Abstract
The complete nucleotide sequence was determined for the putative RNA polymerase (183K protein) gene of tobacco mosaic virus (TMV) OM strain, which differed from the related strain, vulgare, by 51 positions in its nucleotide sequence and 6 residues in its amino acid sequence. Three segments of this 183K protein, each containing the sequence motif of methyltransferase (M), helicase (H), or RNA-dependent RNA polymerase (P), were expressed in Escherichia coli as fusion proteins with hexahistidine tags, and domain-specific antibodies were raised against purified His-tagged M and P polypeptides. By immunoaffinity purification, a template-specific RNA-dependent RNA polymerase containing a heterodimer of the full-length 183K and 126K (an amino-terminal-proximal portion of the 183K protein) viral proteins was isolated. We propose that the TMV RNA polymerase for minus-strand RNA synthesis is composed of one molecule each of the 183- and 126-kDa proteins, possibly together with two or more host proteins.
The genome of tobacco mosaic virus (TMV) consists of a single-stranded RNA molecule of about 6,400 nucleotides in length with positive polarity, which encodes at least four polypeptides: 126- and 183-kDa proteins required for transcription and replication (hereafter referred to as the 126K and 183K proteins, respectively), a 30-kDa (30K) protein for cell-to-cell virus movement in infected plants, and an 18-kDa protein for virus coat formation. The sequence of the 126K protein is encoded by the 5′-proximal region of the viral genome and includes the methyltransferase and RNA helicase motifs, while the 183K protein is a read-through protein of the 126K open reading frame (ORF) and contains, in addition to the above two motifs, the RNA-dependent RNA polymerase motif. The RNA polymerase is considered to be involved in both transcription and replication (8). From sequence analysis, it is believed that the viral RNA polymerase contains the 183K protein as a catalytic subunit, but the precise molecular compositions of transcriptase and replicase have not yet been determined.
In positive-strand RNA virus-infected cells, RNA-dependent RNA polymerases localize on virus-infected cell membrane (27, 31). The membrane fractions of plant tissues, however, contain the activities of cellular RNA-dependent RNA polymerase (3, 20) and terminal nucleotidyl transferase (20, 37). Even though the physiological functions have not yet been identified, these cellular enzymes interfere with the detection and purification of viral RNA polymerases from plant tissues.
Recently, Osman and Buck (21) succeeded in the solubilization of TMV RNA polymerase from virus-infected membrane fractions by using sodium taurodeoxycholate (TDC) (20) and in the separation of the viral RNA polymerase from cellular enzyme activities of RNA-dependent RNA synthesis by conventional protein purification (21). Since the purified viral RNA polymerase preparation contained six major and four to five minor protein components, including the TMV-encoded 126K and 183K proteins, the molecular composition of viral RNA polymerase remained undetermined. To overcome the difficulty in the purification of TMV RNA polymerase, we cloned cDNAs for each domain of the 183K protein (the putative RNA polymerase), expressed them in Escherichia coli, and raised antibodies against the purified proteins. Using immunoaffinity column chromatography, we isolated an enzyme complex containing a 126K/183K protein heterodimer that was virtually free from cellular RNA polymerase activities. The isolated complex exhibited template-dependent and template-specific RNA polymerase activity. We propose that the TMV RNA polymerase for minus-strand RNA synthesis is composed of one molecule each of the 183K and 126K proteins, possibly together with two or more host proteins.
MATERIALS AND METHODS
Preparation of TMV virus and viral RNA.
Virus particles of TMV strain OM, a widely used common strain, were purified from virus-infected Nicotiana tabacum L. cv. Xanthi according to a published procedure (5). Viral RNA (vRNA) was extracted from purified virus by treatment with phenol and sodium dodecyl sulfate (SDS) (2, 36). vRNA from cucumber mosaic virus (CMV) strain Y was kindly provided by Masashi Suzuki (University of Tokyo).
cDNA synthesis and cloning of the 183K gene.
First-strand cDNA covering the entire sequence of 183K gene was synthesized by reverse transcription of vRNA with a 3′ primer (reverse primer corresponding to TMV-OM sequence from nucleotide positions 4897 to 4916 with an attached 5′-CGCGCG [PstI and XhoI sites] sequence) (see Table 1 for the primer sequences). Double-stranded cDNA was synthesized by using this single-stranded cDNA product as a template and a set of primers, the same reverse primer as a 3′ primer and primer 1B corresponding to nucleotide positions 69 to 88 of the TMV-OM sequence attached with 5′-GCGCGC (KpnI and NcoI sites) as a 5′ primer. The resulting double-stranded cDNAs were inserted into the pUC119 cloning vector. Two clones, pUC183K-1 and pUC183K-2, derived from two independent reactions of the first-strand cDNA synthesis were used for subcloning and sequencing.
TABLE 1.
Primers for TMV cDNA synthesis
| Primer | Sequence (5′ to 3′) |
|---|---|
| Reverse primer | CGCGCG CTGCAG CTCGAG ACAACTAGAGCCATCTATAA(4897) |
| PstI XhoI | |
| Primer 1B | GCGCGC GGTACC CCATGG CATACACACAGACAGC(88) |
| KpnI NcoI | |
| Primer H | CGCGCG CTGCAG CTCGAG CCAGGTATTAACCCTGGTGAC(966) |
| PstI XhoI | |
| Primer F | GCGCGC GGTACC CCATGG ATATGCAGTTTTACTATG(3510) |
| KpnI NcoI | |
| Primer A | CGCGCG CTGCAG CTCGAG ACAACTAGAGCCATCTATGAA(4895) |
| PstI XhoI | |
| Primer 3′(+)F | CGC AAGCTT TAATACGACTCACTATA GAGAGCTCTTCTGGTTTGGT(6166) |
| HindIII T7 promoter | |
| Primer 3′(+)R | CGC GGATCC CATATG GGCCCCTACCGGGGGTAACGG(6373) |
| BamHI NdeI |
Nucleotide sequence analysis.
The nucleotide sequence of DNA was determined by the dideoxynucleotide sequence technique (30). The sequence covering the entire 183K-ORF was determined for both orientations of pUC183K-1 and in the forward direction of pUC183K-2 by using at least three overlapping deletion clones. For some uncertain regions, direct sequencing of reverse transcription-PCR products was carried out with vRNA as template. Complete nucleotide and predicted amino acid sequences of 126K-183K ORFs were submitted to the DDBJ, EMBL, and GenBank databases (accession number D78608).
Construction of expression plasmids of RNA polymerase proteins.
For construction of the expression plasmids pET-M, pET-H, and pET-P, each encoding C-terminal hexahistidine (H6)-tagged methyltransferase (M; nucleotide positions 69 to 986 or amino acid residues 1 to 306), helicase (H; nucleotide positions 2483 to 3416 or amino acid residues 806 to 1116), and RNA-dependent RNA polymerase (P; nucleotide positions 3492 to 4919 or amino acid residues 1143 to 1616) domain polypeptides, respectively, cDNA fragments were amplified by PCR with pUC183K as a template and the following sets of primers: (i) 5′-primer 1B and 3′-primer H (positions 966 to 987) attached to the sequence 5′-CGCGCG (PstI and XhoI sites) for the synthesis of the M-domain segment and (ii) 5′-primer F (positions 3492 to 3510) attached to the sequence 5′-GCGCGC (KpnI and NcoI sites) and 3′-primer A (positions 4895 to 4916) attached to the sequence 5′-CGCGCG (PstI and XhoI sites) for the synthesis of the P-domain segment. Resulting PCR products were ligated, after digestion with NcoI and XhoI, to pET21d(+), which had been previously treated with NcoI and XhoI.
