3.7. GIGYF2 as an RNA-binding protein (RBP) enhances STAU1 mRNA stability
However, the question still needs to be answered regarding the nature of GIGYF2 up-regulating STAU1 expression. Given the fact that GIGYF2 is an RNA-binding protein (RBP) involved in regulating the mRNA stability [9,29], and our data that GIGYF2 depletion reduces STAU1 mRNA expression level in HUVECs, we thus propose that GIGYF2 as an RBP binding to STAU1 mRNAs enhances STAU1 mRNA stability. Indeed, RNA immunoprecipitation (RIP) confirmed the binding of GIGYF2 protein to STAU1 mRNA (Fig. S7A). To further elucidate the specific binding between GIGYF2 and STAU1 mRNA, we truncated the ORF of human GIGYF2 (1-1320) into three fragments GIGYF2 A (1-495), GIGYF2 B (496-742) and GIGYF2 C (743-1320) to explore their specific binding domain by RIP (Fig. S7B). The RIP assay showed that GIGYF2 WT (1-1320) with full length and GIGYF2 A (1-495), but not GIGYF2 B and GIGYF2 C, bound with STAU1 mRNA (Fig. S7C), which demonstrates that the STAU1 mRNA specifically interacts with the region of GIGYF2 spans residues 1-495 containing the eIF4E-homologous protein (4EHP) and Menin binding motif (MBM) binding motifs. Subsequently, we performed mRNA stability assays to determine the effects of GIGYF2 knockdown or overexpression on the stability of STAU1 mRNA. qRT-PCR analysis showed that silencing GIGYF2 remarkably reduced STAU1 mRNA (Fig. S7D), conversely hyperactive GIGYF2 expression enhances the STAU1 mRNA stability (Fig. S7E), demonstrating a stabilizing effect of GIGYF2 protein on STAU1 mRNAs in human endothelial cells.
3.8. LAMTOR4 mediates STAU1-mTORC1 signaling cascade
Next, we asked how STAU1 activates mTORC1. The Ragulator complex plays a vital role in anchoring Rag GTPase to the lysosomal surface to activate the mTORC1 pathway. A protein encoded by the late endosomal/lysosomal adaptor and MAPK and MTOR activator 4 (LAMTOR4) gene as a novel Ragulator component is essential for the mTORC1 activation [30]. Here, we found that silencing STAU1 down-regulated LAMTOR4 mRNA expression in senescent HUVECs (Fig. 5A). Considering that STAU1 is also an RBP that has been reported to bind to gene RNA to regulate gene expression [31,32], we performed RIP assay to precipitate LAMTOR4 mRNA with STAU1 antibody, confirming that STAU1 as an RBP can bind to LAMTOR4 mRNA (Fig. 5B). POSTAR3 database shows three potential binding motifs of STAU1 (Fig. 5C), and three binding sites and sequences with LAMTOR4 intron regions (Fig. 5D). Accordingly, we presume that STAU1 binding to LAMTOR4 intron region boosts LAMTOR4 expression, which in turn induces mTORC1 activation. Luciferase reporter assays confirmed this presumption that the WT1 region (chr7:100151697-100151717 region) but not WT1-MUT, WT2 regions (chr7:100152017-100152037) and WT3 regions (chr7:100152437-100152457) of LAMTOR4 intron could bind to STAU1 protein (Fig. 5E). Furthermore, the immunoblotting assay demonstrated that LAMTOR4 knockdown protected against the mTORC1-S6K1 activation caused by GIGYF2 overexpression (Fig. 5F). In line with immunofluorescence staining results, we also found that overexpressing GIGYF2 significantly promoted the translocation of mTORC1 to lysosome (LAMP1), which could be blockaded by silencing LAMTOR4 (Fig. 5G). These results suggest that the GIGYF2-STAU1 signaling axis drivers the mTORC1-S6K1 pathway activation through up-regulating the expression of LAMTOR4 that facilitates the translocation of mTORC1 to lysosome.
Fig. 5.
LAMTOR4 mediates STAU1-mTORC1 signaling cascade. Senescent HUVECs were transduced with pLKO.1-control (Con) or pLKO.1-GIGYF2 shRNA. (A) qRT-PCR analysis of LAMTOR4 mRNA expression. (B) RIPs were performed using control immunoglobulin G (IgG) or rabbit polyclonal antibody directed against STAU1. qRT-PCR analysis of co-precipitated LAMTOR4 mRNAs normalized to GAPDH. (C) POSTAR3 shows the potential binding motifs of LAMTOR4 mRNA with STAU1. (D) Binding regions of STAU1 and LAMTOR4 mRNA predicted by POSTAR3 database. (E) Luciferase reporting assay confirmed STAU1 binding to LAMTOR4 RNA regions chr7:100151697-100151717). (F) Young HUVECs with GIGYF2 overexpression were transduced with pLJM1-EJMP as control (con) or pLJM1-HA-GIGYF2 alone or pLJM1-HA-GIGYF2 plus pLKO.1- LAMTOR4 shRNA. Immunoblotting analysis of S6-S235/236, S6, S6K1-T389 and S6K1 expression levels. (G) Immunofluorescence staining of mTOR (red), and LAMP1 (green) followed by counterstaining with DAPI (blue). The images with white outlines in the corner are the enlargements of the selected area of corresponding pictures. n = 3. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 between the indicated groups. n.s. not significant. Scale bar = 0.1 mm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.9. Endothelial-specific GIGYF2 knockout mice are protected from aging-associated vascular aging and dysfunction
