Abstract
Tardigrades are remarkable in their ability to survive extreme environments. The damage suppressor (Dsup) protein is thought responsible for their extreme resistance to reactive oxygen species (ROS) generated by irradiation. Here we show that expression of Ramazzottius varieornatus Dsup in Saccharomyces cerevisiae reduces oxidative DNA damage and extends the lifespan of budding yeast exposed to chronic oxidative genotoxicity. This protection from ROS requires either the Dsup HMGN-like domain or sequences C-terminal to same. Dsup associates with no apparent bias across the yeast genome, using multiple modes of nucleosome binding; the HMGN-like region interacts with both the H2A/H2B acidic patch and H3/H4 histone tails, while the C-terminal region binds DNA. These findings give precedent for engineering an organism by physically shielding its genome to promote survival and longevity in the face of oxidative damage.
INTRODUCTION
Tardigrades (also termed water bears) are an invertebrate phylum of > 1,200 species with broad-reaching habitats. Many can survive desiccation, extreme temperatures, high pressure, severe irradiation, and exposure to space1. The mechanisms by which tardigrade species resist such extreme stressors are poorly understood. Ramazzottius varieornatus is highly resistant to ionizing radiation (IR); capable of surviving > 48 hours after a dose of 4000 Gy2, compared to the human LD50 of ~ 4.5 Gy3. The R. varieornatus Dsup (Damage suppressor) protein is chromatin associated and predicted to promote IR resistance, being absent from IR sensitive tardigrade species4. Indeed, when expressed in human cells, Dsup localizes to nuclear DNA and confers IR-resistance accompanied by reduced levels of single- and double-strand DNA breaks (SSBs and DSBs)4; it also confers protection from radiation when expressed in tobacco plants5.
While IR can directly induce SSBs and DSBs, much of its genotoxicity is mediated by hydroxyl radicals (OH•), the most powerful oxidant among the reactive oxygen species (ROS) and generated when radiation interacts with water molecules6. Consistent with Dsup protecting against hydroxyl radicals, it also reduces the number of DNA breaks in human cells exposed to hydrogen peroxide (H2O2)4,7. The high-energy hydroxyl radicals react with DNA bases to form lesions (including 8-oxoguanine; 8-oxo-G), while oxidation of the deoxyribose backbone dissociates sugar-phosphate bonds leading to DNA breaks8. Throughout life, oxidative DNA damage is generated from aerobic metabolism, with the resulting mutations thought to contribute to the ageing process9 and the development of age-related diseases10, such as neurodegeneration11 and cancer12,13. Most cancer treatments cause oxidative DNA damage and strand breaks, and thus contributes to long-term side effects in survivors14. As such, the means by which proteins such as R. varieornatus Dsup protect the genome from oxidative damage are of extreme interest.
R. varieornatus Dsup is a 445 amino acid protein predicted to be intrinsically disordered15. Of note, disorder at the N- and C-termini is an important feature of proteins that scan and engage DNA, consistent with a DNA-binding role for Dsup16,17. C-terminal deletion (Δ aa 208–445) abrogates Dsup binding to naked DNA or human chromatin4. Indeed, Dsup binds with higher affinity to reconstituted chromatin over free DNA, and sequences within aa 360–445 are required for the association with chromatin and protection from DSBs caused by hydroxyl radicals18. While Dsup induction in human cells upregulates the expression of DNA repair genes7; the protein also physically prevents DNA damage via chromatin binding, as this ability is observed in a reconstituted system lacking DNA repair factors18.
Within the Dsup C-terminal region, an eight amino acid stretch (aa 363–370, RRSSRLTS) has homology to the core consensus (RSRARLSA) of the nucleosome binding domain of vertebrate High Mobility Group-N (HMGN) proteins18,19,20. The chromatin binding of HMGN proteins influences a wide variety of functions (including embryogenesis, development and disease protection) across diverse cell types and species21. Mutation of the Dsup HMGN-like domain or deletion of its entire C-terminus respectively reduces or ablates binding to reconstituted chromatin and DNA protection from hydroxyl radicals18. As such, in the prevailing model revealed by use of the reconstituted system, Dsup protects the genome from DNA damage by physically shielding chromatin from hydroxyl radicals, involving the Dsup HMGN-like domain within its C-terminal sequences18. Whether the Dsup HMGN-like domain functions in vivo to mediate the interaction with chromatin and protect it from oxidative DNA damage is unknown.
Here, we show that when highly expressed in budding yeast R. varieornatus Dsup uses its HMGN-like domain and an additional region in the adjacent C-terminal sequences to bind chromatin and protect the genome from oxidative DNA damage in a manner dependent on chromatin engagement but independent of scavenging hydroxyl radicals. Dsup expression also extends yeast replicative lifespan in the face of chronic endogenous oxidative DNA damage. A detailed analysis of [Dsup : nucleosome] engagement finds that its HMGN-like domain mediates interaction with both the H2A/H2B acidic patch on the nucleosome surface and the H3/H4 N-terminal tails, while the distal C-terminal sequences binds DNA. Of note such a binding mechanism supports a broad engagement with in vivo chromatin independent of the landscape of histone post-translational modifications (PTMs). Our studies indicate that tardigrade Dsup can be introduced to a heterologous in vivo system and confer viability and longevity. This is achieved by physically coating the chromatinized genome via multivalent interactions to prevent hydroxyl radicals from damaging genomic DNA.
