Summary
Ralstonia solanacearum invades plants through their roots and causes devastating bacterial diseases in multiple crops. Here, we present a versatile inoculation assay in Arabidopsis thaliana seedlings grown in sterile agar plates. We describe steps for plant preparation, bacterial inoculation, tissue sampling, and bacterial quantification. This protocol can be used for accurate assessment of bacterial colonization and observation of plant response to infection.
For complete details on the use and execution of this protocol, please refer to Dindas et al. (2022).1
Subject areas: Microbiology, Model Organisms, Plant Sciences
Graphical abstract

Highlights
-
•
Quantitative evaluation of Arabidopsis susceptibility to R. solanacearum
-
•
Step-by-step instructions for plant preparation and bacterial inoculation
-
•
Versatility to combine plant genotypes, bacterial strains, and chemical treatments
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Ralstonia solanacearum invades plants through their roots and causes devastating bacterial diseases in multiple crops. Here, we present a versatile inoculation assay in Arabidopsis thaliana seedlings grown in sterile agar plates. We describe steps for plant preparation, bacterial inoculation, tissue sampling, and bacterial quantification. This protocol can be used for accurate assessment of bacterial colonization and observation of plant response to infection.
Before you begin
As a soil-borne bacterial pathogen, R. solanacearum penetrates underground root tissues and colonizes xylem vessels in the whole plant, hindering the visualization of disease progression in both temporal and spatial manner. To address this issue, we have recently developed several methods to determine the contribution of either plant genes to host resistance or bacterial genes to virulence and fitness.2,3,4 Although R. solanacearum mainly infects plants from the Solanaceae family, it is also able to infect the model plant Arabidopsis thaliana in laboratory conditions, and this pathosystem has become a powerful tool for genetic and molecular studies of R. solanacearum-plant interaction. To take advantage of the abundant genetic resources in A. thaliana, including mutants, transgenic lines, and fluorescent marker lines, we have developed a method to dissect plant molecular responses and susceptibility to R. solanacearum infection.
This method allows for: 1) observation of fluorescence in reporter or marker lines; 2) biochemical characterization of plant responses to R. solanacearum in various metabolic and signaling transduction pathways; 3) phenotypic observations of plant responses to R. solanacearum, including inhibition of root growth, cell death in the root tip, and formation of lateral roots and root hairs; 4) precise quantification of bacterial growth in different Arabidopsis genotypes; 5) determination of interplays among genotypes, chemical cues, and plant development. This method is performed in fully-controlled sterile conditions, requiring little space of laboratory flow hood and plant growth chamber, which is a significant advantage considering that R. solanacearum is classified as a quarantine microorganism in most countries.
Using this method, we have shown that plant immune responses include an inhibition of phosphate transport to increase R. solanacearum resistance.1 We also showed that R. solanacearum infection enhances plant pyruvate decarboxylase activity, which is involved in tolerance to bacterial wilt disease.5 We have also shown that R. solanacearum infection affects A. thaliana root developmental programs, including the inhibition of primary root growth and a promotion of the emergence of lateral roots and root hairs, in an auxin-dependent and independent manner.6 With the continuous increase of the availability of Arabidopsis genetic materials and genetic manipulation techniques, this protocol will contribute significantly to dissect immune responses to R. solanacearum at the molecular level, elucidate signaling networks, and assess plant susceptibility as an utmost goal.
To simplify the description of this method, we use A. thaliana and R. solanacearum wild-type GMI1000 as examples, and focus on a protocol to quantify bacterial colonization and replication.
Prepare media
Timing: 1 h
-
1.
Prepare MS- medium (MS medium with no sucrose, see recipe) square plates.
-
2.
