Abstract
Glycosylation is a common post-translational modification in eukaryotic cells. It is involved in the production of many biologically active glycoproteins and the regulation of protein structure and function. Core fucosylation plays a vital role in the immune response. Most immune system molecules are core fucosylated glycoproteins such as complements, cluster differentiation antigens, immunoglobulins, cytokines, major histocompatibility complex molecules, adhesion molecules, and immune molecule synthesis-related transcription factors. These core fucosylated glycoproteins play important roles in antigen recognition and clearance, cell adhesion, lymphocyte activation, apoptosis, signal transduction, and endocytosis. Core fucosylation is dominated by fucosyltransferase 8 (Fut8), which catalyzes the addition of α-1,6-fucose to the innermost GlcNAc residue of N-glycans. Fut8 is involved in humoral, cellular, and mucosal immunity. Tumor immunology is associated with aberrant core fucosylation. Here, we summarize the roles and potential modulatory mechanisms of Fut8 in various immune processes of the gastrointestinal system.
Keywords: Fucosyltransferase 8, Core fucosylation, Glycoimmunology, Gastrointestinal tumor immunology, T cell signal pathway
Core Tip: Core fucosylation is driven by fucosyltransferase 8 (Fut8), which catalyzes the addition of α-1,6-fucose to the innermost GlcNAc residue of N-glycans. Core fucosylation plays a vital role in immune responses. Most immune system molecules are core fucosylated glycoproteins that play important roles in antigen recognition and clearance, cell adhesion, lymphocyte activation, apoptosis, signal transduction, and endocytosis. Fut8 is involved in humoral immune responses, cellular immunity, mucosal immunity, and tumor immunology. Here, we summarize the roles and potential modulatory mechanisms of Fut8 in various immune responses of the gastrointestinal system.
INTRODUCTION
Glycosylation is a vital post-transcriptional modification involving the addition of glycans to proteins via chemical bonds[1]. This process occurs in the secretory pathway and affects most intracellular protein folding and trafficking[2]. N-glycosylation and O-glycosylation are the two main types of glycosylation[3]. N-linked glycosylation, also called N-glycosylation, refers to the attachment of an oligosaccharide sugar molecule to a nitrogen atom in the asparagine residue of a protein molecule[4]. In contrast, O-glycosylation refers to the attachment of a sugar molecule to the oxygen atom of a serine or threonine residue[5]. Both N-glycosylation- and O-glycosylation commonly occur during post-translational modifications[6]. Diverse proteins participate in immunological processes, most of which are glycosylated. Glycoimmunology refers to the research on the interactions between glycans and glycan-binding proteins involved in various immune responses and biological/pathological effects[7]. Glycoproteins are also effectors of the immune system, because protein glycosylation promotes immune cell migration throughout the body. Glycosylation is closely associated with pathogen recognition, immune cell homeostasis, and inflammation[8]. In addition, glycosylation is involved in the folding, quality control, maturity, packaging, antigen presentation, assembly of the T cell receptor (TCR) complex and peptide-loaded major histocompatibility complex (p-MHC) antigens, and stability of immune molecules[9]. Previous studies reported that glycans on T cells, immune molecules, and pathogens can influence cellular signal transduction[10-12]. Glycoproteins are key components of the innate and adaptive immune responses[9].
Abnormal glycosylation has been observed in many immune system diseases[1]. O-glycosylation plays a vital role in T cell immunity[13]. Moreover, abnormal O-glycan expression levels in leukomonocytes and cancer cells have been reported in acquired immunodeficiency syndrome, Wiskott–Aldrich syndrome, and T lymphocytic leukemia[3,12,14-17]. Excess core 2 O-glycosylation in T cells reduces primary T cell responses and impairs the interaction between antigen-presenting cells (APC) and T cells, which reduces the production of cytokines, resulting in reduced T cell activation. Core 2 O-glycans serve as a critical backbone for selectin ligand carbohydrate structures[18] and are involved in the adhesion and development of white blood cells[19]. N-Glycosylation mediates innate immune system recognition, inflammation, and autoimmune diseases[20]. Branching and number of N-glycans influence cell proliferation and differentiation[21]. Patients with rheumatoid arthritis (RA) exhibit altered N-glycosylation of immunoglobulin G (IgG). Low N-glycosylation of CD55 (DAF) and CD59 has been observed in RA and inflammation. Furthermore, patients with systemic lupus erythematosus (SLE) display altered N-glycosylation levels in T cells. Decreased synthesis of mannosyl (alpha-1,6-)-glycoproteinbeta-1,6-N-acetyl-glucosaminyltransferase (MGAT5) is observed in autoimmune diseases, whereas it is increased in malignancies[22].
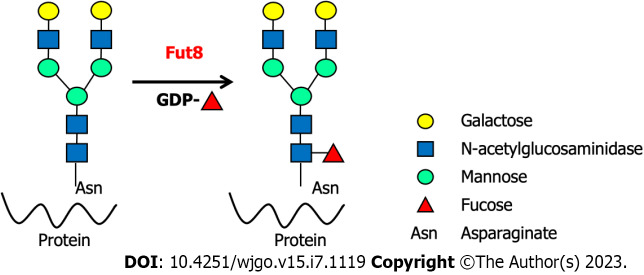
Fucosylation is widely involved in oligosaccharide modifications, which play an essential role in immune-related diseases and cancer, and is often accompanied by the disordered expression of fucosyltransferases (FUTs)[9,23-24,27]. As the sole glycosyltransferase, Fut8 catalyzes the fucose residue transfer via the α1,6-linkage from GDP-fucose (GDP-Fuc) to the innermost N-acetylglucosamine (GlcNAc) residue of N-glycans in the Golgi apparatus of mammalian cells[28] (Figure 1). The core fucose levels of proteins depend on the substrate GDP-Fuc and its transportation to the Golgi apparatus[29]. After core fucosylation, proteins exhibit different spatial structures and biological activities. Core-fucosylated glycoproteins are widely expressed in mammalian tissues, and their aberrant expression is observed under pathological conditions[30]. Fut8 is involved in many types of immune-related diseases and immune responses, such as the humoral immune response, T cell signal transduction, CD4+ T cell activation, CD8+ cytotoxic T cell (CTL) activation, and benign and malignant biological regulation of the gastrointestinal system. Almost all innate and adaptive immune molecules are glycoproteins[31]. Aberrant core fucosylation is one of the most important glycosylation events and has been observed in many immune-related diseases. Most immunoglobulins, clusters of differentiation (CDs), adhesive molecules, soluble and membrane-type lectin receptors, cytokines and their receptors, complement T and B cell receptors (BCRs), and MHCs are core fucosylated proteins. Programmed cell death protein-1 (PD-1, CD279) (NM-005018) is one of the most important immune inhibitory receptors expressed on tumor-infiltrating T cells (TILs). Mass spectrometry has shown that PD-1 is a typical core-fucosylated protein containing four core-fucosylated N-glycans: N49, N58, N74, and N116[32]. Loss of core fucosylation in PD-1 decreases its expression in CTL, leading to more efficient activation, cytotoxicity, and tumor eradication[33].
Figure 1.
The reaction was catalyzed by fucosyltransferase 8. Fucosyltransferase 8 transfers an L-fucose reside from GDP-β-L-fucose onto the innermost GDP-fucose to the innermost N-acetylglucosamine of an N-glycan to form an α-1,6 linkage. Fut8: Fucosyltransferase 8.
Gastrointestinal cancers, mostly colorectal cancer, esophageal cancer (OC), gastric cancer (GC), pancreatic cancer (PC), hepatocellular cancer (HCC), and biliary tract cancers, are the most common causes of cancer mortality worldwide, with significant associated morbidity[34]. Core fucosylation is widely involved in the development of neoplasms of the digestive system. Oral squamous cell carcinoma[35], esophageal squamous cell carcinoma (ESCC)[36], hepatocellular carcinoma[37], and PC[38] were all accompanied with high levels of core fucosylation. A high level of core fucosylation is also observed in colon (also known as bowel) cancer, which is the 3rd most common cancer worldwide (2022)[39,40]. Unlike other tumors, the level of core fucosylation is decreased in GC, and upregulation of core fucosylation can inhibit the proliferation of human GC cells[41]. High levels of Fut8 expression have also been observed in other tumors, such as non-small cell lung cancer, ovarian cancer, prostate cancer, melanoma, and breast cancer, and are associated with unfavorable prognosis[42-46].
Multiple complex mechanisms are involved in Fut8 immunoregulation of gastrointestinal diseases. This review provides an overview of the effect of core fucosylation on glycoimmunology in gastrointestinal diseases.
CORE FUCOSYLATION ON GASTROINTESTINAL CELLULAR IMMUNE MODULATION
T cells can be adoptively transferred from an immunized organism to a native organism to modulate cell-mediated immunity, which involves cell-mediated clearance mechanisms. The cell-mediated immune response is required in the process of elimination during intracellular infectious agents, such as viruses and certain bacteria (e.g., Mycobacterium tuberculosis or Listeria monocytogenes)-induced infection. APC and T helper (Th) cells participate in this response to recognize infected cells. T-cell antigen recognition plays an essential role in both cellular and humoral immunity. After antigen recognition, Th cells release cytokines and chemokines and orchestrate subsequent reactions involving other immune cells, such as natural killer (NK) cells, macrophages, and CTL, to trigger T cell activation.
During T cell recognition and activation, molecules on the surface of CD4+ and CD8+ T cells bind to their respective class II and class I MHC ligands on APC[47]. Moreover, TCR and its co-receptors, CD4 and CD8, participate in the T cell recognition of pMHC ligands. Previous studies have reported that core fucosylation modulates the activation of CD4, CD8, and T-cell receptors.
Core fucosylation on T cell activation via TCR
Proper T cell activation promotes T cell migration to the periphery, where CTLs are found to infect cells and release toxic substances, and Th cells undergo further differentiation into subtypes. However, excessive T cell activation leads to a series of immune system disorders[48]. This abnormal activation was accompanied by changes in core fucosylation levels. SLE is a severe autoimmune disease characterized by the production of autoantibodies and subsequent inflammatory disorders[49]. Gastrointestinal symptoms, including acute abdominal pain (owing to pleurisy and peritonitis), diffuse abdominal pain, epigastric pain, epigastric pain with vomiting and chronic constipation, and diffuse abdominal pain with bleeding from the rectum, probably occur during SLE[50]. The onset and development of SLE pathogenesis are accompanied by excessive activation of CD4+ T cells[51]. Higher core fucosylation levels and IgG antinuclear antibody expression have been observed in the sera of patients with SLE and are related to the severity of SLE[56]. Hyperactivity of B cells in SLE is also T cell-dependent[52]. Increased core fucosylation in patients with SLE has also been correlated with CD4+ T-cell activation[53]. However, the correlation between core fucosylation and SLE-related gastrointestinal symptoms requires further investigation.
Experimental autoimmune encephalomyelitis (EAE), an activated CD4+ T cell-mediated autoimmune disease model[54], shows slower and mild EAE symptoms and recovers body weight reduction after losing core fucosylation in mice[56]. Loss of core fucosylation markedly reduced the proliferation of CD4+ T cells. Viral or bacterial infections and adverse reactions to medications leading to gastrointestinal symptoms are common among patients with SLE. SLE-related gastrointestinal symptoms cannot be ignored since their severity could cause the death of patients if not treated properly. Among all SLE-related gastrointestinal symptoms, lupus mesenteric vasculitis occurs most commonly, accompanied by intestinal pseudo-obstruction, acute pancreatitis protein-losing enteropathy, and other rare complications such as inflammatory bowel diseases and celiac disease[55]. Both SLE and EAE models indicate that the lack of core fucosylation reduces CD4+ T cell activation and consequently ameliorates symptoms[56].
