Significance
SARS-CoV-2, the causative agent of COVID-19, encodes several proteins not present in other coronaviruses. SARS-Unique Domain (SUD), a domain in the nonstructural protein 3 (nsp3) of coronavirus, is present only in Sarbecovirus, a subgenus of Betacoronavirus, including two highly pathogenic human coronaviruses, SARS-CoV and SARS-CoV-2. We report that nsp3-S676T, a single mutation in the SUD, causes SARS-CoV-2 fitness loss through dysregulation of its translational enhancement ability. N-S194L, a compensatory mutation identified in the nucleocapsid (N) protein, reverts viral virulence. Our study reveals a mechanism by which N-nsp3 interactions contribute to SARS-CoV-2 replication and pathogenesis but also identifies an effective antiviral therapeutic target for COVID-19.
Keywords: SARS-CoV-2, N protein, nsp3, SARS-unique domain
Abstract
SARS-CoV-2, the causative agent of COVID-19 encodes at least 16 nonstructural proteins of variably understood function. Nsp3, the largest nonstructural protein contains several domains, including a SARS-unique domain (SUD), which occurs only in Sarbecovirus. The SUD has a role in preferentially enhancing viral translation. During isolation of mouse-adapted SARS-CoV-2, we isolated an attenuated virus that contained a single mutation in a linker region of nsp3 (nsp3-S676T). The S676T mutation decreased virus replication in cultured cells and primary human cells and in mice. Nsp3-S676T alleviated the SUD translational enhancing ability by decreasing the interaction between two translation factors, Paip1 and PABP1. We also identified a compensatory mutation in the nucleocapsid (N) protein (N-S194L) that restored the virulent phenotype, without directly binding to SUD. Together, these results reveal an aspect of nsp3-N interactions, which impact both SARS-CoV-2 replication and, consequently, pathogenesis.
SARS-CoV-2, the causative agent of COVID-19, has resulted in at least 677 million cases and 6.9 million deaths as of March 2023. SARS-CoV-2 belongs to the Betacoronavirus genus and shares 79% genome sequence identity with SARS-CoV (1). Two-thirds of the genome encodes for 16 nonstructural proteins (nsps) essential for viral replication (2). Among them, nsp3 is the largest and plays an essential role in the formation of replication/transcription complexes (RTC). It contains at least eight domains, with specific organization varying in different coronavirus genera (3). Several conserved domains exist within all coronaviruses, (e.g., papain-like protease and ubiquitin-like domain 1) and have similar functions in virus replication (3). In addition, there are also some nonconserved domains, including the SARS-unique domain (SUD), that only exist in the Sarbecovirus (4), which includes two highly pathogenic human coronaviruses, SARS-CoV and SARS-CoV-2. The SUD sequences of SARS-CoV and SARS-CoV-2 have 75% identity (5). The nsp3 SUD consists of three distinct domains, SUD-N (N-terminal; Mac-2), SUD-M (Middle; Mac-3), and SUD-C [C-terminal; DPUP (domain preceding Ubl2 and PL2pro)] (3). SUD-N and SUD-M share structural, but not sequence homology with the macrodomain (Mac), a conserved domain that is essential for coronavirus virulence (Fig. 1B) (3, 6–8). SUD-N/M are capable of binding guanine-quadruplexes formed by RNA and DNA oligonucleotides (6). Further, SUD-M deletion or mutation of residues in SUD-N/M responsible for binding with guanine-quadruplexes diminished replicon replication (9). SUD-C adopts a frataxin-like fold and can regulate SUD-M/C binding to specific RNA regions, such as SARS-CoV Transcription Regulatory Sequence, important for the initiation of subgenomic CoV transcription (10).
Fig. 1.
Nsp3-S676T substitution attenuates SARS-CoV-2 virulence in mice. (A) Schematic showing the generation of SARS2-N501YMA30-Nsp3-S676T in mice. SARS2-N501YMA30-Nsp3-S676T was generated by passage through mouse lungs as previously described (11). After 30 serial passages, the virus was collected from the lungs and then plaque purified three times on Vero E6 cells. Individual plaques were isolated, and viral genomes were sequenced by Nanopore sequencing. (B) Schematic diagram of SARS-CoV-2 genome (①), adaptive mutations in SARS2-N501YMA30-Nsp3-S676T (②), and SUD (③). The viral proteins containing mutations associated with mouse adaptation are shown in the middle panel (②). SUD-M, SUD-N, and SUD-C are colored purple, teal, and pink. Lower panel: The serine to threonine substitution at position 676 in Nsp3 is shown (③). (C) Percentage of initial weight and survival of BALB/c mice infected with 5,000 PFU of SARS2-N501YMA30 (n = 9 mice) or SARS2-N501YMA30-Nsp3-S676T (n = 7 mice). (D) Predicted structures of SUD of SARS-CoV-2 (Left) and SARS-CoV (Right). Globular domains, including SUD-M, SUD-N, and SUD-C, are colored in purple, teal, and pink, respectively. Linker regions between the globular domains are orange colored. Residue S676 is shown as a stick model (Inset). Corresponding residue in SARS-CoV SUD is S652. (E) Representative plaques of recombinant viruses in Vero E6 cells (Left). rSARS-CoV-2Nsp3-S676T, rSARS-CoV-2 (Wuhan-Hu-1), rSARS2-N501YMA30-Nsp3-S676T, and rSARS2-N501YMA30 (named rWuH-1Nsp3-S676T, rWuH-1, rMA30Nsp3-S676T, and rMA30, respectively in text). Comparison of the diameter of plaques from different viruses (Left) (10 plaques each group). (F and G) Weight and survival of BALB/c mice challenged with 5,000 PFU (F) or 500 PFU (G) of rMA30Nsp3-S676T or rMA30 (F. n = 8 mice/group. G. n = 5 mice/group). (H) C57BL/6 mice were infected with 5,000 PFU of rMA30 or rMA30Nsp3-S676T. Weight and survival were monitored daily until 14 dpi (n = 8 mice each group). (I) K18-ACE2 mice were inoculated with 2,000 PFU of rWuH-1 or rWuH-1Nsp3-S676T (n = 4 mice each group). Weight and survival were recorded daily until 14 dpi. P values were determined by a log-rank (Mantel–Cox) test and a two-tailed, unpaired t test with Welch’s correction. Data are combined from two independent experiments.