Purification of H6-tagged RNA polymerase proteins and generation of domain-specific antibodies.
Expression plasmids pET-M, pET-H and pET-P were transformed into E. coli BL21(DE3). The transformants were cultured at 37°C in Luria broth medium containing 100 μg of ampicillin per ml. When the culture reached 40 Klett units, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to 1 mM, and the culture was continued until it reached 100 Klett units. Cells were harvested and suspended in a lysis buffer (50 mM Tris-HCl [pH 8.0], 0.1 M NaCl, 1 mM EDTA, 1 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.1% sodium deoxycholate, and 0.3 mg of lysozyme per ml). After incubation on ice for 20 min, cell lysates were sonicated and centrifuged at 15,000 × g for 15 min at 4°C. The inclusion bodies were collected by centrifugation, resuspended in a binding buffer (20 mM Tris-HCl [pH 8.0], 0.5 M NaCl, 5 mM imidazole, and 6 M guanidine-HCl), and centrifuged at 15,000 × g for 15 min at 4°C. The supernatant was applied onto Ni2+-charged nitrilotriacetic acid-Sepharose (His-Bind Resin; Novagen). The column was washed successively with 6 bed volumes of the binding buffer and 1 bed volume of a washing buffer (20 mM Tris-HCl [pH 8.0], 0.5 M NaCl, 60 mM imidazole, and 6 M guanidine-HCl). The H6-tagged polypeptides, M-H6, H-H6, and P-H6, were eluted by washing with 3 bed volumes of an elution buffer (20 mM Tris-HCl [pH 8.0], 0.5 M NaCl, 1 M imidazole, and 6 M guanidine-HCl) and then fractionated by SDS–12.5% polyacrylamide gel electrophoresis (PAGE). M-H6, H-H6, and P-H6 were then eluted from the corresponding bands of gels. The protein concentration was determined by staining with Coomassie brilliant blue G250 (Protein Assay kit; Bio-Rad).
Polyclonal antibodies against each 183K protein domain polypeptide were raised in rabbits by using these gel-purified His-tagged polypeptides as antigens. The antibodies were affinity purified with the Ampure PA kit (Amersham). The titer of the antibodies against H-H6 was, however, too low to use for Western blot analysis of small amounts of TMV proteins in virus-infected plant tissue extracts.
Preparation of antibody-conjugated Sepharose.
For conjugation of antibodies to Sepharose, 3 g of CNBr-activated Sepharose 6MB (Pharmacia) was suspended in 50 ml of 1 mM HCl and washed a few times with a total of 550 ml of 1 mM HCl each for 15 min at room temperature on a sintered glass filter. The washed CNBr-activated Sepharose was mixed with 8.5 mg of the affinity-purified polyclonal antibodies in 15 ml of coupling buffer (0.1 M NaHCO3 [pH 8.3], 0.5 M NaCl, and 0.5% Tween 80) in a sealed vessel and rotated for 2 h at room temperature. Unconjugated excess antibodies were washed away with 0.2 M glycine dissolved in coupling buffer for 2 h at room temperature. Antibody-conjugated Sepharose was washed for four cycles with alternating pH buffers, a low-pH acetate buffer (0.1 M acetic acid [pH 4.0], 0.5 M NaCl, and 0.5% Tween 80) and a high-pH Tris buffer (0.1 M Tris-HCl [pH 8.0], 0.5 M NaCl, and 0.5% Tween 80), and subsequently washed with an elution buffer (50 mM glycine-HCl [pH 2.5], 0.15 M NaCl, and 0.1% Triton X-100) and stored in Tris buffer at 4°C.
Preparation of TMV-infected tobacco tissue extracts.
Young leaves of tobacco plants (Nicotiana tabacum L. cv. Xanthi.) were inoculated with TMV-OM, and 4 days later the infected leaves (100 g) were homogenized with a blender in 2 volumes of buffer A (50 mM Tris-HCl [pH 7.5], 15 mM MgCl2, 120 mM KCl, 0.1% 2-mercaptoethanol, 3 μg of pepstatin A per ml, 1 mM PMSF, and 20% [vol/vol] glycerol) at 4°C. After it was filtered through two layers of Miracloth (Calbiochem), the extract was centrifuged for 10 min at 500 × g at 4°C. The supernatant was subjected to further centrifugation at 30,000 × g for 30 min at 4°C. The pellet (P30) was resuspended in 20 ml of buffer B (50 mM Tris-HCl [pH 8.2], 10 mM MgCl2, 1 mM dithiothreitol (DTT), 3 μg of pepstatin A per ml, 3 μg of leupeptin per ml, and 1% TDC, stirred gently for 1 h at 4°C, and then centrifuged at 100,000 × g for 1 h at 4°C to give a supernatant (S100) fraction.
Immunoaffinity purification of TMV RNA polymerase proteins.
Fifty microliters of antibody-conjugated Sepharose (Ab-Sepharose) were mixed with 5 bed volumes of S100 and shaken gently on a rotary shaker for 1 h at 4°C. After the supernatant was removed, the Ab-Sepharose was washed four to five times with a total of 30 or 50 bed volumes of Tris-Triton-NaCl buffer (50 mM Tris-HCl [pH 8.0], 0.1% Triton X-100, 0.5 M NaCl, and 0.1% 2-mercaptoethanol). The Ab-Sepharose was directly subjected to SDS-PAGE for analysis of bound proteins and to in vitro transcription assay for the detection of RNA polymerase activity. For the activity assay, proteins were also eluted from the Ab-Sepharose resin with 5 bed volumes of a glycine buffer (50 mM glycine-HCl [pH 2.5], 0.1% Triton X-100, 0.15 M NaCl, and 0.1% 2-mercaptoethanol). The elution fractions were collected into tubes containing 0.15 volume of 1 M Tris-HCl (pH 9.0).
Glycerol gradient centrifugation.
The S100 fraction was mixed with molecular-size marker proteins (yeast alcohol dehydrogenase, 76 kDa; rabbit muscle aldolase, 158 kDa; bovine liver catalase, 240 kDa; E. coli RNA polymerase holoenzyme, 450 kDa) and layered on the top of a 10 to 40% linear gradient of glycerol in 50 mM Tris-HCl (pH 8.2), 10 mM MgCl2, 0.1 N NaCl, and 1 mM DTT. After centrifugation for 8 h at 45,000 rpm in a Beckman SW50.1 rotor, the glycerol gradient was fractionated into 25 tubes. Aliquots were fractionated by SDS-PAGE for detection of the marker proteins by staining with Coomassie brilliant blue and for analysis of the distribution of the 126K and 183K proteins by Western blot analysis.
Western blotting.
Proteins were separated by SDS–7.5% PAGE and electrotransferred from the gel onto a polyvinylidene difluoride membrane by using a wet transfer unit in a transfer buffer (20 mM Tris-base, 150 mM glycine, 0.1% SDS, and 20% methanol) at 0.8 mA/cm2 for 3 h. After being blocked with 5% bovine serum albumin (BSA) in phosphate-buffered saline (PBS), the membrane was incubated at room temperature for 1 h with rabbit antiserum against the M-H6 or P-H6 protein, which was diluted 1,000-fold with 1% BSA-PBS. The membrane was then washed with 0.5% Tween 80-PBS and incubated at room temperature for 1 h with 2,000-fold-diluted horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G serum (Cappel) in 1% BSA-PBS. After a washing with 0.5% Tween 80-PBS, bound antibodies were visualized with 0.5 μg of 3,3′-diaminobenzidine per ml and 0.5% H2O2 in PBS.
