Abstract
CCR5 and CXCR4 are the principal CD4-associated coreceptors used by human immunodeficiency virus type 1 (HIV-1). CXCR4 is also a receptor for the feline immunodeficiency virus (FIV). The rat CXCR4 cannot mediate infection by HIV-1NDK or by FIVPET (both cell line-adapted strains) because of sequence differences with human CXCR4 in the second extracellular loop (ECL2). Here we made similar observations for HIV-189.6 (a strain also using CCR5) and for a primary HIV-1 isolate. It showed the role of ECL2 in the coreceptor activity of CXCR4 for different types of HIV-1 strains. By exchanging ECL2 residues between human and rat CXCR4, we found that several amino acid differences contributed to the inactivity of the rat CXCR4 toward HIV-189.6. In contrast, its inactivity toward HIV-1NDK seemed principally due to a serine at position 193 instead of to an aspartic acid (Asp193) in human CXCR4. Likewise, a mutation of Asp187 prevented usage of CXCR4 by FIVPET. Different mutations of Asp193, including its replacement by a glutamic acid, markedly reduced or suppressed the activity of CXCR4 for HIV-1NDK infection, indicating that the negative charge was not the only requirement. Mutations of Asp193 and of arginine residues (Arg183 and Arg188) of CXCR4 reduced the efficiency of HIV-1 infection for all HIV-1 strains tested. Other ECL2 mutations tested had strain-specific effects or no apparent effect on HIV-1 infection. The ECL2 mutants allowed us to identify residues contributing to the epitope of the 12G5 monoclonal antibody. Overall, residues with different charges and interspersed in ECL2 seem to participate in the coreceptor activity of CXCR4. This suggests that a conformational rather than linear epitope of ECL2 contributes to the HIV-1 binding site. However, certain HIV-1 and FIV strains seem to require the presence of a particular ECL2 residue.
In most situations, the cell entry of the human immunodeficiency virus type 1 (HIV-1) seems to be initiated by the interaction of its surface envelope protein (SU) with two cell surface components, CD4 and a chemokine receptor, often termed the coreceptor (reviewed in references 2, 12, 21, and 31). This interaction is thought to trigger conformational changes eventually activating the transmembrane envelope protein which mediates fusion of the viral envelope with the cell membrane. Several chemokine receptors, or related orphan G-protein-coupled receptors, were found to be capable of mediating HIV-1 infection under particular experimental conditions (21). However, only the chemokine receptors CCR5 and CXCR4 seem to be used by HIV-1 in vivo. The majority of primary HIV-1 strains are CCR5 dependent (R5), while strains that use CXCR4 (X4) or both CCR5 and CXCR4 (R5X4) are less frequently isolated until relatively late stages of infection (4, 10, 43). Their emergence might play a detrimental role in the evolution of the infectious process (29). The resistance of CCR5-deficient individuals to HIV-1 infection (21) might lead one to consider that CCR5 has a prevalent, if not exclusive, role in the transmission and/or establishment of HIV-1 infection. However, cases of AIDS have since been reported among CCR5-deficient individuals (3, 31, 33, 51), and X4 strains were isolated in the only characterized case (28). These elements point to the importance of addressing the role of CXCR4, as well as CCR5, in the process of HIV-1 infection.
Although less information is available, CCR5 and CXCR4 seem to play a major role in the cell entry process of other lentiviruses. Most primary and cell line-adapted HIV-2 strains tested could infect CD4+ cells expressing CCR5 or CXCR4 (48), while CXCR4 was the receptor used by HIV-2 strains adapted to replication in CD4-negative cell lines (16). All of the simian immunodeficiency viruses (SIVs) tested could use CCR5 as a CD4-associated coreceptor but apparently not CXCR4 (21), but the opposite was recently reported for a mandrill SIV isolate (45). A role for CXCR4 in the process of infection with the feline immunodeficiency virus (FIV) has been described (22, 58, 59); this virus is thought to be more related genetically to the ungulate lentiviruses (e.g., visna virus) than to the HIVs or SIVs (34). In these studies, CXCR4 was found to be the primary receptor for strains of FIV that have been selected for the ability to replicate in the Crandell feline kidney (CrFK) cell line (22, 39, 58, 59). We have extended these studies recently and have found that primary FIV isolates that are unable to productively infect CrFK cells could nevertheless be neutralized by the CXCR4 antagonist AMD3100 and other CXCR4 ligands (41). These data suggest a role for CXCR4 in infection with primary strains of FIV and in viral replication in vivo. This model could therefore be of a great interest in evaluating antiviral strategies based on CXCR4 antagonists.
The ability of the HIV-1 SU (gp120) to form a ternary complex with CXCR4 and CD4 was suggested by coprecipitation experiments (26) and by confocal microscopy studies (53). Moreover, the gp120 from X4 or R5X4 strains was found to compete with the CXCR4 ligand, the stromal-cell-derived-factor-1 chemokine, or with anti-CXCR4 monoclonal antibodies (1, 20, 30). Similarly, the gp120 of R5 HIV-1 strains competed with CCR5 ligands (52, 61). While structural studies of HIV-1 gp120 have provided insight on the interaction with CD4, they only gave indirect information on the interaction with coreceptors (24). Different elements suggest that the third variable loop (V3) of gp120 has a direct role in the selectivity for CXCR4 or CCR5, but other domains of gp120 probably contribute to the formation of the coreceptor-binding site (6, 47).
The structural determinants of the HIV-1 coreceptor activity of CCR5 and CXCR4 are not known precisely. Until now, most structure-function studies have used chimeric receptors formed by exchanging homologous domains between CCR5 or CXCR4 and other chemokine receptors devoid of HIV coreceptor activity or deletion mutants. Relatively few studies have used a site-directed mutagenesis approach. In the case of CCR5, the study of chimeras did not allow the identification of an extracellular domain that was absolutely required for HIV-1 coreceptor activity (reviewed in reference 21). More recently, mutation of residues in the amino-terminal domain and in the second extracellular loop (ECL2) of CCR5 were found to impair HIV-1 infection (15, 42). The ability of CXCR4 to mediate infection by certain HIV-1, HIV-2, or FIV strains was found to be determined, at least in a large part, by the ECL2 sequence. We indeed observed that the rat homolog of CXCR4 mediated infection by HIV-1LAI but not by another cell line strain, HIV-1NDK, by HIV-2ROD (37), or by different FIV strains (59). By testing chimeric receptors, we found that the presence of the ECL2 of human CXCR4 was both necessary and sufficient to observe infection by HIV-1NDK and HIV-2ROD (5) or by FIV (59). The role of ECL2 in the HIV-1 coreceptor activity of CXCR4 was also suggested by the properties of chimeras formed with the mouse CXCR4 (35) or with a more distant chemokine receptor CXCR2 (27). Furthermore, we found that the epitope of the 12G5 monoclonal antibody, which can block infection of HIV-1 and HIV-2 strains (16, 50), was at least in part determined by the ECL2 sequence (5) and that mutations in this domain reduced the antiviral efficacy of the AMD3100 bicyclam on HIV-1 infection (25).
Here we show that the ECL2 sequence also determined usage of CXCR4 by primary HIV-1 isolates, and we further explore the role of this domain by a site-directed mutagenesis approach. We sought to identify the residues that were responsible for the distinct HIV-1 and FIV coreceptor activity of the human and rat CXCR4 by reciprocal exchanges in their ECL2 domains. We also tested the effects of a series of amino acid substitutions in ECL2 on the surface expression and coreceptor activity of human CXCR4 and on its reactivity with the 12G5 monoclonal antibody.
MATERIALS AND METHODS
Cell lines and viral strains.