Accumulating studies indicated that endothelial cell senescence is strongly associated with aging-associated vascular dysfunction [33], we therefore further investigated whether the GIGYF2-mediated endothelial cell senescence contributes to the aging-related vascular aging and dysfunction in vivo. Here, we established vascular endothelial GIGYF2-specific knockout mice by crossing GIGYF2flox/flox mice with transgenic mice expressing Cre under the control of the endothelial-specific Cdh5 promoter (Fig. 6A). Homozygous endothelial GIGYF2-specific knockout (GIGYF2flox/flox Cdh-Cre+) mice were screened and validated by PCR genotyping of genomic DNA from tail tissue (Fig. 6B). Co-immunofluorescence staining verified a significantly up-regulated GIGYF2 expression in the aortic endothelial cells of old mice as compared to young mice, and the potent knockout efficiency of GIGYF2 in the aortic endothelial cells of GIGYF2flox/flox Cdh-Cre+ mice (Fig. 6C). Moreover, as compared to young WT mice, we observed that the protein expression levels of GIGYF2, STAU1, S6-S235/236, and S6K1-T389 were significantly upregulated in the aorta of old WT mice (Fig. 6D), which was conversely ablated in the aorta of old GIGYF2flox/flox Cdh-Cre+ mice (Fig. 6D). Also, the aging-associated increases in mRNA expression levels of GIGYF2, STAU1, p21 and p16 (Figs. S8A–C), and the decreases in SIRT1 and SIRT6 mRNA expression (Fig. S8C) were prevented in the aorta of aged GIGYF2flox/flox Cdh-Cre+ mice (Figs. S8A–C). Furthermore, en face fluorescence confocal microscopy demonstrated that the increase in ROS production (Fig. 6E and Fig. S8D) and the decrease in NO generation (Fig. 6F) in old WT mice were also suppressed in old GIGYF2flox/flox Cdh-Cre+ mice. To further assess the loss of endothelial GIGYF2 gene on vascular endothelial function, we isolated mouse aorta for ex vivo functional assay. When compared to young WT mice, the endothelium-dependent relaxations in response to acetylcholine (Ach) were significantly impaired in old WT mice following the stimulation of vascular contractile agent either with KCl (Fig. 6G) or with phenylephrine (Fig. S9A). Strikingly, the old GIGYF2flox/flox Cdh-Cre+ mice exhibited significant amelioration of endothelium-dependent relaxation to Ach (Fig. 6G and Fig. S9A). Whereas, endothelium-independent relaxations in response to the NO donor SNP under endothelial GIGYF2 depletion were not influenced in old mice (Fig. 6H and Fig. S9B). With age, arteries tend to become stiffer with elevated pulse wave velocity (PWV), which can raise blood pressure [34]. As compared to the young WT animal (Young-WT), we observed that PWV, systolic and diastolic blood pressures were significantly elevated in the old mice (Old-WT) (Figs. S9C–E). Of note, endothelial-specific GIGYF2 knockout (Old-GIGYF2flox/flox Cdh-Cre+) remarkably ablated the age-associated increase in PWV (Fig. S9C) rather than blood pressure (
Fig. 6.
Endothelial-specific GIGYF2 knockout ameliorates age-associated vascular dysfunction in mice. (A) Schematic diagram of generating vascular endothelial GIGYF2-specific knockout mice by utilizing the Cre-loxP recombination system. (B) Agarose gel image of PCR-genotyping of genomic DNA obtained from various types of mice. (C) Immunofluorescence staining of GIGYF2 (green) and CD31 (red) in the aortic endothelial cells of young WT, old WT, and old GIGYF2flox/flox Cdh-Cre+ (GIGYF2-CKO) mice (n = 7 per group). (D) Immunoblotting assay detects the protein expression levels of GIGYF2, STAU1, S6-S235/236 and S6K1-T389 in young-WT, old-WT, and old-GIGYF2-CKO mice (n = 5 per group). The bar graphs on the right represent the quantifications of protein level. (E) Representative images of confocal microscopic en face detection of endothelial ROS (DCFH-DA staining) and (F) NO production (DAF-2DA staining) followed by DAPI counterstaining in young WT and GIGYF2-CKO mice and old WT and GIGYF2-CKO mice (n = 9 per group). Quantification of the DCFH-DA and DAF-2DA signals on the right. The cut 3-4 mm aortic rings from each mouse were pre-constricted with KCl (60 mM), and then subject to (G) endothelium-dependent relaxations to acetylcholine in aortas (n = 5 per group), and (H) endothelium-independent relaxations in response to the NO-donor sodium nitroprusside (SNP) in aortas of young WT, young GIGYF2-CKO mice, old WT and old GIGYF2-CKO mice (n = 5 per group). GIGYF2-CKO mice refer to endothelial GIGYF2-specific knockout mice. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 between the indicated groups. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Figs. S9D–E) in old mice.
3.10. Targeting GIGYF2 by ridaifen B alleviates endothelial cells senescence and dysfunction
Ridaifen B (RID-B), a derivative of tamoxifen derivatives, has been reported to bind to the GIGYF2 protein to inhibit AKT-related pathways [35]. Here, we found that RID-B treatment significantly suppressed the protein expression levels of GIGYF2, S6K1-T389 and S6-S235/236 in senescent HUVECs (Fig. S10A). In parallel, the qRT-PCR analysis showed that the mRNA expression levels of GIGYF2 and STAU1 were also reduced in senescent HUVECs with RID-B treatment (Figs. S10B–C). Additionally, upon RID-B treatment in senescent HUVECs, SA-β-gal positive cells (Fig. S10D), p21 and p16 mRNA expression (Fig. S10E), ROS production (Figs. S10F–G), and the mRNA levels of inflammatory cytokines (IL-6, TNF-α, MCP-1, ICAM-1 and VCAM-1) (Fig. S10H) were also significantly decreased, while SIRT1 and SIRT6 mRNA expression (Fig. S10E), and NO production levels were increased (Figs. S10F–G). Moreover, treating the ex vivo isolated aorta of old mice with RID-B was shown to improve endothelial function, i.e., reduced superoxide anion production (Fig. S10I) and enhanced NO generation (Fig. S10J). These results indicate that disruption of GIGYF2 expression by RID-B could attenuate endothelial cells senescence, dysfunction and inflammatory responses, in turn improving aging-related vascular aging.
