Abstract
Hepatitis C virus (HCV) glycoproteins E1 and E2 assemble to form a noncovalent heterodimer which, in the cell, accumulates in the endoplasmic reticulum (ER). Contrary to what is observed for proteins with a KDEL or a KKXX ER-targeting signal, the ER localization of the HCV glycoprotein complex is due to a static retention in this compartment rather than to its retrieval from the cis-Golgi region. A static retention in the ER is also observed when E2 is expressed in the absence of E1 or for a chimeric protein containing the ectodomain of CD4 in fusion with the transmembrane domain (TMD) of E2. Although they do not exclude the presence of an intracellular localization signal in E1, these data do suggest that the TMD of E2 is an ER retention signal for HCV glycoprotein complex. In this study chimeric proteins containing the ectodomain of CD4 or CD8 fused to the C-terminal hydrophobic sequence of E1 were shown to be localized in the ER, indicating that the TMD of E1 is also a signal for ER localization. In addition, these chimeric proteins were not processed by Golgi enzymes, indicating that the TMD of E1 is responsible for true retention in the ER, without recycling through the Golgi apparatus. Together, these data suggest that at least two signals (TMDs of E1 and E2) are involved in ER retention of the HCV glycoprotein complex.
Hepatitis C virus (HCV) is a positive-strand RNA virus which belongs to the Flaviviridae family (18). Its genome contains a long open reading frame of 9,030 to 9,099 nucleotides that is translated into a single polyprotein of 3,010 to 3,033 amino acids (33). Cleavages of this polyprotein are co- and posttranslational and generate at least 10 polypeptides, including 2 glycoproteins, E1 and E2 (50). These glycoproteins are believed to be type I transmembrane proteins with an N-terminal glycosylated ectodomain and a C-terminal hydrophobic anchor. For E2, C-terminal deletions that remove its hydrophobic region result in secretion of the ectodomain (35, 53). This is in accordance with other data proposing that the hydrophobic anchor domain begins at amino acid 718 (position on the polyprotein) (36). For E1, it has been suggested that a second membrane anchor (between amino acids 262 and 290) might exist in addition to its C-terminal hydrophobic domain (34). However, truncated forms ending at amino acid 311 or 334 and containing this internal sequence can also be secreted (23, 35). E1, like E2, is therefore probably anchored by its C-terminal hydrophobic sequence, but the N-terminal limit of this transmembrane domain (TMD) has not been established.
HCV glycoproteins E1 and E2 interact together to form a noncovalent heterodimer (11, 48). The efficiency of HCV glycoprotein assembly is low, and a large portion of them form heterogeneous disulfide-linked aggregates (13, 14). The noncovalent heterodimeric complex is believed to be the prebudding form of HCV glycoprotein oligomer and accumulates in the endoplasmic reticulum (ER) (11). Recently, the mechanism responsible for HCV glycoprotein complex localization in the ER has been analyzed (15). The absence of modifications of HCV glycoprotein glycans by Golgi enzymes indicates that the ER localization of these proteins is not due to their retrieval from the cis-Golgi region. Static retention of HCV glycoprotein complexes in the ER suggests that this compartment plays an important role in the acquisition of the envelope of HCV particles.
HCV glycoprotein E2 expressed in the absence of E1 can fold properly (35), and this has allowed us to analyze the intracellular localization of its properly folded form (9, 55). E2 expressed alone is retained in the ER, as shown by the lack of complex glycans, its intracellular distribution, and the absence of its expression on the cell surface. In addition, replacement of the TMD of E2 with the anchor sequence of CD4 has been shown to be sufficient for its export on the cell surface, and a chimeric protein containing the ectodomain of CD4 fused to the TMD of E2 is retained in the ER (9). This indicates that the TMD of E2 contains the information for its ER localization. This ER localization signal has also been shown to be responsible for true retention of E2 in the ER, without recycling through the Golgi apparatus (15).
The ER retention signal present in E2 could be sufficient to retain E1-E2 complexes in the ER. However, the presence of an ER-targeting signal in E1 as well cannot be excluded. The aim of this study was to look for a potential ER localization signal in E1. By making chimeric proteins containing the ectodomain of CD4 or CD8 fused to the C-terminal hydrophobic sequence of E1, we showed that the TMD of E1 is able to retain these ectodomains in the ER. In addition, these chimeric proteins were not processed by Golgi enzymes. This indicates that the TMD of E1 is responsible for true retention in the ER, without recycling through the Golgi apparatus.
MATERIALS AND METHODS
Cell culture.
The HepG2, HeLa, CV-1, and 143B (thymidine kinase-deficient) cell lines were obtained from the American Type Culture Collection, Rockville, Md. Cell monolayers were grown in Dulbecco modified essential medium (Gibco-BRL) supplemented with 10% fetal bovine serum.
Plasmid constructs.
Plasmids expressing chimeric proteins were constructed by using the standard methodology (52). Briefly, DNA sequences of protein domains were introduced into pTM1 plasmid (38) by PCR. Plasmids expressing chimeric proteins were constructed in two steps by introducing successively the sequences of domains from two different proteins. A unique restriction site was introduced between the sequences of the protein domains. HCV sequences were amplified from H-strain clones (17). Plasmids pTM1/CD4(1–371)-E1(347–383), pTM1/CD4(1–371)-E1(353–383), pTM1/CD8(1–159)-E1(347–383) and pTM1/CD8(1–159)-E1(353–383) contain the signal sequence of CD4 or CD8 followed by the sequence of their ectodomain in fusion with the C-terminal 37 or 31 amino acids of E1. Between these sequences, there is a junction sequence encoding two additional amino acids (Gly and Ser). Plasmid pTM1/CD8 contains the sequence of the entire CD8 glycoprotein (amino acids 1 to 214). Plasmids pTM1/E1(171–346)-CD4(374–435) and pTM1/E1(171–352)-CD4(374–435) contain the signal sequence and the ectodomain of E1 in fusion with the C-terminal 62 amino acids of CD4. Between these two sequences, there is a junction sequence encoding two additional amino acids (Leu and Gln). Plasmids containing sequences amplified by PCR were verified by sequencing.
Generation and growth of viruses.
Vaccinia virus recombinants were generated by homologous recombination essentially as described earlier (25) and plaque purified twice on 143B cells under bromodeoxyuridine selection (50 μg/ml). Stocks of vaccinia virus recombinants were grown and titrated on CV-1 monolayers. Vaccinia virus recombinants vTF7-3 (a vaccinia virus recombinant expressing the T7 DNA-dependent RNA polymerase) (19), vE1E2p7 (a vaccinia virus recombinant expressing HCV glycoproteins E1 and E2 and the p7 polypeptide) (17), vCE1 (a vaccinia virus recombinant expressing HCV proteins C and E1) (35), and vCD4 (a vaccinia virus recombinant expressing full-length CD4) (9) were used in this work.