The human astroglioma cell line U373MG-CD4 stably transfected with Escherichia coli lacZ under the transcriptional control of the HIV-1 long terminal repeat (LTR) (19), the cat kidney cell lines CCC-CD4 (7), and CrFK/FIVPET (persistently infected) (59) have been described. All cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (FCS) and antibiotics. HIV-1LAI (36), HIV-1NDK (49), and HIV-189.6 (9) were produced by transfection of HeLa cells with corresponding molecular clones. The clinical isolates HIV-1OUA and HIV-1ATE (obtained from N. Sol and F. Ferchal, Laboratoire de Virologie, Hôpital Saint-Louis, Paris, France) were propagated in activated peripheral blood mononuclear cells as described earlier (48). The evolutive stages (CDC classification) of the subjects OUA and ATE were A3 (78 CD4+ cells per mm3) and B3 (14 CD4+ cells per mm3), respectively. Subject OUA is a Caucasian from Morocco, while subject ATE is a black African from the Congo. All HIV-1 infectious titers were determined in HeLa P4 cells (LTRlacZ+), by scoring β-galactosidase-positive cells 24 h after infection, as described previously (8).
Chimeric and mutant CXCR4.
The human (H) CXCR4 cDNA (37), the rat (R) CXCR4 cDNA (60) (kindly provided by R. S. Duman), and derived mutants, were expressed from the cytomegalovirus (CMV) immediate-early promoter by standard calcium phosphate transfection techniques. The RRHR and HHRH chimeric constructs correspond to the previously described M and N constructs, respectively (5). Constructs A to D were obtained by blunt-end ligation of two PCR fragments amplified from either human or rat CXCR4 or from the RRHR and HHRH constructs in order to reconstruct a chimeric ECL2 (see Fig. 2A). All other CXCR4 mutants were obtained by site-directed mutagenesis on a single-stranded template. Mutants were screened for the creation of restriction enzyme sites and checked by sequencing the ECL2 region. Except for D182G, D193E, D193N, and D193R, all mutants were created in the epitope-tagged human or rat CXCR4, obtained by subcloning the corresponding cDNA in the pcDNA3-Myc vector (38). The epitope-tagged CXCR4 has a 16-amino-acid sequence from human c-MYC, containing the epitope of the 9E10 monoclonal antibody (MAb) (17) at its amino terminus.
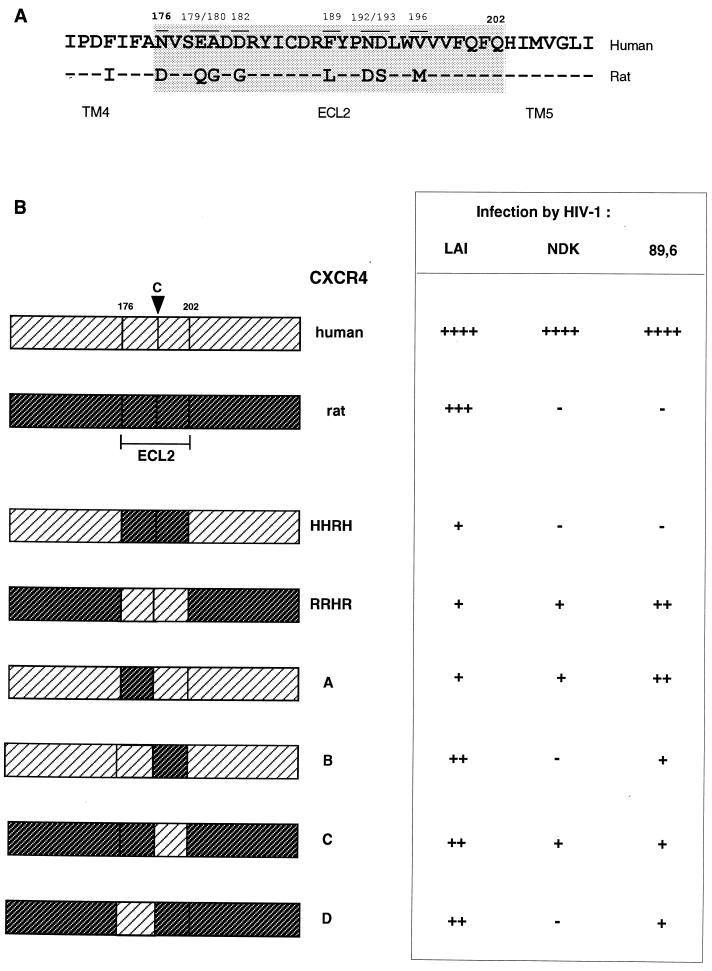
FIG. 2.
Effect of ECL2 substitutions between human and rat CXCR4 on the efficiency of HIV-1 infection. (A) Alignment of the ECL2 domains. The numbering corresponds to the human CXCR4 sequence. (B) Schematic organization of the different chimeric constructs and their efficiency at mediating HIV-1LAI, HIV-1NDK, and HIV-189.6 infection relative to WT human CXCR4. Symbols ++++, 100% (∼103 infected cells per well); +++, >50%; ++, 20 to 50%; +, 10 to 20%; and −, <5%. Infections were performed as described in the legend to Fig. 1. The results are the means of three independent experiments.
HIV-1 infections.
U373MG-CD4 cells were infected in 12-well trays 24 to 48 h after transfection with wild-type (WT) or mutant CXCR4 plasmid. The virus inoculum (104 to 105 infectious units) was left in contact with cells for 36 to 48 h. Supernatant was then harvested, and cells washed and fixed in 0.5% glutaraldehyde. The β-galactosidase activity was revealed by staining with the X-Gal substrate (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Blue-stained foci were scored under 20× magnification. Cell counts of >200 were obtained by extrapolation from randomly selected fields.
Syncytium formation assay.
Cocultures between FIV-infected CrFK cells and CCC-CD4 cells were performed as described earlier (59). Briefly, CCC-CD4 cells were transfected with wild-type or mutant CXCR4 expression vectors and seeded 24 h later in 24-well trays with equivalent numbers of FIV-infected CrFK cells (∼5 × 104 cells per well). Fusion was allowed to proceed for 24 h before cells were fixed and stained with 1% methylene blue–0.2% basic fuschin in methanol. Syncytia (five or more nuclei) were enumerated in three independent fields per well.
Flow cytometry.
COS cells were cotransfected with WT or mutant CXCR4 vectors and with EGFP-N1 (Clontech, Palo Alto, Calif.), a green fluorescent protein (GFP), expression vector, in a 6:1 ratio. Cells were detached with phosphate-buffered saline (PBS) containing 1 mM EDTA at 36 h after transfection and pelleted. Approximately 2 × 105 transfected cells were stained for 1 h at 4°C with 4 μg of the anti-c-MYC MAb 9E10 (17) (Boehringer GmbH, Mannheim, Germany) per ml, 7 μg of the anti-CXCR4 MAb 12G5 (16) (obtained from the NIH AIDS Reagent Program) per ml, or 10 μg of the anti-CXCR4 MAb 6H8 (30) (a gift from A. Amara) per ml in PBS containing 2% FCS. Cells were then washed and further incubated for 1 h with phycoerythrin (PE)-conjugated goat anti-mouse serum (Dako, Glostrub, Denmark) in PBS-FCS. Cells were washed, fixed in 2% formaldehyde, and analyzed on an Epics Elite flow cytometer (Coultronics) for green and red fluorescence.
RESULTS
Strain-dependent activity of rat CXCR4.
