Abstract
Unlike other subclasses of the Retroviridae the Spumavirinae, its prototype member being the so-called human foamy virus (HFV), require the expression of the envelope (Env) glycoprotein for viral particle egress. Both the murine leukemia virus (MuLV) Env and the vesicular stomatitis virus G protein, which efficiently pseudotype other retrovirus capsids, were not able to support export of HFV particles. Analysis of deletion and point mutants of the HFV Env protein revealed that the HFV Env cytoplasmic domain (CyD) is dispensable for HFV particle envelopment, release, and infectivity, whereas deletion of the membrane-spanning-domain (MSD) led to an accumulation of naked capsids in the cytoplasm. Neither alternative membrane association of HFV Env deletion mutants lacking the MSD and CyD via phosphoglycolipid anchor nor domain swapping mutants, with the MSD or CyD of MuLV Env and VSV-G exchanged against the corresponding HFV domains, could restore particle envelopment and the release defect of pseudotypes. However, replacement of the HFV MSD with that of MuLV led to budding of HFV capsids at the intracellular membranes. These virions were of apparently wild-type morphology but were not naturally released into the supernatant and they were noninfectious.
Recent studies have shown that one subclass of retroviruses, the foamy viruses (FVs) or spumaviruses, is unique in regard to virus particle assembly and egress (1, 6). FVs resemble in their capsid assembly strategy those of the type B and D retroviruses which preform capsids (A-type particles) in the cytoplasm of the infected cells before the viral particle buds across cellular membranes (3, 27). Most retroviruses, such as the murine leukemia viruses (MuLVs), the lentiviruses, or the type B and D retroviruses require only the cellular expression of the capsid protein (Gag) for the assembly of viral core structures, their enclosure by lipid membranes of the host cell, and the release of virus-like particles from the expressing cell into the supernatant (reviewed in references 3 and 27). However, unlike all other retroviruses, FVs require the coexpression of the Env protein for viral particle egress (1, 6). Budding occurs into intracellular compartments, presumably the endoplasmic reticulum (ER), but it also takes place at the cell surface (1, 6). In cells transfected with Env-deficient FV proviral clones, no viral particle release could be detected. Instead, an accumulation of naked capsids in the cytoplasm of the cell was observed (6).
Two forms of the human FV (HFV) envelope protein have been described to date (9, 10, 18, 19). The predominant form is expressed as a 130-kDa precursor glycoprotein that is cleaved by a cellular protease into an 80- to 90-kDa surface subunit (SU subunit) and a 47-kDa transmembrane subunit (TM subunit) during its transport to the cell surface and incorporation into the viral particle (11). However, due to a retrieval signal present in the cytoplasmic domain, most of the 130-kDa HFV Env protein is retained in the ER either in the absence of the expression of other HFV structural genes or in the absence of inactivation of the ER retrieval signal (11, 12). The second form is a 170-kDa Env-Bet fusion protein (9, 18). Env-Bet is not essential for particle release and in vitro replication of HFV (18).
The unusual mechanism of FV assembly raised the question as to whether a specific Env protein has to be present to allow budding of FV capsids or whether these capsids can be pseudotyped by other viral glycoproteins. Pseudotyping by heterologous Env proteins has been shown for many other retroviruses (8, 27). Furthermore, it has been demonstrated previously that wild-type HFV Env can pseudotype vesicular stomatitis virus (VSV) and murine leukemia virus (MuLV) (17, 25). The pseudotyping efficiency of MuLV capsids could be enhanced by exchanging the cytoplasmic domain (CyD) of HFV Env for that of MuLV Env (17). In this study we sought to analyze the reverse possibility, i.e., the pseudotyping of HFV capsids with foreign viral Env proteins, and to get a better understanding of the processes of HFV particle release.
MATERIALS AND METHODS
Expression constructs.
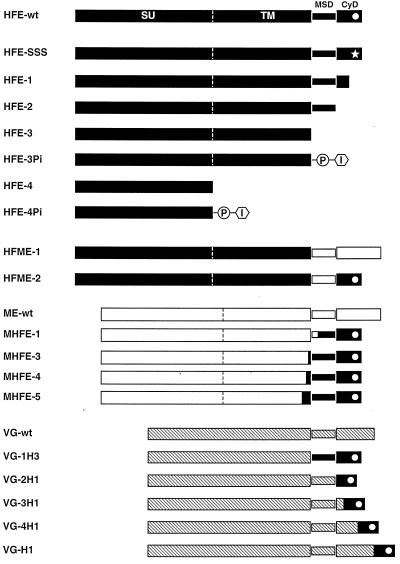
The eukaryotic expression constructs for the different HFV envelope mutants depicted in Fig. 1 are based on the pcHFVenv wild-type plasmid (17) and were generated by standard cloning techniques. All constructs were sequenced to confirm the introduced mutations and to exclude offsite mutations, in particular when using PCR-based techniques. pcHFE-wt contains the complete HFV envelope open reading frame (ORF), starting with the translation start (nucleotide [nt] 5719 relative to the genomic transcription start) up to an EcoRI restriction site (nt 8701) 13 bp downstream of the translation stop, inserted into the expression vector pcDNA3.1/zeo (Invitrogen). This construct, as well as all of the others, includes the EM2 mutation described recently (18). This mutation leads to an inactivation of the splice donor (SD; nt 8530) and splice acceptor (SA; nt 8648) naturally found in the Env ORF and essential for the generation of the Env-Bet