Antibodies.
Monoclonal antibodies (MAbs) A4 (anti-E1 [13]), H53 (anti-E2 [9]), OKT8 (anti-CD8), and OKT4 (anti-CD4) (49) were produced in vitro by using a MiniPerm apparatus (Heraeus) as recommended by the manufacturer. Rabbit antibodies to PDI (SPA-890) and Rab1 were obtained from StressGen (Victoria, British Columbia, Canada) and Zymed (San Francisco, Calif.), respectively. Rabbit polyclonal antibody to mannosidase II (37) was kindly provided by K. Moremen (University of Georgia). The anti-CD4 MAb 13B8.2 was purchased from Immunotech. Rhodamine and cyanogen 2 (Cy2)-conjugated donkey anti-rabbit and anti-mouse immunoglobulin G (IgG) were purchased from Jackson Immunoresearch (West Grove, Pa.).
Metabolic labeling and immunoprecipitation.
Cells expressing HCV proteins were metabolically labeled with 35S-Protein Labeling Mix (100 μCi/ml; DuPont NEN) as previously described (13). Cells were lysed with 0.5% Triton X-100 in Tris-buffered saline (TBS; 50 mM Tris-Cl [pH 7.5], 150 mM NaCl). Immunoprecipitations were carried out as described previously (14). For in vivo labeling of glycan moieties, HepG2 cells were infected with the appropriate vaccinia virus recombinants and pulse-labeled for 30 min with 100 μCi of [2-3H]mannose (Amersham) per ml in α minimal essential medium containing 0.5 mM glucose and 10% dialyzed fetal bovine serum. After 4 h of chase, cells were lysed in TBS–0.5% Triton X-100, and the lysates were used for immunoprecipitation.
Endo H digestions.
Immunoprecipitated proteins were eluted from protein A-Sepharose in 30 μl of dissociation buffer (0.5% sodium dodecyl sulfate [SDS] and 1% 2-mercaptoethanol) by boiling for 10 min. The protein samples were then divided into two equal portions for digestion with endo-β-N-acetylglucosaminidase H (endo H; New England Biolabs) or an undigested control. Digestions were carried out for 1 h at 37°C in the buffer provided by the manufacturer. Digested samples were mixed with an equal volume of 2× Laemmli sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
Analysis of oligosaccharide material.
Immunoprecipitated [2-3H]mannose-labeled proteins were digested overnight at room temperature with 0.2 mg of N-tosyl-l-phenylalanine chloromethyl ketone (TPCK)-treated trypsin in 0.1 M ammonium bicarbonate (pH 7.9). Trypsin-treated proteins were boiled for 10 min to inactivate the trypsin, and the peptides were dried and dissolved in 20 mM sodium phosphate (pH 7.5) containing 50 mM EDTA and 0.2 mg of NaN3 per ml in 50% glycerol. The peptides were incubated overnight at 37°C in the presence of PNGase F (0.5 U; New England Biolabs). For some samples, endo H (10 mU) digestion was performed in 50 mM sodium citrate buffer (pH 5.5). Size analysis of the glycan moieties was achieved by high-pressure liquid chromatography (HPLC) on an amino-derivatized column ASAHIPAK NH2P-50 (250 by 4.6 mm) (Asahi, Kawasaki-ku, Japan) with a solvent system of acetonitrile-water of from 70:30 to 50:50 (vol/vol) at a flow rate of 1 ml/min over 80 min. Oligomannosides were identified as previously described (26) by their retention time. Separation of labeled oligosaccharides was monitored by continuous-flow detection of radioactivity with a Flo-One β detector (Packard). The abbreviations used for the sugars are as follows: GalNAc, N-acetylgalactosamine; Glc, glucose; GlcNAc, N-acetylglucosamine; and Man, mannose.
Affinity chromatography of labeled N glycans.
The lectin column (concanavalin A-Sepharose; 5 by 0.5 cm) was equilibrated at room temperature in 5 mM sodium acetate buffer (pH 5.2) containing 0.1 M NaCl, 1 mM MnCl2, 1 mM CaCl2, and 1 mM MgCl2. Glycan fractions (resulting from PNGase F digestion) were applied to the column which was then eluted with the equilibration buffer (buffer a). Weakly retained glycans were eluted with 10 mM methyl-α-d-glucoside in the equilibration buffer (buffer b) and strongly retained glycans were eluted with 100 mM α-d-mannoside (buffer c).
Indirect immunofluorescence.
Subconfluent HepG2 cells grown on coverslips were infected with the appropriate vaccinia virus recombinants at a multiplicity of infection of 3 PFU/cell. At 8 h postinfection, cells were fixed for 10 min with paraformaldehyde (4% in phosphate-buffered saline [PBS]). Cells were permeabilized or not for 30 min at room temperature with TBS containing 0.1% Triton X-100. Immunofluorescence was carried out as described earlier (11).
Flow-cytometric analysis.
For detection of cell surface expression of chimeric proteins, HeLa cells grown in six-well plates were infected with the appropriate vaccinia virus recombinants at a multiplicity of infection of 10 PFU/cell. At 8 h postinfection, cells were washed twice with PBS at 4°C and resuspended by pipetting. Cells were stained with fluorescein isothiocyanate (FITC)-conjugated anti-CD4 MAb 13B8.2 and fixed in PBS–1% paraformaldehyde for 30 min, resuspended in 1 ml of PBS, and subjected to flow-cytometric analysis with an Epics-Profile II (Coulter). For each sample, 104 cells were analyzed.
RESULTS
Design of chimeric proteins to study the presence of a potential intracellular retention signal in E1.