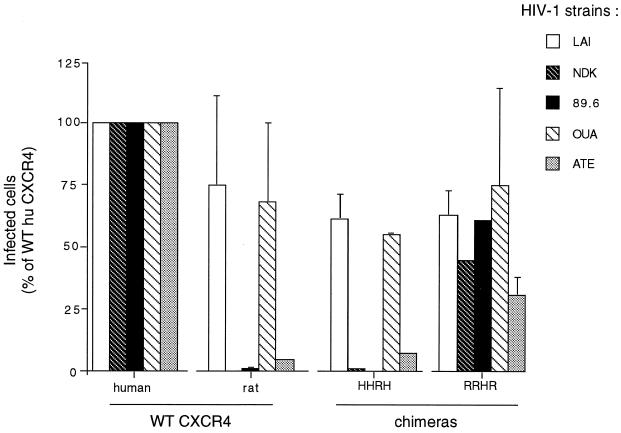
The human astroglioma U373MG-CD4 cells (LTRlacZ+) cannot be infected by HIV-1 or HIV-2 unless they are made to express a functional coreceptor (37, 38). These cells were therefore transfected with different CXCR4 expression vectors and infected in parallel with primary or cell line-adapted HIV-1 strains (Table 1). The efficiency of infection was monitored by scoring β-galactosidase-positive cells in situ. As expected, HIV-1LAI infected cells expressing human CXCR4, rat CXCR4, or the chimeric receptors HHRH and RRHR (corresponding to exchanges of ECL2), while HIV-1NDK only infected cells expressing human CXCR4 or the RRHR chimera (Fig. 1). Likewise, the rat CXCR4 and HHRH chimeras did not allow infection by HIV-189.6 (an R5X4 strain) and HIV-1ATE (a primary X4 strain), whereas they allowed infection by another primary X4 strain, HIV-1OUA (Fig. 1). Differences in ECL2 could therefore prevent usage of CXCR4 by HIV-1 strains belonging to different genetic subtypes (B and D) and by primary and cell-line adapted strains.
TABLE 1.
HIV-1 strains used in this study
| Strain | Type | Molecularly cloned | Coreceptor usage | Genetic subtype |
|---|---|---|---|---|
| LAI | Cell line adapted | Yes | X4 | B |
| NDK | Cell line adapted | Yes | X4 | D |
| 89.6 | Primary | Yes | R5X4 | B |
| ATEa | Primary | No | X4 | Ub |
| OUAa | Primary | No | X4 | U |
The geographical origins for strains ATE and OUA were the Congo and Morocco, respectively.
U, unknown.
FIG. 1.
Infection of U373MG-CD4 cells expressing human or rat CXCR4 or chimeric receptors by the different HIV-1 strains. HIV-1LAI and HIV-1NDK are cell line-adapted X4 strains; HIV-1OUA and HIV-1ATE are primary X4 isolates; and HIV-189.6 is a molecularly cloned R5X4 strain. HHRH is human CXCR4 with the rat CXCR4 ECL2; RRHR is the reciprocal chimera. Bars represent infected cells expressed as a percentage of infection upon transfection with WT human CXCR4. The target cell line bears a Tat-inducible lacZ gene, allowing detection of HIV-infected cells by their high β-galactosidase activity (blue staining with X-Gal). Cells were infected in 12-well trays 24 h after transfection with CXCR4 expression vectors, and X-Gal staining was performed 48 h later. Approximately 1,000 infected cells per well were detected in the case of WT human CXCR4, except for infections with HIV-1ATE (∼200 cells).
The 8-amino-acid differences between human and rat CXCR4 in ECL2 are grouped in two clusters (Fig. 2A). The proximal cluster (residues 176, 179, 180, and 182) at the amino terminus of the loop is separated from the distal cluster (residues 189, 192, 193, and 196) by six residues, one of which is a cysteine conserved in G-protein-coupled receptors, probably forming a disulfide link with the first extracellular loop. The constraints imposed by this link on the spatial structure of ECL2 might bring the two clusters into a relative vicinity.
Homologous fragments of ECL2 were exchanged between the human and rat CXCR4, and the resulting chimeric receptors (constructs A to D) were tested for their ability to mediate infection of U373MG-CD4 cells by HIV-1. Each of these four chimeric receptors mediated infection by HIV-1LAI, although less efficiently than the WT human or rat CXCR4 (Fig. 2B). Infection by HIV-1NDK was observed upon expression of the A and C but not the B and D constructs, suggesting that the proximal cluster of differences had no apparent role for this strain. All four constructs (A to D) allowed infection by HIV-189.6 (Fig. 2B) and by HIV-1ATE (data not shown). Amino acid differences in both clusters must therefore contribute to the lack of coreceptor activity of the rat CXCR4 for these strains. However, construct A was more efficient than construct B at mediating HIV-189.6 and HIV-1ATE infection, which may suggest that amino acid differences in the distal cluster were more important in the lack of activity of the rat CXCR4.
Amino acid exchanges between human and rat CXCR4.
A series of human CXCR4 mutants were obtained by exchanging one or two adjacent residues of ECL2 by the amino acids found at the same position in rat CXCR4. These mutations were created in epitope-tagged forms of the rat or human CXCR4 in order to allow comparisons of their expression at the surface of transfected cells. As previously observed for CCR5 (38), the MYC tag did not affect the HIV-1 coreceptor activity and strain selectivity of the human and rat CXCR4 (Fig. 3 and other data not shown).
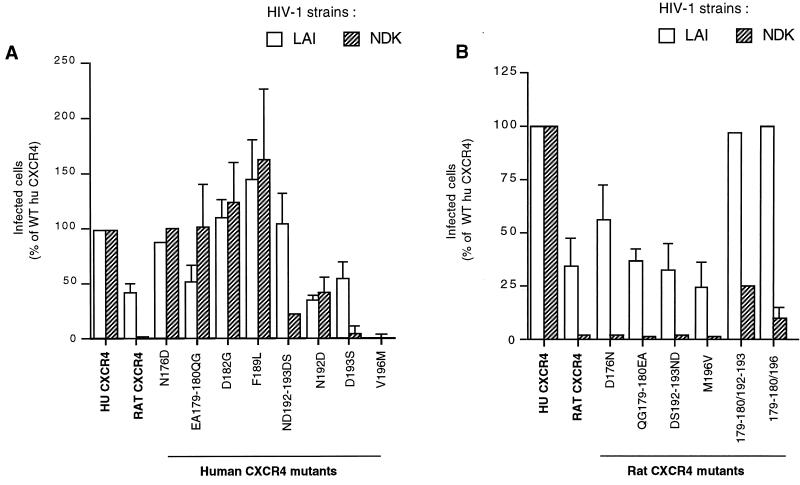
FIG. 3.
Effect of reciprocal exchanges of ECL2 residues between human and rat CXCR4 on infection by HIV-1LAI and HIV-1NDK. Human CXCR4 mutants (A) or rat CXCR4 mutants (B) were expressed in U373MG-CD4 cells, and infections were performed as described in the legend to Fig. 1. Approximately 4,000 infected cells per well were detected for the WT human CXCR4 (100%). The values represent the means of three independent experiments. Mutants are designated by the position of the corresponding residue in the human CXCR4 sequence. For convenience, the same numbering scheme was used for the rat CXCR4 mutants. The WT human and rat CXCR4 and all derived mutants (excepted D182G) bear a MYC epitope tag at their amino terminus.
