Abstract
Background
Epidemics of keratoconjunctivitis may involve various aetiological agents. Microsporidia are an uncommon difficult-to-diagnose cause of such outbreaks.
Aim
During the third quarter of 2022, a keratoconjunctivitis outbreak was reported across Israel, related to common water exposure to the Sea of Galilee. We report a comprehensive diagnostic approach that identified Vittaforma corneae as the aetiology, serving as proof of concept for using real-time metagenomics for outbreak investigation.
Methods
Corneal scraping samples from a clinical case were subjected to standard microbiological testing. Samples were tested by calcofluor white staining and metagenomic short-read sequencing. We analysed the metagenome for taxonomical assignment and isolation of metagenome-assembled genome (MAG). Targets for a novel PCR were identified, and the assay was applied to clinical and environmental samples and confirmed by long-read metagenomic sequencing.
Results
Fluorescent microscopy was suggestive of microsporidiosis. The most abundant species (96.5%) on metagenomics analysis was V. corneae. Annotation of the MAG confirmed the species assignment. A unique PCR target in the microsporidian rRNA gene was identified and validated against the clinical sample. The assay and metagenomic sequencing confirmed V. corneae in an environmental sludge sample collected at the exposure site.
Conclusions
The real-time utilisation of metagenomics allowed species detection and development of diagnostic tools, which aided in outbreak source tracking and can be applied for future cases. Metagenomics allows a fully culture-independent investigation and is an important modality for public health microbiology.
Keywords: keratoconjunctivitis, Microsporidium, PCR, metagenomics, outbreak investigation, unknown aetiology, public health microbiology
Key public health message.
What did you want to address in this study?
We wanted to use advanced diagnostic methods to identify the cause of an elusive national outbreak of keratoconjunctivitis (acute eye infection) and to develop diagnostic tools that can be quickly adapted to a new outbreak investigation.
What have we learnt from this study?
With metagenomic next-generation sequencing and a novel real-time PCR assay, we established that exposure to freshwater containing Vittaforma corneae was the likely source of the keratoconjunctivitis outbreak.
What are the implications of your findings for public health?
So far, this sequencing method has mainly been applied for difficult-to-diagnose clinical cases. Our study provides proof-of-concept that this approach can be used to identify the source of an outbreak in real time and to develop outbreak investigation tools ad hoc.
Introduction
Keratoconjunctivitis is an acute eye infection implicating a wide range of causative pathogens, mainly bacteria and viruses (including staphylococci, Pseudomonas aeruginosa and adenovirus) [1]. Other causes of keratoconjunctivitis include parasites, especially free-living amoeba [2], and fungal pathogens such as Fusarium and Aspergillus [3]. All are readily identified by standard laboratory methods.
Keratoconjunctivitis may be associated with various risk factors, including environmental exposure, eye trauma or surgery, underlying ocular surface disease and use of contact lenses [1]. Epidemics of keratoconjunctivitis can occur and may involve seasonal activity of viruses such as adenovirus [4] or factors affecting amoebal activity such as hot weather, water quality and natural disasters [2].
Microsporidia are ubiquitous degenerate fungi that can cause a wide spectrum of infections in immunocompromised and immunocompetent hosts [5], affecting multiple organs such as the gastrointestinal tract, musculoskeletal system and central nervous system. Reports of microsporidial keratoconjunctivitis have increased in recent years, affecting patients across all age groups, usually with a satisfactory outcome [6]. Whether this increase in incidence is related to changing epidemiology or improved diagnosis is still debated. Because microsporidia are highly ubiquitous in natural environments, identifying an epidemiological link between an infecting species and an environmental source is essential for implementing effective control strategies [7].
Multiple Micorsporidium species have been implicated in sporadic and epidemic keratoconjunctivitis in immunocompetent hosts, especially Encephalitozoon hellem and Vittaforma corneae (previously Nosema corneum) [5]. The main diagnostic procedures for detecting ocular microsporidiosis are microscopy and PCR-based techniques. The accuracy of PCR is limited in light of the different Microsporidium species implicated in keratoconjunctivitis, and thus the definitive identification of the microsporidia at the species level remains challenging. More recently, metagenomic next-generation sequencing (mNGS) enabled the post-mortem identification of an invasive Anncaliia algerae infection in an immunosuppressed kidney transplant recipient after direct PCR failed to detect the microsporidial infection [8].
We report a comprehensive diagnostic approach, including mNGS and ad hoc PCR development and application, which enabled the diagnosis of V. corneae infection in a clinical case of keratoconjunctivitis, as part of a national outbreak of unknown aetiology, and identification of the likely water source of this infection.
Case description
Between August and November 2022, a large number of cases of keratoconjunctivitis were reported across Israel. All patients appeared to have had recreational water exposure in the Sea of Galilee in Northern Israel (Lake Kineret), a popular resort. It is estimated that at least 40 cases have been affected [9]. Standard microbiological investigations (Gram stain and culture) in different institutions failed to detect the aetiology of the outbreak.
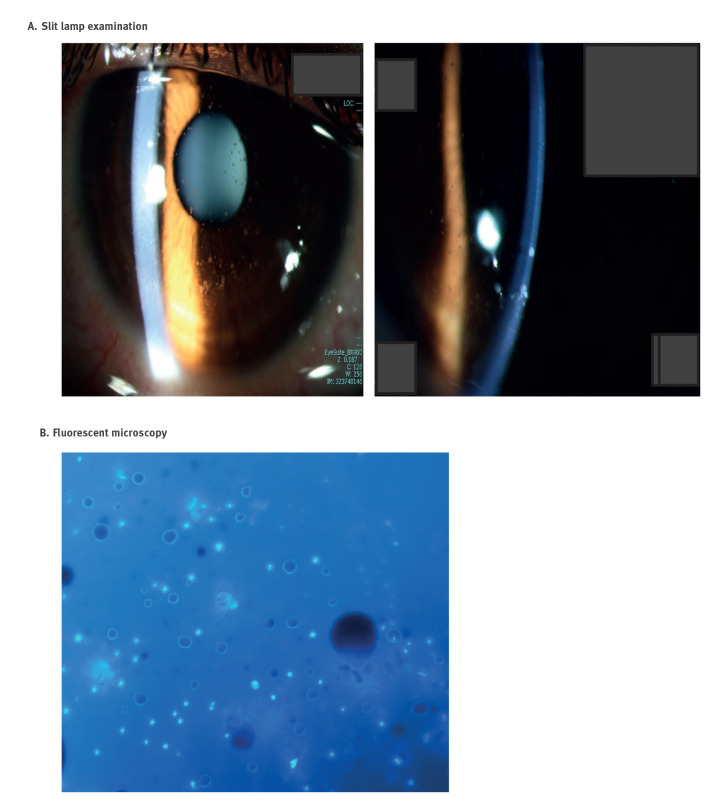
In November 2022, an 18-year-old patient (Jerusalem, Israel) presented with left eye hyperaemia and discomfort for the past 10 days. Fourteen days before the start of symptoms, they swam in the Sea of Galilee. At presentation, the visual acuity was reduced, and they had conjunctival hyperaemia and multiple and diffuse superficial round epithelial and intra-epithelial opacities (Figure 1A). A large 8.0 mm epithelial debridement was performed, and samples were submitted for microbiological investigation. After 4 days, there was a complete epithelialisation, and 11 days post-debridement, the patient was asymptomatic. During follow-up, a clear cornea was evident, with some fading subepithelial haze and no corneal staining, and their vision improved.