fusion protein in the proviral context (18). The amino acid sequences of the various envelope proteins, spanning the C terminus of the extracellular domain, the putative membrane-spanning domain (MSD), and the complete intracellular domain are given in Table 1. The point mutant pcHFE-SSS has the triple lysine motif found near the C terminus, which is responsible for ER retention of the HFV Env protein (12), replaced by a triple serine motif, resulting in an enhanced cell surface expression of the HFV Env protein (11). pcHFE-1, pcHFE-2, pcHFE-3, and pcHFE-4 are constructs for C-terminal deletion mutants leading to the expression of HFV envelope proteins truncated at amino acids (aa) 981, 975, 937, and 571, respectively. Some mutants have one or two additional C-terminal amino acid not encoded by the HFV Env ORF due to the cloning strategy employed (see Table 1). pcHFE-3Pi and pcHFE-4Pi are chimeric envelope proteins based on the deletion mutants pcHFE-3 and pcHFE-4, respectively, which have the signal sequences for a glycophospholipid (GPI) anchor of the human placental alkaline phosphatase (hPLAP) protein (GenBank accession number M12551; aa 497 to 530), which has been shown to result in a membrane anchorage via GPI of otherwise-secreted proteins (28), fused to the C terminus of the protein. pcHFME-1 has the putative MSD and CyD of the HFV Env protein replaced by those of the ecotropic MuLV Env protein (GenBank accession number J02255; aa 604 to 684), whereas the pcHFME-2 mutant has only the putative HFV MSD replaced by that of the MuLV Env protein (aa 604 to 652). Both constructs contain two additional amino acids at the fusion interface, neither of which is encoded by the original HFV Env or by the original MuLV Env sequence (see Table 1). pcVG-wt has an EcoRI fragment of pSVGL1 (24) containing the VSV glycoprotein G (VSV-G) cDNA inserted into the expression vector pcDNA3.1/zeo. Derivatives thereof are pcVG-1H3, with the MSD and CyD of VSV-G replaced by that of HFV, and pcVG-2H1, pcVG-3H1, pcVG-4H1, and pcVG-H1, with various regions of the VSV-G CyD replaced by that of HFV (Table 1). pHIT123 (pcME-wt) is a human cytomegalovirus immediate-early promoter-directed eukaryotic expression vector for the ecotropic MuLV Env protein described previously (26). Derivatives thereof are pcMHFE-1, pcMHFE-3, pcMHFE-4, and pcMHFE-5 lacking the MuLV MSD and CyD and containing various regions of putative HFV MSD and CyD (Table 1). The replication-deficient HFV vector pMH62 expressing the enhanced green fluorescent protein (EGFP) marker gene (Clontech) from an internal spleen focus-forming virus U3 promoter is structurally identical to the HFV vector pMH5 that has been described previously (13).
FIG. 1.
Schematic illustration of the envelope expression constructs. The extracellular domains, MSDs and CyDs of the different envelope proteins are indicated according to previous studies (7, 20, 21). HFV, solid boxes; MuLV, open boxes; VSV-G, cross-hatched boxes. The C-terminal amino acid sequences are given in Table 1. The sequence motif in the cytoplasmic domain of the HFV envelope, which is responsible for ER retention (11), is indicated here by a white dot; the mutated sequence is indicated by a star. The signal sequences for the hPLAP GPI anchor is indicated by PI. SU, surface subunit; TM, transmembrane subunit.
TABLE 1.
Carboxyl amino acid sequences of the individual Env proteins
| Construct | Amino acid sequencea
|
||
|---|---|---|---|
| Ectodomainb | Transmembrane anchor | CyD | |
| 935 | 939 | 976 | |
| HFE-wt | GIGN | FLSGTAQGIFGTAFSLLGYLKPILIGVGVILLVILIF | KIVSWIPTKKKNQ |
| HFE-SSS | GIGN | FLSGTAQGIFGTAFSLLGYLKPILIGVGVILLVILIF | KIVSWIPTSSSNQ |
| HFE-1 | GIGN | FLSGTAQGIFGTAFSLLGYLKPILIGVGVILLVILIF | KIVSWIR |
| HFE-2 | GIGN | FLSGTAQGIFGTAFSLLGYLKPILIGVGVILLVILIF | |
| HFE-3 | GIGS | ||
| 497 | 502 | ||
| HFE-3Pi | GIGSAAGTTD | AAHPGRSVVPALLPLLAGTLLLLETATAP | |
| 570 | |||
| HFE-4 | RRGS | ||
| HFE-4Pi | RRGSAAGTTD | AAHPGRSVVPALLPLLAGTLLLLETATAP | |
| 935 | |||
| HFME-1 | GIGKS | SPWFTTLISTIMGPLIVLLLILLFGPCILN | RLVQFVKDRISVVQALVLTQQYHQLKPIEYEP |
| HFME-2 | GIGKS | SPWFTTLISTIMGPLIVLLLILLFGPCILN | RKIVSWIPTKKKNQ |
| 601 | 604 | 653 | |
| ME-wt | FNR | SPWFTTLISTIMGPLIVLLLILLFGPCILN | RLVQFVKDRISVVOALVLTOOYHOLKPIEYER |
| 947 | |||
| MHFE-1 | FNR | SPWELIFGTAFSLIGYLKPILIGVGVILLVILIP | KIVSWIPTKKKNQ |
| 927 | |||
| MHFE-3 | FNRSPWEPAAASALQGIGN | FLSGTAQGIFGTAFSLLGYLKPILIGVGVILLVILIF | KIVSWIPTKKKNQ |
| 918 | |||
| MHFE-4 | FNRSPWELAAATKDVWPAAASALQGIGN | FLSGTAQGIFGTAFSLLGYLKPILIGVGVILLVILIF | KIVSWIPTKKKNQ |
| 900 | |||
| MHFE-5 | FNRSPWELLRLDIHEGDTPAWIQQLAAATKDVWPAAASALQGIGN | FLSGTAQGIFGTAFSLLGYLKPILIGVGVILLVILIF | KIVSWIPTKKKNQ |
| 457 | 463 | 483 | |
| VG-wt | WFSSWK | SSIASFFFIIGLIIGLFLVL | RVGIHLCIKLKHTKKROITYTKIEMNRLGK |
| VG-1H3 | WFSSWK | SSGTQGIFGTAFSLLGYLKPILIGVGVILLVILIF | KIVSWIPTKKKNQ |
| VG-2H1 | WFSSWK | SSIASFFFIIGLIIGLFLVL | WIPTKKKNQ |
| VG-3H1 | WFSSWK | SSIASFFFIIGLIIGLFLVL | RVGIHLCIKLWIPTKKKNQ |
| VG-4H1 | WFSSWK | SSIASFFFIIGLIIGLFLVL | RVGIHLCIKLKHTKKRQIWQIPTKKKNQ |
| VG-H1 | WFSSWK | SSIASFFFIIGLIIGLFLVL | RVGIHLCIKLKHTKKRQIYTDIEMNRLGKWIPTKKKNQ |
Generation of recombinant HFV supernatants.