4. Discussion
The onset of endothelial cell senescence leading to endothelial dysfunction, is widely recognized as a primary risk factor for cardiovascular disease occurrence. The phenotype of endothelial cell senescence is characterized by a weakened cell proliferation ability, an acquisition of senescence-associated secretory phenotype (SASP), and decreased production of NO and increased ROS caused by eNOS-uncoupling. However, the molecular mechanisms of endothelial cell senescence and aging-associated endothelial dysfunction have yet to be fully understood. Here, we revealed for the first time that GIGYF2 as an RBP promotes endothelial cell senescence, dysfunction, and inflammation through the up-regulation of STAU1 that activates the LAMTOR4-mTORC1-S6K1 signaling cascade in primary endothelial cells, thereby contributing to vascular dysfunction and aging. These findings provide insights into the GIGYF2-STAU1-mTORC1 signaling axis as a potential therapeutic target for the treatment of aging-related cardiovascular disease.
In our current study, we observed hyper-expression of GIGYF2 in senescent human endothelial cells and aortas of aged mice. Silencing GIGYF2 in senescent HUVECs or conditional knockout of GIGYF2 in the endothelial cells of old mice attenuated cellular senescence and rejuvenated vascular function, e.g., reduced endothelial senescence markers, enhanced endothelial or vascular NO generation, decreased endothelial or vascular ROS production and inflammatory responses, and ameliorated arterial stiffness and endothelium-dependent relaxation to acetylcholine. Whereas, endothelial-specific GIGYF2 deficiency failed to prevent age-associated blood pressure elevation in old mice, suggesting that GIGYF2 depletion may affect other blood pressure modulators to counteract the effect of reduced arterial stiffness. Consistently, overexpressing GIGYF2 in young HUVECs resulted in cellular senescence, inflammation as well as endothelial dysfunction. These results from cultured cells and mice models provide conclusive in vitro and in vivo evidence for a causal role of GIGYF2 in promoting endothelial cell senescence, endothelial dysfunction, and vascular aging. Here, for the first time, we disclose the role of GIGYF2 in the modulation of cellular senescence in endothelial cells and vascular aging. Previous studies have demonstrated that the mutations and abnormal expression of GIGYF2 are strongly associated with aging-related diseases such as Parkinson's disease, and diabetes-associated cognitive impairment [10,36]. However, the molecular mechanism that GIGYF2 mediates these neurodegenerative diseases still remains elusive. On the basis of our findings, it is thus tempting to speculate that GIGYF2 might regulate cognitive impairment via the governance of neural cells' senescence. Nevertheless, whether this is true still requires further investigation.
Additionally, our study also demonstrated that GIGYF2 promoting eNOS-uncoupling modulates endothelial senescence and function. eNOS-uncoupling resulting in superoxide anion generation instead of NO causes has previously been reported to contribute to endothelial cell senescence and dysfunction in vascular diseases [20]. Here, we show that exogenous overexpression of GIGYF2 triggers eNOS-uncoupling with concomitant induction of cellular senescence in young endothelial cells. The superoxide anion resulting from GIGYF2-induced eNOS-uncoupling is inhibited by the eNOS activity inhibitor L-NAME. Moreover, in senescent endothelial cells, depleting GIGYF2 is able to recouple eNOS to enhance NO production and alleviate ROS generation, which is also accompanied by inhibition in cellular senescence, inflammatory factors, and endothelial adhesion molecules. Further experiments show that abolishment of ROS with the antioxidant NAC in non-senescent cells upon GIGYF2 overexpression not only restores eNOS function but also suppresses GIGYF2-induced senescence and SASP. In line with in vitro results, endothelial GIGYF2-conditional knockout in aged mice also exhibits improved eNOS-uncoupling compared to WT-aged mice. These findings reveal a positive regulatory crosstalk between eNOS-uncoupling and oxidative stress. The ROS generated from eNOS-uncoupling is required to maintain the persistent eNOS-uncoupling with subsequently reduced NO and augmented ROS generation, which is one of the central underlying mechanisms of endothelial senescence and dysfunction [20,37]. Here, we uncovered a novel function of endothelial GIGYF2 in the regulation of vascular aging linked to eNOS-uncoupling, which was not previously reported.