Assay of RNA-dependent RNA polymerase.
The crude extract (S100) or purified viral RNA polymerase proteins (Ab-Sepharose fractions or eluates) were subjected to an RNA-dependent RNA polymerase assay. The reaction mixture contained in a volume of 150 μl: 50 mM Tris-HCl (pH 8.2); 10 mM MgCl2; 1 mM DTT; 4.8 μg of washed bentonite (2) per ml; 1 mM concentrations each of ATP, CTP, and GTP; 10 μM UTP; 10 μCi of [α-32P]UTP (3,000 Ci/mmol); RNA template, and enzyme. When the crude extract (S100) was used, actinomycin D was added at 100 μg/ml to inhibit endogenous DNA-dependent RNA polymerase activities. Reactions were carried out at 30°C for 1 h. RNA was extracted with phenolchloroform, precipitated with ethanol, dissolved in an RNA sample buffer (10% sucrose, 90% deionized formamide, 0.05% bromophenol blue, and 0.05% xylene cyanol), heat denatured at 70°C for 2 min, and subjected to PAGE in the presence of 8 M urea gel. After electrophoresis, the gel was fixed with 10% methanol–10% acetic acid for 20 min, rinsed with water for 20 min, and subsequently dried with a gel dryer. The dried gel was visualized with a BAS-2000 image analyzer (Fuji).
Template RNA for RNA polymerase assay.
A mini-vRNA of 249 nucleotides with the sequence corresponding to the 3′-terminal portion of TMV RNA was routinely used as a model template for the RNA-dependent RNA polymerase assay. This mini-vRNA was synthesized by transcribing TMV cDNA with T7 RNA polymerase. In brief, cDNA corresponding to a 3′-terminal segment of TMV RNA was PCR-amplified by using pTMV-T7 (a gift from M. Suzuki, University of Tokyo), which includes the full-length cDNA of the TMV-OM genome as a template and a pair of primers: (i) primer 3′(+)F (see Table 1), which includes the sequence 5′-CGC (HindIII site), T7 RNA polymerase promoter, and the viral RNA sequence between nucleotides 6147 and 6166 and (ii) primer 3′(+)R (see Table 1), which includes the sequence 5′-CGC (BamHI and NdeI sites) and the viral RNA sequence between nucleotides 6373 and 6395. After digestion of this PCR-amplified cDNA with HindIII and BamHI, the resulting fragment was ligated into pUC18, which had previously been treated with HindIII and BamHI to give plasmid pTMV3′(+)-T7. For generation of the model viral RNA template, pTMV3′(+)-T7 was linearized by treatment with NdeI and transcribed in vitro by using T7 RNA polymerase. An RNA transcript of 249 nucleotides containing the 3′(+) noncoding sequence of viral RNA, from nucleotide 6147 to 6395 (the 3′ terminus) (see the sequence of 3′-terminal proximal region in Table 1), was used as the template for the detection of the viral RNA polymerase activity. For routine assay, 0.5 pmol of this model RNA was used. A segment of viral RNA containing the coding region for 30K protein was prepared by transcribing plasmid pET30K with T7 RNA polymerase and was kindly provided by Takumi Shimizu (University of Tokyo).
The 53-base v53 RNA is used as a model vRNA template for the influenza virus RNA polymerase assay (22), while the 64-base mRNA2 is used in the functional analysis of the influenza virus RNA polymerase (6a). Both RNAs were synthesized by transcribing the respective DNA templates with T7 RNA polymerase.
RESULTS
Nucleotide sequence of the RNA polymerase gene of TMV-OM.
The entire sequence of the 183K-ORF (nucleotide positions 69 to 4919) of the TMV-OM strain was determined in both orientations by using two sets of at least three overlapping deletion clones prepared from two independently isolated cDNA clones, pUC183K-1 and pUC183K-2. For some unclear regions, direct sequencing of PCR products was carried out by using viral RNA as template. Taken together with the published terminal sequences, i.e., the 5′-terminal sequence from positions 1 to 275 (19) and the 3′-terminal sequence from positions 4768 to 6395 (18, 33) of the TMV-OM genome RNA, the complete nucleotide sequence has been established for the entire genome of TMV strain OM.
The size of the 183K-ORF is identical between two related TMV strains, vulgare and OM. The nucleotide sequence of TMV-OM is similar to that of TMV-vulgare, showing 99.0% homology with a difference of only 51 nucleotides within 4,851 nucleotides (for complete 183K-ORF sequences, see DDBJ-EMBL-GenBank DNA database accession numbers D78608 [OM] and J02415 [vulgare]). The difference in RNA sequence leads to amino acid sequence changes in six amino acid residues within a total of 1,616 residues. The methyltransferase (M), helicase (H), and RNA-dependent RNA polymerase (P) domains are conserved in the same order among alphavirus-like superfamily viruses, each domain containing several conserved sequence segments. The six mutations identified in the OM strain are all located outside the conserved regions of the methyltransferase, helicase, or RNA polymerase polyprotein. At nucleotide positions 624 to 629 from the 5′ terminus of the genome, an A at nucleotide position 624 of vulgare is missing in OM sequence but, instead, an extra A is present at position 629 in OM. These two changes result in a frameshift between nucleotide positions 624 to 629 and amino acid substitution at two residues, Met186Cys and Arg187Glu in the M domain. A single A-to-G substitution at nucleotide 2074 leads to a Lys669Arg change between the M and H domains. Three base substitutions, U-to-C at 3658, U-to-A at 4806, and A-to-U at 4864, result in amino acid substitutions Ile1197Thr, Trp1580Arg and Tyr1599Phe, respectively, in the P domain.
The 5′-terminal sequence between positions 1 and 68 (including the 5′-noncoding region) and the 3′-terminal sequence between positions 4768 and 6395 (including the 30K protein-ORF, the coat protein-ORF, and the 3′-terminal noncoding region) are 100 and 98.9% identical, respectively, between TMV-OM and vulgare.
Expression and purification of M, H, and P domain polypeptides.
The 183K protein consists of three putative functional domains, each carrying the M, H, or P motif. E. coli expression plasmids were constructed for the production of three different polypeptides, M, H and P, each containing one unique functional domain corresponding to the amino acid residues 1 to 306 (306 residues), 806 to 1116 (310 residues), and 1143 to 1616 (473 residues), respectively. For the purification of these polypeptides, a H6 tag was added to the carboxy terminus of each segment polypeptide. All of these polypeptides formed inclusion bodies after cell disruption, which were solubilized in 6 M guanidine-HCl and purified by Ni2+ affinity column chromatography. Each polypeptide was further purified by SDS-PAGE, eluted from the gel, and used to raise antibodies in rabbits. The domain-specific polyclonal antibodies were produced against the M and P polypeptides, but high-titer antibodies were not produced against the H polypeptide.
Isolation of the RNA polymerase from TMV-infected tobacco.
