Abstract
Proteolytic processing of the replicase gene product of mouse hepatitis virus (MHV) is essential for viral replication. In MHV strain A59 (MHV-A59), the replicase gene encodes two predicted papain-like proteinase (PLP) domains, PLP-1 and PLP-2. Previous work using viral polypeptide substrates synthesized by in vitro transcription and translation from the replicase gene demonstrated both cis and trans cleavage activities for PLP-1. We have cloned and overexpressed the PLP-1 domain in Escherichia coli by using a T7 RNA polymerase promoter system or as a maltose-binding protein (MBP) fusion protein. With both overexpression systems, the recombinant PLP-1 exhibited trans cleavage activity when incubated with in vitro-synthesized viral polypeptide substrates. Subsequent characterization of the recombinant PLP-1 revealed that in vitro trans cleavage is more efficient at 22°C than at higher temperatures. Using substrates of increasing lengths, we observed efficient cleavage by PLP-1 requires a substrate greater than 69 kDa. In addition, when PLP-1 was expressed as a polypeptide that included additional viral sequences at the carboxyl terminus of the predicted PLP-1 domain, a fivefold increase in proteolytic activity was observed. The data presented here support previous data suggesting that in vitro and in vivo cleavage of the ORF 1a polyprotein by PLP-1 can occur in both in cis and in trans. In contrast to the cleavage activity demonstrated for PLP-1, no in vitro cleavage in cis or in trans could be detected with PLP-2 expressed either as a polypeptide, including flanking viral sequences, or as an MBP fusion enzyme.
The murine coronavirus mouse hepatitis virus (MHV) is an enveloped virus that belongs to the Coronaviridae family. The genome of MHV strain A59 (MHV-A59) consists of a positive-sense RNA of 31.3 kb. Upon infection, the genome is translated from its 5′ end (gene 1) into a polyprotein, from which an RNA-dependent RNA polymerase is processed; this polymerase then transcribes a nested set of six subgenomic mRNAs as well as negative-sense full-length and subgenomic RNAs. Gene 1, which is 21.7 kb, contains two open reading frames (ORFs), ORF1a and ORF1b. Sequence analysis of ORF1a predicted two papain-like proteinase (PLP) domains, an X domain of unknown function adjacent to PLP-1, and a poliovirus 3C-like proteinase domain. ORF1b, which is expressed via a translational frameshift resulting in readthrough of the ORF1a termination codon, encodes the RNA polymerase domain as well as putative helicase, nucleoside triphosphatase, and zinc-binding domains (5, 16, 22) (Fig. 1A). Similar genome organization and replication strategy are also characteristic of members of the Arteriviridae family. The similarities led to the classification of these two families under the order Nidovirales (12, 28).
FIG. 1.
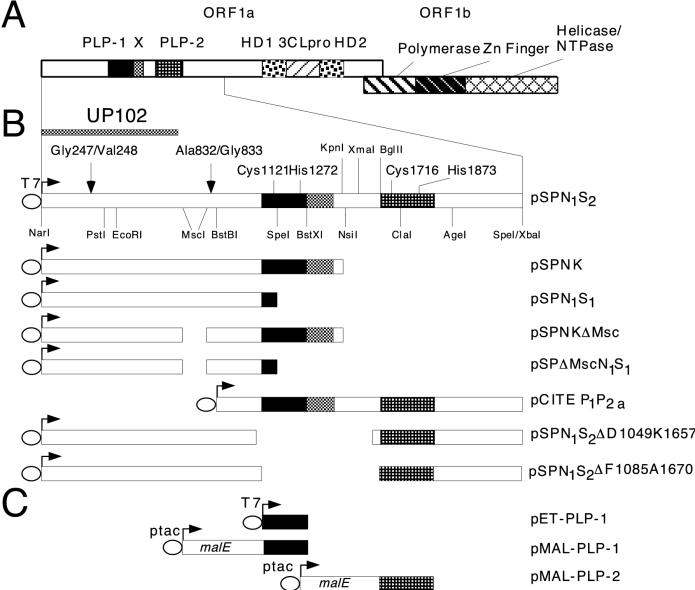
Functional domains of MHV-A59 gene 1 and plasmids encoding ORF1a polypeptides. (A) The two ORFs of gene 1, ORF1a and ORF1b, and their predicted functional domains: PLP-1 and PLP-2, X, picornavirus 3C-like proteinase (3CLpro), and hydrophobic (HD1 and HD2). NTPase, nucleoside triphosphatase. (B) Diagrams of plasmids used for transcription and translation of ORF1a polypeptides, along with relevant functional domains and restriction sites. The catalytic residues of PLP-1, Cys1121 and His1272, and the proposed catalytic residues of PLP-2, Cys1716 and His1873, are indicated. The T7 bacteriophage RNA polymerase promoter is designated T7. The cleavages sites for p28 (Gly247/Val248) and p65 (Ala823/Gly833) and the region of ORF1a polypeptide used to raise antiserum UP102 (11) are also indicated. (C) Plasmids used for expression of the PLP domains in E. coli. T7 and ptac are procaryotic promoters for RNA synthesis, and malE encodes MBP.
Using antisera raised against various regions of the MHV-A59 ORF1a gene product, Denison et al. (8, 11) reported that the ORF1a product was processed in vivo from the amino terminus to produce p28, p65, and p290; p290 was further processed to produce p50 and p240. PLP-1 (also referred to as PCP-1 [13]) has been shown to carry out cleavages of the ORF1a gene product in vitro at the p28 and the predicted p65 sites (2, 3, 5, 6, 20, 27). In vitro studies identified the cleavage site for p28 to be between Gly247 and Val248 (20). The use of deletion constructs identified another in vitro cleavage site, between Ala832 and Gly833; it is likely that this site corresponds to the cleavage site utilized in vivo to generate p65 (5, 6). (It has not yet been proved that the in vitro cleavage site between Ala832 and Gly833 corresponds to the site that is utilized in the synthesis of p65 in vivo. However, since this is very likely to be the same site, we will refer to the second cleavage site as the p65 site.) The cleavage site which generates p50 has not been identified. Deletion studies have mapped the minimal PLP-1 domain to between ORF1a amino acids 1084 and 1316 (5, 6). A role for PLP-1 in proteolytic processing at the amino terminus of ORF1a polypeptide has also been demonstrated in other coronaviruses, including MHV strain JHM (2, 3, 13), human coronavirus strain 229E (19), and infectious bronchitis virus (23). The second PLP domain (PLP-2), predicted to be encoded within amino acids Phe1681 to Ser1936 in the MHV-A59 genome (22), is not found in all coronavirus genomes, and there are no reports to date of an activity for this predicted domain (3, 5, 14).
While the functions of p28 and p65 have yet not been determined, the cleavages that result in their production appear to be a necessary part of MHV life cycle. Addition of the proteinase inhibitors leupeptin and E64d to infected cells resulted in the inhibition of proteolytic processing of the ORF1a gene product, with concomitant reduction in viral RNA synthesis (9, 21). The observation that viral RNA synthesis was inhibited even when the inhibitors were added after viral entry led to the proposal that MHV replication required continuous generation of the replicase; thus, inhibiting this proteolytic processing resulted in inhibition of viral replication. In view of a possible role played by PLP-1 during viral replication, elucidating the mechanism of action of this proteinase could provide insight into the viral life cycle and may help in the development of antiviral drugs. To obtain large quantities of PLP-1 and to characterize its enzymatic properties, we have cloned and overexpressed the predicted PLP-1 domain both as a fusion protein and by itself and have used these recombinant enzymes to further our understanding of the properties of PLP-1. In both expression systems, the recombinant PLP-1 was active on in vitro-synthesized substrates, demonstrating that the previously mapped domain (5) is sufficient to encode a functional enzyme. Further investigation of in vitro trans cleavage by the recombinant PLP-1 showed a strong dependence on the length of substrate, with efficient cleavage detected only with substrates of greater than 69 kDa. In addition, we observed that the presentation of PLP-1 as a polypeptide of 49 kDa or greater, which includes the sequences carboxyl to the proteinase, greatly enhances proteolytic processing. Thus, PLP-1 cleavage activity appears to be optimal when both enzyme and substrate are presented as part of polyproteins, as they are in the virus replication cycle. Expression of the predicted PLP-2 domain either as a polypeptide in vitro or fused to maltose-binding protein (MBP) in Escherichia coli did not result in detectable cleavage.