There is substantial evidence that persistent signaling of mTORC1 accelerates cells senescence and integrates various vascular-related signaling including oxidative stress and pro-inflammatory responses all of which can lead to endothelial dysfunction [27,38]. S6K1, a critical downstream of mTORC1, also has been reported to be constitutively active in senescent endothelial cells and aortas of aged rodents to promote eNOS-uncoupling and endothelial senescence [18]. In a previous study, it was demonstrated that mTORC1-S6K1 activation upregulates eNOS monomer and reduces eNOS dimer/monomer ratio, in turn contributing to mTORC1-induced eNOS uncoupling in endothelial cells [18]. Strikingly, here we found that overexpression of GIGYF2 activates the mTORC1-S6K1 signaling axis in young endothelial cells. In senescent endothelial cells, we previously found that there was more hyperactive mTORC1-S6K1 signaling compared to young cells [20]. Here, we demonstrate that depletion of GIGYF2 markedly blockades this cellular senescence-associated mTORC1 activation, which results in the amelioration of cellular senescence, endothelial function as well as inflammatory response. Consistently, inhibition of the mTORC1 pathway with RAPA has been demonstrated to suppress GIGYF2-induced senescence, pro-inflammatory response, and enhance proliferative capacity. The results for the first time demonstrate a critical role of GIGYF2 in the activation of the mTORC1-S6K1 pathway resulting in the promotion of endothelial cell senescence. Consistent with our data, Giovannone et al. found that the loss of Drosophila CG11148 (ortholog of mammalian GIGYF2) had evident suppressive effects on mTORC1 signaling as evidenced by the down-regulated phosphorylation of mTORC1 substrate S6K1 in Gyf-null mutant tissues [14]. Additionally, the phosphorylation of mTORC2 substrate AKT was also reduced with less extent upon GIGYF2 deficiency, indicating GIGYF2 may mediate mTORC2-AKT signaling axis [14]. A previous study also characterized that the required for cell differentiation1 homolog (RQCD1) protein interacting with GIGYF1 and GIGYF2 is implicated in the regulation of AKT activation in breast cancer cells [39]. In contrast, a most recent study report indicates that GIGYF2 inhibiting pro-survival AKT/Bax/Caspase-3 signaling contributes to MMP-9 mediated cell migration in glioma cells [40]. In mammalian brains, GIGYF2 has been reported to enhance IGF-1-induced extracellular signal-regulated kinase 1/2 (ERK1/2) phosphorylation through modulation of insulin-like growth factor-1 (IGF-1) receptor trafficking [41]. Extensive studies have demonstrated that both AKT and ERK1/2-related pathways are strongly implicated with the regulation of cellular senescence [42,43]. Thus, additional work will be necessary to further explore whether GIGYF2 promotes endothelial cell senescence and dysfunction through the modulation of other pathways.
A further important novel finding of our study is that GIGYF2 as an RBP promoting STAU1 expression results in mTORC1 activation, endothelial cells senescence, dysfunction, and pro-inflammatory responses. Firstly, we demonstrate that GIGYF2 up-regulates STAU1 mRNA expression via enhancing its mRNA stability, which is evidenced by the fact that overexpression of GIGYF2 in young cells promotes the STAU1 mRNA stability, whereas silencing GIGYF2 attenuates the STAU1 mRNA decay in senescent cells. Despite our still incomplete understanding of how GIGYF2 stabilizes STAU1 mRNA, some mechanistic evidence cues are surfacing. The RIP experiment confirmed the binding of GIGYF2 to STAU1 mRNA. Moreover, truncated ORFs of GIGYF2 with diverse motifs demonstrate that GIGYF2 A (1-495) composing 4EHP binding and MBM motifs specifically interacts with STAU1 mRNA. Nevertheless, the exact binding sites between GIGYF2 and STAU1 mRNA still require further investigation. As reported, RPBs mediate mRNA expression through their association with 5′UTR or 3′UTR non-coding regions that have been revealed to modulate pre-mRNA splicing, RNA stability, RNA localization, and translation [44]. It has been reported that GIGYF2 as an RBP interacting with the 5’ cap-binding protein 4EHP mediates post-transcriptional repression of mRNAs [9]. On the other hand, we found that silencing STAU1 prevents GIGYF2-induced mTORC1-S6K1 pathway activation, endothelial cell senescence and dysfunction in young endothelial cells, and overexpressing STAU1 reverses the protective effects that depleting GIGYF2 improves endothelial cells senescence and function in senescent endothelial cells. STAU1, a highly conserved and multifunctional double-stranded RBP, is involved in a variety of biological processes including cell proliferation [45], migration [46], differentiation [47], apoptosis [48], stress response [49], and autophagy [50]. It has been recently reported that exogenous expression of STAU1 sufficiently activates the mTOR pathway in prostate cancer cells and patient-derived cell fibroblast cells, respectively [51]. In non-transformed skeletal muscle cells, STAU1 downregulation can activate autophagy in an mTOR-dependent manner [32]. Also, a clinical study demonstrates that an increased STAU1 expression is strongly associated with the highly phosphorylated mTOR in fibroblast cells and spinal cord tissues from amyotrophic lateral sclerosis (ALS) patients [50]. In our animal model, conditional knockout of GIGYF2 in endothelial cells also shows reduced STAU1 expression, S6K1 and S6 phosphorylation levels. Together, these above findings further support our concept that the GIGYF2 activates mTORC1 signaling pathway via upregulating STAU1, which in turn promotes endothelial cell senescence, endothelial function, and inflammation.
Next, we further answered the question that how STUA1 activates the mTORC1-S6K1 pathway in endothelial cells. Notably, in this study, we uncover another important novel finding that LAMTOR4 is upregulated by STAU1 through its interaction with an intron region of LAMTOR4, which promotes the translocation of mTORC1 to lysosome (LAMP1), ultimately leading to mTORC1 activation in endothelial cells. Our study demonstrates that STAU1 as an RBP with UUUUAA motif binding with the intron region (chr7:100151697-100151717) of LAMTOR4 controls the LAMTOR4 mRNA expression. This is evidenced by the fact that the mutation of binding sites fails to interact with STAU1 and that silencing STAU1 gene in senescent endothelial cells lowers the LAMTOR4 mRNA expression level. STAU1 has been reported to play critical roles in RNA localization [52], splicing [53], stability [54], translation [55], and decay [56]. As the binding sites between STAU1 and LAMTOR4 are located in the intron region of LAMTOR4, we imply that STAU1 controls LAMTOR4 mRNA expression via regulating the splicing of LAMTOR4 mRNA. Nevertheless, this hypothesis warrants further experimental confirmation. It is well accepted that activation of mTORC1 requires the recruitment of mTORC1 to the lysosomal membrane through the Ragulator-Rag complex [57,58]. Thus, LAMTOR4 as one component of the Ragator complex is also involved in the mTORC1 activation [59,60]. In agreement with this notion, here we observe that LAMTOR4 deficiency blocks the activation of the mTORC1-S6K1 pathway and the recruitment of mTORC1 to lysosomal membrane caused by GIGYF2 overexpression. Together, these data point to a role for LAMTOR4 in modulating the GIGYF2-STAU1-mTOC1 signaling axis to regulate endothelial cell senescence and endothelial dysfunction.
