Abstract
Background
TP53 mutations (TP53MT) occur in diverse genomic configurations. Particularly, biallelic inactivation is associated with poor overall survival in cancer. Lesions affecting only one allele might not be directly leukemogenic, questioning the presence of cryptic biallelic subclones in cases with dismal prognosis.
Methods
We have collected clinical and molecular data of 7400 patients with myeloid neoplasms and applied a novel model by identifying an optimal VAF cutoff using a statistically robust strategy of sampling-based regression on survival data to accurately classify the TP53 allelic configuration and assess prognosis more precisely.
Results
Overall, TP53MT were found in 1010 patients. Following the traditional criteria, 36% of the cases were classified as single hits, while 64% exhibited double hits genomic configuration. Using a newly developed molecular algorithm, we found that 579 (57%) patients had unequivocally biallelic, 239 (24%) likely contained biallelic, and 192 (19%) had most likely monoallelic TP53MT. Interestingly, our method was able to upstage 192 out of 352 (54.5%) traditionally single hit lesions into a probable biallelic category. Such classification was further substantiated by a survival-based model built after re-categorization. Among cases traditionally considered monoallelic, the overall survival of those with probable monoallelic mutations was similar to the one of wild-type patients and was better than that of patients with a biallelic configuration. As a result, patients with certain biallelic hits, regardless of the disease subtype (AML or MDS), had a similar prognosis. Similar results were observed when the model was applied to an external cohort. In addition, single-cell DNA studies unveiled the biallelic nature of previously considered monoallelic cases.
Conclusion
Our novel approach more accurately resolves TP53 genomic configuration and uncovers genetic mosaicism for the use in the clinical setting to improve prognostic evaluation.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13045-023-01480-y.
Keywords: TP53 mutations, Allelic inactivation, Myeloid neoplasia, Next-generation sequencing, Single-cell DNA sequencing
Key Points
Patients with single TP53 mutations and variant allele frequency more than 23% act biologically like biallelic TP53 cases.
Traditionally defined single hits might contain subclones with biallelic inactivation, which negatively influences prognosis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13045-023-01480-y.
Introduction
TP53 is a pivotal tumor suppressor gene (TSG) in cancer, including myeloid neoplasia (MN). TP53 can be affected by hypomorphic/loss-of-function (LOF) lesions occurring in diverse configurations. In addition to truncated (frameshifts and stop codons), missense mutations in various hotspots may exert a dominant-negative effect [1, 2]. Poor prognosis has been attributed to TP53 mutations (TP53MT) and loss of heterozygosity (LOH) by 17p deletion (del(17p)) in MN. In particular, this is true for instances as complex karyotype and presumed chemotherapy-related causation [3, 4]. Considering the many variables of TP53MT topography and configuration, the assessment of the prognostic impact of TP53 lesions may be challenging [3, 5].
Congenital heterozygous TP53MT in Li-Fraumeni syndrome (LFS) are the first hits in the oncogenic cascade preceding the evolution of cancers after biallelic inactivation of this gene, a process consistent with a 2-hit theory of recessive TSG inactivation (Knudson’s hypothesis) [6]. Incomplete penetrance in LFS indicates that the residual function of TP53 is at least partially protective [7]. Monoallelic TP53MT may also occur in clonal hematopoiesis without overt leukemia [8]. Thus, somatic monoallelic lesions might not be directly leukemogenic if not accompanied by subsequent hits in trans configuration or affecting other genes. In early SNP-arrays studies, we have shown that some patients with TP53MT may harbor biallelic inactivation by somatic copy-neutral LOH (CN-LOH) or cryptic macro/microdeletions [9]. Presumed monoallelic somatic TP53MT did not affect survival but biallelic hits did confer an unfavorable prognosis [10, 11]. Furthermore, second TP53 hits represent the most associated lesions in patients with primary TP53MT with usually sweeping biallelic subclones, consistent with their functional and clinical impact [12].
The correct assessment of TP53 inactivation status in patients with TP53MT MN in a clinical setting is complicated, as the distinction between two monoallelic hits in a form of subclonal mosaicism vs. true biallelic lesions is not easily possible. Moreover, cases with seemingly monoallelic TP53MT likely contain cryptic clones with biallelic TP53 inactivation, but their detection is not possible using traditional sequencing methods. Even after estimating the clonality of del(17p) or uniparental disomy (UPD), such TP53 configuration analysis is hampered by essential flaws such as the inability to: i) detect and quantify the biallelic fraction in cases with smaller variant allelic frequency (VAF) or ii) prove the presence of subclonal mosaicisms with two different TP53MT clones using traditional bulk DNA sequencing methods. The accuracy of single-cell sequencing technologies although allowing for such delineation is limited by various technical and feasibility problems. For instance, the use of single-cell DNA analysis is clinically impractical and may not always be conclusive due to allelic dropout and other imprecisions [11]. Similarly, SNP-arrays, while precise, are not sensitive in detecting smaller clones (< 20%) containing deletions or CN-LOH [13]. As a result, small clones possibly constituting a significant portion of occult TP53 biallelic inactivation are likely to be overseen. [9, 11, 12, 14].
Most previous reports focused on myelodysplastic syndrome (MDS), and secondary acute myeloid leukemia (sAML) have shown that TP53MT affect the clinical prognostic scoring [4, 11, 15]. Indeed, in the Molecular International Scoring System (IPSS-M) TP53 lesions are the most impactful on clinical outcomes and the different TP53 genomic configuration constitutes an important variable [16]. Based on the assumption that biallelic rather than single TP53 hits are directly leukemogenic, we hypothesized that clinical outcomes such as survival might be used to differentiate cases with biallelic, often cryptic TP53MT clones (and vice versa). Therefore, we investigated TP53MT in a fashion agnostic to the subtypes of MN by devising a new rational method able to predict the impact of these mutations on prognosis in real-life scenarios to further improve the current clinical algorithms.