In addition to these properties, SUD has been demonstrated to interact with additional host factors. Through interaction with ring-finger and CHY zinc-finger domain-containing 1 (RCHY1), SARS-CoV SUD-N/M enhances the E3 ubiquitin ligase activity of RCHY1, inducing p53 degradation, potentially promoting virus replication (12). Further, by interacting with poly(A)-binding protein (PABP)-interacting protein 1 (Paip1), an enhancer of translational initiation, SARS-CoV SUD augments the binding affinity between poly(A)-binding protein (PABP) and Paip1 to stimulate viral translational efficiency (5). Although these studies clarify the role of SUD, the relative importance of each to virus pathogenesis remains unknown.
We addressed the role of SUD by using a set of mouse-adapted SARS-CoV-2. We recently isolated a virulent mouse-adapted virus (SARS2-N501YMA30) by serial mouse lung passage and developed a reverse genetics system for studying this virus (11). During the course of adaptation, we serendipitously isolated a mouse-adapted virus that was attenuated in mice. Viral genome sequencing showed that it contained not only all of the adaptive mutations found in SARS2-N501YMA30 (11) but also an additional mutation at position 676 of nsp3. Further investigation showed that the virus containing this mutation, SARS2-N501YMA30-Nsp3-S676T (called nsp3-S676T herein), replicated to lower titers in cultured cells (Calu-3, Vero-E6), human airway epithelial cells (HAEs), and human macrophages. We also show that the nsp3-S676T mutation diminished the ability of SUD to enhance virus translation.
Results
The Nsp3-S676T Mutation Results in Virus Attenuation.
We isolated nsp3-S676T after serial passage through mouse lungs (Fig. 1A) and showed that it was attenuated in mice (Fig. 1C). Virus genome sequencing revealed that it not only contained all the mutations identified in SARS2-N501YMA30, a lethal virus, but also a single S676T mutation in nsp3 (Fig. 1B). Infection with nsp3-S676T resulted in mild disease, and most of the mice survived until the end of the period of observation (Fig. 1C). In contrast, mice infected with parental SARS2-N501YMA30 developed lethal disease (Fig. 1C). We aligned the SUD sequence from SARS-CoV-2, SARS-CoV, and multiple other SARSr-CoV and found that the amino acid sequence was highly conserved across Sarbecovirus (SI Appendix, Fig. S1). The SARS-CoV SUD structure was determined using X-ray crystallography and NMR (6, 10). Based on these published data and using AlphaFold2 in Colab (13), 3-dimensional structures of SARS-CoV and SARS-CoV-2 SUD were predicted (Fig. 1D). In accordance with earlier reports, the SUD comprises three distinct globular subdomains, SUD-N, SUD-M, and SUD-C, which are connected by two short linkers. S676 sits at the linker region between SUD-M and SUD-C. The corresponding amino acid in SARS-CoV nsp3 is S652 and is also located in the linker between SUD-M and SUD-C. To confirm the role of S676T in virus attenuation, we next introduced the S676T mutation into both MA (rSARS2-N501YMA30) and Wuhan-Hu-1 (rWuH-1) backbones using reverse genetics (11, 14). The resulting recombinant viruses were named rMA30 and rMA30Nsp3-S676T, and rWuH-1 and rWuH-1Nsp3-S676T. Plaque assays showed that plaque sizes after infection with the different recombinant viruses were comparable (Fig. 1E). BALB/c mice and C57BL/6 mice were challenged intranasally with a lethal dose of either rMA30 or rMA30NSP3-S676T (5,000 PFU). Consistent with the data shown above, mice infected with rMA30 died, while mice infected with rMA30NSP3-S676T largely survived (Fig. 1 F and H). Similar results were observed when BALB/c mice were infected with sublethal doses (500 PFU) of the virus (Fig. 1G). To further confirm the attenuating role of S676T, we challenged highly susceptible K18-hACE2 mice intranasally with a lethal dose of rWuH-1 or rWuH-1Nsp3-S676T (2,000 PFU). K18-hACE2 mice infected with rWuH-1 all died by 7dpi, whereas rWuH-1Nsp3-S676T-infected mice developed mild disease and most mice survived (Fig. 1I). Together, these data indicate that the nsp3-S676T mutation significantly decreased the virulence of SARS-CoV-2 in mice.
Diminished Viral Replication and Pathogenesis in rMA30NSP3-S676T-Infected Mice.
We next assessed viral replication and pathogenicity in lungs harvested from BALB/c mice infected with rMA30 or rMA30Nsp3-S676T. Infectious virus titers and levels of sgRNA and gRNA following rMA30Nsp3-S676T infection were significantly lower at 2 and 4 d postinfection (dpi) than was observed in rMA30-infected mice (Fig. 2A). Defects in virus replication were also observed at 16 and 24 h postinfection (hpi) (SI Appendix, Fig. S2A). Gross pathological examination at 4 dpi showed that hyperemia and congestion were present in the lungs of rMA30 but not rMA30Nsp3-S676T-infected mice (Fig. 2B). Histopathological analysis revealed extensive edema in the lungs of rMA30 but not those of rMA30Nsp3-S676T-infected mice (Fig. 2 C and D). Similar results were observed after infection of K18-hACE2 mice. Infectious virus titers and sgRNA levels in rWuH-1Nsp3-S676T infected K18-hACE2 mice at 2 and 6 dpi were lower than those following rWuH-1 infection (SI Appendix, Fig. S2B). Collectively, these results demonstrate that the S676T mutation significantly decreased SARS-CoV-2 replication, alleviating disease in mice.
Fig. 2.
The effect of Nsp3-S676T on viral replication and pathogenesis in mice. BALB/c mice were infected with 5,000 PFU of rMA30 or rMA30Nsp3-S676T. (A) At 2 and 4 dpi, lungs were harvested to evaluate viral titers, levels of subgenomic RNA (sgRNA, N gene) and genomic RNA [gRNA, ORF1 (nsp12)]. LOD: limit of detection. rMA30 (n = 6); rMA30Nsp3-S676T (n = 7). For statistical analysis, in samples with undetectable amounts of virus, we assigned a value of 10 PFU/mL. (B) Gross lung pathology and (C) histopathological analysis of the lungs, harvested at 4 dpi. (n = 3 mice/group). (Scale bars, 430 [Upper] and 86 [Lower] µm, respectively.) (D) Lung edema was quantified as described in Materials and Methods (n = 3 each group). Asterisks indicate pulmonary edema. P values were calculated by a two-tailed, unpaired t test with Welch’s correction. (A) Data are combined from two independent experiments.