CD4+ T cell activation and differentiation are involved in the establishment and control of protective adaptive immune responses establishing and controlling[57]. Core fucosylation participates in all three stages of CD4+ T-cell activation. First, core fucosylation is essential for structural formation of TCRs. Second, core fucosylation of TCR increases the recognition of pMHC and further promotes the CD4+ T cell activation threshold and efficient TCR-pMHC-II contact[56]. De-core fucosylation suppresses IgG class switching by impairing CD4+ T cell activation and signal transduction via TCR[56]. Fucose-specific lectins[58] might participate in T-B cell interactions[56]. Determining the mechanism by which core fucosylation influences CD4+ T cell activation is vital for the treatment of SLE-related gastrointestinal symptoms. De-core fucosylated CD4+ T cell infusion may be a useful intervention for the treatment of SLE-related gastrointestinal symptoms. Future studies should attempt to verify the effects of core fucosylation on SLE using mouse models.
Core fucosylation on T-B cell interaction
Glycosylation of membrane proteins participates in multiple cellular processes, such as immunological recognition, intercellular interactions, and cell signal transduction[59]. The interaction between CD4+ T and B cells is important for optimal responses in adaptive immunity[60]. Fucose-specific lectins may be involved in the T-B cell interactions[56]. Genes related to TCR complex formation (CD3e, CD4, CD8, and CD40L), T cell activation (CD3e, CD4, CD8, and CD40L), and B cell activation (CD79a and CD81) exhibited reduced expression after the loss of core fucosylation in mouse models. A series of cell signaling molecules, such as MAPKKK, Vav1, PIK3, PKC, and Cyclin D3, also show decreased expression after the inhibition of core fucosylation[56]. Inhibition of core fucosylation attenuates T-B cell interactions via TCR-pMHC and consequently reduces CD4+ T cell activation. Core fucosylation is necessary for efficient coalescence of lipid rafts during T-B cell contact. The main characteristics of the lipid raft included high concentrations of cholesterol, glycosphingolipid, GPI-anchored proteins, and parts of signal transduction-related molecules[25,26]. Efficient CD4+ T cell activation requires the membrane compartmentalization of lipid rafts. Lipid raft coalescence and Ag-BCR endocytosis were substantially reduced following inhibition of core fucosylation[61].
Core fucosylation on Toll-like receptors recognition and signaling in macrophages
Receptor fucosylation is critical for BCR antigen recognition and antibody production[61] as well as Toll-like receptor (TLR) recognition and signaling through the scavenger receptor dendritic cell-specific ICAM-grabbing non-integrin[62,63]. CD14 and TLRs are important pattern recognition molecules involved in innate immunity. TLRs serve as bridges between non-specific and specific immunity. CD14, a GPI anchored protein, was involved in governing lipopolysaccharide-induced dimerization of TLR4/myeloid differentiation factor 2 (MD2) complex, subsequent internalization of TLR4 and the activation of the Toll/IL-1R domain-containing adaptor-inducing IFN-β (TRIF)-dependent pathway[64]. CD14 and TLR4 receptor complexes are heavily glycosylated proteins with nine and two potential N-glycosylation sites, respectively[65,66]. Core fucose was essential for CD14-dependent TLR4 internalization and IFN-β production but not TLR4/MD2 activation and dimerization in mouse macrophage activity[64,67].
TLR2, another important component of the TLRs family, mediates the recognition of various G+ bacterial cell wall components. In innate immunity, CD14 cooperates with TLR2/TLR4 to mediate the body's response to various pathogenic components[68]. The immune response of TLR2 and TLR4 is impaired after the loss of core fucosylation in macrophages. Compared to wild-type bone marrow cells, Fut8 knockout exhibited much higher resistance to inflammation in dextran sodium sulfate (DSS)-induced experimental colitis. Core fucose is essential for CD14- dependent TLR4 and TLR2 signaling in murine macrophages, leading to DSS-induced experimental colitis[67]. Screening compounds or small-molecule inhibitors that can inhibit core fucosylation of the CD14/TLR2/TLR4 signaling pathway could be a new strategy for the treatment of colitis. The modes of administration and dosage should first be tested in mouse models.
CORE FUCOSYLATION ON GASTROINTESTINAL MUCOSAL IMMUNE MODULATION
The mucosal immune system is the largest component of the immune system, and protects the main sites of infectious threats[69]. Core fucosylation is involved in the mucosal immune response, which is required for antiserum production during Salmonella enterica subspp. enterica serovar Typhi (S. Typhi) infection[70]. Secretory immunoglobulin A (sIgA) is the first line of defense against bacterial penetration and immunoprotecting against Salmonella infection[71]. The loss of core fucosylation results in suppressed sIgA production, which leads to increased susceptibility to pathogens. During S. Typhi infection, decreased T and B cell activation-related gene expression was observed after the inhibition of core fucosylation. Loss of core fucosylation suppresses the humoral immune response in S. Typhi infection[70].
sIgA is the main antibody of local mucosal anti-infection immunity and is frequently found in body fluids, such as saliva, tears, colostrum, nasal and bronchial secretions, gastric and intestinal fluids, urine, and sweat. In addition to the intestinal mucosal immunity, the influence of core fucosylation on other types of mucosal immunity remains largely unknown. Mucosal immunity differs from systemic immunity at structural, cellular, molecular, and functional levels. The mucosa-associated lymphoid tissues represented a highly compartmentalized immunological system and functioned essentially from the systemic immune apparatus independently[72]. Developing compounds that protect core fucosylation in specific mucosal systems against a variety of microbial pathogens is discussed in the table.
Fucosylated carbohydrate moieties in intestinal epithelial cells (IECs) are involved in creating an environmental niche for commensal and pathogenic bacteria. Core fucosylation is ubiquitously expressed in the intestinal mucosa and has strong effects on gut microbiota. After infection with S. Typhi, the levels of core fucosylation in IECs were upregulated, accompanied by an increased abundance of beneficial microorganisms such as Akkermansia spp. and Lactobacillus. Loss of core fucosylation increases the susceptibility of IECs to S. Typhi. During the infection of S. Typhi, the elevated core fucosylation level of IECs was mediated by the Wnt/β-catenin signaling pathway. Core fucosylation of IECs upregulates the biological antagonism of the intestinal microbiota to protect against S. Typhi infection[73].
CORE FUCOSYLATION ON GASTROINTESTINAL TUMOR IMMUNE MODULATION
Glycoproteomic and microarray analyses have revealed that Fut8 globally modifies surface antigens, receptors, and adhesion molecules and is involved in the regulation of dozens of genes associated with malignancy[74], suggesting that Fut8 contributes to gastrointestinal tumor progression through multiple mechanisms.
Core fucosylation on antibody-dependent cellular cytotoxicity
The antibody-dependent cellular cytotoxicity (ADCC) effector function of antibodies has been applied in oncotherapy and used to increase the clinical efficacy in vivo[75,76]. Therapeutic antibodies may modulate effector functions through the Fc receptor, and ADCC was affected by the Fc oligosaccharide structure[77,78]. In vitro and in vivo experiments have indicated that antibodies lacking core fucosylated Fc receptors exhibit stronger efficacy than core fucosylated antibodies[79-81]. Therapeutic antibodies that lack core fucosylation can evade inhibitory effects to achieve optimal ADCC[78,82-85]. The lack of core fucosylation of Fc oligosaccharides markedly enhances ADCC and is related to advanced antitumor efficacy in vivo[86]. Core fucose-deleted IgGs and N-glycan-reconstructed antibodies can be used in advanced antibody-based cancer immunotherapies. Defucosylated antibodies such as rituximab and trastuzumab, etc., were considered promising next-generation therapeutics that can enhance ADCC activity (https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/fut8). Okazaki et al[87] reported that the loss of core fucosylation in the N-glycans of IgG1 molecules enhanced ADCC activity by 50-100 folds. Core fucosylation can modify physicochemical functions to regulate immunoglobulin function of immunoglobulins[87].
Core fucosylation on the expression of inhibitory receptors
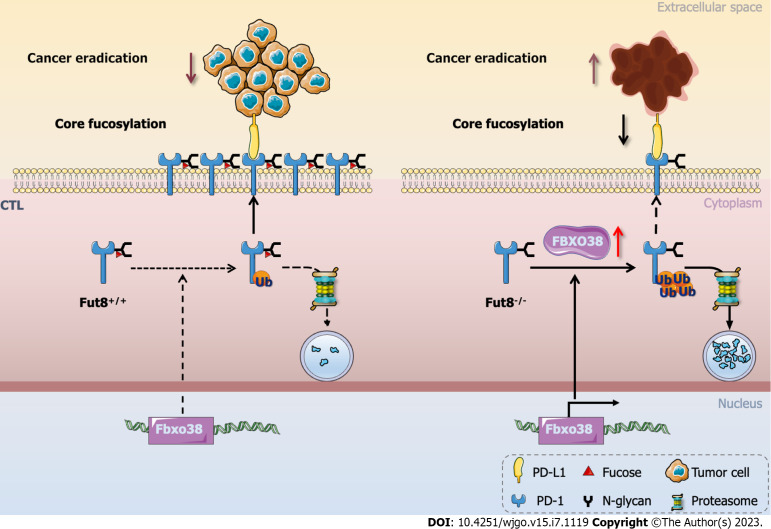
During tumorigenic progression, CTLs experience dysfunction and exhaustion owing to immune-related tolerance and immunosuppression within the tumor microenvironment, favoring adaptive immune resistance. CTLs are key factors in eliminating tumorigenic cells during immune responses[33]. PD-1 was a receptor in the CD28 family of co-stimulatory molecules[88]. Upon interaction with its two ligands, programmed cell death 1 ligand 1 (PD-L1) or PD-L2 to T cells, PD-1 produces inhibitory signals[89]. PD-1 is a core fucosylated glycoprotein with four N-linked glycosylation sites[32], whereas the four N-linked glycosylation sites in PD-L1 (NM-001267706) are N35, N192, N200, and N219[90]. Blocking the core fucosylation of PD-1 inhibits its expression on CTL, which enhances the activation and anti-tumor activity of CTL[33]. Furthermore, the E3 ubiquitin ligase FBXO38 mainly mediates PD-1 polyubiquitination at Lys233[91]. Loss of core fucosylation increases the binding of FBXO38 to PD-1 and subsequent PD-1 degradation through the 26S proteasome to promote PD-1 ubiquitination (Figure 2)[33]. In conclusion, ubiquitination is regulated by core fucosylation, which is involved in tumor immunology[92].
Figure 2.
Programmed cell death 1 core fucosylation in tumorigenesis. Loss of core fucosylation catalyzed by fucosyltransferase 8 significantly enhanced CD8+ cytotoxic T cell activation and cytotoxicity by increasing programmed cell death 1 (PD-1) ubiquitination which in turn led to the degradation of PD-1 in the proteasome and more efficient. PD-1: Programmed cell death 1; PD-L1: Programmed cell death 1 ligand 1; Fut8: Fucosyltransferase 8; CTL: Cytotoxic T cell. This figure was summarized and modified from Zhang et al[33].
PD-1/PD-L1 blockade immunotherapy has been utilized for gastrointestinal malignancies, including biliary tract, esophageal, colorectal, stomach, liver, pancreatic, and anal cancers[93]. The regulation of inhibitory receptor-mediated T cell dysfunction is vital in immunotherapy for gastrointestinal malignancies. The role of the core fucosylation of PD-1 and PD-L1 in the modulation of gastrointestinal cancer remains largely unknown. The effect of core fucosylation on other inhibitory receptors requires further investigation. In addition to PD-1, de-core fucosylation of other receptors, such as antigen 4 (CTLA-4), T-cell immunoglobulin, mucin-domain containing-3 (TIM-3), lymphocyte activation gene-3 (LAG-3), and T-cell immunoglobulin, and ITIM domain (TIGIT), has been investigated. To compare the effects of de-core fucosylation and PD-1/PD-L1 blockade, immunotherapy in a gastrointestinal cancer mouse model is essential.