Prepare solid and liquid phi medium.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| 2-[N-Morpholino] ethanesulfonic acid (MES) | Amresco | Cat#E169 |
| 75% ethanol | Sinopharm Chemical Reagent | Cat#801769610 |
| 100% ethanol | Sinopharm Chemical Reagent | Cat#10009218 |
| Agar | Sinopharm Chemical Reagent | Cat#10000561 |
| Bacto agar | Becton, Dickinson and Co | Cat#214010 |
| Bacto peptone | Becton, Dickinson and Co | Cat#211677 |
| Casein hydrolysate (Casamino acids) | Sigma-Aldrich | Cat#22090-500G |
| Distilled sterile water | N/A | N/A |
| Glucose | Sinopharm Chemical Reagent | Cat#63005518 |
| Murashige & Skoog Basal Medium with Vitamins | PhytoTech Labs | Cat#M519 |
| Potassium hydroxide (KOH) | Sinopharm Chemical Reagent | Cat#10017018 |
| Sucrose | Sinopharm Chemical Reagent | Cat#10021418 |
| Sodium hypochlorite aqueous solution (NaOCl) (11–15% available chlorine) | Sinopharm Chemical Reagent | Cat#80010428 |
| Triphenyltetrazolium chloride (TTC) | Sigma-Aldrich | Cat#T8877-5G |
| Yeast extract | Oxoid | Cat#LP0021 |
| Experimental models: Organisms/strains | ||
| Arabidopsis thaliana: Col-0 | Dindas et al.1 | N/A |
| Ralstonia solanacearum: GMI1000 | Salanoubat et al.7 | N/A |
| Software and algorithms | ||
| GraphPad Prism 9 | GraphPad Software | http://www.graphpad.com |
| Microsoft Excel | Microsoft | |
| Other | ||
| 1.5 mL microcentrifuge tubes | Sangon Biotech | Cat#F601620-0010 |
| 2 mL microcentrifuge tubes | Sangon Biotech | Cat#F601620-0010 |
| Plastic Petri dishes (60 mm diameter) | N/A | N/A |
| Plastic Petri dishes (90 mm diameter) | N/A | N/A |
| Square Petri dishes (10 × 10 cm) | N/A | N/A |
| Pipette tips | AIBIO | Cat#T1040000 |
| Paper towels | Vinda | N/A |
| Metal beads | N/A | N/A |
| Tweezers | Sangon Biotech | Cat#F519021-0001 |
| Scissors | N/A | N/A |
| Spectrophotometer plastic cuvettes | Brand | Cat#759015 |
| Autoclave | SANYO | MLS-3780 |
| Medium- and high-throughput tissue grinder | QIAGEN | Tissue Lyser II |
| Nanodrop spectrophotometer | Thermo Scientific | NanoDrop 2000c |
| Water distiller/sterilizer | Millipore | Milli-Q intergral 10L |
| Vortexer | Scientific Industries | Vortex-Genie 2, Cat#S1-0246 |
| Petri dish incubator at 28°C | Panasonic | I-36VL |
| Flow hood (clean bench) | Shanghai Shangjing Co | CA-1390-1 |
| Plant growth chamber | Percival | I-36VL |
| Electronic balance | Sartorius | BSA224S |
| pH meter | Sartorius | PB-10 |
| Centrifuge | Eppendorf | Centrifuge 5424 |
| Tube incubator (shaker) at 28°C | New Brunswick | Cat#m1324-0006 |
Materials and equipment
| Reagent | Final concentration (w/v%) | Amount |
|---|---|---|
| MS- medium | ||
| Murashige & Skoog (MS) with vitamins | 0.44% | 4.4 g |
| MES | 0.05% | 0.5 g |
| Bacto agar | 0.8% | 8 g |
| ddH2O | N/A | Up to 1 L |
| Total | N/A | 1 L |
| To prepare 1 L of MS- medium, mix 4.4 g MS with Vitamins, 0.5 g MES in 500 mL of double-distilled water (ddH2O) using a magnetic stirrer until the powders are dissolved completely, adjust the pH to 5.8 with KOH, add 8 g bacto agar and make up the volume to 1 L with ddH2O, and autoclave at 121°C for 20 min. Store at room temperature (23°C–25°C) for up to 1 month. | ||
| 1/2 MS medium | ||
| Murashige & Skoog (MS) with vitamins | 0.221% | 2.21 g |
| Sucrose | 1.5% | 15 g |
| Agar | 0.8% | 8 g |
| ddH2O | N/A | Up to 1 L |
| Total | N/A | 1 L |
| To prepare 1 L of 1/2 MS medium, mix 2.21 g MS with Vitamins, 15 g sucrose in 500 mL of double-distilled water (ddH2O) using a magnetic stirrer until the powders are dissolved completely, adjust the pH to 5.8 with KOH, add 8 g agar and make up the volume to 1 L with ddH2O, and autoclave at 121°C for 20 min. Store at room temperature (23°C–25°C) for up to 1 month. | ||