RESULTS
Heterologous expression of R. varieornatus Dsup in budding yeast protects against oxidative damage and promotes longevity in the face of increased oxidative stress
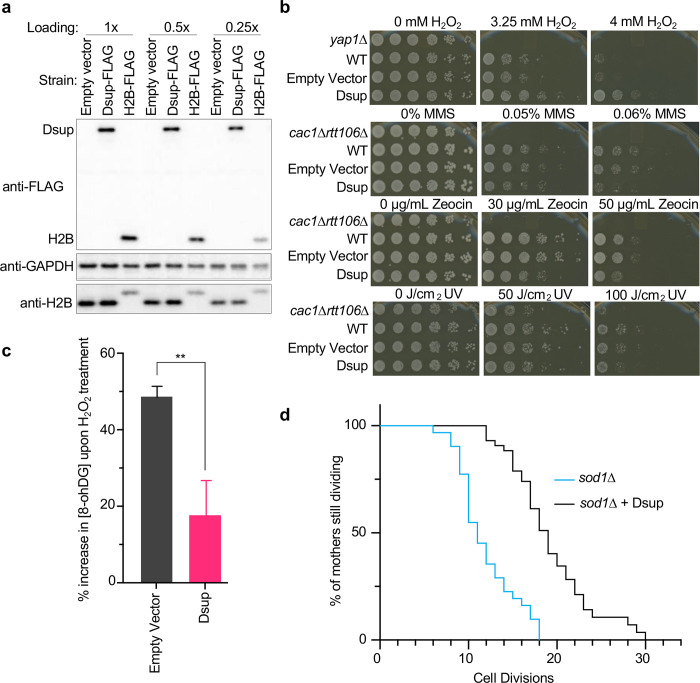
To initiate this study we expressed epitope tagged 6His-Dsup-FLAG (hereafter Dsup-FLAG) in yeast under the constitutive high output TDH3 promoter22, with the goal of achieving in vivo protein levels sufficient to coat the genome. Of note this yielded Dsup-FLAG of similar abundance to H2B-FLAG (Fig. 1a). To investigate the response of Dsup-FLAG yeast to chronic oxidative damage, we performed serial dilution assays on plates containing H2O2, observing a ~25-fold increased survival relative to yeast lacking Dsup (Fig. 1b). This did not extend to general protection from genotoxic insult, since Dsup-FLAG slightly decreased yeast survival in response to non-oxidative DNA-damaging agents such as alkylating methyl methanesulfonate (MMS), radiomimetic Zeocin, or UV (Fig. 1b).
Fig. 1. Heterologous expression of tardigrade Dsup in budding yeast promotes survival after chronic exposure to an oxidative DNA damaging agent, reduces related DNA damage, and extends lifespan upon chronic endogenous oxidative damage.
(a). Comparative immunoblot (three dilutions of protein extracts loaded as indicated) of Dsup-FLAG (from pTDH3–6His-Dsup-FLAG) and H2B-FLAG in yeast strains containing a single integrated copy of each tagged gene (Methods and Suppl. Table 2). (b). Five-fold serial dilutions of strains plated on indicated concentrations of hydrogen peroxide (H2O2), methyl methanesulfonate (MMS), Zeocin, or after exposure to UV. yap1Δ is a positive control for sensitivity to oxidative DNA damage (H2O2); cac1Δrtt106Δ is a positive control for sensitivity to other tested agents. (c). [8-OHdG] increase (mean and standard deviation from three independent experiments; ** = p < 0.01) in response to oxidative DNA damage (120 minutes exposure to 10 mM H2O2). (d). Replicative lifespan of yeast undergoing chronic oxidative damage (sod1Δ −/+ Dsup; n=30 individuals for each background).
In reconstituted assays recombinant Dsup protects chromatin from DSBs caused by hydroxyl radicals18, so we asked if Dsup expression protected the yeast genome from oxidative DNA damage. 8-oxoguanine (8-OHdG) is generated when ROS species react with DNA23, so we quantified the base modification after transient exposure to H2O2 and observed a significant reduction in 8-OHdG in the presence of Dsup (Fig. 1c).
ROS and oxidative damage increase with age, and reducing oxidative damage extends the lifespan of multiple species (yeast, worms, fruit flies, mice24), while elevated ROS production shortens lifespan25. We thus asked if Dsup expression could extend yeast lifespan. In otherwise WT yeast Dsup has a negligible impact on chronological lifespan (the length of time a cell survives in a non-dividing state; Suppl. Fig. 1a), while the replicative lifespan (the maximum number of times a cell can divide), was slightly reduced (Suppl. Fig. 1b). Cells lacking the superoxide dismutase (SOD) genes are deficient in their ability to process both endogenous and exogenous ROS. As a result, they accumulate oxidative stress and damage, such that yeast lacking SOD1 have a shortened replicative lifespan26. When expressed in sod1Δ yeast, Dsup significantly increased their replicative lifespan (Fig. 1d), suggesting enhanced survival and longevity in the face of chronic oxidative damage.