TMV-coded RNA polymerase is known to be associated with membrane fractions of virus-infected plants (6, 26, 29). By immunodetection with antibodies against the M- and P-domain proteins, we confirmed that both the 126 and the 183K proteins are associated with pellet membrane fractions (P30) of TMV-infected tobacco leaf extracts after centrifugation at 30,000 × g (data not shown). For solubilization of the 126K and 183K proteins, the P30 membrane fraction was resuspended in buffer B containing various detergents each at 1% concentration, such as Nonidet P-40, Triton X-100, Triton N-101, sodium deoxycholate, sodium cholate, TDC, Tween 20, Tween 80, and digitonin, followed by centrifugated at 100,000 × g. The supernatant (S100) was treated for immunoprecipitation with the anti-M antibodies, which could cross-react with both the 126K and 183K proteins, and the antigen-antibody complexes thus formed were analyzed by SDS–7.5% PAGE (Fig. 1A). In all cases except for the TDC treatment, the yields of the two RNA polymerase proteins, 126K and 183K, in the S100 fractions were low (data not shown). The yield of the 126K protein in the TDC-treated S100 fraction was at least 10 times more than that of untreated S100, as measured by protein staining with silver stain (Fig. 1A, lanes 2 and 4). These results are in good agreement with the recent finding by Osman and Buck (21).
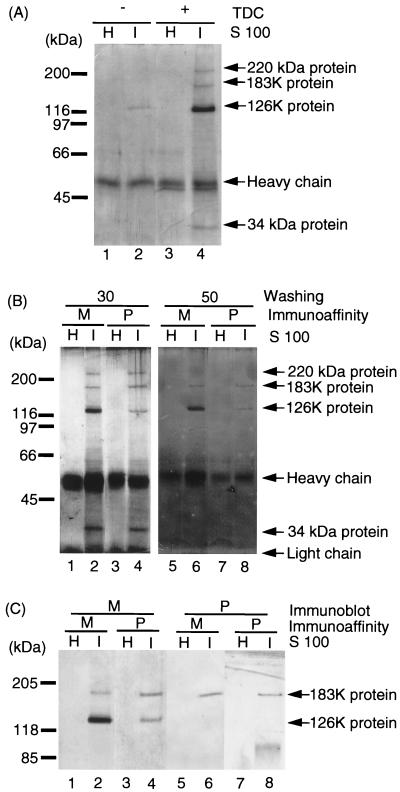
FIG. 1.
Immunoaffinity purification of the 126K and 183K proteins with anti-M and anti-P antibodies. (A) Immunoprecipitation of the 126K and 183K proteins from TDC-treated extracts of TMV-infected tobacco. By starting with 1.25 g of TMV-infected or uninfected tobacco leaves, 250 μl of S100 was obtained in the presence (+) or absence (−) of TDC. The S100 fractions were incubated with anti-M antibody-Sepharose. After a washing with 30 bed volumes of Tris-Triton-NaCl buffer, proteins were eluted from the immunoaffinity resins and subjected to SDS–7.5% PAGE. Proteins in the gel were detected by silver staining. H, uninfected healthy tobacco leaves; I, TMV-infected tobacco leaves. The major protein bands detected are shown on the right, i.e., the 183K and 126K TMV proteins and the 220- and 34-kDa host proteins (see text). The migration positions of the molecular-weight markers are shown on the left. (B) Immunoaffinity purification of the 126K and 183K proteins with anti-M and anti-P antibodies. TDC-treated S100 fractions of TMV-infected and uninfected healthy tobacco leaves were incubated with either anti-M or anti-P antibody-Sepharose. The antigen-bound antibody-Sepharose was washed with 30 (lanes 1 to 4) or 50 (lanes 5 to 8) bed volumes of the Tris-Triton-NaCl buffer. Eluates from these immunoaffinity Sepharose resins were subjected to SDS–7.5% PAGE, and proteins were detected by silver staining. M, anti-M antibody-Sepharose; P, anti-P antibody-Sepharose; H, healthy tobacco leaves; I, TMV-infected tobacco leaves. The proteins identified are indicated on the right, i.e., the 183K and 126K TMV proteins and the 220- and 34-kDa putative host proteins. The migration positions of the molecular-weight markers are shown on the left. (C) Identification of the 126K and 183K proteins in the immunoaffinity-purified RNA polymerase. S100 fractions prepared from healthy tobacco leaves (H) and TMV-infected tobacco leaves (I) were subjected to immunoaffinity purification of the 126K and 183K proteins by using anti-M (M) or anti-P (P) antibody-conjugated Sepharose. After a washing with 50 bed volumes of Tris-Triton-NaCl buffer, the eluates from the immunoaffinity columns were subjected to SDS–7.5% PAGE, and the proteins were detected by immunostaining with polyclonal antibodies against M (lanes 1 to 4) or P (lanes 5 to 8). The positions of the molecular-weight markers are shown on left.
In order to test the protein composition of the RNA polymerase, the S100 fraction was subjected to immunoprecipitation with the anti-M and anti-P antibodies. Both the 126K and 183K proteins were identified in the antigen-antibody complexes formed with not only the anti-M but also the anti-P antibodies (Fig. 1B). Treatment of the same S100 fraction with a preimmune serum did not precipitate the 126K and 183K proteins (data not shown). Identification of the 126K protein by immunoprecipiation with the anti-P antibodies, which did not recognize the isolated 126K protein (see below), itself indicates the presence of complexes between the 126K and 183K proteins.
In addition to the 126K and 183K viral proteins, two additional proteins with the apparent molecular masses of 34 and 220 kDa were identified, which may represent host factors associated with the viral RNA polymerase (Fig. 1B, lanes 2 and 4). These putative host factors were identified with antigen-antibody complexes formed with both anti-M and anti-P antibodies after a washing with 30 bed volumes of Tris-Triton-NaCl buffer (Fig. 1B, “30,” lanes 2 and 4) but were released after a washing with 50 bed volumes of the buffer (Fig. 1B, “50,” lanes 6 and 8), suggesting that the affinities of these putative host factors in the RNA polymerase complexes are weaker than that between the 126K and 183K proteins, the putative subunits of viral RNA polymerase (see below). These putative host factors did not bind to Sepharose without antibodies (data not shown).
Molecular structure of the immunoaffinity purified RNA polymerase.
We next isolated the RNA polymerase proteins by immunoaffinity adsorption with the anti-M or the anti-P antibody-conjugated Sepharose and analyzed the protein compositions of immunopurified fractions by SDS-PAGE. The fraction obtained with the anti-M antibodies contained more 126K protein than 183K protein as detected by both Coomassie brilliant blue staining (data not shown) and silver staining (Fig. 1B, lane 2). The RNA polymerase fraction obtained with the anti-P antibodies, however, contained nearly equal amounts of two components, the 183K and 126K proteins, as detected by silver staining (Fig. 1B, lane 4). Both fractions exhibited the activity of RNA synthesis in vitro when supplemented with a short viral RNA as a model template (see below).