| phi medium | ||
| Bacto peptone | 1% | 10 g |
| Yeast extract | 0.1% | 1 g |
| Casamino acids | 0.1% | 1 g |
| Agar | 1.5% | 15 g |
| ddH2O | N/A | Up to 1 L |
| Total | N/A | 1 L |
| To prepare 1 L of phi liquid medium, mix 10 g Bacto peptone, 1 g yeast extract, and 1 g casamino acids in 500 mL of double-distilled water (ddH2O) using a magnetic stirrer until the powders are dissolved completely, make up the volume to 1 L with ddH2O, and autoclave at 121°C for 20 min. Store at room temperature (23°C–25°C) for up to 1 month. For solid phi medium, prepare phi medium as described above and add 15 g agar to the solution. | ||
| phi TTC plates | ||
| phi medium | N/A | 1 L |
| 1% TTC | 0.05 g/L | 5 mL |
| 20% glucose | 5 g/L | 25 mL |
| Total | N/A | 1 L |
| 1% TTC: Dissolve triphenyltetrazolium chloride (TTC) in distilled water to make 1% (w/v) TTC solution followed by filter sterilization. Store 1% TTC solution at room temperature (23°C–25°C) or 4°C in a dark environment for up to 1 month. | ||
| 20% glucose: Dissolve glucose in distilled water and autoclave to make 20% (w/v) glucose solution. Store 20% glucose at 4°C for up to 1 month. | ||
| Phi agar medium, after cooling down, is supplemented with TTC and glucose, already prepared as sterilized materials as above, to prepare phi TTC plates. To make 1 L phi TTC medium, add 5 mL of 1% TTC and 25 mL of 20% glucose, resulting in a final 0.05 g/L TTC and 5 g/L glucose. Store at 4°C for up to 1 month. | ||
Step-by-step method details
Preparation of plant materials
Timing: 11 days
This section describes the steps for preparation of pant materials in the first step of the experiment. All these steps should be performed in sterile conditions. Growing healthy plants in good conditions is one of the key factors to obtain reliable and reproducible results using this method.
-
1.Sterilize and stratify Arabidopsis seeds.
-
a.Aliquot Arabidopsis seeds in 1.5 mL Eppendorf tubes. Sterilize Arabidopsis seeds using 1 mL 1:20 diluted bleach (we use a commercial sodium hypochlorite aqueous solution with 11%–15% available chlorine; can be replaced by commercial bleach) for 8 min.
-
b.Wash seeds using sterile water 5 times.
-
c.Sow the surface-sterilized Arabidopsis seeds on MS- square plates with a 1 mL pipette tip. If necessary, the seeds can be sown in 3 rows within one 10 × 10 cm sterile plate (Figure 1A). Usually 70%–80% seedlings germinate and grow in perfect conditions for subsequent use.
-
d.Stratify seeds in the dark at 4°C for 3 days.
-
a.
Alternatives: In the step of seed sterilization, it is also possible to use 75% ethanol for 10 min and then 100% ethanol for 3 min. When using Arabidopsis genotypes that cannot grow well on MS- medium, seeds can be sown on 1/2 MS plates to grow for the first 5 days.
-
2.
Grow plants.
Figure 1.
Preparation of plant materials
(A) Sterilize seeds and sow them on MS- medium square plate, then put plates to 4°C and dark.
(B) Transfer 5 day-old seedlings in good conditions to new MS- medium square plate.
(C) Total 15–20 seedlings in one plate.
Move plates to a growth chamber (22 ± 2°C, 16 h light/8 h dark photoperiod, 130 μE m−2 s−1, 65% humidity). Keep plates vertically and let seedlings grow for 5 days.
-
3.Transfer seedlings.
-
a.Transfer seedlings that are in good and consistent growth conditions, such as relatively similar root length and shoot growth, to new MS- square plates, placed as one row, using sterilized tweezers (15–20 seedlings/plate) (Figures 1B and 1C).
