Abstract
Precise diagnosis and treatment of tumors currently still face considerable challenges due to the development of highly degreed heterogeneity in the dynamic evolution of tumors. With the rapid development of genomics, personalized diagnosis and treatment using specific genes may be a robust strategy to break through the bottleneck of traditional tumor treatment. Nevertheless, efficient in vivo gene delivery has been frequently hampered by the inherent defects of vectors and various biological barriers. Encouragingly, spherical nucleic acids (SNAs) with good modularity and programmability are excellent candidates capable of addressing traditional gene transfer-associated issues, which enables SNAs a precision nanoplatform with great potential for diverse biomedical applications. In this regard, there have been detailed reviews of SNA in drug delivery, gene regulation, and dermatology treatment. Still, to the best of our knowledge, there is no published systematic review summarizing the use of SNAs in oncology precision medicine and immunotherapy, which are considered new guidelines for oncology treatment. To this end, we summarized the notable advances in SNAs-based precision therapy and immunotherapy for tumors following a classification standard of different types of precise spatiotemporal control on active species by SNAs. Specifically, we focus on the structural diversity and programmability of SNAs. Finally, the challenges and possible solutions were discussed in the concluding remarks. This review will promote the rational design and development of SNAs for tumor-precise medicine and immunotherapy.
Keywords: Spherical nucleic acids, Tumor, Precision medicine, Molecular imaging, Immunotherapy
Graphical abstract
1. Introduction
Over the past few decades, tremendous progress has been made in exploring cancer treatment, such as discovering new programmed cell death, advancing targeted therapies, and applying immune checkpoint blockade therapies (ICB) [[1], [2], [3]]. Nevertheless, relevant studies have shown that cancer treatment is greatly hampered by intra-tumoral heterogeneity (ITH), which is a reasonable explanation for tumor chemotherapy resistance, and that ITH causes a considerable variation in positive immunotherapy responses of patients [[4], [5], [6], [7]]. Facing the current problems and challenges, we tend to put more emphasis on the application of precision therapy in cancer treatment [8]. The realization of precision medicine relies heavily on accurate diagnosis. Promisingly, with the rapid development of genomics and proteomics, precision medicine is gradually on the right track [[9], [10], [11]]. In the current stage, along with the gradual improvement of gene technology, the shortcomings of tumors are constantly exposed, and gene-level-based diagnosis as well as treatment, especially immunotherapy, can satisfy the requirements for personalized medicine to a certain extent [[12], [13], [14], [15]]. Still, gene-based precision diagnostics and therapies now face significant challenges, mainly in vivo gene delivery that must overcome degradation in peripheral blood, cell membrane-spanning, and intracellular release.
As nanotechnology advances at a rapid pace, non-viral gene delivery vectors are emerging as a powerful alternative to low-security viral vectors [16,17]. Non-viral gene vectors generally have lower toxicity and immunogenicity in comparison to viral vectors, and the raw building materials for non-viral vectors are readily available with low costs for large-scale production. Therefore, these benefits have made non-viral vectors a promising and even better alternative to viral vectors for efficient gene transfer. However, conventional non-viral gene vectors, including lipid nanoparticles (LNPs), synthetic and natural-like cationic polymers, and inorganic nanoparticles, have all faced relevant challenges. For example, LNPs suffer from a single targeting property to the liver [18] and compromised endosomal escape capacity after endocytosis [19,20]. Cationic polymers are likely to affect the cell membrane stability, leading to the potential and pore space changes of the cell membrane for inflammatory response generation [21]. In the case of inorganic nanoparticle-based gene vectors, more consideration should be focused on the low targeting performance, long-term systematic toxicity, and increased metastasis in malignant tumors [22]. All the inherent defects of these typical non-viral gene vectors led to low transfection efficiency for compromised druggability and potential clinical translations. Hopefully, in the last decade, the rapid development of spherical nucleic acids (SNAs) as novel gene vectors in the field of disease diagnosis and immunotherapy has made a distinguished contribution to precision medicine. The SNA is a highly modular and programmable structure with a dense external nucleic acid and a functionally diverse core. This kind of particular spherical structure gives the nucleic acid an unparalleled efficiency [23]. Compared to linear nucleic acids, SNA has some unique physical and chemical properties, mainly due to the dense outer nucleic acid shell of the SNA, independent of the core of the SNA. The negative charge of the dense nucleotide layer of the outer SNA and the resulting high local salt concentration can reduce the risk of nucleic acid degradation by nucleases [24]. Additionally, the high density of charge of the outer SNA can also influence the activity of enzymes that recognize foreign nucleic acids to elicit immune responses, thereby reducing the immunogenicity of SNA [25]. SNA, which is not relying on the conventional controversial EPR effect to penetrate tumor tissue [26,27], has been shown in recent years to be endocytosed into cells via scavenger receptor A [28], allowing SNA to be efficiently taken up by cells in the absence of transfection agents. More surprisingly, in addition to its high transfection efficiency, SNA can also penetrate the blood-brain barrier as well as the skin barrier [[29], [30], [31]].
SNA has demonstrated outstanding results in preclinical and clinical studies, particularly in the treatment of glioblastoma [32], Merkel cell carcinoma (MCC) [33], and COVID-19 [34]. Its multi-modular structure enables it to deliver nucleic acids, proteins [35,36], drugs [37], and fluorescent moieties for gene-based precision diagnostics [38,39]. On top of that, its programmability grants it tumor microenvironment responsiveness [40], passive targeting ability [41], and precise drug release. In this review, we discuss the applications of SNA to precision medicine, focusing on the therapeutic guidance provided by gene-level-based precision fluorescence imaging and how SNA can be used for precise adjuvant and antigen control toward optimum immunomodulation. Initially, we provide a detailed description of the SNA nanostructure, concentrating on the classification and functionalization of the cores that enable multifunctionalities. We then discuss SNA's non-invasive real-time imaging based on the genetic level, summarizing its potential applications for precision diagnosis, precise drug release, and providing precise guidance to surgical operations. Finally, we outline the application of SNA in tumor immunotherapy, mainly summarizing the way in which SNA achieves precise spatiotemporal control of antigens and adjuvants by structure for maximizing immunomodulation (Scheme 1).
Scheme 1.
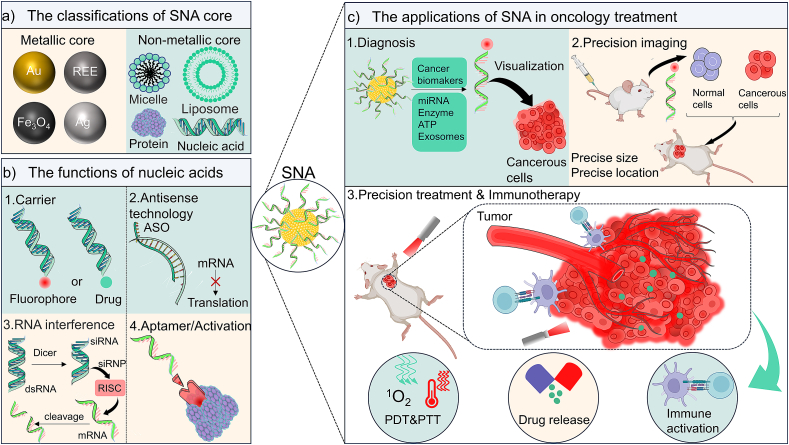
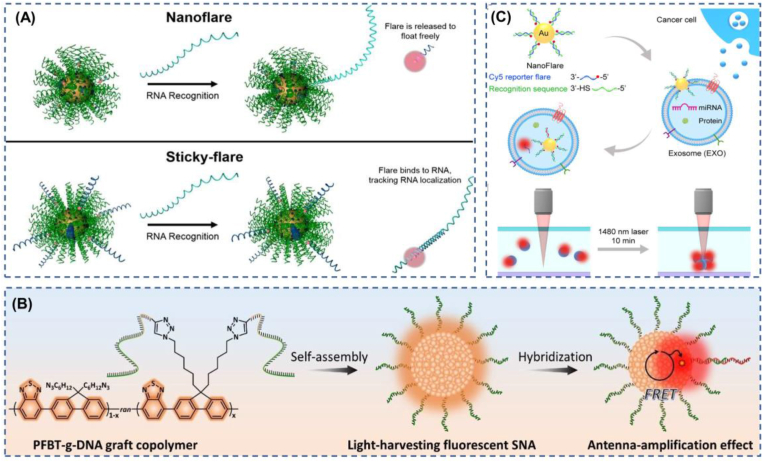
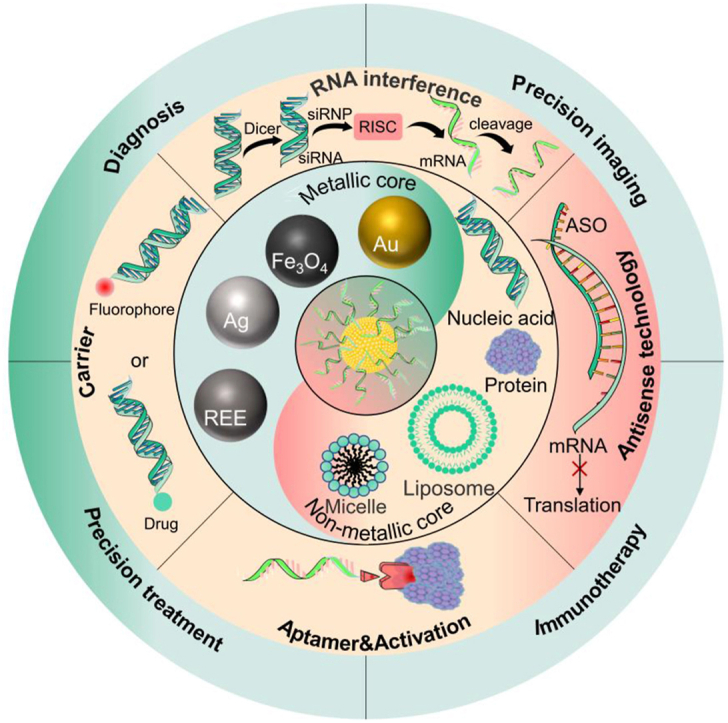
Schematic illustration of the diversity of SNA cores, the functions of nucleic acids, and their applications in treating various diseases. (a) The metallic and nonmetallic types of the SNA cores. (b) The detailed functions achieved by SNA surface nucleic acids, including the “carrier” function often used in diagnostic bio-imaging, as well as RNA interference (RNAi) and antisense technology in the field of gene therapy. (c) The typical ways of SNA are used for tumor diagnosis and treatment.
2. The structure and functionalization of SNA
SNA, in the conventional sense, is a nanoparticle containing two elements, a dense outer layer of nucleic acid and a multi-functionalized inner core. The modular structure of SNA gives it the potential to be prepared as a combined therapeutic platform [42]as well as a theranostics platform [43,44], so the design of SNA nanoparticle structures is of great importance. In addition to the usual roles of suppressing gene expression, artificially increasing gene expression, or gene modification [45], the outer nucleic acid structure can also act as a “bonding agent” through its own base complementary pairing function to create multimodal therapeutic platforms [41]. While some of the unique properties of SNAs arise from the sophisticated design of the sequence, density, and orientation of the outer nucleic acids, the core is often a source of great functional diversity in SNAs. Based on this, we will review the selection of SNA core and explore their structure.
2.1. Exploration of the selection of SNA core and their functionalization
Here we classify the core of conventional SNAs as metallic or non-metallic, as shown in Table 1. The main reason for this classification is that metals have similar structural and physicochemical properties. Metal-containing drugs have many advantages, for instance: 1) the variable coordination number of metals can provide steric structural versatility; 2) the stimulus responsiveness conferred by variable redox states [46]; 3) as well as PTT therapies based on the photothermal conversion of metals and PDT therapies with photodynamic properties [47,48]; 4) metals with luminescence and magnetic capabilities can provide diagnostic guidance. The use of metals in medicine dates back a long time, with records of their use in medicine in China and Egypt more than 2000 years ago. There is growing interest in their new applications in cancer, particularly in three new cell death modes: iron death, calcium death, and copper death. In parallel, an increasing number of metals have been found to inhibit tumor cell proliferation through different mechanisms of action, such as interfering with cellular osmotic pressure homeostasis, activating immune system pathways, and promoting cellular oxidative stress, greatly complementing cancer therapeutics [49].
Table 1.
Summary about the classification of the core in SNA and its advantages and disadvantages.
| Classification | Core | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|
| Metal-SNA | Au; Ag | Diverse physicochemical properties that give SNA systems functional versatility. | Difficult to attach DNA by covalent linkage; potentially toxic to humans | [50,[55], [56], [57],62] |
| Polymers-SNA | PLGA; poloxamer F127 | The use of polymers allows for controllability and greatly improves drug delivery efficiency | Polymeric carriers are generally ineffective on their own and are prone to cause toxic side effects on organs. | [73,74] |
| Protein-SNA | Cas9 protein; glucose oxidase | The nucleic acid shell of SNA is able to reduce the degradation of proteins by proteases, while being able to improve the uptake of proteins by cells. | Requires proteins with sites that allow nucleic acids to attach; the protein needs to be kept active | [36,95] |
| Liposomes-SNA | Lipids | High biocompatibility and wide range of drug delivery. | Does not have a long-term effect in the body; easy to leak drugs; low encapsulation rates | [83,85] |
| Nucleic acid-SNA | DNA Nanoclew; peptide nucleic acid; PSDNA | Capable of self-delivery, avoiding toxicity or immunogenicity of the carrier | Complex preparation; inherent instability | [89,91,93] |
2.1.1. Core of metals and metal compounds
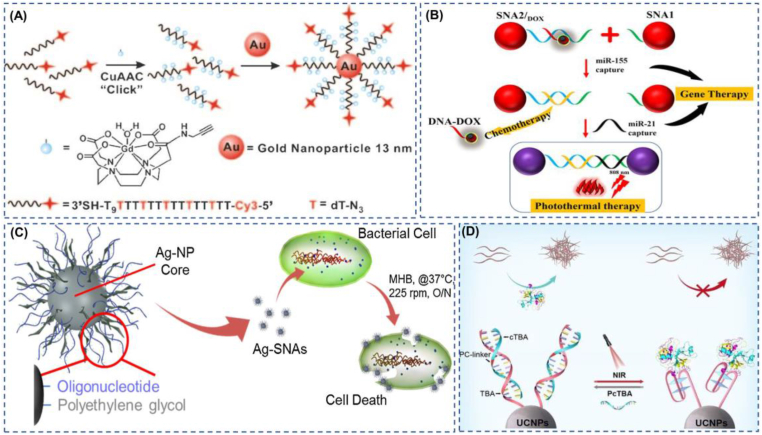
The metal currently most used in SNA core is gold nanoparticles. In 1996, Mirkin et al. reported using oligonucleotide-based methods to assemble supramolecular structures, allowing controlled and reversible gold nanoparticles [50], pioneering the use of gold nanoparticles as SNA core. Many gold nanoparticles based SNAs have been developed for different applications, and gold nanoparticles play different roles in these SNAs. In the early stages of SNA research, Au-SNA was often used as a carrier due to the mature methods for preparing Au-based SNAs and their unique nature of being taken up by cells. For example, the linking of GdIII with alkyne groups to nucleic acids modified with azide groups via click chemistry enables the synthesis of a highly loaded, highly cell-permeable, and high-biocompatible MR contrast agent (DNA-GdIII@AuNPs) [51](Fig. 1a). By functionalizing gold nanoparticles with oligonucleotides containing terminal dodecyl amines, followed by amidation linkage of DNA-AuNPs with cisplatin prodrug (Pt (IV)) containing carboxyl groups, low toxicity, and highly effective cisplatin prodrug complex was developed [52]. In later developments, the therapeutic as well as diagnostic functions of SNAs were gradually emphasized. This further enriched the functions of Au nanoparticle cores as carriers for gene drugs [53], carriers for nucleic acid probes [54], biosensors based on the Localized Surface Plasmon Resonance (LSPR) phenomenon of gold nanoparticles [55], fluorescent bursting agents [56], photothermal therapeutic agents [57] and localization agents for the spatial distribution of nanoparticles in cells or tumors [32]. Using gold nanoparticles as the core of SNA has been a long and progressive development, the most successful example of which is NU-0129. NU-0129 is an Au-SNA drug for the treatment of glioblastoma, which has proven effective in phase 0 clinical trials. This is a significant finding as it is the first time that a nanotherapeutic agent has penetrated the blood-brain barrier by intravenous infusion and altered the genetic mechanism of the tumor to cause tumor cell death [58]. There are many Au–SNAs reported at this stage, of which triple therapy using photothermal, gene therapy, and chemotherapy shows a positive outlook. For example, two SNAs act synergistically in a triple therapy where the nucleotides of SNA1 bind to half of the miRNA-21 sequence, SNA2/Doxorubicin (DOX) can release DNA-DOX when it captures miRNA-155. Then the remaining sequence of the SNA2 nucleic acid shell is able to bind to the remaining half of miRNA-21 to form an SNA aggregate in tumor cells, reducing the side effects of photothermolysis. At the same time, the aggregate prolongs the residence time of SNA in tumor cells [57] (Fig. 1b), which may be a revelation for Au-SNA nanomedicine development, exploiting the modular structure of SNA and the diversity of physicochemical properties of gold nanoparticles for multimodal combination therapy or diagnosis.
Fig. 1.
The typical applications of SNA with metals as the core. (A) The multi-modular structure of SNA as a transport carrier enables accurate multimodal imaging for biomedicine. Reproduced with permission from Ref. [51]. Copyright 2009, Wiley. (B) SNA1 and SNA2 can separately capture the oncogenes miR-21 and miR-155, enabling precision gene therapy. SNA2 can release DOX for precision chemotherapy while capturing miR-155.Then, SNA1 and SNA2 formed aggregates, which increased the retention time of gold nano at the tumor site and improved the efficiency of photothermal treatment. Reproduced with permission from Ref. [57]. Copyright 2022, Wiley. (C) SNAs with a silver core have a more powerful antibacterial effect than a single silver. Reproduced with permission from Ref. [62]. Copyright 2016, Elsevier. (D) SNA based on upconversion nanoparticles performs reversible infrared light modulation of proteins. Reproduced with permission from Ref. [67]. Copyright 2022, Wiley. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In addition to the precious metal gold as the core, silver nanoparticles can also be used as the core of SNA. In the 21st century, with the widespread use of antibiotics, bacterial drug resistance is gradually becoming an urgent problem. Targeting drug resistance genes may be a powerful solution to counter bacterial drug resistance [59]. Plus, Ag nanoparticles act as a safe, environmentally friendly natural bactericide by disrupting bacterial membranes and damaging subcellular structures in multiple ways [60]. Ag-SNA antibacterial drugs have an extensive scope for development. In 2007, a new strategy for preparing silver nanoparticle oligonucleotide adducts based on the modification of DNA by cyclic disulfide anchoring groups was reported, a synthetic approach that gives SNA enhanced stability [61]. In 2016, an SNA with a silver core for use against drug-resistant bacteria was reported [62](Fig. 1c). This Ag-SNA significantly reduces bacterial-resistant gram-negative infections and has lower cytotoxicity and minimum inhibitory concentration compared to Ag-Nps. Except for the well-known antibacterial activity of Ag, it has also been found in a large number of antiviral [63] and anticancer [64] applications, so we believe that Ag-SNA based on silver nanoparticles has a wide range of applications in the future.