To investigate whether reduced replication and pathogenicity correlated with higher expression of Interferon (IFN) and other cytokines, we measured levels of several molecules, including IFN-α, IFN-β, interferon-stimulated gene-15 (ISG-15), tumor necrosis factor (TNF), interleukin-6 (IL-6), and C-X-C motif chemokine ligand 10 (CXCL-10). Levels of these molecules generally correlated with virus loads, with most of them higher in rMA30-infected mice at both 2 and 4 dpi (SI Appendix, Fig. S2C). These data suggest that the attenuated phenotype of rMA30Nsp3-S676T is not caused by increased expression of pro-inflammatory factors but may reflect intrinsic defects in virus replication.
The S676T Mutation Diminishes Viral Replication in Cultured Cells.
To further assess effects on virus replication, we infected Vero E6 and Calu-3 cells with rWuH-1Nsp3-S676T or rWuH-1 (MOI = 8) and measured virus titers. Titers of rWuH-1Nsp3-S676T were significantly lower than those of rWuH-1 in Vero-E6 or Calu-3 cells (Fig. 3A). We further compared the replication of rWuH-1Nsp3-S676T and rWuH-1 in primary HAEs. HAEs from the same donors (n = 13) were inoculated with either rWuH-1Nsp3-S676T or rWuH-1 at 0.1 MOI, and supernatants were harvested from apical surfaces at 24 and 48 hpi. Virus titers from rWuH-1Nsp3-S676T-infected HAEs were significantly lower than those harvested from rWuH-1-infected HAEs (Fig. 3B). We also analyzed virus replication in human monocyte–derived macrophages (MDMs), which are abortively infected with SARS-CoV-2 (15). MDMs, differentiated from four healthy donors, were infected with either rWuH-1Nsp3-S676T or rWuH-1 at 1 MOI, and intracellular RNA was harvested at 12 and 24 hpi. sgRNA and gRNA levels from rWuH-1Nsp3-S676T-infected MDMs were significantly lower than those from rWuH-1-infected MDMs at 12 and 24 hpi (Fig. 3C). Taken together, these data indicate that the S676T mutation attenuated SARS-CoV-2 replication in cultured cells.
Fig. 3.
Nsp3-S676T effect on viral replication in cultured cells. (A) Vero E6 or Calu-3 cells were infected with rWuH-1 or rWuH-1Nsp3-S676T at MOI = 8, and supernatants were harvested at the indicated time points to determine infectious viral titer by plaque assay (Representative of two independent experiments with four replicates/experiment). LOD: limit of detection. (B) Human primary airway epithelial cells (HAEs) from 13 donors were infected with 0.1 MOI of rWuH-1Nsp3-S676T or rWuH-1. At 24 and 48 hpi, PBS was used to elute the released viruses at 37 °C for 30 min. Viral titers were quantified by plaque assay (n = 13). (C) Human MDMs differentiated from human PBMCs (four donors) were infected with rWuH-1 or rWuH-1Nsp3-S676T at an MOI of 1 for 12 or 24 h. The levels of viral sgRNA and gRNA were determined by qRT-PCR (n = 4). A two-tailed, unpaired t test with Welch’s correction (A and C) and a two-tailed, Wilcoxon matched-pairs test (B) were used to assess statistical significance.
Since nsp3 has been shown to counter the innate immune response, we next examined comparative sensitivity to IFN by treating Calu-3 cells with IFN-β (100 U) or PBS for 16h, prior to infection with rWuH-1Nsp3-S676T or rWuH-1. Supernatants and cells were harvested to determine virus titers and sgRNA levels, respectively. rWuH-1Nsp3-S676T and rWuH-1 exhibited equivalent virus titers and sgRNA levels at 24 and 48 hpi in the presence or absence of IFN-β treatment (SI Appendix, Fig. S3A), demonstrating equivalent IFN sensitivity.
S676 Is Required for SUD-Mediated Translational Enhancement.
As described above, multiple functions have been attributed to the SUD. Given the effects that we observed in primary and immortalized cell lines, we chose to focus on the role of the SUD in enhancing virus translation. SARS-CoV SUD interacts with Paip1, thereby enhancing binding affinity between Paip1 and PABP1, and translational efficiency (5). In order to evaluate whether S676T impacts SUD’s ability to enhance translation, we measured its effect on translation using a luciferase-based assay.
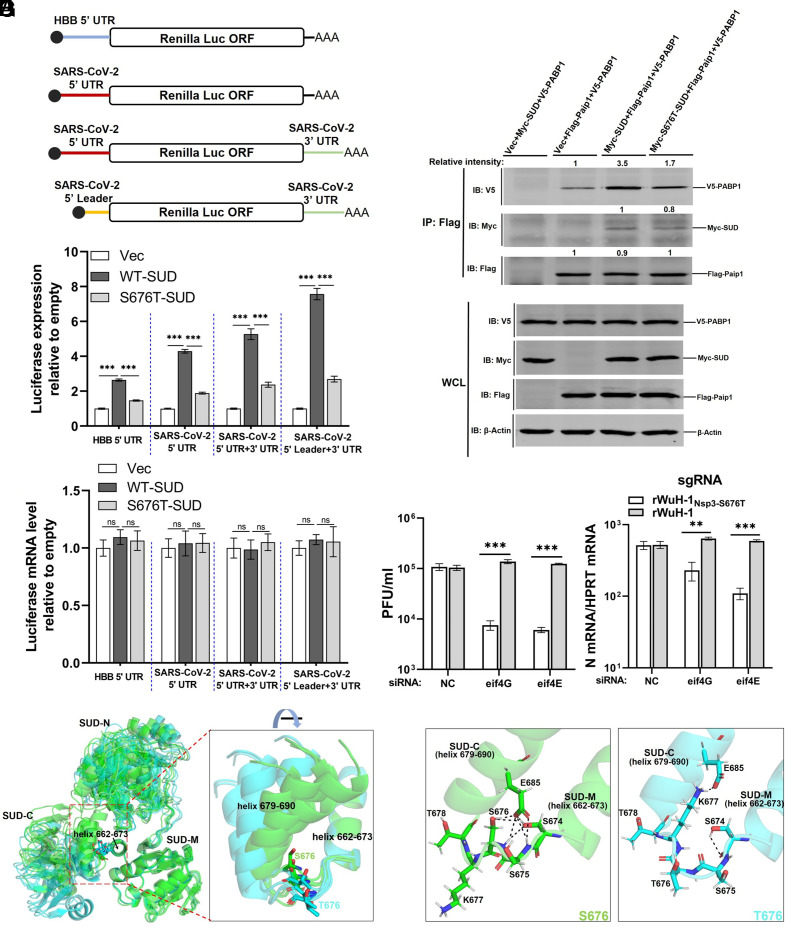
To distinguish host and viral mRNA, we constructed two Renilla luciferase reporters, containing the 5′ UTR of human beta-globin (HBB-5′ UTR) or SARS-CoV-2 5′ UTR (Fig. 4A). Each Renilla luciferase plasmid was cotransfected with plasmid encoding SUD or S676T-SUD into HEK-293T cells. When assessed at 24 h posttransfection, SUD improved the translation of both of these luciferase reporters, whereas S676T-SUD largely lost this enhancing ability (Fig. 4B). Since CoV subgenomic mRNAs each contain a leader, not the full-length 5′ UTR, and a 3′ UTR, we next engineered two additional constructs containing the leader and/or 3′ UTR (Fig. 4A) and used them in the same transfection protocol. Cotransfection with plasmids expressing the SUD but not the S676T SUD improved the translation of these constructs (Fig. 4B). Luciferase mRNA levels were equivalent among all groups (Fig. 4C). We next investigated whether the interaction between Paip1 and PABP1 was affected by the S676T mutation. HEK-293T cells were cotransfected with Paip1 and PABP1 along with vector, SUD or S676T-SUD for 24 h, and cell lysates were harvested for immunoprecipitation. Consistent with a previous study (5), SARS-CoV-2 SUD interacted with Paip1 and formed a ternary complex with PABP1 and Paip1. Additionally, compared to the vector transfected group, SUD increased the interaction between PABP1 and Paip1. In contrast, S676T-SUD was less able to enhance the interaction between PABP1 and Paip1 compared to SUD (Fig. 4D). However, the binding between Paip1 and SUD was not impacted by S676T (Fig. 4D, right-hand lanes), supporting previous results showing that the crucial amino acids in SUD for binding to Paip1 were located within the first 16 amino acids of SUD-N (5).
