Summary
Generation of functional human dopaminergic (DA) neurons from human induced pluripotent stem cells (hiPSCs) is a crucial tool for modeling dopamine-related human diseases and cell replacement therapies. Here, we present a protocol to combine neuralizing transcription factor (NGN2) programming and DA patterning to differentiate hiPSCs into mature and functional induced DA (iDA) neurons. We describe steps from transduction of hiPSCs and neural induction through to differentiation and maturation of near-pure, fully functional iDA neurons within 3 weeks.
For complete details on the use and execution of this protocol, please refer to Sheta et al. (2022).1
Subject areas: Neuroscience, Stem Cells, Cell Differentiation
Graphical abstract

Highlights
-
•
Efficient and rapid differentiation of iPSCs into functional DA neurons
-
•
A step-by-step guide that combines NGN2 programming and DA differentiation kits
-
•
This protocol provides an in vitro platform to study DA neurons
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Generation of functional human dopaminergic (DA) neurons from human induced pluripotent stem cells (hiPSCs) is a crucial tool for modeling dopamine-related human diseases and cell replacement therapies. Here, we present a protocol to combine neuralizing transcription factor (NGN2) programming and DA patterning to differentiate hiPSCs into mature and functional induced DA (iDA) neurons. We describe steps from transduction of hiPSCs and neural induction through to differentiation and maturation of near-pure, fully functional iDA neurons within 3 weeks.
Before you begin
The current protocol for generating functional human induced pluripotent stem cell (hiPSC)-derived dopaminergic (DA) neurons involves programming with the neuralizing transcription factor (NGN2) to induce neuronal cell differentiation, coupled with the use of commercially available midbrain defined differentiation and maturation media (STEMCELL Technologies). This four-step protocol, illustrated in the graphical abstract, begins by delivering the NGN2 overexpression construct via lentivirus infection under the tetracycline-inducible gene expression system (TetO) into hiPSC. The protocol for producing and purifying lentivirus follows the methods described by Tiscornia and colleagues.2 In the second step, hiPSC differentiate into iNeurons upon treatment with doxycycline-induced NGN2 expression, in a rapid and reproducible manner.3,4,5,6,7 Next, in step 3, iNeurons undergo a defined timeline of media supplementation using commercially available midbrain differentiation media, leading to iNeuron patterning towards the DA phenotype (iDA). Finally, in step 4, iDA neurons are incubated in midbrain maturation media to boost their electrical activity and neurotransmitter release. This protocol allows for the efficient and reproducible generation of functional iDA cells within three weeks.
To demonstrate this protocol, we used hiPSC obtained from a healthy male subject and derived from Peripheral Blood Mononuclear Cells (PBMC) via the C-BIG Repository at the Montreal Neurological Institute (cell line name: AIW002-02). This protocol can also be applied to any hiPSC, as demonstrated in our study.1
It is essential to regularly authenticate the cell lines, analyze them for multiple quality control parameters (including pluripotency by immunofluorescence, trilineage differentiation potential, and genomic integrity), and check for mycoplasma contamination. All procedures were carried out under sterile conditions in a Class II biological hood, with the hiPSC cultured in a humidified incubator at 37°C with 5% CO2.
The TetO-NGN2-T2A-puromycin and Ubi-rtTA plasmids used in this protocol for NGN2 forced expression were provided by Dr. Thomas C. Südhof’s laboratory.3,8 It is worth noting that similar plasmids are available from Addgene: pTet-O-Ngn2-puro (Addgene #52047) and the lentiviral plasmid expressing the reverse tetracycline transactivator (rtTA) (Addgene #20342).
Before generating iDA neurons, prepare lentiviral particle stocks of pTetO-NGN2-puro and Ubi rtTA, as well as Matrigel-coated plates for growing hiPSC to an optimal confluency. Finally, prepare and coat coverslips or plates with Poly-L-ornithine and laminin.
It is also essential to ensure good quality requirements for all material and reagents used throughout the protocol. All material and reagents used in the differentiation protocol should be purchased from well-established sources to ensure the integrity and reliability of the protocol for the generation of functional hiPSC-derived dopaminergic neurons.
The protocol described here is based on cell culture procedures that require a strictly controlled environment for cell growth. Sterile distilled water should be used in incubators, in addition to the preparation of sterile buffers described in the protocol.
Additionally, it is necessary to always carefully wipe down media bottles, buffers and cryopreserved vials with 70% ethanol before their transfer to a cell culture hood.
Institutional permissions
If obtaining hiPSC from patients, the collection should be performed in accordance with relevant ethical guidelines. The hiPSC utilized in the study was approved by the CHU de Quebec Research Center (#2022-6079).
Coating tissue culture plates with Matrigel hESC-qualified matrix
Timing: 1 h 30 min
Note: Ensure using aseptic techniques to ensure the sterility of all material used under a cell culture hood.
Note: When aliquoting Matrigel hESC-qualified Matrix, thaw a bottle of Matrigel at 4°C in an ice bucket and place in the backside of the fridge for 16 h. One to two hours before aliquoting Matrigel, cool 1.5-mL microcentrifuge tubes and sterile 200 μL pipette tips at −20°C. While keeping the Matrigel on ice, aliquot 150–200 μL per tube. Store Matrigel aliquots at −80°C until use.
CRITICAL: Matrigel should be aliquoted to prevent freeze/thaw cycles. Matrigel is also very sensitive to heat and can quickly polymerize at 20°C–22°C. Thaw Matrigel on ice and always leave on ice.
Alternatives: There are different variations of the Matrigel available. Alternatives such as GelTrex, or Vitronectin may be used. However the protocol described here present results based on the use of Matrigel basement membrane matrix from Corning.
-
1.
Thaw one aliquot of Matrigel from the −80°C stock on ice for 15–20 min before use.
-
2.
Dispense the desired amount of cold DMEM/F12 + antibiotic-antimycotic into a 15-mL conical tube that is kept on ice, depending on the dilution required (concentration varies depending on the LOT number). Add the desired volume of the thawed Matrigel aliquot and add it to the cold DMEM/F12 + antibiotic-antimycotic medium and mix well.
CRITICAL: The dilution factor of Matrigel basement membrane matrix is LOT-specific and found on the Certificate of Analysis from the supplier. The aliquot (dilution factor) volume is typically between 270 and 350 μL for a 25 mL volume of media and is calculated for each LOT based on the protein concentration.
-
3.Immediately use the diluted Matrigel solution to coat tissue culture dishes. If there were leftovers of non-diluted Matrigel, remaining material can be stored at −20°C until next use.
-
a.The coating volumes of Matrigel in DMEM/F12 + antibiotic-antimycotic used in the study are the following: for a 60-mm plate use 2 mL/plate and for 100-mm plate use 7 mL/plate.
-
a.
Note: Volumes can be adjusted in accordance with the size of the plate or well used.
-
4.
Swirl cell culture plates to spread the Matrigel solution evenly across the surface.
-
5.Incubate at 37°C for a minimum of 1–2 h.
-
a.If Matrigel coated dishes are not used on the same day as coating, they can be left for 16 h or a maximum of 1 week at 37°C before use.
-
b.The coated flasks can be stored for up to 1 week in a 37°C incubator. Incubation at 37°C for long periods of time may result in reagent evaporation and thus uneven covering of the flask surface.
-
c.Alternatively, plates can be sealed with parafilm and stored at 4°C for up to 1 week. Allow stored coated tissue culture plates to come to (20°C–22°C) for 30 min before further use.
-
a.
-
6.
Once ready to use, remove excess Matrigel solution by gently tilting the plate to one side and remove Matrigel solution by aspiration. Ensure that the coated surface is not scratched.
Preparation of acid-washed glass coverslips
Timing: 2 days
-
7.
Pour around 200 coverslips into a 125-mL Erlenmeyer flask.
-
8.
Under a fume hood, pour 70% (vol/vol) nitric acid onto coverslips, adding enough to properly cover all coverslips.
CAUTION: Due to the exothermic nature of the acid reaction, dilution of nitric acid must be done by slowly adding the acid to water to limit the risk of splashing concentrated acid.
-
9.Cover the Erlenmeyer flask with a parafilm and place on a benchtop shaker at 150 rpm for 1 h at 20°C–22°C.
-
a.Under the fume hood slowly discard the nitric acid with a pipette into a glass beaker containing at least 2 L of dH2O, and discard following local security instructions.
-
a.
-
10.Thoroughly wash coverslips four times with dH2O using 100 mL or more per 125-mL Erlenmeyer flask.
-
a.Discard the rinsed volumes into a glass beaker containing at least 2 L of dH2O and pour the mixture down the drain.
-
a.
-
11.Place the Erlenmeyer uncovered, in a sterile 70°C incubator for 16–36 h (dry heat).
-
a.For the first 6 h, tap the back of the Erlenmeyer every hour to detach coverslips that may be found attached to each other due to the humidity of the chamber.
-
a.
-
12.Once coverslips are dry, under a sterile hood, pour out the coverslips from the Erlenmeyer into a sterile 50-mL tube.
-
a.Turn the hood’s UV light for at least 2 h to ensure sterility.