Viral supernatants containing the different recombinant viruses were generated by cotransfection of 293T cells (4) with the HFV vector pMH62 and the respective HFV envelope expression construct essentially as described earlier (6, 17, 18).
Viral infectivity assay.
The infectivity of supernatants of 293T cells cotransfected with the pMH62 HFV vector and the different HFV envelope mutants was analyzed essentially as described previously (13, 17). Cell populations transduced by pMH62 complemented with wild-type HFV Env contained 35 to 50% of the cells expressing the EGFP protein. Transduced cell populations expressing EGFP of as low as 0.2% of the total cells could be reliably detected by this assay.
Radioimmunoprecipitation assay (RIPA).
Transiently transfected 293T cells were metabolically labeled with [35S]methionine and [35S]cysteine for approximately 20 h. At 36 h after the addition of the DNA, the cells were lysed in RIPA buffer (20 mM Tris, pH 7.4; 0.3 M NaCl, 1% [wt/vol] sodium deoxycholate; 1% [vol/vol] Triton X-100; 0.1% [wt/vol] sodium dodecyl sulfate [SDS]) containing protease inhibitors. Viral proteins were precipitated as described earlier (6, 18) using HFV-positive chimpanzee sera (18), rabbit antisera generated against recombinant HFV proteins and specific for SU (16), Env (16) and Gag (2), a rabbit serum specific for VSV-G (a gift of P. Clapham and R. Weiss), or hybridoma supernatants recognizing the SU (ATCC CRL1913) or TM (ATCC CRL1893) subunits of the MuLV envelope protein. Particle-associated proteins were detected after centrifugation through a 20% sucrose cushion as described previously (6, 18).
Pulse-chase analysis and Endo H digestion.
At 36 h posttransfection, 293T cells were washed once with methionine- and cysteine-free medium (labeling medium) and starved in labeling medium for 1 h. Subsequently, the cells were metabolically pulse-labeled for 1 h in labeling medium containing 100 μCi of [35S]methionine and [35S]cysteine per ml and chased for various time periods in normal growth medium containing a 10-fold excess of methionine and cysteine. Cell lysis and radioimmunoprecipitation with a rabbit serum specific for HFV SU was performed as described above. The protein A-Sepharose pellet containing the immunoprecipitates was boiled for 2 min in 5× endoglycosidase H (Endo H) buffer (0.1 M sodium phosphate, pH 6.5; 0.5% SDS; 0.1% sodium azide) and subsequently diluted with 4 volumes of water. The supernatant was divided in two equal aliquots, and 4.5 × 10−3 units of Endo H (ICN) was added to one aliquot. After incubation at 37°C for 11 to 18 h, both aliquots were precipitated overnight at −70°C by the addition of glycogen and ethanol. The precipitate was collected by centrifugation, dried, solubilized in gel sample buffer, and then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) as described above.
Electron microscopy.
At 48 h after transfection, the 293T cells were harvested and processed for electron microscopy analysis as described previously (15).
RESULTS
Analysis of heterologous viral envelope proteins for their ability to pseudotype HFV capsids.
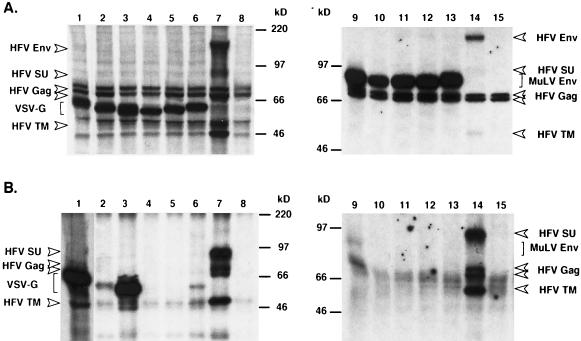
It has long been known that phenotypically mixed viruses, termed pseudotypes, can be observed in cells infected by two different viruses (reviewed in references 14 and 27). Pseudotyping is based on the incorporation of the envelope protein of one virus into viral particles of the other virus, thereby eventually changing the tropism of a given virus. Retroviruses have been shown to incorporate envelope proteins from foreign retroviruses and even those from other classes of viruses, such as VSV or influenza virus (8, 27). We were interested to see whether coexpression of foreign viral Env proteins, together with HFV Gag, would result in the particle incorporation of these glycoproteins and could overcome the block in capsid membrane envelopment and particle release of envelope-deficient HFV. For this purpose, we cotransfected either the Env protein of the ecotropic MuLV or the VSV-G together with the replication-deficient HFV vector pMH62, expressing the EGFP protein from an internal promoter, into 293T cells and analyzed the intracellular protein expression and the release of HFV particles into the supernatant. No HFV proteins could be detected in the supernatant (Fig. 2B, lanes 1 and 9), despite high intracellular expression levels of the foreign envelope proteins and HFV Gag (Fig. 2A, lanes 1 and 9). In line with this a transfer of the EGFP marker gene to susceptible target cells was never observed (Table 2). In the case of cotransfection of the wild-type MuLV Env, faint bands corresponding in size to that of the HFV Gag proteins and the MuLV Env were occasionally observed in the supernatant pellet (Fig. 2B, lane 9). However, electron microscopy analysis of these cells revealed no signs of virus budding, although extracellular aggregates of naked HFV capsids were occasionally detectable (data not shown). Coexpression of the wild-type VSV-G protein lead to the appearance of particulate structures in the supernatant that could be pelleted through 20% sucrose that contained VSV-G but not HFV proteins (Fig. 2B, lane 1). However, this also occurred when expressing only the wild-type VSV-G protein in 293T cells (data not shown) and has been reported in the literature (23). In contrast, cells coexpressing the wild-type HFV Env and pMH62 (Fig. 2A, lanes 7 and 14) after transfection, secreted particle-associated HFV Gag and Env proteins (SU and TM subunits) into the supernatant (Fig. 2B, lanes 7 and 14), whereas cotransfection of the empty expression vector together with pMH62 (Fig. 2A, lanes 8 and 15) resulted in a block of the secretion of HFV capsids into the supernatant (Fig. 2B, lanes 8 and 15). These results indicated that HFV capsids cannot be pseudotyped with the wild-type ecotropic MuLV Env or the wild-type VSV-G protein, despite the fact that these Env proteins have been shown to be incorporated well in other retroviral particles (22, 29).