Fig. 4.
Nsp3-S676T diminishes the translational stimulatory ability of SUD. (A) Schematic diagram of luciferase reporters. Human beta-globin (HBB), SARS-CoV-2 5′ UTR, or 5′ leader was positioned upstream of the Renilla luciferase gene in pRL-TK. The SARS-CoV-2 3′ UTR was inserted downstream of the Renilla luciferase in plasmids containing the 5′ UTR or 5′ leader. (B and C) Quantification of Renilla luciferase expression. HEK-293T cells were transfected with HBB-5’ UTR or SARS-CoV-2 5′ UTR Renilla-Luc and vector, SUD or S676T-SUD. After 24 h, cells were lysed in 1× lysis buffer for measuring luciferase activity (B, n = 5 replicates, representative of three independent experiments) or in TRIzol to evaluate mRNA levels of luciferase using qRT-PCR (C, n = 4 replicates, representative of two independent experiments). (D) HEK293 cells were transfected with vector, myc-SUD, or myc-S676T-SUD, together with V5-PABP1 and Flag-Paip1. As a control, cells were transfected with vector, myc-SUD, and V5-PABP1. After 24 h, transfected cells were lysed and immunoprecipitated with Pierce™ Anti-Flag Affinity Resin. Intensity of PABP1 or SUD was normalized to Paip1 in the same immunoprecipitation. Intensity of Paip1 relative to the vector group is shown (second lane from left). (E) Vero E6 cells were transfected with 40 nM of NC (negative control siRNA) siRNA, eIF4E siRNA or eIF4G siRNA, prior to infection 48 h later with 0.05 PFU/cell of rWuH-1 or WTNsp3-S676T. At 24 hpi, supernatants were collected for virus plaque assay, and cells were harvested with TRIzol to measure viral sgRNA levels by qRT-PCR (n = 3 replicates, representative of two independent experiments). (F) Predicted structures of the SUD domains of nsp3-S676 and nsp3-T676 were superimposed using all five top-ranked structures shown in SI Appendix, Fig. S4, based on helix 662-673. S676 and T676 SUD are colored in green and cyan, respectively. Residues S676 and T676 are shown in a stick model. (G) Predicted hydrogen bonds were identified using each of the rank 1 predicted structures (SI Appendix, Fig. S4), based on analyses of S674, S675, S676/T676, K677, and E678. Left: S676. Right, T676. Hydrogen bonds are shown as black dashed lines. Statistical significance was determined by a one-way ANOVA test with Tukey's multiple comparisons test (B and C) and a two-tailed, unpaired t test with Welch’s correction (E).
In the experiments described in Fig. 3, Vero E6 and Calu-3 cells were infected at a high multiplicity of infection (MOI). These cells, which are immortalized, are expected to express high levels of translation factors such as eIF4G and eIF4E (16). Both of these factors are known to play key roles in rapid cancer cell growth and to contribute to tumorigenesis (17–19). We reasoned that the role of the SUD might be less important in the presence of an excess of translation factors, such as occurs in immortalized cells, after infection at a lower MOI. Therefore, we infected Vero E6 and Calu-3 cells at an MOI of 0.01 with rWuH-1Nsp3-S676T or rWuH-1 and measured virus titers at 12, 24, 48, and 72 hpi. Viral titers after rWuH-1 and rWuH-1Nsp3-S676T, or rMA30Nsp3-S676T and rMA30 infection were comparable at all time points in both cell types (SI Appendix, Fig. S3 B and C). We transfected siRNA targeting eIF4G or eIF4E into Vero E6 cells for 48 h and then infected cells with rWuH-1Nsp3-S676T or rWuH-1 at an MOI of 0.05 [Knockdown efficiency of eIF4G or eIF4E was first evaluated by western blotting before infection (SI Appendix, Fig. S4B)]. At 24 hpi, supernatants and cells lysates were harvested. Virus titers and sgRNA levels were significantly reduced in rWuH-1Nsp3-S676T- compared to rWuH-1-infected eIF4G or eIF4E knockdown cells (Fig. 4E). This suggested that high expression levels of eIF4G and eIF4E in immortalized cells compensated for the loss of SUD-mediated translational enhancement when cells were infected at a low MOI. At higher MOIs, translational factors would be limiting, so that the SUD function was required for maximal translation of viral mRNA.
Effects of the nsp3-S676T on the SUD Structure.