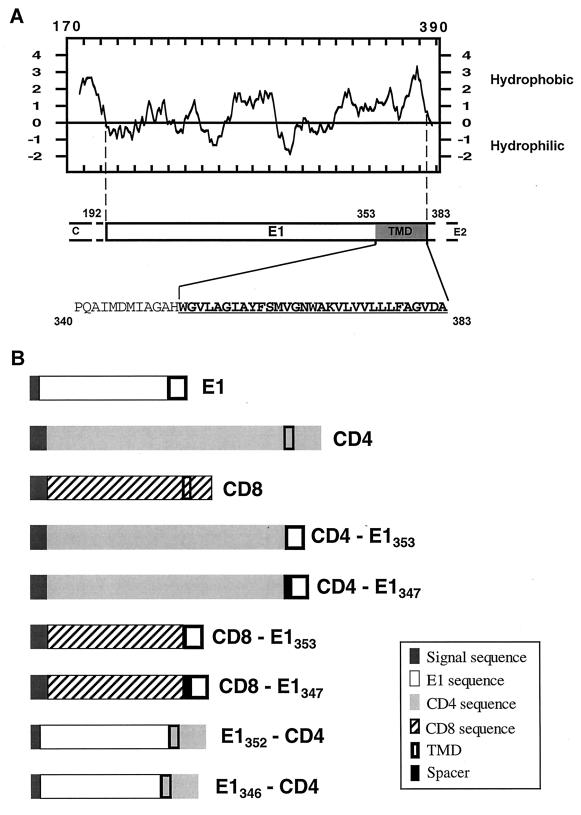
In order to identify the presence of a potential intracellular localization signal in E1, chimeras between proteins normally exported to the cell surface (CD4 or CD8) and E1 were constructed (Fig. 1B) and expressed in HepG2 cells with the vaccinia virus/T7 expression system. However, before designing the constructs, we needed a more precise localization of the limits of the domains in E1. Based on experimental data (23, 35), E1 is believed to be anchored by its C-terminal hydrophobic sequence, but the N-terminal limit of this TMD has not been established. The putative N-terminal border of E1 TMD was therefore deduced from the prediction of transmembrane segments by using the TMAP program (45) based on a multiple sequence alignment carried out with the CLUSTAL W1.6 program (56). Based on these analyses, the N-terminal residue of the TMD of E1 was identified as a tryptophane at position 353 (position on the polyprotein) (Fig. 1A). This limit was therefore taken into account to design the chimeric proteins between E1 and CD4 or CD8 (Fig. 1B). For some constructs (CD4-E1347 and CD8-E1347), 6 amino acids of the C terminus of the E1 ectodomain were added to serve as a spacer between the ectodomain of CD4 or CD8 and the predicted TMD of E1.
FIG. 1.
Schematic representation of the proteins used in this study. (A) Hydropathy plot (28) of HCV glycoprotein E1. The putative ectodomain and TMD of E1 are from amino acids 192 to 352 and amino acids 353 to 383 (position on the polyprotein), respectively. The amino acid sequence of the C-terminal region of E1 is shown, indicated by the single-letter amino acid code. The TMD of E1 is underlined. (B) Schematic representation of the parental (E1, CD4, and CD8) and chimeric proteins. CD4-E1353 and CD8-E1353, ectodomains of CD4 and CD8 fused to the TMD of E1; CD4-E1347 and CD8-E1347, same as CD4-E1353 and CD8-E1353 with an additional 6-amino-acid spacer from the ectodomain of E1 at the N terminus of the TMD of E1; E1352-CD4, ectodomain of E1 fused to the TMD and cytoplasmic domain of CD4; E1346-CD4, same as E1352-CD4 with a 6-amino-acid deletion at the C terminus of the ectodomain of E1. The ectodomain of CD4 is from amino acid 1 to 373, its TMD is from 374 to 395, and its cytosolic domain is from 396 to 435. The ectodomain of CD8 is from amino acid 1 to 160, its TMD is from 161 to 187, and its cytosolic domain is from 188 to 214. Details on the constructions are described in Materials and Methods.
The TMD of HCV glycoprotein E1 functions as an intracellular localization signal.
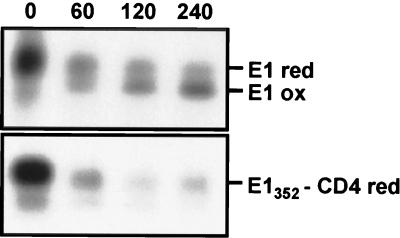
Recently, we have shown that E1 expressed alone is retained in the ER (9). Replacement of the TMD of E1 by the anchor and cytoplasmic domains of CD4 (E1352-CD4 and E1346-CD4) led to ER localization of this protein (data not shown), suggesting that a determinant for ER localization maps in the ectodomain of E1. However, E1 expressed in the absence of E2 does not fold properly (35), and misfolded proteins are usually retained in the ER independently of the presence of a specific retention signal (20). We therefore analyzed the folding of E1352-CD4 and E1346-CD4 to see whether these chimeras would be misfolded like E1 expressed alone. Since we have no conformation-sensitive MAb directed against E1, we monitored disulfide bond formation by SDS-PAGE under nonreducing conditions as previously described (14). This method takes advantage of an increase in mobility as a protein acquires a compact conformation stabilized by the formation of intramolecular disulfide bonds. An oxidized form of E1, which appeared slowly, was clearly detected when E2 was coexpressed with E1 (Fig. 2) as previously observed (14). However, SDS-PAGE analysis of E1346-CD4 (data not shown) and E1352-CD4 (Fig. 2) under nonreducing conditions did not show any shift of their electrophoretic mobilities, indicating that these proteins are not properly folded. Therefore, when E1346-CD4 and E1352-CD4 are expressed alone, we cannot differentiate an ER localization due to a specific signal from a retention caused by misfolding.
FIG. 2.
Analysis of intramolecular disulfide bond formation in E1352-CD4. HepG2 cells were coinfected with vTF7-3 and a vaccinia virus recombinant expressing E1352-CD4 at a multiplicity of infection of 5 PFU/cell. Cells coinfected with vTF7-3 and a vaccinia virus recombinant expressing E1, E2, and p7 were used as a control. At 4.5 h postinfection, infected cells were pulse-labeled for 10 min and chased for the indicated times (in minutes). Cell lysates were immunoprecipitated with MAb A4. Immunoprecipitates were analyzed under nonreducing condition by SDS-PAGE (10% acrylamide). red, reduced; ox, oxidized.
