In the last few years, the study of antivirus defense went through a veritable microrevolution. The diversity of the discovered defense systems that often can be predicted through their colocalization within defense islands in microbial genomes is nothing short of astonishing (1–3). A key unifying theme emerging from these studies is abortive infection (Abi) whereby a broad variety of defense systems cause programmed cell death (PCD) or cell dormancy in response to infection, preventing virus spread across the microbial population. Most of the Abi systems are toxin–antitoxin (TA) modules. In the absence of infection, the toxin is inactive, often, as a result of complex formation with an antitoxin, but infection activates the toxin through a variety of mechanisms, triggering PCD (4). In PNAS, Zhong and colleagues report a novel class of Abi systems that employ a remarkably economical and elegant strategy of antitoxin formation (5).
This new class of Abi systems had been hiding in plain view in numerous bacterial genomes scattered across many phyla and some archaeal genomes, known for years as Rpn (Recombination-Promoting Nucleases). Rpn proteins contain the PD-(D/E)XK (named after the conserved pattern of catalytic amino acids) nuclease domain, which is an extremely abundant type of nuclease in prokaryotes, being present in restriction endonucleases, among others (6). The rpn genes have been considered as a type of transposable element (TE) because their content is highly variable even among closely related bacterial strains, some of which contain numerous rpn copies and because of the demonstrated DNA endonuclease activity of the Rpn protein (7). However, the argument in support of rpn genes being TE is weak as shown by Zhong et al. (5). Indeed, these genes are not flanked by repeats and/or target site duplications like most TE, and PD-(D/E)XK enzymes are not known to function as autonomous transposases. Furthermore, multiple copies of rpn genes appear to be products of tandem duplication rather than transposition. Zhong et al. thus sought to determine the actual function of Rpn and made several notable discoveries. First, it was demonstrated that, in addition to the full-length Rpn protein (denoted RpnL) containing