The Dsup C-terminus is required for protection of yeast from oxidative damage, in a manner not involving ROS scavenging
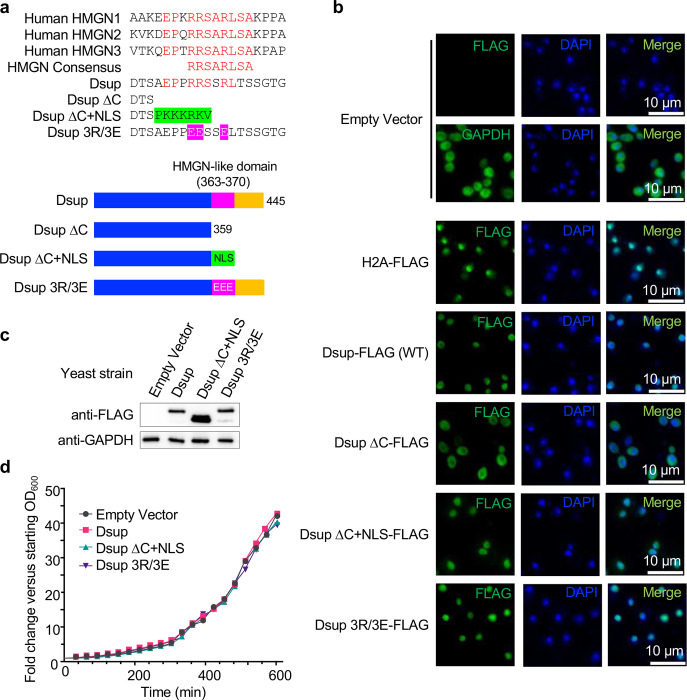
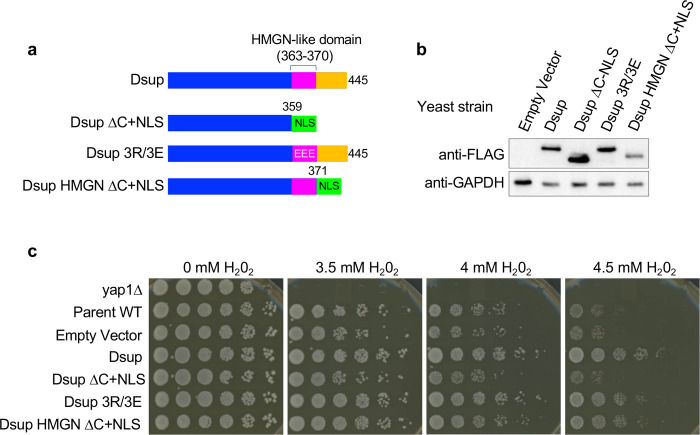
High Mobility Group-N (HMGN) proteins18 contain a conserved HMGN-domain (core consensus RRSARLSA27) required for chromatin binding and protein fuction21 (Fig. 2a). The Dsup C-terminal region contains an eight amino acid stretch with homology to this consensus18 (aa363–370, RRSSRLTS: Fig. 2a), suggesting a physiological relevance. We thus made mutant forms of Dsup by substituting three key arginines with glutamic acid within the motif (Dsup 3R/3E: R363E/R364E/R367E), or by deleting the entire C-terminus including the HMGN-like domain (Dsup ΔC: Δ360–445), alleles previously investigated in vitro18. Since Dsup contains a predicted nuclear localization signal (NLS)28 removed by the ΔC mutation, we added a repeated SV40 NLS (PKKKRKVPKKKRKV)29 C-terminal to Dsup ΔC to make Dsup ΔC+NLS (Fig. 2a). By immunofluorescence Dsup (WT), Dsup 3R/3E and Dsup ΔC+NLS localized to the nucleus, while Dsup ΔC was primarily cytoplasmic, presumably due to removal of the predicted NLS (Fig. 2b). Dsup ΔC was thus omitted from further in vivo study. Importantly, Dsup 3R/3E and Dsup ΔC+NLS proteins were expressed at least as well as Dsup (WT) in yeast (Fig. 2c), and the presence of each did not significantly impact cell growth (Fig. 2d).
Fig. 2. Heterologous Dsup is nuclear localized in yeast and does not negatively impact growth rate.
(a). Alignment of the Dsup HMGN-like domain (aa 363–370, RRSSRLTS), mutants of same from this study, and the HMGN core consensus (RRSARLSA) derived from human HMGN1–3. Residues in red are identical between human HMGN1–3, the HMGN core consensus and Dsup. Green indicates the nuclear localization signal (NLS: PKKKRKVPKKKRKV) added onto the Dsup ΔC construct. Pink indicates three arginine to glutamic acid substitutions (R363E/R364E/R367E) in the HMGN-like sequence to create Dsup 3R/3E (Figure adapted from17). Beneath are schematics of the Dsup wild-type and mutant alleles (all containing N-terminal 6xHIS and C-terminal FLAG tags; not depicted). (b). Immunofluorescence to examine the subcellular location of Dsup alleles (anti-FLAG) in yeast. DAPI co-staining identifies nuclei. H2A-FLAG and GAPDH are respective controls for nuclear and cytoplasmic localization. (c). Western blot showing relative expression of indicated Dsup alleles (anti-FLAG) in yeast. Anti-GAPDH is a loading control for each strain protein extract. (d). Representative growth curves of yeast expressing Dsup alleles.