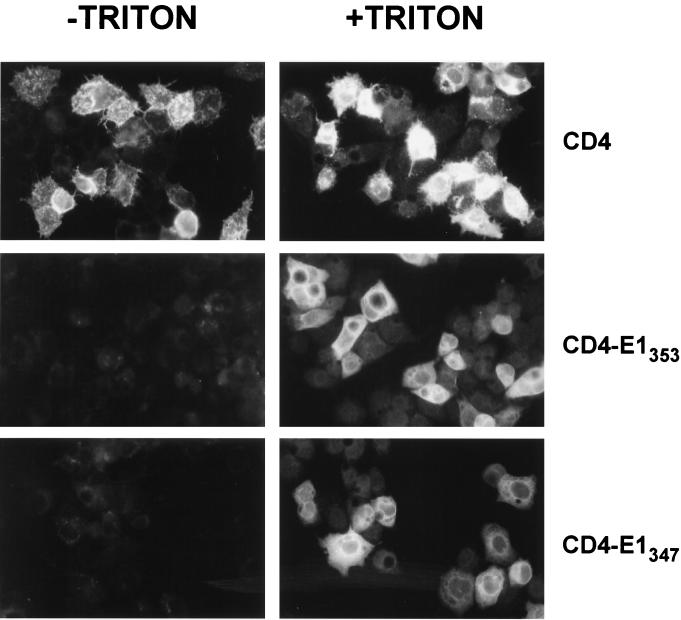
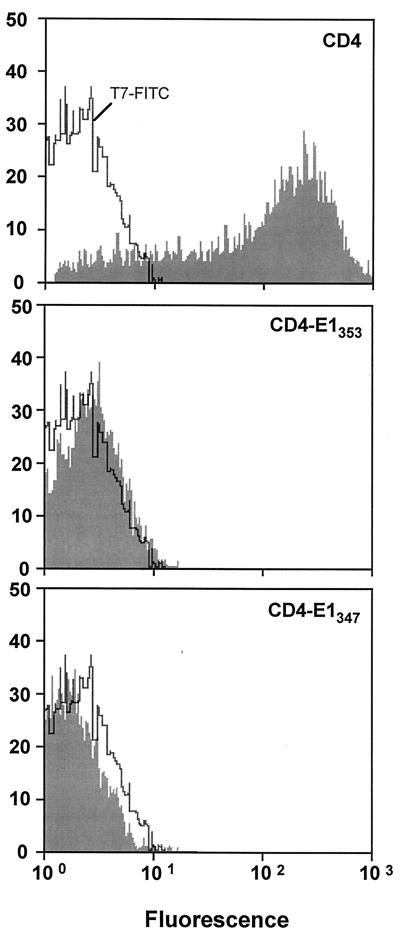
Due to the difficulties in analyzing chimeric proteins containing the ectodomain of E1, we focused our work on the potential role of the TMD of E1 in ER retention. Since transmembrane sequences are supposed to fold autonomously as α-helix structures in the lipid environment of the ER membrane (47), chimeric proteins containing the ectodomain of CD4 in fusion with the TMD of E1 (CD4-E1347 and CD4-E1353) were produced, and their subcellular localization was studied. Cell surface expression of these proteins was analyzed by immunofluorescence and flow cytometry. Cells infected by a vaccinia virus recombinant expressing full-length CD4 (vCD4) were used as a control of cell surface expression. Cells expressing CD4, CD4-E1347, or CD4-E1353 and fixed with paraformaldehyde were all positive after permeabilization with Triton X-100, as determined by immunofluorescence, whereas only CD4-expressing cells were detected in the absence of detergent (Fig. 3). The absence of detectable CD4-E1347 and CD4-E1353 on the surface of cells infected by vaccinia virus recombinants expressing these proteins was confirmed by flow cytometry (Fig. 4). For this approach, HeLa cells were used instead of HepG2 cells because of their better dissociation capacity. Intracellular retention of CD4-E1353 and CD4-E1347 does not seem to be due to misfolding of these chimeric proteins because single mutations of some amino acid residues in the TMD of E1 lead to their cell surface expression (10). Since CD4-E1353 and CD4-E1347 showed similar intracellular retention profiles (Fig. 3 and 4 and data not shown), only results obtained with CD4-E1353 are presented in the following paragraphs.
FIG. 3.
Absence of cell surface expression of CD4-E1353 and CD4-E1347. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 3 PFU/cell. At 8 h postinfection, cells were treated for indirect immunofluorescence light microscopy. Cells were fixed with paraformaldehyde, permeabilized or not with Triton X-100, and immunostained with anti-CD4 MAb OKT4 (secondary donkey anti-mouse IgG-Cy2).
FIG. 4.
Expression of chimeric proteins analyzed by flow cytometry. HeLa cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 10 PFU/cell. At 8 h postinfection, cells were immunostained with FITC-conjugated anti-CD4 MAb 13B8.2. Stained cells were fixed in PBS–1% paraformaldehyde before flow-cytometric analysis. The level of cell surface expression is indicated by the shift of the solid histogram to the right from the open control histogram (T7-FITC, MAb 13B3.2 on cells infected with vTF7-3 alone).
Together, these data indicate that the TMD of E1 functions as an intracellular localization signal.
The TMD of E1 is a signal for ER localization.
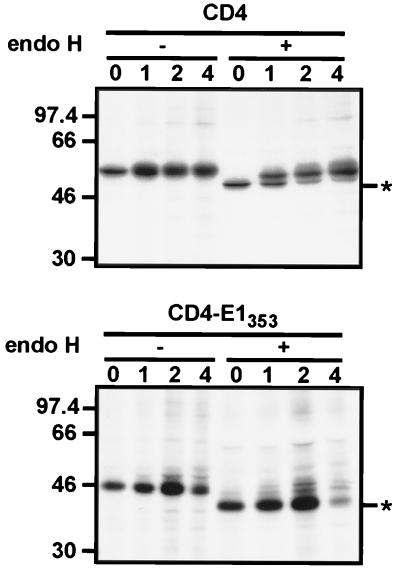
Since CD4-E1353 was retained in an intracellular compartment, we suspected that the TMD of E1 plays a role in ER localization. In a first approach to analyze the intracellular localization of CD4-E1353, its sensitivity to digestion by endo H was analyzed in pulse-chase experiments (Fig. 5). Endo H removes the chitobiose core of high-mannose and some hybrid forms of N-linked sugars but not the complex forms (51). Resistance to digestion with endo H is indicative that glycoproteins have moved from the ER to at least the medial- or trans-Golgi region, where complex sugars are added. The CD4 protein contains two N-linked glycans, and only one of them becomes endo H resistant (54). During the pulse, CD4 was sensitive to endo H treatment, and its resistant form was detected after 1 h of chase or more, whereas the chimeric protein remained endo H sensitive even after 4 h of chase (Fig. 5). This suggests that CD4-E1353 does not reach the trans-Golgi region.
FIG. 5.
Sensitivity of CD4-E1353 to endo H treatment. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. At 4.5 h postinfection, infected cells were pulse-labeled for 10 min and chased for the indicated times (in hours). Cell lysates were immunoprecipitated with MAb OKT4 and then treated or not with endo H. Samples were separated by SDS-PAGE (10% polyacrylamide). Deglycosylated proteins are indicated by asterisks. Sizes (in kilodaltons) of protein molecular-mass markers are indicated on the left.