Core fucosylation on the modulation of the epithelial to mesenchymal transition process
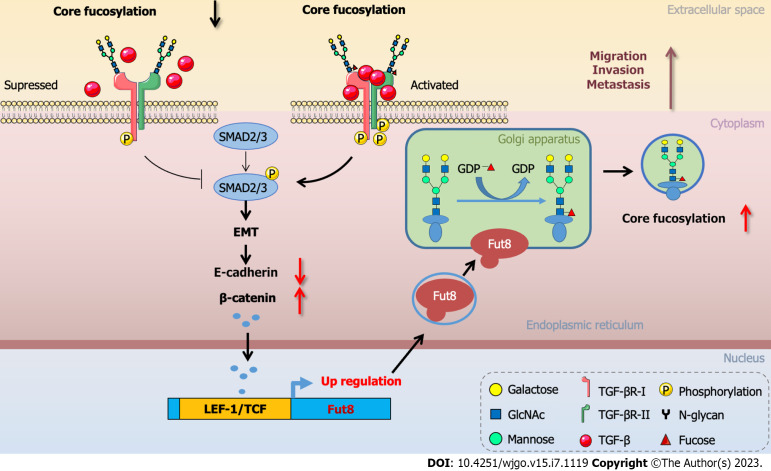
Fut8 inhibition suppresses highly metastatic breast cancer cell invasion and the ability to metastasize to the lung[46]. Epithelial to mesenchymal transition (EMT) is involved in cancer cell metastasis, wherein epithelial cells are transformed into mesenchymal cells to enhance cell dynamics, migration, and invasion[46]. E-cadherin/β-catenin signaling pathway could be activated by transforming growth factor β1 (TGF-β1) to regulate the EMT process. The TGF-β1 signaling pathway led to a deficiency in E-cadherin expression, allowing β-catenin translocation from the plasma membrane into the nucleus, and complexes with lymphoid enhancer-binding factor 1 (LEF-1)/T cell factor (TCF) to promote EMT[94,95]. Fut8 is a direct target of the transcription factor LEF-1, which regulates Fut8 levels. The promoter 1 region of Fut8 5′-UTR included multiple LEF-1/TCF binding sites which participated in the E-cadherin/β-catenin/LEF-1 signaling axis to regulate Fut8 expression during EMT[74,96] (Figure 3). Core fucosylation triggered TGF-β signaling and EMT, which subsequently stimulated the invasion and metastasis of cancer cells[46]. Caveolin-1 (Cav-1), a critical structural protein, could activate Wnt/β-catenin signaling to promote Fut8 expression which led to the proliferation and invasion of HCC[97]. Fut8 was also a driver for the progression of hepatocyte growth factor (HGF)-induced EMT, which was partially blocked by silencing of Fut8 in HCC cells. HGF or TGF-β1 treatment of HCC cells can increase the expression of glycosyltransferase Fut8 to up-regulate the core fucosylation of N-glycans, especially at the glycosylation site Asn-201 on glycoproteins including the folate receptor α (FOLR1)[98]. Core fucosylation of FOLR1 can enhance folate uptake capacity to promote EMT progression in HCC cells[98].
Figure 3.
Core fucosylation modulated epithelial to mesenchymal transition during cancer progression. Loss of core fucosylation impaired the binding of transforming growth factor β to its receptor and inhibited the phosphorylation of regulatory Smad2 and Smad3. During epithelial to mesenchymal transition, the expression of E-cadherin was suppressed, resulting in the accumulation of β-catenin in the nucleus, where it complexed with lymphoid enhancer-binding factor 1 to transactivate fucosyltransferase 8 expression. The core fucosylation level on surface molecules of cancer cells was increased which promoted cancer cell malignant biological properties involving migration, invasion, and metastasis. EMT: Epithelial to mesenchymal transition; TCF: T cell factor; LEF: Lymphoid enhancer-binding factor 1; TGF: Transforming growth factor. This figure was summarized and modified from Tu et al[46], Chen et al[74], and Lin et al[101].
In colorectal cancer, there also existed high levels of Fut8 and E-cadherin protein expression. The enzymatic activity of Fut8 is involved in the appearance of a low-molecular-weight population of E-cadherin and regulates the total amount of E-cadherin[99]. The low-molecular-weight population of E-cadherin significantly increased after the overexpression of Fut8[99]. Core fucosylation participates in changes in N-glycosylation patterns in E-cadherin, stabilization of cell-cell contacts, and regulation of metastatic potential in colorectal cancer[99]. In colorectal cancer, liver metastasis is the most common incident, with a probability of at least 25%, which leads to colorectal liver metastases[100]. Decreased core fucosylation in colorectal cancer may positively influence tumor growth and metastasis. Exploring small molecules, miRNAs, and medicines that could target and downregulate the expression of Fut8 will make sense of the hepatic metastasis of colonic carcinoma.
TGF-β receptors I (TβRI) and II (TβRII) were highly glycosylated proteins. The TGF-β receptor was also modulated by core fucosylation modification[101]. Core fucosylation markedly facilitated functions of the heteromeric complexes of TβRII and TβRI to promote ligand binding and downstream signaling[46]. Loss of core fucosylation impaired the binding of TGF-β to its receptor[46] and inhibited the phosphorylation of regulatory Smad2 and Smad3 (R-Smads) to restrain TGF-β signal transduction(Figure 2). In conclusion, core fucosylation of TGF-β receptors modulated the receptor functions and a series of biological functions[102].
Core fucosylation on the modulation of epidermal growth factor receptor and hepatocyte growth factor receptor and biological functions
Functional growth factor receptors, such as hepatocyte growth factor receptor (HGFR) and epidermal growth factor receptor (EGFR) are regulated by core fucosylation[103,104]. EGFR, a type I transmembrane protein belonging to the ErbB family of receptor tyrosine kinases (RTKs) was activated following binding with peptide growth factors of the EGF family of proteins. Evidence suggests that the high expression of EGFR and EGF-like peptides is associated with the pathogenesis and progression of human carcinomas[37]. De-core fucosylation attenuates responses to EGF and HGF, and blocks EGF-induced phosphorylation of EGFR in hepatocellular carcinoma[37].
Ligand-receptor binding is also essential for core fucosylation between EGF and EGFR, whereas the expression levels of EGFR are not influenced by core fucosylation on the cell surface. Core fucosylation of EGFR is closely related to cancer immunotherapy, which could modulate cellular sensitivity to immunotherapeutic agents. Cells with high Fut8 expression were more sensitive to gefitinib, an EGFR-specific tyrosine kinase inhibitor (EGFR-TKI)[106]. Collectively, core fucosylation plays a vital role in EGFR-mediated biological functions[103].
Fucosylation on the modulation of mitogen-activated protein kinase signal pathway
The mitogen-activated protein kinase (MAPK) pathway participates in the regulation of gene expression, cellular growth, and survival[107]. Abnormal MAPK signaling may induce increased or uncontrolled cell proliferation and apoptosis resistance, leading to oncogenesis[105]. The activation of phosphorylated ERK1/2 and p38 MAPK is involved in tumor cell proliferation and migration[108,109]. Loss of fucosylation suppresses the activation of the ERK1/2 and p38 MAPK signaling pathways, which inhibits carcinoma invasion. Therefore, fucosylation participates in the MAPK signaling pathway activation to modulate biological processes[110].
O-fucosylation is another type of fucosylation modification that is involved in embryonic development. Protein O-fucosyltransferase 1 (poFUT1) is an essential enzyme that catalyzes the synthesis of protein O-fucosylation. poFUT1 promotes trophoblast proliferation by increasing cell cycle progression and promoting cells in the S-phase via activating the MAPK and PI3K/Akt signaling pathways. Activated MAPK and PI3K/Akt signaling pathways are accompanied by increased expression of cyclin D1, cyclin E, CDK 2, CDK 4, and pRb, and decreased levels of the cyclin-dependent kinase inhibitors p21 and p27[111]. However, the influence of core fucosylated N-glycoproteins on the MAPK signaling pathway during tumorigenesis remains largely unknown.
Core fucosylation on the modulation of PI3K-AKT-NF-κB signal pathway
The hepatitis C virus (HCV) is a major cause of chronic liver disease and hepatocellular carcinoma[112]. HCV infection could induce Fut8 expression to promote hepatocellular carcinoma proliferation by activating PI3K-AKT-NF-κB signaling[113]. HCV also stimulates the expression of multidrug resistance-related protein 1 (MRP1) and drug-resistant protein P-glycoprotein (P-GP) and enhances the chemoresistance of HCC to 5-fluorouracil (5-FU)[113]. Inhibition of the PI3K/AKT pathway by its specific inhibitor wortmannin or by small interfering RNA (siRNA) of AKT resulted in decreased MDR of 5-FU chemoresistance in HCC cells, partly through the downregulation of MRP1[114]. In cancer treatment, Fut8 may play a role in reversing chemotherapy resistance and may serve as a therapeutic target.
Core fucosylation on the modulation of cancer cell stemness
Cancer stem cells (CSC) were responsible for cancer reconstitution and propagation. CSCs possess self-renewal, differentiation, and proliferative abilities similar to those of normal stem cells. Cancer stemness, the stem cell-like phenotype of cancer cells, plays an important role in various aspects of carcinogenesis. During anticancer therapies, CSCs exhibited resistant effects, leading to subsequent relapse[115]. Fut8 is required for stemness maintenance in tumorigenic cells[116]. Fut8 deficiency downregulates the stemness of cancer cells, which decreases the expression of CSC-related biomarkers, such as EpCAM, CD133, c-Met, and CXCR4[117]. Fut8-catalyzed α-1,6-fucosylation also promoted stemness induced by fentanyl, an opioid analgesic widely used in cancer patients[118].
Core fucosylation on cancerous radio-resistance
Radioresistance is a major factor affecting the success of radiation therapy for ESCC[119]. There was a high level of core fucosylation in patients with radioresistant ESCC, which led to a poor prognosis. Fut8 inhibition increases the radiosensitivity of radioresistant ESCC cells and suppresses tumor growth and formation[119]. As a key regulatory tumor-associated antigen, native CD147 from human cancer tissue contains a high percentage of core fucosylated structures (28.8%) analyzed via N-linked glycan profiling[120]. In ESCC cells, the inhibition of CD147 partly reversed Fut8-induced radioresistance[119]. Collectively, core fucosylation plays a vital role in the radiosensitivity of ESCC.
Modulation of core fucosylation via microRNA, long non-coding RNA, and circular RNAs on gastrointestinal Tumor
MicroRNAs (miRNAs) and long non-coding RNA (lncRNAs) play vital roles in the post-transcriptional regulation of genes via gene imprinting, chromosome remodeling, and cell cycle regulation. The antisense RNA 1 of LEF1, LEF1-AS1, is a newly identified highly conserved lncRNA encoded on the plus strand of LEF1 on chromosome 4q25, which can modulate LEF1 expression[121]. LEF1-AS1 mediated Fut8 Level through activation of Wnt/β-catenin/LEF1 pathway, thereby resulting in β-catenin nuclear translocation. LEF1-AS1 silencing hinders tumorigenesis and lung and liver metastases of colon cancer cells in vivo, while overexpressed of Fut8 abolished the suppressive effect of LEF1-AS1 repression on the biological behavior of colon cancer cells[121]. Furthermore, miR-198 was shown to target the 3'UTR of Fut8 directly to downregulate Fut8 expression at both mRNA and protein levels and suppressed the proliferation and invasion of colorectal carcinoma[122]. In hepatocarcinoma cells, miR-122 and miR-34a were downregulated in spontaneous human hepatocarcinoma which could specifically interact with and regulate the 3'UTR of Fut8[123]. MiR-122-5p, as a post-transcriptional regulator of Fut8, inhibits the expression of Fut8 and suppresses the proliferation and migration of intrahepatic cholangiocarcinoma cells via the PI3K/AKT signaling pathway[124].