All of these mutants, except V196M (Fig. 3A), mediated infection of U373MG-CD4 cells by HIV-1LAI. As will be seen, this mutant was apparently not expressed at the cell surface. For most of the mutants tested, the efficiency of infection was similar for HIV-1LAI and HIV-1NDK or was even higher for the latter strain, suggesting that the corresponding amino acid differences did not play a major role in the phenotypic differences between human and rat CXCR4. By contrast, mutation of an aspartic acid residue (Asp193) into serine (Ser) almost abolished HIV-1NDK infection, while it reduced HIV-1LAI infection to a lesser extent. When mutations of Asp193 and of the adjacent asparagine residue (Asn192) were combined (ND192-193DS), the efficiency of infection was restored to the WT level for HIV-1LAI but not for HIV-1NDK. These results indicate that Asp193 is crucial for the usage of CXCR4 by HIV-1NDK. Noteworthy, the negative effect of the D193S mutation was compensated for by another mutation (N192D) impairing HIV-1LAI infection.
We next tested rat CXCR4 mutants in which one or two adjacent residues of ECL2 were replaced by their human CXCR4 counterparts. All of these mutants mediated infection of U373MG-CD4 cells by HIV-1LAI with an efficiency similar or even higher than WT rat CXCR4, but none of them, including D192N, S193D, or their combination, mediated detectable HIV-1NDK infection (Fig. 3B and other data not shown). Among the different combinations of mutations tested, two resulted into detectable HIV-1NDK infection. They correspond to the combination of mutations at positions 179/180 and 192/193 or at positions 179/180 and 196. Why the latter combination could mediate HIV-1NDK infection is unclear, since it does not contain the Asp193 residue. It is noteworthy that the same two mutants mediated HIV-1LAI infection markedly more efficiently than the WT rat CXCR4.
Mutations of Asp193.
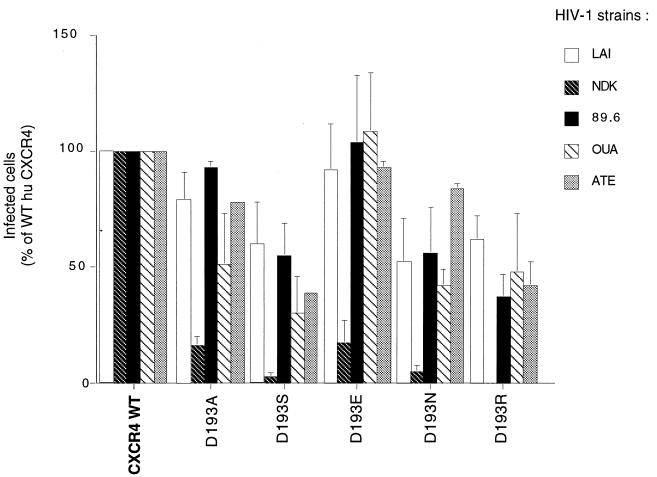
Since the D193S mutation prevented usage of the human CXCR4 coreceptor by HIV-1NDK and impaired its utilization by HIV-1LAI, we have tested the effect of substituting other amino acids for Asp193. Four other HIV-1 strains were included in this experiment. All Asp193 mutants tested could mediate infection of U373MG-CD4 cells by HIV-1LAI, HIV-189.6, HIV-1OUA, and HIV-1ATE with efficiencies ranging from 40 to 100% of that of the WT CXCR4 (Fig. 4). A reduced efficiency of infection was seen upon replacing Asp193 by a serine (D193S), an asparagine (D193N), or an arginine (D193R). Substitutions of a glutamic acid (D193E) or an alanine (D193A) had no apparent effect on HIV-1 infection (or only for HIV-1OUA in the case of D193A). All mutations tested had markedly more important effects on HIV-1NDK. In particular, the D193R mutant was apparently unable to mediate infection by this strain. It confirmed the selective role of Asp193 for a functional interaction between CXCR4 and HIV-1NDK. The results obtained with the D193E mutant indicated that the negative charge was not the only feature supporting the role of Asp193. The coreceptor activity of the D193E was indistinguishable from WT CXCR4 for the three other HIV-1 strains tested, while other mutations were associated with reduced efficiency of infection by one or several of these strains. It suggests that the negative charge of Asp193 could be important for the HIV-1 coreceptor activity of CXCR4 in a non-strain-selective manner.
FIG. 4.
Effect of Asp193 substitutions in human CXCR4 ECL2 on infection by different HIV-1 strains. The experiment was performed and results are represented as described in Fig. 1. The inocula yielded ∼1,000 infected cells per well (∼200 with HIV-1ATE) for WT human CXCR4. The D193A and D193S mutants are MYC tagged.
Effect of other mutations in ECL2.
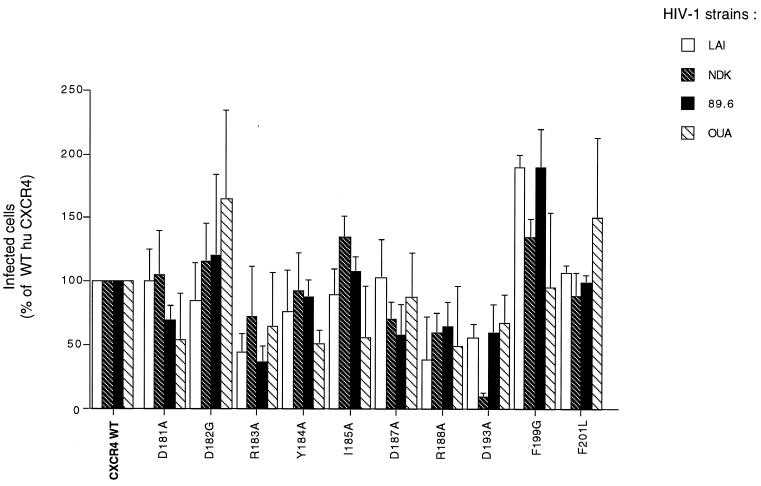
To address the possible role of electrostatic interactions between ECL2 and HIV-1 gp120, we have tested CXCR4 mutants in which a charged residue (Asp181, Asp187, Arg183, or Arg188) was replaced by an alanine, along with the previously described D182G and D193A mutants. We also substituted alanine for tyrosine (Y184A) and isoleucine residues (I185A) and replaced phenylalanine residues by glycine (F199G) or leucine (F201G). These CXCR4 mutants were transiently expressed in U373MG-CD4 cells, and parallel infections were performed with four different HIV-1 strains (Fig. 5). Several mutations resulted in a lower efficiency of infection, either for all of the strains tested (R183A, R188A, and D193A) or for certain strains only (D181A, Y184A, I185A, and D187A). However, the numbers of infected cells were at least 40% of those observed with WT CXCR4, except for HIV-1NDK infection mediated by the D193A mutant. The D182G, I185A, F199G, and F201L mutations increased the efficiency of infection by one or several HIV-1 strains. Overall, this experiment indicated that different types of amino acid substitutions could affect the HIV-1 coreceptor activity of CXCR4, with none being sufficient to completely prevent infection.
FIG. 5.
Effect of single amino acid substitutions in human CXCR4 ECL2 on infection by different HIV-1 strains. The experiment was performed and the results are represented as described in Fig. 1. The inocula yielded 600 to 1,000 infected cells per well for WT human CXCR4. All mutants (excepted D182G) and WT were MYC tagged.
Usage of CXCR4 mutants by FIV.