MATERIALS AND METHODS
Plasmids.
Plasmid pSPNK (20) (Fig. 1B) contains the MHV-A59 genome sequences between nucleotides 182 and 4664 and encodes the amino-terminal 1,484 amino acids of ORF1a. Plasmid pSPNKΔMsc (5) contains an in frame MscI-MscI deletion (ΔMsc) between codons 623 and 869, and pSPNKΔMAG (6), previously referred to as ΔAG, contains in addition an in-frame deletion of the codons encoding the p65 cleavage site, Ala832 to Gly833. Plasmids pSPN1S1, pSPΔMscN1S1, and pSPΔMAGN1S1, which are truncated at the SpeI site within PLP-1, were constructed by digesting pSPNK, pSPNKΔMsc, and pSPNKΔMAG with BglII (within the vector sequence) and SpeI (within the PLP-1 domain, ORF1a nucleotide 3690). The resulting BglII/SpeI fragments were ligated into pSP72 (Promega) previously digested with BglII and XbaI. The resulting plasmids encode viral polypeptides with the same amino termini as authentic viral ORF1a polypeptide; however, the viral sequences in these polypeptides end at Lys1160 (within PLP-1 [Fig. 1B]) and contain 26 plasmid-derived amino acids downstream of the viral polypeptides before termination.
Plasmid pSPN1S2, which contains nucleotides 182 to 6755 and encodes the amino-terminal 2,181 amino acids of ORF1a (including both PLPs [Fig. 1B]), was constructed as follows. A 0.56-kb KpnI/BglII fragment (nucleotides 4664 to 5219 of the genome) from cDNA clone pUC19-K1E2 (4) was ligated into KpnI/BamHI-digested pSP72, which resulted in pSPKBg. This plasmid was then digested with BglII (within the vector sequence) and KpnI (nucleotide 4664) and ligated to a 4.6-kb fragment from pO1aNK1.4 (a plasmid derived from pSPNK containing the same viral sequences as pSPNK) that had been digested with BglII (within the vector sequence) and KpnI (nucleotide 4664). The resulting construct, pSPN1Bg, contains ORF1a nucleotide from the NarI site at nucleotide 182 to the BglII site at nucleotide 5219 (Fig. 1B). Finally, pSPN1Bg was digested with KpnI (nucleotide 4664) and XbaI (within the vector sequence) and then ligated to a KpnI/SpeI fragment (nucleotides 4664 to 6752) from cDNA clone K1E2 (4) to generate pSPN1S2.
For the construction of pCITE P1P2a, encoding both PLP domains (Fig. 1B), pSPN1S2 was digested with BstBI (ORF1a nucleotide 2811), and the ends were filled in with DNA polymerase I Klenow fragment. The blunt-ended plasmid was then digested with SpeI (ORF1a nucleotide 6752), and the fragment containing ORF1a nucleotides 2811 to 6752 was ligated into pCITE (Novagen) previously treated with MscI and XbaI. To construct plasmids in which PLP-2 was placed closer to the p28 cleavage site, pSPN1S2 was treated with XmaI and SpeI and then with exonuclease Bal 31. The recessed ends were filled in with Klenow fragment and ligated. Deletion clones were then screened for (i) retention of the BglII site upstream of the predicted PLP-2 domain and (ii) maintenance of an ORF through PLP-2. Two of the resulting clones, which positioned the predicted amino terminus of PLP-2 domain at 824 and 848 amino acids from the p28 site, are designated pSPN1S2ΔD1049K1657 and pSPN1S2ΔF1085A1670, respectively (Fig. 1B).
Overexpression plasmids.
For the construction of pET-PLP-1 (Fig. 1C), the region corresponding to nucleotides 3393 to 4301, which encodes Ser1062 to Lys1364, was amplified from pSPNK by using PCR primers PLP NS, which introduced an NdeI site at the 5′ end, and PLP CA, which introduced a translational termination codon followed by an XbaI site at the 3′ end (Table 1). Following XbaI and partial NdeI digestion, a 0.9-kb fragment containing the predicted PLP-1 domain was ligated into XbaI/NdeI-cleaved pHB40P (a pET overexpression vector derivative obtained from R. Fletterick, University of California, San Francisco).
TABLE 1.
Primers used for PCR amplifications and mutagenesis
| Primer | Sequence (5′–3′)a | ORF1a positionb |
|---|---|---|
| Cloning primers | ||
| PLP NS | TCGATCGCATATGTCTATCTTGGATGAGCTTCAA | 3393–3413 |
| PLP CA | GAGTCTAGATAACTTTTCAGCTATAGCACCTGC | 4301–4281 |
| PLP-2 FCP | GAAGGAGATCGCGGATCCTTTGATGAACCACAACTGCTG | 5250–5270 |
| PLP-2 RCP | GCCCGCCTGCAGAAGCTTCTACGATAAATCTGGCTTATA | 6017–6000 |
| Mutagenesis primers | ||
| FMPH1272P | GATGTTAATGATTGTCCCTCTATGGCTGTAGTAGAGGG | 4008–4045 |
| RMPH1272P | CCCTCTACTACAGCCATAGAGGGACAATCATTAACATC | 4045–4008 |
| FMPH1873P | GGTGGTAGTGTGGGCCCTTACACGCATGTGAAATG | 5811–5843 |
| RMPH1873P | CATTTCACATGCGTGTAAGGGCCCACACTACCACC | 5843–5811 |
| FMPD904G | CCTTGTAAGGAGCATGGTGTGATAGGCACAAAAG | 2904–2937 |
| RMPD904G | CTTTTGTGCCTATCACACCATGCTCCTTACAAGG | 2937–2904 |
Restriction sites introduced into primers are shown in boldface; mutated codons are underlined.
Reverse position of ORF1a indicates a negative-strand primer.
PLP-1 was also expressed as an MBP–PLP-1 fusion protein. Plasmid pMAL-PLP-1 was constructed by amplifying the same PLP-1 domain as described above for pET-PLP-1 and ligating the fragment into XmnI/EcoRI-digested pMAL-c2 (New England Biolabs) (Fig. 1C). DNA sequencing showed that the construct contained conservative A1084V substitutions 38 and 189 amino acids from the catalytic residues, Cys1121 and His1272, respectively. PLP-2 was also expressed as an MBP-virus enzyme fusion protein. The region encoding Phe1681 through Ser1936 was amplified from plasmid pCITE P1P2a by using primers PLP-2 FCP, which introduced a BamHI site at the 5′ end, and PLP-2 RCP, which added a translational termination signal followed by a HindIII site at the 3′ end (Table 1). The amplified fragment was then digested with BamHI and HindIII and ligated into BamHI/HindIII-cleaved pMAL-c2 (Fig. 1C).
Mutagenesis.
Site-directed mutagenesis was carried out by using PCR technology and the primers listed in Table 1, using a QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s protocol. Mutagenic primers FMPH1272P and RMPH1272P were used to replace the catalytic His1272 of PLP-1 with Pro. Primers FMPH1873P and RMPH1873P were used to substitute Pro for the proposed catalytic His1873 in PLP-2. In this mutagenesis procedure, the entire plasmid is amplified. Thus, to avoid sequencing the entire plasmids to ascertain that no secondary mutation was introduced, restriction fragments containing the desired mutations (which were sequenced and shown not to contain secondary mutations) were introduced into the parental plasmids as indicated. For construction of pMAL-PLP-1 H1272P, a 0.6-kb SpeI/EcoRI fragment containing the mutation was ligated to a 6.9-kb fragment of pMAL-PLP-1 digested with the same enzymes. To introduce H1272P into pCITE P1P2a, a 0.5-kb SpeI/BstXI fragment from pMAL-PLP-1 H1272P was ligated to a 7.2-kb SpeI/BstXI fragment from pCITE P1P2a. For pMAL-PLP-2, the entire insert (BamHI to HindIII) carrying the H1873P mutation was ligated to pMAL-c2 digested with BamHI and HindIII.