-
a.
Note: For long term storage, keep the dry coverslips dry in a sterile 50-mL tube for 3–4 months at 20°C–22°C.
Coating acid-washed glass coverslips with Poly-L-ornithine followed by laminin
Timing: 1 day
Note: Ensure using aseptic techniques to ensure the sterility of all material used under a cell culture hood.
Note: The coating protocol described here also applies for use on plastic tissue culture dishes or plates.
Note: Prepare 10 mg/mL (200×) aliquots of Poly-L-Ornithine (PLO) by reconstituting the PLO in dH2O, filter the stock solution with a 0.22-μm-pore filter. Aliquot the diluted Poly-L-Ornithine and keep the aliquots in −20°C for up to 4 months.
-
13.
Thaw the PLO solution at 20°C–22°C and dilute (1:200) the filtered PLO at 50 μg/mL in sodium borate buffer (1 M, pH 8.5).
-
14.Place glass coverslips from step 12 into 48-well plates, and add 300 μL of PLO into each well of a 48-well plate.
-
a.Incubate the plate or the plate with coverslips at 20°C–22°C for 24 h or a minimum of 4 h before use.
-
a.
Note: Alternatively, the plates or the plate with coverslips can be incubated at 37°C and 5% CO2 for 24 h or a minimum of 4 h before use.
-
15.
When the plate or the plate with coverslips is ready for use, aspirate the PLO solution and wash the PLO coated coverslips or plates four times with generous amounts of sterile water (≥ 1 mL per well).
CRITICAL: PLO and sodium borate buffer are toxic for cells. Thorough washing with generous amounts of dH2O prior to laminin treatment is therefore mandatory.
-
16.
Allow the coverslips to dry at 20°C–22°C for at least 4 h before proceeding with laminin coating.
Note: For extended storage, dried plates or plates containing coated PLO coverslips can be sealed with parafilm and stored at 4°C for up to two weeks.
CRITICAL: The efficiency of coating is relatively low when PLO is prepared in dH2O due to the acidic pH of the solution that can range between pH 3.3 and 6.6. Hence, using borate buffer at pH 8.5 as described above greatly improves cell adherence to the glass coverslips.
-
17.Thaw 1 mg/mL aliquots of laminin on ice.
-
a.Dilute the 1 mg/mL laminin stock with DPBS to a final concentration of 10 μg/mL.
-
a.
CRITICAL: Laminin should be aliquoted to prevent freeze/thaw cycles. Thaw laminin on ice and always leave on ice.
-
18.Coat the coverslips with 150 μL of 10 μg/mL laminin for 24 h or a minimum, of 8 h at 4°C.
-
a.Allow stored coated tissue culture plates to come to 20°C–22°C for 30 min before further use.
-
a.
Note: Alternatively, plates can also be kept for a minimum of 2 h at 37°C and 5% CO2 or otherwise left at 37°C and 5% CO2 for 16 h. Coated PLO/laminin coverslips or plates can be sealed with parafilm and stored at 4°C for up to 1 week.
CRITICAL: Sixteen hour coating at 4°C is recommended for a more reliable coating. Seal the coated plates with parafilm to avoid evaporation. Do not use coverslips or plates if discoloration or web-like structures are observed on the surface of the coated coverslips.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-TH; 1:1000 | Sigma-Aldrich | Cat#MAB318; RRID: AB_2201528) |
| Rabbit polyclonal anti-TH; 1:1000 | PhosphoSolutions | Cat#2025-THRAB; RRID: AB_2492276) |
| Rabbit polyclonal anti-MAP2 polyclonal; 1:1000 | Cell Signaling Technology | Cat#4542S; RRID: AB_10693782 |
| Mouse monoclonal anti-MAP2 (2a+2b); 1:1000 | Sigma-Aldrich | Cat#M1406; RRID: AB_477171 |
| Goat polyclonal anti-mouse IgG AF488; 1:400 | Thermo Fisher | Cat#A-11029; RRID: AB_2534088) |
| Goat polyclonal anti-rabbit IgG AF594, 1:400 | Thermo Fisher | Cat#A11037; RRID: AB_2534088) |
| Plasmids | ||
| TetO-NGN2-T2A-puromycin plasmid | Dr. Thomas C. Südhof laboratory3,8 | N/A |
| Ubi-rtTA plasmid | Dr. Thomas C. Südhof laboratory3,8 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Accutase | STEMCELL Technologies | Cat#07920 |
| Antibiotic-Antimycotic (100×) | Thermo Scientific | Cat#15240062 |
| B-27 Supplement (50×) serum free | Thermo Scientific | Cat#17504044 |
| BDNF Recombinant Human/Murine/Rat | PeproTech | Cat#450-02 |
| Boric acid | Sigma-Aldrich | Cat#B6768 |
| Bovine serum albumin (BSA) | BioShop | Cat#ALB001.500 |
| Cytosine β-D-arabinofuranoside hydrochloride (Ara-C) | Sigma-Aldrich | Cat#C6645 |
| DMEM/F-12 GlutaMAX | Thermo Fisher | Cat#10565018 |
| Doxycycline | Takara Bio Company | Cat#631311 |
| Dulbecco’s phosphate-buffered saline (DPBS) | Thermo Fisher | Cat#14190250 |
| DPBS calcium magnesium (Ca+ Mg+) | Thermo Fisher | Cat#14040182 |
| Ethylenediaminetetraacetic acid (EDTA) | Fisher Scientific | Cat#E478-500 |
| Fetal bovine serum (FBS) | Sigma-Aldrich | Cat#F1051 |
| Fluoromount-G Mounting Medium | Thermo Fisher | Cat#00-4958-02 |
| GDNF Recombinant Human/Murine/Rat | PeproTech | Cat#450-10 |
| GlutaMAX Supplement | Thermo Fisher | Cat#35050061 |
| Human Recombinant Sonic Hedgehog (C24II) (Shh) | STEMCELL Technologies | Cat#78065 |
| Laminin Mouse Protein | Thermo Fisher | Cat#23017015 |
| Matrigel basement membrane matrix LDEV-free | Corning | Cat#354277 |
| MEM Nonessential Amino Acid solution 100× (NEAA) | Wisent Bioproducts | Cat#321-011-EL |
| mFreSR | STEMCELL Technologies | Cat#05854 |
| mTeSRTM Plus | STEMCELL Technologies | Cat#100-0276 |
| N-2 Supplement (100×) | Thermo Scientific | Cat#17502048 |
| Neurobasal Medium | Thermo Scientific | Cat#21103049 |
| Nitric acid ACS reagent 70% | Sigma-Aldrich | Cat#438073 |
| Normal goat serum | Wisent Bioproducts | Cat#053-110 |
| Paraformaldehyde (PFA) | Electron Microscopy Sciences | Cat#19210 |
| PBS 10× solution | Fisher Scientific | Cat#BP399-20 |
| Poly-L-ornithine hydrobromide mol wt 30,000–70,000 | Sigma-Aldrich | Cat#P3655 |
| Potassium chloride (KCl) | Sigma-Aldrich | Cat#P3911 |
| Puromycin 2HCL | Wisent Bioproducts | Cat#400-160-EM |
| STEMdiff Midbrain Neuron Differentiation Kit | STEMCELL Technologies | Cat#100-0038 |
| STEMdiff Midbrain Neuron Maturation Kit | STEMCELL Technologies | Cat#100-0041 |
| Sodium chloride (NaCl) | Sigma-Aldrich | Cat#S7653 |
| Sucrose ≥99.5% (GC) | Sigma-Aldrich | Cat#S9378 |
| Triton X-100 | Sigma-Aldrich | Cat#T8787 |
| Trypan blue | Bio-Rad | Cat#1450022 |
| Y-27632 (ROCK inhibitor) | Tocris | Cat#1254 |
| DAPI | Thermo Fisher Scientific | Cat#D3571 |
| Experimental models: Cell lines | ||
| Human Induced Pluripotent Stem Cells (hiPSC), cell line: AIW002-02. Approved by the CHU de Quebec Research Center (#2022-6079) | Obtained through the C-BIG Repository at the Montreal Neurological Institute (The Neuro) | N/A |
| Critical commercial assays | ||
| qPCR Lentivirus Titer Kit | Applied Biological Materials Inc. | Cat#LV900 |
| Software and algorithms | ||
| ImageJ | N/A | https://imagej.nih.gov/ij/ |
| GraphPad Prism 9.0 | GraphPad Software Inc. | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| German glass coverslips #1.5 (8 mm) | Electron Microscopy Sciences | Cat#72296-08 |
Materials and equipment
Note: Ensure using aseptic techniques to ensure the sterility of all material used under a cell culture hood.
CRITICAL: Sterilize all media and buffers with a 0.22-μm-pore filter or a Stericup vacuum filter unit before use.
-
•
EDTA-based dissociation reagent: Makeup 500 mL of the dissociation reagent by adding 500 μL of 0.5 M EDTA with 0.9 g of NaCl into 500 mL DPBS. The solution can be stored for up to 1 year at 20°C–22°C.