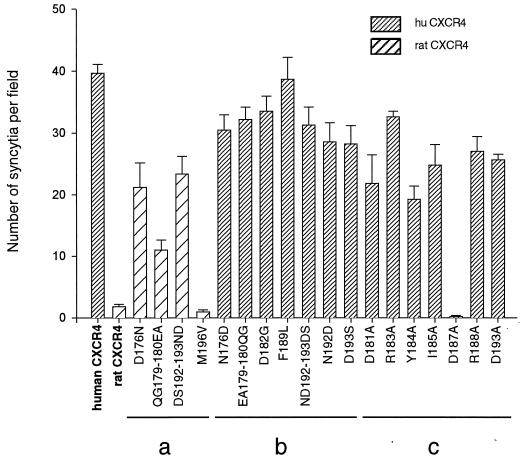
The effect of ECL2 mutations on the usage of CXCR4 by FIV was addressed by syncytium formation assay as described previously (59). As expected, FIV-infected CrFK cells fused with feline CCC cells expressing the human CXCR4 but not with cells expressing the rat homolog (Fig. 6). All human CXCR4 mutants in which ECL2 residues were replaced by their rat CXCR4 counterpart mediated fusion with an efficiency similar to WT human CXCR4 (Fig. 6). Similar to the observations with HIV-189.6, usage of rat CXCR4 as a receptor for FIV was very inefficient. The lack of activity of rat CXCR4 for FIV could not be linked to a single amino acid difference between the ECL2 of rat and human CXCR4. Several rat CXCR4 mutants, in which one or two residues were replaced by the corresponding human CXCR4 residues, allowed fusion with FIV-infected cells, although less efficiently than with WT human CXCR4. The data suggest that the lack of activity of rat CXCR4 as a receptor for FIV may be due to the cumulative effect of several differences in ECL2 between rat and human CXCR4. Furthermore, no single human-to-rat mutation could, by itself, inhibit the usage of human CXCR4 by FIV. In contrast, when a series of human CXCR4 mutants in which residues in ECL2 had been replaced with alanine were assayed for the ability to support fusion mediated by FIV, three mutations were detected that either markedly reduced the efficiency of human CXCR4 to support fusion (40 to 50% with the D181A and Y184A mutants) or abolished receptor function completely (D187A mutant). The Asp187 residue seemed therefore critical for a functional interaction of CXCR4 with FIV but not with HIV-1.
FIG. 6.
Formation of syncytia between FIVPET-infected cells and cells expressing WT or mutant CXCR4. Feline CCC cells were transfected with WT or mutant CXCR4 expression vectors and cocultured (1:1 ratio) with FIVPET-infected CrFKcells in 24-well trays. Cells were fixed after 24 h, and syncytia were scored in five independent fields. Results represent the mean number of syncytia and the standard error in three experiments. The rat CXCR4 mutants (a) and human CXCR4 mutants (b and c) correspond to reciprocal exchanges of ECL2 residues (a and b) or to substitutions of Ala for other human CXCR4 ECL2 residues (c). All mutants (excepted D182G) are MYC tagged.
Surface expression of ECL2 mutants.
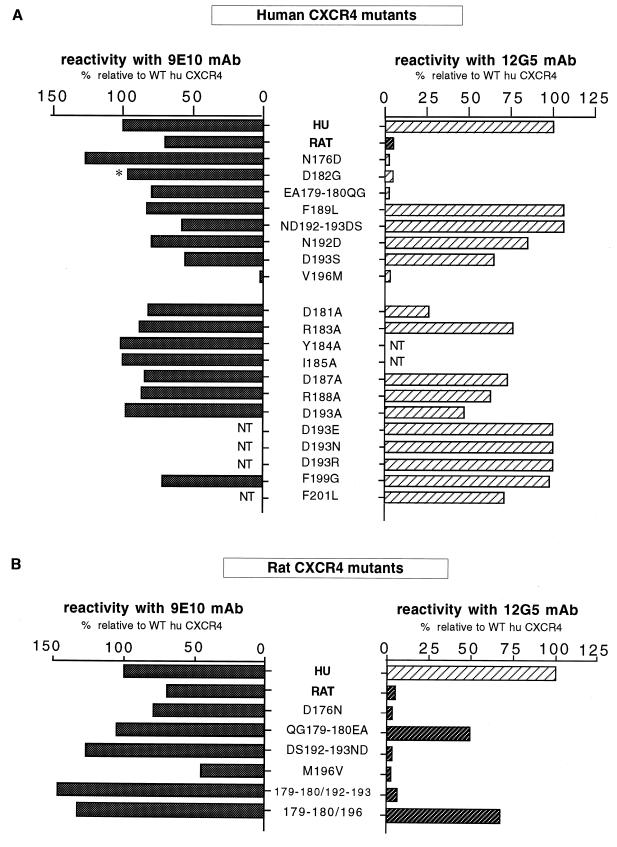
The cell surface expression of the epitope-tagged human and rat CXCR4 mutants was analyzed by indirect immunofluorescence with the 9E10 MAb and by flow cytometry. Some mutants did not have the c-MYC tag (D182G, D193R, D193N, and D193E) or were not tested (F201L) but reacted with the 12G5 MAb-like WT human CXCR4. This was not the case for the D182G mutant, for which the surface expression was tested with the 6H8 MAb raised against the amino-terminal extracellular domain of CXCR4.
Simian COS cells cotransfected with CXCR4 and GFP expression vectors were stained in parallel with the 9E10 (or 6H8) and 12G5 MAbs. The fraction of cells reacting with these antibodies was determined among GFP-positive cells, which correspond to cells that actually expressed the transfected plasmids. The results are presented in Fig. 7.
FIG. 7.
Cell surface expression of human and rat CXCR4 mutants. Flow cytometry analysis was performed on COS cells cotransfected with CXCR4 and GFP expression vectors (6:1) and stained with the 9E10 (anti-c-MYC) or the 12G5 (anti-CXCR4) MAb as indicated. The WT human (HU) and rat CXCR4 and the mutants tested for reactivity with 9E10 bear a c-MYC epitope tag at their amino terminus. The asterisk indicates that the 6H8 anti-CXCR4 MAb was used instead of 9E10. NT, not tested. After the staining with a secondary PE-conjugated antibody, cells were analyzed for green (GFP) and red (PE) fluorescence. The fractions of GFP+ cells, indicating efficient transfection, ranged from 20 to 40%. The bars represent the fractions of GFP+ cells that were stained by 9E10, 12G5, or 6H8 as indicated. The results are expressed relative to cells transfected with WT human CXCR4 (100%).
The V196M mutation prevented expression of CXCR4 at the cell surface, thereby explaining the lack of HIV-1 coreceptor activity of this mutant. The mutation of the corresponding residue in rat CXCR4 (M196V) also markedly reduced expression. All other human and rat CXCR4 mutants yielded fractions of 9E10-positive (or 6H8-positive) cells that were at least 50% relatively to WT human CXCR4. Some human CXCR4 mutants displaying a reduced HIV-1 coreceptor activity, in particular D193S, were expressed at a relatively low level. Conversely, rat CXCR4 mutants mediating HIV-1LAI infection more efficiently than WT rat CXCR4 were also expressed at a higher level (e.g., 179/180 and 192/193). However, there was not an obvious correlation between cell surface expression and the efficiency of infection. For example, the human CXCR4 mutant ND192-193DS mediating HIV-1LAI infection with an efficiency comparable to WT human CXCR4 was expressed at a lower level. The higher efficiency of infection mediated by the F199G mutant was not due to an increased level of expression.
The reactivity of human CXCR4 with the 12G5 MAb was abolished by mutations at position 176 and positions 179 to 180 and was markedly reduced by mutations of Asp181 and Asp182. Certain Asp193 mutations (D193S and D193A) apparently reduced the reactivity with 12G5, but others had no such effect (e.g., D193R), suggesting that this residue is not directly part of the epitope. All other mutants tested (excluding V196M) yielded fractions of 12G5-positive cells comparable to WT human CXCR4. The 12G5 epitope therefore seems critically dependent upon residues of the amino-terminal part of ECL2 at the vicinity of its junction with the fourth membrane-spanning domain. Accordingly, two rat CXCR4 mutants bearing residues 179 and 180 of human CXCR4 (QG179-180EA) reacted with 12G5. However, a rat CXCR4 mutant combining the QG179-180EA and DS192-193ND mutations reacted very weakly with 12G5. The reason for this apparent discrepancy is unclear. Mutations at positions 192 to 193 might modify the ECL2 conformation in the rat CXCR4 context, thereby hampering the access of 12G5.