Overexpression and purification of recombinant PLP-1 and PLP-2.
Overexpression and affinity chromatography purification of MBP–PLP-1 and MBP–PLP-2 fusion proteins were carried out according to the protocols from New England Biolabs. Briefly, the plasmids was transformed into E. coli DH5α, and the transformed cells were grown in LB supplemented with ampicillin (100 μg/ml) at 37°C until the A600 was between 0.4 and 0.7, at which time isopropyl-β-d-thiogalactopyranoside was added to 0.5 mM to induce fusion protein expression. Induction was continued for 3 h before harvesting, and cell pellets were stored frozen at −80°C in column buffer (20 mM Na-HEPES [pH 7.4], 1 mM EDTA, 200 mM NaCl, 2 mM dithiothreitol [DTT]). After thawing and sonication of the cell suspension, the crude extract was centrifuged at 18,000 × g, and the supernatant was applied onto an amylose column (New England Biolabs) previously equilibrated with column buffer. After washing with column buffer to remove contaminants, the fusion enzymes were eluted with column buffer supplemented with 10 mM maltose. The MBP–PLP-1 fusion enzyme was then concentrated in a Centriplus 10 concentrator (Amicon) and stored frozen at −80°C in the presence of 25% glycerol. To release PLP-1, the fusion enzyme was incubated with factor Xa (New England Biolabs) at 4°C overnight according to the manufacturer’s protocol.
For expression of pET-PLP-1 and pET-PLP-1 H1272P, the plasmids were transformed into E. coli BL21(DE3)/pLysS (29). The transformed cells were grown and harvested as described above except that chloramphenicol (25 μg/ml) was included in the medium. For purification of enzyme, the cell pellet was thawed and sonicated on ice for 2 min. Subsequently, 50 to 100 U of DNase I (Boehringer Mannheim) was added, and the mixture was incubated at room temperature for 20 min before another round of sonication. The suspension was centrifuged at 18,000 × g for 30 min, and the inclusion bodies were solubilized in buffer containing 50 mM sodium phosphate (pH 9), 1 mM EDTA, 1 mM DTT, and 8 M urea. To refold the denatured enzyme, the solubilized enzyme was centrifuged at 27,000 × g for 40 min, and the urea in the supernatant was removed stepwise by dialysis at 4°C against buffer containing 50 mM sodium phosphate (pH 9), 1 mM EDTA, 1 mM DTT, and 8 M urea. To refold the denatured enzyme, the solubilized enzyme was centrifuged at 27,000 × g for 40 min, and the urea in the supernatant was removed stepwise by dialysis at 4°C against buffer containing 50 mM sodium phosphate (pH 9), 1 mM EDTA, 1 mM DTT, and 1 mM reduced/0.1 mM oxidized glutathione, and with the urea gradually replaced by 8% glycerol. The refolded enzyme was centrifuged at 27,000 × g for 40 min to remove insoluble aggregates. The supernatant containing the refolded enzyme was then precipitated with ammonium sulfate (10% saturation); the pellet was dissolved in 50 mM sodium phosphate (pH 9)–1 mM EDTA–1 mM DTT–33% glycerol, and stored at 4°C. Protein concentrations of the recombinant enzymes were determined with the Bradford assay, using bovine serum albumin as the standard (1).
In vitro transcription and translations and trans cleavage assays.
In vitro transcription and translation of plasmid DNAs was carried out by using the TnT coupled reticulocyte lysate system (Promega) as described previously (5), using as template 1 μg of plasmid and [35S]methionine to radiolabel the products. For in vitro synthesis of nonradiolabeled enzymes, radiolabeled methionine was replaced with 1 mM methionine. The coupled in vitro transcription-translation reaction (50 μl) was carried out at 30°C for 90 min, and the synthesis was terminated by the addition of 1/10 volume of stop buffer (0.6 U of RQ DNase I per μl, 1.6 μg of RNase A per μl, 20 mM methionine), followed by further incubation for 15 min at 30°C. This procedure has been shown to terminate further incorporation of radiolabel (6). The radiolabeled substrates were then incubated in vitro at 22 to 25°C overnight with either purified recombinant enzyme or nonradiolabeled enzyme synthesized in vitro, as designated. Cleavage reactions were quenched with sodium dodecyl sulfate (SDS)-gel loading buffer, and the samples were then boiled for 2 min before loading onto SDS-polyacrylamide gels (20). After electrophoresis, gels were treated with Autofluor (National Diagnostics), dried at 80°C under vacuum, and exposed to X-ray films at −80°C (6). Immunoprecipitation of cleavage products with polyclonal antibody UP102 (8) was done as described previously (5).
Quantitative analysis of cleavage.
Cleavage activity was determined by quantification of radiolabeled polypeptides after analysis on SDS-polyacrylamide gels, using a Molecular Dynamics Storm 860 PhosphorImager equipped with the ImageQuant software. In vitro trans cleavage efficiency with substrate synthesized from pSPN1S1 was determined by using the equation {1 − [p131/(p28 + p103 + p131)]}, in which p131 is the uncleaved substrate and p28 and p103 are the cleavage products. Cleavage efficiency with substrates of various lengths was determined with a similar equation. In the case of substrate synthesized from pSPΔMscN1S1, cleavage at both the p28 site (between Gly247 and Val248) and the p65 site (between Ala832 and Gly833) produces p28, a downstream p43 (the deleted form of p65), and the carboxyl-terminal p50 (calculated molecular mass is 39 kDa). With partial processing at only the p28 site, p28 and p93 (p43 + p50) are obtained. Cleavage only at the p65 site produces p70 (p28 + p43) in addition to the carboxyl-terminal p50. The in vitro trans cleavage activity of each construct was calculated by subtracting the fraction of unprocessed precursor (p120) radioactivity from total radioactivity, using the equation {1 − [p120/(p28 + p43 + p50 + p70 + p93 + p120)]}. The cleavage activity was expressed as a percentage of cleavage activity obtained for the longest substrate. In determining the effect of substrate length on cleavage efficiency (Fig. 5), the extent of cleavage was determined by subtracting the radioactivity generated with inactive PLP-1 (translated from pET-PLP-1 H1272P) from the radioactivity generated with active PLP-1 (translated from pET-PLP-1).
FIG. 5.
In vitro trans cleavage efficiency of PLP-1 increases with increase substrate length. Plasmid pSPNK H1272P was digested with the enzymes indicated at the top followed by transcription and translation in the coupled system in the presence of [35S]methionine. The radiolabeled viral substrates were incubated with unlabeled mutant PLP-1 H1272P (A) or wild-type PLP-1 (B), also synthesized in the coupled transcription-translation using pET-PLP-1 H1272P or pET-PLP-1, respectively, as the templates. Cleavage products were immunoprecipitated with UP102 and then analyzed by SDS–10% polyacrylamide gel electrophoresis; p28 is indicated by the arrow. The molecular masses of marker proteins are indicated between the panels.
To quantify cleavage efficiency by PLP-1-containing polypeptides of different lengths, cleavages were carried out with enzymes synthesized from pCITE P1P2a. For this purpose, pCITE P1P2a was truncated at various restriction sites, and the linearized plasmids were translated in vitro in the presence of [35S]methionine or with 0.1 mM unlabeled methionine. In the in vitro trans cleavage assays with the viral substrates described above and unlabeled enzymes, the cleavage efficiency was determined by normalizing the amount of cleavage to equal molar amount of enzymes. The amount of enzyme synthesized in vitro from each template was calculated by measuring incorporation of [35S]methionine into the truncated polypeptides after adjusting for the methionine content. Cleavage activity was calculated as described above.