-
b.Move plates to a growth chamber (22 ± 2°C, 16 h light/8 h dark photoperiod, 130 μE m−2 s−1, 65% humidity). Keep plates vertically and let seedlings grow for 3 days.
-
a.
Alternatives: Add chemicals into MS- plates if necessary. In this step, move plates to a growth chamber to grow for 5 days, then transfer seedlings to other MS- plates supplemented with chemicals to grow for 3 more days.
Note:R. solanacearum would grow effortlessly on the surface of MS medium with sucrose, leading to a reduced infection efficiency. Hence, MS- medium is highly recommended.
Preparation of R. solanacearum inoculum (usually starting from 3 days before inoculation)
Timing: 3 days
This section describes the steps for the preparation of R. solanacearum inoculum. All these steps should be performed in sterile conditions.
-
4.
Streak out R. solanacearum strains from the glycerol stock on phi solid medium (with TTC/glucose), and incubate bacteria for 2 days at 28°C (Figures 2A and 2B).
-
5.
Start overnight R. solanacearum culture by picking a pink colony to inoculate phi liquid medium and grow at 28°C (Figure 2C).
-
6.
Pellet bacterial cells at 5,000 × g for 10 min at room temperature (23°C–25°C).
-
7.
Re-suspend the pellet in sterile water, and dilute to a concentration of OD600 = 0.1.
-
8.
Measure the concentration of the bacterial solution using a Nanodrop spectrophotometer (NanoDrop 2000c). Then dilute the bacterial solution to a final concentration of OD600 = 0.0001, corresponding to 105 cfu/mL, before inoculation (Figures 2D and 2E).
Alternatives: Depending on the purpose of different experiments, one might use different inoculation doses. For bacterial quantification, we use 105 cfu/mL as initial inoculation dose. An inoculum of OD600 = 0.0001 or higher can be used for studying transcriptional responses, fluorescent marker line observation using confocal microscopy, or the observation of developmental responses.
Note:R. solanacearum undergoes spontaneous phenotypic conversions.8 Pick pink bacterial colonies instead of the dark red colonies when grown on phi + TTC medium, since this colony color-related phenotypic conversion might correlate with changes in pathogenicity. For R. solanacearum, an OD600 of 0.1 corresponds to approximately 108 cfu/mL.
Figure 2.
Ralstonia solanacearum inoculation of Arabidopsis seedlings
(A) Bacteria growing on solid phi medium with TTC.
(B) Picking a pink colony using a pipette tip.
(C) Bacterial culture after 14 h overnight incubation.
(D) Bacterial suspension in sterile distilled water.
(E) Dilute bacterial solution to a final concentration of OD600 = 0.0001, corresponding to 105 cfu/mL.
(F) Inoculate roots of seedlings (0.5–1 cm from root tip) by placing a 5 μL droplet of R. solanacearum suspension at the inoculation site.
Inoculation of Arabidopsis seedlings
Timing: 2–3 h
-
9.
If there is any water condensation in the plates, remove the water before starting. Mark the inoculation site of each primary root, roughly 0.5–1 cm from the root tip of seedlings on MS- plates, and inoculate roots by placing a 5 μL droplet of R. solanacearum suspension on top of the root tissue at the inoculation site (Figure 2F).
-
10.
Keep the plates under a sterile cabinet for 10 min to let inoculation spots dry.
-
11.
Move the plates to a growth chamber with appropriate conditions for R. solanacearum infection (75% humidity, 12 h light, 130 μE m−2 s−1, 27°C) after inoculation. Keep the plates vertically on a rack.
Note: Always include a mock inoculation as a negative control (for example, H2O) to compare the primary root growth inhibition phenotype in seedlings inoculated with R. solanacearum GMI1000. Take a photograph before and after inoculation to record any interesting phenotypes, if necessary.
Sample collection for bacterial quantification
Timing: 3 days
-
12.Take samples.
-
a.Prepare 2 mL Eppendorf tubes. Label and measure the weight of the empty tubes (ET) on the bench (Figure 3A). Usually collect plant samples for quantification of bacterial growth 2 dpi or 3 dpi.