Figure 1.
Slit lamp and fluorescent microscopy findings, keratoconjunctivitis patient, Israel, 2022
Panel A: Slit lamp examination of the left eye with indirect illumination showing diffuse superficial corneal round opacities (left picture) and the subepithelial location of round corneal opacities (right picture).
Panel B: Fluorescent microscopy of calcofluor white stained preparation of scraped corneal material showing round white homogenous structures compatible with microsporidia. The slide was visualised with a 420 nm filter at 365 nm excitation and 395 nm emission.
Methods
Routine microbiological work-up
Corneal scraping material was subjected to standard microbiological work-up at the Clinical Microbiology Laboratory of Hadassah Medical Center, including Gram stain and microscopy, and attempted bacterial and fungal culture. We extracted DNA using the QIAamp DNA Mini Kit following pre-treatment with ATL buffer and proteinase K (QIAGEN) and performed in-house PCR assays for Acanthamoeba spp. as well as broad-range PCRs targeting the 16S ribosomal RNA (rRNA) gene (pan-bacterial assay) and the internal transcribed spacer of the rRNA gene (pan-fungal assay). Microscopy for suspected microsporidian infection was performed on corneal scraping material using Calcofluor white staining, using 30 µL of 0.1% Fluorescent Brightener 28 mixed with 10% KOH (Sigma-Aldrich). The slide was visualised under a fluorescent Axioskop microscope (Zeiss) with a 420 nm filter at 365 nm excitation and 395 nm emission.
Clinical metagenomic sequencing
We re-extracted the corneal sample using the above method, yielding 5.8 ng/µL of DNA. Whole genome sequencing was performed at the NGS Unit of the Department of Genetics, Hadassah Medical Center. Sequencing libraries were prepared from 500 ng of DNA with the Illumina Nextera Flex Library Prep Kit and sequenced on the Illumina NovaSeq6000 instrument. Sequencing was performed on an S2–300 flowcell yielding a total of 129,074,310 raw reads.
Bioinformatics analysis
Short sequence reads underwent quality control (using fastp, v0.23.2) and taxonomic assignment using kraken2 (v2.0.8) with the Standard-8, Viral and EuPathDB46 databases. The latter includes nearly 400 genomes of eukaryotic pathogens, including microsporidia (available at http://ccb.jhu.edu/data/eupathDB). We removed host (human) reads using kraken2 assignment with the Standard-8 database and KrakenTools (v1.2, extract_kraken_reads.py with parameters ‘--taxid 9606 --exclude’). To assess possible spurious taxonomical assignments, we mapped extracted reads of the taxa relatively abundant in the sample (> 100 reads) against reference genomes of the respective taxa and measured the breadth of coverage was using minimap2 (v2.22-r1101) and samtools (v1.9 with htslib v1.9).
Unclassified reads were subjected to additional analysis at protein level using mmseqs2 (v 14.7e284). The UniRef50 database (https://www.uniprot.org/help/uniref) was downloaded using the mmseqs ‘databases’ command. An mmseqs database was then created with the unclassified reads using the mmseqs ‘createdb’ command and was annotated via similarity search against the UniRef50 database using the mmseqs ‘taxonomy’ command (with the parameter ‘-s 2’). Taxonomic assignment results were summarised using the mmseqs ‘createtsv’ and ‘taxonomyreport’ commands.
Host-filtered reads (ca 1.8 million paired reads) were de novo assembled using Megahit (v1.2.9), and taxonomic assignments of the contigs were assigned using kraken2 with the EuPathDB46 database to identify potential eukaryotic pathogen sequences. Contigs identified as V. corneae in the previous step were further selected for analyses. We compared the resultant metagenome-assembled genome (MAG) with the reference genome of V. corneae ATCC 50505 (National Center for Biotechnology Information (NCBI) GenBank accession GCA_000231115.1) using progressiveMauve (v2.4.0) and mummer (v3.1). The isolated MAG contigs were arranged according to the reference genome of V. corneae ATCC 50505 contigs using the tool jContigSort (v1.3). Plots were generated using mummer2circos (v1.4.2) and pg-mauve (v0.3.1). Depth of coverage of the isolated MAG was calculated using minimap2 (v2.22-r1101) and samtools (v1.9 with htslib v1.9).
After annotating the isolated MAG with Prokka (v1.14.5), species assignment of V. corneae was confirmed by comparing the predicted small-subunit (SSU) 18S rRNA gene to the NCBI nucleotide database using megaBLAST. A multiple sequence alignment (using mafft, v7.475 and trimAl, v1.4) was conducted against V. corneae SSU 18S rRNA gene sequences obtained from the SILVA database (r138.1) and the MAG’s 18S rRNA gene, and a subsequent maximum-likelihood (ML) phylogenetic tree was built using IQ-Tree2 (v2.1.2).