Finally, the above results urge us to assess whether disrupting GIGYF2 by chemical inhibitors or genetic deficiency improves vascular aging and aging-associated endothelial dysfunction. Ridaifen B (RID-B), a novel tamoxifen derivative, has been reported to bind directly with GIGYF2 protein to suppress IGF1R and AKT signaling pathways [15,35]. In this study, we show that RID-B treatment inhibiting GIGYF2-induced mTORC1 activation is accompanied with amelioration of cellular senescence, endothelial dysfunction, and inflammation in senescent endothelial cells with concomitant suppression of mTORC1-S6K1 pathway and reduction of STAU1 expression level. Ischemia is closely associated with endothelial cell dysfunction [61], in the middle cerebral artery occlusion (MCAO) rat model, consistently, tamoxifen treatment has been shown to reduce ischemic damage following middle cerebral artery occlusion via improving endothelial cell function [62]. In the ex vivo model, we confirm that RID-B markedly improves the endothelial function of cultured aortic rings isolated from WT-aged mice as compared to the non-treated group. Furthermore, in comparison with old WT mice, endothelial-specific GIGYF2 knockout mice have also been shown to ameliorate aging-associated vascular endothelium-dependent relaxation. These results substantially support the notion that disrupting GIGYF2 could improve cellular senescence-associated endothelial dysfunction or aging-associated vascular dysfunction. Intriguingly, we found that the expression of GIGYF2 was reduced with RID-B treatment in senescent endothelial cells. In line with previous studies, tamoxifen has been reported to downregulate the expression levels of genes ETV4 and ETV5 in benign breast tissue [63]. Nevertheless, the underlying mechanism of RID-B mediating GIGYF2 expression still deserves further investigation.
5. Conclusion
In summary, our work discloses that GIGYF2 serving as an RBP enhances the mRNA stability of STAU1 that upregulates LAMTOR4 expression through binding with its intron region, which activates the mTORC1-S6K1 signaling pathway via the recruitment of mTORC1 to the lysosomal membrane, ultimately leading to endothelial cell senescence, dysfunction, and vascular aging. Disrupting the GIGYF2-STAU1-mTORC1 signaling axis may represent a promising therapeutic approach against vascular aging and aging-related cardiovascular diseases.
Sources of funding
This work was supported by the National Natural Science Foundation of China (grant no. 82103589, and 82104155), Natural Science Foundation of Shaanxi Province (grant no. 2023-JC-YB-705), Key Research and Development Program of Shaanxi (grant no. 2018SF-223 and 2021SF-096).
Author contributions
Fanglin Niu, Yi Yu, and Yuyan Xiong designed the experiments and drafted the manuscript. Fanglin Niu, Zhuozhuo Li, Yuanyuan Ren, Zi Li, Yang Li, and Yan Zhang performed the experiments. Fanglin Niu, Junle Yang, Lu Qian, Wenzhen Shi, and Xiaobin Fan analyzed the data. Hua Guan, Jinli Li, Lele Shi provided critical appraisal. Fanglin Niu, Zhuozhuo Li, Yi Yu, and Yuyan Xiong revised the manuscript. All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgments
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102824.
Contributor Information
Yi Yu, Email: yiyu@nwu.edu.cn.
Yuyan Xiong, Email: yuyan.xiong@nwu.edu.cn.
Abbreviations
- ECs
vascular endothelial cells
- CVDs
cardiovascular disease
- GIGYF2
the mutation of Grb10-interacting GYF protein 2
- RAPA
rapamycin
- NAC
N-Acetyl-l-cysteine
- STAU1
staufen Double-Stranded RNA Binding Protein 1
- RBP
RNA binding protein
- LAMTOR4
late endosomal/lysosomal adaptor and MAPK and MTOR activator 4
- LAMP1
lysosome
- NO
nitric Oxide
- Ang II
angiotensin II
- ROS
reactive oxygen species
- ICAM-1
intercellular cell adhesion molecule-1
- VCAM-1
vascular cell adhesion molecule-1
- eNOS
endothelial nitric oxide synthase
- PD
Parkinson's disease
- mTORC1
mechanistic target of rapamycin complex 1
- S6K1
ribosome S6 protein kinase
- GIGYF1
Grb10-interacting GYF protein 1
- HUVEC
human umbilical vein endothelial cells
- SNP
sodium nitroprusside
- Ach
acetylcholine
- SASP
senescence-associated secretory phenotype
- RID-B
Ridaifen B
- ALS
amyotrophic lateral sclerosis
- IL-6
interleukin 6
- MCP-1
monocyte chemotactic protein-1
- TNF-α
tumor Necrosis Factor
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- NLRP3
nod-like receptor thermal protein domain associated protein 3
- PPARG
peroxisome proliferative activated receptor
- EMP2
epithelial Membrane Protein 2
- TXNIP
thioredoxin Interacting Protein
- SERP1
stress Associated Endoplasmic Reticulum Protein 1
- UCP2
uncoupling Protein 2
- ESR2
estrogen Receptor 2
- DKK1
dickkopf WNT Signaling Pathway Inhibitor 1
- GBP2
guanylate Binding Protein 2
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Kovacic J.C., Moreno P., Nabel E.G., Hachinski V., Fuster V. Cellular senescence, vascular disease, and aging: part 2 of a 2-part review: clinical vascular disease in the elderly. Circulation. 2011;123(17):1900–1910. doi: 10.1161/circulationaha.110.009118. [DOI] [PubMed] [Google Scholar]