RESULTS
Cloning, overexpression, and purification of recombinant PLP-1.
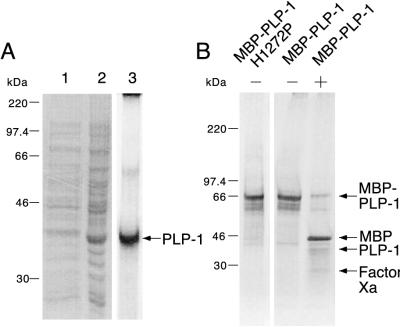
Numerous in vitro studies have suggested a role for coronavirus PLP-1 in proteolytic processing of the ORF1a-encoded polyprotein (2, 3, 5, 6, 10, 13, 19, 20). In the murine coronavirus MHV-A59, deletion of either upstream or downstream sequences flanking the proposed PLP-1 domain defined a minimal region, between Ala1084 and Tyr1316, that when expressed by in vitro transcription-translation was able to cleave in cis and in trans at the p28 and p65 sites (5, 6). To further study the properties of PLP-1 in MHV-A59, we have cloned and overexpressed recombinant PLP-1, using the region from Ser1062 to Lys1364. We initially expressed PLP-1 from a pET vector. However, a majority of the recombinant enzyme was partitioned into insoluble inclusion bodies (Fig. 2A, lanes 1 and 2) and required solubilization with urea followed by refolding and purification. The yield was about 7 mg of purified enzyme per liter of culture (Fig. 2A, lane 3). This insolubility led us to overexpress PLP-1 as an MBP-virus enzyme fusion protein from pMAL-c2. Expression of this plasmid resulted in high levels of a soluble fusion protein and, following affinity chromatography on an amylose column, yielded about 10 mg of fusion enzyme per liter of culture (Fig. 2B, − lanes). However, cleavage by factor Xa, which released PLP-1 from the fusion enzyme, substantially reduced the amount of PLP-1 polypeptide obtained (Fig. 2B, + lane). Therefore in the experiments described below we used the fusion enzyme rather than the factor Xa-cleaved PLP-1.
FIG. 2.
Overexpression and purification of recombinant PLP-1. Proteins were purified as described in Materials and Methods, analyzed by SDS–10% (A) or 4 to 15% (B) polyacrylamide gel electrophoresis, and stained with Coomassie blue. (A) Expression of PLP-1 from pET-PLP-1. Lane 1, supernatant after sonication and centrifugation; lane 2, pellet after sonication and centrifugation; lane 3, refolded PLP-1. PLP-1 is indicated by the arrow. (B) Purified MBP–PLP-1 H1272P and MBP–PLP-1 with (−) and without (+) factor Xa cleavage. MBP–PLP-1 (or MBP–PLP-1 H1272P), MBP, PLP-1, and factor Xa are indicated by arrows. The molecular masses of marker proteins are indicated to the left of each panel.
Proteolytic activity of recombinant PLP-1.
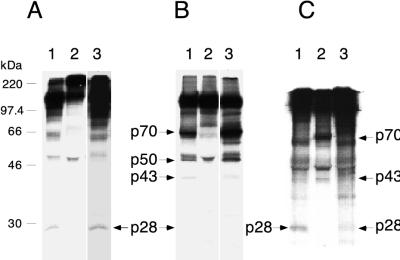
To examine whether the recombinant PLP-1 proteins are enzymatically active, we incubated MBP–PLP-1 with viral substrates, synthesized in transcription-translation reactions from templates pSPN1S1, pSPΔMscN1S1, and pSPΔMAGN1S1. These constructs, derived from pSPNK, pSPNKΔMsc (Fig. 1B), and pSPNKΔMAG, respectively, terminate at the SpeI site within PLP-1, resulting in polypeptides lacking the carboxyl two-thirds of PLP-1, and are unable to catalyze either cis or trans cleavage (6). Results in Fig. 3A and B (lanes 1) demonstrate that the fusion enzyme can catalyze cleavage of these viral substrates. The identities of the products were confirmed by immunoprecipitation using antiserum UP102, which is directed against the first 593 amino acids encoded in gene 1 (8) (Fig. 1B). As expected, the cleavage product, p28, was immunoprecipitated when the viral substrate was translated from pSPN1S1 (Fig. 3C, lane 1). With viral substrate translated from pSPΔMscN1S1, p28, p43, and p70 were immunoprecipitated (Fig. 3C, lane 2). (The p50 polypeptide indicated in Fig. 3B is the carboxyl-terminal product of the cleavage at the p65 site and is therefore not immunoprecipitated by UP102.) When the p65 site is deleted, as in the pSPΔMAGN1S1 construct, neither p43 nor p70 was detected, as expected (Fig. 3C, lane 3) (6). These products are the same as those cleaved in cis, using full-length plasmids as substrates, or those cleaved in trans, from similar substrates by TnT-derived enzyme. Similar to our previous observation, the amount of p28 produced with substrate synthesized from constructs carrying the ΔMsc deletion was considerably less than with substrates synthesized from either a full-length construct or a construct carrying the ΔMAG deletion (6, 20). The same products were obtained with refolded PLP-1 expressed from pET-PLP-1 in vivo (Fig. 3A and B, lanes 3). The authenticity of proteolytic activity was demonstrated by the observation that incubation of these substrates with mutant enzyme containing an inactivating H1272P mutation (6) did not result in cleavage of these polypeptides (Fig. 3A and B, lanes 2). Thus, these results demonstrate that expression of the region of the MHV-A59 genome encoding Ser1062 to Lys1364 does result in a functional proteinase.
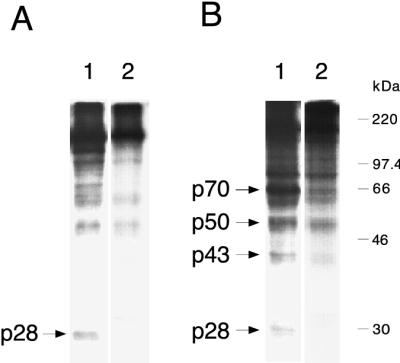
FIG. 3.
trans cleavage of viral substrates by PLP-1. (A) Cleavage of viral substrate translated in vitro from pSPN1S1. Lane 1, with MBP–PLP-1 enzyme; lane 2, with MBP–PLP-1 H1272P enzyme; lane 3, with refolded PLP-1. (B) Cleavage of viral substrate translated in vitro from pSPΔMscN1S1. Lane 1, with MBP–PLP-1 enzyme; lane 2, with MBP–PLP-1 H1272P enzyme; lane 3, with refolded PLP-1. (C) Immunoprecipitation with antiserum UP102 after cleavage of viral substrates translated in vitro from pSPN1S1 (lane 1), pSPΔMscN1S1 (lane 2), and pSPΔMAGN1S1 (lane 3) by MBP–PLP-1 enzyme. Samples were analyzed on SDS–10% polyacrylamide gels. Cleavage products are indicated by arrows. In panel B, p50 represents the carboxyl-terminal cleavage product resulting from cleavage at the p65 site. The molecular masses of marker proteins are indicated to the left of panel A. Much less p28 was produced with substrate translated from pSPΔMscN1S1 than with substrates translated from pSPN1S1 and pSPΔMAGN1S1 (B, lanes 1 and 3; C, lane 2). The p28 protein bands were visible upon prolonged exposure.
In a previous study we reported that cleavage in trans could be carried out with TnT-generated PLP-1 at 22 to 25°C (6). To allow direct comparison with those results, experiments described here were carried out under similar conditions, including length of incubation and reaction temperature (22 to 25°C). Subsequently, we investigated the temperature dependence of trans cleavage by incubating MBP–PLP-1 with viral substrates at 22 and 30°C. Cleavage was significantly more efficient when carried out at 22°C (Fig. 4, lanes 1) than when carried out at 30°C (lanes 2). We further observed that cleavage at both the p28 and p65 sites was even more efficient at 16°C, whereas cleavage at 37°C was even less efficient than cleavage at 30°C (data not shown). With the temperature used in the trans cleavage experiments (22 to 25°C), the proteinase activity was slow, with substantial cleavage at the p28 and p65 sites detected after 6 to 8 h of incubation (data not shown).