-
•
Sodium Borate Buffer (1 M, pH 8.5): Makeup 1 L of a 1 M stock in dH2O by adding 61.83 g of Boric Acid (H3BO3) to 800 mL of dH2O in a graduated glass beaker. Add 10 g of sodium hydroxide to the solution to reach a pH of 8.5. Adjust the volume to 1 L of dH2O. The solution can be stored for up to 1 year at 20°C–22°C.
-
•
DMEM/F12+antibiotic-antimycotic medium: DMEM/F12+antibiotic-antimycotic is used in coating tissue culture plates and all other cell culture stages when working with hiPSC. In a 500-mL bottle of DMEM/F12 add 5 mL of antibiotic antimycotic (100×). DMEM/F12+antibiotic-antimycotic can be stored at 4°C for up to 6 months.
-
•
mTeSR Plus medium: mTeSR Plus from STEMCELL technologies is provided as a kit consisting of a 400 mL bottle of mTeSR Plus basal medium and a frozen 100 mL mTeSR Plus 5× supplement. The media bottle is stored at 4°C and the 100 mL 5× supplement is stored at −20°C. The day before the media is ready to use, place the supplement in a 4°C fridge and allow thawing slowly for 16 h. The next day, add the entire 100 mL supplement to the 400 mL bottle of mTeSR Plus basal medium. Mix well and aliquot 40 mL of the complete mTeSR Plus medium into sterile 50 mL Falcon tubes. Store each aliquot at −20°C for up to 6 months until needed. If not used immediately, store complete and thawed mTeSR Plus medium at 4°C for up to 2 weeks.
CRITICAL: mTeSR Plus medium should be warmed to 20°C–22°C. Warming at 37°C causes the media to denature and it will increase cell differentiation.
Day 0+ Day 1 (D0/1) medium for the differentiation of hiPSC into iNeurons
| Reagent | Stock concentraion | Final concentration | Amount |
|---|---|---|---|
| N-2 | 100× | 1× | 500 μL |
| B-27 | 100× | 1× | 1000 μL |
| NEAA | 100× | 1× | 500 μL |
| BDNF | 20 μg/mL | 10 ng/mL | 25 μL |
| GDNF | 20 μg/mL | 10 ng/mL | 25 μL |
| Laminin | 1 mg/mL | 1 μg/mL | 50 μL |
| Doxycycline | 1 mg/mL | 2 μg/mL | 100 μL |
| DMEM/F12 | N/A | N/A | 47.8 mL |
| Total | N/A | N/A | 50 mL |
Store aliquots at 4°C for up to 1 month until needed.
Note: Aliquot doxycycline into working volumes to avoid repeated freeze-thaw cycles. Aliquots can be stored at −20°C, and should be stable for at least one year. Protect aliquots from prolonged exposure to light.
Note: The DMEM/F12 medium used for D0/1 medium described here does not contain antibiotics. The addition of Penicillin-Streptomycin at 0.5% is possible but not necessary throughout the differentiation protocol. We have not tested the use of antibiotics during the differentiation protocol, as the use of antibiotics is unnecessary when applying good laboratory practice.
Day 2 (D2) medium for the differentiation of hiPSC into iNeurons
| Reagent | Stock concentraion | Final concentration | Amount |
|---|---|---|---|
| N-2 | 100× | 1× | 500 μL |
| B-27 | 100× | 1× | 1000 μL |
| GlutaMAX | 100× | 1× | 500 μL |
| NEAA | 100× | 1× | 500 μL |
| BDNF | 20 μg/mL | 10 ng/mL | 25 μL |
| GDNF | 20 μg/mL | 10 ng/mL | 25 μL |
| Laminin | 1 mg/mL | 1 μg/mL | 50 μL |
| Doxycycline | 1 mg/mL | 2 μg/mL | 100 μL |
| Neurobasal Medium | N/A | N/A | 47.3 mL |
| Total | N/A | N/A | 50 mL |
Store aliquots at 4°C for up to 1 month until needed.
Note: The Neurobasal Medium medium used for D2 medium does not contain antibiotics. The addition of Penicillin-Streptomycin at 0.5% is possible but not necessary throughout the differentiation protocol. We have not tested the use of antibiotics during the differentiation protocol, when applying good laboratory practice, the use of antibiotics is unnecessary.
-
•
STEMdiff Midbrain Neuron Differentiation Medium
STEMdiff Midbrain Neuron Differentiation medium from STEMCELL technologies is provided as a kit consisting of a 20 mL differentiation supplement and 80 mL of STEMdiff Midbrain Neuron Differentiation Basal Medium. The basal media bottle is stored at 4°C and the differentiation supplement is stored at −20°C. When ready for use, thaw the differentiation supplement at 20°C–22°C or at 4°C for 16 h. Before opening, mix the supplement thoroughly. Add 20 mL of the differentiation supplement to 80 mL of the differentiation basal medium. Mix the bottle thoroughly. Add Human Recombinant Shh at a concentration of 200 ng/mL to the 100 mL bottle mixture. Mix thoroughly. If not used immediately, store STEMdiff Midbrain Neuron Differentiation Medium at 4°C for up to 1 month. Before use, aliquot a volume of needed medium, and warm the medium aliquot at 37°C.
Note: To maintain the integrity of the Shh present in STEMdiff Midbrain Neuron Differentiation Medium, it is advised to refrain from subjecting the medium to prolonged exposure to a 37°C water bath or bead bath. This exposure can potentially degrade the Shh. Therefore, it is recommended to only utilize and warm the necessary volumes of medium required for the day.
CAUTION: STEMdiff Midbrain Neuron Differentiation Supplement contains material derived from human plasma. Therefore, this product should be considered potentially infectious and treated in accordance with universal handling precautions as recommended by the manufacturer.
-
•
STEMdiff Midbrain Neuron Maturation Medium
STEMdiff Midbrain Neuron Maturation medium from STEMCELL technologies is provided as a kit consisting of 25 mL of maturation supplement and 100 mL of BrainPhys Neuronal Medium. The neuronal media bottle is stored at 4°C and the maturation supplement is stored at −20°C. When ready for use, thaw the maturation supplement at 20°C–22°C or at 4°C for 16 h. Before opening, mix the supplement thoroughly. Add 25 mL of the maturation supplement to 100 mL of differentiation basal medium. Mix the bottle thoroughly. If not used immediately, store STEMdiff Midbrain Neuron Maturation medium at 4°C for up to 1 month. Before use, medium needs to be warmed to 37°C.
CAUTION: STEMdiff Midbrain Neuron Maturation supplement contains material derived from human plasma. Therefore, this product should be considered potentially infectious and treated in accordance with universal handling precautions as recommended by the manufacturer. BrainPhys Neuronal medium needs to be protected from light.
Alternatives: BrainPhys Neuronal medium alone is also available from STEMCELL technologies for applications such as live neuronal imaging (STEMCELL technologies; cat. no. 05796).
Step-by-step method details
Transduction of hiPSC with NGN2 particles
Timing: 7–8 days
Note: Ensure using aseptic techniques to ensure the sterility of all material used under a cell culture hood.
Step 1 of the protocol is the transduction of hiPSC with lentivirus particles overexpressing NGN2, under the control of doxycycline (TetO).
CAUTION: All steps involved in handling lentivirus particles require a biosafety level 2 laboratory. Handle with extreme care and wear appropriate personal protective equipment when using viruses. Wear a double layer of gloves and wear a lab coat or appropriate personal protective equipment. Any used equipment in contact with the viruses (serological pipettes or tips) must be handled in accordance with institutional viral waste handling procedures.
CRITICAL: It is critical to passage hiPSC at least once post thaw before preparing them for infection and differentiation, since this may affect their infection and differentiation capacity.
Day -7
Timing: 2 h
-
1.To start the transduction procedure (Figure 1A), plate hiPSC as single cells following the procedure described in supplement protocol “passaging hiPSC as single cells”.
-
a.Count cells using a Trypan Blue cell counting method using an automated cell counter or hematocytometer to determine the concentration of the cells in suspension. Only counting live cells when determining the final cell density.
-
a.
-
2.Plate 3.8 × 105 or (0.95 × 105 cells/cm2) of hiPSC per well of a 12-well plate previously coated with Matrigel in mTeSR Plus medium containing Y-27632 at 10 μM.
-
a.Use healthy hiPSC with minimal spontaneously differentiated cells. Working with high quality hiPSC is very important for the successful generation of iDA neurons. Troubleshooting 1.
-
b.Culture the cells for 16 h at 37°C in 5% CO2.
-
a.
Figure 1.
Detailed protocol of the viral transduction of hiPSC and patterning into iNeurons
(A) The viral transduction of hiPSC starts from Day -7 where the cells are passaged as single cells in a 12-well plate. The following day (Day -6), hiPSC are transduced with NGN2 and Ubi rtTA lentiviral particles. After 2 days of incubation with the viral particles (Day -4), transduced hiPSC are passaged as colonies and cultured over three days. One day prior to neuronal induction (Day -1), transduced cells are plated as single cells in mTeSR Plus media. To start the neuronal induction of hiPSC (Day 0 and Day 1), cells are exposed to D0/1 media containing doxycycline and puromycin. Finally, on Day 2, patterned cells are passaged as single cells in D2 media in the presence of doxycycline and Ara-C to continue with the dopaminergic differentiation steps.