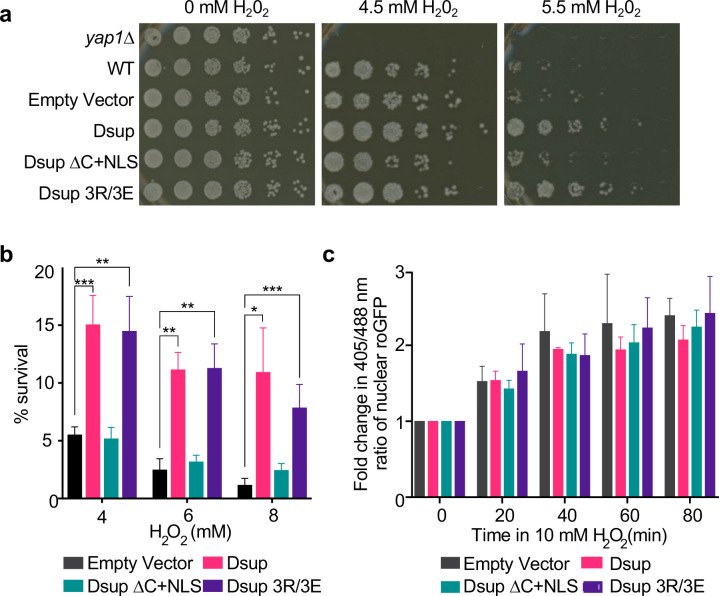
We next examined the ability of Dsup mutants to enhance survival after chronic H2O2 exposure. Mutation of the HMGN-like domain (Dsup 3R/3E) protected cells comparably to Dsup (WT), while Dsup ΔC+NLS yielded no protection, with similar growth to an empty-vector strain (Fig. 3a). As such, the entire C-terminus of Dsup is important for protecting yeast against oxidative DNA damage, while the included HMGN-like domain is dispensable for this function. The observed sensitivity of yeast expressing Dsup (WT) to MMS, Zeocin and UV (Fig. 1b) was not seen upon expression of Dsup 3R/3E or Dsup ΔC+NLS (not shown), indicating that while Dsup 3R/3E can protect from oxidative damage, it does not fully mimic the WT protein.
Fig. 3. Dsup promotes survival after oxidative DNA damage in a manner that requires its chromatin binding C-terminus but is not due to ROS scavenging.
(a). Five-fold serial dilution analysis of yeast expressing Dsup alleles plated on H2O2 (concentrations as indicated). (b). Cell survival after 90-minute exposure to indicated concentrations of H2O2. Shown are average and standard deviation of experiments performed from three independent yeast colonies for each strain. * = p < 0.05, ** = p < 0.01, *** = p < 0.001 by student’s t-test. (c). Nuclear ROS (measured by Redox analysis as in Methods) for indicated yeast strains. Shown are average and standard deviation of experiments performed from three independent colonies. No significant differences were observed between each time point.
To examine whether Dsup expression has any influence on growth following acute oxidative stress, we exposed cells in liquid culture to H2O2 for 1.5 hours, before allowing them to recover on plates with no oxidizing agent. Here Dsup or Dsup 3R/3E expression significantly (and indistinguishably) increased survival following acute oxidative stress, while Dsup ΔC+NLS conferred no protection (Fig. 3b). As such the findings from chronic and acute H2O2 exposure analyses are consistent with expression of Dsup or Dsup 3R/3E, but not Dsup ΔC+NLS, promoting yeast survival in response to oxidative stress.
Free-radical scavengers are effective at protecting yeast from oxidative stress and extending lifespan30. Therefore, we investigated whether Dsup acts as a free-radical scavenger. Redox-sensitive GFPs are excited at 405 nm in an oxidizing environment but 488 nm in reducing conditions, so emissions from excitation at [405/488 nm] allows the measurement of relative changes in redox state. To make a nuclear reporter for this study we added a C-terminal NLS to a roGFP2-Grx1 (glutathione reductase enzyme Grx131) fusion, and confirmed the desired sub-cellular localization (Suppl. Fig. 2). Using this approach, we found that the redox state of the nucleus increased upon H2O2 treatment, but this was not impacted by any Dsup alleles (Fig. 3c). Therefore, Dsup expression had no influence on the yeast nucleus redox state, indicating it uses a mechanism distinct from ROS scavenging to protect the genome from oxidative damage.