- 2.Najjar S.S., Scuteri A., Lakatta E.G. Arterial aging: is it an immutable cardiovascular risk factor? Hypertension. 2005;46(3):454–462. doi: 10.1161/01.Hyp.0000177474.06749.98. [DOI] [PubMed] [Google Scholar]
- 3.Tian X.L., Li Y. Endothelial cell senescence and age-related vascular diseases. J. Genet. Genomics. 2014;41(9):485–495. doi: 10.1016/j.jgg.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 4.North B.J., Sinclair D.A. The intersection between aging and cardiovascular disease. Circ. Res. 2012;110(8):1097–1108. doi: 10.1161/circresaha.111.246876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Versari D., Daghini E., Virdis A., Ghiadoni L., Taddei S. Endothelial dysfunction as a target for prevention of cardiovascular disease. Diabetes Care. 2009;32(Suppl 2):S314–S321. doi: 10.2337/dc09-S330. Suppl 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siragusa M., Fleming I. The eNOS signalosome and its link to endothelial dysfunction. Pflügers Archiv. 2016;468(7):1125–1137. doi: 10.1007/s00424-016-1839-0. [DOI] [PubMed] [Google Scholar]
- 7.Giovannone B., Lee E., Laviola L., Giorgino F., Cleveland K.A., Smith R.J. Two novel proteins that are linked to insulin-like growth factor (IGF-I) receptors by the Grb10 adapter and modulate IGF-I signaling. J. Biol. Chem. 2003;278(34):31564–31573. doi: 10.1074/jbc.M211572200. [DOI] [PubMed] [Google Scholar]
- 8.Morita M., Ler L.W., Fabian M.R., Siddiqui N., Mullin M., Henderson V.C., et al. A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development. Mol. Cell Biol. 2012;32(17):3585–3593. doi: 10.1128/mcb.00455-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amaya Ramirez C.C., Hubbe P., Mandel N., Béthune J. 4EHP-independent repression of endogenous mRNAs by the RNA-binding protein GIGYF2. Nucleic Acids Res. 2018;46(11):5792–5808. doi: 10.1093/nar/gky198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., Sun Q.Y., Yu R.H., Guo J.F., Yan X.X.J.N.S. The contribution of GIGYF2 to Parkinson’s disease: a meta-analysis. Neurol. Sci. 2015;36(11):2073–2079. doi: 10.1007/s10072-015-2316-9. [DOI] [PubMed] [Google Scholar]
- 11.Lautier C., Goldwurm S., Dürr A., Giovannone B., Tsiaras W.G., Pezzoli G., et al. Mutations in the GIGYF2 (TNRC15) gene at the PARK11 locus in familial Parkinson disease. Am. J. Hum. Genet. 2008;82(4):822–833. doi: 10.1016/j.ajhg.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huo Z., Luo X., Zhan X., Chu Q., Xu Q., Yao J., et al. Genetic analysis of indel markers in three loci associated with Parkinson's disease. PLoS One. 2017;12(9) doi: 10.1371/journal.pone.0184269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giovannone B., Tsiaras W.G., de la Monte S., Klysik J., Lautier C., Karashchuk G., et al. GIGYF2 gene disruption in mice results in neurodegeneration and altered insulin-like growth factor signaling. Hum. Mol. Genet. 2009;18(23):4629–4639. doi: 10.1093/hmg/ddp430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim M., Semple I., Kim B., Kiers A., Nam S., Park H.W., et al. Drosophila Gyf/GRB10 interacting GYF protein is an autophagy regulator that controls neuron and muscle homeostasis. Autophagy. 2015;11(8):1358–1372. doi: 10.1080/15548627.2015.1063766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie J., Wei Q., Deng H., Li G., Ma L., Zeng H. Negative regulation of Grb10 Interacting GYF Protein 2 on insulin-like growth factor-1 receptor signaling pathway caused diabetic mice cognitive impairment. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0108559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruvinsky I., Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem. Sci. 2006;31(6):342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Rajapakse A.G., Yepuri G., Carvas J.M., Stein S., Matter C.M., Scerri I., et al. Hyperactive S6K1 mediates oxidative stress and endothelial dysfunction in aging: inhibition by resveratrol. PLoS One. 2011;6(4) doi: 10.1371/journal.pone.0019237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selman C., Tullet J.M., Wieser D., Irvine E., Lingard S.J., Choudhury A.I., et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326(5949):140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yepuri G., Velagapudi S., Xiong Y., Rajapakse A.G., Montani J.P., Ming X.F., et al. Positive crosstalk between arginase-II and S6K1 in vascular endothelial inflammation and aging. Aging Cell. 2012;11(6):1005–1016. doi: 10.1111/acel.12001. [DOI] [PubMed] [Google Scholar]
- 21.Taze C., Drakouli S., Samiotaki M., Panayotou G., Simos G., Georgatsou E., et al. Short-term hypoxia triggers ROS and SAFB mediated nuclear matrix and mRNA splicing remodeling. Redox Biol. 2022;58 doi: 10.1016/j.redox.2022.102545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng R., Cai W.K., Xu W.M., Feng Q., He G.H.J.T.R. Generation and identification of endothelial-specific Hrh2 knockout mice. Transgenic Res. 2021;30(3):251–261. doi: 10.1007/s11248-021-00244-z. [DOI] [PubMed] [Google Scholar]