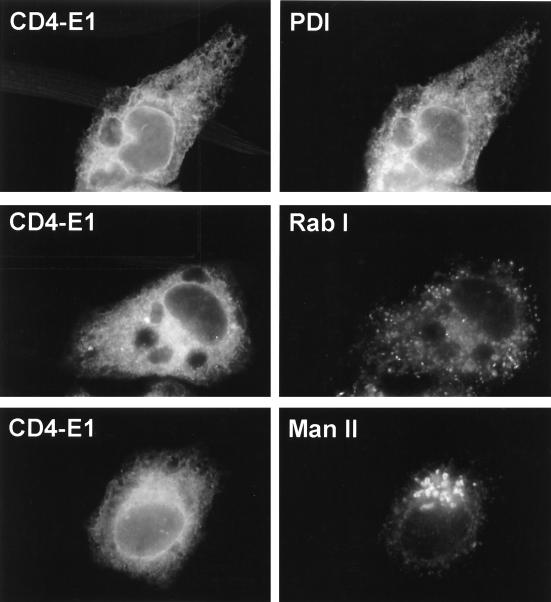
To identify the organelle(s) containing CD4-E1353, we employed double-label immunofluorescence microscopy with antibodies to known ER, intermediate-compartment, and Golgi antigens. The pattern observed after labeling with MAb OKT4 was similar to that revealed by an antibody against PDI (an ER-resident protein) and different from those shown by antibodies directed against mannosidase II (a marker of the Golgi apparatus) or Rab1 (a marker of the ER-to-Golgi intermediate compartment) (Fig. 6). These data indicate that CD4-E1353 is localized in the ER at steady state.
FIG. 6.
Indirect double-label immunofluorescence characterization of the organelle containing CD4-E1353. Subconfluent HepG2 cells grown on coverslips were infected with vTF7-3 and vCD4-E1353 at a multiplicity of infection of 3 PFU/cell. At 8 h postinfection, cells were fixed with paraformaldehyde, permeabilized with Triton X-100, and labeled with anti-CD4 MAb OKT4 (secondary donkey anti-mouse IgG-Cy2) and antibodies to PDI, Rab I, or mannosidase II (Man II) (secondary donkey anti-rabbit IgG-rhodamine red-X).
The TMD of E1 is a determinant for ER retention and not retrieval.
Keeping a protein in a subcellular compartment can be achieved either by a strict retention in this compartment or by retrieval. In the case of HCV glycoproteins, the E1-E2 heterodimer is genuinely retained in the ER without recycling through the Golgi apparatus, and the TMD of E2 has been shown to be responsible for static retention of E2 in the ER (15). Here, we wanted to know whether ER retention by the TMD of E1 would involve a similar mechanism.
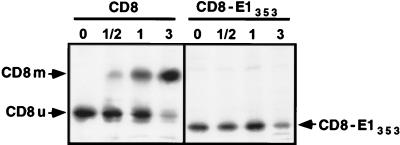
As a first approach to answer this question, chimeric proteins between the ectodomain of CD8 and the TMD of E1 were constructed (CD8-E1353 and CD8-E1347; Fig. 1). The human CD8 protein is a uniquely O-glycosylated type I membrane protein (29, 43). Since the initial step of O-glycosylation occurs in an early Golgi compartment (44), chimeric proteins containing the ectodomain of CD8 (CD8-E1353 and CD8-E1347) should be useful tools for analyzing the mechanism of retention mediated by the TMD of E1. If it involves recycling through the Golgi apparatus, addition of O-linked GalNAc should occur in the CD8 portion of the chimeric proteins. Molecules containing O-glycans should therefore accumulate after several cycles through the cis-Golgi region and back to the ER, and this should be visualized by a shift of the electrophoretic mobility in SDS-PAGE (32, 44). CD8 is synthesized as a 27-kDa species (CD8u), which is converted to a transient and initially glycosylated 29-kDa form, before the full maturation to a completely glycosylated 32- to 34-kDa doublet (CD8m). The intermediate form is generated in an early Golgi compartment, and the mature form in the trans-Golgi/trans-Golgi network region. When expressed in HepG2 cells with the vaccinia virus/T7 expression system, the intermediate 29-kDa form was barely detectable (Fig. 7). This is probably due to a faster processing of the glycans in HepG2 cells. Another characteristic of CD8 expression in HepG2 cells is the longer half-life of the 27-kDa precursor (Fig. 7) compared to previously reported data (44). When the transmembrane and cytosolic domains of CD8 were replaced by the TMD of E1 (CD8-E1353 and CD8-E1347), no shift in the electrophoretic mobility of the molecules was observed in pulse-chase experiments (Fig. 7 and data not shown). The absence of accumulation of an immature O-glycosylated form suggests that neither CD8-E1353 nor CD8-E1347 cycle through the cis-Golgi region. In addition, these data confirm that the TMD of E1 is a signal for ER localization.
FIG. 7.
Expression of CD8 and CD8-E1353 analyzed in pulse-chase experiments. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. At 4.5 h postinfection, infected cells were pulse-labeled for 10 min and chased for the indicated times (in hours). Cell lysates were immunoprecipitated with MAb OKT8 (anti-CD8). Samples were separated by SDS-PAGE (10% polyacrylamide). CD8u, unglycosylated precursor of CD8; CD8m, mature form of CD8.
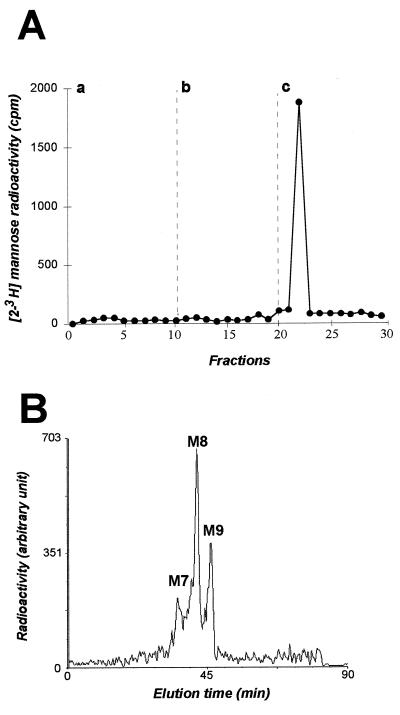
As an additional approach to study the mechanism involved in ER localization, we analyzed the modifications that CD4-E1353 glycans have potentially acquired in the compartment into which they have transited. As shown above, CD4-E1353 is endo H sensitive when analyzed in pulse-chase experiments with chase times of up to 4 h (Fig. 5). The lack of complex-type glycosylation excludes transit through the trans- but not the cis- or medial-Golgi region. In the cis-Golgi region, these proteins would be exposed to Golgi α-mannosidase I, which would process their sugar chains to Man5GlcNAc2 (27). Molecules containing Man5GlcNAc2 should accumulate after several cycles through the cis-Golgi region and back to the ER. To better characterize their potential processing, CD4-E1353 glycans were removed by PNGase F treatment and analyzed by affinity chromatography and HPLC. For this approach, CD4-E1353 was labeled with [2-3H]mannose and immunoprecipitated with MAb OKT4 before PNGase F treatment and characterization of labeled glycans. Affinity chromatography analysis of these glycans showed that 100% of them bound strongly to concanavalin A and eluted in a buffer containing 100 mM α-d-mannoside (buffer c) (Fig. 8A and data not shown), indicating that CD4-E1353 oligosaccharide moieties are of the oligomannoside type only. In addition, HPLC analysis of these glycans demonstrated the presence of three species: Man9, Man8, and Man7GlcNAc2, respectively (Fig. 8B and data not shown). Since the oligosaccharide precursor which is transferred onto nascent proteins is the Glc3Man9GlcNAc2, this reveals the sequential actions of ER glucosidases I and II and at least the action of ER mannosidase yielding Man8 species. The presence of Man7 is probably due to the trimming of mannose residues occurring after prolonged residence in the ER (21). Indeed, both Man9 mannosidase (5) and soluble ER mannosidase (6) have been demonstrated to be able to trim the mannosidic linkage down to Man6GlcNAc2 species. As expected, the affinity chromatography and HPLC profiles of the glycans associated with full-length CD4 revealed the presence of processed species characteristic of the Golgi compartment (data not shown). The nature of the glycans observed in this work confirms the data obtained with CD8-E1353 and CD8-E1347 and indicates that CD4-E1353 is retained in the ER and cannot reach the Golgi vesicles where additional processing takes place.