Materials and methods
Patient cohort
We have compiled molecular and clinical data of a meta-analytic cohort of 1010 patients with TP53 alterations, along with 6390 TP53WT cases. Data were collected from The Cleveland Clinic, (CC, n = 1357), The Munich Leukemia Laboratory (MLL, n = 1962), and publicly available data sets (Memorial Sloan Kettering Cancer Center [11], The Cancer Genome Atlas (TCGA) [17], The BEAT AML master trial, n = 4081 [18] (Additional file 1: Table S1).
Genetic studies
For the data collected at CC, whole exome sequencing (WES) was performed [19–21] on a subset of samples. Paired tumor and germline DNAs were used for WES. Data were validated using a TruSeq or Nextera platform Custom Amplicon Kit (Illumina, San Diego, CA, USA). The targeted sequencing panel is shown in Additional file 1: Table S2. Variants were annotated using Annovar and filtered using an in-house bioanalytical pipeline [14, 19, 21]. The gene sequencing methods of publicly shared data were previously described [22, 23].
Statistical analyses
To identify the optimal VAF cutoff able to delineate TP53MT allelic status, we utilized a statistically robust strategy of subsampling-based regression on survival data. Specifically, using R package randomForestSRC [24], we built a survival model with VAF as a single covariate and minimum node size set to 15. (Further statistical analyses are described in supplementary methods).
Results
Clinical characteristics of patients’ cohort
We screened a cohort of 7400 patients with MN and found that 1010 (14%) of the patients had 1285 TP53MT (Figs. 1A, Additional file 1: Figure S1, Table S3). The median age at diagnosis was 70 years (IQR 61–77). Low-risk MDS (LR-MDS), high-risk MDS (HR-MDS), sAML, and primary AML (pAML) were present in 34%, 17%, 4%, and 27%, respectively (Table 1 and Additional file 1: Figure S1). For this study, HR-MDS patients were defined by a blast count of ≥ 5%. The majority of TP53MT patients harbored complex karyotype at cytogenetic evaluation (704/981; 72%) (Table 1).
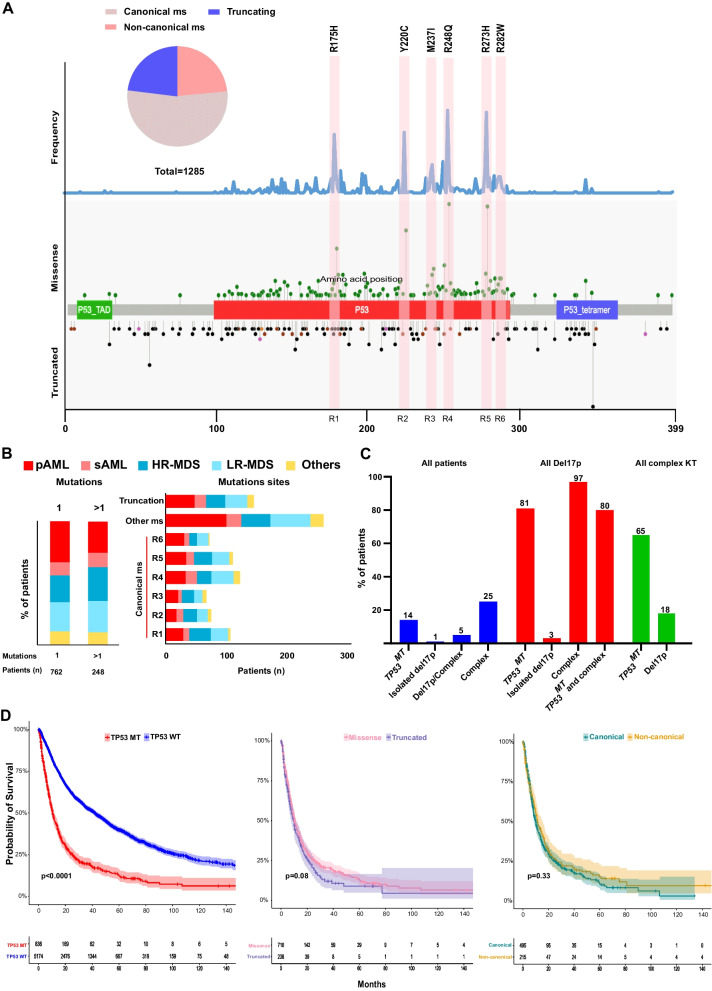
Fig. 1.
Distribution of TP53 mutations with hotspot locations and chromosome 17 deletion. A Schematic drawing of TP53 gene showing the location of mutations, the type of mutations, and canonical sites. Frequency of mutations in all cohort is shown in the upper part. Missense and truncated mutations are indicated in the upper and lower part of the gene structure, respectively. B Bar graphs showing the frequencies of single (1) and multiple TP53 mutations (> 1) and canonical missense locations in each disease subtype. C Percentage of patients with cytogenetics abnormalities in relation to the TP53 mutational status. D Kaplan–Meier survival curves of patients with TP53 mutations vs. wild type, missense vs. truncated mutations, and canonical vs. non-canonical missense mutations
Table 1.