Next, we sought a structural explanation for SUD-T676’s relatively low association with PABP by using AlphaFold2 in Colab (13) to predict the potential effects of nsp3-S676T on the SUD structure. The five highest-ranked structures containing S676 were very similar but different from those containing T676, which, in turn, were internally consistent (SI Appendix, Fig. S4A). The spatial location of S676 and T676 in the linker (residues 674 to 678) was different when the SUDS676 and SUDT676 structures were compared. S676 was in close proximity to the helix (residue 679 to 690 in SUD-C) that follows the linker, whereas T676 was shifted outward (SI Appendix, Fig. S4A). Next, the predicted structures in SI Appendix, Fig. S4A were superposed using the helix (residue 662 to 673 in SUD-M) preceding the linker region as a starting point for alignment. All the T676 structures predicted that the helix at 679 to 690 would move further away from the 662 to 673 helix in comparison to the structures containing S676 (Fig. 4F), resulting in a wider angle between the two helices in T676 compared to S676. Using the top-ranked models, we further assessed the formation of hydrogen bonds across the linker region (Fig. 4G). In SUDS676, seven hydrogen bonds were formed between residues S674, S675, S676, and a residue in the helix 679 to 690, E685, while only two hydrogen bonds were found in SUDT676. The greater number of hydrogen bonds in SUDS676 may have increased the rigidity of the linker and stabilized the local conformation, while the linker in SUDT676 was relatively flexible. Collectively, these structural analyses suggest that loss of hydrogen bonds caused by nsp3-S676T resulted in increased motion of SUD-M relative to SUD-C.
N-S194L Reverses rMA30Nsp3-S676T Attenuation.
To further understand the effects of the nsp3-S676T mutation, we serially passaged rMA30Nsp3-S676T in mouse lungs to identify compensatory mutations that restored virus virulence. After 5 mouse passages, we isolated virus, propagated it for one passage in Calu-3 cells and sequenced the viral genome. We detected only a single nucleotide change, C28854T, resulting in an S194L mutation in the N protein (N-S194L). Remarkably, this second site suppressor was identified in three independent experiments (Fig. 5 A and B); same-site reversion was never detected. Of note, 5,000 PFU of passaged virus encoding N-S194L caused severe clinical disease and weight loss in mice, and all mice succumbed to infection in 6 d, indicating a reversion to lethality (SI Appendix, Fig. S5A). To further validate that N-S194L restored virus fitness, we introduced N-S194L into both rMA30Nsp3-S676T and rWuH-1Nsp3-S676T using reverse genetics (rMA30Nsp3-S676T;N-S194L and rWuH-1Nsp3-S676T;N-S194L). Recombinant virus sequences were confirmed. K18-hACE2 mice were inoculated intranasally with 2,000 PFU of rWuH-1, rWuH-1Nsp3-S676T, or rWuH-1Nsp3-S676T;N-S194L and monitored for weight loss and survival. Mice infected with rWuH-1Nsp3-S676T;N-S194L developed lethal disease and succumbed to infection similarly to rWuH-1 infected-mice. Most of the mice inoculated with rWuH-1Nsp3-S676T survived the infection (SI Appendix, Fig. S5B). Similarly, all BALB/c mice inoculated with rMA30Nsp3-S676T;N-194L or rMA30 developed severe disease and died over the following 6 d, while all rMA30Nsp3-S676T-infected mice survived (Fig. 5C). Lung viral titers and sgRNA levels in rMA30Nsp3-S676T;N-S194L and rMA30 infected mice were comparable and significantly higher than those in mice infected with rMA30Nsp3-S676T (Fig. 5D). Histopathological analyses indicated that levels of edema were comparable in rMA30Nsp3-S676T;N-S194L and rMA30 infected mice lungs (Fig. 5 E and F). In order to further confirm the virulence of rMA30Nsp3-S676T;N-S194L, we infected BALB/c mice with rMA30Nsp3-S676T;N-S194L and rMA30 at sublethal doses. rMA30Nsp3-S676T;N-S194L and rMA30 caused similar clinical disease in these mice (Fig. 5G). Collectively, these results show that N-S194L reverses viral virulence by compensating for the viral fitness loss caused by the nsp3-S676T mutation.
Fig. 5.
The effect of a compensatory N-S194L mutation on viral virulence. (A) Location of S194L mutation in the N protein. NTD: N-terminal domain. CTD: C-terminal domain. SR: Serine/Arginine rich region. (B) Viral genomic RNA sequence analysis showing the proportion of the C28854T base (N-194L) in three mice after five passages. Three BALB/c mice were challenged with 5,000 PFU of rMA30Nsp3-S676T. At 2 dpi, supernatants from lung homogenates from each mouse were serially passaged four more times in uninfected mice. The passaged virus was then propagated in Calu-3 cells. At 24 hpi, supernatants from infected Calu-3 cells were harvested to isolate viral RNA. Viral genomic RNA from each group was sequenced by Nanopore sequencing and compared to unpassaged rMA30Nsp3-S676T. (C) Weight and survival of BALB/c mice infected with 5,000 PFU of rMA30, rMA30Nsp3-S676T or rMA30Nsp3-S676T;N-S194L (n = 3 for rMA30, n = 4 for rMA30Nsp3-S676T, n = 5 for rMA30Nsp3-S676T; N-S194L) (D) Viral titers and sgRNA levels from BALB/c mice after infection with 5,000 PFU rMA30 (n = 5), rMA30Nsp3-S676T (n = 5) or rMA30Nsp3-S676T;N-S194L (n = 5) infection. (E) Histopathological analysis of mouse lungs at 4 dpi after rMA30 or rMA30Nsp3-S676T;N-S194L infection. Asterisks indicate pulmonary edema. (Scale bars, 170 µm.) (F) Summary of histopathological scores of lung edema (rMA30, n = 4. rMA30Nsp3-S676T;N-S194L, n = 5). (G) BALB/c mice were infected with 500 PFU (sublethal dose) of rMA30 or rMA30Nsp3-S676T;N-S194L. Weight and survival were recorded daily until 14 dpi (n = 5 at each group). The log-rank (Mantel–Cox) test (C and G), one-way ANOVA test with Tukey’s multiple comparisons test (D), and two-tailed, unpaired t test with Welch’s correction (F) were used to calculate P values.
N-S194L Enhances N Protein Expression.