FIG. 8.
Analysis of the oligosaccharides bound to CD4-E1353. HepG2 cells were coinfected with vTF7-3 and vCD4-E1353. At 4.5 h postinfection, infected cells were pulse-labeled for 30 min with [2-3H]mannose, chased for an additional 4 h, and lysed with Triton X-100. Cell lysates were used for immunoprecipitation with MAb OKT4, and labeled glycans were removed by PNGase F treatment as described in Materials and Methods. Panel A represents concanavalin A-Sepharose chromatography of glycan fractions obtained after PNGase F treatment with the equilibration buffer alone (a), with 10 mM methyl-α-d-glucoside (b), or with 100 mM methyl-α-d-mannoside (c). Panel B shows HPLC analysis of glycans bound to immunoprecipitated glycoproteins after PNGase F treatment. M7, M8, and M9 indicate the oligosaccharide species possessing two GlcNAc residues at their reducing end and 7, 8, or 9 mannose residues, respectively.
DISCUSSION
Due to their limited genetic capacity, viruses exploit basic cellular mechanisms throughout their replicative cycle. For instance, the maturation of viral proteins in infected cells involves mostly host-cell metabolic pathways, including localization mechanisms, folding proteins, and enzymes that modify the primary translation product. For this reason, viral glycoproteins have often been used as tools for cell biology studies. Viral and cellular proteins in the secretory pathway contain some information in their primary structure for determining their subcellular localization. Keeping proteins in a particular compartment can be achieved either by a strict retention in this compartment or by retrieval. Many luminal and type I transmembrane proteins of the ER contain carboxy-terminal sequences of the prototypes KDEL and KKXX, respectively (39, 40). These sequences act as retrieval signals, returning proteins that have left the compartment in which they reside (41). HCV glycoproteins have been shown to localize in the ER at steady state (11), but they do not cycle between the ER and the Golgi apparatus (15). Recently, we have shown that the TMD of E2 is a signal for retention in the ER (9), and in this report we show that the TMD of E1 can play a similar function.
Immunolocalization of chimeric proteins containing the TMD of E1 and analysis of their glycans showed that this TMD is a signal for static retention in the ER. Proteins are transported from the ER to the Golgi complex by carrier vesicles that are formed from the membrane of the ER and that selectively fuse with the cis-Golgi membrane. These vesicles are coated with a set of proteins known as coatomer protein II (COPII) (4). Partitioning of membrane proteins into these vesicles is now believed to be based on a positive sorting signal in the cargo molecules which could interact with the membrane-proximal surfaces of the COPII coat proteins (24). A number of transmembrane proteins that are transported out of the ER contain the motif Asp-X-Glu (where X is any amino acid) in their cytosolic C-terminal domain. Mutational studies of this diacidic motif in the context of the cytoplasmic tail of the vesicular stomatitis virus glycoprotein G (VSV-G) showed that mutation of either acidic residue to alanine reduced by fivefold the rate of transport of VSV-G from the ER (42). A different motif (paired phenylalanine residues near the C terminus) that also specifies exit from the ER has also been identified in proteins that recycle within the early part of the secretory pathway (12, 16). There is no experimental evidence that HCV glycoproteins contain a cytoplasmic tail or that they possess a positive sorting signal. This could explain some retention of HCV glycoproteins in the ER. However, in the absence of this type of signal, we would expect to detect some slow release of HCV glycoproteins out of the ER, as was observed for mutants of VSV-G deleted of their cytoplasmic tail (42). TMDs have been previously identified as ER localization signals of some proteins (1, 58). For these proteins, it has not been shown whether a mechanism for retrieval or strict retention is involved. Similarly, the transmembrane domain of Golgi proteins and part of their flanking regions contain sufficient information for Golgi retention (41). For these proteins, subcellular localization is not due to retrieval from other compartments but to strict retention. Although the mechanism by which Golgi retention occurs is still unclear, it has been suggested that membrane thickness could play a role (7). Such a “lipid-based” mechanism could also be responsible for the ER retention mediated by the TMDs of E1 and E2.
HCV glycoprotein complex has at least two signals for ER retention. Recently, we have reported that the TMD of E2 is involved in ER localization (9), and here we show that the TMD of E1 plays a similar role. Enveloped viruses acquire their envelope by budding through one of several host cellular membranes. There needs therefore to be an accumulation of viral membrane glycoproteins, which form the spikes, in the appropriate compartment before budding can take place (46). A strategy that most of these viruses have developed is to endow the spike proteins with signals for compartment-specific localization (2, 3, 22, 30, 31, 57). The TMDs of E1 and/or E2 probably play such a role in retaining HCV glycoprotein complex in the ER where budding is supposed to occur (15). Why has the HCV glycoprotein complex evolved two signals for ER retention? One reason could be that it is necessary to maintain both proteins in the ER before they interact together to form a complex. However, these proteins also interact during their folding (8). In addition, only folded proteins are supposed to leave the ER (20). Alternatively, the fact that both TMDs act as retention signals could be due to the constraints imposed by the other functions played by these domains. Besides their role in ER retention, the TMDs of E1 and E2 also are responsible for the membrane anchor, serve as signal sequences, and are involved in E1-E2 interactions. Mutations in the TMD of one of these glycoproteins, which would suppress its ER localization function, would not probably allow this domain to retain the other functions.
In conclusion, the TMDs of E1 and E2 are both involved in static retention in the ER. As multifunctional domains, these TMDs seem to play a crucial role for HCV envelope formation, and further studies will be needed to decipher their different functions.