Clinical and cytogenetic characteristics of the study cohort
| Characteristics | All patients N (%) |
TP53MT N (%) |
TP53WT N (%) |
p-value |
|---|---|---|---|---|
| Number of patients | 7400 (100%) | 1010 (14%) | 6390 (86%) | |
| Median age at diagnosis (IQR) | 70.3 (61–77) | 71 (64–77) | 70 (61–77) | 0.001 |
| Male gender (%) | 58% | 53% | 59% | 0.015 |
| Disease subtypes | ||||
| pAML | 1985 (26.8%) | 316 (31.2%) | 1669 (26.1%) | < 0.001 |
| sAML | 272 (3.6%) | 111 (10.9%) | 161 (2.5%) | < 0.001 |
| HR–MDS | 1273 (17.2%) | 234 (23.1%) | 1038 (16.2%) | < 0.001 |
| LR–MDS | 2558 (34.6%) | 244 (24.1%) | 2314 (36.3%) | < 0.001 |
| MDS/MPN | 1312 (17.8%) | 105 (10.4%) | 1208 (18.9%) | < 0.001 |
| Cytogenetic data* | ||||
| Normal | 2084 (47.8%) | 122 (12.4%) | 1967 (58.2%) | < 0.001 |
| Complex | 1083 (24.8%) | 704 (71.7%) | 344 (10.1%) | < 0.001 |
| Deletion 5q | 211 (4.8%) | 95 (9.6%) | 116 (3.4%) | < 0.001 |
| Deletion 7 | 146 (3.3%) | 20 (2.0%) | 126 (3.7%) | 0.009 |
| Deletion 17p | 207 (4.7%) | 167 (17.0%) | 40 (1.1%) | < 0.001 |
| Deletion 20q | 93 (2.1%) | 8 (0.8%) | 85 (2.5%) | 0.001 |
| Trisomy 8 | 252 (5.7%) | 36 (3.6%) | 216 (6.3%) | 0.001 |
| Deletion Y | 84 (1.9%) | 10 (1.0%) | 74 (2.1%) | 0.018 |
| Median Hb (IQR) | 9.7 (8.5–11.1) | 9.1 (8.2–10.2) | 9.8 (8.6–11.2) | < 0.001 |
| Median WBC (IQR) | 6.8 (3.3–23.6) | 5.1 (2.6–16.3) | 7.1 (3.4–24.9) | < 0.001 |
| Median platelet (IQR) | 105 (50–208) | 65 (38–120) | 113 (54–220) | < 0.001 |
*Cytogenetics were available for total of 4,358 patients, of these 981 TP53MT and 3377 TP53WT
MT mutation, WT wild type, IQR interquartile range, pAML primary acute myeloid leukemia, sAML secondary acute myeloid leukemia, HR-MDS high-risk myelodysplastic syndrome, LR-MDS low-risk myelodysplastic syndrome, MDS/MPN myelodysplastic/myeloproliferative overlap neoplasms, MPN myeloproliferative syndrome, CMML chronic myelomonocytic leukemia, Hb hemoglobin, WBC white blood cells
Distribution of TP53 mutations and 17p deletions
Of TP53MT detected, missense mutations were registered in 74% of patients, truncations in 15%, while 11% had concomitant missense and truncated hits (Fig. 1A). Missense mutations were distributed into 6 main sites, including canonical hotspot lesions (defined here as ± 5 amino acids from the most canonical sites, Fig. 1A). Majority of the missense TP53MT (69%) were detected in the canonical sites (R175H, Y220C, M237I, R248Q, R273H, R282W) [1] (Additional file 1: Figure S2). Notably, we did not observe any differences in the number of TP53MT nor in the location of the lesions among various MN subtypes (Fig. 1B). Overall, 203 patients had del(17p), of whom 118 (58%) had missense TP53MT, and 17% had truncated TP53MT, 6% of the patients had concurrent missense and truncated TP53MT, while in 19% no mutation was found. Mutations often coincided with deletions of TP53 locus either as isolated lesions or more often in the context of complex karyotype, wherein TP53MT was found in 81% of the patients with del(17p) (Fig. 1C). Irrespective of configuration, TP53MT carriers had worse overall survival (OS) compared to TP53WT carriers (HR 2.7 [95%CI 2.53–3.02]). No significant differences in OS between truncated and missense TP53MT or canonical and non-canonical missense TP53MT were observed (Fig. 1D & B).
Different TP53 configurations and disease subtypes impact prognosis
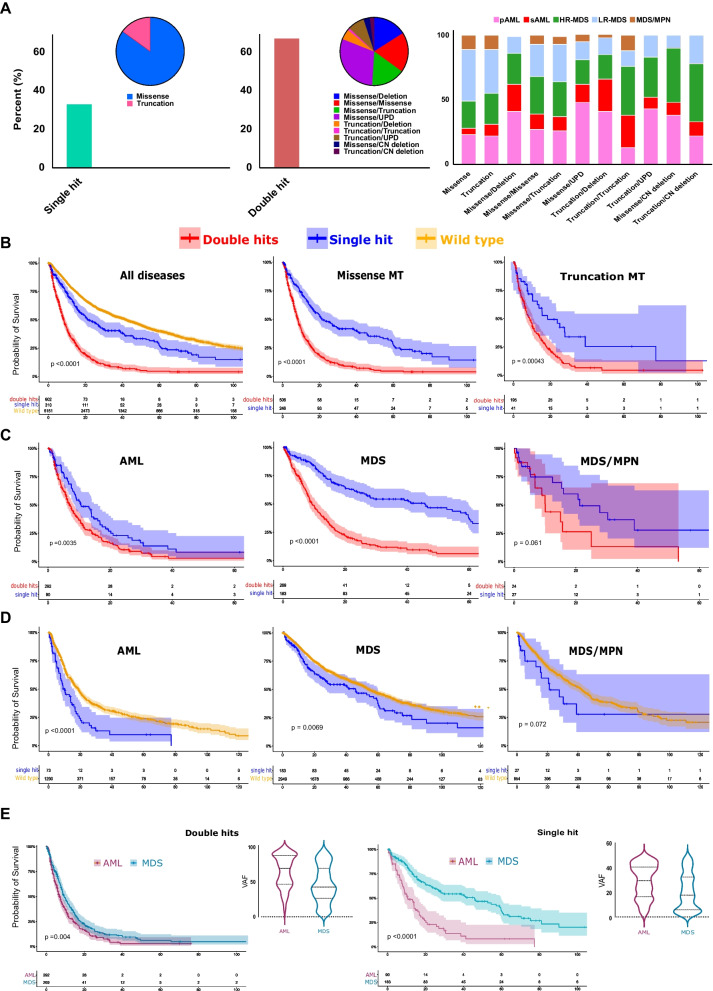
Following the traditional definition [10, 11, 25], we categorized the patients into single- and double-TP53MT hit groups. Briefly, a single TP53 hit was defined as either: (i) one TP53 mutation or (ii) isolated 17p deletion, while double TP53 hits were defined as TP53MT and (i) another TP53MT or (ii) 17p deletion or (iii) TP53 locus UPD. We found that 36% of TP53MT patients had single hits, while 64% exhibited double hits (Fig. 2A). TP53 double hits with different configurations were enriched in p/sAML and HR-MDS cases (Fig. 2A, middle and right panels and Additional file 1: Figure S3). Overall, carriers of TP53 double hits, whether missense and/or truncated had a worse OS than those with single TP53 hits (HR: 2.5 [2.08–3.02], Fig. 2B. Subgroup analysis according to the underlying disease morphology yielded similar results (data not shown). However, the significantly larger difference in OS between single and double TP53 hits was observed in MDS (HR: 3.1 [2.43–4.09]) compared to AML (HR: 1.5 [1.16–2.04]; Fig. 2C). TP53MT was also associated with worse OS when we compared TP53 single hit to TP53WT in both AML (HR: 1.8 [1.42–2.46]) and MDS (HR: 1.3 [1.08–1.67]) but not in MDS/MPN subtype (Fig. 2C and D). Ultimately, while the OS of AML cases was consistently worse compared to that of MDS cases across all patients, we observed that the magnitude of difference was more significant in the patents with presumed monoallelic TP53MT (Fig. 2E). Paralleling these findings, such a worse prognosis was also accompanied by a significantly higher VAF of TP53MT in AML vs. MDS in both double hit (64% vs. 46.5%, p < 0.001) and single hit (median 30 vs. 18%, p < 0.001; Fig. 2E) lesions.