(B) Representative bright-field microscope images illustrating the morphological changes occurring during the neuronal induction, between Day -1 and Day 2. Scale bar, 50 μm.
Day -6
Timing: 2 h
-
3.
Thaw lentivirus stocks from −80°C, by placing the tubes on ice.
-
4.
Based on the viral titer, use a multiplicity of infection (MOI) of 10 for pTetO-NGN2-puro and a MOI of 5 for Ubi rtTA. Troubleshooting 2.
Note: For the viral titration, a titration kit provided by ABM (ABM; cat. no. LV900) has been used. The protocol is based on extracting RNA from viral particles, which is then followed by an RT-qPCR to evaluate the titer. This kit is highly sensitive, and allows for a precise titration of the viral particles against a provided standard. The Ct values are plotted against virus titer, then a logarithmic regression using the standard control DNA dilutions is generated to determine the unknown virus sample titer using y = m ln(x) + b where m is the slope of the line and b is the y-intercept. We always ensure that the R2 coefficient, which allows the determination of the statistical measure, is above 0.95. The final equation follows:
Once the IU/mL is calculated, several MOI are tested based on the number of cells to be infected. Therefore, the cells used to determine the MOI are the single cell plated hiPSCs.
-
5.
Prepare a master mix of the lentiviral particles in 1 mL of mTeSR Plus medium following Table 1 below:
CRITICAL: Make sure to adjust the viral particles volumes based on the viral titer.
-
6.Infect the cells by adding the viral mix dropwise to the cells and incubate the hiPSC for 2 days at 37°C in 5% CO2, until they reach confluency.
-
a.Cells can take up to 3 days to reach confluency, depending on the toxicity of the viral particles added.
-
b.Eventually, cells will form colonies since they are maintained in mTeSR Plus medium.
-
a.
Table 1.
Viral titer and volume per well
| Titer | Volume per 12-well plate | |
|---|---|---|
| pTetO-NGN2-puro | 3.0 × 109 TU/mL | 1 μL |
| Ubi rtTA | 1.5 × 109 TU/mL | 1 μL |
Day -4
Timing: 1 h
-
7.When the hiPSC are confluent, split each well of the 12-well plate following steps 47–49 from supplement protocol “routine maintenance and passaging of hiPSC”.
-
a.Plate the cells as colonies in mTeSR Plus medium into a 10-cm plate previously coated with Matrigel.
-
a.
Day -3 to day -2
Timing: 30 min
-
8.
Starting from day -3 feed cells with 7 mL of mTeSR Plus (every other day).
-
9.When cells are confluent in the 10 cm plates (usually around day -1). Check the cells under a light microscope, you should observe tight colonies with tight edges (Figure 1B).
-
a.Passage the cells for use in both maintenance and expansion for neuronal differentiation as described in supplement protocol “routine maintenance and passaging of hiPSC” following steps 47–51.
-
a.
Pause point: At this stage and after transducing hiPSC with the virus, cells can be expanded for maintenance as described in supplement protocol “routine maintenance and passaging of hiPSC” and cryopreserved using the protocol described in supplement protocol “cryopreservation of hiPSC”. Keeping a stock of already transduced cells will save time and avoid the cost of repeated transductions. It is advisable to cryopreserve cells that exhibit the best response to puromycin selection and doxycycline induction. Cryopreserved and transduced cells can be maintained and used up to 10 passages (the use of cells beyond passage 10 has not been attempted).
Preparing NGN2 expressing hiPSC for neuronal induction
Timing: 4 days
This section describes the treatment of hiPSC with doxycycline to express NGN2 to trigger the neuronal induction of hiPSC to generate iNeurons.
Day -1
Timing: 1 h
-
10.To start the neuronal induction protocol following NGN2 transduction (Figure 1A), on day -1, once hiPSC become confluent, passage the cells, and plate them as single cells following the procedure described in supplement protocol “passaging hiPSC as single cells”.
-
a.Count cells using a Trypan Blue cell counting method using an automated cell counter or hematocytometer to determine the concentration of the cells in suspension. Only take in consideration live cells when determining the final cell density.
-
a.
-
11.
Dilute the cells in the desired volume of mTeSR Plus containing 10 μM of Y-27632, and plate 8.5 × 105 or (1 × 105 cells/cm2) cells per 10-cm plate.
Day 0
Timing: 1 h
-
12.
Prepare the desired volume of D0/1 medium (materials and equipment) containing puromycin.
CRITICAL: Puromycin concentration needs to be optimized depending on the hiPSC line used. We have tested puromycin concentrations ranging between 1 μg/mL to 10 μg/mL. Adding puromycin on day 0 will kill off cells not expressing the NGN2/puromycin resistance gene.
-
13.
Aspirate media from the plate and add 7 mL of D0/1 medium (materials and equipment) containing puromycin.
-
14.
Culture cells for 16 h at 37°C in 5% CO2.
Day 1
Timing: 30 min
-
15.Replace the media on the plates with a fresh 7 mL of D0/1 medium (materials and equipment) containing puromycin.
-
a.On day 1, cells may be surrounded by debris after the puromycin selection. The debris observed at this point will not affect the subsequent steps of the protocol.
-
b.On day 1, cells should start to change morphology (Figure 1B).
-
a.
Day 2
Timing: 1 h 30 min
-
16.Prepare D2 media (materials and equipment), and allow the prepared plates or coverslips coated with PLO/laminin (coating acid-washed glass coverslips with Poly-L-ornithine followed by laminin) to reach 20°C–22°C.
-
a.Aspirate the media from the 10-cm plates and wash the cells with DPBS.
-
b.Aspirate the DPBS and add 3–4 mL of accutase per 10-cm plate.
-
c.Incubate the plate at 20°C–22°C for 2–3 min.
-
d.Check under an inverted microscope for cell detachment, once cells are detached, tap the plate to aid in detaching the cells.
-
a.
CRITICAL: It is highly recommended that plates or coverslips are coated with PLO/laminin on day 0 and day 1 of the protocol respectively. Troubleshooting 3.
-
17.Without aspirating the accutase, add 5–6 mL of DMEM/F12 medium containing antibiotic-antimycotic (materials and equipment).
-
a.Using a 5-mL pipette, pipette up and down to ensure the collection of all the cells.
-
b.Transfer the cell suspension to a 15- or 50-mL centrifuge tube.
-
a.
-
18.Centrifuge the suspended cells at 300 × g for 3 min.
-
a.Aspirate media and carefully resuspend cells in 2–5 mL (depending on the size of the cell pellet) of D2 medium (materials and equipment).
-
b.Using a 1-mL filter pipette tip gently triturate the cells, gently pipette again using a filtered 200-μL pipette tip.
 CRITICAL: Do not over-pipette, cells are more fragile at this stage, over-pipetting can reduce cell viability.
CRITICAL: Do not over-pipette, cells are more fragile at this stage, over-pipetting can reduce cell viability. -
c.Count cells using a Trypan Blue cell counting method using an automated cell counter or hematocytometer to determine the concentration of the cells in suspension. Only consider live cells when determining the final cell density.
-
a.
-
19.
Dilute the cells in the desired volume of D2 medium containing 10 μM of Y-27632 and 2 μM of Ara-C and plate 300 μL of 10–12 × 105 cells (2.4–3 × 105 cells/cm2) per well of a 48-well plate.
CRITICAL: The recommended medium volume is 0.2–0.5 mL/cm2 , a volume range suggested to allow for adequate gas exchange in the culture plate. In this protocol we use 300 μL media volumes for cells grown in 48 well plates, and this volume is in the right range recommended for adequate cell growth.
CRITICAL: The plating number here is suggested for a 48-well plate, cell numbers should be adjusted based on the plate type used (scale the number of plated cells to the surface area used).
CRITICAL: Ara-C is recommended as an anti-mitotic agent to eliminate proliferating non-neuronal cells.9Troubleshooting 4.
CAUTION: Ara-C is a hazardous substance, and it must be handled with caution.
-
20.
Culture the cells for 16 h at 37°C in 5% CO2.
Differentiation of NGN2 expressing hiPSC to iDA cells
Timing: 6 days
This section describes the forced differentiation of iNeurons towards the DA phenotype and generation of iDA neurons.
Day 3
Timing: 1 h 30 min
-
21.Dopaminergic neuronal differentiation starts on day 3 (Figure 2A).
-
a.Start by gently taping the plate and carefully wash the cells with DPBS Ca+ Mg+ to wash off any dead cells.
-
b.Gently tap the plate again and replace media with 150 μL of warm (37°C) STEMdiff Midbrain Neuron Differentiation medium containing 1 μM of Ara-C + 2 μg/mL of doxycycline per well of a 48-well plate.
-
a.
Note: Day 3 is the last day when neuronal media contains Ara-C. Cells start to exhibit neuronal morphology (Figure 2B).
Figure 2.