Real-time PCR development and validation
We performed in silico PCR on the isolated MAG and the V. corneae ATCC 50505 reference genome using primer sequences available from the literature (Table) with seqkit (v2.3.0, using the ‘amplicon’ subcommand). An amplicon was generated from both the isolated MAG and reference genomes using the primer set LPW20469-LPW20470 while other primer sets failed (Table). Since the former primers are pan-microsporidian, we sought to utilise a V. corneae-specific primer pair. Upon close inspection of the failed primers LPW20475-LPW20476, we found a number of base variations in the MAG primer binding sites. We therefore redesigned the primers LPW20475-LPW20476 by selecting a conserved downstream region with Primer3 (using the ‘check primers’ module with the isolated MAG 18S rRNA gene as template sequence). The optimised primers (Forward -5’-GGTTGACGGGAAATTAGGGT-3’; Tm = 57.5 °C; Reverse -5’-CTGCAGCATCTCTCATACACAC-3’; Tm = 59.1 °C) were confirmed in silico against the MAG and then tested in vitro using an ad hoc SYBR green-based assay. The full real-time PCR protocol can be accessed in the Supplement. The PCR products were Sanger sequenced and analysed against the NCBI nucleotide database using megaBLAST.
Table. Primers used for in silico PCR development and validation, keratoconjunctivitis outbreak, Israel, 2022.
| Primer specificity | Primer set label | Forward (5’ → 3’) | Reverse (5’ → 3’) | Amplicon size (bases) | Reference |
|---|---|---|---|---|---|
| Pan-microsporidial | LPW20469-LPW20470 | CACCAGGTTGATTCTGCC | GTGACGGGCGGTGTGTAC | 1,200 | [27] |
| Microspo | TTCCGGAGAGGGAGCCTGAGAG | CCACTCCTTGTGGTGTCCTTCCGTCAA | 558 | [13] | |
| Vittaforma corneae species- specific | Vc-specific | CTACCAAGACAGTGACGGTTGA | GGCATCTTTTACTGCTGGAACT | 472 | [14] |
| LPW20475-LPW20476 | ACAGTGACGGTTGACGGGAA | TGAGACCTCGCATCTCTCTTT | 280 | [27] | |
| HMC-Vc | GGTTGACGGGAAATTAGGGT | CTGCAGCATCTCTCATACACAC | 224 | This study |
Environmental sampling, molecular testing and metagenomic sequencing
To identify a potential environmental source, three freshwater (à 1,000 mL) and three underwater sludge samples (à 50 mL) were obtained about 6 weeks following clinical presentation, from the specific coastline to which the patient reported exposure. Sampling involved three beach locations along a continuous coastline on a single day. Samples were processed following vigorous shaking and filtration of 20 mL using a 0.45 μm filter, followed by extraction using the DNeasy PowerWater kit (QIAGEN) per manufacturer’s instructions. DNA extracts were then tested using the novel real-time PCR (HMC-Vc) and amplicons were Sanger-sequenced for confirmation. Sequence traces showing double peaks underwent deconvolution using Tracy (v0.7.3), using the ‘decompose’ subcommand using the isolated MAG’s 18S rRNA gene as reference. An in-house 16S rRNA broad-range TaqMan qPCR assay was performed in parallel as process control.
Positive samples were further analysed using long-read metagenomic sequencing. Briefly, ca 1 µg of DNA was prepared with the SQK-LSK109 Ligation Sequencing Kit (ONT). Sequencing was carried out using the Minion Oxford Nanopore (ONT) with an R9.4.1 Flow cell (FLO-MIN106). Long reads were basecalled using Guppy (v6.0.1 + 652ffd1, with the HAC mode, and parameters ‘--trim_adapters --kit SQK-LSK109 --flowcell FLO-MIN106’). Reads longer than 1 kb were analysed using kraken2 (v2.0.8) with the Standard-8, Viral and EuPathDB46 databases, as above, to identify potential pathogens. Reads that were assigned to the phylum Microsporidia (taxid 6029) were filtered using KrakenTools (v1.2, extract_kraken_reads.py with parameters ‘--taxid 6029 --include-children’) and compared with the isolated MAG and V. corneae ATCC 50505 reference genome using BLASTn. To explore strain-level relatedness of environmental V. corneae reads, host-removed short reads of the clinical metagenome described above were mapped to all long reads that were assigned to the taxon V. corneae (taxid 42399) using minimap2 and samtools. The mapped reads were analysed for breadth and depth of coverage.
Analysis of historical samples from the Sea of Galilee
We downloaded a publicly available metagenomic sequencing data of 61 samples from the Sea of Galilee from the NCBI SRA database (https://www.ncbi.nlm.nih.gov/sra; accessed in December 2022), under the bioprojects PRJNA488159, PRJNA497963 and PRJNA637457. Taxonomic assignments of each sample’s reads were performed using kraken2 with the EuPathDB46 database to identify potential eukaryotic pathogen sequences, and summarised using KrakenTools (v1.2, combine_kreports.py). Reads that were assigned to the taxon V. corneae ATCC 50505 (taxid 993615) for each sample were mapped to the isolated MAG using minimap2. For the samples harbouring the greatest number of reads assigned to V. Corneae (SRR8088693, SRR7764488, SRR7764487, and SRR7764492), reads were mapped to the SSU of the isolated MAG. Consensus sequences were generated from these samples using samtools consensus and aligned to the amplicons generated in silico from the optimised primers as described above.
Results
Routine microbiological analyses
Routine microscopy and bacterial and fungal culture were unremarkable, yielding no pathogen. The broad-range PCR assays and the specific PCR for free-living amoeba were also negative. Calcofluor White staining analysed by fluorescent microscopy demonstrated small round fluorescent cells suggestive of Microsporidium cells (Figure 1B).
Metagenomic sequencing of the clinical sample
Sequencing of the mNGS short reads (Illumina) produced 129 million high-quality paired-end reads with an average size of 147 bp. Taxonomic assignment of the mNGS reads assigned 86.12% of the reads to human origin, while 0.01% were assigned to either bacterial or viral sequences, and the remaining 13.87% were unassigned; for the exact number of reads see Supplementary Table S1. Assigned bacterial sequences included 3,759 taxa of which only several taxa were assigned more than 50 reads. When mapped against the reference genomes of the respective genera (Salmonella, Bartonella, Klebsiella, Streptomyces, Bacillus), the reads covered only a small fraction of their genomes (between 0 and 2,000 bases), suggesting spurious assignments. Analysis against the Virus database did not reveal any pathogenic viruses; a list of these viruses is appended in Supplementary Table S2.