FIG. 2.
Expression analysis of various MuLV Env and VSV-G proteins and HFV particle release. 293T cells were cotransfected with pMH62 and the individual Env expression constructs depicted in Fig. 1 and were metabolically labeled. (A) Cellular lysates were precipitated with a mixture of antisera against HFV Env, HFV Gag, and VSV-G (lanes 1 to 8) or MuLV (lanes 9 to 15). (B) Virus particles secreted into the supernatant were pelleted by centrifugation through 20% sucrose and resuspended in sample buffer, and proteins were separated by SDS-PAGE. (Lane 1 was exposed for a five-fold shorter period than the other lanes). All samples were cotransfected with pMH62 and the following expression vectors: lane 1, pcVG-wt; lane 2, pcVG-1H3; lane 3, pcVG-2H1; lane 4, pcVG-3H1; lane 5, pcVG-4H1; lane 6, pcVG-H1; lane 7, pcHFE-wt; lane 8, pcDNA3.1+zeo; lane 9, pcME-wt; lane 10, pcMHFE-1; lane 11, pcMHFE-3; lane 12, pcMHFE-4; lane 13, pcMHFE-5; lane 14, pcHFE-wt; and lane 15, pCDNA3.1/zeo.
TABLE 2.
Relative transduction efficiencies of the pMH62 HFV vector cotransfected with different HFV envelope expression constructs
| Envelope protein | Relative infectivity (% of wild type) | No. of expts |
|---|---|---|
| HFE-wt | 100 | 14 |
| HFF-SSS | 63 ± 20 | 14 |
| HFE-1 | 54 ± 31 | 14 |
| HFE-2 | 53 ± 31 | 14 |
| HFE-3 | <0.5 | 8 |
| HFE-3Pi | <0.5 | 8 |
| HFE-4 | <0.5 | 5 |
| HFE-4Pi | <0.5 | 5 |
| HFME-1 | <0.5 | 4 |
| HFME-2 | <0.5 | 3 |
| ME-wt | <0.5 | 5 |
| MHFE-1 | <0.5 | 4 |
| MHFE-3 | <0.5 | 2 |
| MHFE-4 | <0.5 | 2 |
| MHFE-5 | <0.5 | 2 |
| VG-wt | <0.5 | 4 |
| VG-1H3 | <0.5 | 2 |
| VG-2H1 | <0.5 | 2 |
| VG-3H1 | <0.5 | 2 |
| VG-4H1 | <0.5 | 2 |
| VG-H1 | <0.5 | 2 |
Therefore, several chimeric envelope proteins containing various portions of the HFV MSD and CyD were generated and examined in regard to the ability to support HFV particle egress. The analysis of particulate material in supernatants of 293T cells cotransfected with pMH62 and various VSV-G chimeras containing both the MSD and CyD (VG-1H3) or only the CyD of HFV (VG-2H1, VG-3H1, VG-4H1, and VG-H1) (see Fig. 1 and Table 1) revealed that none of them was able to restore HFV particle release (Fig. 2B, lanes 2 to 6) despite high intracellular expression levels (Fig. 2A, lanes 2 to 6). We extended our analysis by generating several chimeric envelope proteins containing the extracellular domains of the ecotropic MuLV and various C-terminal portions of the HFV Env. Cotransfection of none of these mutants together with the pMH62 vector permitted the release of HFV particles into the supernatant (Fig. 2A, lanes 10 to 13), even those chimeras that contained, in addition to the HFV MSD and CyD, further sequences of the extracellular domain of the HFV TM subunit. Furthermore, cotransfection of none of the VSV-G and MuLV Env chimeras resulted in a transfer of the marker gene of the pMH62 HFV vector to susceptible target cells (Table 2). Taken together, these results indicated that the FV MSD and CyD regions tested are not sufficient to confer FV particle release in the context of fusion proteins containing extracellular domains of foreign envelope proteins.
C-terminal deletion and mutagenesis analysis of the HFV envelope protein.
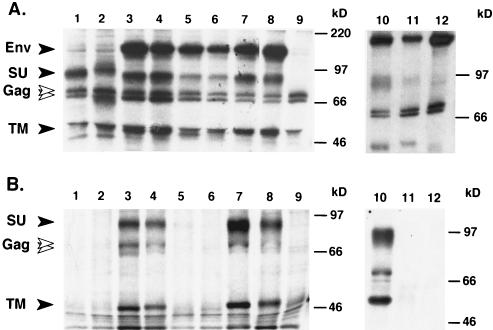
To determine which parts of the HFV envelope protein are required for capsid envelopment, particle release, and infectivity, several C-terminal truncation mutants and point mutants of the HFV Env protein were generated (Fig. 1 and Table 1). Intracellular viral protein expression was monitored by RIPA with a chimpanzee serum recognizing all major HFV proteins (18) (Fig. 3A). The protein composition of metabolically labeled HFV particles released into the supernatant was analyzed by SDS-PAGE after purification through a 20% sucrose cushion (Fig. 3B). This analysis showed that all HFV envelope mutants were expressed at roughly comparable levels intracellularly (Fig. 3A). However, cells expressing the HFE-3 (Fig. 3B, lane 5) or the HFE-4 (Fig. 3B, lane 1) envelope protein and cells cotransfected with the empty expression vector (Fig. 3B, lane 9) failed to release detectable amounts of particle-associated viral proteins into the supernatants. In contrast, in supernatants of cells coexpressing the HFE-wt (Fig. 3B, lanes 8 and 10), HFE-SSS (Fig. 3B, lane 7), HFE-1 (Fig. 3B, lane 4), or HFE-2 (Fig. 3B, lane 3) envelope proteins viral-particle-associated Gag proteins, as well as the respective envelope SU and TM subunits, were readily detectable.