-
b.Collect the aerial part of seedlings by using a sterilized blade or scissors and place samples in plate with sterile water on the bench (Figures 3B and 3C). Take 4 seedlings as one sample.
-
c.Wash sample with sterile water 3 times. Dry the samples with tissue paper and weigh the seedling materials within the tubes as total weight (TW) (Figure 3D). For the collection of root tissues, wash 5 times wash with sterile water.
-
d.Calculate fresh weight (FW) of the seedling materials. Fresh weight of seedling tissue for each sample is calculated as FW = TW – ET.
-
a.
Note: To minimize the variation in each replicate, the sample volumes could be n = 8–10 per genotype. After plant sample collection, all the following procedures can be performed on the bench.
-
13.Grind plant samples.
-
a.Add 200 μL of sterile water to each Eppendorf tube containing plant samples, and three small metal beads or one big metal bead on the bench.
-
b.Grind the samples using a Tissue Lyzer at 25 cycles for 1 min with the adapters at room temperature (23°C–25°C) (Figure 3E). Afterward, add 800 μL of sterile water to obtain 1 mL of suspension in each tube.
-
a.
Alternatives: To grind samples using a drill, put each sample in a 2 mL Eppendorf tube with 200 μL of sterile water. Grind the tissue with a grinder placed in the drill stand. Afterwards, add 800 μL of sterile water.
-
14.
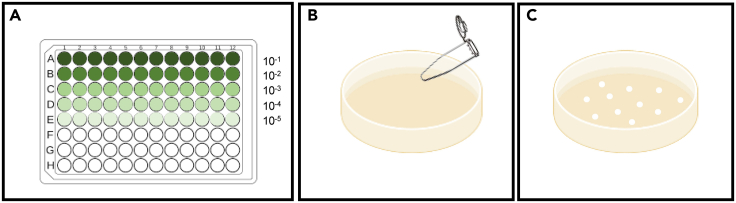
Make a series of dilutions from 10−1 to 10−5. Use a 96-well 200 μL plate for serial dilution (Figure 4A). Fill the plate with 180 μL sterile water in each well, then add 20 μL bacterial solution (10-fold dilution) from step 13 in the first dilution (10−1 dilution).
-
15.
For samples from 3 days post-bacterial inoculation, plate 10−4 and 10−5 dilutions on phi plates by hand-spreading (Figure 4B) or using any other automated system. Turn the plate upside-down after the suspension is spread and dry.
-
16.
Incubate 2 days at 28°C and count colonies (Figure 4C).
Note: After each experiment, make sure that all materials containing R. solanacearum (e.g., plates, gloves, bottles, etc.) are autoclaved to kill the bacteria (bleach is also acceptable to kill the bacteria).
Figure 3.
Sample collection for bacterial quantification after 3 days post inoculation
(A) Prepare empty 2 mL tubes and weigh; record as empty tube (ET).
(B) Take shoots of seedlings by using a sterilized blade or scissors; usually take 4 shoots as a sample.
(C) Wash shoots in sterile water 3 times and dry shoots with tissue paper.
(D) Weigh samples, record as total weight (TW).
(E) Grind samples with 200 μL sterile water and metal beads (3 small beads or 1 big beads), then add 800 μL sterile water to 1 mL.
Figure 4.
Spread bacterial solution on phi plates
(A) Make a series of dilutions by 96-well 200 μL plate. Add 180 μL sterile water each well, and add 20 μL bacterial solution from Figure 3 in the first row and record as 10−1 dilution.
(B) Spread bacterial solution (usually spread 2 different dilutions) by using 1.5 mL Eppendorf tube with hand on phi plates. Put plates to 28°C for 2 days.
(C) Count bacterial number after 2 dpi.
Expected outcomes
This protocol allows for the quantification of bacterial cells (colony-forming units) in shoot tissues after inoculation in roots, which provides a quantitative assessment of bacterial colonization in Arabidopsis tissues (Figure 5). The versatility of this method allows researchers to compare the efficiency of bacterial colonization in different plant genotypes and different compositions of growth media.1 The final quantitative outcomes of bacterial numbers can be subjected to statistical analysis to determine statistically significant differences, as we have published before.1 This protocol also allows for observations of physiological changes in plant development, including root length, the formation of lateral roots, and the number and length of root hairs.6 Combined with confocal microscopy and the use of tissue staining and/or fluorescent markers, this protocol can also be used to monitor molecular changes after Ralstonia infection in a time-dependent manner.6
Figure 5.