Circular RNAs (circRNAs) regulate cancer development via multiple mechanisms, including miRNA sponges, modulation of the Wnt signaling pathway, and EMT[125]. CircRNA cFUT8 promotes HCC development by binding free miR-548c and inhibiting the miR-548c/FUT8 regulatory axis[126].
MiRNAs are promising therapeutic targets for cancer treatment[127]. CircRNAs, such as miRNA sponges, protein scaffolding, or autophagy regulators, interact with RNA-binding proteins (RBP) to act as potential biomarkers for cancer prevention, diagnosis, and therapeutic targets[128]. Screening out more miRNAs and circRNAs targeting Fut8 both in scientific research and in the clinical treatment of gastrointestinal diseases, could be a directional research area.
MODULATION OF CORE FUCOSYLATION ON T CELL IMMUNOREGULATION
The results of the current study showed that Fut8 played a pivotal role in T and B cell activation. CD4+ T cells represent a unique branch of the adaptive immune system that is crucial for achieving a regulated and effective immune response against pathogens and their proper functioning is vital for survival[129]. CD4+ T cells perform multiple functions, including activation of cells of the innate immune system, B-lymphocytes, CTLs, and non-immune cells, and play a critical role in the suppression of immune reactions[129]. However, several uncertainties remain. Fut8 participates in the contact between TCR and pMHC and subsequent CD4+ T cell activation. Owing to the complexity of the in vivo environment, we cannot simply assume that malfunction of the TCR signaling pathway is the only reason for T cell activation. Functions of Fut8 on the interaction of CD4+ T cells with other APCs (such as dendritic cells and macrophages) are also important issues to be explored. SLE is characterized by the overproduction of autoantibodies, mainly IgG[130]. The pathogenesis of SLE is accompanied by hyper CD4+ T cell metabolism[49,131,132]. Core fucosylation is associated with SLE severity and is significantly increased in the CD4+ T cells of patients with SLE. Hyper core fucosylation-induced CD4+ T cell activation could be involved in the development of SLE[56]. To further explore how core fucosylation modulates other cell types in the adaptive immune system, the entire function of SLE therapy may be an urgent project.
Core fucosylation is essential for CD4+ T cell activation via TCR and affects the geometry and conformation of TCR-pMHC clusters in T-B cell interactions. The loss of core fucosylation enhances the anticancer activity of CD8+ CTLs in lung adenocarcinoma. The IL-2 pathway is involved in immune cell activation and downstream effects[133], which may pave the way for further therapeutic developments. In the IL-2/ IL-2 receptor (IL-2R) pathway, the low-affinity IL-2 receptor, IL-2Rβγ, tended to be expressed on CD8+ T cells and NK cells, while the high-affinity heterotrimeric IL-2-receptor alpha beta gamma subunits (IL-2Rαβγ) were constitutively expressed by Tregs[134]. Researchers have determined that IL-2 has pleiotropic effects; at high concentrations, IL-2 causes the expansion and activation of cytotoxic CD8+ effector T cells, whereas at low concentrations, IL-2 expands and activates Tregs[135] which is the dual and seemingly opposing activity during tumor eradication. NKTR-214 is a CD122-biased IL-2R agonist that stimulates the proliferation of anticancer immune cells in vivo by targeting CD122-specific receptors on the surfaces of NK, CD4+ T, and CD8+ T cells. CD122, also known as the IL-2Rβ subunit, was an important signaling receptor known to increase the proliferation of these effector T cells. In preclinical and clinical studies, NKTR-214 treatment led to rapid expansion and mobilization of these cells into the microenvironment. Given that the mRNA expression of IL-2 was upregulated in Fut8-/-OT-I CTLs, IL-2-induced FBXO38 expression resulted in enhanced PD-1 ubiquitination, which was correlated with lower PD-1 protein abundance on the surface of activated CTLs. We hypothesize Fut8 may also participate in IL-2R subunit regulation and function, which subsequently influences CTL activation.
Physiologically, there may be an equilibrium of core fucosylation on the TCR and the inhibitory receptor PD-1 to regulate T cell activation. Our study is the first revealed that Fut8 inhibition promoted CD8+ T cell activation while inhibiting TCR-pMHC contact and subsequent CD4+ T cell activation. The complexity of in vivo conditions makes it impossible to determine the final influence of Fut8 on tumor immunology. CD8+ T cells mainly participate in tumor eradication, whereas CD4+ T cells are involved in immunoregulation. Although the study identified the role of Fut8 in CD4+ T-cell activation during SLE and the anticancer activity of CTL in lung adenocarcinoma, the influence of Fut8 on CD4+ T-cells during cancer progression remains largely unknown. The balance between specific and degenerate T cell recognition by pMHC-II has important implications for protective immunity vs autoimmunity.
FUT8 INHIBITOR AND GASTROINTESTINAL ONCOTHERAPY
Fucosylation, which regulates the biological functions of adhesion molecules and growth factor receptors[136,137]. Many types of fucosyltransferases (FucTs), the GDP-Fuc synthesis pathway, and GDP-Fuc transporters participate in the regulation of fucosylation. The inhibition of FucT activity by FucT inhibitors represents a potential strategy for cancer immunotherapy. The metabolic fucosylation inhibitor 2-fluoro-L-fucose (2F-Fuc) inhibits natural GDP-Fuc production by blocking fucosylation via endogenous salvage pathways[138]. 2F-Fuc affects the functions of both immune and tumor cells. Blocking core fucosylation of CD8+ CTL via 2F-Fuc downregulates PD-1 expression and enhances CTL activation and tumor eradication[32]. Beyond this mechanism, 2F-Fuc inhibited the binding affinity of galectin-3 to glycoproteins, which could promote TCR signaling pathways and dendritic cell maturation by reducing the threshold for lattice disruption by peptide-MHC with easier TCR engagement (Abstract 4005: Understanding the mechanism of 2FF-induced immune modulation). 2F-Fuc enhances the immune-dependent protective effects of lymphoma vaccines[139].
2F-Fuc can also inhibit fucosylation and modulate tumor cell function. Core fucosylation of a series of membrane glycoproteins, integrin β1, and EGFR could be inhibited by 2F-Fuc treatment, which suppressed the phosphorylation of downstream signal molecules like EGFR, AKT, ERK, and FAK. Proliferation, migration, and tumor formation in human cancer cells are consequently suppressed by 2F-Fuc treatment[140].
Core fucosylated proteins are crucial components of the gastrointestinal immune system that modulate the immunoregulation of various immune processes and immune-related gastrointestinal diseases. Abnormal core fucosylation has been observed in several immune processes. Thus, given the unfavorable regulation of immune-related diseases by core fucosylation, Fut8 inhibitors should be developed. 2F-Fuc inhibits fucosylation of proteins. In total, 13 FUT genes have been identified in the human genome. Different genes encode different fucosyltransferases with various functions. Core fucosylation is an important modification of the N-glycan core structure, forming the α-1,6 fucosylation of the GlcNAc residue linked to asparagine, which is catalyzed by Fut8. The inhibition of core fucosylation has shown great therapeutic potential in a series of diseases, and is an important first step towards the development of future inhibitors. Compared to 2F-Fuc, the specific Fut8 inhibitor had a more precise function towards immunoregulation, with minor side effects on the entire gastrointestinal immune system.
Chimeric antigen receptor (CAR) T-cell therapy is an innovative form of cancer immunotherapy in which autologous T cells are genetically modified to express chimeric receptors encoding antigen-specific single-chain variable fragments and co-stimulatory molecules[141]. Although numerous advances have been made in CAR-T cell therapy for hematological tumors, this technology remains ineffective for treating solid cancers, especially gastrointestinal tumors. These shortcomings include on-target, off-tumor toxicity, a paucity of tumor-specific antigen targets, T cell exhaustion in the gastrointestinal tumor microenvironment, and low infiltration of immune cells into gastrointestinal solid tumor niches[142]. Thus, progress in gastrointestinal tumor immunology and improvements in the manufacturing of cell products have promoted the development of cellular immunotherapies. High expression of inhibitory receptors, such as PD-1, on the surface of T cells is modulated by core fucosylation. Inhibitors with smaller molecules can penetrate to the inner tumor tissue more easily. In comparison to monotherapy, CAR-T cell therapy combined with a Fut8 inhibitor may improve the therapeutic effect, especially in solid tumors such as gastrointestinal tumors.
CONCLUSION
Core fucosylation plays a major role in gastrointestinal glycoimmunology (Table 1). Owing to the complicated regulation of core fucosylation in a variety of immune-related gastrointestinal diseases and cancers, other regulatory mechanisms involved in core fucosylation modifications also need to be explored. Future studies should aim to identify the detailed and complex regulatory mechanisms and therapeutic effects of Fut8 on more kinds of gastrointestinal system-related immune cells and immune diseases.
Table 1.
Abnormal core fucosylation in gastrointestinal diseases
|
Types of immune modulation
|
Mechanism
|
Patterns of modulation
|
Disease
|
Ref.
|
| Core fucosylation on gastrointestinal cellular immune modulation | Core fucosylation modulated T cell activation via TCR | Higher core fucosylation level existed in the sera of SLE patients and related to the severity of SLE. Increased core fucosylation in SLE patients was also correlated with CD4+ T cell activation | SLE | [51,53] |
| Core fucosylation modulated TLRs recognition and signaling in macrophages | Core fucose was essential for CD14- dependent TLR4 and TLR2 signaling in murine macrophage activity, leading to DSS-induced experimental colitis | DSS-induced experimental colitis | [67] | |
| Core fucosylation on gastrointestinal humoral immune modulation | Core fucosylation modulated humoral immune response | Loss of core fucosylation suppressed the humoral immune response in S. Typhi infection and resulted in suppressed sIgA production, which led to increased susceptibility to pathogens | S. Typhi infection | [70] |
| Core fucosylation on gastrointestinal tumor immune modulation | Core fucosylation modulated EMT | Caveolin-1 (Cav-1) could activate Wnt/β-catenin signaling to promote Fut8 expression which led to the proliferation and invasion of HCC | HCC | [97] |
| Fut8 was a driver for the progress of hepatocyte growth factor (HGF)-induced EMT which was partially blocked by the silencing of Fut8 in HCC cells | HCC | [98] | ||
| Core fucosylation on FOLR1 could enhance the folate uptake capacity to finally promote the EMT progress of HCC cells | HCC | [98] | ||
| The low molecular weight population of E-cadherin was significantly increased after overexpression of Fut8, which resulted in an enhancement in cell–cell adhesion | Colorectal cancer | [99] | ||
| Core fucosylation modulated EGFR and HGFR and biological functions | De-core fucosylation attenuated responses to EGF and HGF and blocked the EGF-induced phosphorylation of the EGFR in hepatocellular carcinoma | HCC | [37] | |
| Core fucosylation modulated PI3K-AKT-NF-κB signal pathway | HCV infection induced Fut8 expression to promote hepatocellular carcinoma proliferation by activating PI3K-AKT-NF-κB signaling | HCC | [113] | |
| Core fucosylation modulated cancerous radio-resistance | Fut8 inhibition increased the radiosensitivity of radioresistant ESCC cells and suppressed the growth and formation of tumors | ESCC | [119] | |
| Modulation of core fucosylation via microRNA, long non-coding RNA, and circular RNAs | LEF1-AS1 (lncRNA) silence hindered the tumorigenesis, and lung and liver metastasis of colon cancer cells in vivo, while overexpressed Fut8 abolished the suppressive impact of LEF1-AS1 repression on the biological behavior of colorectal cancer cells | Colorectal cancer | [121] | |
| MiR-198 targeted the 3'UTR of Fut8 directly to downregulate Fut8 expression at both mRNA and protein levels and suppressed the proliferation and invasion of colorectal carcinoma | Colorectal cancer | [122] | ||
| MiR-122 and miR-34a were downregulated in spontaneous human hepatocarcinoma which could specifically interact with and regulate the 3'UTR of Fut8 | HCC | [123] | ||
| MiR-122-5p inhibited the expression of Fut8 and suppressed the proliferation and migration ability of the intrahepatic cholangiocarcinoma cell line via PI3K/AKT signaling pathway | Intrahepatic cholangiocarcinoma | [124] | ||
| CircRNA cFUT8 promoted HCC development by binding free miR-548c and inhibiting the miR-548c/FUT8 regulatory axis | HCC | [126] |
Mechanisms and patterns of different types of gastrointestinal immune modulation included cellular, humoral, and tumor immune modulation after modification with core fucosylation. The related diseases were also listed with the references. HCC: Hepatocellular cancer; EMT: Epithelial to mesenchymal transition; Fut8: Fucosyltransferase 8; CTL: Cytotoxic T cell; HCV: Hepatitis C virus; EGFR: Epidermal growth factor receptor; HGFR: Hepatocyte growth factor receptor; SLE: Systemic lupus erythematosus; ESCC: Esophageal squamous cell carcinoma. TLRs: Toll-like receptors; S. typhimurium: Salmonella typhimurium.