Detailed protocol of the iDA differentiation and maturation steps
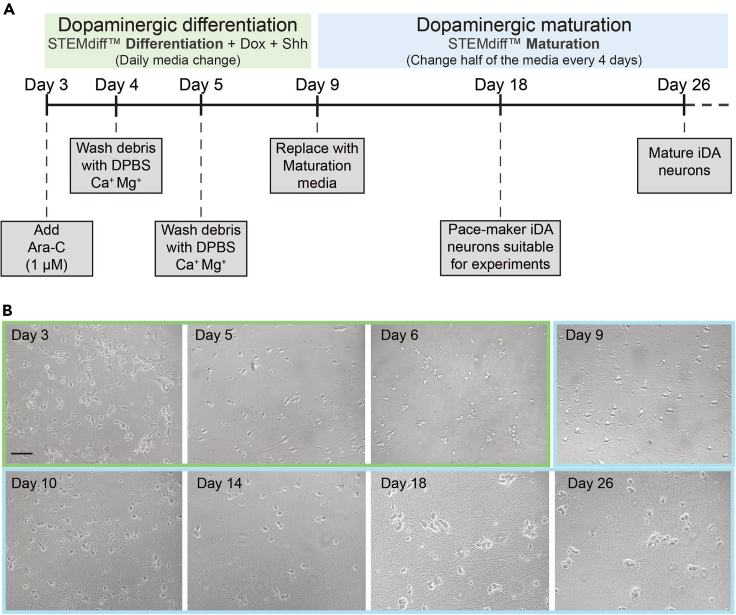
(A)The dopaminergic differentiation protocol (Day 3 to Day 8) is based on daily media changes with STEMdiff differentiation media supplemented with doxycycline and Shh. On Day 3, culture media is supplemented with Ara-C (1 μM) to further eliminate any remaining proliferating cells. To ensure good quality of the cells, cellular debris are thoroughly washed off using DPBS Ca+ Mg+ prior to media change on Day 4 and Day 5. Between Day 6 and Day 8, daily media changes are performed using STEMdiff differentiation media supplemented with doxycycline and Shh. Starting from Day 9, the maturation protocol starts by replacing full media with STEMdiff Maturation media. Following Day 9, half of the culture media is replaced every 3–4 days.
(B) Representative bright-field microscope images illustrating the morphological changes occurring during the iDA differentiation and maturation, between Day 3 and Day 26. Scale bar, 50 μm.
Day 4
Timing: 1 h
-
22.Gently tap the plate and carefully wash the cells with DPBS Ca+ Mg+ to wash off any dead cells.
-
a.Gently tap the plate again and replace media with warmed 150 μL (37°C) STEMdiff Midbrain Neuron Differentiation Medium containing 2 μg/mL of doxycycline per well of a 48-well plate.
-
b.Incubate the cells for 16 h at 37°C and 5% CO2.
-
a.
Day 5–8
Timing: 20 min
-
23.
Perform daily media changes (300 μL) with warm (37°C) STEMdiff Midbrain Neuron Differentiation Medium containing 2 μg/mL of doxycycline.
Note: Day 8 will be the last day when neuronal media contains doxycycline.
Establishment of mature iDA cells
Timing: 15 days
Day 9
Timing: 30 min
-
24.Dopaminergic neuronal maturation starts on day 9 (Figure 2A).
-
a.Replace STEMdiff Midbrain Neuron Differentiation Medium with 300 μL of STEMdiff Midbrain Neuron Maturation medium per well of a 48-well plate.
-
b.Culture the cells for 16 h at 37°C in 5% CO2.
-
a.
Day 10–25
Timing: 30 min
-
25.
Starting from this step onwards, perform half-medium changes (150 μL) with warm (37°C) STEMdiff Midbrain Neuron Maturation medium every 3–4 days.
Note: The cells will start to exhibit a mature neuronal morphology (Figure 2B).
CRITICAL: Avoid daily monitoring of cells, since this may affect their viability.
Note: This protocol highlights the rapidity of the approach we have developed1; however, cells can be cultured for longer periods of time (50–60 days) with medium changes every 4 days. Troubleshooting 5.
Day 26 onwards
-
26.
On day 26, or later, iDA cells can be characterized; proceed to “characterization of mature iDA cells” and follow the appropriate option. Troubleshooting 6.
CRITICAL: This protocol was established using hiPSC that have been used in different studies in the laboratory,1,10 which have also been routinely karyotyped, characterized and checked for mycoplasma contamination. In our published paper,1 we used multiple hiPSC lines, and we show highly similar levels of successful DA neuronal phenotype.1 Nonetheless, we cannot eliminate the possibility of differences in the differentiation kinetics of hiPSC from different donors. Therefore, optimization of MOI requirements for viral transduction, response to puromycin selection and starting cell number plating may be required. Moreover, programming with the NGN2 system, induces a rapid and efficient differentiation of hiPSC into iNeurons, if no neuronal induction is observed at the early time points as presented in Figure 1 on day 1, refer to troubleshooting 1 for possible solutions.
Characterization of mature iDA cells
Immunofluorescence analysis to confirm the identity of iDA neurons
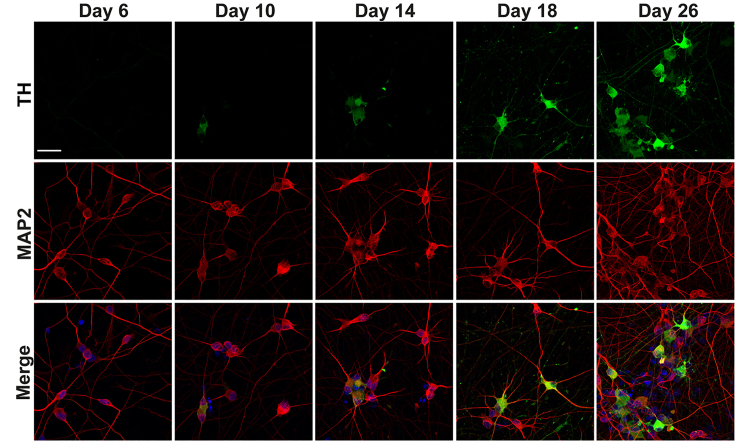
Note: To confirm successful DA neural induction, perform immunofluorescence analysis using a list of mature neuronal markers such as Microtubule Associated Protein 2 (MAP2), and the DA neuronal marker, the tyrosine hydroxylase (TH), as we previously reported1 (Figure 3). The neuronal marker (MAP2) is visible as early as day 4 after neuronal induction and the DA marker (TH) is visible as early as day 10 (Figure 3).
Note: The present protocol is an optimized version of the protocol we previously published.1 This optimization is based on improvements in Ara-C treatment time, and in the maintenance of cells through rigorous washing to eliminate any debris which may affect neuronal health. Most importantly, avoiding cell clumps from accumulating on the plates and/or coverslips has shown to improve the efficiency of the differentiation and maturation protocol allowing for higher TH+ positive cells, and users can expect to achieve a yield of almost 80% of TH + cells as early as day 18 (Figure 4). Troubleshooting 7.
CRITICAL: As we previously reported,1 we recommend including negative controls during the programming protocol, these controls can include (non-expressing NGN2 hiPSC or NPCs, and NGN2 expressing hiPSC not exposed to the dopaminergic differentiation/maturation protocols). Including such controls will allow for a clear comparison for the phenotypic characteristics associated with DA midbrain neurons, including morphological phenotypes, as well as an expression of midbrain-specific genes and proteins throughout the protocol timeline.
Note: For the growth and maintenance of NGN2 expressing hiPSC not exposed to the dopaminergic differentiation/maturation protocols follow this: Starting from day 2 onwards, NGN2 expressing hiPSC can be grown in D2 media for the duration of the protocol. Performing half media changes on the cells every 2–3 days.
-
27.
Fix cells in culture with 4% (wt/vol) paraformaldehyde (PFA) and 3% (wt/vol) sucrose for 15 min at 20°C–22°C.
-
28.
Perform three 5-min washes with DPBS Ca+ Mg+ and permeabilize the cells with 0.3% (vol/vol) Triton X-100 in 1× PBS for 10 min at 20°C–22°C.
-
29.
Rinse the cells with 1× PBS to remove remaining Triton.
-
30.
Block cells for 1 h at 20°C–22°C with 5% (vol/vol) normal goat serum (NGS), and 2% BSA (vol/vol) in 0.1% (vol/vol) Triton X-100–1× PBS.
-
31.
Stain with primary antibodies against MAP2, and TH (key resources table) diluted in blocking solution at 4°C for 16 h.
-
32.
Perform three 10-min washes with fresh blocking buffer.
-
33.
Stain with secondary antibodies (key resources table) diluted at 1:400 for 1 h 30 min at 20°C–22°C.
-
34.
Perform two 10-min washes with 1× PBS.
-
35.
Counterstain the cells with DAPI at 1:5000 in 1× PBS for 2 min.
-
36.
Perform two quick washes with 1× PBS.
-
37.
Before mounting, quickly rinse the coverslips with dH2O to remove salts.
-
38.
Mount the 8-mm coverslips using 3.5 μL of mounting media.
-
39.
Evaluate the protein expression by performing high-resolution microscopy as used in our original report.1
Figure 3.