Generation of graphs using GraphPad Prism
(A) Open GraphPad Prism and choose type column.
(B) Record log10 of bacterial number calculated in Excel.
(C) Change the graph type to individual values.
(D and F) Analyze data by using t-test.
(G) Add statistical analysis results to the graph. Ns indicates no statistically significant difference.
Quantification and statistical analysis
Timing: 1–2 h
Note: FW = TW – ET. cfu/g = (Bacterial number) × (10ˆ3 or 4) × 2 × 50/ FW.
-
2.
Generate graphs using GraphPad Prism (Figure 5) or a similar program for statistical analysis. To compare individual results, perform a Student’s t-test, or an appropriate statistical analysis method for multiple comparisons, and present the mean value, standard error, and p-value to represent the statistical significance of data difference.
Table 1.
Calculation of bacterial numbers in plant tissues in genotype A
|
Ralstonia infection, 3dpi | ||||||
|---|---|---|---|---|---|---|
| Genotype: A |
Dilution |
cfu/g | Log10 cfu/g | |||
| Empty tube (ET)/g | Total weight (TW)/g | Fresh weight (FW)/g | 10−4 (cfu) | 10−5 (cfu) | ||
| 0.915 | 0.936 | 0.021 | 52 | 1238095238 | 9.09275405 | |
| 0.8681 | 0.884 | 0.0159 | 74 | 23270440252 | 10.3668046 | |
| 0.9163 | 0.9301 | 0.0138 | 58 | 21014492754 | 10.3225189 | |
| 0.9027 | 0.9193 | 0.0166 | 40 | 12048192771 | 10.0809219 | |
| 0.8742 | 0.8888 | 0.0146 | 47 | 16095890411 | 10.206715 | |
| 0.9273 | 0.9432 | 0.0159 | 146 | 4591194969 | 9.66192574 | |
| 0.8748 | 0.8871 | 0.0123 | 77 | 31300813008 | 10.4955556 | |
| 0.908 | 0.9198 | 0.0118 | 60 | 25423728814 | 10.4052392 | |
| 0.8939 | 0.9091 | 0.0152 | 167 | 5493421053 | 9.73984289 | |
| 0.8935 | 0.9059 | 0.0124 | 49 | 19758064516 | 10.2957444 | |
Table 2.
Calculation of bacterial numbers in plant tissues in genotype B
|
Ralstonia infection, 3dpi | ||||||
|---|---|---|---|---|---|---|
| Genotype: B |
Dilution |
cfu/g | Log10 cfu/g | |||
| Empty tube (ET)/g | Total weight (TW)/g | Fresh weight (FW)/g | 10−4 (cfu) | 10−5 (cfu) | ||
| 0.9228 | 0.9288 | 0.006 | 49 | 4083333333 | 9.61101483 | |
| 0.891 | 0.8978 | 0.0068 | 37 | 2720588235 | 9.43466282 | |
| 0.9159 | 0.9228 | 0.0069 | 21 | 15217391304 | 10.1823402 | |
| 0.9024 | 0.9099 | 0.0075 | 221 | 14733333333 | 10.168301 | |
| 0.9112 | 0.9198 | 0.0086 | 162 | 9418604651 | 9.97398657 | |
| 0.9155 | 0.9226 | 0.0071 | 31 | 21830985915 | 10.3390733 | |
| 0.9405 | 0.9473 | 0.0068 | 24 | 17647058824 | 10.2466723 | |
| 0.8942 | 0.901 | 0.0068 | 27 | 19852941176 | 10.2978249 | |
| 0.8711 | 0.8787 | 0.0076 | 21 | 13815789474 | 10.1403757 | |
| 0.8599 | 0.8692 | 0.0093 | 87 | 46774193548 | 10.6700063 | |
Optional step: Phenotypic observation of developmental alterations
R. solanacearum alters Arabidopsis development, particularly in root tissues,9 which involves both molecular and physiological reprograming. To dissect those changes at different levels, this inoculation system provides an excellent platform to make use of the available plant genetic materials, molecular tools and techniques, and pharmacological approaches. Here we mention a few examples that could be suitable for this setup.