Dsup binds chromatin throughout the yeast genome, in a manner dependent on sequences within the C terminus
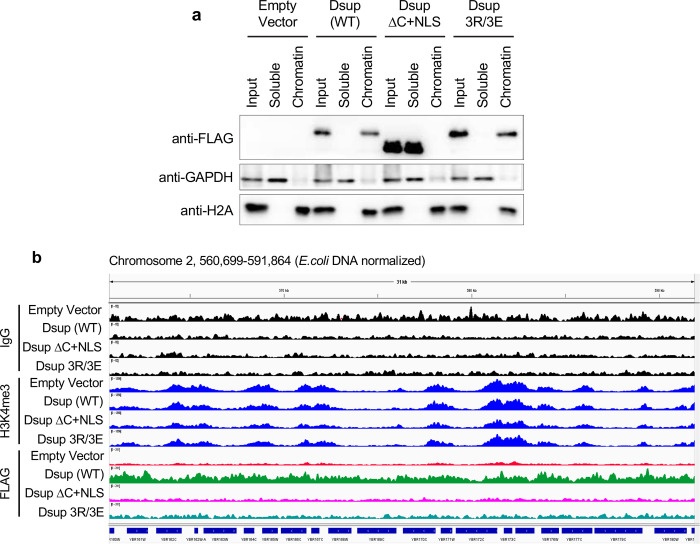
Dsup was first isolated from the chromatin fraction of Tardigrade cells4, and shown to bind preferentially to nucleosomes over free DNA in vitro18. Therefore, we investigated if Dsup binds yeast chromatin in vivo. After cellular fractionation to separate chromatin-bound from soluble proteins, Dsup and Dsup 3R/3E were enriched in the chromatin-bound fraction (Fig. 4a). By contrast, Dsup ΔC+NLS was entirely in the soluble fraction (Fig. 4a), suggesting that despite nuclear localization (Fig. 2b), it does not bind chromatin. Of note, the chromatin localization of Dsup and Dsup 3R/3E, but not Dsup ΔC+NLS, parallels their ability to promote cell survival in the face of oxidative damage (Fig. 3a,b), suggesting that chromatin binding is key.
Fig. 4. Dsup fractionates with yeast chromatin and associates across the yeast genome without apparent bias.
(a). The Dsup C-terminus is required for stable association with chromatin in vivo. Total protein extracts (input) from indicated strains were separated to chromatin (H2A control) and soluble (GAPDH control) fractions and immunoblotted as indicated. (b). CUT&RUN analysis of Dsup allele interactions across the yeast genome in vivo. For each strain anti-IgG (assay background) and anti-H3K4me3 (transcriptionally active gene promoters) were respectively included as negative and positive controls. Each target is group-scaled (after normalization to E. coli spike-in) to the highest signal in the depicted representative IGV window (IgG (82); H3K4me3 (1356) or Dsup-FLAG (311)).
Tardigrade Dsup expression in human and plant cells alters transcription factor binding and gene expression in response to DNA damage5,7. This suggests that Dsup may bind preferentially to certain areas of the genome to influence gene expression. Alternatively, to have the largest physically protective effect from oxidative DNA damage, Dsup might uniformly coat the genome. To investigate these possibilities, we used Cleavage Under Targets & Release Using Nuclease (CUT&RUN)32 to map 6His-Dsup-FLAG localization (by anti-FLAG) across the yeast genome, and observed that Dsup (WT) associated with all regions, with little noticeable bias or selectivity (i.e. without forming peaks / domains; Fig. 4b). Of note, the ability of CUT&RUN to map transcriptionally active promoters with anti-H3K4me3 was unaffected by Dsup (compare Empty vector and Dsup (WT)), indicating a minimal impact on local chromatin structure (Fig. 4b). In agreement, on titrated MNase digestion of yeast cells we observed no significant difference in chromatin accessibility between strains −/+ Dsup expression (not shown).
We next compared CUT&RUN across Dsup alleles, first noting that the relative DNA yield post MNase digestion (prior to adapter ligation) was consistently Dsup (WT) >> Dsup 3R/3E > Dsup ΔC+NLS > Empty vector (EV) (Suppl. Fig. 3). This suggests Dsup 3R/3E has weaker (or higher turnover) binding relative to Dsup (WT) during the CUT&RUN steps prior to MNase activation. Their relative yield is mirrored in the CUT&RUN data, where Dsup 3R/3E showed less enrichment than Dsup (WT) across all genomic regions, while Dsup ΔC+NLS resembled empty vector (Fig. 4b; all data group scaled after normalizing to E. coli spike-in to allow comparisons of global changes in factor binding). It would appear CUT&RUN is a more stringent analysis of chromatin interaction (presumably due [at least in part] to the long incubation times) as compared to chromatin fractionation where Dsup (WT) and Dsup 3R/3E were indistinguishable (Fig. 4a). Taken together, these data indicate that Dsup binds without obvious bias across the genome in a manner that is dependent on its C-terminus (which includes the HMGN-like domain), while mutation within the HMGN-like domain (3R/3E) reduces chromatin binding relative to wild type Dsup, but not enough to confer loss of protection from oxidative DNA damage (Fig. 3a,b).