Footnotes
Conflict-of-interest statement: The authors declare no commercial or financial conflict of interest.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Peer-review started: January 18, 2023
First decision: February 24, 2023
Article in press: May 8, 2023
Specialty type: Immunology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Miyoshi E, Japan; Wang Q, China S-Editor: Yan JP L-Editor: A P-Editor: Zhang XD
Zhang XD
Contributor Information
Nian-Zhu Zhang, Clinical Laboratory, The Second Hospital of Dalian Medical University, Dalian 116023, Liaoning Province, China.
Li-Fen Zhao, Clinical Laboratory, The Second Hospital of Dalian Medical University, Dalian 116023, Liaoning Province, China.
Qian Zhang, Department of Cell Therapy, Shanghai Tianze Yuntai Biomedical Co., Ltd., Shanghai 200100, China.
Hui Fang, Graduate School of Comprehensive Human Sciences, University of Tsukuba, Tsukuba 305-0005, Ibaraki, Japan.
Wan-Li Song, Guangdong Provincial Key Laboratory of Infectious Diseases and Molecular Immunopathology, Shantou University Medical College, Shantou 515041, Guangdong Province, China.
Wen-Zhe Li, Guangdong Provincial Key Laboratory of Infectious Diseases and Molecular Immunopathology, Shantou University Medical College, Shantou 515041, Guangdong Province, China.
Yu-Song Ge, Department of Neurology, The Second Hospital of Dalian Medical University, Dalian 116023, Liaoning Province, China.
Peng Gao, Clinical Laboratory, The Second Hospital of Dalian Medical University, Dalian 116023, Liaoning Province, China. gaop@dicp.ac.cn.
References
- 1.Zhu Y, Hart GW. Nutrient regulation of the flow of genetic information by O-GlcNAcylation. Biochem Soc Trans. 2021;49:867–880. doi: 10.1042/BST20200769. [DOI] [PubMed] [Google Scholar]
- 2.Hülsmeier AJ, Welti M, Hennet T. Glycoprotein maturation and the UPR. Methods Enzymol. 2011;491:163–182. doi: 10.1016/B978-0-12-385928-0.00010-9. [DOI] [PubMed] [Google Scholar]
- 3.Essentials of Glycobiology. Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, Editors. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press, 2009. [PubMed] [Google Scholar]
- 4.Jefferis R. Recombinant Proteins and Monoclonal Antibodies. Adv Biochem Eng Biotechnol. 2021;175:281–318. doi: 10.1007/10_2017_32. [DOI] [PubMed] [Google Scholar]
- 5.Khairnar A, Sunsunwal S, Babu P, Ramya TNC. Novel serine/threonine-O-glycosylation with N-acetylneuraminic acid and 3-deoxy-D-manno-octulosonic acid by bacterial flagellin glycosyltransferases. Glycobiology. 2021;31:288–306. doi: 10.1093/glycob/cwaa084. [DOI] [PubMed] [Google Scholar]
- 6.Abu-Qarn M, Eichler J, Sharon N. Not just for Eukarya anymore: protein glycosylation in Bacteria and Archaea. Curr Opin Struct Biol. 2008;18:544–550. doi: 10.1016/j.sbi.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 7.Colomb F, Giron LB, Trbojevic-Akmacic I, Lauc G, Abdel-Mohsen M. Breaking the Glyco-Code of HIV Persistence and Immunopathogenesis. Curr HIV/AIDS Rep. 2019;16:151–168. doi: 10.1007/s11904-019-00433-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Kooyk Y, Rabinovich GA. Protein-glycan interactions in the control of innate and adaptive immune responses. Nat Immunol. 2008;9:593–601. doi: 10.1038/ni.f.203. [DOI] [PubMed] [Google Scholar]
- 9.Rudd PM, Wormald MR, Dwek RA. Glycosylation and the immune system. J Protein Chem. 1998;17:519. [PubMed] [Google Scholar]
- 10.Garg A, Barnes PF, Roy S, Quiroga MF, Wu S, García VE, Krutzik SR, Weis SE, Vankayalapati R. Mannose-capped lipoarabinomannan- and prostaglandin E2-dependent expansion of regulatory T cells in human Mycobacterium tuberculosis infection. Eur J Immunol. 2008;38:459–469. doi: 10.1002/eji.200737268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Y, Chen H, Wang Q, Luo F, Yan J, Zhang XL. IL-24 protects against Salmonella typhimurium infection by stimulating early neutrophil Th1 cytokine production, which in turn activates CD8+ T cells. Eur J Immunol. 2009;39:3357–3368. doi: 10.1002/eji.200939678. [DOI] [PubMed] [Google Scholar]
- 12.Zhang XL. Roles of glycans and glycopeptides in immune system and immune-related diseases. Curr Med Chem. 2006;13:1141–1147. doi: 10.2174/092986706776360897. [DOI] [PubMed] [Google Scholar]
- 13.Chang YH, Weng CL, Lin KI. O-GlcNAcylation and its role in the immune system. J Biomed Sci. 2020;27:57. doi: 10.1186/s12929-020-00648-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefebvre JC, Giordanengo V, Limouse M, Doglio A, Cucchiarini M, Monpoux F, Mariani R, Peyron JF. Altered glycosylation of leukosialin, CD43, in HIV-1-infected cells of the CEM line. J Exp Med. 1994;180:1609–1617. doi: 10.1084/jem.180.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brockhausen I, Kuhns W, Schachter H, Matta KL, Sutherland DR, Baker MA. Biosynthesis of O-glycans in leukocytes from normal donors and from patients with leukemia: increase in O-glycan core 2 UDP-GlcNAc:Gal beta 3 GalNAc alpha-R (GlcNAc to GalNAc) beta(1-6)-N-acetylglucosaminyltransferase in leukemic cells. Cancer Res. 1991;51:1257–1263. [PubMed] [Google Scholar]
- 16.Huang MM, Tsuboi S, Wong A, Yu XJ, Oh-Eda M, Derry JM, Francke U, Fukuda M, Weinberg KI, Kohn DB. Expression of human Wiskott-Aldrich syndrome protein in patients' cells leads to partial correction of a phenotypic abnormality of cell surface glycoproteins. Gene Ther. 2000;7:314–320. doi: 10.1038/sj.gt.3301085. [DOI] [PubMed] [Google Scholar]
- 17.Essentials of Glycobiology. Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, Seeberger PH, Editors. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press, 2015. [PubMed] [Google Scholar]
- 18.Ellies LG, Tsuboi S, Petryniak B, Lowe JB, Fukuda M, Marth JD. Core 2 oligosaccharide biosynthesis distinguishes between selectin ligands essential for leukocyte homing and inflammation. Immunity. 1998;9:881–890. doi: 10.1016/s1074-7613(00)80653-6. [DOI] [PubMed] [Google Scholar]
- 19.Tsuboi S, Fukuda M. Roles of O-linked oligosaccharides in immune responses. Bioessays. 2001;23:46–53. doi: 10.1002/1521-1878(200101)23:1<46::AID-BIES1006>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 20.Green RS, Stone EL, Tenno M, Lehtonen E, Farquhar MG, Marth JD. Mammalian N-glycan branching protects against innate immune self-recognition and inflammation in autoimmune disease pathogenesis. Immunity. 2007;27:308–320. doi: 10.1016/j.immuni.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 22.Li D, Li Y, Wu X, Li Q, Yu J, Gen J, Zhang XL. Knockdown of Mgat5 inhibits breast cancer cell growth with activation of CD4+ T cells and macrophages. J Immunol. 2008;180:3158–3165. doi: 10.4049/jimmunol.180.5.3158. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Hsu HC, Mountz JD, Allen JG. Unmasking Fucosylation: from Cell Adhesion to Immune System Regulation and Diseases. Cell Chem Biol. 2018;25:499–512. doi: 10.1016/j.chembiol.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Miyoshi E, Noda K, Yamaguchi Y, Inoue S, Ikeda Y, Wang W, Ko JH, Uozumi N, Li W, Taniguchi N. The alpha1-6-fucosyltransferase gene and its biological significance. Biochim Biophys Acta. 1999;1473:9–20. doi: 10.1016/s0304-4165(99)00166-x. [DOI] [PubMed] [Google Scholar]
- 25.Dykstra M, Cherukuri A, Sohn HW, Tzeng SJ, Pierce SK. Location is everything: lipid rafts and immune cell signaling. Annu Rev Immunol. 2003;21:457–481. doi: 10.1146/annurev.immunol.21.120601.141021. [DOI] [PubMed] [Google Scholar]
- 26.Cheng PC, Brown BK, Song W, Pierce SK. Translocation of the B cell antigen receptor into lipid rafts reveals a novel step in signaling. J Immunol. 2001;166:3693–3701. doi: 10.4049/jimmunol.166.6.3693. [DOI] [PubMed] [Google Scholar]
- 27.Zhang K, Wang H. [Role of Fucosylation in Cancer] Zhongguo Fei Ai Za Zhi. 2016;19:760–765. doi: 10.3779/j.issn.1009-3419.2016.11.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson JR, Williams D, Schachter H. The control of glycoprotein synthesis: N-acetylglucosamine linkage to a mannose residue as a signal for the attachment of L-fucose to the asparagine-linked N-acetylglucosamine residue of glycopeptide from alpha1-acid glycoprotein. Biochem Biophys Res Commun. 1976;72:909–916. doi: 10.1016/s0006-291x(76)80218-5. [DOI] [PubMed] [Google Scholar]
- 29.Norton PA, Mehta AS. Expression of genes that control core fucosylation in hepatocellular carcinoma: Systematic review. World J Gastroenterol. 2019;25:2947–2960. doi: 10.3748/wjg.v25.i23.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Inoue S, Gu J, Miyoshi E, Noda K, Li W, Mizuno-Horikawa Y, Nakano M, Asahi M, Takahashi M, Uozumi N, Ihara S, Lee SH, Ikeda Y, Yamaguchi Y, Aze Y, Tomiyama Y, Fujii J, Suzuki K, Kondo A, Shapiro SD, Lopez-Otin C, Kuwaki T, Okabe M, Honke K, Taniguchi N. Dysregulation of TGF-beta1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc Natl Acad Sci U S A. 2005;102:15791–15796. doi: 10.1073/pnas.0507375102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the immune system. Science. 2001;291:2370–2376. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- 32.Okada M, Chikuma S, Kondo T, Hibino S, Machiyama H, Yokosuka T, Nakano M, Yoshimura A. Blockage of Core Fucosylation Reduces Cell-Surface Expression of PD-1 and Promotes Anti-tumor Immune Responses of T Cells. Cell Rep. 2017;20:1017–1028. doi: 10.1016/j.celrep.2017.07.027. [DOI] [PubMed] [Google Scholar]
- 33.Zhang N, Li M, Xu X, Zhang Y, Liu Y, Zhao M, Li P, Chen J, Fukuda T, Gu J, Jin X, Li W. Loss of core fucosylation enhances the anticancer activity of cytotoxic T lymphocytes by increasing PD-1 degradation. Eur J Immunol. 2020;50:1820–1833. doi: 10.1002/eji.202048543. [DOI] [PubMed] [Google Scholar]
- 34.Abdul-Latif M, Townsend K, Dearman C, Shiu KK, Khan K. Immunotherapy in gastrointestinal cancer: The current scenario and future perspectives. Cancer Treat Rev. 2020;88:102030. doi: 10.1016/j.ctrv.2020.102030. [DOI] [PubMed] [Google Scholar]