- 23.Viswambharan H., Seebeck T., Yang Z. Enhanced endothelial nitric oxide-synthase activity in mice infected with Trypanosoma brucei. Int. J. Parasitol. 2003;33(10):1099–1104. doi: 10.1016/s0020-7519(03)00180-2. [DOI] [PubMed] [Google Scholar]
- 24.Di Lascio N., Kusmic C., Stea F., Faita F. Ultrasound-based pulse wave velocity evaluation in mice. J. Vis. Exp. 2017;120 doi: 10.3791/54362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng M., Whitesall S., Zhang Y., Beibel M., D'Alecy L., DiPetrillo K. Validation of volume-pressure recording tail-cuff blood pressure measurements. Am. J. Hypertens. 2008;21(12):1288–1291. doi: 10.1038/ajh.2008.301. [DOI] [PubMed] [Google Scholar]
- 26.Whitesall S.E., Hoff J.B., Vollmer A.P., D'Alecy L.G. Comparison of simultaneous measurement of mouse systolic arterial blood pressure by radiotelemetry and tail-cuff methods. Am. J. Physiol. Heart Circ. Physiol. 2004;286(6):H2408–H2415. doi: 10.1152/ajpheart.01089.2003. [DOI] [PubMed] [Google Scholar]
- 27.Carroll B., Nelson G., Rabanal-Ruiz Y., Kucheryavenko O., Dunhill-Turner N.A., Chesterman C.C., et al. Persistent mTORC1 signaling in cell senescence results from defects in amino acid and growth factor sensing. J. Cell Biol. 2017;216(7):1949–1957. doi: 10.1083/jcb.201610113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y.M., Huang A., Kaley G., Sun D. eNOS uncoupling and endothelial dysfunction in aged vessels. Am. J. Physiol. Heart Circ. Physiol. 2009;297(5):H1829–H1836. doi: 10.1152/ajpheart.00230.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber R., Chung M.Y., Keskeny C., Zinnall U., Landthaler M., Valkov E., et al. 4EHP and GIGYF1/2 mediate translation-coupled messenger RNA decay. Cell Rep. 2020;33(2) doi: 10.1016/j.celrep.2020.108262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bar-Peled L., Schweitzer L.D., Zoncu R., Sabatini D.M. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150(6):1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paul S., Dansithong W., Gandelman M., Tao Z., Ranum L., Figueroa K.P., et al. bioRxiv; 2019. Staufen Blocks Autophagy in Neurodegeneration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almasi S., Crawford Parks T.E., Ravel-Chapuis A., MacKenzie A., Côté J., Cowan K.N., et al. Differential regulation of autophagy by STAU1 in alveolar rhabdomyosarcoma and non-transformed skeletal muscle cells. Cell. Oncol. 2021;44(4):851–870. doi: 10.1007/s13402-021-00607-y. [DOI] [PubMed] [Google Scholar]
- 33.Li Z., Wang L., Ren Y., Huang Y., Liu W., Lv Z., et al. Arginase: shedding light on the mechanisms and opportunities in cardiovascular diseases. Cell Death Dis. 2022;8(1):413. doi: 10.1038/s41420-022-01200-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliveira A.C., Cunha P., Vitorino P.V.O., Souza A.L.L., Deus G.D., Feitosa A., et al. Vascular aging and arterial stiffness. Arq. Bras. Cardiol. 2022;119(4):604–615. doi: 10.36660/abc.20210708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsukuda S., Kusayanagi T., Umeda E., Watanabe C., Tosaki Y.T., Kamisuki S., et al. Ridaifen B, a tamoxifen derivative, directly binds to Grb10 interacting GYF protein 2. Bioorg. Med. Chem. 2013;21(1):311–320. doi: 10.1016/j.bmc.2012.10.037. [DOI] [PubMed] [Google Scholar]
- 36.Xie J., Wei Q., Deng H., Li G., Ma L., Zeng H.J.P.O. Negative regulation of Grb10 interacting GYF protein 2 on insulin-like growth factor-1 receptor signaling pathway caused diabetic mice cognitive impairment. PLoS One. 2014;9 doi: 10.1371/journal.pone.0108559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kietadisorn R., Juni R.P., Moens A.L. Tackling endothelial dysfunction by modulating NOS uncoupling: new insights into its pathogenesis and therapeutic possibilities. Am. J. Physiol. Endocrinol. Metab. 2012;302(5):E481–E495. doi: 10.1152/ajpendo.00540.2011. [DOI] [PubMed] [Google Scholar]
- 38.Sciarretta S., Forte M., Frati G., Sadoshima J. New insights into the role of mTOR signaling in the cardiovascular system. Circ. Res. 2018;122(3):489–505. doi: 10.1161/circresaha.117.311147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ajiro M., Katagiri T., Ueda K., Nakagawa H., Fukukawa C., Lin M.L., et al. Involvement of RQCD1 overexpression, a novel cancer-testis antigen, in the Akt pathway in breast cancer cells. Int. J. Oncol. 2009;35(4):673–681. doi: 10.3892/ijo_00000379. [DOI] [PubMed] [Google Scholar]
- 40.Yang W., Yuan Q., Zhang S., Zuo M., Li T., Li J., et al. Elevated GIGYF2 expression suppresses tumor migration and enhances sensitivity to temozolomide in malignant glioma. Cancer Gene Ther. 2022;29(6):750–757. doi: 10.1038/s41417-021-00353-1. [DOI] [PubMed] [Google Scholar]
- 41.Higashi S., Iseki E., Minegishi M., Togo T., Kabuta T., Wada K. GIGYF2 is present in endosomal compartments in the mammalian brains and enhances IGF-1-induced ERK1/2 activation. J. Neurochem. 2010;115(2):423–437. doi: 10.1111/j.1471-4159.2010.06930.x. [DOI] [PubMed] [Google Scholar]
- 42.Minamino T., Miyauchi H., Tateno K., Kunieda T., Komuro I. Akt-induced cellular senescence: implication for human disease. Cell Cycle. 2004;3(4):449–451. [PubMed] [Google Scholar]