In 2022, Liang et al. developed a NaGdF4 SNA, an SNA formed by siRNA coordinates with NaGdF4 nanoparticles [65]. Gd3+ can bind to phospholipids of endosomal membranes, which disrupts the integrity of membrane structure and promotes endosomal escape of NaGdF4-SNA. SNA multivalent binding with DNA as the nucleic acid shell can be utilized for nanomaterials that are difficult to functionalize by DNA covalent attachment, such as metals, metal oxides, metal-organic frameworks, and nanocarbon. SNA-mediated DNA adsorption is up to a thousand times more efficient compared to free DNA adsorption, with the DNA in the SNA shell acting partly as an attachment and partly as a function, the structure also having the specific properties of SNA [66]. Rare earth metal ions can also act as a core for SNAs, and upconversion materials containing rare earth element ions (REE), UCNPs SNAs, can be programmed to control protein activity in a spatiotemporally controllable manner by infrared light excitation [67](Fig. 1d).
2.1.2. Polymer-based core
Polymeric materials, as the main components of new drug delivery systems, have the following characteristics: 1) good biocompatibility, degradability, low toxicity, and low immunogenicity; 2) powerful drug delivery capacity; 3) controlled drug release behavior using physical, chemical, and biological parameters as well as improved target distribution of drugs; 4) improved efficacy of drugs and reduced toxic side effects [68].
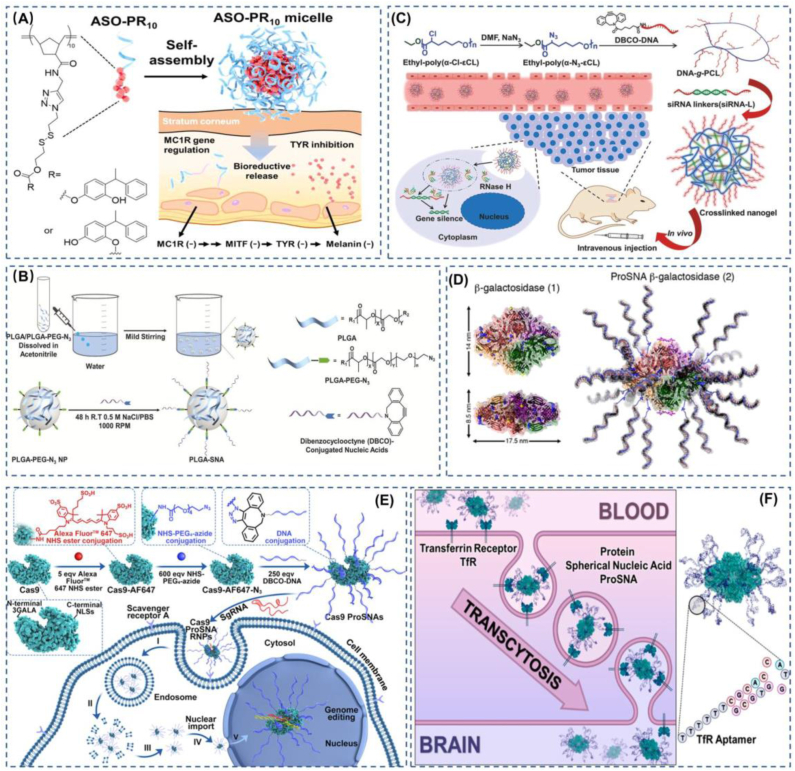
SNA inherently has excellent modularity and efficient transfection efficiency, and thus it can be used as a delivery vector for small molecule chemotherapeutic drugs. Nanoparticles made of soft materials can encapsulate many chemotherapeutic drugs, and further developed nanoparticles for the co-delivery of chemotherapeutic drugs and nucleic acid drugs have shown good performance [69]. Inspired by amphiphilic copolymer fragments forming nanomicelles, polymer-DNA conjugates self-assembled into SNAs with a hydrophobic core capable of encapsulating the hydrophobic small molecule chemotherapeutic drug BKM120 were developed, and the nano-SNAs were able to increase uptake by Hela cells, prolong circulation time and reduce leakage through the blood-brain barrier [37]. There are also micellar SNAs targeting specific tissues [70], where biocompatible polypropylene oxide (PPO) is selected as the hydrophobic portion of an amphiphilic copolymer fragment to encapsulate the hydrophobic chemotherapeutic drug DOX. The DNA portion is then conjugately linked to a complementary fragment to which folic acid is attached, enabling the nanoparticles to target the highly expressed folic acid receptor in tumor cells. The assembly of amphiphilic molecules into micelle-like SNAs allows for the covalent coupling of hydrophobic drugs with polymers to form nucleic acid-drug couples, other than encapsulating hydrophobic drugs as a core. Nucleic acid-drug couples (NADCs) hold broad promise for combination therapy yet are often underestimated. SNA, as a modular tool, can provide an ideal strategy for nucleic acid and drug co-delivery, and as a proof of concept, an SNA nanoparticle reported in 2016 is strong evidence of this. By linking paclitaxel and functional nucleic acids to a norbornene acyl polymer to form an amphiphilic molecule, which in turn assembles into a micelle SNA, the results show that the biocompatibility of oligonucleotides is an ideal carrier for hydrophobic drugs and that they can functionally complement each other [71]. Not only that, but this research team also used azidized tyrosine kinase inhibitor phenethyl resorcinol (PR) as the hydrophobic part, modified with antisense oligonucleotides targeting a key gene (MC1R) in the process of melanin synthesis in human skin via a click reaction. These couples were then able to assemble into micelle SNAs, and this SNA nanoparticle had a significant effect in inhibiting localized skin pigmentation [72](Fig. 2a). However, there are often many challenges with co-delivery vehicles, including how to mitigate low drug loading rates and the inability to control the release of drugs separately effectively. Based on this, Mirkin et al. designed PLGA-SNA to enable independent control of the surface nucleic acid shell and independent release of the encapsulated drug by changing the composition of the PLGA (PLA: PGA) [73](Fig. 2b). What is more, Mirkin et al. developed a thermally responsive micellar SNA in 2017. The SNA uses Poloxamer F127 as the core of the SNA, and lipid tail-modified nucleic acids can be inserted into the hydrophobic region of the core; this temperature-sensitive SNA can be easily assembled and disassembled using temperature changes [74].
Fig. 2.
Some exquisite examples of SNA with polymers and proteins as functional cores in practical applications. (A) A poly-PR with a bioreducible disulfide bond was used as the core, and the shell was modified with ASO to deregulate the gene for synthesizing tyrosinase. This SNA demonstrates a strong ability to treat skin diseases. Reproduced with permission from Ref. [72]. Copyright 2021, American Chemical Society. (B) The use of PLGA as the core of SNA enables the encapsulation of hydrophobic drugs and the controlled release of the drugs. Reproduced with permission from Ref. [73]. Copyright 2018, Wiley. (C) Nanogel-like SNA not only protects functional nucleic acids from nuclease degradation but also has size-controllable properties. Reproduced with permission from Ref. [75]. Copyright 2018, Wiley. (D) SNA with protein as the core is capable of efficiently transfecting proteins into cells. Reproduced with permission from Ref. [80]. Copyright 2015, American Chemical Society. (E) The conversion of Cas9 proteins into SNA structures through chemical design and synthesis allows efficient access to the nucleus while maintaining the biological activity of Cas9 proteins. This provides a powerful strategy for a wide range of CRISPR-Cas9-based therapeutic applications. Reproduced with permission from Ref. [36]. Copyright 2022, American Chemical Society. (F) Modification of SNA with TfR aptamers enables the delivery of functional proteins across the blood-brain barrier into the brain or CNS. Reproduced with permission from Ref. [81]. Copyright 2022, American Chemical Society.
In our general understanding, SNA as gene therapy is achieved through its outer dense oligonucleotide, but the exposure of the oligonucleotide to the physiological environment cannot avoid causing damage to the therapeutic oligonucleotide, so it is an excellent option to bury the therapeutic nucleotide within the outer nucleic acid. Based on this, Ding et al. developed an SNA-like nanoparticle [75](Fig. 2c). This SNA nanoparticle is a spherical nanohydrogel formed by cross-linking siRNA with that DNA which is grafted onto PCL, and this nanoparticle also shows physiological stability and resistance to enzymatic degradation similar to SNA, as well as good siRNA release properties. Acidic mucopolysaccharide hyaluronic acid, which is widely available in the human extracellular matrix, has also been used as a carrier for SNA for the treatment of hyperplastic scarring (HS) and has shown significant results in both in vivo and in vitro experiments [76]. Polymer-based SNAs are attractive because they can be programmed to achieve the multifunctional nature of polymers. Although polymer based SNAs have been developed at this stage, there is still a gap in translating marketed products. Therefore, further optimization of polymeric SNA delivery systems is still needed.
2.1.3. Protein-like core
Protein therapies have shown an increasingly important role in recent medical developments, which often utilize antibodies and a variety of enzymes that can regulate cellular functions to manipulate normal cellular processes or replace abnormal proteins in cells [77]. Recently, protein-based oncology therapies have been rapidly evolving [78]. But protein agents acting from in vitro to in vivo are often subject to three barriers, 1) low stability, low permeability, and low aggregation due to extracellular barriers, 2) low uptake due to cell membrane barriers, and 3) intracellular barriers, main susceptibility to enzymatic degradation, which seriously hinder the prospects of protein therapies [79]. In 2015, protein delivery mediated with a DNA oligonucleotide shell that not only protected the natural conformation of the protein but also allowed a 280-fold increase in cellular uptake caught the eye [80](Fig. 2d). Inspired by this report, Huang et al. developed an SNA based on the function of the CRISPR-Cas9 gene editing system. This Pro SNA, with the Cas9 protein as its core, can be effectively taken up by cells without the support of transfection agents as well as electroporation techniques [36](Fig. 2e). Similarly, since SNAs can be modularly programmed, modification of SNAs can achieve different functions, such as increasing the in vivo blood-brain barrier targeting of Pro-SNA using transferrin aptamers [81](Fig. 2f). This work shows that the appropriate addition of transferrin (Tfr) aptamers to the nucleic acid shell of ProSNA to target binding to transferrin receptors present on blood-brain barrier (BBB) endothelial cells can in turn increase the penetration of the blood-brain barrier to protein SNAs, allowing greater access of protein SNAs to the brain and central nervous system (CNS). ProSNAs are modular structures that show promise in disease diagnosis due to the high uptake of SNA by a large number of cells. At the same time, due to the protection of the SNA shell and the ability to regulate biodistribution, ProSNAs are able to achieve long circulation and wide distribution in vivo compared to ordinary proteins [35].
2.1.4. Liposomal spherical nucleic acid (LSNA)
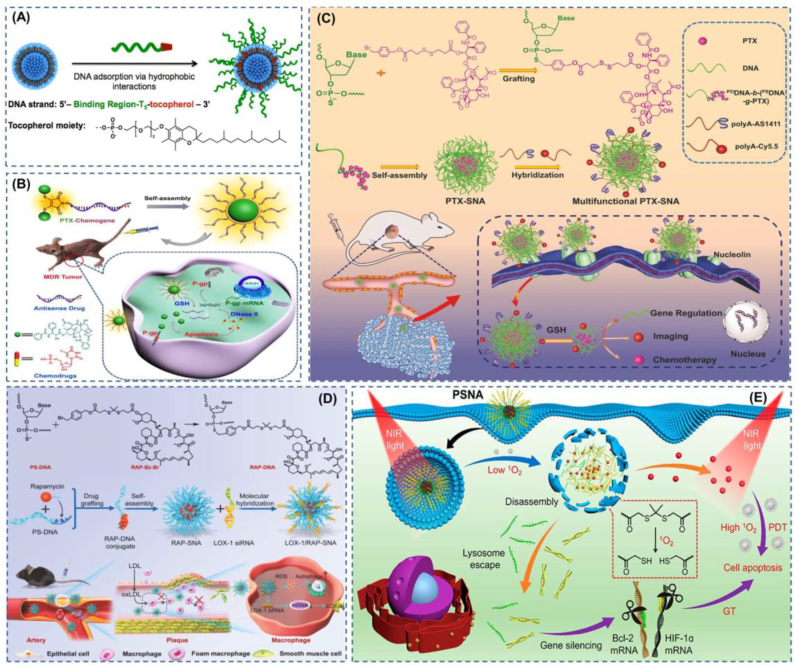
It is not hard to understand the broad application of liposomes in SNA as a common nanocarrier based on their biocompatibility and low toxicity [82]. In 2014, Mirkin et al. developed a metal-free liposomal SNA (LSNA), a novel SNA that is less prone to aggregation and precipitation due to the charged nature of the nucleic acid shell compared to its liposomal core. The liposomal SNA can also be highly taken up by cells like Au-SNA and has little toxicity to cells at high concentrations [83](Fig. 3a). In the subsequent development of liposomal SNAs, a liposomal SNA with a non-phospholipid nucleolipid was developed, which was synthesized by coupling a dibenzocyclooctyne (DBCO)-modified oligonucleotide with an azide-modified 1,3-hexadecylpropan-2-ol (DHP)-free copper click reaction [84]. Based on the preparation of nucleolipids by conventional click chemistry reactions, where the metal catalysts and radical catalysts used may introduce toxicity to the nanocarrier system, thiol-ene click reactions can be used to synthesize nucleolipids to prepare liposomal SNA with less toxicity [85]. In addition to the conventional liposomal SNA with nucleic acid as the shell and liposomes as the nucleus, liposomal encapsulated SNA with targeted functionalization can be used to achieve better therapeutic results. For example, liposomes can be modified with the brain-targeting peptide apolipoprotein E (ApoE) or rabies virus glycoprotein (RVG29), and then the liposomes encapsulate the SNA for RNAi treatment of glioblastoma. This approach may offer a viable strategy for brain diseases and central neurological disorders [86].
Fig. 3.
Liposomal SNAs and Carrier-free self-delivery SNAs. (A) Liposomal SNA with stable multi-modular structure. Reproduced with permission from Ref. [83]. Copyright 2014, American Chemical Society. (B) FdU is integrated into the gene to form a chemogene, and then two PTXs are modified at the oligonucleotide ends to form a conjugate that self-assembles into an SNA, which has potent anti-tumor effects. Reproduced with permission from Ref. [90]. Copyright 2020, Wiley. (C) The use of thio-modified nucleic acids as carriers for hydrophobic chemotherapeutic drugs has high drug loading rates, and the unloaded portion of the nucleic acid can also be modified with probes or aptamers by Watson-Crick pairing, etc. Reproduced with permission from Ref. [91]. Copyright 2019, Wiley. (D) Co-delivery of siRNA and RAP to atherosclerotic plaques and implementation of synergistic therapy can significantly slow down the progression of atherosclerosis. Reproduced with permission from Ref. [92]. Copyright 2022, Wiley. (E) Self-delivery SNA that encapsulates photosensitizers in a hydrophobic core enable passive targeting of light responsiveness. Reproduced with permission from Ref. [93]. Copyright 2021, American Chemical Society.
2.1.5. Nucleic acid-based core
In order to reduce the long-lasting and various safety and efficiency problems associated with SNAs such as liposomes, inorganic nanoparticles, micelles, and other cores [87], R&D workers have turned their attention to natural genetic materials. Among these, DNA is a good choice as an inherently biocompatible substance and has excellent properties such as low immunogenicity, biodegradability, and targeting using aptamers. Kim et al. used amphiphilic lipid DNA-loaded siRNA to derepress USE1 expression in lung tumors [88]. Ruan et al. have developed an SNA with DNA Nanoclew (DC) as the core. The new SNA shows significant advantages over metals and polymers due to the excellent biocompatibility of DNA, and the SNA also exhibits promising biological effects [89]. In an effort to minimize the side effects of SNA vectors, a vectorless SNA has emerged that is formed by paclitaxel (PTX)-chemical gene self-assembly. This coupling is formed by the binding of two PTXs to antisense nucleotides (chemical genes) integrated with FdU via fluorescent disulfide-maleimide (DTM) [90](Fig. 3b). To apply the superiority of DNA nucleic acid as a carrier to a greater extent, the researchers cleverly designed the DNA into a diblock DNA strand containing a standard phosphodiester fragment (PODNA) and a phosphorothiolate fragment (PSDNA). PSDNA is a double-bonded oxygen on the phosphodiester group on DNA modified by a thio reagent to form a double-bonded sulfur. This nucleophilic site greatly enhances the ability of DNA to be modified, for example, by attaching the hydrophobic drug PTX so that the DNA becomes an amphiphilic DNA hybrid copolymer that can then self-assemble into a nanomicellar SNA. The normal PODNA as the outer shell of the nanosphere nucleic acid can be modified with a complement fragment attached to the targeting aptamer or fluorescence for functionalization, and the results show both the tumor targeting and tumor suppression effects of the nanoparticles were greatly enhanced [91](Fig. 3c). Zhang et al. prepared rapamycin-containing SNA using PSDNA molecules as carrier materials for plaque targeting imaging and synergistic treatment studies of atherosclerosis [92](Fig. 3d). The self-assembled SNA structure was prepared by thio-modifying a portion of the nucleic acid molecule and then grafting benzyl-bromide-modified rapamycin onto the modified nucleic acid molecule to form an amphiphilic molecule, with the ungrafted portion of the nucleic acid serving as a dense shell for the SNA and the ungrafted portion also modified with Gd-DOTA by base complementary pairing for imaging or with LOX- 1 siRNA to achieve synergistic anti-atherosclerosis. More interestingly, Zhang et al. have developed a carrier-free, near-infrared (NIR) light-controlled, self-delivered SNA (PSNA). The SNA shell is a hydrophilic therapeutically functional siRNA, and the core is a hydrophobic peptide nucleic acid-based antisense oligonucleotide (pASO) and a photosensitizer (PS), which generates single linear oxygen 1O2 upon irradiation with near-infrared light. 1O2 cleaves the linker between the siRNA and pSNA, which allows the nanocarrier to be used in tumor site-specific passive targeting for a perfectly integrated therapy [93](Fig. 3e).
A typical multinodular structure enables self-delivery SNAs with precise spatiotemporal control of multiple therapeutic reagents, but such a structure fails to achieve simultaneous implementation of multiple gene therapies. The nucleic acid amplification method based on rolling circles amplification (RCA) may provide a solution strategy. A notable example is Shi's vectorless self-delivery SNA, capable of aggregating multiple genes, which was self-assembled by rolling up and replication of customized ring templates, including caspase-3 gene-inhibiting antisense oligonucleotide (ASO) and transferrin (TfR) aptamers. The resulting SNA has higher treatment effects on ischemic stroke due to high blood-brain barrier penetration and caspase-3 gene silencing competency [94].
2.2. Structural modification of SNA and modification of its synthesis method
With the booming development of nanomedicine, researchers began to pursue more efficient and high-quality nano-design ideas. SNA binds to serum proteins in the blood to form protein crowns [96], which is one of the main problems affecting the efficacy of SNA. Because protein crown affects the speed of phagocytosis, reduces blood circulation time, affects absorption, effect, etc. With the widespread use of PEGylated nano in nanomedicine, it may provide a solution to this problem, and its improvement is noticeable, but some inherent disadvantages of PEGylated nanoparticles limit its development. In SNA, although PEGylated SNA can prolong their circulation time in vivo, they may also reduce cellular uptake, and the amount of PEG needs to be adjusted to achieve prolonged circulation time and not affect endocytosis [97]. Although the formation of protein crowns is challenging to avoid, there are two sides to everything. A strategy that uses specific protein crowns to reduce immune system phagocytosis of nanocarriers opens up research ideas [98], and specific protein crowns applied to SNAs may have active targeting as well as improved biodistribution, in addition to reducing nonspecific uptake [99,100]. To balance the cycle time as well as cellular uptake, Mirkin et al. modified the EK peptide with a linker that is cleavable by matrix metalloproteinases (MMPs) upregulated in tumors. EK peptide as an amphoteric ionic peptide can play a similar function to PEG in prolonging blood circulation time, and when EK peptide-modified SNA enters MMP enzyme-upregulated tumor cells, it can be cleaved by the enzyme, preventing the peptide from blocking the binding of SNA to the uptake receptor [40]. Besides using PEGylation or protein crown to improve the stability and uptake efficiency of SNA, glycosylation to modify the SNA shell of nucleic acids can also cloak SNA and facilitate its effect [101,102].