After removal of the host (human) reads, we compared the remaining 3.7 million reads with the eukaryotic pathogens database (EuPathDB46) using kraken2, where 96.5% of the assigned reads (34,665/35,895) matched to V. corneae, thus establishing the diagnosis of ocular microsporidiosis; we provide a list of the matched taxa in Supplementary Table S3. Another 396 reads were assigned to several other closely related microsporidia while 834 reads were assigned to 19 other eukaryotic taxa at negligible abundance (20–65 reads each), probably representing misclassified V. corneae reads.
We subsequently analysed the 3,650,413 unclassified reads at protein level. Of these, 1,921,058 reads passed the mmseqs pre-filtering steps, while 1,031,665 reads remained unclassified. An additional 90,654 reads were ambiguously assigned as either root or cellular organisms. Of the classified reads, the most abundant taxonomic assignment was V. corneae ATCC 50505 (515,515 reads; 26.8%) or other microsporidia at different taxonomical levels (69,389 reads; 3.6%). The remaining reads were assigned to several thousand different taxa across different kingdoms, at abundancies of less than 0.1%.
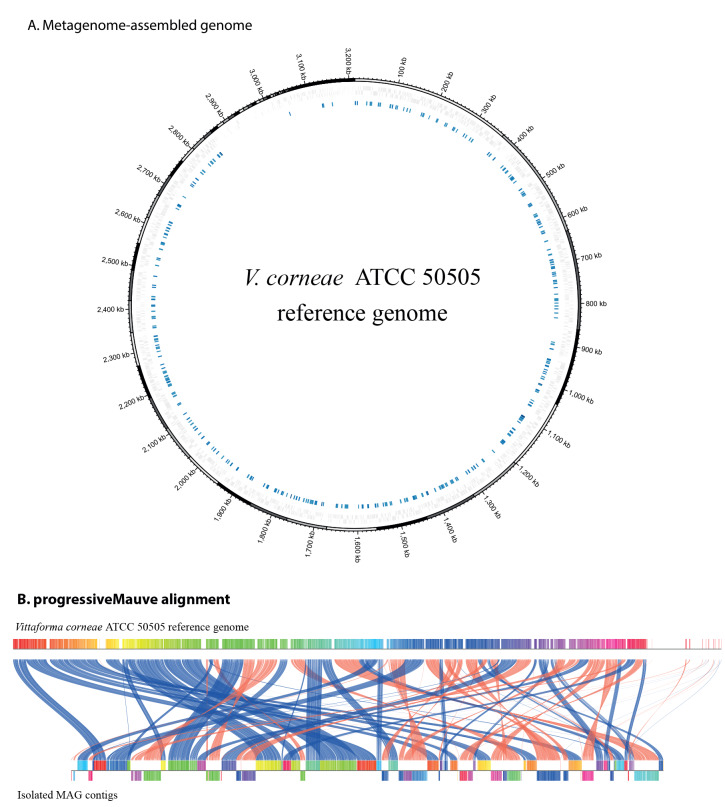
The host (human)-removed mNGS reads underwent de novo assembly, generating a 3.6 Mb metagenomic assembly consisting of 1,645 contigs (N50 47,773). Taxonomic assignments of the metagenomic assembly contigs to the eukaryotic pathogens database (EuPathDB46) using kraken2 assigned 69 contigs to V. corneae, totalling 2.67 Mb, with a 61-fold mean depth of coverage and a contiguity of N50 of 64,665. Comparison of the isolated MAG against the V. corneae ATCC 50505 reference genome showed 71.3% breadth of coverage (Figure 2). After filtering the complete metagenome assembly by size, only 116 of the unassigned contigs (7.3%) were > 1 kb. Repeated analysis classified one contig as a related microsporidium (Enterocytozoon bieneusi H348) and another 107 contigs as eukaryotic or root or cellular organism.
Figure 2.
Comparison of the isolated MAG contigs with the Vittaforma corneae ATCC 50505 reference genome, keratoconjunctivitis patient, Israel, 2022
ATCC: American Type Culture Collection; MAG: metagenome-assembled genome.
Panel A: The outer ring (contigs coloured black, grey or white) of the mummer2circos plot shows the V. corneae ATCC 50505 reference genome sequence, the middle ring (grey) shows the coding regions in both directions in the reference genome, the innermost ring (blue) shows the isolated MAG’s contigs that aligned to the reference genome (using mummer).
Panel B: progressiveMauve alignment between the V. corneae ATCC 50505 reference genome (top) and the isolated MAG contigs (bottom). Aligned contigs are coloured according to their arrangement in the reference genome. Connecting lines (coloured blue for same direction alignment, or orange for opposing direction alignment) between the reference genome and the isolated MAG contigs show the aligned regions.
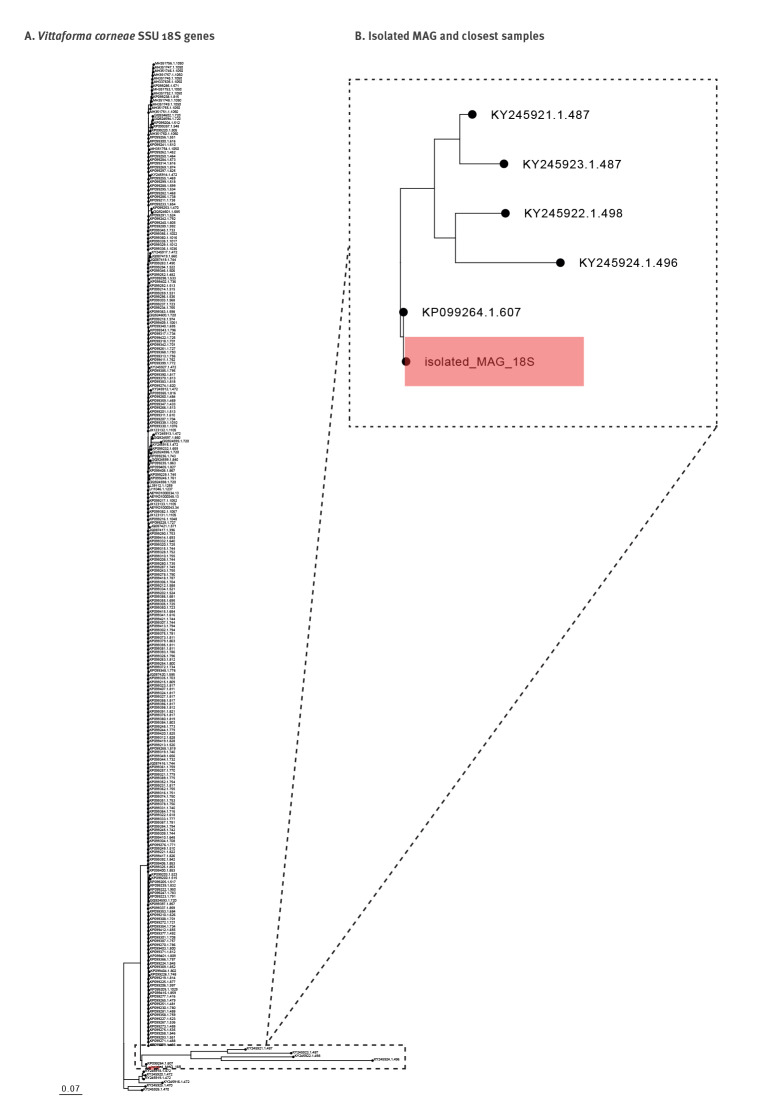
The isolated MAG’s annotated 18S rRNA gene was aligned against 278 V. corneae SSU 18S rRNA genes obtained from the SILVA rRNA database. The derived ML phylogenetic tree (Figure 3) shows the isolated MAG clustered outside of the main cluster of V. corneae samples obtained from the SILVA database. The nearest sample to the isolated MAG (KP099264.1.607) involved a human ocular sample from India, dated 2015, at 99.5% identity.