The N protein interacts with the ubiquitin-like domain of nsp3 (Ubl1) to enhance the initiation of viral RNA replication (20–23). To test whether nsp3-S676T impacted the formation of the N-nsp3 complex, we performed coimmunoprecipitation assays. A set of truncated nsp3s, including the N terminal domain of nsp3 from Ubl1 to SUD (Ubl1-SUD), Ubl1-SUD with the S676T mutation (Ubl1-SUDS676T), and Ubl1 and SUD (SI Appendix, Fig. S6A) were constructed. HEK-293 cells were transfected with Flag-N or vector, along with one of the nsp3 constructs, and cell lysates were analyzed by coIP after 24 h. The result demonstrated interactions between Ubl1 or Ubl1-SUD and N (SI Appendix, Fig. S6 B and C), as expected (22). The nsp3-S676T mutation did not affect Ubl1-SUDS676T binding to N, suggesting that nsp3-S676T had no direct effect on N-nsp3 interactions (SI Appendix, Fig. S6C). Notably, SUD alone did not interact with N (SI Appendix, Fig. S6D). To further dissect the compensatory role of N-S194L, we next analyzed the expression of N protein encoding S194 or L194 after transfection of HEK-293 cells. S194L-mutated N was expressed at higher levels than WT N (Fig. 6A), without enhancing cellular protein synthesis (SI Appendix, Fig. S6E). To assess these results in the context of infectious virus, we inserted N-S194L mutation into the rWuH-1 backbone to generate rWuH-1N-S194L. We then compared the replication of rWuH-1, rWuH-1Nsp3-S676T, rWuH-1Nsp3-S676T;N-S194L, and rWuH-1N-194L in Vero E6 or Calu-3 cells at 8 and 24 hpi after infection at high MOIs. rWuH-1N:S194L virus grew to the highest titers, while rWuH-1 and rWuH-1Nsp3-S676T;N-S194L replicated equivalently (Fig. 6B). As expected, rWuH-1Nsp3-S676T replicated least well (Fig. 6B). We also measured N protein expression in infected cells. N protein expression in cells infected with the rWuH-1N-S194L virus was higher than that detected in cells infected with the other three viruses (Fig. 6C). To determine whether translation was impacted in cells infected at lower MOIs, Vero E6 cells were transfected with siRNA targeting eIF4G or eIF4E for 48 h, and infected with rWuH-1, rWuH-1Nsp3-S676T, rWuH-1Nsp3-S676T;N-S194L, or rWuH-1N-S194L for 24 h. The results showed that in the presence or absence of eIF4G/E knockdown, rWuH-1N-S194L replicated to higher levels than the other viruses (Fig. 6D). These results indicate that N-L194S enhanced N protein expression, independent of SUD effects on protein translation.
Fig. 6.
N-S194L enhances N protein expression. (A) Expression of N protein. HEK-293T cells were transfected with plasmids encoding Flag-N, Flag-N-S194L, or vector, and cell lysates were subject to analysis by western blotting at 24 h posttransfection. (B) Vero E6 or Calu-3 cells were inoculated with rWuH-1, rWuH-1Nsp3-S676T, rWuH-1Nsp3-S676T;N-S194L, or rWuH-1N-S194L at MOI = 8. Supernatants were collected from cells at 8 and 24 hpi to determine virus titers by plaque assay (n = 4 replicates). (C) Vero E6 or Calu-3 cells infected with rWuH-1, rWuH-1Nsp3-S676T, rWuH-1Nsp3-S676T;N-S194L, or rWuH-1N-S194L were lysed at 8 and 24 hpi and then subject to western blotting to detect N protein. (D) Vero E6 cells were transfected with NC siRNA or siRNA targeting eIF4G or eIF4E; after 48 h, the transfected cells were infected with 0.05 PFU/cell rWuH-1, rWuH-1Nsp3-S676T, rWuH-1Nsp3-S676T;N-S194L, or rWuH-1N-S194L. Supernatants were harvested at 24 h to measure virus titers by plaque assay, and cell lysates were harvested for detecting viral N gene expression by qRT-PCR (n = 3 replicates). A one-way ANOVA test with Tukey’s multiple comparisons test was used for assessing statistical significances in (B) and (D). (A–D) Representative of two to five independent experiments.
Discussion
The data shown here indicate that nsp3-S676T of the SUD results in SARS-CoV-2 attenuation. Unexpectedly, reversion of rMA30Nsp3-S676T to virulence resulted in a single compensatory mutation in the N protein, supporting previous reports that demonstrated an important role of nsp3-N interactions in coronavirus replication (20, 21). Specifically, among other functions, SARS-CoV-2 SUD is capable of enhancing translation activity (Fig. 4B), as was previously shown for the SARS-CoV SUD (5). The alteration from serine to threonine represents a modest change; however, this mutation changes the predicted structure and profoundly changes the phenotype of SARS-CoV-2 in mice. The predicted structure of SARS-CoV-2 SUD showed that S676T was located in the short linker region, Ser–Ser–Ser–Lys–Thr, found between SUD-M and SUD-C. The amino acid sequence of this short linker is identical among SARS-CoV, SARS-CoV-2, and other SARSr-CoVs (SI Appendix, Fig. S1), which implies that it is important for SUD function. It is also notable that the compensatory N-S194L mutation is located in a linker region of the N protein (24). Previous work has shown that other mutations in this linker region (R203K/G204R) accelerated viral replication and virulence of SARS-CoV-2 (25) or assembly (S202R/R203M) in the context of virus-like particles (26). Our results identify another mutation in the same region that has the same effect.
The SUD preferentially enhances viral RNA translation by increasing the interaction between PABP1 and Paip1 (5). In conjunction with several other coronavirus proteins, such as nsp1, that inhibit host RNA translation (27, 28), this allows the virus to co-opt cells for viral protein synthesis. Consistent with this report (5), we also observed that SARS-CoV-2 SUD bound to Paip1, forming a ternary complex with PABP1 and Paip1 and further augmenting the binding between PABP1 and Paip1. The nsp3-S676T mutation largely abrogated SUD’s ability to increase PABP1-Paip1 interactions, explaining the loss of translational enhancing ability of nsp3-S676T SUD (SI Appendix, Fig. S7). A striking result was the differential effects of the nsp3-S676T mutation on virus replication, when low and high MOIs of the virus were used to infect Vero E6 and Calu-3 cells (Fig. 3A and SI Appendix, Fig. S3 B and C). At low MOIs, rWuH-1 and rWuH-1S676T displayed similar replication kinetics, whereas rWuH-1S676T showed decreased replication compared to rWuH-1 when high MOIs were used. In addition, rWuH-1S676T virus also grew to lower titers than rWuH-1 in primary cell culture, including HAEs and MDMs (human monocyte-derived macrophages). We speculate that the basis for these differences is different levels of translation factors in primary and immortalized cell lines. Many studies indicate that aberrant protein synthesis is a key driver of tumorigenesis, and that the translation machinery in immortalized cells is hyperactive (16, 17). Two translation initiations, eIF4E and eIF4G, are amplified to high levels in cancer and are important for promoting cellular transformation (17–19). Knockdown of eIF4G or eIF4E levels in Vero E6 cells reduced rWuH-1S676T replication at low MOI compared to WT, supporting the notion that the abundance of translation factors is critical for SUD-enhancing ability. In the context of immortalized cells, like Vero E6 and Calu-3 cells, SARS-CoV-2 at low MOI can rely on the presence of abundant translational factors to produce viral proteins for replication, even in the absence of the enhancing SUD effects. SUD-mediated enhancement would be more apparent after infection at high MOIs in these cells because there is increased competition for translation factors. In addition, in the presence of normal levels of translation factors, such as occurs in HAEs and MDMs, the enhancing activity of SUD becomes more apparent.