FIG. 3.
Expression analysis of HFV Env mutants and chimeras. 293T cells were cotransfected with pMH62 and the individual Env expression constructs depicted in Fig. 1 and were metabolically labeled. (A) Cellular lysates were precipitated with a chimpanzee serum recognizing all major HFV proteins. (B) Virus particles secreted into the supernatant were pelleted by centrifugation through 20% sucrose and resuspended in sample buffer, and the proteins were separated by SDS-PAGE. Lanes 1, pcHFE-4; 2, pcHFE-4Pi; 3, pcHFE-2; 4, pcHFE-1; 5, pcHFE-3; 6, pcHFE-3Pi; 7, pcHFE-SSS; 8, pcHFE-wt; 9, mock, pcDNA3.1/zeo; 10, pcHFE-wt; 11, pcHFME-1; 12, pcHFME-2.
We also examined the infectivity of the different viral particles by incubating NIH 3T3 cells with cell-free supernatants of the transfected 293T cells and determined the transduction efficiency by measuring the percentage of EGFP-expressing cells. As can been seen in Table 2, only supernatants that contained particle-associated viral proteins, as determined above, were able to transduce target cells. The relative transduction efficiency of the different infectious mutant viruses (HFE-1 and HFE-2) was, at maximum, two- to four-fold lower than that of wild type. Taken together, these results showed that the MSD of the HFV envelope protein, but not the CyD, is essential for the release of viral particles. Furthermore, these results indicated that the short CyD of the HFV envelope protein has only a minor influence on the infectivity of the HFV particle.
Alternative membrane anchorage of the HFV Env protein.
The results presented above indicated that membrane anchorage of the HFV envelope protein via the HFV MSD domain is essential for viral particle release. This raised the question of whether the MSD is involved specifically in this process of whether simply membrane anchorage of the HFV envelope extracellular domains is required. To test this, additional envelope expression constructs (as shown in Fig. 1 and Table 1) were analyzed. All chimeric proteins were expressed intracellularly, although the HFE-3Pi (Fig. 3A, lane 6) and HFE-4Pi (Fig. 3A, lane 2) proteins were expressed at a somewhat lower level than the other proteins. Cell surface anchorage of these mutants was confirmed by cell surface biotinylation (data not shown). Interestingly, whereas all other HFV envelope proteins, including the HFME-1 chimera, displayed a normal SU-TM cleavage (Fig. 3A, lane 11), the HFME-2 mutant clearly showed an accumulation of envelope precursor protein and almost no cleavage products (Fig. 3A, lane 12). Analysis of viral-particle-associated proteins (Fig. 3B) and measurement of infectivity (Table 2) in the supernatant showed that none of these additional mutant envelope proteins supported HFV particle release. Furthermore, the cell-associated infectivity of all mutants was determined after transfected cells were subjected to a freeze-thaw cycle to release intracellular trapped FV particles and the supernatant was passed the supernatant through a 0.45 μm (pore size) filter. This analysis gave results similar to that for the supernatants, although the infectivity of all of the infectious mutants was increased in general by a factor of three to five (data not shown). These results indicated that the HFV MSD cannot be substituted by the corresponding domain of another retroviral envelope protein or by other forms of membrane anchorage, such as a glycophospholipid, and that it may be specifically involved in the particle release process.
Pulse-chase analysis of Env expression.
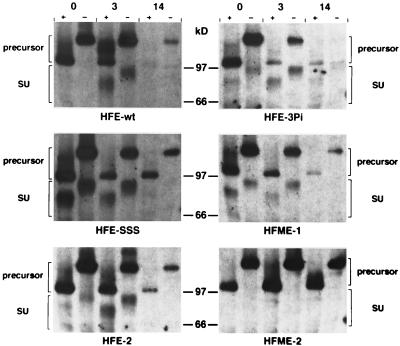
One possible explanation for the failure of some of the HFV Env mutants to facilitate secretion of FV particles upon Gag coexpression could be a decreased stability or changes in the intracellular transport of these proteins. To address these questions, a pulse-chase analysis combined with digestion by Endo H of the mutants was performed. An example of this analysis for some of the mutants is shown in Fig. 4. In this assay the HFE-SSS, HFE-2, and HFME-1 mutants had a protein stability similar to that of the wild-type Env, whereas the HFE-3Pi, HFE-4, and HFE-4Pi mutants seemed to be degraded somewhat faster (Fig. 4 and data not shown). In contrast to wild-type Env, all of these mutants showed a faster processing of the gp130 into SU and TM subunits. This result was most prominent for the HFE-SSS protein, where large amounts of SU could be detected already during the 1-h labeling period (Fig. 4). Furthermore, for most mutants Endo H-resistant forms of gp130 were only weakly detectable after the pulsing period (see HFE-SSS, 0-h time point). In contrast, in wild-type HFV Env-expressing cells, Endo H-resistant forms of gp130 were more prominent but were first detectable in the 3-h chase period. A possible explanation for this could be a reshuttling of Endo H-resistant wild-type gp130 to the ER before cleavage into the subunits occurred, as a result of the cytoplasmic ER retrieval signal, which is missing in all of the other mutants. The HFME-2 protein differed completely from all other mutants, since its gp130 precursor was much more stable (compare the 14-h time points in Fig. 4). Furthermore, no cleavage of HFME-2 gp130 into SU and TM subunits or Endo H-resistant forms of gp130 could be observed (Fig. 4), indicating a block in the intracellular trafficking of this mutant. Similar results were observed in cells coexpressing HFV Gag in addition to the individual Env proteins (data not shown).
FIG. 4.