FIG. 4.
In vitro trans cleavage by recombinant PLP-1 is more efficient at room temperature. Radiolabeled viral polypeptides were synthesized in a coupled transcription-translation system using as templates pSPN1S1 (A) and pSPΔMscN1S1 (B). Viral substrates were incubated with recombinant MBP–PLP-1 fusion enzyme overnight at either 22°C (lane 1) or 30°C (lane 2), followed by electrophoresis on SDS–10% polyacrylamide gels. Cleavage products are indicated by the arrows. In panel B, cleavage at the p65 site generates the carboxyl-terminal p50 polypeptide. The molecular masses of marker proteins are indicated to the right of panel B.
Effect of substrate length on p28 cleavage efficiency.
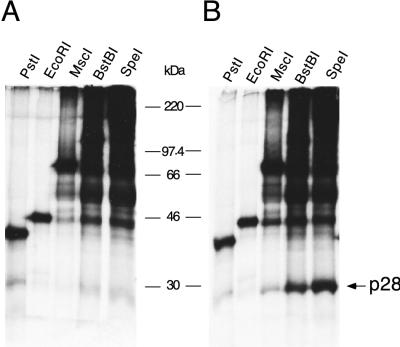
Baker et al. (2) reported that in vitro-translated MHV-JHM PLP-1 failed to cleave in trans a 65-kDa polypeptide including the p28 cleavage site and downstream sequences. In MHV-A59, deletions of amino acid sequences downstream of the cleavage sites reduced cleavage efficiency from 22 to 63%, depending on the region and amount deleted (5). This suggests that PLP-1 may prefer long substrates, probably above 65 kDa. To examine this hypothesis, plasmid pSPNK H1272P was linearized with various enzymes, followed by in vitro transcription and translation. This resulted in the synthesis of polypeptides of increasing lengths: 301 amino acids (34.7 kDa), generated by PstI digestion; 368 amino acids (40.9 kDa), generated by EcoRI digestion; 622 amino acids (69.4 kDa), generated by MscI digestion; 867 amino acids (96.2 kDa), generated by BstBI digestion; and 1,160 amino acids (128 kDa), generated by SpeI digestion. The polypeptide products were then incubated in a trans cleavage assay with PLP-1 synthesized in vitro from pET-PLP-1 as described in Materials and Methods. (Substrates were also incubated with mutant PLP-1 containing the H1272P substitution; the low level of radioactivity present in the p28 region of the gel following incubation with mutant PLP-1 was considered background and was subtracted in the calculation of the cleavage efficiency shown in Table 2.) PhosphorImager quantifications showed that cleavage activity was not detectable with substrates of less than 301 amino acids (34.7 kDa) (Fig. 5 and Table 2). As substrate length increased to 622 amino acids (69.4 kDa), substantial cleavage (44%) could be detected. Further increasing the substrate length to 867 amino acids (96.2 kDa) resulted in maximal cleavage. The results presented here thus demonstrate that efficient trans cleavage by PLP-1 in vitro is strongly dependent on substrate length. Consistent with this finding, we failed to detect cleavage of synthetic peptides containing either the p28 or the p65 sites by recombinant PLP-1 (data not shown).
TABLE 2.
Cleavage efficiency at the p28 site, using substrates of increasing length
| Enzymea | Length (amino acids)b | Mass (kDa)c | Cleavage efficiency (%)d |
|---|---|---|---|
| PstI | 301 | 34.7 | 0 |
| EcoRI | 368 | 40.9 | 5 |
| MscI | 622 | 69.4 | 44 |
| BstBI | 867 | 96.2 | 116 |
| SpeI | 1,160 | 128.0 | 100 |
Plasmid pSPNK H1272P (6) was linearized with the enzymes listed; each linearized plasmid was used as template in in vitro-coupled transcription-translation reactions to produce polypeptides of increasing size.
Lengths of polypeptides synthesized by using the truncated plasmid templates.
Predicted molecular mass of corresponding viral polypeptides.
Cleavage efficiency (p28 production) was calculated as described in Materials and Methods; the cleavage efficiency with the longest substrate was arbitrarily set to 100%.
Effect of flanking regions on p28 cleavage efficiency.
Results from earlier studies and those described above demonstrated that expression of the region between Ala1084 and Tyr1316 is sufficient to produce an enzymatically active PLP-1. However, the presence of additional sequences on either the amino or carboxyl side of the mapped domain result in more efficient cleavage of p28 (5). Furthermore, the X domain, adjacent to the carboxyl side of the PLP-1 in MHV (15), is conserved among different viruses and has been suggested to play a role in the proteinase activity (5, 17). In view of the observation that the major polypeptide, detected in infected cells, containing the PLP-1 domain is p290 (11), it is possible that the proteolytic activity exhibited by PLP-1 is most effective when synthesized as part of a very large polyprotein. To further examine possible roles for the X domain, PLP-2, and the intervening sequences in affecting the efficiency of PLP-1 cleavage, we used plasmid pCITE P1P2a as a source of PLP-1; this plasmid contains the viral sequences encoding Lys869 to Leu2182 (including both proteinase domains), downstream of a T7 RNA polymerase promoter (Fig. 1B). Plasmid pCITE P1P2a was linearized with SpeI (between the catalytic Cys1121 and His1272 of PLP-1), BstXI (at the carboxyl terminus of PLP-1), NsiI (including the entire PLP-1 and the X domain), ClaI (including in addition the amino one-third of PLP-2), and AgeI (including in addition the rest of PLP-2), followed by in vitro transcription and translation in the presence of unlabeled methionine as described in Materials and Methods. The products were then incubated overnight at 22°C with viral substrates synthesized in vitro in the presence of [35S]methionine, using either pSPN1S1 or pSPΔMscN1S1 as the template (as described in Materials and Methods). The results demonstrated that the presence of the downstream sequence enhanced cleavage efficiency (Fig. 6 and Table 3). As the inclusion of the X domain increased cleavage from 16 to 18% to 29 to 33%, addition of the amino one-third of the PLP-2 domain further enhanced the extent of cleavage of p28 to 48%. Interestingly, increasing the downstream sequence to include the complete PLP-2 doubled the cleavage efficiency (Table 3), suggesting that regions in additional to the X domain may play a role in regulating PLP-1 activity (see Discussion).
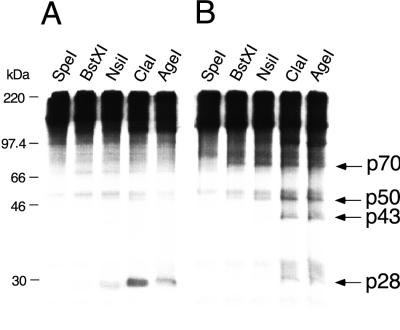
FIG. 6.
PLP-1 cleavage efficiency at the p28 and p65 sites increases with increased length of proteinase polypeptide. Plasmid pCITE P1P2a was digested with the enzymes listed at the top and used as templates in coupled transcription-translation reactions in the presence of unlabeled methionine. The translated polypeptides, serving as a source of PLP-1, were incubated in trans cleavage assays with viral substrates synthesized in coupled transcription-translation reactions in the presence of [35S]methionine (as described in Materials and Methods), using pSPN1S1 (A) and pSPΔMscN1S1 (B) as templates. Cleavage reactions were analyzed by SDS–10% polyacrylamide gel electrophoresis; cleavage products p28, p43, p50, and p70 are indicated by the arrows. p50 is the carboxyl-terminal product derived by cleavage at the p65 site. The molecular masses of marker proteins are indicated to the left of panel A.
TABLE 3.