Fig. 2.
Diverse TP53 configurations and disease subtypes impact prognosis. A patients with single hit (left panel) vs. double hits (middle panel) TP53 lesions and the type of configuration for each hit class and various constellations among different myeloid neoplasms (right panel). B Kaplan–Meier survival estimates of patients with TP53 wild type, single hits, and double hits, all patients (left panel) and cases with missense (middle panel) and truncated mutations (right panel). C Kaplan–Meier survival estimates of patients with TP53 single hits and double hits distributed according to different disease subtypes: AML (left panel), MDS (middle panel), and MDS/MPN (right panel). D Kaplan–Meier survival estimates of patients with TP53 wild type and single hit distributed according to different disease subtypes: AML (left panel), MDS (middle panel), and MDS/MPN (right panel). E Kaplan–Meier survival estimates of TP53 double hits (left panel) and single hit (right panel) in acute myeloid leukemia (AML) vs. myelodysplastic syndrome (MDS) in correlation with variant allelic frequency (VAF) for each subtype
A new approach to resolve the dilemma of biallelic vs. monoallelic TP53MT
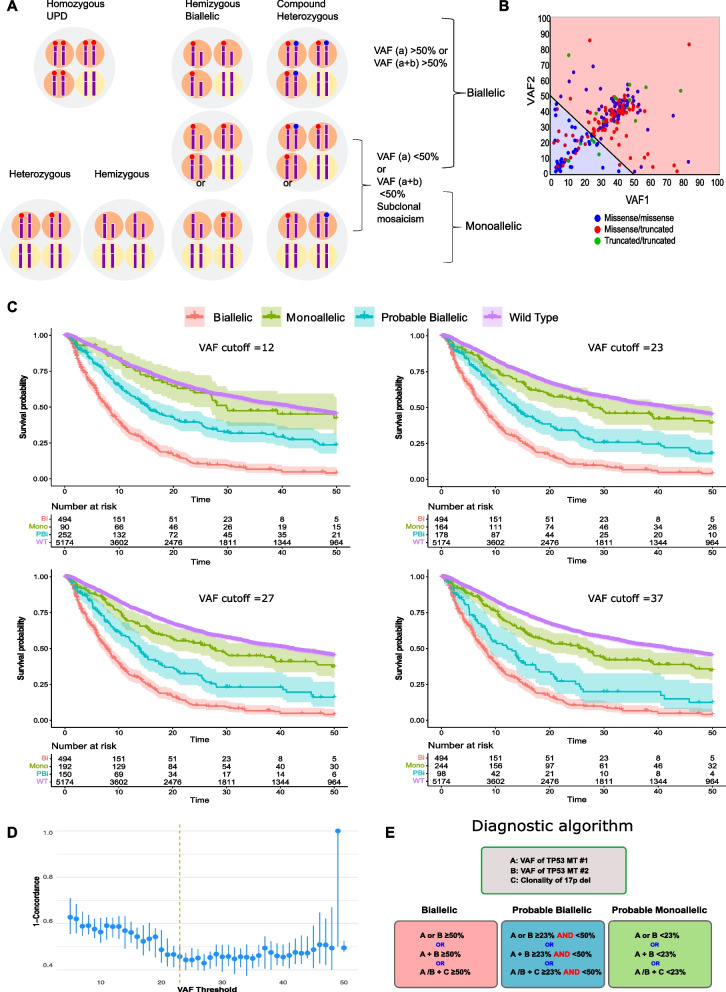
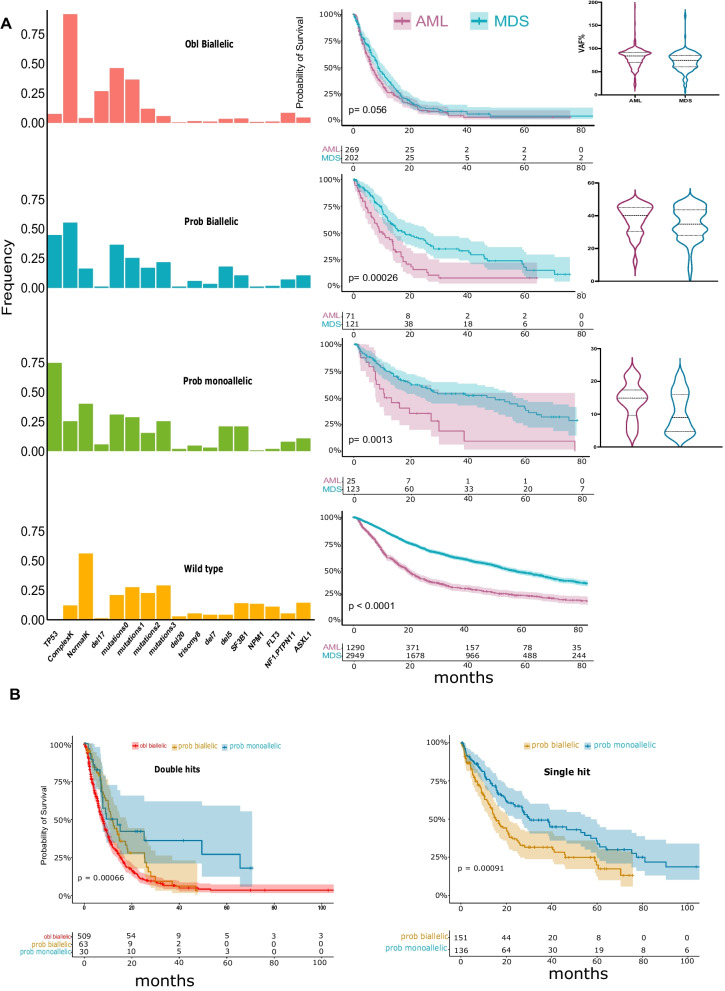
To resolve the uncertainty regarding the precise TP53MT allelic status, we developed a new approach to accurately predict the TP53 allelic configuration. Based on VAF values available from clinical sequencing, obligatory biallelic TP53MT were identified among patients with: i) one TP53 hit and VAF > 50% or ii) two TP53 hits with a combined VAF > 50% or iii) TP53MT VAF + clonality of del(17p) > 50% (Fig. 3A, B & Additional file 1: Fig. S6). However, the presence of subclonal mosaicism vs. biallelic TP53 hits (deletion and homozygous mutation) may not be discriminated with one TP53MT and VAF < 50%. Similarly, patients with ≥ 2 TP53 hits and a combined VAF < 50% may have subclonal mosaicism of several purely monoallelic or biallelic hits (Additional file 1: Fig. S7). Previously presumed monoallelic cases by traditional classification should be differentiated into those likely true monoallelic TP53MT and/or those likely containing a subclonal cryptic biallelic TP53 lesions. Since the presence of biallelic TP53MT clones is associated with poor OS, we applied a random forest regression analysis, with survival as a surrogate marker for TP53 allelic status, to fine-tune the VAF cutoff in order to separate the questionable cases into likely monoallelic vs. likely biallelic. We cross-validated VAF cutoff values by randomly splitting the data into test/train sets with 20%/80% ratios and calculating the Harrell’s C-index (Concordance) in the test set over 30 runs (Fig. 3D, Additional file 1: Table S4). A VAF cutoff of 23% was found to be optimal for separating these monoallelic and biallelic TP53MT (Additional file 1: Figures S8 & Fig. 3D). Accordingly, we classified the TP53MT into three main groups: A) “obligatory” biallelic, B) “probable biallelic”, and C) “probable monoallelic” groups (Fig. 3E). Based on this approach, 579 (57%) patients had obligatory biallelic TP53MT, 239 (24%) had probable biallelic TP53MT, and 192 (19%) had probable monoallelic TP53MT (Additional file 1: Table S5). The OS of patients with probable monoallelic TP53MT (median OS: 29 [10–77]) was similar to the TP53WT group (median: 