Pulse-chase analysis of various HFV env mutants. 293T cells were transfected with the individual HFV Env expression constructs as indicated. The pulse-chase analysis was performed as described in Materials and Methods. The time periods (in hours) of the chase are indicated at the top. +, Incubated with Endo H; −, mock incubated.
Electron microscopy analysis of viral particle maturation.
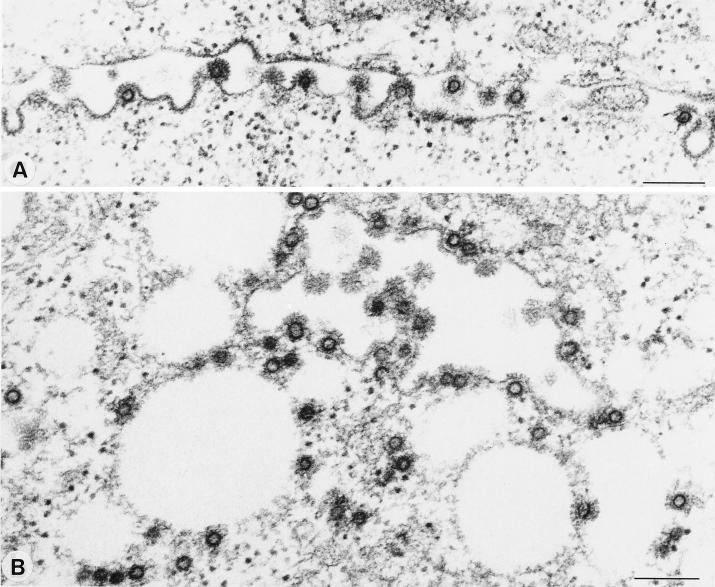
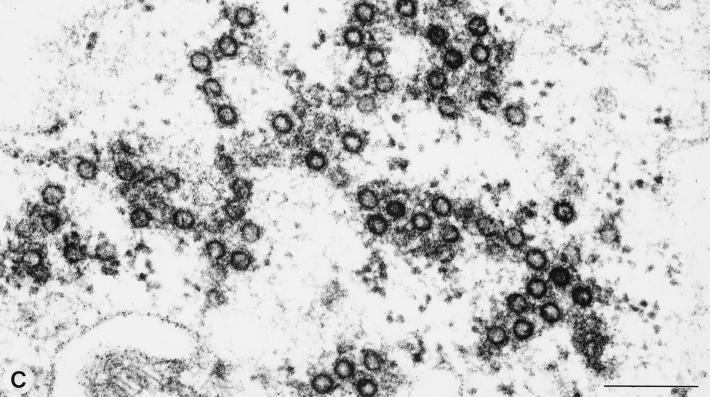
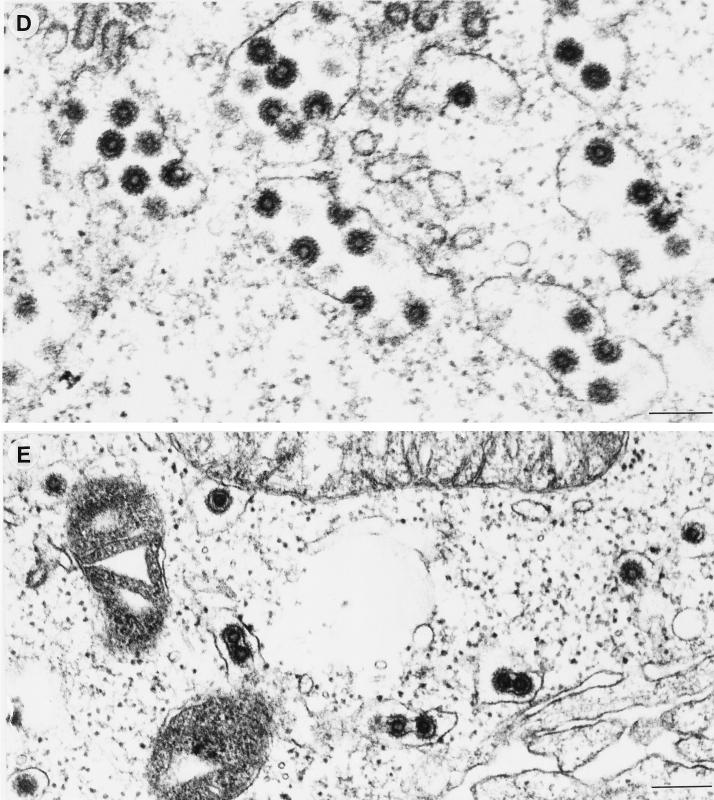
Whereas for all other subgroups of retroviruses the Gag expression is sufficient for the membrane envelopment and release of viral capsids, FVs require the coexpression of the FV Env protein for this process. To distinguish the two steps of membrane envelopment and viral particle release, we employed electron microscopy analysis of 293T cells cotransfected with the pMH62 HFV retroviral vector and different envelope mutants. The analysis of cells cotransfected with the pMH62 vector and the wild-type HFV envelope expression construct pcHFE-wt revealed the release of enveloped HFV capsids at extracellular membranes (Fig. 5A) and into intracellular compartments (Fig. 5B) as described previously (1, 6). The individual HFV envelope mutants examined could be grouped into three different classes according to their phenotype in the electron microscopy analysis. The first class comprises all envelope mutants that upon cotransfection with pMH62 lead to the appearance of infectious HFV particles in the supernatant (HFE-SSS, HFE-1, and HFE-2), as determined earlier. They showed a phenotype indistinguishable from that of the HFE-wt described above (data not shown). In contrast, the second class showed a phenotype identical to that of cells that were cotransfected with pMH62 and a control plasmid, resulting in the intracellular accumulation of naked HFV capsids. To this class belong the secreted mutants HFE-3 and HFE-4, as well as their PI membrane-anchored forms HFE-3Pi and HFE-4Pi, respectively, and the HFME-1 chimeric envelope containing the MSD and CyD of the ecotropic MuLV envelope. A representative example of the naked capsids seen in HFE-3Pi-expressing cells is shown in Fig. 5C. Interestingly, a third class could be observed, its only member being the HFME-2 chimeric envelope with the HFV MSD replaced by that of the ecotropic MuLV Env. The unique phenotype of this mutant was that no budding of viral particles at the extracellular membrane could be observed, whereas budding into intracellular compartments was readily detectable (Fig. 5D). These enveloped particles also contained spike-like structures on their lipid membrane (Fig. 5D and E), most probably representing envelope protein multimeric structures, similar to those observed on wild-type particles (Fig. 5A+B). Another unique feature of this mutant was the appearance of small membrane-enclosed vesicles containing single enveloped particles (Fig. 5E). This was not observed in cells cotransfected with any other HFV envelope mutant, including the wild-type protein. Taken together, these observations supported the results of the biochemical analysis presented above indicating that the HFV MSD is essential for HFV particle release. However, the requirement of the HFV MSD for capsid envelopment can be complemented by that of the MuLV Env, at least when the HFV CyD is still present.