Dsup binds nucleosomes via multivalent interactions with the histone tails, acidic patch, and DNA
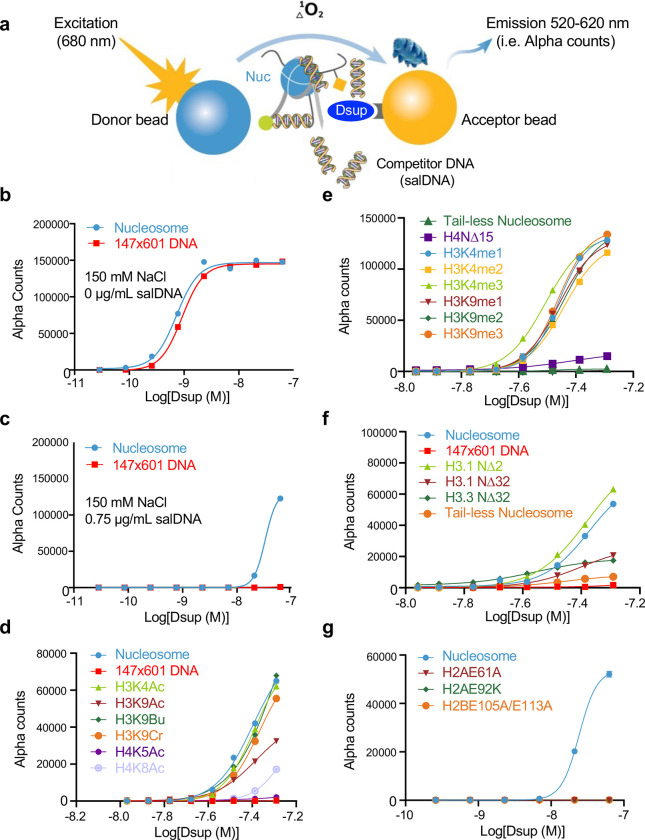
To rigorously interrogate the mode of interaction of Dsup with nucleosomes or free DNA, we used the dCypher in vitro chemiluminescent assay33. Here the biotinylated target (e.g., free DNA or fully defined mononucleosome) couples to streptavidin-donor beads while epitope-tagged query (here WT or mutant forms of 6His-Dsup-FLAG (Suppl. Fig. 4)18) couples to anti-tag acceptor beads. After mixing potential reactants the donor beads are excited at 680 nm, releasing a singlet oxygen that causes emission (520–620 nm) in proximal acceptor beads: this luminescent signal is directly correlated to interaction / binding affinity (Fig. 5a). To compare across each [Query : Target], data is presented as their relative concentration effective in producing 50% of the maximal response (EC50rel) by plotting Alpha Counts (fluorescence) as a function of protein concentration (see Suppl. Table 3 for all EC50rel from this study).
Fig. 5. Dsup binds DNA, nucleosomal histone tails and the nucleosome acidic patch.
(a). Schematic of the dCypher assay (Methods) to measure the interaction between epitope-tagged Dsup queries (Suppl. Fig. 4) and biotinylated nucleosome or 147×601 free DNA targets (former is depicted). The assay can be performed under a variety of conditions (e.g., ionic strength) or −/+ modulators (here salmon sperm DNA (salDNA) as a competitor). (b). At 150mM NaCl, Dsup binds free DNA and (unmodified) nucleosome with equivalent affinities (EC50rel : calculated as in Methods33; see also Suppl. Table 3 for all generated in this study). (c). Competitor salDNA diminishes the interaction of Dsup with nucleosomes, but ablates that with free DNA (147×601). (d-g). Interaction of Dsup with mononucleosomes containing defined lysine acylations (d); lysine methylations (e); histone tail truncations (f); or acid patch mutations (g). All assays performed under optimized conditions (62.5 nM Dsup (from WT), 10nM unmodified nucleosome (or 2.5 nM 147×601 (free) DNA), 150 mM NaCl, 0.75 μg/ml salDNA competitor).
To begin these studies, we titrated salt (sodium chloride) to examine the potential complication of non-specific ionic interactions (Suppl. Fig. 5). At lower salt (50 mM) Dsup showed slightly stronger binding to mononucleosomes over naked DNA (EC50rel 0.6 nM vs. 1.1 nM; Suppl. Fig. 5a), but as salt increased, the apparent affinity of Dsup for both targets gradually declined to undetectable (Suppl. Fig. 5b-f). We chose to move forward with approximately physiological salt (150 mM NaCl) where Dsup binding to nucleosomes and DNA was equivalent (Fig. 5b and Suppl Fig. 5), and next tested the impact of adding salmon sperm DNA (salDNA) as a non-specific competitor to isolate multivalent nucleosome engagement (Suppl. Fig. 5g)33,34. This identified an optimized assay condition (150 mM NaCl, 0.75 μg/ml salDNA) where nucleosome binding was retained over free-DNA (EC50rel 24.6 nM vs. Not Determined (ND); Fig. 5c); demonstrated that the Dsup-DNA association is a significant part of its interaction with chromatin; and further suggested multiple co-operative interactions between Dsup and the nucleosome.