42 [15–103]), p = 0.070. However, patients with probable biallelic TP53MT (median OS: 14 [7–37]) had worse outcomes as compared to TP53WT (p < 0.001; Fig. 3C and Additional file 1: Fig. S9) at all VAF cutoffs. While the OS was similar when we compared AML to MDS within the obligatory biallelic TP53MT group; however, AML showed a worse prognosis than comparable MDS, coinciding with higher TP53MT VAFs in AML patients (Fig. 4A). Additionally, in comparison with monoallelic and probable biallelic groups, obligatory biallelic TP53MT status was found to be associated more with pAML (OR: 1.84, [1.44–2.96]), sAML (OR: 2.50 [1.25–4.99]), complex karyotype (OR: 10.70 [5.20–21.9]), or carriers of del(17p) (OR: 5.42 [2.80–10.5]) (Fig. 4A, Additional file 1: Table S6 and S7).
Fig. 3.
Assessment of allelic status of TP53 lesions. A possibilities of different TP53 configurations and del(17p) presence resulting in monoallelic vs. biallelic lesions. For illustration purpose, four cells scheme is presented. For biallelic mutations, the sum VAF% is more than 50% in hemizygous, homozygous uniparental disomy (UPD) or compound heterozygous configurations. For monoallelic mutations, the sum VAF% is less than 50% in different configurations including heterozygous, hemizygous, or subclonal mosaicism. B The variant allele frequency (VAF) of two TP53 mutations of patients in our cohort was plotted in two colors (red area, obligatory biallelic) with the sum of VAF1 and VAF2 exceeding 50% and (blue area, non-obligatory biallelic) with sum of VAF1 and VAF2 less than 50. C Kaplan–Meier survival estimates of obligatory biallelic, probable biallelic, probable monoallelic, and wild-type TP53 patients after applying different VAF cutoffs based on random forest analysis separating probable monoallelic from probable biallelic mutations. D VAF cutoffs cross-validation by randomly splitting the data into test/train sets with %20/%80 ratios and calculating the Harrell’s C-index (Concordance-index) in the test set over 30 runs. A VAF of 23% resulted optimal for separating the monoallelic and biallelic TP53 mutations. E A novel algorithm for the precise classification of TP53MT into obligatory biallelic, probable biallelic, or probable monoallelic groups
Fig. 4.
Identifying the probable allelic involvement in relation to clinical and cytogenetic factors. A Frequency and importance of number of concurrent somatic mutations and cytogenetic abnormalities with Kaplan–Meier survival estimates comparing acute myeloid leukemia (AML) vs. myelodysplastic syndrome (MDS) for each group. Median variant allele frequency (VAF) for AML vs. MDS. B Kaplan–Meier survival estimates comparing obligatory biallelic, probable biallelic, and probable monoallelic mutations in double hits and single hit groups
To further highlight the limitations of the traditional classification methods, we applied our new algorithm to the previously classified single and double TP53 hit groups. Indeed, we found significant OS differences between the new subgroups with the better prognostic resolution of the monoallelic/biallelic cases. Moreover, patients with probable biallelic TP53 hits had survival rates between probable monoallelic TP53MT and obligatory biallelic TP53MT cases (Fig. 4B, Additional file 1: Fig. S10). This survival difference was also validated in an external confirmatory cohort (Additional file 1: Fig. S11).