Figure 3.
Phylogenetic analysis of Vittaforma corneae from a keratoconjunctivitis patient, Israel, 2022
MAG: metagenome-assembled genome; ML: maximum likelihood; SSU: small subunit.
Panel A: ML phylogenetic tree based on the SSU 18S rRNA gene from the isolated MAG (highlighted in red) and 278 V. corneae SSU 18S genes obtained from the SILVA database.
Panel B: Enlargement of the isolated MAG and closest samples in the phylogenetic tree. The nearest sample to the isolated MAG (KP099264.1.607) involved a human ocular sample from India, dated 2015.
Molecular analysis of the clinical sample
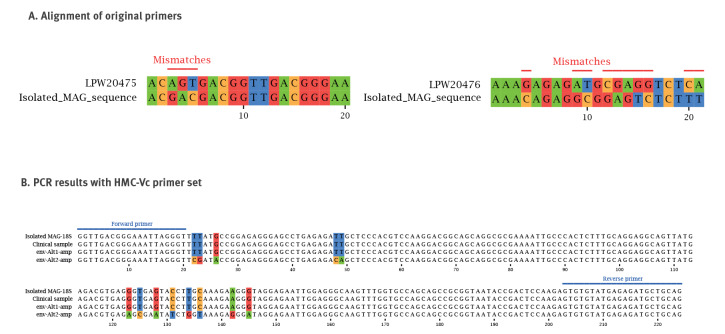
Due to a number of base variations in the isolated MAG primer binding sites (Figure 4A), we redesigned and optimised the V. corneae-specific primers (LPW20475-LPW20476 primer set). The new primers (HMC-Vc, Table) were applied in triplicate on a DNA extract from the original corneal scraping material and yielded a clear positive result with a melting temperature of 85.8 °C. The PCR product was subjected to Sanger sequencing and demonstrated 100% identity to the predicted MAG-derived amplicon and 96.43% identity to V. corneae deposited in NCBI.
Figure 4.
Development and application of real-time PCR for Vittaforma corneae, Israel, 2022
MAG: metagenome-assembled genome.
Panel A: LPW20475-LPW20476 PCR primer set (see Table) alignment to isolated MAG sequence for mismatch identification. Several forward and reverse base variations, necessitating optimisation, are highlighted.
Panel B: Optimised PCR primer set testing results, with the forward and reverse primers binding sequences highlighted in blue. PCR testing of the clinical sample (second row) produced an amplicon with 100% match to the 18S rRNA MAG target (first row). PCR testing of the environmental sample produced two potential amplicons (after deconvolution of trace files): Alt1-amp (third row) showed an exact match to the 18S rRNA MAG target, while Alt2-amp (fourth row) showed 13 mismatches.
Molecular and metagenomic analysis of environmental samples
We tested six environmental samples; three samples contained 1,000 mL of freshwater, whereas three others contained ca 50 mL of underwater solids (sludge, pebbles and gravel) mixed in freshwater from each of the sampled sites. All samples were positive for 16S rRNA in the qPCR performed as process control, reflecting a concentration of ca 109 bacterial cfu/L. One of the six samples was positive for V. corneae by means of the novel real-time PCR. Sanger sequencing of the PCR product yielded the expected amplicon size (224 bases) with multiple evident double peaks, suggesting a possible mixture of several microsporidian amplicons. The Sanger sequences were deconvoluted, producing two possible closely related amplicon sequences (Figure 4B). The two amplicon sequences were aligned with the MAG, showing 100% (Alt1-amp) and 94.2% (Alt2-amp) identity. We also queried the deconvoluted amplicons against the NCBI nucleotide database using megaBLAST, with top matches being V. corneae and Microsporidium sp., respectively; the BLAST search details can be accessed in Supplementary Table S4.
Long read sequencing produced 1.42 million reads with an average size of 4,536 bases (range: 1,000–77,496). Taxonomic assignment using kraken2 identified human (4.71%), archaeal (0.19%), bacterial (30.78%) and viral (0.03%) sequences; we refer to Supplementary Table S5 for the exact numbers. We also compared the long reads with the eukaryotic pathogens database (EuPathDB46) using kraken2, with 11,992 reads (0.84%) assigned a phylum (listed in Supplementary Table S6), of which 117 reads (ranging between 1,088 and 37,669 bases in size) were assigned to microsporidia. These included three long reads assigned to V. corneae (17,485, 6,710 and 6,671 bases in size) and other reads assigned to non-Vittaforma species; for the complete list see Supplementary Table S7. This was consistent with the PCR results, indicating the presence of several microsporidian species in the sample and the presence of V. corneae at the site of reported exposure. Analysis using the kraken Virus database revealed non-pathogenic viruses and analysis for bacteria revealed mostly environmental taxa but also several species of enteric bacteria at different quantities; these species and read numbers can be accessed in Supplementary Tables S8 and S9.