FIG. 5.
Electron micrographs showing representative thin sections of transiently transfected 293T cells. Cells cotransfected with the wild-type HFV env showed budding at the plasma membrane (A) and into intracellular compartments (B). (C) An accumulation of naked intracellular HFV capsids was observed in cells cotransfected with the HFE-3Pi mutant. (D) In cells coexpressing the HFME-2 Env protein, only budding of capsids into intracellular compartments could be detected. (E) A unique feature of these cells was the appearance of vesicles containing single enveloped virus particles. Magnifications: A and D ×63,400; B, ×69,500; C, ×99,400; E, ×60,900. Bar size, 200 nm.
DISCUSSION
Most retroviruses require only the expression of the Gag protein for the assembly of capsid structures, their membrane envelopment, and the release of viral particles into the supernatant (reviewed in reference 27). FVs are unique in regard to these steps, since particle egress is dependent on the coexpression of the gp130 Env protein (1, 6). Using C-terminal envelope deletion mutants, we have shown that the CyD of gp130 containing an ER retrieval signal is dispensable but that membrane anchorage by the HFV MSD is essential for these events in FV particle maturation. The HFV MSD seems to be specifically involved in this process since cells expressing chimeric envelope mutants that were alternatively membrane anchored, by using a GPI moiety or the MSD and CyD of a foreign retroviral envelope, failed to release HFV particles into the supernatant. However, from our experiments it is not clear whether the HFV MSD participates at the amino acid or the structural level. Furthermore, structural changes of the deletion or chimeric HFV envelope mutants could potentially account for their inability to support HFV particle egress, although no major differences in precursor processing compared to the still infectious mutants was observed. Interestingly, we have identified one envelope chimera, HFME-2, with the HFV MSD replaced by the corresponding domain of the MuLV Env, that still showed efficient capsid membrane envelopment and envelope incorporation at the intracellular compartments. However, this mutant, like all of the others lacking the HFV MSD, failed to support HFV particle release into the supernatant. Therefore, it is tempting to speculate that for the first step, i.e., the envelopment of the capsid structure by the lipid bilayer, only certain structural requirements are necessary that can be complemented by the MSD of a foreign Env protein in the context of gp130, whereas in the second step, i.e., the transport of an enveloped HFV particle to the cell surface and its release into the supernatant, a specific amino acid sequence motif of the HFV MSD may be involved. The small vesicles in the cytoplasm containing single, membrane-enveloped HFV particles with spike-like structures, which were observed with the HFME-2 mutant, may represent HFV particles blocked at some point of their export from the cell. A transport block of this particular mutant was also suggested by the pulse-chase Endo H analysis.
In addition, we found that neither the wild-type ecotropic MuLV envelope nor the wild-type VSV-G, which have been shown previously to efficiently pseudotype other retroviral capsids (22, 29), was able to overcome the block in particle release of Env-deficient HFV vectors. This indicates that the HFV gp130 contains unique information required for the HFV budding process. Furthermore, different chimeric envelope proteins containing the extracellular domains of MuLV or VSV and the HFV MSD and/or CyD of various lengths were also unable to restore HFV budding into the supernatant when cotransfected with an HFV vector. These results imply that the HFV MSD and CyD, although essential, are not sufficient to support HFV particle egress, at least to the extent analyzed. Furthermore, they suggest that additional extracellular domains of gp130 are required for these steps in HFV maturation. A factor that might have an influence on this is the oligomeric state of the HFV Env proteins. For some retroviral Env proteins, such as the MuLV Env or the Rous sarcoma virus Env, it has been shown that they form trimers and that sequences within the extracellular domain of the TM subunit are required for oligomerization (5). However, until now nothing has been known about the oligomeric structure of HFV gp130 or which regions of the HFV Env are required for proper oligomerization. It is possible that the chimeric Env proteins do not oligomerize correctly and therefore fail to support HFV particle egress. Experiments are in progress to address this question.
Baldwin and Linial (1) have reported the occurrence of occasional membrane enveloped or budding HFV capsids in BHK-21-derived FAB cells transfected with proviral clones expressing a truncated Env protein. Using 293T cells transfected with comparable Env-deficient constructs, we were unable to detect such HFV particles in previous experiments, despite extensive examination (6). In addition, in the current study with Env-negative HFV vectors alone or together with various Env mutants defective in supporting HFV particle release into the supernatant, we were again unable to detect HFV capsid membrane envelopment or intracellular budding. The only exception was the HFME-2 Env, although in samples cotransfected with this mutant numerous membrane-enveloped and intracellularly budding HFV particles could be observed (Fig. 5D and E). Furthermore, these particles had a morphology similar to that of wild-type virus, including the prominent surface spike structures.
ACKNOWLEDGMENTS
We thank P. Clapham and R. Weiss for the VSV-G-specific antiserum, Birgit Hub for excellent technical assistance, and Ulrike Ackermann for photographic work.
This work was supported by the EU (BMH4-CT97-2010), Bayerische Forschungsstiftung, and DFG (SFB 165 and Li-621/2-1). D.L. is supported by the virology fellowship program of the BMBF, Bonn, Germany.