- 35.Chang SC, Lin WL, Chang YF, Lee CT, Wu JS, Hsu PH, Chang CF. Glycoproteomic identification of novel plasma biomarkers for oral cancer. J Food Drug Anal. 2019;27:483–493. doi: 10.1016/j.jfda.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadeghzadeh Z, Khosravi A, Jazi MS, Asadi J. Upregulation of Fucosyltransferase 3, 8 and protein O-Fucosyltransferase 1, 2 genes in esophageal cancer stem-like cells (CSLCs) Glycoconj J. 2020;37:319–327. doi: 10.1007/s10719-020-09917-z. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Fukuda T, Isaji T, Lu J, Im S, Hang Q, Gu W, Hou S, Ohtsubo K, Gu J. Loss of α1,6-fucosyltransferase inhibits chemical-induced hepatocellular carcinoma and tumorigenesis by down-regulating several cell signaling pathways. FASEB J. 2015;29:3217–3227. doi: 10.1096/fj.15-270710. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe K, Ohta M, Yada K, Komori Y, Iwashita Y, Kashima K, Inomata M. Fucosylation is associated with the malignant transformation of intraductal papillary mucinous neoplasms: a lectin microarray-based study. Surg Today. 2016;46:1217–1223. doi: 10.1007/s00595-015-1299-8. [DOI] [PubMed] [Google Scholar]
- 39.Muinelo-Romay L, Villar-Portela S, Cuevas Alvarez E, Gil-Martín E, Fernández-Briera A. α(1,6)Fucosyltransferase expression is an independent prognostic factor for disease-free survival in colorectal carcinoma. Hum Pathol. 2011;42:1740–1750. doi: 10.1016/j.humpath.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 40.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 41.Zhao YP, Xu XY, Fang M, Wang H, You Q, Yi CH, Ji J, Gu X, Zhou PT, Cheng C, Gao CF. Decreased core-fucosylation contributes to malignancy in gastric cancer. PLoS One. 2014;9:e94536. doi: 10.1371/journal.pone.0094536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lv X, Song J, Xue K, Li Z, Li M, Zahid D, Cao H, Wang L, Song W, Ma T, Gu J, Li W. Core fucosylation of copper transporter 1 plays a crucial role in cisplatin-resistance of epithelial ovarian cancer by regulating drug uptake. Mol Carcinog. 2019;58:794–807. doi: 10.1002/mc.22971. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Chen J, Li QK, Peskoe SB, Zhang B, Choi C, Platz EA, Zhang H. Overexpression of α (1,6) fucosyltransferase associated with aggressive prostate cancer. Glycobiology. 2014;24:935–944. doi: 10.1093/glycob/cwu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agrawal P, Fontanals-Cirera B, Sokolova E, Jacob S, Vaiana CA, Argibay D, Davalos V, McDermott M, Nayak S, Darvishian F, Castillo M, Ueberheide B, Osman I, Fenyö D, Mahal LK, Hernando E. A Systems Biology Approach Identifies FUT8 as a Driver of Melanoma Metastasis. Cancer Cell. 2017;31:804–819.e7. doi: 10.1016/j.ccell.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Honma R, Kinoshita I, Miyoshi E, Tomaru U, Matsuno Y, Shimizu Y, Takeuchi S, Kobayashi Y, Kaga K, Taniguchi N, Dosaka-Akita H. Expression of fucosyltransferase 8 is associated with an unfavorable clinical outcome in non-small cell lung cancers. Oncology. 2015;88:298–308. doi: 10.1159/000369495. [DOI] [PubMed] [Google Scholar]
- 46.Tu CF, Wu MY, Lin YC, Kannagi R, Yang RB. FUT8 promotes breast cancer cell invasiveness by remodeling TGF-β receptor core fucosylation. Breast Cancer Res. 2017;19:111. doi: 10.1186/s13058-017-0904-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miceli MC, Parnes JR. The roles of CD4 and CD8 in T cell activation. Semin Immunol. 1991;3:133–141. [PubMed] [Google Scholar]
- 48.Dimeloe S, Burgener AV, Grählert J, Hess C. T-cell metabolism governing activation, proliferation and differentiation; a modular view. Immunology. 2017;150:35–44. doi: 10.1111/imm.12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin Y, Choi SC, Xu Z, Perry DJ, Seay H, Croker BP, Sobel ES, Brusko TM, Morel L. Normalization of CD4+ T cell metabolism reverses lupus. Sci Transl Med. 2015;7:274ra18. doi: 10.1126/scitranslmed.aaa0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fawzy M, Edrees A, Okasha H, El Ashmaui A, Ragab G. Gastrointestinal manifestations in systemic lupus erythematosus. Lupus. 2016;25:1456–1462. doi: 10.1177/0961203316642308. [DOI] [PubMed] [Google Scholar]
- 51.Ohmes J, Comdühr S, Akbarzadeh R, Riemekasten G, Humrich JY. Dysregulation and chronicity of pathogenic T cell responses in the pre-diseased stage of lupus. Front Immunol. 2022;13:1007078. doi: 10.3389/fimmu.2022.1007078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mao L, Hou H, Wu S, Zhou Y, Wang J, Yu J, Wu X, Lu Y, Mao L, Bosco MJ, Wang F, Sun Z. TIGIT signalling pathway negatively regulates CD4(+) T-cell responses in systemic lupus erythematosus. Immunology. 2017;151:280–290. doi: 10.1111/imm.12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Y, Li Z, Liang W, Zhang Y, Song W, Song J, Xue K, Wang M, Sun W, Gu J, Li M, Li W. A novel immunochromatographic strips assay for rapid and simple detection of systemic lupus erythematosus. Sci Rep. 2020;10:14178. doi: 10.1038/s41598-020-71137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCarthy DP, Richards MH, Miller SD. Mouse models of multiple sclerosis: experimental autoimmune encephalomyelitis and Theiler's virus-induced demyelinating disease. Methods Mol Biol. 2012;900:381–401. doi: 10.1007/978-1-60761-720-4_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tian XP, Zhang X. Gastrointestinal involvement in systemic lupus erythematosus: insight into pathogenesis, diagnosis and treatment. World J Gastroenterol. 2010;16:2971–2977. doi: 10.3748/wjg.v16.i24.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liang W, Mao S, Sun S, Li M, Li Z, Yu R, Ma T, Gu J, Zhang J, Taniguchi N, Li W. Core Fucosylation of the T Cell Receptor Is Required for T Cell Activation. Front Immunol. 2018;9:78. doi: 10.3389/fimmu.2018.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reed J, Wetzel SA. CD4(+) T Cell Differentiation and Activation. Methods Mol Biol. 2018;1803:335–351. doi: 10.1007/978-1-4939-8549-4_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lehrman MA, Hill RL. The binding of fucose-containing glycoproteins by hepatic lectins. Purification of a fucose-binding lectin from rat liver. J Biol Chem. 1986;261:7419–7425. [PubMed] [Google Scholar]
- 59.Chen R, Seebun D, Ye M, Zou H, Figeys D. Site-specific characterization of cell membrane N-glycosylation with integrated hydrophilic interaction chromatography solid phase extraction and LC-MS/MS. J Proteomics. 2014;103:194–203. doi: 10.1016/j.jprot.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 60.Tay C, Kanellakis P, Hosseini H, Cao A, Toh BH, Bobik A, Kyaw T. B Cell and CD4 T Cell Interactions Promote Development of Atherosclerosis. Front Immunol. 2019;10:3046. doi: 10.3389/fimmu.2019.03046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li W, Yu R, Ma B, Yang Y, Jiao X, Liu Y, Cao H, Dong W, Liu L, Ma K, Fukuda T, Liu Q, Ma T, Wang Z, Gu J, Zhang J, Taniguchi N. Core fucosylation of IgG B cell receptor is required for antigen recognition and antibody production. J Immunol. 2015;194:2596–2606. doi: 10.4049/jimmunol.1402678. [DOI] [PubMed] [Google Scholar]
- 62.Gringhuis SI, Kaptein TM, Wevers BA, van der Vlist M, Klaver EJ, van Die I, Vriend LE, de Jong MA, Geijtenbeek TB. Fucose-based PAMPs prime dendritic cells for follicular T helper cell polarization via DC-SIGN-dependent IL-27 production. Nat Commun. 2014;5:5074. doi: 10.1038/ncomms6074. [DOI] [PubMed] [Google Scholar]
- 63.Gringhuis SI, Kaptein TM, Wevers BA, Mesman AW, Geijtenbeek TB. Fucose-specific DC-SIGN signalling directs T helper cell type-2 responses via IKKε- and CYLD-dependent Bcl3 activation. Nat Commun. 2014;5:3898. doi: 10.1038/ncomms4898. [DOI] [PubMed] [Google Scholar]
- 64.Iijima J, Kobayashi S, Kitazume S, Kizuka Y, Fujinawa R, Korekane H, Shibata T, Saitoh SI, Akashi-Takamura S, Miyake K, Miyoshi E, Taniguchi N. Core fucose is critical for CD14-dependent Toll-like receptor 4 signaling. Glycobiology. 2017;27:1006–1015. doi: 10.1093/glycob/cwx075. [DOI] [PubMed] [Google Scholar]
- 65.da Silva Correia J, Ulevitch RJ. MD-2 and TLR4 N-linked glycosylations are important for a functional lipopolysaccharide receptor. J Biol Chem. 2002;277:1845–1854. doi: 10.1074/jbc.M109910200. [DOI] [PubMed] [Google Scholar]
- 66.Meng J, Parroche P, Golenbock DT, McKnight CJ. The differential impact of disulfide bonds and N-linked glycosylation on the stability and function of CD14. J Biol Chem. 2008;283:3376–3384. doi: 10.1074/jbc.M707640200. [DOI] [PubMed] [Google Scholar]
- 67.Nakayama K, Wakamatsu K, Fujii H, Shinzaki S, Takamatsu S, Kitazume S, Kamada Y, Takehara T, Taniguchi N, Miyoshi E. Core fucose is essential glycosylation for CD14-dependent Toll-like receptor 4 and Toll-like receptor 2 signalling in macrophages. J Biochem. 2019;165:227–237. doi: 10.1093/jb/mvy098. [DOI] [PubMed] [Google Scholar]
- 68.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 69.Russell MW, Moldoveanu Z, Ogra PL, Mestecky J. Mucosal Immunity in COVID-19: A Neglected but Critical Aspect of SARS-CoV-2 Infection. Front Immunol. 2020;11:611337. doi: 10.3389/fimmu.2020.611337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zahid D, Zhang N, Fang H, Gu J, Li M, Li W. Loss of core fucosylation suppressed the humoral immune response in Salmonella typhimurium infected mice. J Microbiol Immunol Infect. 2021;54:606–615. doi: 10.1016/j.jmii.2020.02.006. [DOI] [PubMed] [Google Scholar]
- 71.Jia S, Zhang W, Tan X, He W, Wang W. The distribution of SIgA and IgG antibody-secreting cells in the palatine tonsils of Bactrian camels (Camelus bactrianus) of different ages. Histol Histopathol. 2017;32:511–521. doi: 10.14670/HH-11-808. [DOI] [PubMed] [Google Scholar]
- 72.Holmgren J, Czerkinsky C. Mucosal immunity and vaccines. Nat Med. 2005;11:S45–S53. doi: 10.1038/nm1213. [DOI] [PubMed] [Google Scholar]