-
1.
To observe the molecular changes induced by R. solanacearum, it is possible to use available florescent marker lines to visualize changes in gene expression in a dynamic manner.10 Usually, significant fluorescent changes can be observed 48 h after R. solanacearum inoculation by using confocal microscopy.
-
2.
To measure transcriptional changes of specific target genes during R. solanacearum infection, it is possible to use RT-qPCR after extraction of RNA from root and shoot tissues collected in the step 12b.
-
3.
Seedling tissues collected in the step 12b can also be used for large-scale ’omics studies, after the appropriate assessment of the necessary amount of tissue and time point(s) post-inoculation.
-
4.
Significant root developmental reprograming after R. solanacearum inoculation has been recently reported.9,11 It is possible to apply the observation of root-associated phenotypes in time-course experiments as indices for a successful inoculation, since infection by R. solanacearum in Arabidopsis usually results in several visible phenotypes around the inoculation spot: 1) primary root growth inhibition at 24–48 hpi; 2) root hair formation closes to the root tip at 24–48 hpi; 3) formation of lateral root structures at 72 hpi.
Limitations
Although this protocol allows for a sensitive detection of differences in resistance/susceptibility between plant genotypes and/or chemical treatments, in our experience, it is not the most appropriate method to compare the virulence of bacterial mutant strains. We are currently trying to understand the reasons why the virulence attenuation of bacterial mutant strains is not so obvious upon direct inoculation in young seedling tissues using this method. For this purpose, we recommend more traditional inoculation methods, such as soil-drenching followed by the observation of disease symptoms.4 Alternatively, host plants with thicker stems also allow other alternative inoculation methods, such as stem injection, which also provide quantitative results,2 or leaf infiltration.3
Although root tissues collected after R. solanacearum inoculation can be used for molecular studies, we found it very difficult to remove bacteria from the root surface, including the inoculation site. Therefore, the quantification of bacterial numbers in root tissues may not reflect bacterial colonization or replication inside plant tissues.
Finally, it is worth reminding that the mechanisms observed in a model plant like Arabidopsis may not necessarily resemble those taking place in crop plants infected by R. solanacearum in natural systems.
Troubleshooting
Problem 1
Specific plant genotypes cannot germinate and grow well on MS- medium (related to step 1).
Potential solution
Sow and grow these plants, together with any control plants, on ½ MS plates for the first 5 days.
Problem 2
Some seedlings die after transfer to a new MS- plates for another 3-day growth (related to step 3).
Potential solution
Use minimal pressure when handling the seedlings using tweezers, since young seedlings may be very susceptible to mechanical damage.
Problem 3
Bacterial solution droplet may flow down right after bacterial inoculation (related to step 9).
Potential solution
Dry plates in a sterile cabinet or flow chamber before bacterial inoculation.
Problem 4
Contamination issues arise in the plant growth and bacterial inoculation steps (related to step 1 and step 9).
Potential solution
Perform seed sterilization, sowing, seedling transfer, and bacterial inoculation in sterile flow hood.
Problem 5
R. solanacearum strains cannot grow well in liquid phi medium (related to step 5).
Potential solution
Always streak out fresh R. solanacearum strains from a glycerol stock, and grow the liquid culture with freshly growing colonies. Do not use R. solanacearum colonies growing on plates for more than 3 days.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Alberto P. Macho (alberto.macho@icloud.com).
Materials availability
This study did not generate new unique reagents. All the non-commercial materials described in this study are available upon request.
Data and code availability
This study did not generate/analyze datasets.
Acknowledgments
We thank Xinyu Jian and Fangyuan Wu for technical and administrative assistance during this work, Hao Xue for using this protocol for the observation of florescent marker lines, and all the members of the Macho laboratory for the constant improvement of this protocol and helpful discussions. This work was supported by the National Natural Science Foundation of China (NSFC; grants 32170289 to A.P.M. and 32270284 to G.Y.) and the Shanghai Center for Plant Stress Biology (Chinese Academy of Sciences).