REFERENCES
- 1.Baldwin D N, Linial M L. The roles of Pol and Env in the assembly pathway of human foamy virus. J Virol. 1998;72:3658–3665. doi: 10.1128/jvi.72.5.3658-3665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baunach G, Maurer B, Hahn H, Kranz M, Rethwilm A. Functional analysis of human foamy virus accessory reading frames. J Virol. 1993;67:5411–5418. doi: 10.1128/jvi.67.9.5411-5418.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boulanger P, Jones I. Use of heterologous expression systems to study retroviral morphogenesis. Curr Top Microbiol Immunol. 1996;214:237–260. doi: 10.1007/978-3-642-80145-7_8. [DOI] [PubMed] [Google Scholar]
- 4.DuBridge R B, Tang P, Hsia H C, Leong P-M, Miller J H, Calos M P. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Einfeld D. Maturation and assembly of retroviral glycoproteins. Curr Top Microbiol Immunol. 1996;214:133–176. doi: 10.1007/978-3-642-80145-7_5. [DOI] [PubMed] [Google Scholar]
- 6.Fischer N, Heinkelein M, Lindemann D, Enssle J, Baum C, Werder E, Zentgraf H, Müller J G, Rethwilm A. Foamy virus particle formation. J Virol. 1998;72:1610–1615. doi: 10.1128/jvi.72.2.1610-1615.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flügel R M, Rethwilm A, Maurer B, Darai G. Nucleotide sequence analysis of the env gene and its flanking regions of the human spumaretrovirus reveals two novel genes. EMBO J. 1987;6:2077–2084. doi: 10.1002/j.1460-2075.1987.tb02473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedmann T, Yee J K. Pseudotyped retroviral vectors for studies of human gene therapy. Nat Med. 1995;1:275–277. doi: 10.1038/nm0395-275. [DOI] [PubMed] [Google Scholar]
- 9.Giron M L, de The H, Saib A. An evolutionarily conserved splice generates a secreted Env-Bet fusion protein during human foamy virus infection. J Virol. 1998;72:4906–4910. doi: 10.1128/jvi.72.6.4906-4910.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giron M-L, Rozain F, Debons-Guillemin M-C, Canivet M, Peries J, Emanoil-Ravier R. Human foamy virus polypeptides: identification of env and bel gene products. J Virol. 1993;67:3596–3600. doi: 10.1128/jvi.67.6.3596-3600.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goepfert P A, Shaw K L, Ritter G D, Jr, Mulligan M J. A sorting motif localizes the foamy virus glycoprotein to the endoplasmic reticulum. J Virol. 1997;71:778–784. doi: 10.1128/jvi.71.1.778-784.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goepfert P A, Wang G, Mulligan M J. Identification of an ER retrieval signal in a retroviral glycoprotein. Cell. 1995;82:543–544. doi: 10.1016/0092-8674(95)90026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinkelein M, Schmidt M, Fischer N, Moebes A, Lindemann D, Enssle J, Rethwilm A. Characterization of a cis-acting sequence in the pol region required to transfer human foamy virus vectors. J Virol. 1998;72:6307–6314. doi: 10.1128/jvi.72.8.6307-6314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter E. Viral entry and receptors. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 71–119. [PubMed] [Google Scholar]
- 15.Kräusslich H G, Fäcke M, Heuser A M, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehrmann, H., D. Lindemann, S. Bräutigam, and A. Rethwilm. Expression of foamy virus envelope proteins by recombinant baculoviruses and generation of protein-specific antisera. Submitted for publication.
- 17.Lindemann D, Bock M, Schweizer M, Rethwilm A. Efficient pseudotyping of murine leukemia virus particles with chimeric human foamy virus envelope proteins. J Virol. 1997;71:4815–4820. doi: 10.1128/jvi.71.6.4815-4820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindemann D, Rethwilm A. Characterization of a human foamy virus 170-kilodalton Env-Bet fusion protein generated by alternative splicing. J Virol. 1998;72:4088–4094. doi: 10.1128/jvi.72.5.4088-4094.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Netzer K O, Rethwilm A, Maurer B, ter Meulen V. Identification of the major immunogenic structural proteins of human foamy virus. J Gen Virol. 1990;71:1237–1241. doi: 10.1099/0022-1317-71-5-1237. [DOI] [PubMed] [Google Scholar]
- 20.Odell D, Wanas E, Yan J, Ghosh H P. Influence of membrane anchoring and cytoplasmic domains on the fusogenic activity of vesicular stomatitis virus glycoprotein G. J Virol. 1997;71:7996–8000. doi: 10.1128/jvi.71.10.7996-8000.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ragheb J A, Anderson W F. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J Virol. 1994;68:3220–3231. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rizvi T A, Panganiban A T. Simian immunodeficiency virus vectors: replication and pseudotyping. J Med Primatol. 1992;21:69–73. [PubMed] [Google Scholar]
- 23.Rolls M M, Webster P, Balba N H, Rose J K. Novel infectious particles generated by expression of the vesicular stomatitis virus glycoprotein from a self-replicating RNA. Cell. 1994;79:497–506. doi: 10.1016/0092-8674(94)90258-5. [DOI] [PubMed] [Google Scholar]
- 24.Rose J K, Bergmann J E. Altered cytoplasmic domains affect intracellular transport of the vesicular stomatitis virus glycoprotein. Cell. 1983;34:513–524. doi: 10.1016/0092-8674(83)90384-7. [DOI] [PubMed] [Google Scholar]
- 25.Schnitzer T J. Simian foamy virus pseudotypes of vesicular stomatitis virus: production and use in sero-epidemiological investigations. J Gen Virol. 1982;59:203–206. doi: 10.1099/0022-1317-59-1-203. [DOI] [PubMed] [Google Scholar]
- 26.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 28.Udenfriend S, Kodukula K. How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu Rev Biochem. 1995;64:563–591. doi: 10.1146/annurev.bi.64.070195.003023. [DOI] [PubMed] [Google Scholar]
- 29.Yee J K, Friedmann T, Burns J C. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 1994;43:99–112. doi: 10.1016/s0091-679x(08)60600-7. [DOI] [PubMed] [Google Scholar]