- 73.Hao S, Fan Q, Bai Y, Fang H, Zhou J, Fukuda T, Gu J, Li M, Li W. Core Fucosylation of Intestinal Epithelial Cells Protects Against Salmonella Typhi Infection via Up-Regulating the Biological Antagonism of Intestinal Microbiota. Front Microbiol. 2020;11:1097. doi: 10.3389/fmicb.2020.01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen CY, Jan YH, Juan YH, Yang CJ, Huang MS, Yu CJ, Yang PC, Hsiao M, Hsu TL, Wong CH. Fucosyltransferase 8 as a functional regulator of nonsmall cell lung cancer. Proc Natl Acad Sci U S A. 2013;110:630–635. doi: 10.1073/pnas.1220425110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Clynes RA, Towers TL, Presta LG, Ravetch JV. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nat Med. 2000;6:443–446. doi: 10.1038/74704. [DOI] [PubMed] [Google Scholar]
- 76.Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, Watier H. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood. 2002;99:754–758. doi: 10.1182/blood.v99.3.754. [DOI] [PubMed] [Google Scholar]
- 77.Carter P. Improving the efficacy of antibody-based cancer therapies. Nat Rev Cancer. 2001;1:118–129. doi: 10.1038/35101072. [DOI] [PubMed] [Google Scholar]
- 78.Kanda Y, Yamada T, Mori K, Okazaki A, Inoue M, Kitajima-Miyama K, Kuni-Kamochi R, Nakano R, Yano K, Kakita S, Shitara K, Satoh M. Comparison of biological activity among nonfucosylated therapeutic IgG1 antibodies with three different N-linked Fc oligosaccharides: the high-mannose, hybrid, and complex types. Glycobiology. 2007;17:104–118. doi: 10.1093/glycob/cwl057. [DOI] [PubMed] [Google Scholar]
- 79.Niwa R, Sakurada M, Kobayashi Y, Uehara A, Matsushima K, Ueda R, Nakamura K, Shitara K. Enhanced natural killer cell binding and activation by low-fucose IgG1 antibody results in potent antibody-dependent cellular cytotoxicity induction at lower antigen density. Clin Cancer Res. 2005;11:2327–2336. doi: 10.1158/1078-0432.CCR-04-2263. [DOI] [PubMed] [Google Scholar]
- 80.Niwa R, Natsume A, Uehara A, Wakitani M, Iida S, Uchida K, Satoh M, Shitara K. IgG subclass-independent improvement of antibody-dependent cellular cytotoxicity by fucose removal from Asn297-linked oligosaccharides. J Immunol Methods. 2005;306:151–160. doi: 10.1016/j.jim.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 81.Suzuki E, Niwa R, Saji S, Muta M, Hirose M, Iida S, Shiotsu Y, Satoh M, Shitara K, Kondo M, Toi M. A nonfucosylated anti-HER2 antibody augments antibody-dependent cellular cytotoxicity in breast cancer patients. Clin Cancer Res. 2007;13:1875–1882. doi: 10.1158/1078-0432.CCR-06-1335. [DOI] [PubMed] [Google Scholar]
- 82.Iida S, Misaka H, Inoue M, Shibata M, Nakano R, Yamane-Ohnuki N, Wakitani M, Yano K, Shitara K, Satoh M. Nonfucosylated therapeutic IgG1 antibody can evade the inhibitory effect of serum immunoglobulin G on antibody-dependent cellular cytotoxicity through its high binding to FcgammaRIIIa. Clin Cancer Res. 2006;12:2879–2887. doi: 10.1158/1078-0432.CCR-05-2619. [DOI] [PubMed] [Google Scholar]
- 83.Mori K, Iida S, Yamane-Ohnuki N, Kanda Y, Kuni-Kamochi R, Nakano R, Imai-Nishiya H, Okazaki A, Shinkawa T, Natsume A, Niwa R, Shitara K, Satoh M. Non-fucosylated therapeutic antibodies: the next generation of therapeutic antibodies. Cytotechnology. 2007;55:109–114. doi: 10.1007/s10616-007-9103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Preithner S, Elm S, Lippold S, Locher M, Wolf A, da Silva AJ, Baeuerle PA, Prang NS. High concentrations of therapeutic IgG1 antibodies are needed to compensate for inhibition of antibody-dependent cellular cytotoxicity by excess endogenous immunoglobulin G. Mol Immunol. 2006;43:1183–1193. doi: 10.1016/j.molimm.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 85.Nechansky A, Schuster M, Jost W, Siegl P, Wiederkum S, Gorr G, Kircheis R. Compensation of endogenous IgG mediated inhibition of antibody-dependent cellular cytotoxicity by glyco-engineering of therapeutic antibodies. Mol Immunol. 2007;44:1815–1817. doi: 10.1016/j.molimm.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 86.Imai-Nishiya H, Mori K, Inoue M, Wakitani M, Iida S, Shitara K, Satoh M. Double knockdown of alpha1,6-fucosyltransferase (FUT8) and GDP-mannose 4,6-dehydratase (GMD) in antibody-producing cells: a new strategy for generating fully non-fucosylated therapeutic antibodies with enhanced ADCC. BMC Biotechnol. 2007;7:84. doi: 10.1186/1472-6750-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Okazaki A, Shoji-Hosaka E, Nakamura K, Wakitani M, Uchida K, Kakita S, Tsumoto K, Kumagai I, Shitara K. Fucose depletion from human IgG1 oligosaccharide enhances binding enthalpy and association rate between IgG1 and FcgammaRIIIa. J Mol Biol. 2004;336:1239–1249. doi: 10.1016/j.jmb.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 88.Dai S, Jia R, Zhang X, Fang Q, Huang L. The PD-1/PD-Ls pathway and autoimmune diseases. Cell Immunol. 2014;290:72–79. doi: 10.1016/j.cellimm.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 89.Cho H, Kang H, Lee HH, Kim CW. Programmed Cell Death 1 (PD-1) and Cytotoxic T Lymphocyte-Associated Antigen 4 (CTLA-4) in Viral Hepatitis. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18071517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, Khoo KH, Chang SS, Cha JH, Kim T, Hsu JL, Wu Y, Hsu JM, Yamaguchi H, Ding Q, Wang Y, Yao J, Lee CC, Wu HJ, Sahin AA, Allison JP, Yu D, Hortobagyi GN, Hung MC. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun. 2016;7:12632. doi: 10.1038/ncomms12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meng X, Liu X, Guo X, Jiang S, Chen T, Hu Z, Liu H, Bai Y, Xue M, Hu R, Sun SC, Zhou P, Huang X, Wei L, Yang W, Xu C. FBXO38 mediates PD-1 ubiquitination and regulates anti-tumour immunity of T cells. Nature. 2018;564:130–135. doi: 10.1038/s41586-018-0756-0. [DOI] [PubMed] [Google Scholar]
- 92.Chamoto K, Yaguchi T, Tajima M, Honjo T. Insights from a 30-year journey: function, regulation and therapeutic modulation of PD1. Nat Rev Immunol. 2023 doi: 10.1038/s41577-023-00867-9. [DOI] [PubMed] [Google Scholar]
- 93.Long J, Lin J, Wang A, Wu L, Zheng Y, Yang X, Wan X, Xu H, Chen S, Zhao H. PD-1/PD-L blockade in gastrointestinal cancers: lessons learned and the road toward precision immunotherapy. J Hematol Oncol. 2017;10:146. doi: 10.1186/s13045-017-0511-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Medici D, Hay ED, Goodenough DA. Cooperation between snail and LEF-1 transcription factors is essential for TGF-beta1-induced epithelial-mesenchymal transition. Mol Biol Cell. 2006;17:1871–1879. doi: 10.1091/mbc.E05-08-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim K, Lu Z, Hay ED. Direct evidence for a role of beta-catenin/LEF-1 signaling pathway in induction of EMT. Cell Biol Int. 2002;26:463–476. doi: 10.1006/cbir.2002.0901. [DOI] [PubMed] [Google Scholar]
- 96.Lee HS, Park MH, Yang SJ, Park KC, Kim NS, Kim YS, Kim DI, Yoo HS, Choi EJ, Yeom YI. Novel candidate targets of Wnt/beta-catenin signaling in hepatoma cells. Life Sci. 2007;80:690–698. doi: 10.1016/j.lfs.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 97.Zhang C, Wu Q, Huang H, Chen X, Huang T, Li W, Zhang J, Liu Y. Caveolin-1 upregulates Fut8 expression by activating the Wnt/β-catenin pathway to enhance HCC cell proliferative and invasive ability. Cell Biol Int. 2020;44:2202–2212. doi: 10.1002/cbin.11426. [DOI] [PubMed] [Google Scholar]
- 98.Jia L, Li J, Li P, Liu D, Shen J, Zhu B, Ma C, Zhao T, Lan R, Dang L, Li W, Sun S. Site-specific glycoproteomic analysis revealing increased core-fucosylation on FOLR1 enhances folate uptake capacity of HCC cells to promote EMT. Theranostics. 2021;11:6905–6921. doi: 10.7150/thno.56882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Osumi D, Takahashi M, Miyoshi E, Yokoe S, Lee SH, Noda K, Nakamori S, Gu J, Ikeda Y, Kuroki Y, Sengoku K, Ishikawa M, Taniguchi N. Core fucosylation of E-cadherin enhances cell-cell adhesion in human colon carcinoma WiDr cells. Cancer Sci. 2009;100:888–895. doi: 10.1111/j.1349-7006.2009.01125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martin J, Petrillo A, Smyth EC, Shaida N, Khwaja S, Cheow HK, Duckworth A, Heister P, Praseedom R, Jah A, Balakrishnan A, Harper S, Liau S, Kosmoliaptsis V, Huguet E. Colorectal liver metastases: Current management and future perspectives. World J Clin Oncol. 2020;11:761–808. doi: 10.5306/wjco.v11.i10.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin H, Wang D, Wu T, Dong C, Shen N, Sun Y, Xie H, Wang N, Shan L. Blocking core fucosylation of TGF-β1 receptors downregulates their functions and attenuates the epithelial-mesenchymal transition of renal tubular cells. Am J Physiol Renal Physiol. 2011;300:F1017–F1025. doi: 10.1152/ajprenal.00426.2010. [DOI] [PubMed] [Google Scholar]
- 102.Venkatachalam MA, Weinberg JM. New wrinkles in old receptors: core fucosylation is yet another target to inhibit TGF-β signaling. Kidney Int. 2013;84:11–14. doi: 10.1038/ki.2013.95. [DOI] [PubMed] [Google Scholar]
- 103.Wang X, Gu J, Ihara H, Miyoshi E, Honke K, Taniguchi N. Core fucosylation regulates epidermal growth factor receptor-mediated intracellular signaling. J Biol Chem. 2006;281:2572–2577. doi: 10.1074/jbc.M510893200. [DOI] [PubMed] [Google Scholar]
- 104.Wang Y, Fukuda T, Isaji T, Lu J, Gu W, Lee HH, Ohkubo Y, Kamada Y, Taniguchi N, Miyoshi E, Gu J. Loss of α1,6-fucosyltransferase suppressed liver regeneration: implication of core fucose in the regulation of growth factor receptor-mediated cellular signaling. Sci Rep. 2015;5:8264. doi: 10.1038/srep08264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Anjum J, Mitra S, Das R, Alam R, Mojumder A, Emran TB, Islam F, Rauf A, Hossain MJ, Aljohani ASM, Abdulmonem WA, Alsharif KF, Alzahrani KJ, Khan H. A renewed concept on the MAPK signaling pathway in cancers: Polyphenols as a choice of therapeutics. Pharmacol Res. 2022;184:106398. doi: 10.1016/j.phrs.2022.106398. [DOI] [PubMed] [Google Scholar]