By immunostaining, the 126K protein cross-reacted with the anti-M antibodies (Fig. 1C, lanes 2 and 4) but not with the anti-P antibodies (Fig. 1C, lanes 6 and 8), while the 183K protein cross-reacted against both the anti-M and the anti-P antibodies (Fig. 1C, lanes 2, 4, 6, and 8). By Coomassie brilliant blue staining (data not shown) or silver staining (see Fig. 1B, lane 2), the content of the 126K protein in the 126K/183K fraction obtained with the anti-M antibodies was more than 10-fold that of the 183K protein. In good agreement with the patterns of protein staining, the intensity of immunostaining of the 126K protein was approximately 30-fold higher than that of the 183K protein (Fig. 1C, lane 2). On the other hand, the fraction obtained with the anti-P antibodies contained almost equal amounts of both the 126K and 183K proteins as detected by silver staining (Fig. 1B, lanes 4 and 8) or immunostaining with anti-M antibodies (Fig. 1C, lane 4). Taken together, these findings lead us to propose that the RNA polymerase protomer of TMV is composed of one molecule each of the 126K and 183K proteins and that the virus-infected cells contain excess free 126K protein.
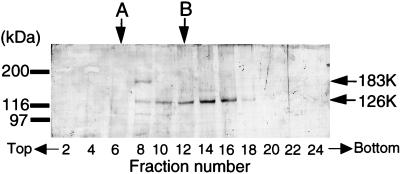
To confirm this prediction, we next subjected the S100 fraction, together with several size-marker proteins, to glycerol gradient centrifugation. As shown in Fig. 2, the 183K protein formed a single peak between 240-kDa bovine liver catalase and 450-kDa E. coli RNA polymerase holoenzyme. In the 183K peak fraction, an almost equimolar amount of the 126K protein was identified. Treatment of this fraction with anti-P and anti-M antibodies precipitated both the 126K and the 183K proteins (data not shown). This finding supported our conclusion that the TMV RNA polymerase is composed of one molecule each of the 126K and the 183K protein, with the calculated molecular mass of 309 kDa for the 126K/183K heterodimer. On the other hand, the unbound 126K protein alone formed a diffuse peak, sedimenting faster than the 126K-183K heterodimer, suggesting that the 126K alone formed oligomers or it was associated with an as-yet-unidentified host protein(s).
FIG. 2.
Glycerol gradient centrifugation of the 126K-183K complex. The S100 fraction from TMV-infected tobacco leaves was mixed with the molecular-size marker proteins as described in Materials and Methods and centrifuged on a 10 to 40% linear gradient of glycerol in Tris-MgCl2-DTT buffer for 8 h at 45,000 rpm. Aliquots of the glycerol fractions were fractionated by SDS-PAGE. The gel was treated for Western blot analysis against the anti-M antibodies. The location of the marker proteins is shown by arrows: A, bovine liver catalase (240 kDa); B, E. coli RNA polymerase (450 kDa).
Viral RNA-dependent RNA synthesis by immunoaffinity-purified RNA polymerase.
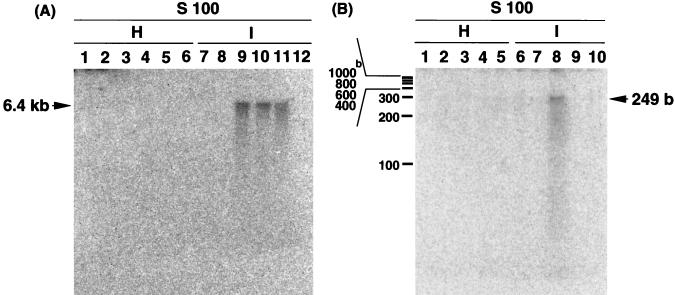
The TMV RNA polymerase is considered to recognize the 3′-terminal sequence of genome RNA as the promoter for transcription (or the origin for the negative-strand template synthesis in replication). We then constructed a model mini-vRNA template of 249 nucleotides, which contained the 3′-terminal sequence of vRNA between nucleotide positions 6147 and 6395 (the 3′ terminus). When model mini-vRNA template-dependent RNA synthesis in vitro was examined by using the antigen-antibody complexes as the enzyme, we detected the synthesis of RNA which migrated as fast as the 249-nucleotide template RNA as analyzed by urea–6% PAGE (Fig. 3, lanes 5 and 7).
FIG. 3.
Model mini-vRNA template-directed RNA synthesis in vitro by TMV RNA polymerase. The model mini-vRNA template of 249 nucleotides, prepared by transcribing pTMV3′(+)-T7 template by T7 RNA polymerase, corresponds to 3′-terminal noncoding positive-strand RNA between positions 6147 and 6395 (the 3′ terminus). The immunoaffinity-purified RNA polymerase was added to the reaction mixture of the standard in vitro transcription assay, including the model mini-vRNA template and [α-32P]UTP as a labeled substrate. The enzymes used were as follows: lanes 1 and 2, S100 fractions from healthy (H) and TMV-infected (I) tobacco; lane 3, no added enzyme; lanes 4 and 5, the RNA polymerase fractions isolated with anti-M antibodies after a washing with 30 bed volumes of the Tris-Triton-NaCl buffer; and lanes 6 and 7, the RNA polymerase fractions isolated with anti-P antibodies after a washing with a 30 bed volumes of the Tris-Triton-NaCl buffer. RNA products were analyzed by urea–6% PAGE, and the gel was visualized with a BAS-2000 image analyzer (Fuji). The positions of single-stranded RNA molecular-size markers are shown on the left.
When the S100 fraction from uninfected tobacco was used as an enzyme source, the synthesis of model template-sized small RNA was also detected (Fig. 3, lane 1), suggesting that a cellular RNA polymerase(s) is able to utilize the model RNA as a template or a primer for RNA synthesis in vitro. However, the activity of model mini-vRNA-dependent RNA synthesis became negligible after immunoaffinity purification of the enzyme from uninfected healthy plants (Fig. 3, lanes 4 and 6), indicating that the activity of the 126K/183K protein fraction purified with immunoaffinity resins is exclusively due to the viral RNA-dependent RNA polymerase. The activity of virus-specific RNA synthesis was observed for both of the immunopurified proteins by using anti-M and anti-P antibodies (Fig. 3, lanes 5 and 7). The S100 fraction from not only TMV-infected tobacco but also uninfected healthy plants produced not only the model template-sized RNA but also RNA that was more than 1,000 nucleotides long (Fig. 3, lanes 1 and 2). This activity also disappeared after immunoaffinity purification of the viral RNA polymerase (Fig. 3, lanes 4 to 7).
The activity of model RNA-directed RNA synthesis in vitro on the basis of the equivalent quantity of the 183K protein was almost equal between the RNA polymerase preparations obtained with anti-M and anti-P antibodies (Fig. 3, lanes 5 and 7). As noted above, the fraction obtained with the anti-M antibodies contained the 126K protein at least 10-fold more than the 183K protein. Thus, the catalytic site of RNA polymerization may be located on the 183K protein subunit in the 126K/183K heterodimer.
Template selectivity of the TMV RNA polymerase.
For detection of the TMV RNA polymerase, we used a model mini-vRNA template carrying the 3′-terminal sequence of TMV RNA. Next we tested whether the full-length TMV RNA can be transcribed by the immunoaffinity-purified 126K/183K heterodimeric form of RNA polymerase. As shown in Fig. 4A, both viral RNA prepared from TMV virions (lanes 5 and 11) and an RNA transcript of full-length TMV-cDNA with (lanes 3 and 9) or without 5′ cap structure (lanes 4 and 10) served as a template and produced RNA products as long as the 6.4-kb TMV RNA.
FIG. 4.