3. Application of SNA in tumor diagnosis and imaging
The leading cause of death in cancer patients is delayed diagnosis as well as treatment. By diagnosing and treating cancer in the preinvasive stage, early cancer detection may significantly influence the control of the disease and thus improve the chances of successful treatment. However, traditional cancer screening methods, such as low-dose computed tomography screening for lung cancer [103], mammography for breast cancer [104], prostate-specific antigen (PSA) blood tests [105], and colonoscopy for colon cancer [106], are not accurate enough for early detection due to over-diagnosis issues. Using tumor biomarkers in diagnosis can help reduce the risk of overdiagnosis. For example, risk stratification biomarkers may help to distinguish low-risk populations from high-risk populations prone to develop advanced malignant tumors. Biomarkers may be less likely to detect precancerous and painless tumors during early tumor detection and prognosis than imaging and help to distinguish tumors with inert or aggressive behavior. In this area, continued advances may reduce the burden and harm of overdiagnosis [107]. Cancer biomarkers are highly selective, specific, and reproducible, which play an essential role in the diagnosis, prognosis as well as efficacy prediction of cancer patients. However, the sensitivity and specificity of currently clinically applied biomarker assays are low and need to be improved. Nanoparticle-based assays offer ultra-high sensitivity and specificity as a promising method in cancer biomarker detection [108]. Furthermore, the detection is still simplified by technologies focusing on improving the detection sensitivity and building nanosensors for biomarker amplification systems [109,110]. However, facing the complex physiological environment, the main challenge in the development of nanotechnology-based assays at this stage is to overcome the precise detection of ultra-low biomarker concentrations as well as point-of-care testing [111].
3.1. Benefits of SNA for diagnosis
3.1.1. Outstanding contribution of SNA structure in biodiagnostics
DNA-based probes constitute a versatile biometric platform due to their ability to recognize nucleic and non-nucleic acid targets, ease of synthesis and chemical modification, ease of interfacing with signal amplification schemes, and inherent biocompatibility. However, in live cell analysis, such linear DNA probes do not efficiently cross cell membranes without transfection reagents and are susceptible to rapid degradation by nucleases in the cellular environment. The arrangement of nucleic acids around the nanoparticle core in an SNA structure at high density leads to active cellular uptake, enhanced resistance to nuclease degradation, and enhanced binding to complementary targets [112](Fig. 4a). In addition, DNA-functionalized gold nanoparticle probes have higher binding constants compared to molecular fluorescent probes of the same sequence, which greatly increases their sensitivity in analytical assays [113]. In 1997, Mirkin et al. reported a colorimetric biosensor based on the aggregation of AuNPs. They modified oligonucleotides on the surface of AuNPs as probes, and when target DNA is present, the probes hybridize to form a multimeric network, the AuNPs aggregate, and the color of the solution is visible to the naked eye from red to blue, which is one of the most typical biospecific reaction to make plasma nanoparticles aggregation of the sensor, since then began a new generation of medical diagnostic technology based on nanomaterial colorimetric method [114]. In 1999, a method for assembling semiconductor quantum dots of cadmium selenide into macroscopic materials in three or two dimensions using the principle of DNA base complementary pairing was reported [115]. This method using DNA nucleic acid shells as cross-linkers to connect nanomaterials assembled into macroscopic materials enables a sensitizing effect on the materials [116]. SNA probes have better performance in comparison to nucleic acid molecular probes in terms of cellular uptake properties, resistance ability to enzymatic degradation, and sensitivity. Moreover, SNA nanoprobes can perform self-structure optimization for improved detection effects due to their own modular structure and programmable performance. The traditional nucleic acid functionalization of AuNPs using Au–S bonds has been extensively investigated together with the exploration of detection applications. Nevertheless, biological thiols in the body (e.g.a high intracellular GSH concentration) are generally susceptible to any interferences with the Au–S bond for a high likelihood of false-positive events [117,118]. Additionally, the electrostatic repulsions between negative charges and steric hindrance result in a compromised DNA density on the surface of gold particles [119]. To address the aforementioned critical issues, the nucleic acid functionalization methods for AuNPs have been continuously innovated. A notable example is the development of SNAs based on the Au–Se bond with a more resistant ability to any interferences with biothiols than Au–S-based analogs. Interestingly, a tighter connection between the nucleic acid and the gold nano core could be realized for more stable and improved accuracy of the gold-SNA-based nanoprobe via replacing the Au–S bonds with Au–Se bonds due to the greater sensitivity of Se than that of S [120,121]. Yet, the compromised stability of Au–Se due to the high reactivity of Se is also unbeneficial for the potential practical applications. Qing et al. later developed Pt–S bond-based nanoflares with enhanced SNA stability in biological conditions for high-fidelity detection of miRNA-21 microRNAs in experiments [122]. Besides, nucleic acid functionalization of AuNPs using a non-covalent binding approach has been confirmed to be a useful strategy. The use of polyA as a linker between DNA and AuNPs can achieve SNA probe sensitivity control by adjusting the length of the polyA tail, thus enabling reliable detection with a lower detection limit [123]. PolyA-mediated SNAs have been shown to be highly sensitive for imaging the tumor-associated nucleic acid marker miRNA-155 in living cells [124], because polyA20 tails as attachment blocks for nucleic acid probes offer programable modulation of the DNA loading density on the gold nanointerface for a greatly reduced detection limit of SNA probes compared to that of thiolated probes without surface density modulation [125]. Programmable DNA detection in a serum environment for simultaneous multicolor detection of pancreatic cancer-associated three different microRNAs can be facilitated by modulating the length of the diblock DNA probe on gold nanoparticles as the anchoring spacer of the polyadenine [126]. However, the Poly-A bond has a low adsorption affinity relative to thiols and is susceptible to biothiols, which limits its utility. Still further, Zeng et al. developed a method for modifying AuNPs using 2′-Fluorinated DNA (FDNA), leading to Au/FxDNA with good colloidal stability at high salt concentrations as well as in a wide pH range [127].
Fig. 4.
Some common design concepts and applications of SNA-based bio-diagnostics and imaging. (A) Mode of nanoflare and sticky-flare action on fluorescence imaging. Reproduced with permission from Ref. [112]. Copyright 2020, American Chemical Society. (B) Based on fluorescent π-conjugated polymers with light-trapping capability, it enables amplified detection of low concentrations of nucleic acids. Reproduced with permission from Ref. [130]. Copyright 2022, Wiley. (C) Nanoflares can precisely identify microRNAs in tumor exosomes and then combine them with thermophoresis to amplify the sensitivity of the detection. Reproduced with permission from Ref. [138]. Copyright 2020, American Chemical Society.
The core of SNAs can also be selected to achieve assay diversity. For example, the valence tunability of SNAs (FNA-mSNA) using framework nucleic acids (FNAs) that provide high valence controllability as the core can modulate biosensing performance to be suitable for bioassays in different situations [128]. A new strategy for chemical analysis of living cells using protein SNAs, known as ProSNAs constructs, allows for the analytical detection of targets through highly programmable nucleic acid shells or functional protein cores. As a proof-of-concept, i-base sequences are utilized as nucleic acid recognition elements to detect pH in living cells. By interfacing the i-base sequence with a forced insertion readout, a quencher-free approach against false-positive signals is introduced, overcoming the limitations of conventional fluorophore/quencher-based gold nanoflare correlation. Moreover, this study demonstrates activity-based amplified sensing of glucose using glucose oxidase as a functional protein core [95]. Additionally, SNA probes based on the upconversion material LRET effect and catalytic hairpin assembly (CHA) can simultaneously achieve the robustness, sensitivity, and uniformity required to achieve the ideal probe [129].
3.1.2. SNA for ultra-low concentration molecular detection
For low-concentration nucleic acid detection, DNA strand cleavage by nucleases to achieve recovery of target miRNAs, as well as recruitment of large amounts of electro-chemicals [Ru(NH3)6]3+ by hybridization of SNAs with DNA1 on electrodes, enables double-amplified signal detection. Experiments have shown that this strategy is highly sensitive to detecting down-regulated microRNAs in influenza virus-infected cells [54].To detect deficient concentrations of nucleic acids in complex biological samples requiring highly sensitive and specific fluorescent probes, Xiao et al. developed an amplified in situ nucleic acid detection and imaging SNA tool that can be applied at the single cell level, using FRET technology with light-collecting antennae fluorescent π-conjugated polymers (FCPs) as hydrophobic cores to enhance signal transduction for nucleic acid detection [130](Fig. 4b). Qu et al. used SNA to design a DNA Walker with directional response capability, using exonuclease III as a driving force to provide a nucleic acid detection tool with autonomous movement and cascade signal amplification recognition [131].
3.2. Major pathways of SNA in enabling biodiagnostics and imaging
In 2007, Seferos et al. developed the nanoflare, the only new intracellular probe that combines cell transfection, enzymatic degradation protection, and oligonucleotide acid detection [132], whose main conformation is an oligonucleotide capture sequence probe attached to gold nanoparticles and a short nucleotide sequence with a fluorescent molecule hybridization, in this conformation, the proximity of the fluorophore to the gold nanosurface causes fluorescence quenching [133]. Furthermore, when the target binds to the capture sequence, a fluorescent chain is released, and a corresponding increase in fluorescence is detected by the displacement of the fluorescent chain [134]. In later developments, nanoflare has been refined; for example, light-activated nanoflare has now been developed [135], which has high spatiotemporal control performance and make up for the shortage of target recognition response triggered by traditional nanoflare once they enter the cell, which can improve the detection sensitivity to a certain extent. Furthermore, there are multiple application nanoflare [136], which can detect multiple sequences simultaneously using a single structure. These advances have led to multiple applications of nanoflare, such as the identification of skin scars [38], identification and enrichment of skeletal stem cells [137], identification of cancer cells [138](Fig. 4c), and assessment of drug nephrotoxicity to human kidney tubules [139]. Traditional nanoflare is only able to detect the “presence” or “absence” of a target, but since the quantification and location of RNA are critical for the diagnosis of some diseases, sticky-flare has been developed to study the dynamics of RNA in cells and to perform quantitative and spatial localization studies [140]. Sticky-flare differs from nanoflare in that sticky-flare bind specifically to the target with a fluorescent reporter strand rather than SNA. Sticky-flare has also been used in several applications, such as in situ monitoring of telomerase in tumor cells [141] and tracking of p21 mRNA in living cells for visual assessment of tumor treatment efficacy [142]. In addition to nanoflare and sticky-flare, metal aggregation or dispersion with localized surface plasmon resonance phenomenon is also used to achieve visual detection, which is discernible to the naked eye and less costly. A colorimetric detection method for HIV-1 nucleic acid based on iambic structure-controlled gold nanoparticle SNA assembly and dispersion is reported to be able to easily detect target DNA with picomole amounts of target DNA observed to the naked eye, which may be able to be extended to the detection of other viruses [55].
3.3. SNA for precise diagnosis and treatment of tumors
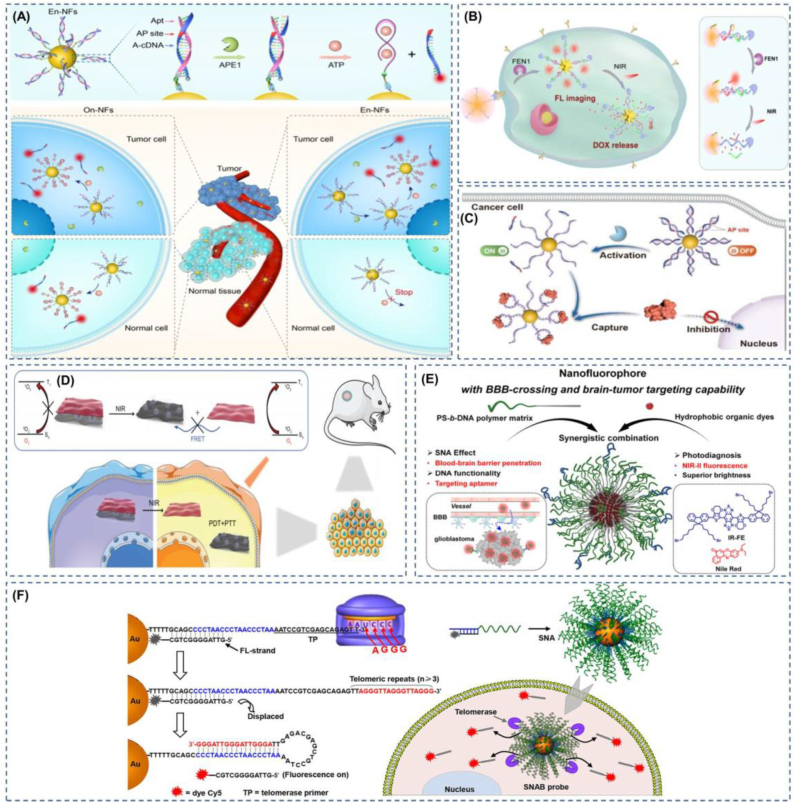
Fluorescent probes are ideal tools for the detection of tumor-related biomarkers and the visualization of tumors for in situ imaging. This non-invasive detection can provide immediate, highly specific, and accurate imaging [143], and, in addition to its active role in early tumor diagnosis, intraoperative imaging, and prognosis, its application in providing precise drug release at the tumor site during treatment is also promising [144]. At this stage, with the rapid development of SNA, its application area is becoming more and more extensive, and the detection method is becoming more and more novel. In tumor biomarker diagnosis, SNA can provide accurate detection of tumor-specific mRNA, miRNA, exosomes, enzymes, ATP, and other biomarkers. Nanoflares are capable of qualitatively detecting “yes” or “no”. Furthermore, sticky-flares provide quantitative and spatiotemporal resolution [145]. Pathogenic nucleic acids or disease nucleic acid markers have an indelible role in the diagnosis of some diseases. However, this type of nucleic acid is often only quantitatively up- or down-regulated. It is also present in healthy individuals to a greater or lesser extent, so a single test for the presence or absence of nucleic acid may give false positives. Exploiting the diversity of nucleic acid sequences may provide a solution to this dilemma. Jou et al. designed a nucleic acid functionalized with CdSe/ZnS quantum dots, and the nucleic acid includes two sequences; sequence one is the recognition sequence of prostate cancer biomarker miRNA-141, which can detect the presence of miRNA-141, and sequence two is the telomerase primer sequence, which can detect the effect more accurately [146]. In situ detection of cancer cells is vital for early tumor screening, but conventional SNA nanoflares probes are often limited by off-target interference. Designing enzyme-triggered nanoflares by adding sites activated by apurinic/apyrimidinic endonuclease 1 (APE1) overexpressed in the cytoplasm of cancer cells enables the development of tumor-specific molecular nanoflares [147](Fig. 5a). Using FRET nanoflares, false positive signals generated by biological or thermodynamic systems can be minimized when detecting mRNA in living cells [117].
Fig. 5.
The figure shows the precise tumor diagnosis based on biological responsiveness as well as physical stimulus responsiveness. Apart from that, there is also precision imaging of tumors. (A) Designing tumor-specific enzyme-triggered sites in nucleic acid sequences that can specifically identify tumor cells. Reproduced with permission from Ref. [147]. Copyright 2022, American Chemical Society. (B) Schematic of SNA application on theranostics. Reproduced with permission from Ref. [150]. Copyright 2021, American Chemical Society. (C) SNA activated by intracellular specific enzymes is used for the selective regulation of proteins. Reproduced with permission from Ref. [151]. Copyright 2023, Wiley. (D) Photothermal mode modulation within situ targeted photodynamic therapy. Reproduced with permission from Ref. [41]. Copyright 2022, Wiley. (E) Non-invasive imaging of glioblastoma using SNA-transported NIR–II–Emitting Dye. Reproduced with permission from Ref. [157]. Copyright 2022, Wiley. (F) The use of telomerase enables specific diagnostic imaging of tumor cells, overcoming the challenge of precise diagnosis due to tumor heterogeneity. Reproduced with permission from Ref. [158]. Copyright 2018, American Chemical Society.
The ability to detect tumor biomarkers more accurately using SNA provides excellent conditions for precise treatment at the tumor site. For example, the unique oligonucleotide stem-loop structure of the fluorescent DOX inserted into the molecular beacon (MB) leads to fluorescence quenches. When the MB encounters the target DNA or RNA, it can bind to the target to release DOX and restore the cytotoxicity of DOX [148]. Subsequently, the group designed a dual-targeted nano-SNA for tumor cell imaging and therapy, capable of detecting tumor cell-specific mRNA and releasing DOX in an mRNA concentration-dependent manner [149]. Using gold nanostars (AUNs) functionalized with reporter chain substrates that can be cleaved by the tumor biomarker flap-like endonuclease (FEN1), tumor cells can be precisely identified, and then the AUNs can be made to generate heat at the desired site by near-infrared to release DOX [150](Fig. 5b). The use of enzyme-activated SNA can now also precisely and reversibly regulate the location of proteins in tumor cells. Li et al. modified the activatable site of APE1 enzyme on the nucleic acid sequence, which is widely present in the cytoplasm of tumor cells and in the nucleus of normal cells, thus achieving tumor cell-specific protein control [151](Fig. 5c). Apart from enzyme activation, there is also ATP-activated SNA for tumor imaging and therapy, using BHQ2 to quench the fluorescence of Ce6 and inhibit the ability to produce singlet oxygen. After targeting tumor cells through AS1411, ATP turns on the fluorescence of Ce6 and the ability to produce singlet oxygen and releases DOX [152]. Nevertheless, in their complex biological systems, such stimulus-responsive targeting is easily disturbed, and targeting using external stimuli can well avoid such disturbances. Du et al. used SNA containing an NMR imaging agent as a linker to connect the fluorescent photosensitizer PPF-1 and black phosphorus sheets and were able to achieve photothermally triggered precision photodynamic therapy by changing the ratio of nucleic acid sequences (A + T) and (C + G) [41](Fig. 5d).
3.4. SNA guides precision surgery
At this stage, surgical resection remains the primary treatment for primary and regional solid tumors. However, in surgery, physicians subjectively evaluate tumor tissue versus normal tissue primarily through textural palpation and visual tissue differences, which may lead to incomplete surgical resection or accidental removal of normal tissue [153]. Many imaging techniques have been applied to surgical procedures for tumors to date, but there are some drawbacks, such as the use of MRI causing surgical interruptions and perhaps fluorescent optical contrast agents more appropriate for the intraoperative setting [154]. Molecular fluorescence imaging of tumors enables non-invasive visualization of tumor-associated marker molecules at different stages, thus facilitating precise diagnosis and treatment of cancer [155,156]. Bladder cancer is challenging to precisely outline tumor margins due to the lack of effective tumor margin recognition techniques, and it is still extremely difficult for surgical treatment. SNA nanoprobes modified with R11 peptide-promoting molecular beacons are used to achieve precise determination of bladder tumor boundaries through the endosomal escape pathway [39]. Besides, the researchers used the amphiphilic DNA block copolymer PS-b-DNA as a polymer vector, loaded with nanofluorophores that can emit near-infrared-II rays and cross the blood-brain barrier, which showed superior performance to the PS-b-PEG polymer-based nanofluorophores in the diagnostic imaging of glioblastoma [157](Fig. 5e). The diagnostic and imaging identification of tumors at this stage is diverse, as the heterogeneity among tumors and the lack of tumor-specific biomarkers for all tumors have led to a lack of methods to identify all tumors with high accuracy at the molecular, cellular, and systemic scales. The detection of telomerase may be a breakthrough point to solve this problem. The uncontrollable growth of tumor cells is a significant feature of tumors, and uncontrollable cell growth requires high telomerase activation. However, the activity is inhibited in normal human cells. Therefore, using an SNA with a telomerase primer (Tp) long strand and a short strand carrying a fluorescent group complementary to the long strand, when telomerase is present, telomerase will extend the TTAGGG sequence of Tp and then form a thermodynamically more stable hairpin-like DNA structure, displacing the fluorescent strand and restoring fluorescence for easy detection, which is expected to affect clinical diagnosis as well as surgical guidance [158] (Fig. 5f). Studies have shown that the choice of excision thickness for skin tumors significantly affects patient mortality [159]. SNA nanoflares can be used for rapid, real-time monitoring of skin scar types and their severity. This method avoids traditional invasive biopsies of scar tissue that result in new scars or worsening of original scars. This high specificity and sensitivity of nanoflares as a non-invasive diagnostic tool can be extended to the diagnosis of skin diseases characterized by overexpression of other nucleic acid markers and to the screening of dermatologically based drugs with high effectiveness; in particular, we should note that this tool can determine the extent of skin tumor excision and provide guidance for the surgical excision of skin tumors [38], improving the efficiency of diagnosis and treatment.