Strain-level analysis of Vittaforma corneae from short and long reads
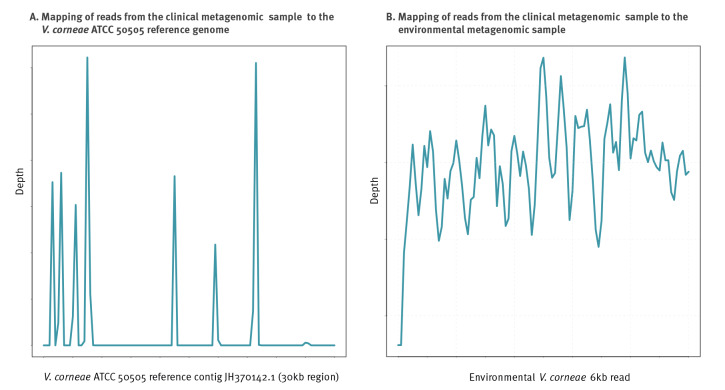
We attempted mapping the host-removed mNGS short reads from the clinical sample to each of the three V. corneae-assigned long reads (3.7 million reads, of which 1,131,712 mapped to the MAG). This resulted in mapping of 2,798 short reads to one long read (6,671 bases in size), with 98.91% breadth of coverage and 59.91-fold depth of coverage. When these short reads were mapped to the V. corneae ATCC 50505 reference genome, only 2,239 bases in a 30,000-base region in contig JH370142.1 were covered, with 7.46% breadth of coverage and 1.97-fold mean depth of coverage (Figure 5); details are appended in Supplementary Table S10.
Figure 5.
Mapping of clinical metagenomic sample to environmental metagenomic sample and to Vittaforma corneae ATCC 50505, keratoconjunctivitis patient, Israel, 2022
ATCC: American Type Culture Collection.
Host-removed reads from clinical metagenomic sample (3.7 million) covered 2,239 bases in a 30 kb region of the V. corneae ATCC 50505 contig JH370142.1 (at a mean depth of 2 ×) and 6,598 bases (at a mean depth of 60 ×) in the environmental 6 kb V. corneae read.
Analysis of historical metagenomes from the Sea of Galilee
We analysed a total of 61 metagenomic short reads from samples taken between 2013 and 2017 in the Sea of Galilee, totalling 1,459,047,627 reads (mean: 23,918,814, median: 21,977,956; range: 401,405–510,04,644). After taxonomic assignment of the reads using kraken2 with the EuPathDB46 database, 27,198 reads were assigned to the phylum Microsporidia, of which 460 reads were assigned to the species V. corneae. The distribution of V. corneae reads across historical samples is appended in Supplementary Figure S1. For each of the four most abundant samples, mapping of the reads to the MAG provided a consensus sequence for the SSU. When aligned to our 224 bp PCR amplicon (including the isolated MAG, reference genome and environmental sample amplicons as shown in Figure 4B), regions of variation were observed for all samples; the alignment for these is appended in Supplementary Figure S2.
Discussion
We describe the establishment of an aetiological diagnosis and attempted source attribution of a nationwide keratoconjunctivitis outbreak using mNGS and development of complementary molecular diagnostics. The aetiology of this outbreak remained unknown for several months; therefore, the confirmation of V. corneae, a microsporidian pathogen known to cause keratoconjunctivitis, had an important public health impact. Following the confirmation of the case reported herein, several cases have been reported as confirmed V. corneae infection using NGS or PCR elsewhere in Israel [10].
The main clinical manifestations of microsporidiosis among immunocompetent patients are usually self-limited gastroenteritis and keratoconjunctivitis, and V. corneae is a predominant aetiology of the latter, causing either sporadic cases or outbreaks. Nine cases were reported in Taiwan in 2011 and linked to two types of non-disinfected hot springs [11]. Another outbreak in Taiwan in 2017 involved 13 teenagers following exposure to a local swimming pool [12]. Another outbreak was reported from Singapore following a rugby tournament; it involved 47 cases, attributed to exposure to muddy field conditions following local floods [7]. Interestingly, only five cases in that outbreak were microbiologically confirmed, and the authors mention that liberal corneal scraping could not be justified.
There is a wide range of pathogens that could be considered as difficult to diagnose. Such pathogens may include unculturable microorganisms (e.g. Tropheryma whipplei or rickettsial organisms), organisms that are very difficult to culture such a spirochetes (e.g. Leptospira or Borrelia) or organisms for which routine diagnostics such as validated PCR assays are not widely available. The epidemiology of such pathogens and their scarcity often results in a lack of developed commercial or in-house diagnostic solutions. Microsporidia can be considered such difficult-to-diagnose organisms. In addition, obtaining high-quality sample material might not be trivial for some types of infection and can become a limiting factor. In that respect, corneal scrapings could be considered such samples that may not be widely available. Our case confirms the usefulness of fluorescent microscopy in diagnosing ocular microsporidiosis in cases where corneal samples are obtained. Nevertheless, expertise in microscopic diagnosis of microsporidia is lacking in many laboratories. Moreover, in the context of an elusive outbreak, microscopy may be insufficient for establishing the aetiology for a sentinel case but could be used in further cases once the aetiology has been unequivocally confirmed using molecular or genomic tools.
Several diagnostic PCR assays for Microsporidium detection in keratoconjunctivitis have been published, primarily targeting the SSU ribosomal 18S rRNA gene [13]. Another approach is a dual PCR assay comprised of SSU PCR as a target for microsporidia detection and a complementary target specific to Vittaforma [14]. Due to the notable diversity of Microsporidium spp., the accuracy of PCR in this setting is far from optimal, creating the need for a more comprehensive approach, such as mNGS.
Use of mNGS is increasingly being reported in clinical microbiology as a cutting-edge diagnostic modality [15] having impact on the diagnosis of infections across all organ systems, such as the central nervous, musculoskeletal or respiratory system infections. mNGS has been used successfully for the diagnosis of serious eye infection such as ocular Whipple’s disease [16], bacterial endophthalmitis [17,18] and uveitis [19] but also of more superficial eye infections such as acute conjunctivitis, infectious keratitis and corneal ulcers [20-22]. Several challenges still surround the application of mNGS in the diagnosis of eye infection, including the order by which samples are obtained [23], difficulties in validation and interpretation, the limit of detection, reagent contamination, regulatory aspects and costs, to name a few [24]. While the reports of mNGS in ophthalmology have not focused on microsporidiosis, a recent report from China describes an invasive fatal microsporidial infection in which A. algerae was detected by deep sequencing of respiratory and muscle tissue samples [8]. Light and transmission electron microscopy yielded compatible findings. This report exemplifies the utility of mNGS in the diagnosis of elusive cases.