Confirming biallelic TP53 inactivation and clonal mosaicism in selected cases
Our novel VAF-based method allowed a re-classification of 192 (19%) patients with single hits to probable biallelic class and 32 (3%) patients with double hits to probable monoallelic class of TP53MT compared to the traditional classification (Additional file 1: Fig. S12). Accordingly, VAF < 50% in single or double TP53 hits could be indeed associated with the presence of a subclone that acquired biallelic inactivation. To further confirm these results and characterize the subclonal configurations, we selected four patients (3 likely monoallelic and one likely biallelic TP53MT) and applied single-cell DNA mutational and copy number analysis. We found that some cells contained monoallelic TP53MT, while others had biallelic lesions in all cases studied. For example, in a pAML case (UPN13), the bulk NGS showed TP53MT with a VAF of 8%; however, 6% of the cells were actually biallelic. In another HR-MDS case (UPN125), the bulk NGS showed TP53MT with a VAF of 33%, while 4% were biallelic. In case (UPN423), NGS detected two TP53MT with a combined VAF of 20%, but this sample contained 32% biallelic cells by single-cell DNA analysis. Finally, a case (UPN875) of LR-MDS had two TP53MT (missense and truncated); however, the single-cell DNA analysis showed three TP53MT, and the percentage of biallelic cells was 34% (Additional file 1: Fig. S13A–D).
Frequency of concurrent somatic mutations and factors associated with TP53MT
We found that 41% of cases had TP53MT as a sole molecular lesion, while the remaining 59% harbored additional somatic events. In particular, complex karyotype was more frequent among patients with isolated TP53MT (p < 0.001). As to disease associations, LR-MDS cases had a lower burden of co-mutations, while the highest percentages were registered in MDS/MPN group (p = 0.0015; Additional file 1: Fig. S14). Interestingly, the rate of co-occurring events varied according to the TP53MT status. The TP53WT group had a rate of 2.10 co-mutation per patient. In contrast, this rate was lower in other groups with rates of 0.80, 1.42, and 1.76 co-mutations per patient in obligatory biallelic, probable biallelic, and probable monoallelic groups, respectively (Additional file 1: Fig. S15). We then applied our new classification scheme to investigate whether any difference was notable in the mutational configuration between patients with obligatory vs. probable monoallelic and probable biallelic TP53MT. In univariate analysis, we found that IDH1, IDH2, EZH2, SUZ12, ASXL1, DNMT3A, JAK2, RUNX1, SF3B1, SRSF2, TET2, and U2AF1 mutations were less common in obligatory biallelic patients compared to probable monoallelic/probable biallelic cases; however, this correlation was not significant in a multivariate setting (Additional file 1: Table S8).
Discussion
The prognostic impact of TP53MT depends on the allelic configuration of the TP53 hits reversely engineered a clinically applicable system enabling the imputation of allelic status that allows for a more precise and clinically applicable assignment of prognosis using routinely available molecular tools. The underlying hypothesis for our strategy was that traditionally defined single hit cases [11, 25] might also contain subclones with a biallelic TP53 inactivation, which negatively influences prognosis.
Since in clinical situation the direct clonality measure is not available, we have proposed an algorithm that approximated allelic burden/copy number based on the cutoffs benchmarked according to the survival by applying a rationally developed strategy to our large and well-annotated cohort and provide a method for a more precise assessment of prognosis in carriers of TP53 lesions under the assumption that the higher the clonal burden, the more likely is the presence of a cryptic biallelic subclone. Using a newly devised bioinformatics approach, we established a VAF cutoff for prognostic diversification of traditionally considered monoallelic TP53MT groups. Conversely, we have also shown that double hits are not necessarily biallelic but may constitute a subclonal TP53 mosaicism in a branching evolution mode. A more intricate method such as single-cell DNA sequencing, including the simultaneous analysis of mutations and CN-LOH, demonstrated the above-described points, in agreement with the observation in LFS. Indeed, using single-cell DNA sequencing we were able to discover the presence of biallelic clones carrying TP53 lesions in cases with likely monoallelic hits, demonstrating that the analysis of clonal architecture at single-cell level is able to identify cryptic alterations not always detected by bulk sequencing. The critical size of the biallelic clone to affect the prognosis would be impossible to be precisely estimated, but as demonstrated by us a combined VAF may give away the presence and size of the double hit clones. Of note is that to date none of the studies systematically and directly addressed this issue in clinical setting due to obvious feasibility issues.
TP53MT was common in MDS and AML patients in agreement with other studies [26, 27], while our earlier analyses have shown that a second TP53 lesion was the most common hit associated with TP53MT [14] and that second TP53 hits are likely sweeping lesions [12]. Previous studies in MDS [11, 25] have already demonstrated that monoallelic lesions have no clinical impact on prognosis vs. TP53WT cases, but biallelic clones defined by the presence of two TP53 lesions have more aggressive phenotypes. We initially confirmed this finding according to traditional methods, but our current strategy, as described, further refined both double- and single-hit cases so that the latter could be sub-stratified according to the probability of the presence of truly biallelic subclones and thereby distinguished by worse outcomes and vice versa. For instance, patients with TP53MT, whether single or double hits and combined VAF of < 23% rarely harbor biallelic subclones and indeed show WT-like survival. Conversely, TP53MT patients with a VAF of > 23% have more aggressive disease.
In our cohort, MDS/MPN has lower frequency of TP53MT and lower clonal burdens and thus by inference the double hits should be less common. Indeed, other studies have shown that TP53MT (independently from single or double hits) are relatively infrequent in MDS/MPN (< 5%) especially CMML accounting for 1–5% of the cases [28–30]. Although TP53MT frequency increases in MDS/MPN-U [21] and therapy-related CMML (< 2%) [31], the presence of such lesions remains uncommon most likely due to the lower rate of transformation of patients with MDS/MPN compared to MDS. However, one can also stipulate that the initiation and progression of MN with myeloproliferative phenotype are more likely driven by RAS pathway alterations possibly mutually exclusive with TP53MT.