Cleavage efficiency at the p28 and p65 sites, using polypeptides of increasing length as sources of PLP-1
| Enzymea | Length (amino acids)b | Mass (kDa)c | Cleavage efficiency (%)d
|
|
|---|---|---|---|---|
| pSPN1S1 | pSPΔMscN1S1 | |||
| SpeI | 294 | 31.9 | 0 | 1 |
| BstXI | 448 | 49.2 | 18 | 16 |
| NsiI | 638 | 69.3 | 33 | 29 |
| ClaI | 904 | 99.2 | 48 | 48 |
| AgeI | 1,161 | 128.0 | 100 | 100 |
To synthesize PLP-1-containing polypeptides, plasmid pCITE P1P2a (Fig. 1B) was linearized with the enzymes listed, followed by in vitro transcription-translation in the presence of 0.1 mM unlabeled methionine.
Lengths of polypeptides synthesized by using the truncated plasmid templates.
Predicted molecular mass of corresponding viral polypeptides.
Substrates were synthesized in similar reactions using [35S]methionine and either pSPN1S1 or pSPΔMscN1S1 as the template. Cleavage efficiency was calculated as described in Materials and Methods; the cleavage efficiency with the longest enzyme polypeptide was arbitrarily set to 100%.
PLP-2 does not cleave viral substrates under the same conditions under which PLP-1 activity is observed.
Despite the prediction of a second conserved PLP domain (PLP-2) in several, but not all, coronaviruses (12, 16, 17, 22), there have not yet been any reports of detection of an in vitro cleavage activity associated with this second proteinase. Therefore, we examined the PLP-2 domain for a proteinase activity in several ways. Based on the results presented previously (5) and in Fig. 6, which show that PLP-1 cleavage is more efficient in the presence of adjacent regions, we hypothesized that PLP-2 cleavage might also require the presentation of the proteinase as a larger polypeptide. To examine this hypothesis, we introduced the H1272P mutation into pCITE P1P2a and then used the wild-type and mutant plasmids to produce polypeptides in a coupled transcription-translation reaction. The polypeptides were then incubated with substrate translated from pSPΔMscN1S1 in a trans cleavage assay. The results in Fig. 7A show that inactivation of PLP-1 abolishes cleavage at both the p28 and p65 sites.
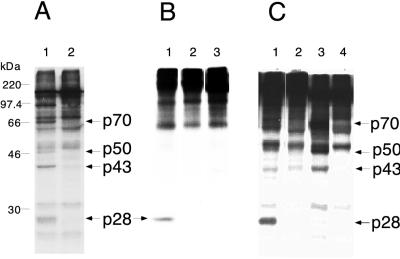
FIG. 7.
Proteolytic activity could not be detected for PLP-2. (A) trans cleavage of [35S]methionine-labeled viral substrates encoded by pSPΔMscN1S1 with enzymes synthesized by coupled transcription-translation using as the template pCITE P1P2a (lane 1) or pCITE P1P2a H1272P (lane 2). (B) cis cleavage with [35S]methionine-labeled viral polypeptides synthesized by transcription-translation using as templates pSPNK (lane 1), pSPN1S2ΔD1049K1657 (lane 2), and pSPN1S2ΔF1085A1670 (lane 3), followed by immunoprecipitation with UP102. (C) trans cleavage carried out with MBP fusion enzymes using [35S]methionine-labeled viral polypeptides as substrates. Lane 1, MBP–PLP-1 (enzyme) and pSPN1S1-encoded polypeptide (substrate); lane 2, MBP–PLP-2 (enzyme) and pSPN1S1-encoded polypeptide (substrate); lane 3, MBP–PLP-1 (enzyme) and pSPΔMscN1S1-encoded polypeptide (substrate); lane 4, MBP–PLP-2 (enzyme) and pSPΔMscN1S1-encoded polypeptide (substrate). All cleavage reactions were analyzed by SDS–10% polyacrylamide gel electrophoresis; cleavage products p28, p43, p50, and p70 are indicated by the arrows. p50 is the carboxyl-terminal product derived by cleavage at the p65 site. The molecular masses of marker proteins are indicated to the left of panel A. The p28 protein bands were visible upon prolonged exposure.
To examine whether the position of PLP-2 can affect its activity, or whether the presence of PLP-1 can inhibit PLP-2 cleavage activity, we constructed plasmids pSPN1S2ΔD1049K1657 and pSPN1S2ΔF1085A1670, in which the PLP-1 domain is deleted and the predicted PLP-2 domain is moved to the approximate position of PLP-1 (Fig. 1B). As shown in Fig. 7B, cis cleavage was not observed at either the p28 or p65 sites when the plasmids pSPN1S2ΔD1049K1657 (lane 2) and pSPN1S2ΔF1085A1670 (lane 3) were transcribed and translated in vitro. However, it was still possible that the activity of PLP-2 synthesized in vitro from these constructs was too low for detection. To address this possibility, we subcloned the predicted region for PLP-2 (Phe1681 to Ser1936) (16) into pMAL-c2, and overexpressed PLP-2 as a fusion protein. As with PLP-1, the fusion protein was soluble, and about 10 mg of fusion enzyme per liter of culture could be isolated following affinity chromatography. Results in Fig. 7C show that under conditions with which cleavage by MBP–PLP-1 could be detected (lanes 1 and 3), incubation of MBP–PLP-2 with polypeptides generated from either pSPN1S1 or pSPΔMscN1S1 did not result in detectable cleavage (lanes 2 and 4). Therefore, the results presented suggest that PLP-2 may not be active in cleavage at the p28 and p65 sites (see Discussion).
DISCUSSION
To further our understanding of the MHV-A59 replicase processing events, we have cloned and overexpressed the predicted PLP-1 domain. We previously mapped the minimal functional domain of PLP-1 to between Ala1084 and Tyr1316 (5), nearly the same span of residues as originally predicted by Lee et al. (16) based on sequence analysis. However, we also found that activity of the polypeptide containing this minimal domain was not optimal. Starting with plasmid pSPNK (Fig. 1B), which encodes the first 1,484 amino acids of ORF1a, deletion of various sequences between the enzyme and cleavage sites, or truncation of the ORF1a polypeptide to eliminate the X domain, reduced cleavage by 22 to 63% (5). For expression of the proteinase in E. coli, we have cloned a region slightly larger than the minimal domain, from Ser1062 to Lys1364; this was a compromise between cloning a region large enough to optimize the efficiency of proteinase activity and cloning a region that was not too large for efficient polypeptide expression in E. coli.
Initial expression of the region from Ser1062 to Lys1364, using a pET-based vector (Fig. 1C), resulted in the majority of the recombinant enzyme partitioning into inclusion bodies (Fig. 2A, lane 2), which required solubilization and refolding in order to reconstitute an active enzyme (Fig. 3A and B, lanes 3). This led us to express this region as an MBP–PLP-1 fusion protein (Fig. 1C), which was soluble (Fig. 2B, − lanes) and enzymatically active (Fig. 3A and B, lanes 1). Our results, therefore, demonstrate that the region between Ser1062 and Lys1364 encodes a functional proteinase. However, incubation of MBP–PLP-1 with factor Xa results in greatly reduced yield of the amount of PLP-1-containing polypeptide (Fig. 2B, + lane). This is likely due to either nonspecific cleavage of PLP-1 by factor Xa, as reported for other MBP fusion proteins (25, 26), or insolubility of the PLP-1 domain alone after cleavage. The poor recovery of PLP-1 after factor Xa digestion thus led us to perform the subsequent characterizations with the MBP–PLP-1 fusion enzyme.