The dCypher platform allowed us to query a diversity of fully defined mononucleosomes (Suppl. Table 1D) to ascertain which surfaces are most important for Dsup binding to chromatin (Fig. 5d–g). Here the apparent affinity of Dsup for mononucleosomes was minimally impacted by a variety of lysine acylations or methylations (Fig. 5d,e and not shown). However, individual deletion of the H4 N-terminal tail (NΔ15) greatly reduced the apparent affinity of Dsup (EC50rel 38.2 nM WT to NΔ15 ND), as did individual deletion of the H3.1 or H3.3 tails (NΔ32; both to ND), or parallel deletion of all histone tails (H2A, H2B, H3 and H4 by trypsin digestion of a nucleosome: tail-less; again to ND) (Fig. 5e,f). Of particular interest mutations (H2AE61A, H2AE92K and H2BE105A/E113A) within the nucleosome acidic patch, a hub of interaction for many nucleosome binding proteins35,36, profoundly impacted Dsup binding to mononucleosomes (EC50rel 22.8 nM WT to ND for each acid patch mutant; Fig. 5g). Taken together, these experiments indicate that Dsup interaction with chromatin is mediated by the N-terminal tails of H3 and H4, the acidic patch of H2A and H2B, and DNA.
The Dsup HMGN-like domain mediates interactions with histones while the region C-terminal to the HMGN-like domain binds DNA
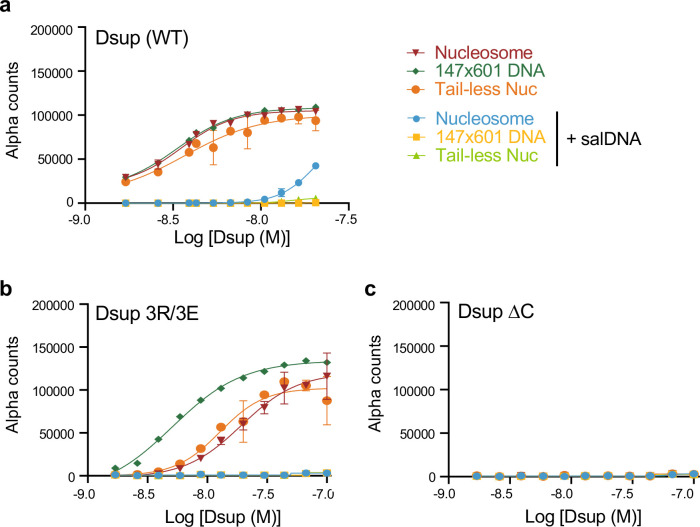
We next used dCypher to examine the contribution of the Dsup HMGN-like domain and C-terminus for nucleosome and DNA engagement. In the absence of competitor DNA, 3R/3E showed noticeably reduced binding to intact and tail-less nucleosome relative to free DNA (compare to WT: Fig. 6a,b). The addition of competitor DNA (to conditions optimized for Dsup WT) then reduced nucleosome binding by Dsup 3R/3E below the level of detection (Fig. 6a,b). In profound contrast, Dsup ΔC (which lacks the HMGN-like domain and C-terminal sequences) showed no interaction with nucleosomes or free DNA under conditions optimized for Dsup (WT) (Fig. 6c), only binding at reduced NaCl (Suppl. Fig. 6). Such non-physiological salt concentrations facilitate nonspecific interactions between proteins and DNA37, so it is unlikely that the observed binding of Dsup ΔC has any functional relevance for Dsup in vivo. Together, these studies indicate that the Dsup interaction with nucleosomal histone tails and the acidic patch are mediated by the HMGN-like domain, while C-terminal sequences may mediate the interaction with DNA in physiological conditions.
Fig. 6. Mutation of the Dsup HMGN-like domain (3R/3E) reduces nucleosome / spares DNA binding, while C-terminal deletion (ΔC) ablates nucleosome and DNA binding.
(a-c). Binding of Dsup alleles (WT (a); 3R/E (R363E/R364E/R367E) (b); or ΔC (Δ360–445) (c)) to nucleosome (−/+ histone tails) or free DNA (147×601) under optimized conditions (Fig. 3d–g) −/+ salDNA competitor.
Dsup interaction with either the histones or DNA is sufficient to survive oxidative damage
Our finding that the Dsup-DNA interaction remained intact after mutation of the HMGN-like domain (aa 363–370: Fig. 6b), but was lost on deletion of the entire C-terminus (Δ360–445: Fig. 6c), suggests that Dsup nucleosome and DNA interactions are mediated by distinct elements within the region previously defined as the Dsup C-terminal domain (aa 208–445)4. To determine the relative functional importance of these elements we created a Dsup construct that retained the HMGN-like domain, and only removed the DNA-binding C-terminus (Dsup HMGN ΔC+NLS: (Δ371–445 + NLS [PKKKRKVPKKKRKV]) (Fig. 7a). This allele is expressed in yeast at slightly reduced levels relative to the other forms of Dsup (Fig. 7b) but was notably able to promote yeast survival in the face of chronic H2O2 exposure (Fig. 7c). These data show that either an intact HMGN-like domain or intact C-terminal downstream sequences, by respectively binding to the nucleosome or DNA, are sufficient for Dsup to protect the genome against oxidative damage (Figure 8).