Waterborne microsporidiosis can be linked to a putative source of exposure based on conventional epidemiological methods elucidating behavioural risk factors. Nevertheless, microbiological confirmation of an outbreak source may be challenging. Based on PCR testing of water and soil samples, a follow-up study of the 2017 Taiwan outbreak has stipulated that environmental contamination following rainfall resulted in secondary swimming pool contamination following human activity [25]. Notably, sequencing of PCR products revealed a high sequence diversity with only a few amplicons being similar to those derived from clinical samples, suggesting that environmental contamination with microsporidia involved multiple clones. In a further ecological study, molecular surveillance of Vittaforma-like organisms in rivers and water reservoirs in Taiwan identified positive samples in 47 of 50 tested sites [26]. That study also reported a notable phylogenetic diversity based on sequenced PCR amplicons, with only 32 of 82 amplicons analysed showing a very close identity to V. corneae. Moreover, most samples yielded two or three bands on DNA electrophoresis, suggesting a mixture of different microsporidia, a finding confirmed by cloning and aligning sequences. These environmental findings coincide with our real-time PCR results, which showed evidence of two microsporidia in the environmental samples, one being V. corneae identical to the clinical sample and the other a closely related unidentified Microsporidium sp. Furthermore, our analysis of historical samples also confirmed the abundance of different microsporidia in the Sea of Galilee, including different strains of V. corneae, which were closely related but not identical to the outbreak strain, testifying to the complexity of this ecosystem.
The long-read metagenomic sequencing results further supported this finding. Sequencing of the environmental sample identified several microsporidia co-existing in the sample. While a complete strain-level metagenomic analysis could not be carried out, due to the paucity of public genomes and the small number of high-quality long reads identified, mapping of the short reads of the clinical sample to a 6 kb read, resulted in very high depth and coverage, a finding highly suggestive of the infection source in the environmental sample.
Use of mNGS enables a range of microbiological applications in a single test, including the detection of known pathogens in an unbiased manner (as opposed to the hypothesis-driven utilisation of PCR), the study of microbial populations (such as the microbiome) or pathogen discovery in case of an emerging or new pathogens. With the appropriate computational approach, mNGS could also allow interrogation of identified genomes within a metagenome to perform strain-level analysis (e.g. for resistance and virulence determinants), determination of phylogeny, and identification of diagnostic targets for specific PCR development. In the context of an outbreak lacking a clear aetiology, mNGS is especially powerful since it allows relatively rapid culture-independent detection of the causative pathogens while facilitating comparison to previous or other cases and the ad hoc development of diagnostics, which could enable further case finding or source tracking. Notably, in the case presented herein, mNGS was also useful in searching for alternative aetiologies such as common bacterial and viral causes of keratoconjunctivitis, further supporting the established diagnosis.
Our study has several limitations. Corneal samples from additional patients were not available and therefore a strain-level comparison of V. corneae between outbreak cases was not possible. Moreover, in the absence of additional samples, clinical validation of the ad hoc PCR assay could not be performed beyond the current case. The depth of long-read sequencing of the environmental sample was limited, thus not allowing a complete comparison of the MAG between the short-reads and long-reads metagenome. Despite using several analytical tools, many sequencing reads remained unclassified or resulted in taxonomic assignments which were considered spurious. This reflects limitations in public databases, and in particular, the paucity of V. corneae genomes in such databases.
Conclusion
As deep sequencing is becoming increasingly available across health systems, its application is expected to improve individual case management but also the public health response to outbreaks and emerging infections. Further development and validation of mNGS are needed to overcome existing diagnostic and operational challenges associated with routine mNGS use.
Ethical statement
The use of anonymous clinical and microbiological data has been approved by the Research Ethics Committee of the Hadassah Medical Center, Jerusalem, Israel (#HMO-0019-16). Written informed consent was obtained from the patient reported in this study.
Data availability
Anonymised sequence data have been deposited in the European Nucleotide Archive (ENA) database under bioProjects PRJEB58700 and PRJEB58701.
Supplementary Data
Note
This study has been presented in part at the 33rd European Congress of Clinical Microbiology and Infectious Diseases (ECCMID), Copenhagen, Denmark, April 2023.
Conflict of interest: None declared.
Authors’ contributions: DW collected data and specimens and performed clinical diagnosis; VT and AM-G performed microscopy and clinical diagnostic work; SM, RBMM and IS performed sample processing and experimental diagnostic work; CR performed sequencing work; YM and JM-G performed bioinformatics and data analyses; YM, DW, JS and JM-G drafted the paper.