Molecular characterization of iDA cells
Immunostaining analysis of iDA cells with Tyrosine hydroxylase (TH, green) and Microtubule-associated protein 2 (MAP2, red) at different timepoints. Nuclei are stained with DAPI. These representative images show the increase in the number of TH+ cells overtime. Scale bar, 20 μm.
Figure 4.
Efficiency of the optimized protocol to generate TH+ neurons
(A) Immunostaining analysis of iDA cells after D18 in culture showing a high number of TH+ cells and extensions. The cells were stained for TH (green), MAP2 (red) and DAPI (blue). Scale bar, 20 μm.
(B) Histograms representing the percentage of DA neurons (TH = ) relative to the total cells in culture (DAPI) or to neuronals cells (MAP2+) at Day 18 (n = 3). Data are represented as mean ± SEM.
Supplement protocol for culturing hiPSC
Routine maintenance and passaging of hiPSC
Timing: 6–7 days
-
40.
Thaw a vial of frozen hiPSC in a 37°C water bath. Prepare a 15-mL tube containing 5 mL of mTeSR Plus medium. Add the hiPSC mixture dropwise into the 5 mL medium containing tube.
CRITICAL: Do not pipette since it can risk breaking-up the hiPSC colonies.
-
41.
Centrifuge for 3 min at 300 × g at 20°C–22°C.
-
42.Resuspend the pelleted cells in 2 mL of mTeSR Plus medium containing 10 μM of Y-27632.
-
a.Add the 2 mL of medium containing cells to a 60 mm Matrigel-coated plate.
-
b.Rock the plate forward and backward and from side to side to ensure an even plating of the hiPSC on the plate.
-
a.
CRITICAL: Do not expose cells to Y-27632 for more than 24 h since long-term exposure to Y-27632 negatively affects the proliferation and survival of hiPSC.
-
43.
Culture cells for 16 h at 37°C in 5% CO2.
-
44.
On the following day, remove the mTeSR Plus medium and replace with fresh mTeSR Plus medium lacking Y-27632.
Note: From this point, continue to incubate the cells, changing the medium every second day until hiPSC form colonies with diameters ranging from 1 mm-2 mm, reaching around 70% confluency. Depending on the cell line, this can take between 5 to 7 days.
CRITICAL: Do not overgrow the hiPSC colonies; otherwise, spontaneous differentiation is observed.
-
45.
Regularly monitor hiPSC under a light microscope to assess the morphology and health of the colonies.
Note: If spontaneous differentiation is observed, manually remove these areas using the tip of a pipet tip. Troubleshooting 8.
-
46.
When cells reach more than 50% confluency, change media every day since it will oxidize quickly.
Note: From this step onwards, floating cells may be observed in the media, this is due to normal cell death induced by the cells growing as 3D colonies. Troubleshooting 9.
-
47.Once visible colonies are at 70% confluency, passage the hiPSC colonies with EDTA dissociation buffer (materials and equipment).
-
a.First remove the mTeSR Plus medium, rinse the plate with 2 mL of DPBS.
-
b.Add 2 mL of EDTA dissociation buffer per 60-mm plate.
-
c.Keep the plate at 20°C–22°C for 5–6 min, checking for the detachment of the colonies every 2 min on an inverted microscope.
-
a.
-
48.Once the edges of the colonies start to detach, the cells are ready for harvesting.
-
a.Aspirate the EDTA dissociation buffer by gently aspirating the buffer.
-
a.
CRITICAL: Do not over-incubate the cells with the EDTA dissociation buffer since over-incubation may favor the dissociation of hiPSC as single-cells, which will reduce their viability and promote spontaneous differentiation.
CRITICAL: Using EDTA as part of the dissociation protocol does not require the need to add Y-27632. Therefore, for routine maintenance of hiPSC, the addition of Y-27632 treatment is optional. Troubleshooting 10.
-
49.Add 2 mL of mTeSR Plus medium and tap the dish to slightly detach the colonies.
-
a.Gently pipetting up and down 2 to 3 times with a 5-mL pipette.
-
b.Detach and collect the colonies from the plate.
-
a.
CRITICAL: Do not over pipette; if the colonies fragment excessively, the yield of hiPSC will be affected. If the colonies are too small, or too big, the colonies will spontaneously differentiate.
CRITICAL: Make sure to use a 5-mL pipette, smaller pipettes or micropipettes will excessively fragment the colonies.
-
50.Plate the hiPSC colonies at a ratio of 1:20 (relative to the volume of starting media suspension; it’s usually one to two drops of the media containing cells) into fresh Matrigel-coated pre-filled with 2.5 mL of mTeSR Plus medium.
-
a.Rock the plate forward and backward and from side to side to ensure an even plating of the hiPSC colonies on the plate.
-
a.
-
51.Incubate the cells for 5–7 days and change the medium every second day with mTeSR Plus medium until hiPSC form colonies with diameters ranging from 1 mm-2 mm.
-
a.Regularly monitor hiPSC under a light microscope to assess the morphology and health of the colonies.
-
a.
Note: If spontaneous differentiation is observed, manually remove these areas using the tip of a pipet tip by scratching.
Alternatives: Here we use mTeSR Plus medium, other media such as mTeSR™1, StemFlex, Essential 8 Medium can be used for the maintenance and culture of hiPSC. It is highly recommended to use one kind of media, and not change the type of mTeSR the hiPSC cells were first exposed to.
Pause point: Expanded hiPSC need to be passaged at least one before starting the NGN2 reprogramming protocol. Likewise, hiPSC colonies cells can be cryopreserved following the procedure described in supplement protocol “cryopreservation of hiPSC” after at least 1 passage post thawing.
Cryopreservation of hiPSC
Timing: 0.5–1 h
-
52.
For the cryopreservation of hiPSC, collect the cells following steps 47-49 from supplement protocol “routine maintenance and passaging of hiPSC”.
-
53.
Thaw a mFreSR aliquot at 4°C or on ice.
Note: After thawing aliquots, use immediately. Do not refreeze.
-
54.
Resuspend the collected cell pellets in cooled mFreSR medium while maintaining cells as colonies.
-
55.
Aliquot 1 mL of the cell suspension (keeping cells as colonies) into prelabeled cryovials.
-
56.
Transfer the cryovials into a Corning CoolCell LX Cell Freezing Vial Container, and immediately put at −80°C.
CRITICAL: Steps 52–56 above must be performed quickly not to compromise cell viability.
CRITICAL: To maximize cell viability, alternative freezing approaches such as utilizing a controlled rate freezer can also be performed.
-
57.
Twenty four hours later, transfer the cryovials into a liquid nitrogen tank for long-term storage.
Passaging hiPSC as single cells
Timing: 0.5–1 h
-
58.
Passage hiPSC following steps 47–49 from supplement protocol “routine maintenance and passaging of hiPSC”.
-
59.To the plate containing slightly detached hiPSC colonies, add 2 mL of mTeSR Plus medium.
-
a.Collect the colonies by pipetting up and down 5 to 7 times with a 5-mL pipette to detach the colonies from the plate.
-
b.Gently pipette the cell suspension with a 1-mL filter pipette tip 3 to 5 times.
-
a.
Note: If clusters of cells are still visible, gently pipette again, using a filtered 200-μL pipette tip, and confirm that cells have been dissociated into single cells. Troubleshooting 11.
-
60.Harvest the cell suspension and centrifuge the cells at 300 × g for 3 min at 22°C with maximum acceleration and deceleration.
-
a.Aspirate the media and resuspend cells in 2–3 mL of mTeSR Plus medium containing 10 μM of Y-27632.
-
b.Gently triturate the cell suspension with a 1-mL filtered pipette tip 3–5 times to further confirm that the cells have been dissociated into single cells.
-
a.
-
61.
Count cells using a Trypan Blue cell counting method using an automated cell counter or hematocytometer to determine the concentration of the cells in suspension.
CRITICAL: Using Trypan Blue to accurately estimate the number of viable cells is highly recommended.
-
62.
Examine the plated cells under the microscope making sure the cells are not clumped. Troubleshooting 4.
-
63.
Culture the cells for 16 h at 37°C in 5% CO2.
Expected outcomes
The present protocol has been validated in multiple differentiation experiments, using at least three different hiPSC lines from three different donors. We do not report high variability amongst the different programming experiments, and we are able to successfully derive iDA cells with more than 70–80% TH+ cells (Figure 4). The efficiency of the protocol mainly relies on the quality of the hiPSC and the procedure of their maintenance, which we have optimized as described in the present protocol.
Furthermore, the DA phenotype of iDA neurons should be validated by assessing the expression of key dopaminergic markers such as TH, G-protein-regulated inward-rectifier potassium channel 2 (GIRK2), Forkhead Box A2 (FOXA2), the vesicular monoamine transporter (VMAT) and dopamine receptor D2 (D2R), using immunocytochemistry and RT-qPCR analysis. Likewise, successful iDA differentiation should also be associated with the loss of the expression of pluripotency genes, such as SRY-Box transcription factor 2 (SOX2) and the octamer-binding transcription factor 4 (Oct4).