Template activity of various RNAs in RNA synthesis in vitro by TMV RNA polymerase. (A) Template activity was tested for viral RNA from CMV strain Y (lanes 1 and 7), CMV RNA segment 3 (lanes 2 and 8), transcript of the full-length cDNA of TMV strain OM added with the 5′-cap structure (lanes 3 and 9), the same RNA without the 5′-cap structure (lanes 4 and 10), and viral RNA from TMV strain OM (lanes 5 and 11). Lanes 6 and 12, no added template. The 5′-cap structure of the transcript of TMV cDNA was added by using the vaccinia virus capping enzyme. To 0.06 pmol each of these templates, the immunoaffinity-purified RNA polymerase from healthy (H, lanes 1 to 6) or virus-infected (I, lanes 7 to 12) was added as described in Fig. 3. In vitro transcription assay was carried out under the standard conditions with [α-32P]UTP as a labeled substrate. RNA products were analyzed by urea–4% PAGE, and the gel was visualized with a BAS-2000 image analyzer (Fuji). The migration position of full-length TMV RNA is shown on the right. (B) Template activity was tested for the 53-base model mini-vRNA for influenza virus RNA polymerase (lanes 1 and 6); 64 base mRNA2 used in the analysis of influenza virus RNA polymerase (lanes 2 and 7); the 249-base model mini-vRNA for TMV RNA polymerase (lanes 3 and 8); 807-base TMV RNA, including the 30K protein coding sequence (lanes 4 and 9); and no RNA control (lanes 5 and 10). All of these RNAs were prepared by transcribing the respective template DNAs with T7 RNA polymerase. In vitro transcription assay was carried out by using 0.5 pmol each of these templates and under the standard reaction conditions as above. RNA products were analyzed by urea–8% PAGE.
As an attempt to reveal the template selectivity of TMV RNA polymerase, we examined RNA synthesis in vitro with various RNA templates. The genome of CMV includes four RNA segments, RNA 1 (3,361 bases), RNA 2 (3,051 bases), RNA 3 (2,217 bases), and RNA 4 (1,034 bases) (for the sequences, see references 15 to 17). As shown in Fig. 4A, neither the four vRNA segment mixture (lanes 1 and 7) nor the vRNA segment 3 alone (lanes 2 and 8) produced detectable transcripts.
To test the requirement of the 3′-terminal proximal sequence of TMV RNA for the template activity, an 807 base-long internal segment of TMV RNA from nucleotides, 4903 to 5709 corresponding to the 30K protein coding region was prepared by transcribing the cDNA, but it did not give any specific transcript (Fig. 4B, lanes 4 and 9). We also tested the model 53-base vRNA template for the influenza virus RNA polymerase (22) and a 64 base mRNA2 used in functional analyses of the influenza virus RNA polymerase (6a). Both RNAs were inactive in directing RNA synthesis by the TMV RNA polymerase (Fig. 4B, lanes 1 and 6 and lanes 2 and 7). Thus, we concluded that the TMV RNA polymerase complex, containing 126K/183K protein heterodimer, is template specific.
DISCUSSION
Molecular composition of the TMV RNA polymerase.
Model templates carrying transcription promoter and/or replication origin sequences of the viral genomes were established and widely used for detection of viral RNA polymerases from negative-strand animal viruses (22; also reviewed in reference 8). RNA polymerases from negative-strand RNA viruses require ribonucleoprotein templates for the efficient transcription of the genome-sized RNA, but short model templates can be transcribed in the absence of RNA-bound nucleoprotein (7). Here we demonstrated that a model viral RNA of short size can be used as a template by the RNA-free viral RNA polymerase from positive-strand virus-infected cells. Up to the present time, however, it was not known whether the full-length TMV RNA can serve as a template for the solubilized RNA polymerase and be used to generate the template-sized transcript. For detection of the TMV-specific RNA polymerase activity, we then constructed a model mini-vRNA template 249 nucleotides in length containing the 3′-terminal proximal sequence of the TMV genome RNA and used it as a template during the purification of TMV RNA polymerase.
The model RNA constructed in this study was, however, transcribed by extracts from not only virus-infected plants but also uninfected healthy plants (see Fig. 3, lanes 1 and 2), indicating that host RNA-dependent RNA polymerases and/or terminal nucleotidyltransferase recognize this model RNA as either a template or a primer of RNA synthesis in vitro. Recently, Osman and Buck (20) succeeded in removing the cellular RNA-dependent RNA polymerase activity from the viral enzyme by sucrose density gradient centrifugation in the absence of Mg2+. Here we succeeded in separating the cellular RNA polymerase activities by using immunoaffinity isolation of the viral RNA polymerase with anti-M and anti-P antibodies (see Fig. 3, lanes 5 and 7). The TMV RNA polymerase free from the cellular enzyme activities was able to transcribe the model RNA.
The RNA polymerase obtained by immunoaffinity absorption with anti-P antibodies contained both the 126K and 183K proteins at approximately the molar ratio of 1:1 (see Fig. 1B, lanes 4 and 8). Thus, the most likely model is that the molecular composition of TMV RNA polymerase is a heterodimer consisting of one molecule each of 126K and 183K proteins. This prediction was indeed confirmed by glycerol gradient centrifugation (see Fig. 2). Previously, Saito et al. (28) detected only the 183K protein by immunoprecipitation of TMV-infected tobacco protoplasts with antibodies against a 183K-specific region. In their experiments, however, the protoplast samples were treated with SDS, which might dissociate the 126K/183K heterodimer.
On the other hand, not only the 126K/183K heterodimer but also the free 126K protein could be purified using anti-M antibodies. The affinity-purified protein fraction isolated with the anti-M antibodies contained at least 10-fold more 126K protein than the 183K (see Fig. 1B, lane 2, and Fig. 1C, lane 2). Since we detected a large excess of free 126K protein in virus-infected cell extracts, there should be a large excess of unassembled free 126K proteins in virus-infected plants. The biological function(s) of the free form(s) of 126K protein remains to be determined. Mutant TMV-L with substitution of its amber codon at the junction between the 126K and 183K ORFs for a tyrosine codon was shown to replicate in vivo in the absence of 126K protein (9), indicating that the 126K protein is not essential for RNA synthesis. However, the growth rate of this mutant virus was about one-tenth the rate of wild-type virus. Thus, the 126K protein may control the formation of 126K/183K heterodimer or it may also be needed at a step in transcription and/or replication after the synthesis of minus-strand RNA.
The 126K/183K heterodimer isolated by immunoaffinity chromatography with either anti-M or anti-P antibodies exhibited the activity of model RNA-directed RNA synthesis even in the simultaneous presence of antibodies, indicating that the major epitopes recognized by the antibodies generated in this study do not include the catalytic sites for RNA polymerization. In contrast, the antibodies generated by Osman and Buck (21) against M, H, and P domains all inhibited the exogenous template-dependent RNA synthesis by the membrane-bound RNA polymerase. This difference may be due to (i) the difference in the protein segments of the TMV RNA polymerase used as antigens; (ii) the different effect of tags, either the His tag (this study) or the maltose-binding protein (MBP) tag (20), on the antigenicity; (iii) the difference in the protein purity on the enzyme activity, i.e., immunopurified proteins (this study) or membrane-bound enzyme complexes (20); and (iv) the difference in templates used for activity detection with model RNA (this study) or full-sized vRNA (20).
Template selectivity of the TMV RNA polymerase.