DISCUSSION
This study confirms the importance of a discrete domain of CXCR4, the ECL2, in the process of HIV-1 and FIV entry. We had shown that amino acid differences in ECL2 accounted for the inability of the rat CXCR4 to mediate infection by HIV-1NDK, a cell line-adapted strain belonging to the D genetic subtype, while it mediated infection by HIV-1LAI, a subtype B cell line-adapted strain (5). These HIV-1 strains therefore had different requirements for a functional interaction with CXCR4, a finding consistent with the genetic divergence of their surface envelope proteins (SU). Likewise, human but not rat CXCR4 could mediate infection by HIV-2 and FIV strains (5, 40, 56). Here we report that the rat CXCR4 did not allow infection of CD4+ cells by two primary HIV-1 strains, one from the B subtype. Again, this lack of activity was due to the ECL2 sequence. This result directly shows the role of ECL2 for HIV-1 strains with different properties (primary or cell line-adapted X4 or R5X4) and from different subtypes. Likewise, HIV-1LAI, another primary HIV-1 strain, could infect cells via the rat CXCR4. These HIV-1 strains either could not depend upon ECL2 for usage of CXCR4 or could have different sequence requirements in this domain. The latter possibility seems supported by the inhibitory effects of several mutations in ECL2 on HIV-1LAI infection and by the properties of chimeras formed between CXCR4 and CXCR2. Only chimeras with ECL2 from CXCR4 could indeed support fusion with cells expressing HIV-1LAI envelope proteins (27).
Different experiments have shown the interaction of CXCR4 with the HIV-1 surface envelope protein (SU) gp120, usually in the presence of CD4 (1, 20, 26, 30, 53), and with the FIV SU (22). Interaction of CXCR4 with the HIV-2 SU is also likely, but it has not yet been directly demonstrated. Since the ability of CXCR4 to mediate infection appears to be dependent upon the ECL2 sequence, the most straightforward explanation is that this domain interacts directly with SU. Other domains of CXCR4 probably also contribute to the interaction with gp120. Indeed, the deletion of most of the amino-terminal extracellular domain (NT) of CXCR4 reduced the efficiency of HIV-1LAI infection and almost abolished HIV-1NDK infection (5). However, CXCR4 chimeras bearing the NT domain or the third extracellular loop (ECL3) from a different receptor (CXCR2) retained HIV-1 coreceptor activity (27). The NT and ECL3 domains of CXCR4 could therefore tolerate very important changes. They could interact with gp120 in a relatively nonstringent way. Alternatively, their role could be to maintain ECL2 in a conformation compatible with gp120 binding.
The study of the strain selectivity of the rat CXCR4 could provide insight into the role of ECL2 in the process of HIV or FIV entry. We have examined the effects of exchanges of ECL2 residues between human and rat CXCR4 on HIV-1 infection and fusion with FIV-infected cells. The inability of rat CXCR4 to mediate infection by HIV-189.6 was due to several differences at nonadjacent residues with human CXCR4. In contrast, the Asp193 of human CXCR4 (replaced by Ser in rat CXCR4) was apparently crucial for infection by HIV-1NDK. The other differences in ECL2 apparently had a smaller role in the lack of activity of the rat CXCR4 for this strain. Like all of the mutations of Asp193 tested, the D193E mutation markedly reduced the efficiency of HIV-1NDK infection. This suggests that the negative charge of Asp193 is not the only feature required for a functional interaction with this strain. Interestingly, most Asp193 mutations, but not D193E, also reduced the efficiency of infection by HIV-1LAI and other strains. The negative charge of Asp193 might therefore be of a general importance for the HIV-1 coreceptor activity of CXCR4.
Different elements could suggest that negatively charged residues of ECL2 had a direct role in the HIV-1 coreceptor activity of CXCR4, possibly mediating an electrostatic interaction with the third variable loop (V3) of gp120. Indeed, usage of CXCR4 by both HIV and FIV seems determined at least in part by the V3 sequence (6, 37, 54) and by the accumulation of basic residues in this domain (18, 23, 54). Also, HIV-1 and FIV infection is blocked by different positively charged compounds interacting with CXCR4, such as the AMD3100 bicyclam (13, 46, 57), the ALX40-4C poly-Arg peptide (14, 57), or the T22 peptide (32). We found that replacing any of the four Asp residues of ECL2 by Ala markedly reduced the efficiency of HIV-1 neutralization by AMD3100 (25).
While the Asp193 mutations reduced the efficiency of infection by all strains tested, no consistent pattern emerged for mutations resulting in a loss of net negative charge (D181A, D182G, and D187A) or their effects were strain selective. Mutations of Asp193 and Asp187 prevented usage of CXCR4 by HIV-1NDK and by FIVPET, respectively. However, as was seen before, the negative charge of Asp193 did not seem crucial, while the effect of other Asp187 substitutions has not been tested. These results do not support the view that negatively charged residues are particularly important for the function of CXCR4. Also, the gain of a negative charge (N192D) was associated with a reduced efficiency of infection by HIV-1LAI and HIV-1NDK, as were mutations resulting in the loss of a positive charge (R183A and R188A). Both positively and negatively charged residues of ECL2 seem therefore to contribute to the function of CXCR4. Since residues important for HIV-1 coreceptor activity or supporting the strain selectivity of rat CXCR4 were located in distinct areas of ECL2, this domain is more likely to contribute to the gp120 binding site of CXCR4 as a conformational structure rather than as a linear epitope.
In a recent study, Wang et al. (55) found that mutations of charged ECL2 residues in CXCR4 had no effect on cell-cell fusion mediated by HIV-1IIIB, an HIV-1LAI variant. It is possible that the highly efficient vaccinia virus-based system used to express HIV-1 envelope proteins and to monitor cell fusion did not allow detection of a partial loss of CXCR4 activity. Interestingly, the D187A mutation allowed fusion with an R5 HIV-1 strain (55), suggesting that the chemokine receptor binding site in HIV gp120 is a relatively conserved structure and that minor changes in either the chemokine receptor or the viral gp120 determine the specificity of coreceptor usage. In this study, we observed that the D187A mutation completely ablated the usage of CXCR4 as a receptor by FIV, suggesting that the conservation of gp120 structure may extend to the feline lentiviruses.
We do not know whether the inability of the human CXCR4 mutants or of rat CXCR4 to mediate infection by some HIV-1 strains is due simply to their lack of interaction with the corresponding gp120 or rather to an inadequate interaction, one insufficient to trigger either the molecular events leading to membrane fusion, or an intracellular signal potentially involved in postentry steps (11, 44). It will be of interest to compare the ability of recombinant SU from different HIV-1 strains to bind to WT and mutant CXCR4 (and to induce signalling via the receptor), keeping in mind that different interactions might take place with oligomeric SU at the surface of the virions. Further characterization of the CXCR4-gp120 interaction may provide valuable information regarding the process of viral entry and for planning future antiviral approaches.
ACKNOWLEDGMENTS
We thank our colleagues L. Picard for helpful discussion, N. Sol and F. Ferchal (Hôpital Saint-Louis, Paris, France), R. Duman (Yale University, New Haven, Conn.), and A. Amara (Institut Pasteur, Paris, France) for gifts of reagents, and I. Bouchaert, F. Letourneur, and C. Tréboute (ICGM) for technical help.