ACKNOWLEDGMENTS
We thank Françoise Jacob-Dubuisson for critical reading of the manuscript, F. Penin for help in the sequence analyses of E1, and Sophana Ung for excellent technical assistance. We are grateful to D. R. Littman, B. Moss, and K. Moremen for the gifts of plasmids containing the sequence of CD8, the vaccinia virus recombinant vTF7-3, and the anti-mannosidase II antibody, respectively.
This work was supported by the CNRS, the Institut Pasteur de Lille, and grant 9736 from the ARC.
REFERENCES
- 1.Ahn K, Szczesna-Skorupa E, Kemper B. The amino-terminal 29 amino acids of cytochrome P450 2C1 are sufficient for retention in the endoplasmic reticulum. J Biol Chem. 1993;268:18726–18733. [PubMed] [Google Scholar]
- 2.Andersson A M, Melin L, Bean A, Pettersson R F. A retention signal necessary and sufficient for Golgi localization maps to the cytoplasmic tail of a Bunyaviridae (Uukuniemi virus) membrane glycoprotein. J Virol. 1997;71:4717–4727. doi: 10.1128/jvi.71.6.4717-4727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong J, Patel S. The Golgi sorting domain of coronavirus E1 protein. J Cell Sci. 1991;98:567–575. doi: 10.1242/jcs.98.4.567. [DOI] [PubMed] [Google Scholar]
- 4.Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach M F, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 5.Bause E, Breuer W, Schweden J, Roeser R, Geyer R. Effect of substrate structure on the activity of Man9-mannosidase from pig liver involved in N-linked oligosaccharide processing. Eur J Biochem. 1992;208:451–457. doi: 10.1111/j.1432-1033.1992.tb17207.x. [DOI] [PubMed] [Google Scholar]
- 6.Bischoff J, Kornfeld R. The use of 1-deoxymannojirimycin to evaluate the role of various α-mannosidases in oligosaccharide processing in intact cells. J Biol Chem. 1986;261:4758–4765. [PubMed] [Google Scholar]
- 7.Bretcher M S, Munro S. Cholesterol and Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 8.Choukhi A, Ung S, Wychowski C, Dubuisson J. Involvement of endoplasmic reticulum chaperones in folding of hepatitis C virus glycoproteins. J Virol. 1998;72:3851–3858. doi: 10.1128/jvi.72.5.3851-3858.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocquerel L, Meunier J-C, Pillez A, Wychowski C, Dubuisson J. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J Virol. 1998;72:2183–2191. doi: 10.1128/jvi.72.3.2183-2191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocquerel, L., C. Wychowski, F. Minner, F. Penin, and J. Dubuisson. Unpublished data. [DOI] [PMC free article] [PubMed]
- 11.Deleersnyder V, Pillez A, Wychowski C, Blight K, Xu J, Hahn Y S, Rice C M, Dubuisson J. Formation of native hepatitis C virus glycoprotein complexes. J Virol. 1997;71:697–704. doi: 10.1128/jvi.71.1.697-704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominguez M, Dejgaard K, Fullekrug J, Dahan S, Fazel A, Paccaud J P, Thomas D Y, Bergeron J J, Nilsson T. gp25L/emp24/p24 protein family members of the cis-Golgi network bind both COP I and II coatomer. J Cell Biol. 1998;140:751–765. doi: 10.1083/jcb.140.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubuisson J, Hsu H H, Cheung R C, Greenberg H B, Russell D G, Rice C M. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol. 1994;68:6147–6160. doi: 10.1128/jvi.68.10.6147-6160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubuisson J, Rice C M. Hepatitis C virus glycoprotein folding: disulfide bond formation and association with calnexin. J Virol. 1996;70:778–786. doi: 10.1128/jvi.70.2.778-786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duvet S, Cocquerel L, Pillez A, Cacan R, Verbert A, Moradpour D, Wychowski C, Dubuisson J. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J Biol Chem. 1998;273:32088–32095. doi: 10.1074/jbc.273.48.32088. [DOI] [PubMed] [Google Scholar]
- 16.Fiedler K, Veit M, Stamnes M A, Rothman J E. Bimodal interaction of coatomer with the p24 family of putative cargo receptors. Science. 1996;273:1396–1399. doi: 10.1126/science.273.5280.1396. [DOI] [PubMed] [Google Scholar]
- 17.Fournillier-Jacob A, Cahour A, Escriou N, Girard M, Wychowski C. Processing of the E1 glycoprotein of hepatitis C virus expressed in mammalian cells. J Gen Virol. 1996;77:1055–1064. doi: 10.1099/0022-1317-77-5-1055. [DOI] [PubMed] [Google Scholar]
- 18.Francki R I B, Fauquet C M, Knudson D L, Brown F. Classification and nomenclature of viruses. Fifth report of the International Committee on Taxonomy of Viruses. Arch Virol. 1991;2(Suppl.):223. [Google Scholar]
- 19.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 21.Hebert D N, Foellmer B, Helenius A. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell. 1995;81:425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- 22.Hobman T C, Woodward L, Farquahr M G. Targeting of a heterodimeric membrane complex to the Golgi complex: rubella virus E2 glycoprotein contains a transmembrane Golgi retention signal. Mol Biol Cell. 1995;6:7–20. doi: 10.1091/mbc.6.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussy P, Schmid G, Mous J, Jacobsen H. Purification and in vitro-phospholabeling of secretory envelope proteins E1 and E2 of hepatitis C virus expressed in insect cells. Virus Res. 1996;45:45–57. doi: 10.1016/0168-1702(96)01365-2. [DOI] [PubMed] [Google Scholar]
- 24.Kaiser C, Ferro-Novick S. Transport from the endoplasmic reticulum to the Golgi. Curr Opin Cell Biol. 1998;10:477–482. doi: 10.1016/s0955-0674(98)80062-8. [DOI] [PubMed] [Google Scholar]
- 25.Kieny M-P, Lathe R, Drillien R, Spehner D, Skory S, Schmitt D, Wiktor T, Koprowski H, Lecocq J-P. Expression of rabies virus glycoprotein from a recombinant vaccinia virus. Nature. 1984;312:163–166. doi: 10.1038/312163a0. [DOI] [PubMed] [Google Scholar]
- 26.Kmiécik D, Herman V, Stroop C J M, Michalski J C, Mir A M, Labiau O, Verbert A, Cacan R. Catabolism of glycan moieties of lipid intermediates leads to a single Man5GlcNAc oligosaccharide isomer: a study with permeabilized CHO cells. Glycobiology. 1995;5:483–494. doi: 10.1093/glycob/5.5.483. [DOI] [PubMed] [Google Scholar]
- 27.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 28.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 29.Littman D R, Thomas Y, Maddon P J, Chess L, Axel R. The isolation and sequence of the gene encoding T8: a molecule defining functional classes of T lymphocytes. Cell. 1985;40:237–246. doi: 10.1016/0092-8674(85)90138-2. [DOI] [PubMed] [Google Scholar]