Author contributions
G.Y., L.Z., and A.P.M. wrote the manuscript; G.Y. and L.Z. prepared figures and tables; K.W. helped initial setup of inoculation assay.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Gang Yu, Email: gangyu@sjtu.edu.cn.
Alberto P. Macho, Email: alberto.macho@icloud.com.
References
- 1.Dindas J., DeFalco T.A., Yu G., Zhang L., David P., Bjornson M., Thibaud M.-C., Custódio V., Castrillo G., Nussaume L., et al. Direct inhibition of phosphate transport by immune signaling in Arabidopsis. Curr. Biol. 2022;32:488–495.e5. doi: 10.1016/j.cub.2021.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y., Xian L., Yu G., Macho A.P. Tomato stem injection for the precise assessment of Ralstonia solanacearum fitness in planta. Bio. Protoc. 2021;11 doi: 10.21769/BioProtoc.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu W., Macho A.P. A fast and easy method to study Ralstonia solanacearum virulence upon transient gene expression or gene silencing in Nicotiana benthamiana leaves. Bio. Protoc. 2021;11:e4116. doi: 10.21769/BioProtoc.4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morcillo R.J.L., Zhao A., Tamayo-Navarrete M.I., García-Garrido J.M., Macho A.P. Tomato root transformation followed by inoculation with Ralstonia Solanacearum for straightforward genetic analysis of bacterial wilt disease. JoVE. 2020 doi: 10.3791/60302. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y., Zhao A., Morcillo R.J.L., Yu G., Xue H., Rufian J.S., Sang Y., Macho A.P. A bacterial effector protein uncovers a plant metabolic pathway involved in tolerance to bacterial wilt disease. Mol. Plant. 2021;14:1281–1296. doi: 10.1016/j.molp.2021.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Xue H., Yu G., Zhang L., Li M., Lozano-Durán R., Macho A.P. Ralstonia solanacearum alters root developmental programs in auxin-dependent and independent manners. bioRxiv. 2022 doi: 10.1101/2022.06.22.497157. Preprint at. [DOI] [Google Scholar]
- 7.Salanoubat M., Genin S., Artiguenave F., Gouzy J., Mangenot S., Arlat M., Billault A., Brottier P., Camus J.C., Cattolico L., et al. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature. 2002;415:497–502. doi: 10.1038/415497a. [DOI] [PubMed] [Google Scholar]
- 8.Poussier S., Thoquet P., Trigalet-Demery D., Barthet S., Meyer D., Arlat M., Trigalet A. Host plant-dependent phenotypic reversion of Ralstonia solanacearum from non-pathogenic to pathogenic forms via alterations in the phcA gene. Mol. Microbiol. 2003;49:991–1003. doi: 10.1046/j.1365-2958.2003.03605.x. [DOI] [PubMed] [Google Scholar]
- 9.Lu H., Lema A S., Planas-Marquès M., Alonso-Díaz A., Valls M., Coll N.S. Type III secretion-dependent and -independent phenotypes caused by Ralstonia solanacearum in Arabidopsis roots. Mol. Plant Microbe Interact. 2018;31:175–184. doi: 10.1094/mpmi-05-17-0109-fi. [DOI] [PubMed] [Google Scholar]
- 10.Marquès-Bueno M.D.M., Morao A.K., Cayrel A., Platre M.P., Barberon M., Caillieux E., Colot V., Jaillais Y., Roudier F., Vert G. A versatile multisite gateway-compatible promoter and transgenic line collection for cell type-specific functional genomics in Arabidopsis. Plant J. 2016;85:320–333. doi: 10.1111/tpj.13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao C., Wang H., Lu Y., Hu J., Qu L., Li Z., Wang D., He Y., Valls M., Coll N.S., et al. Deep sequencing reveals early reprogramming of Arabidopsis root transcriptomes upon Ralstonia solanacearum infection. Mol. Plant Microbe Interact. 2019;32:813–827. doi: 10.1094/mpmi-10-18-0268-r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze datasets.