This work was supported by the Agence Nationale de Recherches sur le SIDA and The Wellcome Trust.
REFERENCES
- 1.Bandres J C, Wang Q F, O’Leary J, Baleux F, Amara A, Hoxie J A, Zolla-Pazner S, Gorny M K. Human immunodeficiency virus (HIV) envelope binds to CXCR4 independently of CD4, and binding can be enhanced by interaction with soluble CD4 or by HIV envelope deglycosylation. J Virol. 1998;72:2500–2504. doi: 10.1128/jvi.72.3.2500-2504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger E A. HIV entry and tropism: the chemokine receptor connection. AIDS. 1997;11(Suppl. A):S3–S16. [PubMed] [Google Scholar]
- 3.Biti R, Ffrench R, Young J, Bennetts B, Stewart G, Liang T. HIV-1 infection in an individual homozygous for the CCR-5 deletion allele. Nat Med. 1997;3:252–253. doi: 10.1038/nm0397-252. [DOI] [PubMed] [Google Scholar]
- 4.Björndal A, Deng H, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyö E M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brelot A, Heveker N, Pleskoff O, Sol N, Alizon M. Role of the first and third extracellular domains of CXCR-4 in human immunodeficiency virus coreceptor activity. J Virol. 1997;71:4744–4751. doi: 10.1128/jvi.71.6.4744-4751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 7.Clapham P R, Blanc D, Weiss R A. Specific cell surface requirements for the infection of CD4-positive cells by human immunodeficiency virus types 1 and 2 and by simian immunodeficiency virus. Virology. 1991;181:703–715. doi: 10.1016/0042-6822(91)90904-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clavel F, Charneau P. Fusion from without directed by human immunodeficiency virus particles. J Virol. 1994;68:1179–1185. doi: 10.1128/jvi.68.2.1179-1185.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collman R, Balliet J, Gregory S, Friedman H, Kolson D, Nathanson N, Srinivasan A. An infectious clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J Virol. 1992;66:7517–7521. doi: 10.1128/jvi.66.12.7517-7521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis C B, Dikic I, Unumatz D, Hill C M, Arthos J, Siani J A, Thompson D A, Schlessinger J, Littman D R. Signal transduction due to HIV-1 envelope interactions with chemokine receptors CXCR4 or CCR5. J Exp Med. 1997;186:1793–1798. doi: 10.1084/jem.186.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doms R W, Peiper S C. Unwelcomed guests with master keys: how HIV uses chemokine receptors for cellular entry. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- 13.Donzella G A, Schols D, Lin S W, Esté J A, Nagashima K A, Maddon P J, Allaway G P, Sakmar T P, Henson G, De Clercq E, Moore J P. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4:72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 14.Doranz B J, Grovit-Ferbas K, Sharron M P, Mao S-H, Goetz M B, Daar E S, Doms R W, O’Brien W A. A small-molecule inhibitor directed against the chemokine receptor CXCR4 prevents its use as an HIV-1 coreceptor. J Exp Med. 1997;186:1395–1400. doi: 10.1084/jem.186.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dragic T, Trkola A, Lin S W, Nagashima K A, Kajumo F, Zhao L, Olson W C, Wu L, Mackay C R, Allaway G P, Sakmar T P, Moore J P, Maddon J P. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J Virol. 1998;72:279–285. doi: 10.1128/jvi.72.1.279-285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 17.Evan G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freed E O, Martin M A. The role of human immunodeficiency virus type 1 envelope glycoproteins in virus infection. J Biol Chem. 1995;270:23883–23886. doi: 10.1074/jbc.270.41.23883. [DOI] [PubMed] [Google Scholar]
- 19.Harrington R D, Geballe A P. Cofactor requirements for human immunodeficiency virus type 1 entry into a CD4-expressing human cell line. J Virol. 1993;67:5939–5947. doi: 10.1128/jvi.67.10.5939-5947.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heselgesser J, Halks-Miller M, DelVecchio V, Peiper S, Hoxie J, Kolson D L, Taub D, Horuk R. CD4-independent association between HIV-1 gp120 and CXCR4: functional chemokine receptors are expressed in human neurons. Curr Biol. 1997;7:112–121. doi: 10.1016/s0960-9822(06)00055-8. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman T L, Doms R W. Chemokines and coreceptors in HIV/SIV-host interactions. AIDS. 1998;12(Suppl. A):S17–S26. [PubMed] [Google Scholar]
- 22.Hosie M J, Broere N, Hesselgesser J, Turner J D, Hoxie J A, Neil J C, Willett B J. Modulation of feline immunodeficiency virus infection by stromal cell-derived factor (SDF-1) J Virol. 1998;72:2097–2104. doi: 10.1128/jvi.72.3.2097-2104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuiken C L, de Jong J J, Baan E, Keulen W, Tersmette M, Goudsmit J. Evolution of the V3 envelope domain in proviral sequences and isolates of human immunodeficiency virus type 1 during transition of the viral biological phenotype. J Virol. 1992;66:4622–4627. doi: 10.1128/jvi.66.7.4622-4627.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J G, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labrosse B, Brelot A, Heveker N, Sol N, Schols D, De Clercq E, Alizon M. Determinants for sensitivity of human immunodeficiency virus coreceptor CXCR4 to the bicyclam AMD3100. J Virol. 1998;72:6381–6388. doi: 10.1128/jvi.72.8.6381-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Evidence for cell-surface association between fusin and the CD4-gp120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 27.Lu Z-H, Berson J F, Chen Y-H, Turner J D, Zhang T-Y, Sharron M, Jenk M H, Wang Z-X, Kim J, Rucker J, Hoxie J A, Peiper S C, Doms R W. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc Natl Acad Sci USA. 1997;94:6426–6431. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michael N L, Nelson J A E, Kewalramani V N, Chang G, O’Brien S J, Mascola J R, Volsky B, Louder M, White II G C, Littman D R, Swanstrom R, O’Brien T R. Exclusive and persistent use of the entry coreceptor CXCR4 by human immunodeficiency virus type 1 from a subject homozygous for CCR5 Δ32. J Virol. 1998;72:6040–6047. doi: 10.1128/jvi.72.7.6040-6047.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miedema F, Meyaard L, Koot M, Klein M R, Roos M T L, Groenink M, Fouchier R A M, Van’t Wout A B, Tersmette M, Schellekens P T A, Schuitemaker H. Changing virus-host interactions in the course of HIV-1 infection. Immunol Rev. 1994;140:35–72. doi: 10.1111/j.1600-065x.1994.tb00864.x. [DOI] [PubMed] [Google Scholar]
- 30.Mondor I, Moulard M, Ugolini S, Klasse P-J, Hoxie J, Amara A, Delaunay T, Wyatt R, Sodroski J, Sattentau Q J. Interactions among HIV gp120, CD4, and CXCR4: dependence on CD4 expression level, gp120 viral origin, conservation of the gp120 COOH- and NH2-termini and V1/V2 and V3 loops, and sensitivity to neutralizing antibodies. Virology. 1998;248:394–405. doi: 10.1006/viro.1998.9282. [DOI] [PubMed] [Google Scholar]
- 31.Moore J P, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 32.Murakami T, Nakajima T, Koyanagi Y, Tachibana K, Fujii N, Tamamura H, Yoshida N, Waki M, Matsumoto A, Yoshie O, Kishimoto T, Yamamoto N, Nagasawa T. A small molecule CXCR4 inhibitor that blocks T cell line-tropic HIV-1 infection. J Exp Med. 1997;186:1389–1393. doi: 10.1084/jem.186.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Brien T, Winkler C, Dean M, Nelson J A E, Carrington M, Michael N L, White G C., II HIV-1 infection in a man homozygous for CCR-5Δ32. Lancet. 1997;349:1219. doi: 10.1016/s0140-6736(97)24017-1. [DOI] [PubMed] [Google Scholar]