To highlight the limitations of previous traditional classifications of TP53MT, the International Consensus Classification of myeloid neoplasms and acute leukemias group recently published new classification schemes of MN with TP53MT. [32] Although the new classifications addressed the constraints of the previous schemes, the issue of possible subclonal mosaicism vs. truly biallelic hits and the possibility of cryptic biallelic hits in seemingly monoallelic cases remained unresolved. In our algorithm, we showed that patients with single TP53MT and VAF more than 23% are acting biologically like biallelic TP53MT cases. We think that TP53 VAF is more accurate to determine the biallelic involvement than the presence of complex karyotypes unless the latter contain del(17p).
According to our proposed algorithm, MDS and AML with obligatory biallelic mutations had similar survival, thus upstaging MDS irrespective of the blast count. Recent findings that patients with AML have similar OS compared to those with MDS with excess blasts [25] overlapping with the presented results. Although probable biallelic group had worse outcome compared to probable monoallelic group, their OS was better compared to the obligatory biallelic TP53MT. This finding can be explained by the small biallelic clone among these patients contributing to a better OS when compared to the obligatory biallelic group.
Monoallelic TP53MT are not unimportant as they constitute the first step in establishing a dominant clone characterized by biallelic TP53 loss. Unexpectedly, we could not differentiate the prognosis between frameshift and dominant-negative missense mutations in any of the possible configurations.
Because our method relies on parameter optimization alone, while improving previous approaches, it has clear limitations. For instance, we did not consider the clonality of all samples since these data were not readily available for all the cohorts in our analysis. However, such consideration will be necessary for future studies to estimate allelic involvement accurately.
In sum, our study demonstrates the importance of delineating the subclonal mosaicism of TP53MT for modeling disease progression in MN. In addition to the genetic context, the role of TP53MT may also vary in different disease subtypes (e.g., AML vs. MDS), and individual (single hit) VAF levels can shape patients' trajectories differently. In the future, our proposed approach could be incorporated into prognostication systems [16], such as IPSS-M to improve its precision with regard to patients’ outcomes. Ultimately, resolution of TP53 inactivation status may proof a valuable tool for identifying the most suitable candidates for TP53-targeted therapeutic strategies.
Supplementary Information
Additional file 1. Novel scheme for defining the clinical implications of TP53 mutations in myeloid neoplasia.
Author contributions
WB, TK collected, annotated, and analyzed clinical and molecular data, interpreted results, and wrote the manuscript. CG collected clinical and molecular data and edited the manuscript. AD applied the algorithm for TP53 genomic re-classification, analyzed the data, interpreted the results, and wrote the statistical methods. BP helped in the analysis of single-cell DNA sequencing. YK, ODO, MZ analyzed data and edited the manuscript. IP collected and prepared samples for single-cell DNA sequencing. YM, TB, and SKB provided annotated validation cohorts and helped in editing the manuscript. HA, RA, MM collected data and edited the manuscript. MM, TH provided data and significant insights to the manuscript. VV and JPM offered invaluable help to the manuscript preparation, generated, and conceived the study design, designed figures and tables, and wrote the manuscript. All authors participated in the critical review of the final paper and submission.
Funding
Source of funding: the HENRY & MARILYN TAUB FOUNDATION (J.P.M.), grants R01HL118281 (to J.P.M.), R01HL123904 (to J.P.M., R.A.P.), R01HL132071 (to J.P.M., R.A.P.), R35HL135795 (all to J.P.M), AA&MDSIF (to V.V., S.P., J.P.M), The Leukemia & Lymphoma Society TRP Award 6645–22 (to J.P.M), VeloSano 9 Pilot Award, Vera and Joseph Dresner Foundation–MDS (to V.V.). C.G. was supported by a grant from the Edward P. Evans Foundation. This work used the High-Performance Computing Resource in the Core Facility for Advanced Research Computing at Case Western Reserve University. We thank the Mission Bio team for their technical expertise in single-cell DNA analysis.
Data availability
All the data used to support our results are available in this article. NGS data of The Cleveland Clinic cohort can be requested by contacting the corresponding author (maciejj@ccf.org).
Declarations
Ethics approval and consent to participate
Samples were collected after obtaining written informed consent according to the protocols approved by the institutional review boards of the participating institutions under the regulations set forth in the Declaration of Helsinki.
Consent for publication
Consent was obtained from patients.
Competing interests
Y.M has received honoraria/consulting fees from BluePrint Medicines, GERON, OncLive and MD Education participated in advisory boards and received honoraria from Sierra Oncology, Stemline Therapeutics, Blueprint. Medicines, Morphosys, Taiho Oncology, Rigel Pharmaceuticals and Novartis. YFM received travel reimbursement from Blueprint Medicines, MD Education, and Morphosys. None of these relationships were related to this work.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Waled Bahaj and Tariq Kewan have equally contributed to the work.
Contributor Information
Valeria Visconte, Email: visconv@ccf.org.