Characterization of the enzymatic properties of MBP–PLP-1 revealed a requirement for low temperature for in vitro trans cleavage (Fig. 4). Similar temperature requirements were also observed with the refolded enzyme and PLP-1 synthesized in vitro from pET-PLP-1 and pCITE P1P2a (data not shown), indicating that this unusual cleavage condition is a characteristic of PLP-1 activity and is not specific to MBP–PLP-1. Taking into consideration that PLP-1 has to be functionally active at in vivo temperatures, the dependence of low temperature for cleavage could reflect an intrinsic property specific to the in vitro trans cleavage conditions. A possible explanation for this temperature dependence could be that the ORF1a polypeptide must adopt a specific conformation in order to present the cleavage sites to the active site of PLP-1, and under the in vitro conditions used, a low temperature (16 to 25°C) favors the adoption of this conformation. Substrate conformation has also been proposed to explain the lack of in vitro cleavage by PLP-1 at the p65 site in polypeptide encoded by pSPNK (5).
The cleavage pattern observed for PLP-1 is quite different from those of some other viral papain-like proteinases, including the nonstructural nsP2 protein of Sindbis virus, the leader proteinase of foot-and-mouth disease virus, and the PCPα and PCPβ from porcine reproductive and respiratory syndrome virus and lactate dehydrogenase-elevating virus (7, 18, 24). In those cases, the fully functional proteinases are generated via cis and/or trans cleavages of the precursor polypeptides at the amino and carboxyl termini of the proteinase domains. The data to date suggest, however, that (i) MHV PLP-1 does not have cleavage sites flanking the amino and carboxyl termini of the predicted domain and (ii) PLP-1 is probably more active as part of a larger polypeptide. An in vivo study with infected cells showed that PLP-1 exists as part of the p290 polypeptide (9, 11). Kinetic studies in which viral genomic RNA was translated in vitro demonstrated that p28 cleavage is initially detectable at about the time that the PLP-1 domain is synthesized (2, 10). This led to the speculation that PLP-1 acts in cis to cleave p28 from the amino terminus of a nascent ORF1a polyprotein. Subsequent in vitro studies using deletion constructs demonstrated that a second cleavage could occur at the p65 site (5). Furthermore, both cleavage events have been demonstrated to occur in vitro in trans (reference 6 and this study). There is also evidence that cleavage at the p28 and p65 sites can occur in trans in vivo as well as in vitro. Previous in vivo kinetic studies show that production of both p28 and p65 is rapid, and no precursors containing p28 or p65 sequences could be detected (11). If the cleavages at these two sites were to occur exclusively in cis, then p28 and p65 should not be detectable until translation of a functional PLP-1 occurs. The lack of detection of a precursor polypeptide containing p28 and p65 suggests that cleavage may occur in vivo, at least at late times after infection when these studies were carried out, in trans.
We demonstrate here that efficient cleavage in trans is dependent on the lengths of both the polypeptides containing substrate and enzyme. By measuring the in vitro trans cleavage efficiency by using substrates and enzyme presented as polypeptides of increasing lengths, we observed that substrates of greater than 69 kDa show substantial cleavage, with the highest cleavage efficiency achieved with substrates of greater than 90 kDa (Table 2). The observation that maximal cleavage was detected when PLP-1 was expressed with the downstream X and PLP-2 domains, regardless of the whether the cleavage was at the p28 or the p65 site (Table 3), suggests that the presentation of the enzyme within a polyprotein also enhances cleavage. In results not presented here, we observed that substituting the proposed catalytic His1873 of PLP-2 (in pCITE P1P2a [Fig. 1B]) with a Pro did not reduce in vitro trans cleavages at the p28 and p65 sites significantly, suggesting that the presence of adjacent residues contributes more to efficient PLP-1 cleavage rather than the presence of a functional PLP-2 domain. Thus, in vitro trans cleavage by PLP-1 is most efficient when both the enzyme and substrate are presented as long polypeptides. Thus, taken together the in vivo and in vitro data are most consistent with a model in which PLP-1 cleavage may occur in cis and in trans, providing that the ORF1a polypeptide folds into a conformation which exposes the p28 and p65 sites to the active site of PLP-1.
As proposed by Bonilla et al. (6), the consensus sequence for PLP-1 cleavage recognition is [P5-(Arg,Lys)XXX(Gly,Ala)↓(Gly,Ala,Val)-P1′], with the residues at P5, P1, and P1′ shown to be essential for cleavage recognition. We previously proposed that the sequence Lys1258-Val-Phe-Arg-Ala↓Ala1262 in MHV-A59 ORF1a was the sequence closest to a consensus sequence within ORF1a (other than the p28 and p65 cleavage sites) (6). Cleavage at this site, which is between the catalytic dyad of PLP-1, would generate a 50-kDa protein, which could be the p50 detected in vivo as a result of processing of the p290 polypeptide (6, 11, 30). Nevertheless, in vitro transcription and translation of the full-length construct, pSPNK, did not result in cleavage at either the p65 or p50 site (5, 6, 20). Similarly, when pCITE P1P2a was in vitro transcribed and translated, products corresponding to cleavage at this potential p50 site were not detected (data not shown), indicating that the production of p50 (as well as p65) was not determined simply by the length of the substrate. Based on the in vitro cleavage experiments described previously (5, 6, 20) and here, which show that cleavage at the p65 site is observed only with the ΔMsc deletion construct; we have hypothesized that only substrates translated in vitro from the deletion constructs could adopt specific conformation to allow access of the p65 site to the active site of PLP-1. The lack of processing at the potential p50 site may be similarly explained. Interestingly, if cleavage between Ala1262 and Ala1263 were to occur in vivo, that could suggest a possible mechanism for regulating the proteolytic processing of the replicase gene product during infection, possibly via a trans-acting autocatalytic inactivation of PLP-1.
Recently, Schiller et al. (27) proposed a PLP-1 cleavage site (to generate p72) in the MHV-JHM ORF1a-encoded polyprotein, not detected in the MHV-A59 ORF1a polyprotein. They speculated that cleavage occurred within the sequence [P5-Cys900-Lys-Glu-His-Gly↓Val905-P1′]. It should be noted that this proposed JHM p72 cleavage sequence differs from the proposed consensus PLP-1 cleavage sequence discussed above (6) at the P5 residue; the corresponding region in MHV-A59 ORF1a is [P5-Cys900-Lys-Glu-His-Asp-Val905-P1′], which differs from the consensus sequence at P1 as well as P5 residues. Moreover, Schiller et al. (27) suggested that the lack of cleavage in MHV-A59 ORF1a at this site was due to the presence of an Asp, rather than a Gly (as in MHV-JHM), at the P1 site (residue 904) in the MHV-A59 sequence. To examine their hypothesis, we mutated Asp904 in pSPNKΔMAG (6) to Gly, which results in the sequence [P5-Cys900-Lys-Glu-His-Gly-Val905-P1′]. In vitro transcription and translation of this construct, pSPNKΔMAGD904G, did not result in any new cleavage products (data not shown). The lack of cleavage between Gly904 and Val905 in this mutant construct provides further support for the important roles of P5, P1, and P1′ residues in determining cleavage by PLP-1. However, we cannot rule out the possibility that the lack of cleavage was due to the translated polypeptide adopting an unfavorable conformation in vitro, or the lack of cellular cofactors essential for cleavage at this mutated site, similar reasons as those proposed to explain the lack of cleavage at the p65 site in the pSPNK-encoded polypeptide (5, 6).
Based on sequence comparisons, it has been proposed that MHV ORF1a encodes a second PLP domain, located downstream of PLP-1 (16, 22). However, no proteolytic activity for this domain has yet been demonstrated (3, 5, 14). In this study, we used several approaches to investigate a possible proteolytic role for PLP-2: (i) expression of PLP-2 as part of a polypeptide adjacent to an inactive PLP-1, (ii) removal of PLP-1 and placing PLP-2 into the approximate location of PLP-1, and (iii) expression of PLP-2 as an MBP fusion. In all cases, no cleavage activity could be detected (Fig. 7). The lack of cleavage with PLP-2 was at least not due to the incubation temperature, as overnight incubation from 22 to 37°C did not result in cleavage at either the p28 or the p65 site (data not shown). Thus, the PLP-2 domain may be nonfunctional in mediating cleavage at the p28 and p65 sites. Alternatively, cellular factors not present in our in vitro transcription-translation system, or a certain conformation adopted by the substrate in vivo, may be essential for the PLP-2-mediated catalysis. Lastly, PLP-2 may be involved in cleavage other than the p28 and p65 sites within the ORF1a polypeptide. Further experiments will be required to examine these hypotheses.