- 106.Matsumoto K, Yokote H, Arao T, Maegawa M, Tanaka K, Fujita Y, Shimizu C, Hanafusa T, Fujiwara Y, Nishio K. N-Glycan fucosylation of epidermal growth factor receptor modulates receptor activity and sensitivity to epidermal growth factor receptor tyrosine kinase inhibitor. Cancer Sci. 2008;99:1611–1617. doi: 10.1111/j.1349-7006.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Knight T, Irving JA. Ras/Raf/MEK/ERK Pathway Activation in Childhood Acute Lymphoblastic Leukemia and Its Therapeutic Targeting. Front Oncol. 2014;4:160. doi: 10.3389/fonc.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen L, Mayer JA, Krisko TI, Speers CW, Wang T, Hilsenbeck SG, Brown PH. Inhibition of the p38 kinase suppresses the proliferation of human ER-negative breast cancer cells. Cancer Res. 2009;69:8853–8861. doi: 10.1158/0008-5472.CAN-09-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhou X, Liu Y, You J, Zhang H, Zhang X, Ye L. Myosin light-chain kinase contributes to the proliferation and migration of breast cancer cells through cross-talk with activated ERK1/2. Cancer Lett. 2008;270:312–327. doi: 10.1016/j.canlet.2008.05.028. [DOI] [PubMed] [Google Scholar]
- 110.Carrascal MA, Silva M, Ramalho JS, Pen C, Martins M, Pascoal C, Amaral C, Serrano I, Oliveira MJ, Sackstein R, Videira PA. Inhibition of fucosylation in human invasive ductal carcinoma reduces E-selectin ligand expression, cell proliferation, and ERK1/2 and p38 MAPK activation. Mol Oncol. 2018;12:579–593. doi: 10.1002/1878-0261.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu C, Liang X, Wang J, Zheng Q, Zhao Y, Khan MN, Liu S, Yan Q. Protein O-fucosyltransferase 1 promotes trophoblast cell proliferation through activation of MAPK and PI3K/Akt signaling pathways. Biomed Pharmacother. 2017;88:95–101. doi: 10.1016/j.biopha.2017.01.026. [DOI] [PubMed] [Google Scholar]
- 112.Yeung CY, Lee HC, Chan WT, Jiang CB, Chang SW, Chuang CK. Vertical transmission of hepatitis C virus: Current knowledge and perspectives. World J Hepatol. 2014;6:643–651. doi: 10.4254/wjh.v6.i9.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li S, Liu XY, Pan Q, Wu J, Liu ZH, Wang Y, Liu M, Zhang XL. Hepatitis C Virus-Induced FUT8 Causes 5-FU Drug Resistance in Human Hepatoma Huh7.5.1 Cells. Viruses. 2019;11 doi: 10.3390/v11040378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cheng L, Luo S, Jin C, Ma H, Zhou H, Jia L. FUT family mediates the multidrug resistance of human hepatocellular carcinoma via the PI3K/Akt signaling pathway. Cell Death Dis. 2013;4:e923. doi: 10.1038/cddis.2013.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Najafi M, Farhood B, Mortezaee K. Cancer stem cells (CSCs) in cancer progression and therapy. J Cell Physiol. 2019;234:8381–8395. doi: 10.1002/jcp.27740. [DOI] [PubMed] [Google Scholar]
- 116.Ma M, Guo D, Tan Z, Du J, Guan F, Li X. Fucosyltransferase 8 regulation and breast cancer suppression by transcription factor activator protein 2γ. Cancer Sci. 2021;112:3190–3204. doi: 10.1111/cas.14987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liang C, Fukuda T, Isaji T, Duan C, Song W, Wang Y, Gu J. α1,6-Fucosyltransferase contributes to cell migration and proliferation as well as to cancer stemness features in pancreatic carcinoma. Biochim Biophys Acta Gen Subj. 2021;1865:129870. doi: 10.1016/j.bbagen.2021.129870. [DOI] [PubMed] [Google Scholar]
- 118.Yang HF, Yu M, Jin HD, Yao JQ, Lu ZL, Yabasin IB, Yan Q, Wen QP. Fentanyl Promotes Breast Cancer Cell Stemness and Epithelial-Mesenchymal Transition by Upregulating α1, 6-Fucosylation via Wnt/β-Catenin Signaling Pathway. Front Physiol. 2017;8:510. doi: 10.3389/fphys.2017.00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shen L, Xia M, Deng X, Ke Q, Zhang C, Peng F, Dong X, Luo Z. A lectin-based glycomic approach identifies FUT8 as a driver of radioresistance in oesophageal squamous cell carcinoma. Cell Oncol (Dordr) 2020;43:695–707. doi: 10.1007/s13402-020-00517-5. [DOI] [PubMed] [Google Scholar]
- 120.Huang W, Luo WJ, Zhu P, Tang J, Yu XL, Cui HY, Wang B, Zhang Y, Jiang JL, Chen ZN. Modulation of CD147-induced matrix metalloproteinase activity: role of CD147 N-glycosylation. Biochem J. 2013;449:437–448. doi: 10.1042/BJ20120343. [DOI] [PubMed] [Google Scholar]
- 121.Qi Y, Shan Y, Li S, Huang Y, Guo Y, Huang T, Zhao X, Jia L. LncRNA LEF1-AS1/LEF1/FUT8 Axis Mediates Colorectal Cancer Progression by Regulating α1, 6-Fucosylationvia Wnt/β-Catenin Pathway. Dig Dis Sci. 2022;67:2182–2194. doi: 10.1007/s10620-021-07051-w. [DOI] [PubMed] [Google Scholar]
- 122.Wang M, Wang J, Kong X, Chen H, Wang Y, Qin M, Lin Y, Xu J, Hong J, Chen YX, Zou W, Fang JY. MiR-198 represses tumor growth and metastasis in colorectal cancer by targeting fucosyl transferase 8. Sci Rep. 2014;4:6145. doi: 10.1038/srep06145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bernardi C, Soffientini U, Piacente F, Tonetti MG. Effects of microRNAs on fucosyltransferase 8 (FUT8) expression in hepatocarcinoma cells. PLoS One. 2013;8:e76540. doi: 10.1371/journal.pone.0076540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen F, Li Y, Aye L, Wu Y, Dong L, Yang Z, Gao Q, Zhang S. FUT8 is regulated by miR-122-5p and promotes malignancies in intrahepatic cholangiocarcinoma via PI3K/AKT signaling. Cell Oncol (Dordr) 2023;46:79–91. doi: 10.1007/s13402-022-00736-y. [DOI] [PubMed] [Google Scholar]
- 125.He J, Xie Q, Xu H, Li J, Li Y. Circular RNAs and cancer. Cancer Lett. 2017;396:138–144. doi: 10.1016/j.canlet.2017.03.027. [DOI] [PubMed] [Google Scholar]
- 126.Li C, Xin Z, He L, Ning J, Lin K, Pan J, Rao J, Wang G, Zhu H. cFUT8 promotes liver cancer progression by miR-548c/FUT8 axis. Signal Transduct Target Ther. 2021;6:30. doi: 10.1038/s41392-020-00393-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mollaei H, Safaralizadeh R, Rostami Z. MicroRNA replacement therapy in cancer. J Cell Physiol. 2019;234:12369–12384. doi: 10.1002/jcp.28058. [DOI] [PubMed] [Google Scholar]
- 128.Tang Q, Hann SS. Biological Roles and Mechanisms of Circular RNA in Human Cancers. Onco Targets Ther. 2020;13:2067–2092. doi: 10.2147/OTT.S233672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Luckheeram RV, Zhou R, Verma AD, Xia B. CD4⁺T cells: differentiation and functions. Clin Dev Immunol. 2012;2012:925135. doi: 10.1155/2012/925135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jost SA, Tseng LC, Matthews LA, Vasquez R, Zhang S, Yancey KB, Chong BF. IgG, IgM, and IgA antinuclear antibodies in discoid and systemic lupus erythematosus patients. ScientificWorldJournal. 2014;2014:171028. doi: 10.1155/2014/171028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Perl A. Oxidative stress in the pathology and treatment of systemic lupus erythematosus. Nat Rev Rheumatol. 2013;9:674–686. doi: 10.1038/nrrheum.2013.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Morel L. Immunometabolism in systemic lupus erythematosus. Nat Rev Rheumatol. 2017;13:280–290. doi: 10.1038/nrrheum.2017.43. [DOI] [PubMed] [Google Scholar]
- 133.Osinalde N, Moss H, Arrizabalaga O, Omaetxebarria MJ, Blagoev B, Zubiaga AM, Fullaondo A, Arizmendi JM, Kratchmarova I. Interleukin-2 signaling pathway analysis by quantitative phosphoproteomics. J Proteomics. 2011;75:177–191. doi: 10.1016/j.jprot.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 134.Doberstein SK. Bempegaldesleukin (NKTR-214): a CD-122-biased IL-2 receptor agonist for cancer immunotherapy. Expert Opin Biol Ther. 2019;19:1223–1228. doi: 10.1080/14712598.2019.1685489. [DOI] [PubMed] [Google Scholar]
- 135.Boyman O, Kovar M, Rubinstein MP, Surh CD, Sprent J. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science. 2006;311:1924–1927. doi: 10.1126/science.1122927. [DOI] [PubMed] [Google Scholar]
- 136.Miyoshi E, Moriwaki K, Nakagawa T. Biological function of fucosylation in cancer biology. J Biochem. 2008;143:725–729. doi: 10.1093/jb/mvn011. [DOI] [PubMed] [Google Scholar]
- 137.Kondo J, Sakata N, Morishita K, Hayashibara A, Sakon D, Takamatsu S, Asakura N, Suzuki T, Miyoshi E. Transcription factor SP1 regulates haptoglobin fucosylation via induction of GDP-fucose transporter 1 in the hepatoma cell line HepG2. Biochem Biophys Rep. 2022;32:101372. doi: 10.1016/j.bbrep.2022.101372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rillahan CD, Antonopoulos A, Lefort CT, Sonon R, Azadi P, Ley K, Dell A, Haslam SM, Paulson JC. Global metabolic inhibitors of sialyl- and fucosyltransferases remodel the glycome. Nat Chem Biol. 2012;8:661–668. doi: 10.1038/nchembio.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Okeley NM, Alley SC, Anderson ME, Boursalian TE, Burke PJ, Emmerton KM, Jeffrey SC, Klussman K, Law CL, Sussman D, Toki BE, Westendorf L, Zeng W, Zhang X, Benjamin DR, Senter PD. Development of orally active inhibitors of protein and cellular fucosylation. Proc Natl Acad Sci U S A. 2013;110:5404–5409. doi: 10.1073/pnas.1222263110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhou Y, Fukuda T, Hang Q, Hou S, Isaji T, Kameyama A, Gu J. Inhibition of fucosylation by 2-fluorofucose suppresses human liver cancer HepG2 cell proliferation and migration as well as tumor formation. Sci Rep. 2017;7:11563. doi: 10.1038/s41598-017-11911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Toulouie S, Johanning G, Shi Y. Chimeric antigen receptor T-cell immunotherapy in breast cancer: development and challenges. J Cancer. 2021;12:1212–1219. doi: 10.7150/jca.54095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xiao BF, Zhang JT, Zhu YG, Cui XR, Lu ZM, Yu BT, Wu N. Chimeric Antigen Receptor T-Cell Therapy in Lung Cancer: Potential and Challenges. Front Immunol. 2021;12:782775. doi: 10.3389/fimmu.2021.782775. [DOI] [PMC free article] [PubMed] [Google Scholar]