Nsp3 is a major component of the double-membrane vesicle (DMV), the site of viral RNA synthesis. Two studies recently showed that nsp3 of two betacoronaviruses, MHV and SARS-CoV-2, is a major component of the pore that acts as a channel for transporting viral mRNA from the DMV lumen to the cytoplasm (29, 30). Other studies found that N protein interacted with the Ubl1 domain of nsp3 to regulate RNA binding in the context of MHV and SARS-CoV-2 (20, 22, 31). Together these reports suggest that by binding the Ubl1 domain, N protein localizes to the spanning pore to facilitate viral RNA transport from the DMV.
N-S194L may reverse the effects of nsp3-S676T, and enhance virus growth by one or more mechanisms as follows. First, N-S194L increased viral protein expression and thereby compensated for the diminished translation occurring in the presence of nsp3-S676T (Fig. 7A). Second, N-S194L may enhance virus RNA exit from the site of synthesis, the DMV (Fig. 7B). The DMV spanning pore is formed by nsp3-4, which contains a crown-like-shaped gate on the surface of the DMV (30). Deletion of the N-terminal domain of nsp3 from Ubl1 to Ubl2 destroyed this crown-like structure (see SI Appendix, Fig. S6A for nsp3 organization), but nsp3 lacking Ubl1-Mac still maintained a crown-like structure with collapsed, shortened prongs (30), suggesting that the SUD domain was a main component of the crown-like structure. Nsp3-S676T had no effect on the interaction between N and Ubl1, suggesting that nsp3-S676T might affect the stability of the crown-like gate and thereby diminish viral mRNA and genomic RNA export. The T676 was predicted to make the linker more flexible compared to S676, resulting in increased mobility of the SUD-M relative to the SUD-C (Fig. 4G). Increased amounts of the N protein may have stabilized the crown-like gate. Alternatively, Nsp3-N interactions at the pore may be critical for entry of viral genomic RNA into the DMV, to initiate RNA replication and transcription, as proposed by Koetzner et al (32). Third, N-S194L may enhance replication by augmenting virion transcription or assembly (26, 33, 34). These seemingly disparate effects of the nsp3-S676T may result from a single role for the nsp3-SUD, although given the small number of pores in DMVs and the large number of translation complexes, this seems unlikely. Rather, we think that it is more likely that these different effects reflect the widespread distribution of nsp3 within infected cells. Nsp3 is present in 1) DMVs (35–38); 2) DMV pores (29, 30); 3) Virions (39); 4) Other cellular membranes (35, 36, 40). Nsp3 in the endoplasmic reticulum may impact protein translation, separate from any effects on viral RNA transit through DMV pores. Additional structural and ultrastructural studies will be required to determine the relative importance of these postulated nsp3 functions.
Fig. 7.
Two putative models of the effect of Nsp3-S676T on virus replication and compensatory role of N-S194L. (A) Coronaviruses employ Nsp3 SUD to increase the binding between Paip1 and PABP1 to enhance viral mRNA translation. SARS-CoV-2 with the Nsp3-S676T mutation decreases translational stimulation, resulting in diminished virus infectivity. The N-S194L mutation augments N protein expression to compensate for the loss of viral fitness caused by S676T and reverts virus virulence. (B) Nsp3 and nsp4 are the minimal components required for the formation of the DMV spanning pore (30). The N-terminal domains of nsp3, including Ubl-1, HVR, Mac, and SUD, are critical for forming the crown-like shaped gate of the pore at the convex side of DMV. The SUD is a major component of this gate (30). N protein, serving as an RNA chaperone in CoV, is located at the gate and binds to Ubl1 to regulate viral RNA export. T676 in SUD is postulated to increase linker flexibility (Fig. 4F), resulting in increased mobility of the SUD-M relative to the SUD-C and leading to instability of the gate and diminished viral mRNA transport. S194L augments expression of the N protein. Increased levels of N are postulated to stabilize the gate, restoring viral RNA exit from the DMV. Note that the DMV contains two bilayers, but for the sake of simplicity, only one layer is shown.
Collectively, these results identify key roles for the linker regions of SUD and the N protein in replication. Notably, N-S194L has also been identified in 0.8% of SARS-CoV-2 on the GISAID website (122,543/14,904,508 sequences as of 21 March 2023) and was associated with more severe disease (41–43). However, even though this mutation appears to offer a replication advantage to the virus, this mutation has not been selected to a significant extent in human isolates during the pandemic, probably reflecting the complex selective forces present in human but not mouse or in vitro infections.
Materials and Methods
Cells and Mice.
Vero E6 (ATCC CRL-1586) and HEK-293T (ATCC CRL-3216) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO) supplemented with 10% fetal bovine serum (FBS, Atlanta Biologicals). Calu-3 2B4 (ATCC HTB-55) cells were grown in DMEM containing 20% FBS. Primary HAEs obtained from deidentified donors were grown in air–liquid interface cultures with DMEM/F12 containing 2% Ultroser G as previously described (44).
Six- to ten-week-old BALB/c and 6- to 8-mo-old C57BL/6 mice were purchased from Charles River Laboratories. Seven- to eight-week-old K18-hACE2 mice were obtained from the Jackson Laboratory [034860-B6.Cg-Tg(K18-ACE2)2Prlman/J]. All mice were housed in the Animal Care Facility at the University of Iowa prior to infection. All animal studies were approved by the University of Iowa Animal Care and Use Committee and met stipulations of the Guide for the Care and Use of Laboratory Animals.
Additional figures (SI Appendix, Figs. S1–S7) supporting the main text are provided in SI Appendix. A full overview of the methods and materials referred to in this study is available in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Dr. Tom Gallagher for careful review of the manuscript, Jianqiang Shao for help with histological staining, and Dr. Jian Zheng for help with culturing human monocyte–derived macrophages. This study was supported by grants from the NIH [P01 AI060699 and R01 A129269 (S.P.)]. We thank the Center for Gene Therapy of Cystic Fibrosis for providing primary human airway epithelial cells.