- 34.Olmsted R A, Hirsch V M, Purcell R H, Johnson P R. Nucleotide sequence analysis of feline immunodeficiency virus: genome organization and relationship to other lentiviruses. Proc Natl Acad Sci USA. 1989;86:8088–8092. doi: 10.1073/pnas.86.20.8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parolin C, Borsetti A, Choe H, Farzan M, Kolchinsky P, Heesen M, Ma Q, Gerard C, Palu G, Dorf M E, Springer T, Sodroski J. Use of murine CXCR-4 as a second receptor by some T-cell-tropic human immunodeficiency viruses. J Virol. 1998;72:1652–1656. doi: 10.1128/jvi.72.2.1652-1656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1lai, HIV-1mal, and HIV-1eli. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 37.Pleskoff O, Sol N, Labrosse B, Alizon M. Human immunodeficiency virus strains differ in their ability to infect CD4+ cells expressing the rat homolog of CXCR-4 (fusin) J Virol. 1997;71:3259–3262. doi: 10.1128/jvi.71.4.3259-3262.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pleskoff O, Tréboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- 39.Poeschla E M, Looney D J. CXCR4 is required by a nonprimate lentivirus/heterologous expression of feline immunodeficiency virus in human, rodent, and feline cells. J Virol. 1998;72:6858–6866. doi: 10.1128/jvi.72.8.6858-6866.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reeves J D, Heveker N, Brelot A, Alizon M, Clapham P, Picard L. The second extracellular loop of CXCR4 is involved in CD4-independent entry of human immunodeficiency virus type 2. J Gen Virol. 1998;79:1793–1799. doi: 10.1099/0022-1317-79-7-1793. [DOI] [PubMed] [Google Scholar]
- 41.Richardson, J., G. Pancino, R. Merat, T. Teste-Lasserre, A. Moraillon, J. Schneider-Mergener, M. Alizon, P. Sonigo, and N. Heveker. Shared usage of the chemokine receptor CXCR-4 by primary and laboratory-adapted strains of the feline immunodeficiency virus. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 42.Ross T M, Bieniasz P D, Cullen B R. Multiple residues contribute to the inability of murine CCR-5 to function as a coreceptor for macrophage-tropic human immunodeficiency virus type 1 isolate. J Virol. 1998;72:1918–1924. doi: 10.1128/jvi.72.3.1918-1924.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scarlatti G, Tresoldi E, Björndal A, Frederiksson R, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, Fenyö E M, Lusso P. In vivo evolution of HIV-1 coreceptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 44.Schmidtmayerova H, Alfano M, Nuovo G, Bukrinsky M. Human immunodeficiency virus type 1 T-lymphotropic strains enter macrophages via a CD4- and CXCR4-mediated pathway: replication is restricted at postentry level. J Virol. 1998;72:4633–4642. doi: 10.1128/jvi.72.6.4633-4642.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schols D, De Clercq E. The simian immunodeficiency virus mnd(GB-1) strain uses CXCR4, not CCR5, as coreceptor for entry in human cells. J Gen Virol. 1998;79:2203–2205. doi: 10.1099/0022-1317-79-9-2203. [DOI] [PubMed] [Google Scholar]
- 46.Schols D, Struyf S, Van Damme J, Esté J A, Henson G, De Clercq E. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J Exp Med. 1997;186:1383–1388. doi: 10.1084/jem.186.8.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smyth R J, Yi Y, Singh A, Collman R G. Determinants for entry cofactor utilization in a dual tropic human immunodeficiency virus type 1 primary isolate. J Virol. 1998;72:4478–4484. doi: 10.1128/jvi.72.5.4478-4484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sol N, Ferchal F, Braun J, Pleskoff O, Tréboute C, Ansard I, Alizon M. Usage of the coreceptors CCR-5, CCR-3, and CXCR-4 by primary and cell line-adapted human immunodeficiency virus type 2. J Virol. 1997;71:8237–8244. doi: 10.1128/jvi.71.11.8237-8244.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spire B, Sire J, Zachar V, Rey F, Barré-Sinoussi F, Galibert F, Hampe A, Chermann J-C. Nucleotide sequence of HIV1-NDK, a highly cytopathic strain of the human immunodeficiency virus, HIV1. Gene. 1989;81:275–284. doi: 10.1016/0378-1119(89)90188-1. [DOI] [PubMed] [Google Scholar]
- 50.Strizki J M, Turner J D, Collman R G, Hoxie J, Gonzalez-Scarano F. A monoclonal antibody (12G5) directed against CXCR-4 inhibits infection with the dual-tropic human immunodeficiency virus type 1 isolate HIV-189.6, but not the T-tropic isolate HIV-1HxB. J Virol. 1997;71:5678–5683. doi: 10.1128/jvi.71.7.5678-5683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Theodorou I, Meyer L, Magierowska M, Katlama C, Rouzioux C. HIV-1 infection in an individual homozygous for CCR-5Δ32. Lancet. 1997;349:1219–1220. [PubMed] [Google Scholar]
- 52.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G, Martin S R, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its coreceptor CCR5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 53.Ugolini S, Moulard M, Mondor I, Barois N, Demandolx D, Hoxie J, Brelot A, Alizon M, Davoust J, Sattentau Q. HIV-1 gp120 from T-cell tropic viruses induces an association between CD4 and the chemokine receptor CXCR4. J Immunol. 1997;159:3000–3008. [PubMed] [Google Scholar]
- 54.Verschoor E V, Boven L A, Blaak H, van Vliet A R W, Horzinek M C, de Ronde A. A single mutation within the V3 envelope neutralization domain of feline immunodeficiency virus determines its tropism for CRFK cells. J Virol. 1995;69:4752–4757. doi: 10.1128/jvi.69.8.4752-4757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z, Berson J F, Zhang T, Cen Y-H, Sun Y, Sharron M, Lu Z, Peiper S-C. CXCR4 sequences involved in coreceptor determination of human immunodeficiency virus type-1 tropism. J Biol Chem. 1998;273:15007–15015. doi: 10.1074/jbc.273.24.15007. [DOI] [PubMed] [Google Scholar]
- 56.Willett B J, Adema K, Heveker N, Brelot A, Picard L, Alizon M, Turner J D, Hoxie J A, Peiper S, Neil J C, Hosie M J. The second extracellular loop of CXCR4 determines its function as a receptor for feline immunodeficiency virus. J Virol. 1998;72:6475–6481. doi: 10.1128/jvi.72.8.6475-6481.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willett, B. J., and M. J. Hosie. The role of CXCR4 in infection with feline immunodeficiency virus. Mol. Membr. Biol., in press. [DOI] [PubMed]
- 58.Willett B J, Hosie M J, Neil J C, Turner J D, Hoxie J A. Common mechanism of infection by lentiviruses. Nature. 1997;385:587. doi: 10.1038/385587a0. [DOI] [PubMed] [Google Scholar]
- 59.Willett B J, Picard L, Hoxie M J, Turner J D, Adema K, Clapham P R. Shared usage of the chemokine receptor CXCR4 by the feline and human immunodeficiency viruses. J Virol. 1997;71:6407–6415. doi: 10.1128/jvi.71.9.6407-6415.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong M-L, Xin W W, Duman R S. Rat LCR1: cloning and cellular distribution of a putative chemokine receptor in brain. Mol Psychiatry. 1996;1:133–140. [PubMed] [Google Scholar]
- 61.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]