The 3′-terminal segment of TMV RNA of 249 nucleotides was found to serve as the template for the solubilized, endogenous template-free RNA polymerase, indicating that the promoter for the first-step reaction of transcription is located within this sequence. This model mRNA contains both the 3′-terminal tRNA-like structure and the pseudoknot region immediately upstream from the tRNA-like structure, which are required for the efficient replication of TMV RNA in tobacco and protoplasts (34). Since the template-sized transcripts are produced by using both the intact TMV RNA and the model mini-vRNA, the initiation site of transcription in the in vitro transcription system by the TMV RNA polymerase must be located near the extreme 3′ termini of both RNA templates, indicating that the promoter on the plus-stand RNA is located within the 249-base sequence and the extreme 3′-terminal sequence of ---CCCGGUAGGGGCCCA(3′) provide the site of transcription initiation.
The non-TMV RNAs examined in this study were all inactive in directing RNA synthesis by the TMV RNA polymerase. All four RNA segments of CMV carry the 3′-terminal sequence of ---UCCAAAAGGAGACCA(3′). The primary sequences of CMV RNAs, including the 3′-terminal CCA sequence, are similar to that of TMV RNA but were virtually inactive as the template for the TMV RNA polymerase. This finding supports the prediction that an as-yet-unidentified sequence or RNA conformation other than the 3′-terminal sequence is also needed for promoter recognition by the TMV RNA polymerase. The 53-base influenza virus model mini-vRNA carries the 3′-terminal promoter sequence of ---CACCCUGCUUUUGCU(3′), which is recognized by the influenza virus RNA polymerase (22), while the 64 base nontemplate control M-RNA2 carries the 3′-terminal sequence of ---UUUUCCAACCCGGGA(3′). The 30K protein RNA carries the 3′-terminal sequence of ---CGACUCUAGAGGAUC(3′). All of these RNAs were inactive as the template for the TMV RNA polymerase. These RNAs do not carry the tRNA- or pseudoknot-like structures or the TMV-like 3′-terminal sequence. These data altogether suggest that the TMV RNA polymerase carries a high level of selectivity for the promoter sequence. Details of the RNA sequence or structure requirement for the promoter activity remain to be determined.
Host factors associated with the TMV RNA polymerase.
RNA viruses encode viral RNA-dependent RNA polymerases, the basic apparatus in both transcription and replication of the virus genomes. In most cases so far analyzed, however, host proteins are additionally required for complete cycles of either transcription or replication. Host factors are supposed to modulate the activity or specificity of viral RNA polymerases. For instance, the RNA polymerase of positive-strand RNA bacteriophage Qβ contains three host-encoded proteins as the enzyme subunits (1, 35) and one host protein for activity control (14). So far several host proteins have been identified which are associated with viral RNA polymerases from positive-strand plant RNA viruses. A protein of the molecular mass of about 50 kDa was copurified with CMV RNA polymerase (4). The 41-kDa subunit of wheat germ eukaryotic initiation factor 3 (eIF-3) was copurified with brome mosaic virus (BMV) RNA-dependent RNA polymerase, and the addition of 41-kDa eIF-3 subunit exhibited a stimulation of negative-strand RNA synthesis in vitro (23, 24). Janda and Ahlquist (13) observed the replication of BMV-RNA in yeast cells expressing viral 1a and 2a proteins (RNA polymerase proteins), suggesting that yeast cells contain the cellular factors necessary for BMV RNA replication. Moreover, the BMV RNA polymerase purified from the transformed yeast exhibited the activity of viral RNA replication in vitro (25).
The initial step of TMV replication is the synthesis of minus-strand template. The cessation of minus-strand RNA accumulation at an early stage of TMV infection was considered to be due to a decrease in an essential host component(s) needed for this step reaction (10). In this study, two putative host proteins, which were 34 and 220 kDa, were found to be copurified with the viral 126K/183K proteins. Since these two proteins were associated with the RNA polymerase fraction obtained not only with anti-M antibodies (126K/183K complex plus 126K protein) but also with anti-P antibodies (126K-183K complex), these proteins are associated with the 126K-183K heterodimeric form of TMV RNA polymerase. Preliminary experiments suggested that the level of model RNA-dependent RNA synthesis activity by the 126K/183K heterodimer polymerase is correlated to a certain extent with the amount of associated 34K and 220K proteins (35a). Possible involvement of these putative host factors in transcription and/or replication of the TMV genome RNA is being examined.
The TMV RNA polymerase purified by Osman and Buck (20) contained a variable number of host proteins; of these only the proteins of 56, 54, and 50 kDa were consistently found. The 56-kDa protein cross-reacted with antibodies against GCD-10 subunit of yeast eIF-3 (21). Moreover, the RNA polymerase activity was inhibited by anti-yeast GCD-10 antibodies, suggesting that this protein is an integral part of the TMV RNA polymerase complex (21). Both of our putative host factors should be different from GCD-10 simply because of the size difference. Since the migration position of the 55-kDa GCD-10 on SDS-PAGE overlaps with the immunoglobulin heavy chain, it is not excluded yet that our RNA polymerase preparation also contains the GCD-10 protein. The observation that the amplification of TMV-related RNA is defective in a tom1 mutant Arabidopsis thaliana (11, 12) suggests the involvement of the tom1 gene product in TMV replication. The molecular nature of tom1 protein is not determined yet. A possible relationship between the tom1 protein and the 34K and 220K proteins in our enzyme preparations remains to be determined.
ACKNOWLEDGMENTS
We thank Masashi Suzuki (University of Tokyo) for pTMV-T7 and CMV RNA and Takumi Shimizu (University of Tokyo) for pET30K.
This work was supported by Grants-in-Aid from the Ministry of Education, Science, and Culture of Japan, and CREST (Core Research for Evolutional Science and Technology) of Japan Science and Technology Corporation.
REFERENCES
- 1.Blumenthal T, Landers T A, Weber K. Bacteriophage Qβ replicase contains the protein biosynthesis elongation factors EFTu and EFTs. Proc Natl Acad Sci USA. 1972;69:1313–1317. doi: 10.1073/pnas.69.5.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fraenkel-Conrat H, Singer B, Tsugita A. Purification of viral RNA by means of bentonite. Virology. 1961;14:54–58. doi: 10.1016/0042-6822(61)90131-3. [DOI] [PubMed] [Google Scholar]
- 3.Fraenkel-Conrat H. RNA-directed RNA polymerases of plants. CRC Crit Rev Plant Sci. 1986;4:213–226. [Google Scholar]
- 4.Hayes R J, Buck K W. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell. 1990;63:363–368. doi: 10.1016/0092-8674(90)90169-f. [DOI] [PubMed] [Google Scholar]
- 5.Hebert T T. Precipitation of plant viruses by polyethylene glycol. Phytopathology. 1963;53:362. [Google Scholar]
- 6.Hills G J, Plaskitt K A, Young N D, Dunigan D D, Watts J W, Wilson T M A, Zaitlin M. Immunogold localization of the intracellular sites of structural and nonstructural tobacco mosaic virus proteins. Virology. 1987;161:488–496. doi: 10.1016/0042-6822(87)90143-7. [DOI] [PubMed] [Google Scholar]
- 6a.Honda, A. Unpublished results.