- 43.Zou J., Lei T., Guo P., Yu J., Xu Q., Luo Y., et al. Mechanisms shaping the role of ERK1/2 in cellular senescence (Review) Mol. Med. Rep. 2019;19(2):759–770. doi: 10.3892/mmr.2018.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grzybowska E.A., Wakula M. Protein binding to cis-motifs in mRNAs coding sequence is common and regulates transcript stability and the rate of translation. Cells. 2021;10(11):2910. doi: 10.3390/cells10112910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghram M., Bonnet-Magnaval F., Hotea D.I., Doran B., Ly S., DesGroseillers L. Staufen1 is essential for cell-cycle transitions and cell proliferation via the control of E2F1 expression. J. Mol. Biol. 2020;432(13):3881–3897. doi: 10.1016/j.jmb.2020.04.016. [DOI] [PubMed] [Google Scholar]
- 46.Ramasamy S., Wang H., Quach H.N., Sampath K. Zebrafish Staufen1 and Staufen2 are required for the survival and migration of primordial germ cells. Dev. Biol. 2006;292(2):393–406. doi: 10.1016/j.ydbio.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 47.Gautrey H., McConnell J., Hall J., Hesketh J. Polarised distribution of the RNA-binding protein Staufen in differentiated intestinal epithelial cells. FEBS Lett. 2005;579(10):2226–2230. doi: 10.1016/j.febslet.2005.02.074. [DOI] [PubMed] [Google Scholar]
- 48.Gandelman M., Dansithong W., Figueroa K.P., Paul S., Scoles D.R., Pulst S.M. Staufen 1 amplifies proapoptotic activation of the unfolded protein response. Cell Death Differ. 2020;27(10):2942–2951. doi: 10.1038/s41418-020-0553-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas M.G., Martinez Tosar L.J., Desbats M.A., Leishman C.C., Boccaccio G.L. Mammalian Staufen 1 is recruited to stress granules and impairs their assembly. J. Cell Sci. 2009;122(Pt 4):563–573. doi: 10.1242/jcs.038208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paul S., Dansithong W., Figueroa K.P., Gandelman M., Scoles D.R., Pulst S.M. Staufen1 in human neurodegeneration. Ann. Neurol. 2021;89(6):1114–1128. doi: 10.1002/ana.26069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marcellus K.A., Parks T.E.C., Almasi S., Jasmin B.J. Distinct roles for the RNA-binding protein Staufen1 in prostate cancer. BMC Cancer. 2021;21(1):120. doi: 10.1186/s12885-021-07844-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Broadus J., Fuerstenberg S., Doe C.Q. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature. 1998;391(6669):792–795. doi: 10.1038/35861. [DOI] [PubMed] [Google Scholar]
- 53.Ravel-Chapuis A., Bélanger G., Yadava R.S., Mahadevan M.S., DesGroseillers L., Côté J., et al. The RNA-binding protein Staufen1 is increased in DM1 skeletal muscle and promotes alternative pre-mRNA splicing. J. Cell Biol. 2012;196(6):699–712. doi: 10.1083/jcb.201108113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu T.P., Liu X.X., Xia R., Yin L., Kong R., Chen W.M., et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34(45):5648–5661. doi: 10.1038/onc.2015.18. [DOI] [PubMed] [Google Scholar]
- 55.Dugré-Brisson S., Elvira G., Boulay K., Chatel-Chaix L., Mouland A.J., DesGroseillers L. Interaction of Staufen1 with the 5' end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005;33(15):4797–4812. doi: 10.1093/nar/gki794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim Y.K., Furic L., Desgroseillers L., Maquat L.E. Mammalian Staufen1 recruits Upf1 to specific mRNA 3'UTRs so as to elicit mRNA decay. Cell. 2005;120(2):195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 57.Roberto Zoncu, Liron, Bar-Peled, Alejo Efeyan, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., Sabatini D.M. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(2):290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang T., Wang R., Wang Z., Wang X., Wang F., Ding J. Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1. Nat. Commun. 2017;8(1):1394. doi: 10.1038/s41467-017-01567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Araujo M.E.G., Naschberger A., Fürnrohr B.G., Stasyk T., Dunzendorfer-Matt T., Lechner S., et al. Crystal structure of the human lysosomal mTORC1 scaffold complex and its impact on signaling. Science. 2017;358(6361):377–381. doi: 10.1126/science.aao1583. [DOI] [PubMed] [Google Scholar]
- 61.Keep R.F., Andjelkovic A.V., Stamatovic S.M., Shakui P., Ennis S.R. Ischemia-induced endothelial cell dysfunction. Acta Neurochir. Suppl. 2005;95:399–402. doi: 10.1007/3-211-32318-x_81. [DOI] [PubMed] [Google Scholar]
- 62.Kimelberg H.K., Feustel P.J., Jin Y., Paquette J., Boulos A., Keller R.W., Jr., et al. Acute treatment with tamoxifen reduces ischemic damage following middle cerebral artery occlusion. Neuroreport. 2000;11(12):2675–2679. doi: 10.1097/00001756-200008210-00014. [DOI] [PubMed] [Google Scholar]
- 63.Euhus D., Bu D., Xie X.-J., Sarode V., Ashfaq R., Hunt K., et al. Tamoxifen downregulates ets oncogene family members ETV4 and ETV5 in benign breast tissue: implications for durable risk reduction. Cancer Prev. Res. 2011;4(11):1852–1862. doi: 10.1158/1940-6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.