4. Application of SNA in immunotherapy
The human innate immune recognition receptors for nucleic acids can be divided into two categories, DNA and RNA sensors. DNA immune recognition sensors are mainly Toll-like receptors (TLRs) located in endolysosomes and cyclic GMP-AMP synthases (CGAS) located in the cytoplasm, depending on their location [160]. RNA immune recognition sensors are another important intermediate of the innate immune response, mainly endolysosomal receptors (TLR3, TLR7/8) and cytoplasmic receptors with retinoic-acid inducible gene I -like receptors [161]. To date, Toll-like pattern recognition receptors have been relatively well studied, and four nucleic acid immune recognition sensing receptors have been reported, namely TLR3, which recognizes dsRNA; TLR7, TLR8, and TLR13, which recognize ssRNA; TLR9, which recognizes ssDNA containing CpG motifs [162]. The interferon gene cGAS stimulator (cGAS-STING), a cGAS recognition receptor mainly in the cytoplasm, can activate the catalytic activity of cGAS when dSDNA or other Y-type DNA and hybrid DNA are elevated in the cytoplasm, generating a second messenger, cGAMP, which triggers a conformational change in the STING ligand binding region on the endoplasmic reticulum and further initiates downstream signaling cascade to express pro-inflammatory large-scale type I interferons (IFN) [163]. RLRs, widely expressed in human immune and non-immune cells, primarily recognize RNA viruses or short-stranded RNAs that are abnormally present in cells, ultimately leading to the upregulation of type I interferons [164]. There are three main ways in which a group of nucleic acids and oligonucleotides can function in the innate immune system: 1) they can stimulate pattern recognition receptors to activate innate immune cells, 2) they encode proteins or peptides with immunostimulatory effects, 3) they silence specific genes to block negative regulators of immune cells [165].
4.1. SNA as immunotherapeutic vaccines
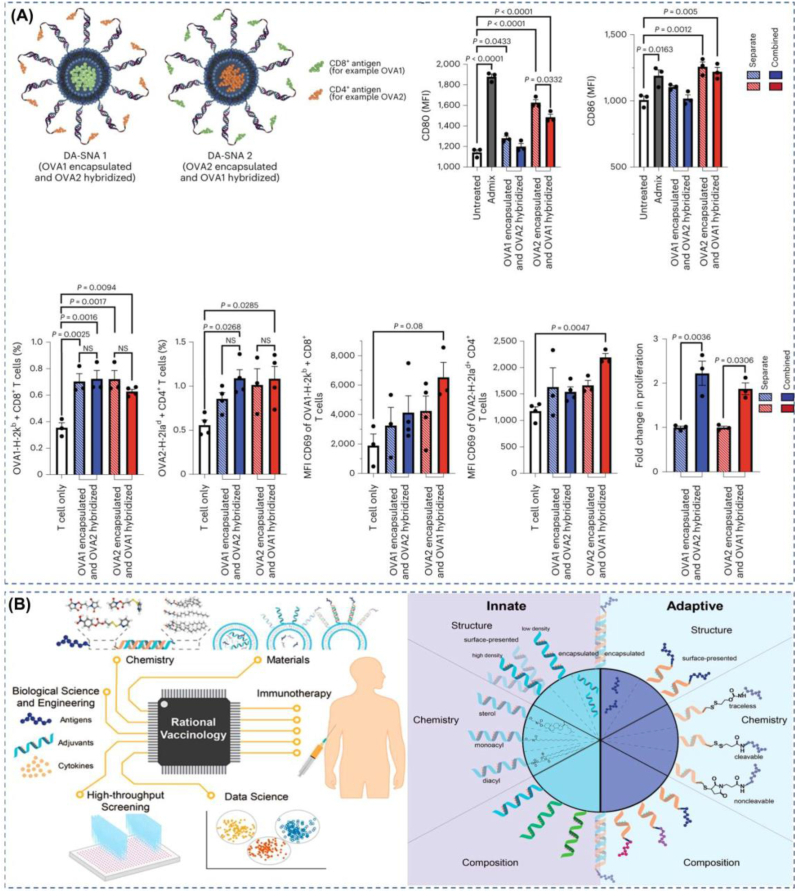
Vaccination has significantly contributed to global human health and has an irreplaceable role in controlling viruses such as poliovirus [166] and measles [167]. Although smallpox and rinderpest viruses have been eliminated [168], vaccination still has a bright future in the control of new infectious diseases and the fight against tumors. The main types of vaccines are live attenuated vaccines, inactivated vaccines, antitoxins, subunit vaccines (including peptide vaccines), vector vaccines, nucleic acid vaccines, etc. In the last decade, mRNA vaccines are gaining momentum. The development of nucleic acid vaccines appears to have been a long process from a historical perspective, with researchers beginning to think about their use as early as 1995 [169]. But it was not until COVID-19 that nucleic acid vaccines showed an unprecedented rate of development [170]. The nucleic acids in nucleic acid vaccines include common DNA and RNA. However, the nucleic acids have different functions, including those that translate antigenic peptides and those that act as adjuvants to stimulate immune responses. Compared with traditional attenuated vaccines, inactivated vaccines, subunit vaccines, etc., mRNA vaccines have high safety, high yield, and high efficacy [171], but in vivo, delivery of nucleic acids is easily degraded by nucleases, which is an essential driving force to promote the application of nanomaterials in vaccines. Of course, the need for better spatiotemporal coordination and higher biosafety of vaccines is also a significant driving force to promote the application of nanomaterials in vaccines [172]. Nanovaccines can increase the stability of antigens as well as adjuvants, target in situ to improve immunogenicity, control release rates, and target delivery, among other advantages [173]. Nanovaccines have also shown excellent results in studies; for example, glycosylated nano-HIV immunogens trigger more potent immunity compared to monomers because glycosylated nano antigens trigger mannose-binding lectin-mediated innate immune mechanisms, leading to their aggregation in germinal centers in vivo [174]. In nanovaccines, designing vaccines based on SNA has considerable advantages because SNA has a highly modular architecture. In addition to vaccine composition, vaccine structure is a key determinant of vaccine efficacy. Mirkin et al. recently demonstrated that even the right target could produce an ineffective vaccine if it's processing by immune cells is not taken into account. The researchers demonstrated that vaccine structure in antigen placement is a design criterion that strongly affects vaccine efficacy because nanoscale antigen placement changes the time scale of intracellular antigen processing and the residence time of antigen in different cellular compartments, and rational design parameters achieved through modular architectures (e.g., SNA) can greatly improve vaccine efficacy [175](Fig. 6a). In past studies, developers have often spent a great deal of time on the composition and combination of adjuvants and tumor-associated antigens, neglecting the impact of structural and spatiotemporal coordination of antigen and adjuvant presentation on immune stimulation [176], when in fact the manner and point in time at which adjuvants and antigens meet the immune system and the size of the nanocarrier are pivotal in exerting immune effects [177]. Researchers have shown that it is not only the structure of the carrier that affects the immune response but also differences in the structure of the antigen and adjuvant combination, even within the same material [178]. SNAs are capable of rapid uptake by antigen-presenting cells through receptor-mediated endocytosis. For innate immune responses, the unique adjuvant nucleic acid three-dimensional structure of the SNA shell can provide better recognition of TLRs [179](Fig. 6b). For adaptive immune responses, some nucleic acid adjuvants or antigenic peptides that are not clinically effective may exhibit the opposite result when delivered using nanostructures. It is also an example of rational vaccinology, such as prostate-specific membrane antigen, which does not do its job well clinically, but when utilized with SNAs, it can show a robust immune response [180].
Fig. 6.
SNA's programmable properties enable optimal immunotherapy results. (A) Precise spatial and temporal control of antigen and adjuvant by designing the structural positions of antigen and adjuvant can enhance the immune effect of the SNA vaccine. Reproduced with permission from Ref. [175]. Copyright 2023, Springer Nature. (B) Some of the altered approaches and combinations of options shown in the figure provide robust strategies for the implementation of rational vaccinology. Reproduced with permission from Ref. [179]. Copyright 2022, American Chemical Society.
TLR9 is the first pattern recognition receptor (PRR) we have identified that can sense nucleic acids, and it is capable of sensing bacterial DNA through CpG sequences, as well as being specifically activated by synthetic non-methylated CpG oligonucleotides [181]. TLR9 receptor is different from other immunotherapeutic target receptors; the development of TLR9 agonist is influenced by its structure and other inherent properties. On the one hand, it is located in the intracellular endosome of the cell membrane; TLR9 agonist needs to enter inside the cell to be effective; therefore, the process of TLR9 agonist entering the cell through endocytosis is the key step for its efficacy. On the other hand, TLR9 agonists are unmethylated CpG oligonucleotides, and oligonucleotides are easily degraded by nucleases in plasma. Hence, the choice of TLR9 carrier or the mode of administration is quite important. Therefore, contemporary TLR9 agonists are mainly optimized in terms of preventing oligonucleotides from being rapidly degraded and enhancing endocytosis. Nowadays, nucleic acids are widely used as adjuvants in the development of cancer vaccines, but they are easily degraded by enzymes in vivo and have poor cellular uptake, so the application of nucleic acids as immune adjuvants should first consider how to maintain the stability of nucleic acids and thus play an immune role. SNA seems to be the best choice for TLR9. SNA can not only protect oligonucleotides from degradation by nucleases but also transport TLR9 nucleic acid agonists to the intracellular side of the cell membrane through receptor endocytosis, which greatly improves transport efficiency. The application of SNA vaccines is not only limited to tumor vaccines, but it has also received a lot of attention in the field of anti-infectious diseases, especially the SNA vaccine against novel coronavirus (SARS-CoV-2) [34]. Using the receptor-binding domain (RBD protein) of the novel coronavirus spike-in protein wrapped in liposomes as an antigen and DNA with an immunostimulatory CpG sequence modified on the liposome shell, a novel vaccine with a high degree of modularity was constructed, and only one dose of the SNA nano vaccine was sufficient for 100% protection against the novel coronavirus in animal studies.
4.2. SNA as modulators of tumor immunotherapy
Cancer treatment mainly consists of chemotherapy, radiotherapy, and surgery, but these traditional methods have corresponding disadvantages. For example, in chemotherapy, the toxic side effects of chemotherapeutic drugs are too strong. Radiotherapy and surgery can only be localized and cannot be used to control the metastasis of tumor cells. Along with the development of nanotechnology, nanomedicines have shown promising applications in improving cancer chemotherapy [182]. Nevertheless, some studies have found that after systemic administration, only 0.7% of nanomedicines reach solid tumors [183]. The high attrition rate is a critical factor in promoting the development of nanomedicine, and these nanomedicines are often eliminated due to low penetration aggregation rate and high toxic side effects. In order to deal with the severe threat of tumors to human life and health, immunotherapy is a good choice. First, with immunotherapy, a small amount of nanomedicine can activate the immune system, reducing the off-target toxicity caused by a large amount of commonly used nano chemotherapy drugs, and the activated systemic immune system can be localized to distant tumor cells [184] (Table 2).
Table 2.
Summary of SNA working strategies for tumor immunotherapy.
| Adjuvant | Antigens or drugs | Disease model | Innovative Strategies | Modes of action | Outcomes | Ref. |
|---|---|---|---|---|---|---|
| CpG | Pembrolizumab/cemiplimab | Advanced MCC/CSCC patients | AST-OO8, an SNA that activates TLR9, in combination with pembrolizumab for the treatment of MCC or with cemiplimab for the treatment of CSCC. | AST-OO8 increases th1-type cytokines, which in turn activate T cells and NK cells; pembrolizumab and cemiplimab bind specifically to PD-1 | Tumor regression was observed and, at the site of the lesion, increased infiltration of toxic immune cells | [189] |
| dsDNA | None | Glioblastoma | Nucleic acids can be delivered to intracranial tumor sites through the nasal cavity using SNA transported dsDNA. | Binding of dsDNA to cGAS induces endogenous CDN production, which in turn stimulates interferon gene-stimulating proteins to activate innate immunity for tumor treatment. | Tumors have good tolerance to STING-SNA, and fewer doses can delay tumor growth and improve survival. | [196] |
| CpG | Prostate peptide antigens (PSA; PSMA; PAP) | C57BL6 mouse prostate cancer transplantation tumor model | Co-delivery of SNA to adjuvants and peptide antigens enhances DC activation, spanning the current limitations of immunotherapy against weakly immunogenic tumors using only antigens such as OVA. | Class B CpG matures DCs to produce Th1 cytokines such as IL-12; peptide antigens activate antigen-specific T-cell immune responses in vivo | Co-delivery of CpG and antigen using SNA produced a stronger immune response than delivery of a simple mixture of CpG and antigen, and structural activation of immunity by antigen and adjuvant on the surface of SNA was most effective. | [194] |
| CpG-1826 | Lysates from TNBC cells (Oxidized or non-oxidized) | EMT6 model of TNBC; Py230 and Py8119 models of TNBC | Use of oxidized tumor cell lysates as antigens in SNA | SNA co-delivery of lysates and adjuvants can increase cellular uptake and bioavailability | Lys-SNAs were more potent than CpG and lysates co-mixtures in delaying tumor growth in Py230 and Py8119 models; OxLys-SNAs showed potent anti-tumor effects in EMT6 model of TNBC. | [193] |
| CpG | DOX | Tumor-bearing E.G7-OVA mice |
Ability to generate tumor-specific antigens, effectively avoiding off-target effects | Using DOX to generate ICD effects and CpG to enhance ICD-generated immune responses | Enhances CD8+ and CD4+ amplification, delays tumor growth and prolongs survival of tumor-bearing mice | [192] |
| CpG | Au&αPD-L1 | Tumor-bearing 4T1 mice |
SNA acts as both a radiosensitizer and an immunotherapeutic agent. | Gold nanoparticles of SNA act as radiosensitizers; RT causes tumor cells to produce ICD; SNA captures antigen to form an in situ vaccine and works synergistically with αPD-L1. | A potent anti-tumor effect was obtained, completely inhibiting tumor growth. | [198] |
In recent years, researchers have been encouraged by advances in tumor immunotherapy, which can not only initiate the immune cycle in response to primary as well as distant tumor cells but can also use a strong immune response to reduce the likelihood of future tumor formation [185]. The main exogenous immunostimulants used for immunotherapy include small chemical molecules, therapeutic proteins, and nucleic acids. Among the therapeutic nucleic acids (TNAs), CpG oligonucleotides acting on Toll-like receptors have received much attention. However, CpG oligonucleotides often cause a strong inflammatory response. Not only that, the vectors delivering the therapeutic nucleic acids can often be immunostimulatory and cause strong side effects [186], so the choice of carriers is very urgent. In the selection of vectors to deliver therapeutic nucleic acids, the 3D structure of nucleic acid nanoparticles (NANPs) has been shown to be the key factor influencing immune recognition rather than the sequence and particle size of the nucleic acid. The connectivity and composition of nucleic acids can also influence immune effects, and the spherical structure of RNA nanoparticles tends to be more effective than RNA rings and RNA fibers [187].
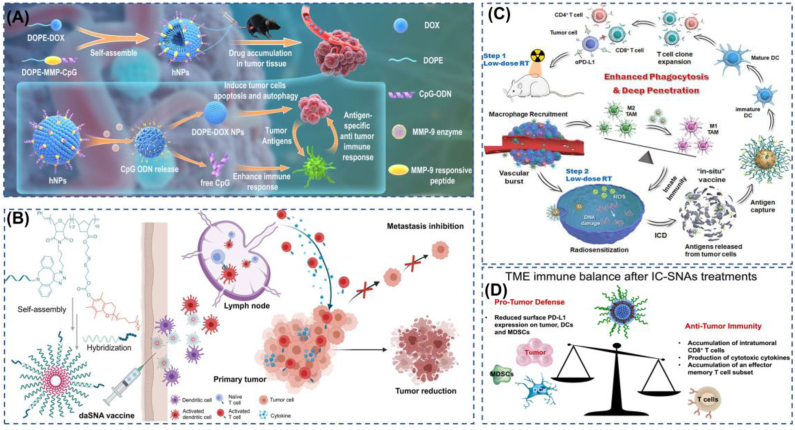
Due to the modular nature of SNAs and their specific three-dimensional structure in cellular uptake and anti-nucleic acid enzymatic cleavage, both immunostimulatory and immunomodulatory SNA can exhibit better immunotherapeutic effects [188]. Among these, Cavrotolimod (AST-008), a Toll-like receptor 9 (TLR-9) agonist designed to activate the innate immune system and induce an effective anti-cancer immune response, is highly effective in combination with immune checkpoint inhibitors. Currently, cavrotolimod is being tested in a Phase 1b/2 clinical trial to evaluate the safety, tolerability, pharmacokinetics, pharmacodynamics, and preliminary efficacy of cavrotolimod alone or in combination with the anti-PD-1 antibody pembrolizumab in the treatment of patients with Merkel cell carcinoma (MCC) or squamous cell carcinoma (cSCC) of the skin [33,189]. There are many excellent results of SNA on the bench here at the proof-of-concept stage. The first SNA conjugate that co-delivers nucleic acids and small molecules to the same cells, with small molecules inhibiting TLR4 and specific nucleic acids inhibiting TLR9, showed that this co-delivery structure has a significant advantage in TLR inhibition compared to oligonucleotide sequences as well as small molecules [190]. Related work has shown that maximum immune responses are possible when co-delivering antigen and adjuvant to the same targeted immune cells [191] and that encapsulating antigen in the core of an SNA increases the co-delivery of antigen and nucleic acid adjuvant to immune cells. In order to avoid the off-target effect of immune stimulation as much as possible, co-delivery of DOX with CpG via SNA, using DOX to induce immunogenic death (ICD) to produce tumor-specific antigens, and activation of the immune system with CpG, can achieve better specific immune effects [192](Fig. 7a). In a triple-negative breast cancer model, the use of SNAs with CpG-1826 as adjuvant and tumor cell lysate as antigen significantly increased the co-delivery of both immune components to the same immune cells in the lymph nodes, providing better tumor control compared to a simple mixture of tumor lysate antigen and nucleic acid adjuvant in vitro as well as in vivo experiments [193]. In a mouse model of prostate tumor, immunostimulant SNA with CpG oligonucleotides as the nucleic acid shell and as an immune adjuvant followed by an empty shell core wrapped around the antigen demonstrated a significant tumor suppressive effect. Further studies showed that chemical coupling of prostate cancer peptide antigen to the shell of CpG SNAs resulted in a stronger immune response, which also demonstrated that the modular programmable structure of SNA can well meet the high dependence of cytotoxic T lymphocyte (CTL) immune response on the carrier structure [194]. For being able to avoid the stimulation of the immune system by SNA carriers, Chen et al. used CpG co-loaded with vitamin E to construct a dual adjuvant self-delivery system that could well meet the requirements of immunostimulants for carrier-free delivery [195](Fig. 7b).