In conclusion, the data presented here provide support for a model in which the ORF1a polypeptide adopts a conformation which allows PLP-1 to catalyze cleavage at the p28 and p65 sites. While early studies of in vitro translation of genome RNA or ORF1a-encoded plasmids suggested that cleavage was in cis only, studies of ORF1a processing in vivo and studies reported here using recombinant PLP-1 demonstrate that PLP-1 can function in trans at both cleavage sites. The question of whether the adjacent X domain and PLP-2, or even other cofactors, may play a regulatory role cannot yet be answered. It should be noted that even though the cloned region, from Ser1062 to Lys1364, may not encode a fully active PLP-1, the proteolytic activity demonstrated in this study suggests that this region may constitute the catalytic domain of the native, or in vivo, form of PLP-1. Thus, studying the cloned region can provide us with useful structural information and can contribute to the development of antiviral therapy.
ACKNOWLEDGMENTS
This work was supported by NIH grant AI-17418.
We acknowledge Ravi R. Mayreddy for construction of plasmid pMAL-PLP-1, Sidhartha Chandela for construction of pMAL-PLP-2, and Pedro J. Bonilla for construction of many ORF1a plasmids used in this study and for valuable advice and critical evaluation of the manuscript.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1987. pp. 10.1.1–10.1.3. [Google Scholar]
- 2.Baker S C, Shieh C K, Chang M F, Vannier D M, Lai M M C. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J Virol. 1989;63:3693–3699. doi: 10.1128/jvi.63.9.3693-3699.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker S C, Yokomori K, Dong S, Carlisle R, Gorbalenya A E, Koonin E V, Lai M M C. Identification of the catalytic sites of a papain-like proteinase of murine coronavirus. J Virol. 1993;67:6056–6063. doi: 10.1128/jvi.67.10.6056-6063.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonilla P J, Gorbalenya A E, Weiss S R. Mouse hepatitis virus strain A59 RNA polymerase gene ORF1a: heterogeneity among MHV strains. Virology. 1994;198:736–740. doi: 10.1006/viro.1994.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonilla P J, Hughes S A, Piñón J D, Weiss S R. Characterization of the leader papain-like proteinase of MHV-A59: identification of a new in vitro cleavage site. Virology. 1995;209:489–497. doi: 10.1006/viro.1995.1281. [DOI] [PubMed] [Google Scholar]
- 6.Bonilla P J, Hughes S A, Weiss S R. Characterization of a second cleavage site and demonstration of activity in trans by the papain-like proteinase of the murine coronavirus MHV-A59. J Virol. 1997;71:900–909. doi: 10.1128/jvi.71.2.900-909.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.den Boon J A, Faaberg K S, Meulenberg J J M, Wassenaar A L M, Plagemann P G W, Gorbalenya A E, Snijder E J. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: identification of two papain-like cysteine proteases. J Virol. 1995;69:4500–4505. doi: 10.1128/jvi.69.7.4500-4505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denison M R, Hughes S A, Weiss S R. Identification and characterization of a 65-kDa protein processed from the gene 1 polyprotein of the murine coronavirus MHV-A59. Virology. 1995;207:316–320. doi: 10.1006/viro.1995.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denison M R, Kim J C, Ross T. Inhibition of coronavirus MHV-A59 replication by proteinase inhibitors. In: Talbot P J, Levy G A, editors. Corona- and related viruses. New York, N.Y: Plenum Press; 1995. pp. 391–397. [DOI] [PubMed] [Google Scholar]
- 10.Denison M R, Perlman S. Translation and processing of mouse hepatitis virus virion RNA in a cell-free system. J Virol. 1986;60:12–18. doi: 10.1128/jvi.60.1.12-18.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denison M R, Zoltick P W, Hughes S A, Giangreco B, Olson A L, Perlman S, Leibowitz J L, Weiss S R. Intracellular processing of the N-terminal ORF1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology. 1992;189:274–284. doi: 10.1016/0042-6822(92)90703-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Vries A A F, Horzinek M C, Rottier P J M, de Groot R J. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong S, Baker S C. Determinants of the p28 cleavage site recognized by the first papain-like cysteine proteinase of murine coronavirus. Virology. 1994;204:541–549. doi: 10.1006/viro.1994.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao H-Q, Schiller J J, Baker S C. Identification of the polymerase polyprotein products p72 and p65 of the murine coronavirus MHV-JHM. Virus Res. 1996;45:101–109. doi: 10.1016/S0168-1702(96)01368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorbalenya A E, Koonin E V. Comparative analysis of amino-acid sequences of key enzymes of replication and expression of positive-strand RNA viruses: validity of approach and functional and evolutionary implications. Sov Sci Rev Sect D. 1993;11:1–84. [Google Scholar]
- 16.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Coronavirus genome: prediction of putative functional domains in the nonstructural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorbalenya A E, Snijder E J. Viral cysteine proteinases. Perspect Drug Discov Des. 1996;6:64–86. doi: 10.1007/BF02174046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy W R, Strauss J H. Processing the nonstructural polypeptides of Sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and trans. J Virol. 1989;63:4653–4664. doi: 10.1128/jvi.63.11.4653-4664.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herold J, Gorbalenya A E, Thiel V, Schelle B, Siddell S G. Proteolytic processing at the amino terminus of human coronavirus 229E gene 1-encoded polyproteins: identification of a papain-like proteinase and its substrate. J Virol. 1998;72:910–918. doi: 10.1128/jvi.72.2.910-918.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes S A, Bonilla P J, Weiss S R. Identification of the murine coronavirus p28 cleavage site. J Virol. 1995;69:809–813. doi: 10.1128/jvi.69.2.809-813.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J C, Spence R A, Currier P F, Lu X, Denison M R. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64d. Virology. 1995;208:1–8. doi: 10.1006/viro.1995.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee H J, Shieh C K, Gorbalenya A E, Koonin E V, LaMonica N, Tuler J, Bagdzhadzhyan A, Lai M M C. The complete sequence of the murine coronavirus gene 1 encoding the putative protease and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim K P, Liu D X. Characterization of the two overlapping papain-like proteinase domains encoded in gene 1 of the coronavirus infectious bronchitis virus and determination of the C-terminal cleavage site of an 87-kDa protein. Virology. 1998;245:303–312. doi: 10.1006/viro.1998.9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medina M, Domingo E, Brangwyn J K, Belsham G J. The two species of the foot-and-mouth disease virus leader protein, expressed individually, exhibit the same activities. Virology. 1993;194:355–359. doi: 10.1006/viro.1993.1267. [DOI] [PubMed] [Google Scholar]
- 25.Nagal K, Thøgersen H C. Synthesis and sequence-specific proteolysis of hybrid proteins produced in Escherichia coli. Methods Enzymol. 1987;153:461–481. doi: 10.1016/0076-6879(87)53072-5. [DOI] [PubMed] [Google Scholar]
- 26.Rodríguez P L, Carrasco L. Improved factor Xa cleavage of fusion proteins containing maltose binding protein. BioTechniques. 1995;18:238–243. [PubMed] [Google Scholar]
- 27.Schiller J J, Kanjanahaluethai A, Baker S C. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology. 1998;242:288–302. doi: 10.1006/viro.1997.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snijder E J, Meulenberg J J M. The molecular biology of arteriviruses. J Gen Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 29.Studier W F, Rosenberg A H, Dunn J J, Dubendorf J W. Use of T7 polymerase to direct the expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 30.Weiss S R, Hughes S A, Bonilla P J, Leibowitz J L, Denison M R. Coronavirus polyprotein processing. Arch Virol. 1994;9:349–358. doi: 10.1007/978-3-7091-9326-6_35. [DOI] [PubMed] [Google Scholar]