In addition to the molecular characterization described in this protocol, functional validation of the iDA neurons can be assessed using high-performance liquid chromatography (HPLC) analysis to examine the production and release of the dopamine and its metabolites.1,11,12 Electrophysiological analysis can also be used to confirm the functionality and electrical activity of the iDA neurons.1 Using this protocol, we have previously shown that iDA neurons exhibit a pace-making activity as early as day 18.1,13 We also reported on a correlation between electrical activity and the release of the dopamine in the culture media, without the need for external stimulus.1 These patterns can be detected by patch-clamp recordings of single neurons or visualized using engineered calcium detectors like GCamP series. Since iDA neurons show such firing patterns, they are also suitable for synaptic activity analysis using markers such as PSD95 and Homer-1.14 These features can be further developed to study dopamine pathways dysfunction and their role in disease development.
Likewise, iDA neurons can be used to investigate the molecular mechanisms underlying the selective DA neuronal vulnerability observed in some neurodegenerative disorders including Parkinson’s disease,15 which can help identify potent inhibitors of neuronal degeneration in these diseases.
Limitations
Although the present protocol has shown a remarkable efficacy in producing DA neurons differentiated from hiPSC, it presents with some limitations. The first limitation consists of the use of the viral delivery method for NGN2 overexpression in hiPSC. While this approach results in high transgene expression levels, viral integration can alter some of the normal gene functions.16 Today, mRNA delivery is regarded as the more suitable approach for disease modeling and transplantation studies since they present with the advantage of absence of long-term expression and integration of genetic material into the genome.17,18
The second limitation is the suitability of the generated iDA to cryopreservation. The challenge comes with increasing the viability of cells after thawing frozen banks of DA neurons. Low cell viability means the need for higher scaling of neuronal production which is often tedious and complex. It has been established that these criteria are dependent on the stage of maturity of DA neurons, on the capacity of the cells to differentiate post thawing, parameters that we need to further optimize for the present protocol.19,20,21
Troubleshooting
Problem 1
Poor neuronal induction and early cell death, related to step 2 and step 26.
Potential solution
-
•High quality of hiPSC is essential for the success of neural induction. It is important to regularly perform quality control checks to ensure healthy hiPSCs colonies, which include the following:
-
○Colonies should have a good round shape with dense centers.
-
○Colonies should have tightly packed cells with a prominent nucleoli. Loss of uniformity, border integrity and cellular packing are indicative of unhealthy hiPSCs.
-
○Check for areas of differentiation, it is normal for hiPSC to show a low percentage (5%) of spontaneous differentiation which can be manually removed before differentiation. However, an increase in areas of differentiation (over 10%) is indicative of poor-quality hiPSCs.
-
○
If the hiPSCs do not pass this quality control thaw a new batch before starting the differentiation protocol.
-
•
Ensure proper puromycin selection.
-
•
Proper counting of hiPSCs used for induction is critical since plating too low or too high cell confluency can affect induction efficiency. The recommended plating density for induction is 0.9 × 105 cells/cm2.
-
•
Ensure that you maintain cells in D0/1 media for 48 h. Cells need to be exposed to D0/1 media on day 0 and day 1 of the protocol. This media contains doxycycline which will allow for the initiation of neuronal induction.
-
•
On day 2 Ara-C is added to D2 media at the recommended concentration of 1 μM. Adding Ara-C at earlier timepoints and/or at higher concentrations can induce a high number of cell death, and may kill off cells that are at the stage of differentiating into neurons. If you observe too much of cell death after treating with Ara-C, then delay treatment of Ara-C to day 3 of the protocol. We would recommend performing a control test to determine how the hiPSC lines used in the study respond to Ara-C treatment.
Problem 2
Low transduction efficiency, related to step 4.
Potential solution
-
•
The transduction efficiency depends on the viral titer. To ensure proper titer, check the titer by transducing hiPSCs with serial dilutions (MOI) of the lentiviral vector.
-
•
To determine the best MOI, use one well for each MOI to test, and test a range of puromycin concentrations (ranging between 1 μg/mL to 10 μg/mL). Include an additional well as an untransduced control, this will allow determine the positive selection of successfully transduced cells.
-
•
Ensure that Lentivector expression construct DNA preparation is of good quality.
Problem 3
iNeurons attach poorly after passaging, which may happen from inappropriate coating of tissue culture plates or coverslips, related to step 16.
Potential solution
-
•
Ensure that the tissue culture plates, or coverslips used to have been coated properly with PLO/Laminin.
-
•
Make sure that the Sodium Borate Buffer has been properly prepared, and that PLO in Sodium Borate Buffer has been properly washed.
Problem 4
Too many neuronal clusters on day 4 onwards, related to step 19 and 62. This may happen when iNeurons were not dissociated properly into single cells during passaging steps on day 2. Likewise, this may occur from having too many proliferating cells, and in turn Ara-C treatment will result in inducing high cell death, and as a result this can create clusters with too many dead cells.
Potential solution
-
•
Ensure that the cells have been dissociated into single cells before plating. To ensure single-cell dissociation, possible options may include increasing the incubation time with the dissociation solution such as accutase or alternative enzymatic based solutions such as ACCUMAX Cell Detachment Solution. This will help reduce the number of proliferating cells.
-
•
Addition of Ara-C at lower concentrations is possible see “problem 1”.
-
•
When choosing a lower concentration of Ara-C, it is possible to prolong the incubation time with Ara-c, however longer exposure times may create clusters. The amount of non-neuronal dead cells may indirectly affect the state of the neurons in culture, since the presence of cell debris in the culture medium is a common cause of cell clustering.
Problem 5
Notable cell death from day 20 of maturation protocol onward, related to step 25. This may occur from the following reasons:
-
•
Starting with poor quality hiPSC can affect the differentiation protocol in the long run.
-
•
Dense cultures leading to a lack of nutrients.
-
•
Low culture number leading to lack of neuronal extensions.
-
•
Cells are out of the incubator for prolonged periods.
-
•
Cells are detaching on the edge of the coverslips, thus wrapping the layer of cells leading to a detachment of the cells.
Potential solution
-
•
Use high quality hiPSC, refer to problem 1.
-
•
Increase the volume of StemCell™ medium added to each well and replace media more often.
-
•
Ensure plating a good number of cells per well. Optimize cell number plating prior to differentiation. A range between 7.5 × 104 and 12 × 104 cells or (1.8 × 104–3 × 104 cells/cm2) per well of a 48-well plate is usually a good starting point.
-
•
hiPSC-derived neurons are sensitive cells. Working with such cells requires the user to change the media as quickly as possible when the cells are out of the incubator while maintaining the sterility of the cells. Do not leave cells out of the incubator for longer than 10-min periods.
-
•
We always recommend using micropipettes to gently aspirate the media at all steps of the neuronal culture. Never touch the layer of cells present on the coverslip. Tilt the plate at a 45° angle to aspirate the culture medium. Don’t allow the coverslip and/or the well or plate to air-dry, and directly add fresh medium on the edge of the well, so the medium drops do not directly fall on the layer of cells.
Problem 6
Inefficient iDA differentiation, where you observe that not enough cells differentiated into neurons on day 3 onwards, related to step 26.
Potential solution
-
•
Be sure to use iNeurons that are healthy, by checking them on D0/1, this will allow for proper differentiation.
-
•
Make sure to select the appropriate puromycin concentration before proceeding with the dopaminergic differentiation and maturation protocols, refer to problem 2.
-
•
Make sure that the media includes the appropriate concentration of doxycycline, which will ensure activation of NGN2 expression. Review doxycycline handling and storage conditions in “materials and equipment” section.
Problem 7
Low number of TH+ iDA neurons when evaluated by ICC, related to “characterization of mature iDA cells”. This may occur from the following reasons.
-
•
Poor quality of hiPSC used for reprogramming, refer to problem 1.
-
•
Expired StemCell™ differentiation or maturation medium.
-
•
Too many cells plated, or too many cell clusters.
-
•
Poor antibody quality.
Potential solution
-
•
Use high quality hiPSC, refer to problem 1, and do not over passage the cells.
-
•
Once reconstituted, StemCell™ medium is usually consumed within 4 weeks. Check that media are not expired and use fresh StemCell™ medium.
-
•
To prevent the degradation of heat-sensitive components of the media, pre-warm small aliquots of StemCell™ medium before each use.
-
•
Having too many cells on the coverslips means more proliferating cells, more dead cells, hence resulting in the clustering of the neurons. To avoid clusters, plates should not be kept out of the incubator for extended periods of time, since exposure to prolonged cold, changes in pH, or changes in O2/CO2 are harmful to the neurons. These stresses can lead to cell detachment and clustering.
Problem 8
Inefficient neuronal differentiation that may be caused from either reason: hiPSC used have a high passage number or from the presence of too many spontaneous differentiating hiPSC, related to step 45.
Potential solution
-
•
Use early passage hiPSC for reprogramming. Do not use overgrown hiPSC for reprogramming.
-
•
Always check for spontaneous differentiating hiPSC in the proliferating hiPSC and try to manually remove these cells before differentiation.