Fig. 7.
Some representative cases of SNA in immune regulation. (A) SNAs with DOX and CpG achieve synergistic functions of chemotherapy and immunotherapy with specific enzymatic activation, which can significantly retard tumor growth. Reproduced with permission from Ref. [192]. Copyright 2022, Springer Nature. (B) The use of vector-free immune SNA with a dual adjuvant can greatly maximize the TLR9 activation effect. Reproduced with permission from Ref. [195]. Copyright 2022, American Chemical Society. (C) Radiotherapy enhances the infiltration accumulation of SNAs, while SNAs act as radiosensitizers and in situ vaccines for tumors, enabling a synergistic treatment strategy of radiotherapy and immunotherapy. Reproduced with permission from Ref. [198]. Copyright 2022, Elsevier B.V. (D) The use of antisense DNA that specifically blocks PD-L1 can effectively block the PD-1/PD-L1 signaling pathway, providing a powerful approach to this precision immunotherapy. Reproduced with permission from Ref. [201]. Copyright 2022, American Chemical Society.
In glioblastoma treatment, Stegh et al. used the Bcl2-Like 12 (Bcl2L12) oncogene as a target for SNA-mediated gene silencing and showed that siRNAs carrying a target for Bcl2L12 were able to cross the blood-brain barrier without transfection agents, effectively reducing tumor burden and progression [29]. And the SNAs (NU-0129) were obtained in a phase 0 clinical trial in rodents and non-human primates with similar results, showing significant inhibition against both recurrent glioblastoma, indicating that SNA is a safe and precise medical treatment with the ability to penetrate the blood-brain barrier. SNA is a safe and precise medical tool that can deliver siRNA oligonucleotide systems to intracranial tumor sites for RNAi therapy [32]. More recently, Mirkin et al. have developed STING-SNA for glioblastoma, targeting cGAS by loading high-density dsDNA, developing STING agonistic immunotherapy, slowing tumor growth with just one dose of intranasal STING-SNA in a homozygous mouse glioma model, and addressing the challenge of delivering nucleic acids to intracranial tumor sites via the intranasal route [196].
Interestingly, in addition to its prominent application in immunotherapy, the immunomodulatory function of SNAs can also be used as an adjuvant tool in combination therapy. For example, there is evidence that treatment with radiotherapy (RT) can alter the tumor microenvironment, in which nanoparticles can increase its infiltration rate using macrophage-mediated vascular rupture. However, the macrophages recruited by RT are mainly M2-type macrophages that suppress inflammation, which can inhibit the tumor microenvironment and thus promote tumor infiltration and invasion [197]. Perhaps SNA may change this status quo, using CpG-modified gold nano SNA, which not only can reshape the tumor immune microenvironment, changing M2 type macrophages to M1 type macrophages but also can act as a radiosensitizer. Moreover, RT can again provide conditions for the infiltration of CpG-AuSNA, increasing the aggregation as well as infiltration of SNA with immune function in tumors [198](Fig. 7c). Nanogel SNA nanoparticles, as mentioned earlier, also have similar physiological stability and resistance to enzymatic degradation as SNAs. A major challenge in the clinical application of CpG is its ability to activate not only M1-type macrophages but also M2-type macrophages, so it is worth considering blocking M2 activation when CpG is used as immunotherapy. Based on this, a nanogel with a highly loaded CpG motif that can activate the immune system and, at the same time, block M2 activation using siRNA has been developed [199]. Photodynamic therapy (PDT) not only kills tumor cells through reactive oxygen species generated by photosensitizers but also theoretically induces tumor immunogenic cell death and thus activates effector T cells to generate an anti-tumor immune response, so the combined application of PDT and ICB therapy can produce a good synergistic anti-tumor effect. Guo et al. have developed a novel self-assembled nucleic acid nanogel platform using PD-L1 siRNA as a cross-linking agent, co-loaded with siRNA and demagnetized chlorophyll A (PPA) photosensitizer, enabling a perfect combination of photodynamic therapy and immune checkpoint blockade therapy [200]. The PD1/PD-L1 signaling pathway is an important target for tumor immunotherapy because the combination of PD1 and PD-L1 will initiate a negative signaling cascade that will further inhibit T-cell activation, which will help the immune escape mechanism in the tumor immune microenvironment. In recent years, ICB therapies with therapeutic antibodies against PD1 (as competitive inhibitors of PD-L1) have been heavily used. However, the effectiveness of such applied antibody immunotherapy is not obvious due to some inherent drawbacks of antibodies. At this point, the development of immune checkpoint inhibition therapy with gene regulation of PD-L1 expression has obvious advantages [201](Fig. 7d), using SNA multi-modular structure to co-deliver anti-PD-L1 expression antisense DNA and immune stimulation antigen into tumor cells, which can reduce PD-L1 expression in all cells in the tumor microenvironment, and thus enhance combined immune stimulation, and we expect that this combined therapy using SNA to achieve ICB and immune stimulation will become a new milestone in tumor immunotherapy.
In the study, it was found that altering the length of the liposomal diacyl lipid tail can change the biological properties of liposomal SNA as well as the immunomodulatory ability of tumors [202], suggesting that liposomal SNA can influence the properties of the SNA system by altering the structure of the liposome. SNA co-delivery of antigens with immune adjuvants shows great promise for application. However, it has been shown that the linker between antigen and complement nucleic acids should not only consider the convenience of the synthesis method, as the coupling chemistry between them can affect the effectiveness of T cell activation [203]. Altering the chemical linkage of antigens to the SNA surface can also affect the immune ability of immunologically applied SNA, as the linkage of antigens to the nucleic acid adjuvant complement controls the rate of antigen release, which was found to strongly influence the efficacy of therapeutic SNA in vivo [204]. In addition to the impact of the internal composition of the nucleic acid on the application of SNA, the diversity of nucleic acid sequences also affects the effect of SNA [205]. It was found that double-sequence (CpG-a and CpG-b) SNA are better delivered compared to those delivered as a mixture of two linear oligonucleotides or as a mixture of single oligonucleotide SNA. Nucleic acid sequences on SNA also affect the immune response of SNA. G-rich SNA induces higher levels of macrophage activation than polyT SNA or single G- or T-rich DNAs, and although no long-term cellular activation is observed in vivo with G-rich SNA, they have short-term macrophage activation in vitro and short-term cytokine release in vivo [206]. It is suggested that SNA used for immunostimulation can be enhanced by adding G-sequences to the shell nucleic acid or by directly coupling G-rich nucleic acids to the SNA surface. Table 3 shows the effect of structural or component variations in SNA on immunomodulation, providing a design strategy for maximizing activated immunity.
Table 3.
Summary of design strategies for SNA's programmable structure on maximizing immune system activation.
| Structure | Variables in the structure | Adjuvant | Antigen | modol | Outcomes | Ref |
|---|---|---|---|---|---|---|
| L -SNA | Differential hydrophobic anchors between nucleic acids and liposomes (lipid-tail or cholesterol-tail) | CpG-1826 | None | RAW-Blue macrophages | The structures utilizing lipid tail-anchored nucleic acids in LSNA have greater stability as well as greater ability to activate TLR receptors than those utilizing cholesterol-anchored nucleic acids. | [207] |
| L -SNA | Diacyl lipid tail | CpG-1826 | Py8119 TNBC cells lysates | Orthotopic mouse models of 4T1 & Py8119 | The use of DPPC with high Tc value as a lipid tail could improve the serum stability of L-SNA as well as the ability to activate immunity, with obvious anti-tumor effects, and inhibit the lung metastasis of TNBC cells. | [202] |
| L -SNA | The Linker between antigen and SNA(Whether or not it can be cracked) | CpG | Gp100 (KVPRNQDWL) | DC cells derived from bone marrow | A cleavable traceless linker between antigen and SNA enhances activation of T cells. | [203] |
| L -SNA | The Linker between peptide antigens and SNAs(The rate of cleavage) | CpG | OVA-1 | C57BL/6&OT-1 mouse splenocytes | The cleavable Linker and the ability to rapidly release antigen can enhance the immunizing effect of SNA. | [204] |
| L -SNA | Location of antigen and adjuvant | CpG | Prostate cancer peptide | Syngeneic mouse models of prostate cancer | HM SNAs, which can simultaneously present antigen and adjuvant on the surface, are capable of inducing stronger anti-tumor immune responses than EM SNAs, in which the SNA core encapsulates the antigen. | [194] |
| L -SNA | Location of OVA1 and OVA2 on the SNA & the way of delivery for both antigens OVA1 and OVA2. | CpG | OVA1&OVA2 | DC cells derived from bone marrow& C57BL/6 | Simultaneous delivery of both antigens with the same SNA induced stronger T cell activation than delivery of two SNAs separately. Moreover, the structure of core-coated OVA2 and externally crosslinked OVA1 significantly improved the tumor suppression effect than the structure of core-coated OVA1 and externally crosslinked OVA2. | [175] |
| L -SNA | Categories of nucleic acids loaded on the surface of LSNA | CpG-A&CpG-B | None | DC cells derived from bone marrow | LSNA containing two-component CpG activated the maturation of DC cells more than mixtures of CpG-A and CpG-B and single-component LSNA containing CpG-A or CpG-B. | [205] |
As a widespread substance, it appears important for the human organism to accurately identify self- and non-self-directed nucleic acids, and it is now well understood that nucleic acid sensors that trigger innate immune responses are associated with the development of diseases, such as systemic lupus erythematosus and arthritis [208]. Imanishi et al. found that nucleic acids, such as DNA and RNA (released by dead cells), can trigger the differentiation of immune naïve T cells into T helper type 2 (Th2 cells) cells [209], which can mediate allergic responses or anti-parasitic infections, all of which demonstrated a strong link between nucleic acids and disease in vivo and provide a solid foundation and direction for the development of research on SNA. Besides SNA as an immunostimulant or immunomodulator, we conjecture that it is feasible to use the nucleic acid sequences of SNA as a tool in immunotherapy, which was recently brought to our attention by a study in which bispecific nanobonders transformed the immune function of solid tumors [210]. The study noted that hematologic immune malignancies are more sensitive to immune responses than solid tumors, primarily because hematologic malignancy cells express a family of molecules capable of expressing targeted activation of lymphatic immune cells member 7 (SLAMF7). The investigators used nano-adherents to modify solid tumors with SLAMF7, which in turn induced a strong phagocytic immune response and greater sensitivity to CD47 immune checkpoint inhibitors. This report is actually similar to the PROTAC technology, both of which enable the functional integration of “targeting” and “recruitment”. The nucleic acid sequence of SNA is mainly used as an immune adjuvant or to silence specific genes to block the negative regulatory factors of immune cells in immunotherapy at this stage, and for a large number of modifications to the structure of the shell nucleic acid sequence of SNA, for example, the complement of the nucleic acid sequence can be modified with different structures, which opens up the application of SNA, and we can also consider SNA as an application of linker.
5. Summary and outlook
Definitely, the applications of SNA in the biomedical field have been a promising prospect. The unique nucleic acid arrangement structure has been reported to enable SNA with sufficient colloidal stability in the extracellular circulation, promoted intracellular transfection efficiency, and even the ability to cross the blood-brain and skin barriers. More importantly, this multi-modular structure, very similar to a core-shell nanoparticulate structure, provides not only a rapidly developed nanoplatform for the target treatment of diverse diseases but also great opportunities for rational design and optimization of therapeutic combinations toward greater synergistic effects. In this review, we made a comprehensive summary on the applications of SNA in precision medicine and immunotherapy from the perspective of the structure-activity relationship, which is believed to inspire some fresh ideas for researchers and physicians, such as non-invasive diagnostics, precise treatment at the lesion site, and intraoperative imaging guidance. We also provided an in-depth discussion on precise spatiotemporal control of adjuvants and antigens or antigens and antigens via SNA for maximizing the immune system activation to fight against diseases.
Looking to the future, we are confident that more and more SNA-based drugs will break through the preclinical research stage, which will require the concerted efforts of multidisciplinary researchers in multiple fields. Of note, most SNA research is currently limited to the proof-of-concept stage, with only a very few studies entering clinical research [32,33,58,211]. There are still critical challenges to overcome for most SNA clinical translations. As a novel gene delivery vector, SNA can somewhat overcome the challenges of enzymatic degradation and toxicity faced by traditional gene delivery vectors, including cationic vectors, and greatly improve the transfection efficiency. Nonetheless, SNAs are similarly confronted with the degradation of endosomes and the formation of protein crowns in the extracellular circulation for severely compromised transfection efficiency. At the current stage, there are solutions to the endosomal escape of SNA [65,212], but these efforts remain insufficient for the general applicability of SNA. Furthermore, only a few examples of SNA nanoplatforms have been constructed based on carrier-free self-delivery strategies, and most of the reported SNAs still utilize conventional nanomaterials for construction, thus requiring more efforts to examine the toxicity and immunogenicity issues brought by the materials. Beyond that, there exist limitations in SNA preparation. The kinds of SNAs explored at this stage seem to be limited despite the multimodular and programmable properties of SNA. The widely investigated SNAs based on gold nanoparticles or liposomes provide numerous opportunities for further SNA exploration. The study on the structure-property relationship of SNA should be performed with the expansion of the core domain materials and application fields. For example, the current organic core-based SNAs, especially polymer-based SNAs, have attracted increasing attention. However, the polymers used for polymeric core construction are limited to linear polymers [73,74], which apparently limits the universal preparation as well as the potential clinical translations of SNAs.
Last but not least, more attention should be paid to self-delivering SNAs that can avoid carrier-associated toxicity or immunogenicity since a stabilized simple structure is considered the most important requirement for druggability. The development of SNA is currently facing great opportunities as well as challenges. The excellent results of SNA in bench, as well as some clinical trial results, including precise diagnostic imaging and drug release; precise intraoperative imaging guidance and prognosis; and the activation of more potent immune effects based on precise structural control and precise spatial-temporal control of components, make us convinced that SNA will be a powerful nanotool that can be eventually clinically translatable in the field of precision medicine and immunotherapy.
Credit author statement
Songbin Liu: Writing-original draft, Writing-review & editing. Cui-Yun Yu: Writing-original draft, Writing-review & editing. Hua Wei: Writing-original draft, Writing-review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors acknowledge the financial support from the Key R&D Program of Hunan province (2021SK2036), the Thousand Young Talent Program, the Hunan Science and Technology Innovation Leading Talent Project (2022RC3080), and the Hunan Provincial Natural Science Foundation (2022JJ40381).
Contributor Information
Cui-Yun Yu, Email: yucuiyunusc@hotmail.com.
Hua Wei, Email: weih@usc.edu.cn.
Data availability
No data was used for the research described in the article.