- 7.Honda A, Ueda K, Nagata K, Ishihama A. RNA polymerase of influenza virus: role of NP in RNA chain elongation. J Biochem. 1988;104:1021–1026. doi: 10.1093/oxfordjournals.jbchem.a122569. [DOI] [PubMed] [Google Scholar]
- 8.Ishihama A, Barbier P. Molecular anatomy of viral RNA-directed RNA polymerase. Arch Virol. 1994;134:235–258. doi: 10.1007/BF01310564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishikawa M, Meshi T, Motoyoshi F, Takamatsu N, Okada Y. In vitro mutagenesis of the putative replicase genes of tobacco mosaic virus. Nucleic Acids Res. 1986;14:8291–8305. doi: 10.1093/nar/14.21.8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikawa M, Meshi T, Ohno T, Okada Y. Specific cessation of minus-strand RNA accumulation at an early stage of tobacco mosaic virus infection. J Virol. 1991;65:861–868. doi: 10.1128/jvi.65.2.861-868.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishikawa M, Obata F, Kumagai T, Ohno T. Isolation of mutants of Arabidopsis thaliana in which accumulation of tobacco mosaic virus coat protein is reduced to low levels. Mol Gen Genet. 1991;230:33–38. doi: 10.1007/BF00290647. [DOI] [PubMed] [Google Scholar]
- 12.Ishikawa M, Naito S, Ohno T. Effects of the tom1 mutation of Arabidopsis thaliana on the multiplication of tobacco mosaic virus RNA in protoplasts. J Virol. 1993;67:5328–5338. doi: 10.1128/jvi.67.9.5328-5338.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janda M, Ahlquist P. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-d. [DOI] [PubMed] [Google Scholar]
- 14.Kajitani M, Ishihama A. Identification and sequence determination of the host factor gene for bacteriophage Qβ. Nucleic Acids Res. 1991;19:1063–1066. doi: 10.1093/nar/19.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kataoka J, Masuda C, Takanami Y. Complete nucleotide sequence of cucumber mosaic virus (Y strain) RNA 2. Ann Phytopathol Soc Jpn. 1990;56:495–500. [Google Scholar]
- 16.Kataoka J, Masuta C, Takanami Y. Complete nucleotide sequence of cucumber mosaic virus (Y strain) RNA 1 and evolutionary relationships among genome RNAs of other CMV strains. Ann Phytopathol Soc Jpn. 1990;56:501–507. [Google Scholar]
- 17.Nitta N, Masuda C, Kuwata S, Takanami Y. Comparative studies of the nucleotide sequence of cucumber mosaic virus RNA 3 between Y strain and O strain. Ann Phytopathol Soc Jpn. 1991;54:516–522. [Google Scholar]
- 18.Meshi T, Ohno T, Okada Y. Nucleotide sequence and its character of cistron coding for the 30K protein of tobacco mosaic virus (OM strain) J Biochem. 1982;91:1441–1444. doi: 10.1093/oxfordjournals.jbchem.a133833. [DOI] [PubMed] [Google Scholar]
- 19.Meshi T, Ishikawa M, Takamatsu N, Ohno T, Okada Y. The 5′-terminal sequence of TMV RNA. Question on the polymorphism found in the vulgare strain. FEBS Lett. 1983;162:282–285. doi: 10.1016/0014-5793(83)80772-8. [DOI] [PubMed] [Google Scholar]
- 20.Osman T A M, Buck K W. Complete replication in vitro of tobacco mosaic virus RNA by a template-dependent, membrane-bound RNA polymerase. J Virol. 1996;70:6227–6234. doi: 10.1128/jvi.70.9.6227-6234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osman T A M, Buck K W. The tobacco mosaic virus RNA polymerase complex contains a plant protein related to the RNA-binding subunit of yeast eIF-3. J Virol. 1997;71:6075–6082. doi: 10.1128/jvi.71.8.6075-6082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parvin J D, Palese P, Honda A, Ishihama A, Krystal M. Promoter analysis of the influenza virus RNA polymerase. J Virol. 1989;63:5142–5152. doi: 10.1128/jvi.63.12.5142-5152.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quadt R, Jaspars E M J. Purification and characterization of brome mosaic virus RNA-dependent RNA polymerase. Virology. 1990;178:189–194. doi: 10.1016/0042-6822(90)90393-6. [DOI] [PubMed] [Google Scholar]
- 24.Quadt R, Kao C C, Browning K S, Hershberger R P. Characterization of a host protein associated with brome mosaic virus RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 1993;90:1498–1502. doi: 10.1073/pnas.90.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quadt R, Ishikawa M, Janda M, Ahlquist P. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc Natl Acad Sci USA. 1995;92:4892–4896. doi: 10.1073/pnas.92.11.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ralph R K, Bullivant S, Wojcik S J. Cytoplasmic membranes, a possible site of tobacco mosaic virus RNA replication. Virology. 1971;43:713–716. doi: 10.1016/0042-6822(71)90295-9. [DOI] [PubMed] [Google Scholar]
- 27.Restrepo-Hartwig M A, Ahlquist P. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito T, Watanabe Y, Meshi T, Okada Y. Preparation of antibodies that react with the large non-structural proteins of tobacco mosaic virus by using Escherichia coli expressed fragments. Mol Gen Genet. 1986;205:82–89. [Google Scholar]
- 29.Saito T, Hosokawa D, Meshi T, Okada Y. Immunocytochemical localization of the 130K and 180K proteins (putative replicase components) of tobacco mosaic virus. Virology. 1987;160:477–481. doi: 10.1016/0042-6822(87)90020-1. [DOI] [PubMed] [Google Scholar]
- 30.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaad M C, Jensen P E, Carrington J C. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 1997;16:4049–4059. doi: 10.1093/emboj/16.13.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki M, Kuwata S, Kataoka J, Masuta C, Nitta N, Takanami J. Functional analysis of deletion mutants of cucumber mosaic virus RNA 3 using an in vitro transcription assay. Virology. 1991;183:106–113. doi: 10.1016/0042-6822(91)90123-s. [DOI] [PubMed] [Google Scholar]
- 33.Takamatsu N, Ohno T, Meshi T, Okada Y. Molecular cloning and nucleotide sequence of the 30K and the coat protein cistron of TMV (tomato strain) genome. Nucleic Acids Res. 1983;11:3767–3778. doi: 10.1093/nar/11.11.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takamatsu N, Watanabe Y, Meshi T, Okada Y. Mutational analysis of the pseudoknot region in the 3′ noncoding region of tobacco mosaic virus RNA. J Virol. 1990;64:3686–3693. doi: 10.1128/jvi.64.8.3686-3693.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wahba A J, Miller M J, Niveleau A, Landers T A, Carmichael G G, Weber K, Hawley D A, Slobin L I. Subunit I of Qβ replicase and 30S ribosomal protein S1 of Escherichia coli. Evidence for the identity of the two proteins. J Biol Chem. 1974;249:3314–3316. [PubMed] [Google Scholar]
- 35a.Watanabe, T. Unpublished data.
- 36.Wilcockson J, Hull R. The rapid isolation of plant virus RNAs using sodium perchlorate. J Gen Virol. 1974;23:107–111. doi: 10.1099/0022-1317-23-1-107. [DOI] [PubMed] [Google Scholar]
- 37.Zabel P, Dorssers L, Wernars K, Kammen A V. Terminal uridylyl transferase of Vigna unguiculata: purification and characterization of an enzyme catalyzing the addition of a single UMP residue to the 3′-end of an RNA primer. Nucleic Acids Res. 1981;11:2433–2453. doi: 10.1093/nar/9.11.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]