the nuclease domain, the rpn genes direct the synthesis of small proteins (RpnS) that correspond to the C-terminal portions of RpnL proteins and are translated separately using an internal translation initiation site (iTIS) (Fig. 1). Previously, it has been shown that Rpn is toxic when overexpressed in Escherichia coli (7), but in those experiments, RpnL and RpnS were coexpressed, potentially, confounding the results. In the new study, Zhong et al. decouple RpnL from RpnS and show substantial toxicity of RpnL that can be abrogated by RpnS (5). Furthermore, RpnS was shown to form a complex with RpnL and inhibit the endonuclease activity of the latter (Fig. 1).
Fig. 1.
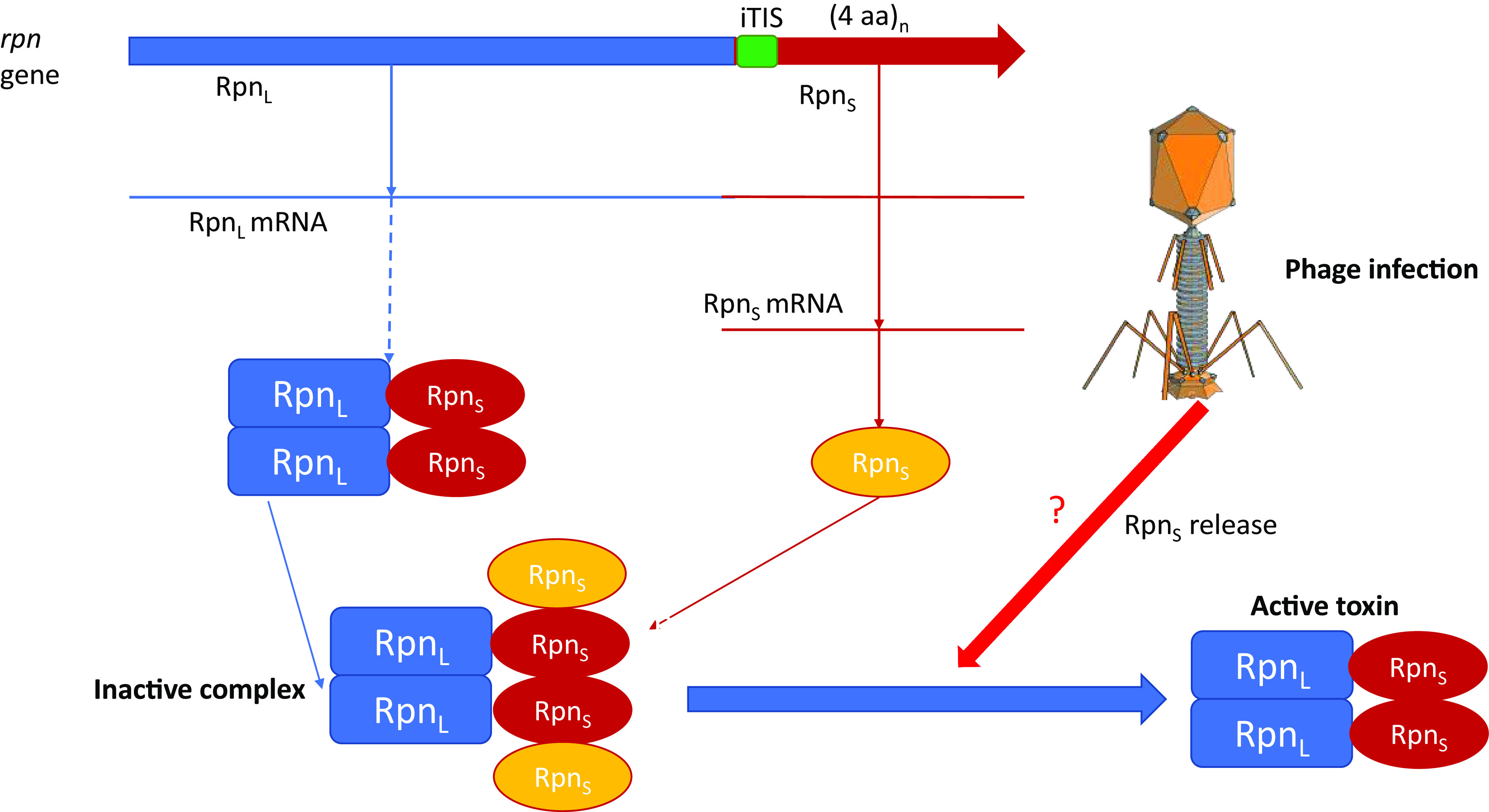
A gene within gene yields the antitoxin in a bacterial TA system. iTIS, internal translation initiation site; (4 aa)n denotes the hypervariable four amino acid repeats in RpnS. The C-terminal domain of RpnL corresponding to RpnS is shown in dark red, and the free RpnS translated from the iTIS is shown in yellow. The active toxin appears to be an RpnL dimer, and the inhibition of the nuclease activity involves RpnS binding to each of the monomers. The mechanism of RpnS release triggered by phage infection (?) remains to be elucidated.
“Zhong et al. report a novel class of Abi systems that employ a remarkably economical and elegant strategy of antitoxin formation.”
The inhibitory effect of RpnS is specific to the particular RpnL from which the RpnS comes, and Zhong et al revealed a remarkable structural basis of such specificity. It turned out that the C-terminal region of Rpn that is shared by RpnL and RpnS contains a hypervariable region composed of four-amino acid repeats such that a mismatch in the number of repeats prevents the formation of the RpnL–RpnS complex. The crystal structure of RpnS was solved, and the hypervariable tetrad repeat region was shown to belong to a long α-helix that forms a dimerization interface.
The hypervariability of Rpn and the patchy distribution of the rpn genes amongst bacteria prompted the hypothesis that these genes comprise a distinct antiphage defense system. Indeed, Zhong et al. showed that expression of RpnLS inhibited the reproduction of several T-even phages in E. coli (5).