References
- 1.Liu X., Xia S., Zhang Z., Wu H., Lieberman J. Channelling inflammation: gasdermins in physiology and disease. Nat. Rev. Drug Discov. 2021;20:384–405. doi: 10.1038/s41573-021-00154-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herbst R.S., Morgensztern D., Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 3.Ribas A., Wolchok J.D. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almendro V., Marusyk A., Polyak K. Cellular heterogeneity and molecular evolution in cancer. Ann. Review Paleopathol. Mech. Disease. 2013;8:277–302. doi: 10.1146/annurev-pathol-020712-163923. [DOI] [PubMed] [Google Scholar]
- 5.Wolf Y., Bartok O., Patkar S., Eli G.B., Cohen S., Litchfield K., Levy R., Jiménez-Sánchez A., Trabish S., Lee J.S., Karathia H., Barnea E., Day C.P., Cinnamon E., Stein I., Solomon A., Bitton L., Pérez-Guijarro E., Dubovik T., Shen-Orr S.S., Miller M.L., Merlino G., Levin Y., Pikarsky E., Eisenbach L., Admon A., Swanton C., Ruppin E., Samuels Y. UVB-induced tumor heterogeneity diminishes immune response in melanoma. Cell. 2019;179:219–235.e221. doi: 10.1016/j.cell.2019.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vitale I., Shema E., Loi S., Galluzzi L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat. Med. 2021;27:212–224. doi: 10.1038/s41591-021-01233-9. [DOI] [PubMed] [Google Scholar]
- 7.Lim Z.F., Ma P.C. Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J. Hematol. Oncol. 2019;12:134. doi: 10.1186/s13045-019-0818-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senft D., Leiserson M.D.M., Ruppin E., Ronai Z.A. Precision oncology: the road ahead. Trends Mol. Med. 2017;23:874–898. doi: 10.1016/j.molmed.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinker K., Chin J., Melsaether A.N., Morris E.A., Moy L. Precision medicine and radiogenomics in breast cancer: new approaches toward diagnosis and treatment. Radiology. 2018;287:732–747. doi: 10.1148/radiol.2018172171. [DOI] [PubMed] [Google Scholar]
- 10.Kumar-Sinha C., Chinnaiyan A.M. Precision oncology in the age of integrative genomics. Nat. Biotechnol. 2018;36:46–60. doi: 10.1038/nbt.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel S.J., Sanjana N.E., Kishton R.J., Eidizadeh A., Vodnala S.K., Cam M., Gartner J.J., Jia L., Steinberg S.M., Yamamoto T.N., Merchant A.S., Mehta G.U., Chichura A., Shalem O., Tran E., Eil R., Sukumar M., Guijarro E.P., Day C.P., Robbins P., Feldman S., Merlino G., Zhang F., Restifo N.P. Identification of essential genes for cancer immunotherapy. Nature. 2017;548:537–542. doi: 10.1038/nature23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mai D., June C.H., Sheppard N.C. In vivo gene immunotherapy for cancer. Sci. Transl. Med. 2022;14 doi: 10.1126/scitranslmed.abo3603. eabo3603. [DOI] [PubMed] [Google Scholar]
- 13.Nissim L., Wu M.R., Pery E., Binder-Nissim A., Suzuki H.I., Stupp D., Wehrspaun C., Tabach Y., Sharp P.A., Lu T.K. Synthetic RNA-based immunomodulatory gene circuits for cancer immunotherapy. Cell. 2017;171:1138–1150.e1115. doi: 10.1016/j.cell.2017.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mardis E.R. The emergence of cancer genomics in diagnosis and precision medicine. Nat. Can. (Que.) 2021;2:1263–1264. doi: 10.1038/s43018-021-00305-6. [DOI] [PubMed] [Google Scholar]
- 15.Sahin U., Türeci Ö. Personalized vaccines for cancer immunotherapy. Science. 2018;359:1355–1360. doi: 10.1126/science.aar7112. [DOI] [PubMed] [Google Scholar]
- 16.Picanço-Castro V., Pereira C.G., Covas D.T., Porto G.S., Athanassiadou A., Figueiredo M.L. Emerging patent landscape for non-viral vectors used for gene therapy. Nat. Biotechnol. 2020;38:151–157. doi: 10.1038/s41587-019-0402-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H., Qin L., Zhang X., Guan J., Mao S. Mechanisms and challenges of nanocarriers as non-viral vectors of therapeutic genes for enhanced pulmonary delivery. J. Contr. Release. 2022;352:970–993. doi: 10.1016/j.jconrel.2022.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akinc A., Querbes W., De S., Qin J., Frank-Kamenetsky M., Jayaprakash K.N., Jayaraman M., Rajeev K.G., Cantley W.L., Dorkin J.R., Butler J.S., Qin L., Racie T., Sprague A., Fava E., Zeigerer A., Hope M.J., Zerial M., Sah D.W., Fitzgerald K., Tracy M.A., Manoharan M., Koteliansky V., Fougerolles A., Maier M.A. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu S., Cheng Q., Wei T., Yu X., Johnson L.T., Farbiak L., Siegwart D.J. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR-Cas gene editing. Nat. Mater. 2021;20:701–710. doi: 10.1038/s41563-020-00886-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C., Stoter M., Epstein-Barash H., Zhang L., Koteliansky V., Fitzgerald K., Fava E., Bickle M., Kalaidzidis Y., Akinc A., Maier M., Zerial M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 21.Vaidyanathan S., Anderson K.B., Merzel R.L., Jacobovitz B., Kaushik M.P., Kelly C.N., van Dongen M.A., Dougherty C.A., Orr B.G., Banaszak Holl M.M. Quantitative measurement of cationic polymer vector and polymer-pDNA polyplex intercalation into the cell plasma membrane. ACS Nano. 2015;9:6097–6109. doi: 10.1021/acsnano.5b01263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng F., Setyawati M.I., Tee J.K., Ding X., Wang J., Nga M.E., Ho H.K., Leong D.T. Nanoparticles promote in vivo breast cancer cell intravasation and extravasation by inducing endothelial leakiness. Nat. Nanotechnol. 2019;14:279–286. doi: 10.1038/s41565-018-0356-z. [DOI] [PubMed] [Google Scholar]
- 23.Mirkin C.A., Laramy C., Skakuj K. The power of spheres. Nature. 2019;576 doi: 10.1038/d41586-019-03713-y. S3-S3. [DOI] [PubMed] [Google Scholar]
- 24.Young K.L., Scott A.W., Hao L., Mirkin S.E., Liu G., Mirkin C.A. Hollow spherical nucleic acids for intracellular gene regulation based upon biocompatible silica shells. Nano Lett. 2012;12:3867–3871. doi: 10.1021/nl3020846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massich M.D., Giljohann D.A., Seferos D.S., Ludlow L.E., Horvath C.M., Mirkin C.A. Regulating immune response using polyvalent nucleic acid-gold nanoparticle conjugates. Mol. Pharm. 2009;6:1934–1940. doi: 10.1021/mp900172m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda H. Toward a full understanding of the EPR effect in primary and metastatic tumors as well as issues related to its heterogeneity. Adv. Drug Deliv. Rev. 2015;91:3–6. doi: 10.1016/j.addr.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Sindhwani S., Syed A.M., Ngai J., Kingston B.R., Maiorino L., Rothschild J., MacMillan P., Zhang Y., Rajesh N.U., Hoang T., Wu J.L.Y., Wilhelm S., Zilman A., Gadde S., Sulaiman A., Ouyang B., Lin Z., Wang L., Egeblad M., Chan W.C.W. The entry of nanoparticles into solid tumours. Nat. Mater. 2020;19:566–575. doi: 10.1038/s41563-019-0566-2. [DOI] [PubMed] [Google Scholar]
- 28.Choi C.H., Hao L., Narayan S.P., Auyeung E., Mirkin C.A. Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc. Natl. Acad. Sci. U.S.A. 2013;110:7625–7630. doi: 10.1073/pnas.1305804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jensen S.A., Day E.S., Ko C.H., Hurley L.A., Luciano J.P., Kouri F.M., Merkel T.J., Luthi A.J., Patel P.C., Cutler J.I., Daniel W.L., Scott A.W., Rotz M.W., Meade T.J., Giljohann D.A., Mirkin C.A., Stegh A.H. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3006839. 209ra152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng D., Giljohann D.A., Chen D.L., Massich M.D., Wang X.Q., Iordanov H., Mirkin C.A., Paller A.S. Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation. Proc. Natl. Acad. Sci. U.S.A. 2012;109:11975–11980. doi: 10.1073/pnas.1118425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kouri F.M., Hurley L.A., Daniel W.L., Day E.S., Hua Y., Hao L., Peng C.Y., Merkel T.J., Queisser M.A., Ritner C., Zhang H., James C.D., Sznajder J.I., Chin L., Giljohann D.A., Kessler J.A., Peter M.E., Mirkin C.A., Stegh A.H. miR-182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes Dev. 2015;29:732–745. doi: 10.1101/gad.257394.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumthekar P., Ko C.H., Paunesku T., Dixit K., Sonabend A.M., Bloch O., Tate M., Schwartz M., Zuckerman L., Lezon R., Lukas R.V., Jovanovic B., McCortney K., Colman H., Chen S., Lai B., Antipova O., Deng J., Li L., Tommasini-Ghelfi S., Hurley L.A., Unruh D., Sharma N.V., Kandpal M., Kouri F.M., Davuluri R.V., Brat D.J., Muzzio M., Glass M., Vijayakumar V., Heidel J., Giles F.J., Adams A.K., James C.D., Woloschak G.E., Horbinski C., Stegh A.H. A first-in-human phase 0 clinical study of RNA interference-based spherical nucleic acids in patients with recurrent glioblastoma. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abb3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milhem M.M., Perez C.A., Hanna G.J., Wise-Draper T.M., Bhatia S., Bexon A.S., Daniel W.L., O'Day S. AST-008: a novel approach to TLR9 agonism with PD-1 blockade for anti-PD-1 refractory Merkel cell carcinoma (MCC) and cutaneous squamous cell carcinoma (CSCC) J. Clin. Oncol. 2020;38 doi: 10.1200/JCO.2020.38.15_suppl.TPS3164. TPS3164-TPS3164. [DOI] [Google Scholar]
- 34.Teplensky M.H., Distler M.E., Kusmierz C.D., Evangelopoulos M., Gula H., Elli D., Tomatsidou A., Nicolaescu V., Gelarden I., Yeldandi A., Batlle D., Missiakas D., Mirkin C.A. Spherical nucleic acids as an infectious disease vaccine platform. Proc. Natl. Acad. Sci. U.S.A. 2022;119 doi: 10.1073/pnas.2119093119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ebrahimi S.B., Samanta D., Kusmierz C.D., Mirkin C.A. Protein transfection via spherical nucleic acids. Nat. Protoc. 2022;17:327–357. doi: 10.1038/s41596-021-00642-x. [DOI] [PubMed] [Google Scholar]
- 36.Huang C., Han Z., Evangelopoulos M., Mirkin C.A. CRISPR spherical nucleic acids. J. Am. Chem. Soc. 2022;144:18756–18760. doi: 10.1021/jacs.2c07913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bousmail D., Amrein L., Fakhoury J.J., Fakih H.H., Hsu J.C.C., Panasci L., Sleiman H.F. Precision spherical nucleic acids for delivery of anticancer drugs. Chem. Sci. 2017;8:6218–6229. doi: 10.1039/c7sc01619k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeo D.C., Wiraja C., Paller A.S., Mirkin C.A., Xu C. Abnormal scar identification with spherical-nucleic-acid technology. Nat. Biomed. Eng. 2018;2:227–238. doi: 10.1038/s41551-018-0218-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma M., Zhang P., Liang X., Cui D., Shao Q., Zhang H., Zhang M., Yang T., Wang L., Zhang N., Jing M., Zhang L., Dan W., Song R., Liu X., Hao J., Chen Y., Gu L., Wang L., Fan J. R11 peptides can promote the molecular imaging of spherical nucleic acids for bladder cancer margin identification. Nano Res. 2022;15:2278–2287. doi: 10.1007/s12274-021-3807-z. [DOI] [Google Scholar]
- 40.Zhang W., Callmann C.E., Meckes B., Mirkin C.A. Tumor-associated enzyme-activatable spherical nucleic acids. ACS Nano. 2022;16:10931–10942. doi: 10.1021/acsnano.2c03323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du T., Shi Z., Qin Z., Hu Y., Zhu Y., Jiang H., Wang X. Tailoring photothermally triggered phase transition of multimodal cascade theranostics platform by spherical nucleic acids. Adv. Funct. Mater. 2022;32 doi: 10.1002/adfm.202207410. [DOI] [Google Scholar]
- 42.Huang S., Liu Y., Xu X., Ji M., Li Y., Song C., Duan S., Hu Y. Triple therapy of hepatocellular carcinoma with microRNA-122 and doxorubicin co-loaded functionalized gold nanocages. J. Mater. Chem. B. 2018;6:2217–2229. doi: 10.1039/c8tb00224j. [DOI] [PubMed] [Google Scholar]
- 43.Yan R., Chen J., Wang J., Rao J., Du X., Liu Y., Zhang L., Qiu L., Liu B., Zhao Y.D., Jiang P., Chen C., Li Y.Q. A NanoFlare-based strategy for in situ tumor margin demarcation and neoadjuvant gene/photothermal therapy. Small. 2018;14 doi: 10.1002/smll.201802745. [DOI] [PubMed] [Google Scholar]
- 44.Zheng J., Zhu G., Li Y., Li C., You M., Chen T., Song E., Yang R., Tan W. A spherical nucleic acid platform based on self-assembled DNA biopolymer for high-performance cancer therapy. ACS Nano. 2013;7:6545–6554. doi: 10.1021/nn402344v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sayed N., Allawadhi P., Khurana A., Singh V., Navik U., Pasumarthi S.K., Khurana I., Banothu A.K., Weiskirchen R., Bharani K.K. Gene therapy: comprehensive overview and therapeutic applications. Life Sci. 2022;294 doi: 10.1016/j.lfs.2022.120375. [DOI] [PubMed] [Google Scholar]
- 46.Wang X., Wang X., Jin S., Muhammad N., Guo Z. Stimuli-responsive therapeutic metallodrugs. Chem. Rev. 2019;119:1138–1192. doi: 10.1021/acs.chemrev.8b00209. [DOI] [PubMed] [Google Scholar]
- 47.Lan G., Ni K., Lin W. Nanoscale metal-organic frameworks for phototherapy of cancer. Coord. Chem. Rev. 2019;379:65–81. doi: 10.1016/j.ccr.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao T., Chen Z., Kuang Y., Ren Z., Yu W., Rao W., Li L., Liu Y., Xu Z., Jiang B., Li C. Small-size Ti(3)C(2)Tx MXene nanosheets coated with metal-polyphenol nanodots for enhanced cancer photothermal therapy and anti-inflammation. Acta Biomater. 2023;159:312–323. doi: 10.1016/j.actbio.2023.01.049. [DOI] [PubMed] [Google Scholar]
- 49.Hu H., Xu Q., Mo Z., Hu X., He Q., Zhang Z., Xu Z. New anti-cancer explorations based on metal ions. J. Nanobiotechnol. 2022;20:457. doi: 10.1186/s12951-022-01661-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mirkin C.A., Letsinger R.L., Mucic R.C., Storhoff J.J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 51.Song Y., Xu X., MacRenaris K.W., Zhang X.Q., Mirkin C.A., Meade T.J. Multimodal gadolinium-enriched DNA-gold nanoparticle conjugates for cellular imaging. Angew. Chem. Int. Ed. 2009;48:9143–9147. doi: 10.1002/anie.200904666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dhar S., Daniel W.L., Giljohann D.A., Mirkin C.A., Lippard S.J. Polyvalent oligonucleotide gold nanoparticle conjugates as delivery vehicles for platinum(IV) warheads. J. Am. Chem. Soc. 2009;131:14652–14653. doi: 10.1021/ja9071282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nemati H., Ghahramani M.H., Faridi-Majidi R., Izadi B., Bahrami G., Madani S.H., Tavoosidana G. Using siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation in psoriasis. J. Contr. Release. 2017;268:259–268. doi: 10.1016/j.jconrel.2017.10.034. [DOI] [PubMed] [Google Scholar]
- 54.Miao P., Tang Y., Wang B., Jiang C., Gao L., Bo B., Wang J. Nuclease assisted target recycling and spherical nucleic acids gold nanoparticles recruitment for ultrasensitive detection of microRNA. Electrochim. Acta. 2016;190:396–401. doi: 10.1016/j.electacta.2016.01.034. [DOI] [Google Scholar]
- 55.Karami A., Hasani M. A palindromic-based strategy for colorimetric detection of HIV-1 nucleic acid: single-component assembly of gold nanoparticle-core spherical nucleic acids. Anal. Chim. Acta. 2020;1102:119–129. doi: 10.1016/j.aca.2019.12.050. [DOI] [PubMed] [Google Scholar]
- 56.Xu J., Tang Q., Zhang R., Chen H., Khoo B.L., Zhang X., Chen Y., Yan H., Li J., Shao H., Liu L. Sensitive detection of microRNAs using polyadenine-mediated fluorescent spherical nucleic acids and a microfluidic electrokinetic signal amplification chip. J. Pharm. Anal. 2022;12:808–813. doi: 10.1016/j.jpha.2022.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X., Yang T., Yu Z., Liu T., Jin R., Weng L., Bai Y., Gooding J.J., Zhang Y., Chen X. Intelligent gold nanoparticles with oncogenic microRNA-dependent activities to manipulate tumorigenic environments for synergistic tumor therapy. Adv. Mater. 2022;34 doi: 10.1002/adma.202110219. [DOI] [PubMed] [Google Scholar]
- 58.Kumthekar P., Rademaker A., Ko C., Dixit K., Schwartz M.A., Sonabend A.M., Sharp L., Lukas R.V., Stupp R., Horbinski C., McCortney K., Stegh A.H. A phase 0 first-in-human study using NU-0129: a gold base spherical nucleic acid (SNA) nanoconjugate targeting BCL2L12 in recurrent glioblastoma patients. J. Clin. Oncol. 2019;37 doi: 10.1200/JCO.2019.37.15_suppl.3012. 3012-3012. [DOI] [Google Scholar]
- 59.MacLean R.C., San Millan A. The evolution of antibiotic resistance. Science. 2019;365:1082–1083. doi: 10.1126/science.aax3879. [DOI] [PubMed] [Google Scholar]
- 60.Zheng K., Setyawati M.I., Leong D.T., Xie J. Antimicrobial silver nanomaterials. Coord. Chem. Rev. 2018;357:1–17. doi: 10.1016/j.ccr.2017.11.019. [DOI] [Google Scholar]
- 61.Lee J.S., Lytton-Jean A.K., Hurst S.J., Mirkin C.A. Silver nanoparticle-oligonucleotide conjugates based on DNA with triple cyclic disulfide moieties. Nano Lett. 2007;7:2112–2115. doi: 10.1021/nl071108g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rische C.H., Goel A., Radovic-Moreno A.F., Gryaznov S.M. Antibacterial silver core spherical nucleic acids. Mater. Today Commun. 2016;9:30–40. doi: 10.1016/j.mtcomm.2016.09.003. [DOI] [Google Scholar]
- 63.Das C., Paul S.S., Saha A., Singh T., Saha A., Im J., Biswas G. Silver-based nanomaterials as therapeutic agents against coronaviruses: a review. Int. J. Nanomed. 2020;15:9301–9315. doi: 10.2147/ijn.S280976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ratan Z.A., Haidere M.F., Nurunnabi M., Shahriar S.M., Ahammad A.J.S., Shim Y.Y., Reaney M.J.T., Cho J.Y. Green chemistry synthesis of silver nanoparticles and their potential anticancer effects. Cancers. 2020;12 doi: 10.3390/cancers12040855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu C., Li K., Xu L., Li B., Li C., Guo S., Li Z., Zhang Y., Hussain A., Tan H., Zhang M., Zhao Y., Huang Y., Liang X.-J. siRNA-functionalized lanthanide nanoparticle enables efficient endosomal escape and cancer treatment. Nano Res. 2022;15:9160–9168. doi: 10.1007/s12274-022-4573-2. [DOI] [Google Scholar]
- 66.Liu B., Huang Z., Liu J. Polyvalent spherical nucleic acids for universal display of functional DNA with ultrahigh stability. Angew. Chem. Int. Ed. 2018;57:9439–9442. doi: 10.1002/anie.201805532. [DOI] [PubMed] [Google Scholar]
- 67.Zhang J., Zhao P., Li W., Ye L., Li L., Li Z., Li M. Near-infrared light-activatable spherical nucleic acids for conditional control of protein activity. Angew. Chem. Int. Ed. 2022;61 doi: 10.1002/anie.202117562. [DOI] [PubMed] [Google Scholar]