Problem 9
hiPSC attach poorly after passaging and do not proliferate and this may be due to either the inappropriate coating of tissue culture plates or due to having hiPSC overgrown before passaging, related to step 46.
Potential solution
-
•
Ensure that the tissue culture plates used have been coated with Matrigel for at least 1 h.
-
•
Make sure that Matrigel has been handled properly following the recommended guidelines.
-
•
Make sure plates are not dry before plating cells.
-
•
Do not let hiPSC overgrow; passage the colonies when they reach 70% confluency.
-
•
Instead of changing media every other day, if media color changes are noticed, suggestive of oxidation, start changing media every day until hiPSC culture reaches 70% confluency.
Problem 10
The Majority of the hiPSC are coming off when removing EDTA-based buffer or inversely hiPSC do not detach from the plate after EDTA treatment. This may be due to long EDTA incubation time or that the EDTA pH/concentration is not correct, related to step 48.
Potential solution
Check the pH and concentration of the solution. If the problem persists, reduce, or increase the incubation time. If the majority of the hiPSC are still coming off, add mTeSR Plus directly in the EDTA buffer. EDTA action should be quenched by mTeSR Plus.
Problem 11
Too many spontaneously differentiated cells, this can also happen when differentiating neuronal culture become over-confluent, related to step 59. The main reason this can happen is that when plating cells, the number of cells plated on day -1 is too high, thus increasing the number of proliferating cells. Another reason can be due to that the hiPSC cells were not dissociated properly into single cells during passaging steps on day -1.
Potential solution
-
•
Ensure that the cells have been dissociated into single cells before plating. Properly count cells, and check under the microscope to make sure cells are not clumped together to ensure accurate counting.
-
•
To ensure better cell dissociation, increase the incubation time with the EDTA-based buffer.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Abid Oueslati (Abid.Oueslati.1@ulaval.ca).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (CIHR). A.O. is supported by Junior 1 and Junior 2 salary awards from the Fonds de Recherche du Québec – Santé (FRQS) and la Société Parkinson du Québec. M.T. is supported by the FRQS doctoral fellowship.
Dr. Thomas Drurcan and Dr. Edward Fon from The Neuro’s Early Drug Discovery Unit (EDDU), McGill University, provided us with the hiPSC. Dr. Thomas C. Südhof provided us with the TetO-NGN2-T2A-puromycin and Ubi-rtTA plasmids.
Author contributions
Conceptualization, A.O. and R.S.; data analysis, A.O. and R.S.; manuscript preparation, A.O. and R.S.; collection and analysis of the cell culture data, R.S., M.T., and W.I.; generation and optimization of the protocol of hiPSC culture, R.S., M.T., and W.I.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Razan Sheta, Email: razan.sheta.1@ulaval.ca.
Abid Oueslati, Email: abid.oueslati.1@ulaval.ca.
Data and code availability
This study did not generate/analyze data and/or code.
References
- 1.Sheta R., Teixeira M., Idi W., Pierre M., de Rus Jacquet A., Emond V., Zorca C.E., Vanderperre B., Durcan T.M., Fon E.A., et al. Combining NGN2 programming and dopaminergic patterning for a rapid and efficient generation of hiPSC-derived midbrain neurons. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-22158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tiscornia G., Singer O., Verma I.M. Production and purification of lentiviral vectors. Nat. Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y., Pak C., Han Y., Ahlenius H., Zhang Z., Chanda S., Marro S., Patzke C., Acuna C., Covy J., et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nehme R., Zuccaro E., Ghosh S.D., Li C., Sherwood J.L., Pietilainen O., Barrett L.E., Limone F., Worringer K.A., Kommineni S., et al. Combining NGN2 Programming with Developmental Patterning Generates Human Excitatory Neurons with NMDAR-Mediated Synaptic Transmission. Cell Rep. 2018;23:2509–2523. doi: 10.1016/j.celrep.2018.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng Y.H., Chanda S., Janas J.A., Yang N., Kokubu Y., Südhof T.C., Wernig M. Efficient generation of dopaminergic induced neuronal cells with midbrain characteristics. Stem Cell Rep. 2021;16:1763–1776. doi: 10.1016/j.stemcr.2021.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmid B., Holst B., Poulsen U., Jørring I., Clausen C., Rasmussen M., Mau-Holzmann U.A., Steeg R., Nuthall H., Ebneth A., Cabrera-Socorro A. Generation of two gene edited iPSC-lines carrying a DOX-inducible NGN2 expression cassette with and without GFP in the AAVS1 locus. Stem Cell Res. 2021;52 doi: 10.1016/j.scr.2021.102240. [DOI] [PubMed] [Google Scholar]
- 7.Schörnig M., Ju X., Fast L., Ebert S., Weigert A., Kanton S., Schaffer T., Nadif Kasri N., Treutlein B., Peter B.M., et al. Comparison of induced neurons reveals slower structural and functional maturation in humans than in apes. Elife. 2021;10 doi: 10.7554/eLife.59323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gan K.J., Südhof T.C. Specific factors in blood from young but not old mice directly promote synapse formation and NMDA-receptor recruitment. Proc. Natl. Acad. Sci. USA. 2019;116:12524–12533. doi: 10.1073/pnas.1902672116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mañáková S., Puttonen K.A., Raasmaja A., Männistö P.T. Ara-C induces apoptosis in monkey fibroblast cells. Toxicol. Vitro. 2003;17:367–373. doi: 10.1016/s0887-2333(03)00024-9. [DOI] [PubMed] [Google Scholar]
- 10.Bérard M., Sheta R., Malvaut S., Rodriguez-Aller R., Teixeira M., Idi W., Turmel R., Alpaugh M., Dubois M., Dahmene M., et al. A light-inducible protein clustering system for in vivo analysis of alpha-synuclein aggregation in Parkinson disease. PLoS Biol. 2022;20 doi: 10.1371/journal.pbio.3001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terraf P., Babaloo H., Kouhsari S.M. Directed Differentiation of Dopamine-Secreting Cells from Nurr1/GPX1 Expressing Murine Embryonic Stem Cells Cultured on Matrigel-Coated PCL Scaffolds. Mol. Neurobiol. 2017;54:1119–1128. doi: 10.1007/s12035-016-9726-4. [DOI] [PubMed] [Google Scholar]
- 12.Roy N.S., Cleren C., Singh S.K., Yang L., Beal M.F., Goldman S.A. Functional engraftment of human ES cell-derived dopaminergic neurons enriched by coculture with telomerase-immortalized midbrain astrocytes. Nat. Med. 2006;12:1259–1268. doi: 10.1038/nm1495. [DOI] [PubMed] [Google Scholar]
- 13.Poulin H., Martineau L., Racine V., Puymirat J., Chahine M. Differentiation of lymphoblastoid-derived iPSCs into functional cardiomyocytes, neurons and myoblasts. Biochem. Biophys. Res. Commun. 2019;516:222–228. doi: 10.1016/j.bbrc.2019.05.176. [DOI] [PubMed] [Google Scholar]
- 14.Shi Y., Kirwan P., Livesey F.J. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat. Protoc. 2012;7:1836–1846. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- 15.Bloem B.R., Okun M.S., Klein C. Parkinson's disease. Lancet. 2021;397:2284–2303. doi: 10.1016/S0140-6736(21)00218-X. [DOI] [PubMed] [Google Scholar]
- 16.Moiani A., Paleari Y., Sartori D., Mezzadra R., Miccio A., Cattoglio C., Cocchiarella F., Lidonnici M.R., Ferrari G., Mavilio F. Lentiviral vector integration in the human genome induces alternative splicing and generates aberrant transcripts. J. Clin. Invest. 2012;122:1653–1666. doi: 10.1172/JCI61852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue Y., Zhan X., Sun S., Karuppagounder S.S., Xia S., Dawson V.L., Dawson T.M., Laterra J., Zhang J., Ying M. Synthetic mRNAs Drive Highly Efficient iPS Cell Differentiation to Dopaminergic Neurons. Stem Cells Transl. Med. 2019;8:112–123. doi: 10.1002/sctm.18-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirkeby A., Grealish S., Wolf D.A., Nelander J., Wood J., Lundblad M., Lindvall O., Parmar M. Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Rep. 2012;1:703–714. doi: 10.1016/j.celrep.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Drummond N.J., Singh Dolt K., Canham M.A., Kilbride P., Morris G.J., Kunath T. Cryopreservation of Human Midbrain Dopaminergic Neural Progenitor Cells Poised for Neuronal Differentiation. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.578907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiramatsu S., Morizane A., Kikuchi T., Doi D., Yoshida K., Takahashi J. Cryopreservation of Induced Pluripotent Stem Cell-Derived Dopaminergic Neurospheres for Clinical Application. J. Parkinsons Dis. 2022;12:871–884. doi: 10.3233/JPD-212934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leitner D., Ramamoorthy M., Dejosez M., Zwaka T.P. Immature mDA neurons ameliorate motor deficits in a 6-OHDA Parkinson's disease mouse model and are functional after cryopreservation. Stem Cell Res. 2019;41 doi: 10.1016/j.scr.2019.101617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze data and/or code.