Author contributions
P.L. and S.P. designed research; P.L. and B.X. performed research; P.L. and S.P. contributed new reagents/analytic tools; P.L., B.X., N.J.S., L.-Y.R.W., D.K.M., and S.P. analyzed data; S.P. supervised the project; and P.L. and S.P. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix. All data are available upon request to the scientific community.
Supporting Information
References
- 1.Lu R., et al. , Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 395, 565–574 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.V’Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V., Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 19, 155–170 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lei J., Kusov Y., Hilgenfeld R., Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res. 149, 58–74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snijder E. J., et al. , Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331, 991–1004 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lei J., et al. , The SARS-unique domain (SUD) of SARS-CoV and SARS-CoV-2 interacts with human Paip1 to enhance viral RNA translation. Embo J. 40, e102277 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan J., et al. , The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog 5, e1000428 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fehr A. R., et al. , The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J. Virol. 89, 1523–1536 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fehr A. R., et al. , The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus. Infection. mBio 7, e01721–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kusov Y., Tan J., Alvarez E., Enjuanes L., Hilgenfeld R., A G-quadruplex-binding macrodomain within the “SARS-unique domain” is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology 484, 313–322 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson M. A., Chatterjee A., Neuman B. W., Wuthrich K., SARS coronavirus unique domain: Three-domain molecular architecture in solution and RNA binding. J. Mol. Biol. 400, 724–742 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong L. R., et al. , Eicosanoid signalling blockade protects middle-aged mice from severe COVID-19. Nature 605, 146–151 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma-Lauer Y., et al. , p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proc. Natl. Acad. Sci. U.S.A. 113, E5192–E5201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirdita M., et al. , ColabFold: Making protein folding accessible to all. Nat. Methods 19, 679–682 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fehr A. R., Bacterial artificial chromosome-based lambda red recombination with the I-SceI homing endonuclease for genetic alteration of MERS-CoV. Methods Mol. Biol. 2099, 53–68 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng J., et al. , Severe acute respiratory syndrome coronavirus 2-induced immune activation and death of monocyte-derived human macrophages and dendritic cells. J. Infect Dis. 223, 785–795 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhat M., et al. , Targeting the translation machinery in cancer. Nat. Rev. Drug. Discov. 14, 261–278 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Xu Y., Ruggero D., The role of translation control in tumorigenesis and its therapeutic implications. Annu. Rev. Cancer Biol. 4, 437–457 (2020). [Google Scholar]
- 18.Furic L., et al. , eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl. Acad. Sci. U.S.A. 107, 14134–14139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaiswal P. K., Koul S., Shanmugam P. S. T., Koul H. K., Eukaryotic Translation Initiation Factor 4 Gamma 1 (eIF4G1) is upregulated during Prostate cancer progression and modulates cell growth and metastasis. Sci. Rep. 8, 7459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurst K. R., Ye R., Goebel S. J., Jayaraman P., Masters P. S., An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J. Virol. 84, 10276–10288 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurst K. R., Koetzner C. A., Masters P. S., Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J. Virol. 87, 9159–9172 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bessa L. M., et al. , The intrinsically disordered SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a. Sci. Adv. 8, eabm4034 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cong Y., et al. , Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J. Virol. 94, e01925–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cubuk J., et al. , The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 12, 1936 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu H., et al. , Nucleocapsid mutations R203K/G204R increase the infectivity, fitness, and virulence of SARS-CoV-2. Cell Host Microbe 29, 1788–1801.e1786 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Syed A. M., et al. , Rapid assessment of SARS-CoV-2-evolved variants using virus-like particles. Science 374, 1626–1632 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schubert K., et al. , SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 27, 959–966 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Vora S. M., et al. , Targeting stem-loop 1 of the SARS-CoV-2 5’ UTR to suppress viral translation and Nsp1 evasion. Proc. Natl. Acad. Sci. U.S.A. 119, e2117198119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolff G., et al. , A molecular pore spans the double membrane of the coronavirus replication organelle. Science 369, 1395–1398 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimmermann L., et al. , SARS-CoV-2 nsp3-4 suffice to form a pore shaping replication organelles. bioRxiv [Preprint] (2022). 10.1101/2022.10.21.513196 (Accessed 21 October 2022). [DOI]
- 31.Keane S. C., Giedroc D. P., Solution structure of mouse hepatitis virus (MHV) nsp3a and determinants of the interaction with MHV nucleocapsid (N) protein. J. Virol. 87, 3502–3515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koetzner C. A., Hurst-Hess K. R., Kuo L., Masters P. S., Analysis of a crucial interaction between the coronavirus nucleocapsid protein and the major membrane-bound subunit of the viral replicase-transcriptase complex. Virology 567, 1–14 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zúñiga S., et al. , Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 84, 2169–2175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson G. W., Stohlman S. A., Tahara S. M., High affinity interaction between nucleocapsid protein and leader/intergenic sequence of mouse hepatitis virus RNA. J. Gen. Virol. 81, 181–188 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Angelini M. M., Akhlaghpour M., Neuman B. W., Buchmeier M. J., Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio 4, e00524-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oudshoorn D., et al. , Expression and cleavage of middle east respiratory syndrome coronavirus nsp3-4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with coronaviral RNA replication. mBio 8, e01658-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Twu W. I., et al. , Contribution of autophagy machinery factors to HCV and SARS-CoV-2 replication organelle formation. Cell Rep. 37, 110049 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knoops K., et al. , SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6, e226 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neuman B. W., et al. , Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 82, 5279–5294 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clementz M. A., Kanjanahaluethai A., O’Brien T. E., Baker S. C., Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology 375, 118–129 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maurya R., et al. , SARS-CoV-2 mutations and COVID-19 clinical outcome: Mutation global frequency dynamics and structural modulation hold the key. Front. Cell Infect. Microbiol. 12, 868414 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narayanan S., et al. , SARS-CoV-2 genomes from Oklahoma, United States. Front Genet. 11, 612571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barona-Gómez F., et al. , Phylogenomics and population genomics of SARS-CoV-2 in Mexico during the pre-vaccination stage reveals variants of interest B.1.1.28.4 and B.1.1.222 or B.1.1.519 and the nucleocapsid mutation S194L associated with symptoms. Microb. Genom. 7, 000684 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karp P. H., et al. , An in vitro model of differentiated human airway epithelia. Methods for establishing primary cultures. Methods Mol. Biol. 188, 115–137 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix. All data are available upon request to the scientific community.