- 68.Kamaly N., Yameen B., Wu J., Farokhzad O.C. Degradable controlled-release polymers and polymeric nanoparticles: mechanisms of controlling drug release. Chem. Rev. 2016;116:2602–2663. doi: 10.1021/acs.chemrev.5b00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen A.M., Zhang M., Wei D., Stueber D., Taratula O., Minko T., He H. Co-delivery of doxorubicin and Bcl-2 siRNA by mesoporous silica nanoparticles enhances the efficacy of chemotherapy in multidrug-resistant cancer cells. Small. 2009;5:2673–2677. doi: 10.1002/smll.200900621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alemdaroglu F.E., Alemdaroglu N.C., Langguth P., Herrmann A. DNA block copolymer micelles – a combinatorial tool for cancer nanotechnology. Adv. Mater. 2008;20:899–902. doi: 10.1002/adma.200700866. [DOI] [Google Scholar]
- 71.Tan X., Lu X., Jia F., Liu X., Sun Y., Logan J.K., Zhang K. Blurring the role of oligonucleotides: spherical nucleic acids as a drug delivery vehicle. J. Am. Chem. Soc. 2016;138:10834–10837. doi: 10.1021/jacs.6b07554. [DOI] [PubMed] [Google Scholar]
- 72.Fang Y., Lu X., Wang D., Cai J., Wang Y., Chen P., Ren M., Lu H., Union J., Zhang L., Sun Y., Jia F., Kang X., Tan X., Zhang K. Spherical nucleic acids for topical treatment of hyperpigmentation. J. Am. Chem. Soc. 2021;143:1296–1300. doi: 10.1021/jacs.0c12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu S., Xing H., Gordiichuk P., Park J., Mirkin C.A. PLGA spherical nucleic acids. Adv. Mater. 2018;30 doi: 10.1002/adma.201707113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Banga R.J., Meckes B., Narayan S.P., Sprangers A.J., Nguyen S.T., Mirkin C.A. Cross-linked micellar spherical nucleic acids from thermoresponsive templates. J. Am. Chem. Soc. 2017;139:4278–4281. doi: 10.1021/jacs.6b13359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ding F., Mou Q., Ma Y., Pan G., Guo Y., Tong G., Choi C.H.J., Zhu X., Zhang C. A crosslinked nucleic acid nanogel for effective siRNA delivery and antitumor therapy. Angew. Chem. Int. Ed. 2018;57:3064–3068. doi: 10.1002/anie.201711242. [DOI] [PubMed] [Google Scholar]
- 76.Jiang K., Zhao D., Ye R., Liu X., Gao C., Guo Y., Zhang C., Zeng J., Wang S., Song J. Transdermal delivery of poly-hyaluronic acid-based spherical nucleic acids for chemogene therapy. Nanoscale. 2022;14:1834–1846. doi: 10.1039/d1nr06353g. [DOI] [PubMed] [Google Scholar]
- 77.Krejsa C., Rogge M., Sadee W. Protein therapeutics: new applications for pharmacogenetics. Nat. Rev. Drug Discov. 2006;5:507–521. doi: 10.1038/nrd2039. [DOI] [PubMed] [Google Scholar]
- 78.Cheng L., Yang L., Meng F., Zhong Z. Protein nanotherapeutics as an emerging modality for cancer therapy. Adv. Healthcare Mater. 2018;7 doi: 10.1002/adhm.201800685. [DOI] [PubMed] [Google Scholar]
- 79.Gu Z., Biswas A., Zhao M., Tang Y. Tailoring nanocarriers for intracellular protein delivery. Chem. Soc. Rev. 2011;40:3638–3655. doi: 10.1039/c0cs00227e. [DOI] [PubMed] [Google Scholar]
- 80.Brodin J.D., Sprangers A.J., McMillan J.R., Mirkin C.A. DNA-mediated cellular delivery of functional enzymes. J. Am. Chem. Soc. 2015;137:14838–14841. doi: 10.1021/jacs.5b09711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kusmierz C.D., Callmann C.E., Kudruk S., Distler M.E., Mirkin C.A. Transferrin aptamers increase the in vivo blood-brain barrier targeting of protein spherical nucleic acids. Bioconjugate Chem. 2022;33:1803–1810. doi: 10.1021/acs.bioconjchem.2c00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xing H., Hwang K., Lu Y. Recent developments of liposomes as nanocarriers for theranostic applications. Theranostics. 2016;6:1336–1352. doi: 10.7150/thno.15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Banga R.J., Chernyak N., Narayan S.P., Nguyen S.T., Mirkin C.A. Liposomal spherical nucleic acids. J. Am. Chem. Soc. 2014;136:9866–9869. doi: 10.1021/ja504845f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dimitrov E., Toncheva-Moncheva N., Bakardzhiev P., Forys A., Doumanov J., Mladenova K., Petrova S., Trzebicka B., Rangelov S. Nucleic acid-based supramolecular structures: vesicular spherical nucleic acids from a non-phospholipid nucleolipid. Nanoscale Adv. 2022;4:3793–3803. doi: 10.1039/d2na00527a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dimitrov E., Toncheva-Moncheva N., Bakardzhiev P., Forys A., Doumanov J., Mladenova K., Petrova S., Trzebicka B., Rangelov S. Original synthesis of a nucleolipid for preparation of vesicular spherical nucleic acids. Nanomaterials. 2022;12 doi: 10.3390/nano12203645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grafals-Ruiz N., Rios-Vicil C.I., Lozada-Delgado E.L., Quiñones-Díaz B.I., Noriega-Rivera R.A., Martínez-Zayas G., Santana-Rivera Y., Santiago-Sánchez G.S., Valiyeva F., Vivas-Mejía P.E. Brain targeted gold liposomes improve RNAi delivery for glioblastoma. Int. J. Nanomed. 2020;15:2809–2828. doi: 10.2147/ijn.S241055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yin H., Kanasty R.L., Eltoukhy A.A., Vegas A.J., Dorkin J.R., Anderson D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 88.Kim H., Jeong I.H., Choi Y.K., Lee Y.K., Moon E., Huh Y.H., Im W., Jin J.O., Kwak M., Lee P.C. Suppression of lung cancer malignancy by micellized siRNA through cell cycle arrest. Adv. Healthcare Mater. 2023 doi: 10.1002/adhm.202202358. [DOI] [PubMed] [Google Scholar]
- 89.Ruan W., Zheng M., An Y., Liu Y., Lovejoy D.B., Hao M., Zou Y., Lee A., Yang S., Lu Y., Morsch M., Chung R., Shi B. DNA nanoclew templated spherical nucleic acids for siRNA delivery. Chem. Commun. 2018;54:3609–3612. doi: 10.1039/c7cc09257a. [DOI] [PubMed] [Google Scholar]
- 90.Zhu L., Guo Y., Qian Q., Yan D., Li Y., Zhu X., Zhang C. Carrier-free delivery of precise drug-chemogene conjugates for synergistic treatment of drug-resistant cancer. Angew. Chem. Int. Ed. 2020;59:17944–17950. doi: 10.1002/anie.202006895. [DOI] [PubMed] [Google Scholar]
- 91.Guo Y., Zhang J., Ding F., Pan G., Li J., Feng J., Zhu X., Zhang C. Stressing the role of DNA as a drug Carrier: synthesis of DNA-drug conjugates through grafting chemotherapeutics onto phosphorothioate oligonucleotides. Adv. Mater. 2019;31 doi: 10.1002/adma.201807533. [DOI] [PubMed] [Google Scholar]
- 92.Guo Y., Qin J., Zhao Q., Yang J., Wei X., Huang Y., Xie M., Zhang C., Li Y. Plaque-targeted rapamycin spherical nucleic acids for synergistic atherosclerosis treatment. Adv. Sci. 2022;9 doi: 10.1002/advs.202105875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen L., Li G., Wang X., Li J., Zhang Y. Spherical nucleic acids for near-infrared light-responsive self-delivery of small-interfering RNA and antisense oligonucleotide. ACS Nano. 2021;15:11929–11939. doi: 10.1021/acsnano.1c03072. [DOI] [PubMed] [Google Scholar]
- 94.Yu W., Xuan C., Liu B., Zhou L., Yin N., Gong E., Zhang Z., Li Y., Zhang K., Shi J. Carrier-free programmed spherical nucleic acid for effective ischemic stroke therapy via self-delivery antisense oligonucleotide. Nano Res. 2023;16:735–745. doi: 10.1007/s12274-022-4402-7. [DOI] [Google Scholar]
- 95.Samanta D., Ebrahimi S.B., Kusmierz C.D., Cheng H.F., Mirkin C.A. Protein spherical nucleic acids for live-cell chemical analysis. J. Am. Chem. Soc. 2020;142:13350–13355. doi: 10.1021/jacs.0c06866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chinen A.B., Guan C.M., Ko C.H., Mirkin C.A. The impact of protein corona formation on the macrophage cellular uptake and biodistribution of spherical nucleic acids. Small. 2017;13 doi: 10.1002/smll.201603847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chinen A.B., Ferrer J.R., Merkel T.J., Mirkin C.A. Relationships between poly(ethylene glycol) modifications on RNA-spherical nucleic acid conjugates and cellular uptake and circulation time. Bioconjugate Chem. 2016;27:2715–2721. doi: 10.1021/acs.bioconjchem.6b00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schöttler S., Landfester K., Mailänder V. Controlling the stealth effect of nanocarriers through understanding the protein corona. Angew. Chem. Int. Ed. 2016;55:8806–8815. doi: 10.1002/anie.201602233. [DOI] [PubMed] [Google Scholar]
- 99.Zhang W., Meckes B., Mirkin C.A. Spherical nucleic acids with tailored and active protein coronae. ACS Cent. Sci. 2019;5:1983–1990. doi: 10.1021/acscentsci.9b01105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang S., Zhang J., Zhou H., Lu Y.C., Jin X., Luo L., You J. The role of protein corona on nanodrugs for organ-targeting and its prospects of application. J. Contr. Release. 2023;360:15–43. doi: 10.1016/j.jconrel.2023.06.014. [DOI] [PubMed] [Google Scholar]
- 101.Tähtinen V., Gulumkar V., Maity S.K., Yliperttula A.M., Siekkinen S., Laine T., Lisitsyna E., Haapalehto I., Viitala T., Vuorimaa-Laukkanen E., Yliperttula M., Virta P. Assembly of bleomycin saccharide-decorated spherical nucleic acids. Bioconjugate Chem. 2022;33:206–218. doi: 10.1021/acs.bioconjchem.1c00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Song W.C., Kim K.R., Park M., Lee K.E., Ahn D.R. Backbone-modified oligonucleotides for tuning the cellular uptake behaviour of spherical nucleic acids. Biomater. Sci. 2017;5:412–416. doi: 10.1039/c6bm00792a. [DOI] [PubMed] [Google Scholar]
- 103.Patz E.F., Jr., Pinsky P., Gatsonis C., Sicks J.D., Kramer B.S., Tammemägi M.C., Chiles C., Black W.C., Aberle D.R., N.O M.W.T. For the, Overdiagnosis in low-dose computed tomography screening for lung cancer. JAMA Intern. Med. 2014;174:269–274. doi: 10.1001/jamainternmed.2013.12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Welch H.G., Prorok P.C., O'Malley A.J., Kramer B.S. Breast-cancer tumor size, overdiagnosis, and mammography screening effectiveness. N. Engl. J. Med. 2016;375:1438–1447. doi: 10.1056/NEJMoa1600249. [DOI] [PubMed] [Google Scholar]
- 105.Etzioni R., Penson D.F., Legler J.M., di Tommaso D., Boer R., Gann P.H., Feuer E.J. Overdiagnosis due to prostate-specific antigen screening: lessons from U.S. prostate cancer incidence trends. J. Natl. Cancer Inst. 2002;94:981–990. doi: 10.1093/jnci/94.13.981. [DOI] [PubMed] [Google Scholar]
- 106.Brenner H., Altenhofen L., Stock C., Hoffmeister M. Prevention, early detection, and overdiagnosis of colorectal cancer within 10 years of screening colonoscopy in Germany. Clin. Gastroenterol. Hepatol. 2015;13:717–723. doi: 10.1016/j.cgh.2014.08.036. [DOI] [PubMed] [Google Scholar]
- 107.Rich N.E., Singal A.G. Overdiagnosis of hepatocellular carcinoma: prevented by guidelines? Hepatology. 2022;75:740–753. doi: 10.1002/hep.32284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ye F., Zhao Y., El-Sayed R., Muhammed M., Hassan M. Advances in nanotechnology for cancer biomarkers. Nano Today. 2018;18:103–123. doi: 10.1016/j.nantod.2017.12.008. [DOI] [Google Scholar]
- 109.Kwong G.A., von Maltzahn G., Murugappan G., Abudayyeh O., Mo S., Papayannopoulos I.A., Sverdlov D.Y., Liu S.B., Warren A.D., Popov Y., Schuppan D., Bhatia S.N. Mass-encoded synthetic biomarkers for multiplexed urinary monitoring of disease. Nat. Biotechnol. 2013;31:63–70. doi: 10.1038/nbt.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jiang Z., Le N.D.B., Gupta A., Rotello V.M. Cell surface-based sensing with metallic nanoparticles. Chem. Soc. Rev. 2015;44:4264–4274. doi: 10.1039/c4cs00387j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lewis J.M., Heineck D.P., Heller M.J. Detecting cancer biomarkers in blood: challenges for new molecular diagnostic and point-of-care tests using cell-free nucleic acids. Expert Rev. Mol. Diagn. 2015;15:1187–1200. doi: 10.1586/14737159.2015.1069709. [DOI] [PubMed] [Google Scholar]
- 112.Ebrahimi S.B., Samanta D., Mirkin C.A. DNA-based nanostructures for live-cell analysis. J. Am. Chem. Soc. 2020;142:11343–11356. doi: 10.1021/jacs.0c04978. [DOI] [PubMed] [Google Scholar]
- 113.Lytton-Jean A.K., Mirkin C.A. A thermodynamic investigation into the binding properties of DNA functionalized gold nanoparticle probes and molecular fluorophore probes. J. Am. Chem. Soc. 2005;127:12754–12755. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]
- 114.Elghanian R., Storhoff J.J., Mucic R.C., Letsinger R.L., Mirkin C.A. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 115.Mitchell G.P., Mirkin C.A., Letsinger R.L. Programmed assembly of DNA functionalized quantum dots. J. Am. Chem. Soc. 1999;121:8122–8123. doi: 10.1021/ja991662v. [DOI] [Google Scholar]
- 116.Cai Q., Wu D., Li H., Jie G., Zhou H. Versatile photoelectrochemical and electrochemiluminescence biosensor based on 3D CdSe QDs-DNA nanonetwork-SnO(2) nanoflower coupled with DNA walker amplification for HIV detection. Biosens. Bioelectron. 2021;191 doi: 10.1016/j.bios.2021.113455. [DOI] [PubMed] [Google Scholar]
- 117.Yang Y., Huang J., Yang X., Quan K., Wang H., Ying L., Xie N., Ou M., Wang K. FRET nanoflares for intracellular mRNA detection: avoiding false positive signals and minimizing effects of system fluctuations. J. Am. Chem. Soc. 2015;137:8340–8343. doi: 10.1021/jacs.5b04007. [DOI] [PubMed] [Google Scholar]
- 118.Rosi N.L., Giljohann D.A., Thaxton C.S., Lytton-Jean A.K., Han M.S., Mirkin C.A. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 119.Jacobson D.R., McIntosh D.B., Stevens M.J., Rubinstein M., Saleh O.A. Single-stranded nucleic acid elasticity arises from internal electrostatic tension. Proc. Natl. Acad. Sci. U.S.A. 2017;114:5095–5100. doi: 10.1073/pnas.1701132114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gao P., Liu B., Pan W., Li N., Tang B. A spherical nucleic acid probe based on the Au-Se bond. Anal. Chem. 2020;92:8459–8463. doi: 10.1021/acs.analchem.0c01204. [DOI] [PubMed] [Google Scholar]
- 121.Hu B., Kong F., Gao X., Jiang L., Li X., Gao W., Xu K., Tang B. Avoiding thiol compound interference: a nanoplatform based on high-fidelity Au-Se bonds for biological applications. Angew. Chem. Int. Ed. 2018;57:5306–5309. doi: 10.1002/anie.201712921. [DOI] [PubMed] [Google Scholar]
- 122.Qing Z., Luo G., Xing S., Zou Z., Lei Y., Liu J., Yang R. Pt-S bond-mediated nanoflares for high-fidelity intracellular applications by avoiding thiol cleavage. Angew. Chem. Int. Ed. 2020;59:14044–14048. doi: 10.1002/anie.202003964. [DOI] [PubMed] [Google Scholar]
- 123.Wang C., Zhang H., Zeng D., Sun W., Zhang H., Aldalbahi A., Wang Y., San L., Fan C., Zuo X., Mi X. Elaborately designed diblock nanoprobes for simultaneous multicolor detection of microRNAs. Nanoscale. 2015;7:15822–15829. doi: 10.1039/c5nr04618a. [DOI] [PubMed] [Google Scholar]
- 124.Qian Q., He G., Wang C., Li S., Zhao X., Xu Y., Mi X. Poly-adenine-mediated spherical nucleic acid probes for live cell fluorescence imaging of tumor-related microRNAs. Mol. Biol. Rep. 2022;49:3705–3712. doi: 10.1007/s11033-022-07210-w. [DOI] [PubMed] [Google Scholar]
- 125.Zhu D., Zhao D., Huang J., Zhu Y., Chao J., Su S., Li J., Wang L., Shi J., Zuo X., Weng L., Li Q., Wang L. Poly-adenine-mediated fluorescent spherical nucleic acid probes for live-cell imaging of endogenous tumor-related mRNA. Nanomed. Nanotechnol. Biol. Med. 2018;14:1797–1807. doi: 10.1016/j.nano.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 126.Wang L., Zhang H., Wang C., Xu Y., Su J., Wang X., Liu X., Feng D., Wang L., Zuo X., Shi J., Ge Z., Fan C., Mi X. Poly-adenine-mediated spherical nucleic acids for strand displacement-based DNA/RNA detection. Biosens. Bioelectron. 2019;127:85–91. doi: 10.1016/j.bios.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 127.Zeng T., Fang J., Jiang Y., Xing C., Lu C., Yang H. Spherical nucleic acid probe based on 2'-fluorinated DNA functionalization for high-fidelity intracellular sensing. Anal. Chem. 2022;94:18009–18016. doi: 10.1021/acs.analchem.2c04294. [DOI] [PubMed] [Google Scholar]
- 128.Hu X., Ke G., Liu L., Fu X., Kong G., Xiong M., Chen M., Zhang X.B. Valency-controlled molecular spherical nucleic acids with tunable biosensing performances. Anal. Chem. 2019;91:11374–11379. doi: 10.1021/acs.analchem.9b02614. [DOI] [PubMed] [Google Scholar]
- 129.Huo M., Li S., Zhang P., Feng Y., Liu Y., Wu N., Ju H., Ding L. Nanoamplicon comparator for live-cell microRNA imaging. Anal. Chem. 2019;91:3374–3381. doi: 10.1021/acs.analchem.8b04661. [DOI] [PubMed] [Google Scholar]
- 130.Xiao F., Fang X., Li H., Xue H., Wei Z., Zhang W., Zhu Y., Lin L., Zhao Y., Wu C., Tian L. Light-harvesting fluorescent spherical nucleic acids self-assembled from a DNA-grafted conjugated polymer for amplified detection of nucleic acids. Angew. Chem. Int. Ed. 2022;61 doi: 10.1002/anie.202115812. [DOI] [PubMed] [Google Scholar]
- 131.Qu X., Zhu D., Yao G., Su S., Chao J., Liu H., Zuo X., Wang L., Shi J., Wang L., Huang W., Pei H., Fan C. An exonuclease III-powered, on-particle stochastic DNA walker. Angew. Chem. Int. Ed. 2017;56:1855–1858. doi: 10.1002/anie.201611777. [DOI] [PubMed] [Google Scholar]
- 132.Seferos D.S., Giljohann D.A., Hill H.D., Prigodich A.E., Mirkin C.A. Nano-flares: probes for transfection and mRNA detection in living cells. J. Am. Chem. Soc. 2007;129:15477–15479. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dubertret B., Calame M., Libchaber A.J. Single-mismatch detection using gold-quenched fluorescent oligonucleotides. Nat. Biotechnol. 2001;19:365–370. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- 134.Prigodich A.E., Lee O.-S., Daniel W.L., Seferos D.S., Schatz G.C., Mirkin C.A. Tailoring DNA structure to increase target hybridization kinetics on surfaces. J. Am. Chem. Soc. 2010;132:10638–10641. doi: 10.1021/ja104859j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lin M., Yi X., Huang F., Ma X., Zuo X., Xia F. Photoactivated nanoflares for mRNA detection in single living cells. Anal. Chem. 2019;91:2021–2027. doi: 10.1021/acs.analchem.8b04434. [DOI] [PubMed] [Google Scholar]
- 136.Prigodich A.E., Randeria P.S., Briley W.E., Kim N.J., Daniel W.L., Giljohann D.A., Mirkin C.A. Multiplexed nanoflares: mRNA detection in live cells. Anal. Chem. 2012;84:2062–2066. doi: 10.1021/ac202648w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xavier M., Kyriazi M.E., Lanham S., Alexaki K., Matthews E., El-Sagheer A.H., Brown T., Kanaras A.G., Oreffo R.O.C. Enrichment of skeletal stem cells from human bone marrow using spherical nucleic acids. ACS Nano. 2021;15:6909–6916. doi: 10.1021/acsnano.0c10683. [DOI] [PubMed] [Google Scholar]
- 138.Zhao J., Liu C., Li Y., Ma Y., Deng J., Li L., Sun J. Thermophoretic detection of exosomal microRNAs by nanoflares. J. Am. Chem. Soc. 2020;142:4996–5001. doi: 10.1021/jacs.9b13960. [DOI] [PubMed] [Google Scholar]