References
- 1. Ting DSJ, Ho CS, Deshmukh R, Said DG, Dua HS. Infectious keratitis: an update on epidemiology, causative microorganisms, risk factors, and antimicrobial resistance. Eye (Lond). 2021;35(4):1084-101. 10.1038/s41433-020-01339-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ibrahim YW, Boase DL, Cree IA. Factors affecting the epidemiology of Acanthamoeba keratitis. Ophthalmic Epidemiol. 2007;14(2):53-60. 10.1080/09286580600920281 [DOI] [PubMed] [Google Scholar]
- 3. Ahmadikia K, Aghaei Gharehbolagh S, Fallah B, Naeimi Eshkaleti M, Malekifar P, Rahsepar S, et al. Distribution, prevalence, and causative agents of fungal keratitis: a systematic review and meta-analysis (1990 to 2020). Front Cell Infect Microbiol. 2021;11:698780. 10.3389/fcimb.2021.698780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martin C, Löw U, Quintin A, Schießl G, Gärtner B, Heim A, et al. Epidemic keratoconjunctivitis: efficacy of outbreak management. Graefes Arch Clin Exp Ophthalmol. 2022;260(1):173-80. 10.1007/s00417-021-05344-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Han B, Pan G, Weiss LM. Microsporidiosis in Humans. Clin Microbiol Rev. 2021;34(4):e0001020. 10.1128/CMR.00010-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Das S, Sharma S, Sahu SK, Nayak SS, Kar S. Diagnosis, clinical features and treatment outcome of microsporidial keratoconjunctivitis. Br J Ophthalmol. 2012;96(6):793-5. 10.1136/bjophthalmol-2011-301227 [DOI] [PubMed] [Google Scholar]
- 7. Tan J, Lee P, Lai Y, Hishamuddin P, Tay J, Tan AL, et al. Microsporidial keratoconjunctivitis after rugby tournament, Singapore. Emerg Infect Dis. 2013;19(9):1484-6. 10.3201/eid1909.121464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu C, Chen Q, Fu P, Shi YY. Anncaliia algerae microsporidiosis diagnosed by metagenomic next-generation sequencing, China. Emerg Infect Dis. 2022;28(7):1466-70. 10.3201/eid2807.212315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Health Ministry suspects parasite in Sea of Galilee causing eye infections. Jerusalem: The Times of Israel; 8 Nov 2022. Available from: https://www.timesofisrael.com/health-ministry-suspects-parasite-in-sea-of-galilee-causing-eye-infections
- 10.Lubitz Greenbaum I, Barequet I, Cohen G, Ninio S, Sukenik A, Gihon I, et al. In the public eye - the story of an elusive aquatic predator. Annual meeting of the Israel Society for Parasitology, Protozoology and Tropical Diseases, Ramat Gan, Israel, March 2023. [Google Scholar]
- 11. Fan NW, Wu CC, Chen TL, Yu WK, Chen CP, Lee SM, et al. Microsporidial keratitis in patients with hot springs exposure. J Clin Microbiol. 2012;50(2):414-8. 10.1128/JCM.05007-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang WY, Chu HS, Lin PC, Lee TF, Kuo KT, Hsueh PR, et al. Outbreak of microsporidial keratoconjunctivitis associated with water contamination in swimming pools in Taiwan. Am J Ophthalmol. 2018;194:101-9. 10.1016/j.ajo.2018.07.019 [DOI] [PubMed] [Google Scholar]
- 13. Sanpool O, Thanathanee O, Laummuanwai P, Maleewong W, Intapan PM. Molecular identification of microsporidian species in patients with epithelial keratitis. J Med Microbiol. 2020;69(3):414-8. 10.1099/jmm.0.001164 [DOI] [PubMed] [Google Scholar]
- 14. Jayahar Bharathi M, Murugan N, Ramesh Kumar G, Ramakrishnan R, Anitha V, Ramesh S. Vittaforma corneae keratitis in southern India: role of a novel duplex PCR. J Med Microbiol. 2013;62(Pt 4):553-9. 10.1099/jmm.0.051722-0 [DOI] [PubMed] [Google Scholar]
- 15. Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet. 2019;20(6):341-55. 10.1038/s41576-019-0113-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gonzales JA, Doan T, VanZante A, Stewart JM, Sura A, Reddy A, et al. Detection of Tropheryma whipplei genome from the aqueous humor by metagenomic sequencing. Ann Intern Med. 2021;174(9):1329-30. 10.7326/L20-1470 [DOI] [PubMed] [Google Scholar]
- 17. Zhu J, Xia H, Tang R, Ng TK, Yao F, Liao X, et al. Metagenomic next-generation sequencing detects pathogens in endophthalmitis patients. Retina. 2022;42(5):992-1000. 10.1097/IAE.0000000000003406 [DOI] [PubMed] [Google Scholar]
- 18. Low L, Nakamichi K, Akileswaran L, Lee CS, Lee AY, Moussa G, et al. Deep metagenomic sequencing for endophthalmitis pathogen detection using a nanopore platform. Am J Ophthalmol. 2022;242:243-51. 10.1016/j.ajo.2022.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Valdes L, Bispo P, Sobrin L. Application of metagenomic sequencing in the diagnosis of infectious uveitis. Semin Ophthalmol. 2020;35(5-6):276-9. 10.1080/08820538.2020.1818795 [DOI] [PubMed] [Google Scholar]
- 20. Seitzman GD, Hinterwirth A, Zhong L, Cummings S, Chen C, Driver TH, et al. Metagenomic deep sequencing for the diagnosis of corneal and external disease infections. Ophthalmology. 2019;126(12):1724-6. 10.1016/j.ophtha.2019.06.013 [DOI] [PubMed] [Google Scholar]
- 21. Bountogo M, Sié A, Coulibaly B, Ruder K, Chen C, Zhong L, et al. Deep sequencing analysis of acute conjunctivitis in Burkina Faso, Africa. Int Health. 2023;15(1):101-3. 10.1093/inthealth/ihac001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lalitha P, Prajna NV, Sikha M, Gunasekaran R, Hinterwirth A, Worden L, et al. Evaluation of metagenomic deep sequencing as a diagnostic test for infectious keratitis. Ophthalmology. 2021;128(3):473-5. 10.1016/j.ophtha.2020.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Redd TK, Lalitha P, Prajna NV, Sikha M, Gunasekaran R, Hinterwirth A, et al. Impact of sample collection order on the diagnostic performance of metagenomic deep sequencing for infectious Keratitis. Cornea. 2022;41(1):39-44. 10.1097/ICO.0000000000002766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ung L, Bispo PJM, Doan T, Van Gelder RN, Gilmore MS, Lietman T, et al. Clinical metagenomics for infectious corneal ulcers: Rags to riches? Ocul Surf. 2020;18(1):1-12. 10.1016/j.jtos.2019.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen JS, Hsu TK, Hsu BM, Chao SC, Huang TY, Ji DD, et al. Swimming pool-associated Vittaforma-like microsporidia linked to microsporidial keratoconjunctivitis outbreak, Taiwan. Emerg Infect Dis. 2019;25(11):2100-3. 10.3201/eid2511.181483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen JS, Hsu BM, Tsai HC, Chen YP, Huang TY, Li KY, et al. Molecular surveillance of Vittaforma-like microsporidia by a small-volume procedure in drinking water source in Taiwan: evidence for diverse and emergent pathogens. Environ Sci Pollut Res Int. 2018;25(19):18823-37. 10.1007/s11356-018-2081-4 [DOI] [PubMed] [Google Scholar]
- 27. Kwok AKH, Tong JMK, Tang BSF, Poon RWS, Li WWT, Yuen KY. Outbreak of microsporidial keratoconjunctivitis with rugby sport due to soil exposure. Eye (Lond). 2013;27(6):747-54. 10.1038/eye.2013.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.