- 30.Locker J K, Klumperman J, Oorschot V, Horzinek M C, Geuze H J, Rottier P J M. The cytoplasmic tail of mouse hepatitis virus M protein is essential but not sufficient for its retention in the Golgi complex. J Biol Chem. 1994;269:28263–28269. [PubMed] [Google Scholar]
- 31.Machamer C E, Rose J K. A specific transmembrane domain of a coronavirus E1 glycoprotein is required for its retention in the Golgi region. J Cell Biol. 1987;105:1205–1214. doi: 10.1083/jcb.105.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martire G, Mottola G, Pascale M C, Malagolini N, Turrini I, Serafini-Cessi F, Jackson M R, Bonatti S. Different fate of a single reporter protein containing KDEL or KKXX targeting signals stably expressed in mammalian cells. J Biol Chem. 1996;271:3541–3547. doi: 10.1074/jbc.271.7.3541. [DOI] [PubMed] [Google Scholar]
- 33.Matsuura Y, Miyamura T. The molecular biology of hepatitis C virus. Semin Virol. 1993;4:297–304. [Google Scholar]
- 34.Matsuura Y, Suzuki T, Suzuki R, Sato M, Aizaki H, Saito I, Miyamura T. Processing of E1 and E2 glycoproteins of hepatitis C virus expressed in mammalian and insect cells. Virology. 1994;205:141–150. doi: 10.1006/viro.1994.1629. [DOI] [PubMed] [Google Scholar]
- 35.Michalak J-P, Wychowski C, Choukhi A, Meunier J-C, Ung S, Rice C M, Dubuisson J. Characterization of truncated forms of hepatitis C virus glycoproteins. J Gen Virol. 1997;78:2299–2306. doi: 10.1099/0022-1317-78-9-2299. [DOI] [PubMed] [Google Scholar]
- 36.Mizushima H, Hijikata M, Asabe S-I, Hirota M, Kimura K, Shimotohno K. Two hepatitis C virus glycoprotein E2 products with different C termini. J Virol. 1994;68:6215–6222. doi: 10.1128/jvi.68.10.6215-6222.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moremen K W, Touster O, Robbins P W. Novel purification of the catalytic domain of Golgi α-mannosidase II: characterization and comparison with the intact enzyme. J Biol Chem. 1991;266:16876–16885. [PubMed] [Google Scholar]
- 38.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 39.Munro S, Pelham H R B. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 40.Nilsson T, Jackson M R, Peterson P A. Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic reticulum. Cell. 1989;58:707–718. doi: 10.1016/0092-8674(89)90105-0. [DOI] [PubMed] [Google Scholar]
- 41.Nilsson T, Warren G. Retention and retrieval in the endoplasmic reticulum and the Golgi apparatus. Curr Opin Cell Biol. 1994;6:517–521. doi: 10.1016/0955-0674(94)90070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishimura N, Balch W E. A di-acidic signal required for selective export from the endoplasmic reticulum. Science. 1997;277:556–558. doi: 10.1126/science.277.5325.556. [DOI] [PubMed] [Google Scholar]
- 43.Nitsch L, Tramontano D, Ambesi-Impiombato F S, Quarto N, Bonatti S. Morphological and functional polarity of an epithelial thyroid cell line. Eur J Cell Biol. 1985;38:57–66. [PubMed] [Google Scholar]
- 44.Pascale M C, Erra M C, Malagolini N, Serafini-Cessi F, Leone A, Bonatti S. Post-translational processing of an O-glycosylated protein, the human CD8 glycoprotein, during the intracellular transport to the plasma membrane. J Biol Chem. 1992;267:25196–25201. [PubMed] [Google Scholar]
- 45.Persson B, Argos P. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol. 1994;237:182–192. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- 46.Pettersson R F. Protein localization and virus assembly at intracellular membranes. Curr Top Microbiol Immunol. 1991;170:67–104. doi: 10.1007/978-3-642-76389-2_3. [DOI] [PubMed] [Google Scholar]
- 47.Popot J-L. Integral membrane protein structure: transmembrane α-helices as autonomous folding domains. Curr Opin Struct Biol. 1993;3:532–540. [Google Scholar]
- 48.Ralston R, Thudium K, Berger K, Kuo C, Gervase B, Hall J, Selby M, Kuo G, Houghton M, Choo Q-L. Characterization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia viruses. J Virol. 1993;67:6753–6761. doi: 10.1128/jvi.67.11.6753-6761.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reinherz E L, Kung P C, Goldstein G, Schlossman S F. Separation of functional subsets of human T cells by a monoclonal antibody. Proc Natl Acad Sci USA. 1979;76:4061–4065. doi: 10.1073/pnas.76.8.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rice C M. Flaviviridae: viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 931–959. [Google Scholar]
- 51.Robbins P W, Timble R B, Wirth D F, Hering C, Maley F, Maley G F, Das R, Gibson B W, Royal N, Biemann K. Primary structure of the Streptomyces enzyme endo-β-N-acetylglucosaminidase H. J Biol Chem. 1984;259:7577–7583. [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 53.Selby M J, Glazer E, Masiarz F, Houghton M. Complex processing and protein:protein interactions in the E2:NS2 region of HCV. Virology. 1994;204:114–122. doi: 10.1006/viro.1994.1515. [DOI] [PubMed] [Google Scholar]
- 54.Shin J, Dunbrack R L, Lee S, Strominger J L. Signals for retention of transmembrane proteins in the endoplasmic reticulum studied with CD4 truncation mutants. Proc Natl Acad Sci USA. 1991;88:1918–1922. doi: 10.1073/pnas.88.5.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spaete R R, Alexander D, Rugroden M E, Choo Q-L, Berger K, Crawford K, Kuo C, Leng S, Lee C, Ralston R, Thudium K, Tung J W, Kuo G, Houghton M. Characterization of the hepatitis E2/NS1 gene product expressed in mammalian cells. Virology. 1992;188:819–830. doi: 10.1016/0042-6822(92)90537-y. [DOI] [PubMed] [Google Scholar]
- 56.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weisz O A, Swift A M, Machamer C E. Oligomerization of a membrane protein correlates with its retention in the Golgi complex. J Cell Biol. 1993;122:1185–1196. doi: 10.1083/jcb.122.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang M, Ellenberg J, Bonifacino J S, Weissman A M. The transmembrane domain of a carboxy-terminal anchored protein determines localization to the endoplasmic reticulum. J Biol Chem. 1997;272:1970–1975. doi: 10.1074/jbc.272.3.1970. [DOI] [PubMed] [Google Scholar]