- 139.Wiraja C., Mori Y., Ichimura T., Hwang J., Xu C., Bonventre J.V. Nephrotoxicity assessment with human kidney tubuloids using spherical nucleic acid-based mRNA nanoflares. Nano Lett. 2021;21:5850–5858. doi: 10.1021/acs.nanolett.1c01840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Briley W.E., Bondy M.H., Randeria P.S., Dupper T.J., Mirkin C.A. Quantification and real-time tracking of RNA in live cells using Sticky-flares. Proc. Natl. Acad. Sci. U.S.A. 2015;112:9591–9595. doi: 10.1073/pnas.1510581112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wu Q., Liu Z., Su L., Han G., Liu R., Zhao J., Zhao T., Jiang C., Zhang Z. Sticky-flares for in situ monitoring of human telomerase RNA in living cells. Nanoscale. 2018;10:9386–9392. doi: 10.1039/c8nr01260a. [DOI] [PubMed] [Google Scholar]
- 142.Zhao T., Dong F., Hu X., Xu Y., Wei W., Liu R., Yu F., Fang W., Shen Y., Zhang Z. Dynamic tracking of p21 mRNA in living cells by sticky-flares for the visual evaluation of the tumor treatment effect. Nanoscale. 2022;14:1733–1741. doi: 10.1039/d1nr05418j. [DOI] [PubMed] [Google Scholar]
- 143.Li Y., Chen Q., Pan X., Lu W., Zhang J. New insight into the application of fluorescence platforms in tumor diagnosis: from chemical basis to clinical application. Med. Res. Rev. 2023;43:570–613. doi: 10.1002/med.21932. [DOI] [PubMed] [Google Scholar]
- 144.Gao M., Yu F., Lv C., Choo J., Chen L. Fluorescent chemical probes for accurate tumor diagnosis and targeting therapy. Chem. Soc. Rev. 2017;46:2237–2271. doi: 10.1039/c6cs00908e. [DOI] [PubMed] [Google Scholar]
- 145.Li J., Li Y., Pan L., Pan W., Li N., Tang B. Spherical nucleic acids-based biosensors for cancer biomarkers detection. Trends Anal. Chem. 2022;157 doi: 10.1016/j.trac.2022.116807. [DOI] [Google Scholar]
- 146.Jou A.F., Lu C.H., Ou Y.C., Wang S.S., Hsu S.L., Willner I., Ho J.A. Diagnosing the miR-141 prostate cancer biomarker using nucleic acid-functionalized CdSe/ZnS QDs and telomerase. Chem. Sci. 2015;6:659–665. doi: 10.1039/c4sc02104e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhao Y., Zhao J., Zhang J., Sun Y., Li L., Li Z., Li M. Enzymatically controlled nanoflares for specific molecular recognition and biosensing. Anal. Chem. 2022;94:8883–8889. doi: 10.1021/acs.analchem.2c00166. [DOI] [PubMed] [Google Scholar]
- 148.Qiao G., Zhuo L., Gao Y., Yu L., Li N., Tang B. A tumor mRNA-dependent gold nanoparticle-molecular beacon carrier for controlled drug release and intracellular imaging. Chem. Commun. 2011;47:7458–7460. doi: 10.1039/c1cc11490e. [DOI] [PubMed] [Google Scholar]
- 149.Pan W., Yang H., Zhang T., Li Y., Li N., Tang B. Dual-targeted nanocarrier based on cell surface receptor and intracellular mRNA: an effective strategy for cancer cell imaging and therapy. Anal. Chem. 2013;85:6930–6935. doi: 10.1021/ac401405n. [DOI] [PubMed] [Google Scholar]
- 150.Li S., Jiang Q., Liu Y., Wang W., Yu W., Wang F., Liu X. Precision spherical nucleic acids enable sensitive FEN1 imaging and controllable drug delivery for cancer-specific therapy. Anal. Chem. 2021;93:11275–11283. doi: 10.1021/acs.analchem.1c02264. [DOI] [PubMed] [Google Scholar]
- 151.Liu Q., Huang Y., Li L., Li Z., Li M. Endogenous enzyme-operated spherical nucleic acids for cell-selective protein capture and localization regulation. Angew. Chem. Int. Ed. 2023 doi: 10.1002/anie.202214958. [DOI] [PubMed] [Google Scholar]
- 152.Li N., Xiang M.H., Liu J.W., Tang H., Jiang J.H. DNA polymer nanoparticles programmed via supersandwich hybridization for imaging and therapy of cancer cells. Anal. Chem. 2018;90:12951–12958. doi: 10.1021/acs.analchem.8b03253. [DOI] [PubMed] [Google Scholar]
- 153.Hu S., Kang H., Baek Y., El Fakhri G., Kuang A., Choi H.S. Real-time imaging of brain tumor for image-guided surgery. Adv. Healthcare Mater. 2018;7 doi: 10.1002/adhm.201800066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rosenthal E.L., Warram J.M., Bland K.I., Zinn K.R. The status of contemporary image-guided modalities in oncologic surgery. Ann. Surg. 2015;261:46–55. doi: 10.1097/sla.0000000000000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Weissleder R. Molecular imaging in cancer. Science. 2006;312:1168–1171. doi: 10.1126/science.1125949. [DOI] [PubMed] [Google Scholar]
- 156.Zhou Z., Lu Z.R. Molecular imaging of the tumor microenvironment. Adv. Drug Deliv. Rev. 2017;113:24–48. doi: 10.1016/j.addr.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 157.Xiao F., Lin L., Chao Z., Shao C., Chen Z., Wei Z., Lu J., Huang Y., Li L., Liu Q., Liang Y., Tian L. Organic spherical nucleic acids for the transport of a NIR-II-emitting dye across the blood-brain barrier. Angew. Chem. Int. Ed. 2020;59:9702–9710. doi: 10.1002/anie.202002312. [DOI] [PubMed] [Google Scholar]
- 158.Liu Z., Zhao J., Zhang R., Han G., Zhang C., Liu B., Zhang Z., Han M.Y., Gao X. Cross-platform cancer cell identification using telomerase-specific spherical nucleic acids. ACS Nano. 2018;12:3629–3637. doi: 10.1021/acsnano.8b00743. [DOI] [PubMed] [Google Scholar]
- 159.Hayes A.J., Maynard L., Coombes G., Newton-Bishop J., Timmons M., Cook M., Theaker J., Bliss J.M., Thomas J.M. Wide versus narrow excision margins for high-risk, primary cutaneous melanomas: long-term follow-up of survival in a randomised trial. Lancet. 2016;17:184–192. doi: 10.1016/s1470-2045(15)00482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chan Y.K., Gack M.U. Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 2016;14:360–373. doi: 10.1038/nrmicro.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Schlee M., Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 2016;16:566–580. doi: 10.1038/nri.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lind N.A., Rael V.E., Pestal K., Liu B., Barton G.M. Regulation of the nucleic acid-sensing Toll-like receptors. Nat. Rev. Immunol. 2022;22:224–235. doi: 10.1038/s41577-021-00577-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Barber G.N. STING: infection, inflammation and cancer. Nat. Rev. Immunol. 2015;15:760–770. doi: 10.1038/nri3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Desmet C.J., Ishii K.J. Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination. Nat. Rev. Immunol. 2012;12:479–491. doi: 10.1038/nri3247. [DOI] [PubMed] [Google Scholar]
- 165.Meng F., Wang J., Yeo Y. Nucleic acid and oligonucleotide delivery for activating innate immunity in cancer immunotherapy. J. Contr. Release. 2022;345:586–600. doi: 10.1016/j.jconrel.2022.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Blake I.M., Pons-Salort M., Molodecky N.A., Diop O.M., Chenoweth P., Bandyopadhyay A.S., Zaffran M., Sutter R.W., Grassly N.C. Type 2 poliovirus detection after global withdrawal of trivalent oral vaccine. N. Engl. J. Med. 2018;379:834–845. doi: 10.1056/NEJMoa1716677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bester J.C. Measles and measles vaccination: a review. JAMA Pediatr. 2016;170:1209–1215. doi: 10.1001/jamapediatrics.2016.1787. [DOI] [PubMed] [Google Scholar]
- 168.The D. Lancet Infectious, Rinderpest, smallpox, and the imperative of destruction. Lancet Infect. Dis. 2019;19:789. doi: 10.1016/s1473-3099(19)30358-5. [DOI] [PubMed] [Google Scholar]
- 169.Vogel F.R., Sarver N. Nucleic acid vaccines. Clin. Microbiol. Rev. 1995;8:406–410. doi: 10.1128/cmr.8.3.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rando Halie M., Lordan R., Kolla L., Sell E., Lee Alexandra J., Wellhausen N., Naik A., Kamil Jeremy P., Gitter A., Greene Casey S. The coming of age of nucleic acid vaccines during COVID-19. mSystems. 2023;0 doi: 10.1128/msystems.00928-22. 00928-00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Luo M., Samandi L.Z., Wang Z., Chen Z.J., Gao J. Synthetic nanovaccines for immunotherapy. J. Contr. Release. 2017;263:200–210. doi: 10.1016/j.jconrel.2017.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Azharuddin M., Zhu G.H., Sengupta A., Hinkula J., Slater N.K.H., Patra H.K. Nano toolbox in immune modulation and nanovaccines. Trends Biotechnol. 2022;40:1195–1212. doi: 10.1016/j.tibtech.2022.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tokatlian T., Read B.J., Jones C.A., Kulp D.W., Menis S., Chang J.Y.H., Steichen J.M., Kumari S., Allen J.D., Dane E.L., Liguori A., Sangesland M., Lingwood D., Crispin M., Schief W.R., Irvine D.J. Innate immune recognition of glycans targets HIV nanoparticle immunogens to germinal centers. Science. 2019;363:649–654. doi: 10.1126/science.aat9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Teplensky M.H., Evangelopoulos M., Dittmar J.W., Forsyth C.M., Sinegra A.J., Wang S., Mirkin C.A. Multi-antigen spherical nucleic acid cancer vaccines. Nat. Biomed. Eng. 2023 doi: 10.1038/s41551-022-01000-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhu M. Immunological perspectives on spatial and temporal vaccine delivery. Adv. Drug Deliv. Rev. 2021;178 doi: 10.1016/j.addr.2021.113966. [DOI] [PubMed] [Google Scholar]
- 177.Zhang Y.N., Lazarovits J., Poon W., Ouyang B., Nguyen L.N.M., Kingston B.R., Chan W.C.W. Nanoparticle size influences antigen retention and presentation in lymph node follicles for humoral immunity. Nano Lett. 2019;19:7226–7235. doi: 10.1021/acs.nanolett.9b02834. [DOI] [PubMed] [Google Scholar]
- 178.Wang S., Qin L., Yamankurt G., Skakuj K., Huang Z., Chen P.C., Dominguez D., Lee A., Zhang B., Mirkin C.A. Rational vaccinology with spherical nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 2019;116:10473–10481. doi: 10.1073/pnas.1902805116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Huang Z., Callmann C.E., Wang S., Vasher M.K., Evangelopoulos M., Petrosko S.H., Mirkin C.A. Rational vaccinology: harnessing nanoscale chemical design for cancer immunotherapy. ACS Cent. Sci. 2022;8:692–704. doi: 10.1021/acscentsci.2c00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Teplensky M.H., Dittmar J.W., Qin L., Wang S., Evangelopoulos M., Zhang B., Mirkin C.A. Spherical nucleic acid vaccine structure markedly influences adaptive immune responses of clinically utilized prostate cancer targets. Adv. Healthcare Mater. 2021;10 doi: 10.1002/adhm.202101262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Krieg A.M., Yi A.K., Matson S., Waldschmidt T.J., Bishop G.A., Teasdale R., Koretzky G.A., Klinman D.M. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 182.Wei G., Wang Y., Yang G., Wang Y., Ju R. Recent progress in nanomedicine for enhanced cancer chemotherapy. Theranostics. 2021;11:6370–6392. doi: 10.7150/thno.57828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wilhelm S., Tavares A.J., Dai Q., Ohta S., Audet J., Dvorak H.F., Chan W.C.W. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016;1 doi: 10.1038/natrevmats.2016.14. [DOI] [Google Scholar]
- 184.Pandit S., Dutta D., Nie S. Active transcytosis and new opportunities for cancer nanomedicine. Nat. Mater. 2020;19:478–480. doi: 10.1038/s41563-020-0672-1. [DOI] [PubMed] [Google Scholar]
- 185.Milling L., Zhang Y., Irvine D.J. Delivering safer immunotherapies for cancer. Adv. Drug Deliv. Rev. 2017;114:79–101. doi: 10.1016/j.addr.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 187.Hong E., Halman J.R., Shah A.B., Khisamutdinov E.F., Dobrovolskaia M.A., Afonin K.A. Structure and composition define immunorecognition of nucleic acid nanoparticles. Nano Lett. 2018;18:4309–4321. doi: 10.1021/acs.nanolett.8b01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Radovic-Moreno A.F., Chernyak N., Mader C.C., Nallagatla S., Kang R.S., Hao L., Walker D.A., Halo T.L., Merkel T.J., Rische C.H., Anantatmula S., Burkhart M., Mirkin C.A., Gryaznov S.M. Immunomodulatory spherical nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 2015;112:3892–3897. doi: 10.1073/pnas.1502850112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.O'Day S., Perez C., Wise-Draper T., Hanna G., Bhatia S., Kelly C., Medina T., Laux D., Daud A., Chandra S., Shaheen M., Gao L., Burgess M., Hernandez-Aya L., Yeung C., Smythe K., DeGoma E., Daniel W., Feltner D., Sindelar L., Michel R., Bexon A., Bexon M., Milhem M. 423 Safety and preliminary efficacy of intratumoral cavrotolimod (AST-008), a spherical nucleic acid TLR9 agonist, in combination with pembrolizumab in patients with advanced solid tumors. J. Immunotherapy Cancer. 2020;8:A257. doi: 10.1136/jitc-2020-SITC2020.0423. [DOI] [Google Scholar]
- 190.Ferrer J.R., Wertheim J.A., Mirkin C.A. Dual Toll-Like receptor targeting liposomal spherical nucleic acids. Bioconjugate Chem. 2019;30:944–951. doi: 10.1021/acs.bioconjchem.9b00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Krishnamachari Y., Salem A.K. Innovative strategies for co-delivering antigens and CpG oligonucleotides. Adv. Drug Deliv. Rev. 2009;61:205–217. doi: 10.1016/j.addr.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Deng B., Ma B., Ma Y., Cao P., Leng X., Huang P., Zhao Y., Ji T., Lu X., Liu L. Doxorubicin and CpG loaded liposomal spherical nucleic acid for enhanced Cancer treatment. J. Nanobiotechnol. 2022;20:140. doi: 10.1186/s12951-022-01353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Callmann C.E., Cole L.E., Kusmierz C.D., Huang Z., Horiuchi D., Mirkin C.A. Tumor cell lysate-loaded immunostimulatory spherical nucleic acids as therapeutics for triple-negative breast cancer. Proc. Natl. Acad. Sci. U.S.A. 2020;117:17543–17550. doi: 10.1073/pnas.2005794117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Qin L., Wang S., Dominguez D., Long A., Chen S., Fan J., Ahn J., Skakuj K., Huang Z., Lee A., Mirkin C., Zhang B. Development of spherical nucleic acids for prostate cancer immunotherapy. Front. Immunol. 2020;11:1333. doi: 10.3389/fimmu.2020.01333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Chen P., Wang D., Wang Y., Zhang L., Wang Q., Liu L., Li J., Sun X., Ren M., Wang R., Fang Y., Zhao J.J., Zhang K. Maximizing TLR9 activation in cancer immunotherapy with dual-adjuvanted spherical nucleic acids. Nano Lett. 2022;22:4058–4066. doi: 10.1021/acs.nanolett.2c00723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mahajan A., Hurley L., Tommasini-Ghelfi S., Dussold C., Stegh A., Mirkin C. IMMU-53. STING-ing glioblastoma with spherical nucleic acids. Neuro Oncol. 2021;23:vi104–vi105. doi: 10.1093/neuonc/noab196.412. [DOI] [Google Scholar]
- 197.Ricketts T.D., Prieto-Dominguez N., Gowda P.S., Ubil E. Mechanisms of macrophage plasticity in the tumor environment: manipulating activation state to improve outcomes. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.642285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Liu J., Guo L., Mi Z., Liu Z., Rong P., Zhou W. Vascular bursts-mediated tumor accumulation and deep penetration of spherical nucleic acids for synergistic radio-immunotherapy. J. Contr. Release. 2022;348:1050–1065. doi: 10.1016/j.jconrel.2022.06.030. [DOI] [PubMed] [Google Scholar]
- 199.Zhang Q., Guo Y., Zhu L., Liu X., Yang J., Li Y., Zhu X., Zhang C. A nucleic acid nanogel dually bears siRNA and CpG motifs for synergistic tumor immunotherapy. Biomater. Sci. 2021;9:4755–4764. doi: 10.1039/d1bm00531f. [DOI] [PubMed] [Google Scholar]
- 200.Guo Y., Zhang Q., Zhu Q., Gao J., Zhu X., Yu H., Li Y., Zhang C. Copackaging photosensitizer and PD-L1 siRNA in a nucleic acid nanogel for synergistic cancer photoimmunotherapy. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abn2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Chou L., Callmann C.E., Dominguez D., Zhang B., Mirkin C.A. Disrupting the interplay between programmed cell death protein 1 and programmed death ligand 1 with spherical nucleic acids in treating cancer. ACS Cent. Sci. 2022;8:1299–1305. doi: 10.1021/acscentsci.2c00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Callmann C.E., Kusmierz C.D., Dittmar J.W., Broger L., Mirkin C.A. Impact of liposomal spherical nucleic acid structure on immunotherapeutic function. ACS Cent. Sci. 2021;7:892–899. doi: 10.1021/acscentsci.1c00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Skakuj K., Wang S., Qin L., Lee A., Zhang B., Mirkin C.A. Conjugation chemistry-dependent T-cell activation with spherical nucleic acids. J. Am. Chem. Soc. 2018;140:1227–1230. doi: 10.1021/jacs.7b12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Skakuj K., Teplensky M.H., Wang S., Dittmar J.W., Mirkin C.A. Chemically tuning the antigen release kinetics from spherical nucleic acids maximizes immune stimulation. ACS Cent. Sci. 2021;7:1838–1846. doi: 10.1021/acscentsci.1c00779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Huang Z.N., Cole L.E., Callmann C.E., Wang S., Mirkin C.A. Sequence multiplicity within spherical nucleic acids. ACS Nano. 2020;14:1084–1092. doi: 10.1021/acsnano.9b08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Guan C.M., Chinen A.B., Ferrer J.R., Ko C.H., Mirkin C.A. Impact of sequence specificity of spherical nucleic acids on macrophage activation in vitro and in vivo. Mol. Pharm. 2019;16:4223–4229. doi: 10.1021/acs.molpharmaceut.9b00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Meckes B., Banga R.J., Nguyen S.T., Mirkin C.A. Enhancing the stability and immunomodulatory activity of liposomal spherical nucleic acids through lipid-tail DNA modifications. Small. 2018;14 doi: 10.1002/smll.201702909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Elkon K.B. Review: cell death, nucleic acids, and immunity: inflammation beyond the grave. Arthritis Rheumatol. 2018;70:805–816. doi: 10.1002/art.40452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Imanishi T., Ishihara C., Badr Mel S., Hashimoto-Tane A., Kimura Y., Kawai T., Takeuchi O., Ishii K.J., Taniguchi S., Noda T., Hirano H., Brombacher F., Barber G.N., Akira S., Saito T. Nucleic acid sensing by T cells initiates Th2 cell differentiation. Nat. Commun. 2014;5:3566. doi: 10.1038/ncomms4566. [DOI] [PubMed] [Google Scholar]
- 210.Lu Y., Huntoon K., Lee D., Wang Y., Ha J., Qie Y., Li X., Schrank B.R., Dong S., Gallup T.D., Kang M., Zhao H., An Y., Yang Z., Li J., Kim B.Y.S., Jiang W. Immunological conversion of solid tumours using a bispecific nanobioconjugate for cancer immunotherapy. Nat. Nanotechnol. 2022;17:1332–1341. doi: 10.1038/s41565-022-01245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Daniel W.L., Lorch U., Mix S., Bexon A.S. A first-in-human phase 1 study of cavrotolimod, a TLR9 agonist spherical nucleic acid, in healthy participants: evidence of immune activation. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1073777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Shi L., Wu W., Duan Y., Xu L., Xu Y., Hou L., Meng X., Zhu X., Liu B. Light-induced self-escape of spherical nucleic acid from endo/lysosome for efficient non-cationic gene delivery. Angew. Chem. Int. Ed. 2020;59:19168–19174. doi: 10.1002/anie.202006890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.