This study reveals a novel strategy for antitoxin formation in TA systems whereby the antitoxin is a portion of the toxin produced from a gene-within-gene. In principle, this is a variation on the general theme of dominant-negative inhibition of biological activities that is, in particular, a widespread antidefense strategy employed by viruses, in particular, large DNA viruses of eukaryotes. For example, many animal viruses encode derivatives of DEATH (named for their role in PCD) adaptor domains that form unproductive complexes with effector components of PCD networks and thus prevent apoptosis (8). However, this principle so far has not been known to apply to the regulation of toxin activity in TA modules, and the “ingenious” utilization of a gene within gene adds another unique feature. It is notable, in this context, that some rpn loci contain a second copy of RpnS indicating that the toxin and the antitoxin can be decoupled, perhaps, enhancing the flexibility of regulation (5).
As befits a truly novel discovery, additional questions abound. In particular, what is the mechanism of the release of the RpnL toxin upon infection? Zhong et al. provide a hint by showing that the RpnL–RpnS complex dissociates at alkaline pH, but obviously, definitive experiments remain to be performed. Further, what is the evolutionary driver of the fine-tuning of the RpnL inhibition by RpnS through the variable number of the amino acid tetrad repeats? It is natural to interpret this phenomenon within the context of the arms race between viruses and defense systems. In the many bacteria that harbor multiple rpn copies, repeat variation will prevent RpnL inhibition in trans, suggesting the intriguing possibility that different rpn copies specifically target distinct phages. And, perhaps, the most important and intriguing question: Is this discovery a tip of a proverbial iceberg, that is, are there many other TA and perhaps other systems that employ iTIS to produce dominant negative regulators? It would appear quite incongruous if such a simple and elegant regulation mechanism was unique to Rpn. Zhong et al. point to ribosome profiling data suggesting that expression of RpnS from iTIS indeed could be a widespread phenomenon in bacteria including at least one more TA module in E. coli, yhaV-prlF. Clearly, however, extensive additional work will be required to assess the generality of the gene within gene principle. While the new studies are eagerly anticipated, it is already clear that the Rpn system is a notable addition to the burgeoning armory of microbial mechanisms.
Acknowledgments
E.V.K’s research is supported through the Intramural Research Program of the NIH (National Library of Medicine).
Author contributions
E.V.K. wrote the paper.
Competing interests
The author declares no competing interest.
Footnotes
See companion article, “Toxic antiphage defense proteins inhibited by intragenic antitoxin proteins,” 10.1073/pnas.2307382120.
References
- 1.Doron S., et al. , Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359, eaar4120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao L., et al. , Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 369, 1077–1084 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Millman A., et al. , An expanded arsenal of immune systems that protect bacteria from phages. Cell Host Microbe. 30, 1556–1569.e5 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Jurenas D., Fraikin N., Goormaghtigh F., Van Melderen L., Biology and evolution of bacterial toxin-antitoxin systems. Nat. Rev. Microbiol. 20, 335–350 (2022). [DOI] [PubMed] [Google Scholar]
- 5.Zhong A., et al. , Toxic antiphage defense proteins inhibited by intragenic antitoxin proteins. Proc. Natl. Acad. Sci. U.S.A. [Preprint] (2023). 10.1101/2023.05.02.539157 (Accessed 12 July 2023). [DOI] [PMC free article] [PubMed]
- 6.Steczkiewicz K., Muszewska A., Knizewski L., Rychlewski L., Ginalski K., Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res. 40, 7016–7045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kingston A. W., Ponkratz C., Raleigh E. A., Rpn (YhgA-Like) proteins of Escherichia coli K-12 and their contribution to RecA-independent horizontal transfer. J. Bacteriol. 199, e00787-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koonin E. V., Dolja V. V., Krupovic M., The logic of virus evolution. Cell Host Microbe. 30, 917–929 (2022). [DOI] [PubMed] [Google Scholar]