Jaroslaw P. Maciejewski, Email: maciejj@ccf.org
References
- 1.Boettcher S, Miller PG, Sharma R, et al. A dominant-negative effect drives selection of. Science. 2019;365(6453):599–604. doi: 10.1126/science.aax3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohgami RS, Ma L, Merker JD, et al. Next-generation sequencing of acute myeloid leukemia identifies the significance of TP53, U2AF1, ASXL1, and TET2 mutations. Mod Pathol. 2015;28(5):706–714. doi: 10.1038/modpathol.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2(1):1001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haase D, Stevenson KE, Neuberg D, et al. TP53 mutation status divides myelodysplastic syndromes with complex karyotypes into distinct prognostic subgroups. Leukemia. 2019;33(7):1747–1758. doi: 10.1038/s41375-018-0351-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hainaut P, Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- 6.Baker SJ, Kinzler KW, Vogelstein B. Knudson's hypothesis and the TP53 revolution. Genes Chromosomes Cancer. 2003;38(4):329. doi: 10.1002/gcc.10249. [DOI] [PubMed] [Google Scholar]
- 7.Achatz MI, Zambetti GP. The inherited p53 mutation in the Brazilian population. Cold Spring Harb Perspect Med. 2016 doi: 10.1101/cshperspect.a026195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen S, Wang Q, Yu H, et al. Mutant p53 drives clonal hematopoiesis through modulating epigenetic pathway. Nat Commun. 2019;10(1):5649. doi: 10.1038/s41467-019-13542-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jasek M, Gondek LP, Bejanyan N, et al. TP53 mutations in myeloid malignancies are either homozygous or hemizygous due to copy number-neutral loss of heterozygosity or deletion of 17p. Leukemia. 2010;24(1):216–219. doi: 10.1038/leu.2009.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malcikova J, Smardova J, Rocnova L, et al. Monoallelic and biallelic inactivation of TP53 gene in chronic lymphocytic leukemia: selection, impact on survival, and response to DNA damage. Blood. 2009;114(26):5307–5314. doi: 10.1182/blood-2009-07-234708. [DOI] [PubMed] [Google Scholar]
- 11.Bernard E, Nannya Y, Hasserjian RP, et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat Med. 2020;26(10):1549–1556. doi: 10.1038/s41591-020-1008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makishima H, Yoshizato T, Yoshida K, et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet. 2017;49(2):204–212. doi: 10.1038/ng.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gondek LP, Tiu R, Haddad AS, et al. Single nucleotide polymorphism arrays complement metaphase cytogenetics in detection of new chromosomal lesions in MDS. Leukemia. 2007;21(9):2058–2061. doi: 10.1038/sj.leu.2404745. [DOI] [PubMed] [Google Scholar]
- 14.Nagata Y, Makishima H, Kerr CM, et al. Invariant patterns of clonal succession determine specific clinical features of myelodysplastic syndromes. Nat Commun. 2019;10(1):5386. doi: 10.1038/s41467-019-13001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cazzola M, Della Porta MG, Malcovati L. The genetic basis of myelodysplasia and its clinical relevance. Blood. 2013;122(25):4021–4034. doi: 10.1182/blood-2013-09-381665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elsa Bernard PDea. Molecular International Prognostic Scoring System for Myelodysplastic Syndromes. NEJM evidence; 2022. [DOI] [PubMed]
- 17.Yan H, Qu J, Cao W, et al. Identification of prognostic genes in the acute myeloid leukemia immune microenvironment based on TCGA data analysis. Cancer Immunol Immunother. 2019;68(12):1971–1978. doi: 10.1007/s00262-019-02408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burd A, Levine RL, Ruppert AS, et al. Precision medicine treatment in acute myeloid leukemia using prospective genomic profiling: feasibility and preliminary efficacy of the Beat AML Master Trial. Nat Med. 2020;26(12):1852–1858. doi: 10.1038/s41591-020-1089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meggendorfer M, Haferlach C, Kern W, Haferlach T. Molecular analysis of myelodysplastic syndrome with isolated deletion of the long arm of chromosome 5 reveals a specific spectrum of molecular mutations with prognostic impact: a study on 123 patients and 27 genes. Haematologica. 2017;102(9):1502–1510. doi: 10.3324/haematol.2017.166173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delic S, Rose D, Kern W, et al. Application of an NGS-based 28-gene panel in myeloproliferative neoplasms reveals distinct mutation patterns in essential thrombocythaemia, primary myelofibrosis and polycythaemia vera. Br J Haematol. 2016;175(3):419–426. doi: 10.1111/bjh.14269. [DOI] [PubMed] [Google Scholar]
- 21.Palomo L, Meggendorfer M, Hutter S, et al. Molecular landscape and clonal architecture of adult myelodysplastic/myeloproliferative neoplasms. Blood. 2020;136(16):1851–1862. doi: 10.1182/blood.2019004229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bersanelli M, Travaglino E, Meggendorfer M, et al. classification and personalized prognostic assessment on the basis of clinical and genomic features in myelodysplastic syndromes. J Clin Oncol. 2021;39(11):1223–1233. doi: 10.1200/JCO.20.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562(7728):526–531. doi: 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishwaran H, and U. B. Kogalur. Fast unified random forests for survival, regression, and classification (RF-SRC); R package version 2.1 2019.
- 25.Grob T, Al Hinai AS, Sanders MA, et al. Molecular characterization of mutant Tp53 acute myeloid leukemia and high-risk myelodysplastic syndrome. Blood. 2022 doi: 10.1182/blood.2021014472. [DOI] [PubMed] [Google Scholar]
- 26.Bejar R, Stevenson K, Abdel-Wahab O, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364(26):2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W, Routbort MJ, Loghavi S, Tang Z, Medeiros LJ, Wang SA. Characterization of chronic myelomonocytic leukemia with TP53 mutations. Leuk Res. 2018;70:97–99. doi: 10.1016/j.leukres.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Patnaik MM, Lasho TL, Vijayvargiya P, et al. Prognostic interaction between ASXL1 and TET2 mutations in chronic myelomonocytic leukemia. Blood Cancer J. 2016;6(1):e385. doi: 10.1038/bcj.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Itzykson R, Kosmider O, Renneville A, et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013;31(19):2428–2436. doi: 10.1200/JCO.2012.47.3314. [DOI] [PubMed] [Google Scholar]
- 31.Patnaik MM, Vallapureddy R, Yalniz FF, et al. Therapy related-chronic myelomonocytic leukemia (CMML): molecular, cytogenetic, and clinical distinctions from de novo CMML. Am J Hematol. 2018;93(1):65–73. doi: 10.1002/ajh.24939. [DOI] [PubMed] [Google Scholar]
- 32.Arber DA, Orazi A, Hasserjian RP, et al. international consensus classification of myeloid neoplasms and acute Leukemias: integrating morphologic, clinical, and genomic data. Blood. 2022;140(11):1200–1228. doi: 10.1182/blood.2022015850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Novel scheme for defining the clinical implications of TP53 mutations in myeloid neoplasia.
Data Availability Statement
All the data used to support our results are available in this article. NGS data of The Cleveland Clinic cohort can be requested by contacting the corresponding author (maciejj@ccf.org).