Fig. 7. The Dsup HMGN-like domain and downstream C-terminal sequences redundantly contribute to survival during oxidative damage.
(a). Schematic of Dsup alleles (all containing N-terminal 6xHIS and C-terminal FLAG tags (not depicted; see also Fig. 2a), including Dsup HMGN ΔC+NLS (Δ374–445 followed by an exogenous NLS (PKKKRKVPKKKRKV)). (b). Western blotting of expression levels (see also Fig. 2c). (c). Sensitivity to oxidative DNA damage (H2O2 ; see also Fig. 1b).
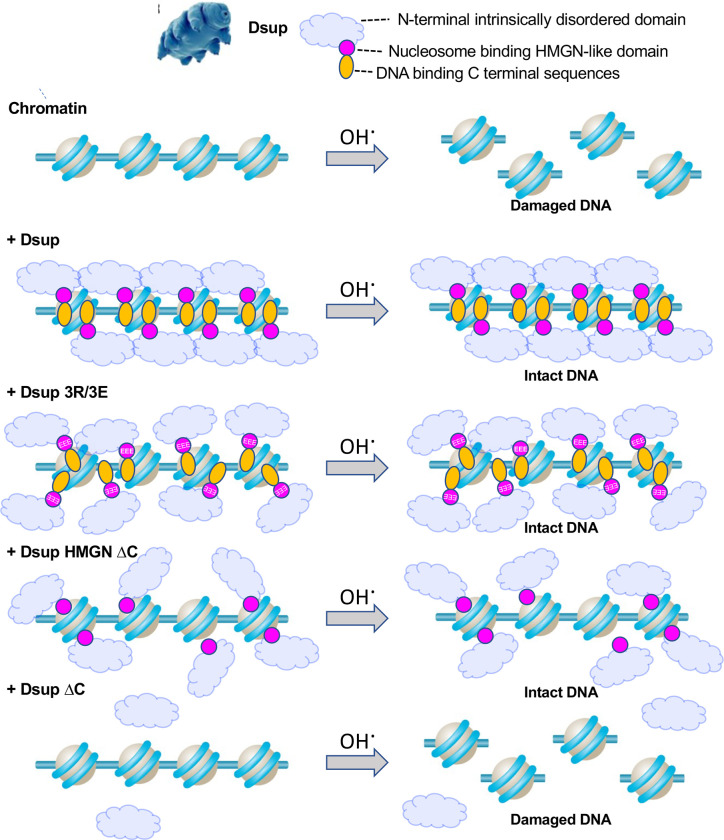
Fig. 8. Model for multivalent association of Dsup with the genome to protect from oxidative DNA damage.
Multivalent binding of Dsup to the chromatinized genome protects against oxidative DNA damage (as exogenously induced by H2O2). Dsup mutations that independently diminish interaction with the nucleosome acidic patch / histone tails (HMGN-like domain; pink), or DNA (C-terminal distal sequences; orange) have reduced chromatin interaction but are still capable of protecting the genome against H2O2-mediated DNA damage. However, loss of both interacting regions ablates the Dsup interaction with chromatin, and thus its ability to protect from oxidative DNA damage.
DISCUSSION
To understand the molecular basis of the extreme radioresistance of tardigrades, we investigated if, and how, their Dsup protein protects against oxidative damage in vivo. When expressed in budding yeast, Dsup coated the entire genome without apparent bias, using two C-terminal regions to associate with chromatin via multivalent interactions involving several nucleosome surfaces and DNA. Functionally, this engagement prevents oxidative DNA damage in a manner independent of ROS scavenging. Our data supports a model where Dsup mediates multivalent interactions with chromatin to protect the underlying genome from oxidative DNA damage (Fig. 8), thus promoting yeast survival and longevity after exposure to elevated levels of hydroxyl radicals (Fig. 1b,d).
HMGN proteins are primarily described in vertebrates38, but whether the Tardigrade Dsup HMGN-like domain is functionally important in vivo was unknown. Human HMGN2 (and likely the other family members) binds nucleosomes at the H2A/H2B acidic patch39. Dsup also binds the acidic patch, with this interaction lost upon mutation of all three arginines in its HMGN-like domain (3R/3E; R363E/R364E/R367E: Fig. 5g). However, we additionally find that the Dsup HMGN-like domain binds histone tails, as this interaction is again lost in Dsup 3R/3E (Fig. 5e,f). Deletion of the histone tails or mutations within the H2A/H2B acidic patch are each sufficient to abolish the Dsup interaction with nucleosomes (Fig. 5e,f), suggesting its HMGN-like domain may bind these regions cooperatively. Despite the popular conception that the histone tails usually extend from the globular nucleosomal core40, recent work instead suggests their default high affinity interaction is with nucleosomal DNA41, which could potentially place the tails close to the nucleosomal acidic patch to facilitate interactions of both entities with a single HMGN-like domain. Alternatively, the HMGN-like domains of different Dsup protein molecules may bind to the histone tails and the acidic patch of the nucleosome.
Dsup is predicted to be intrinsically disordered15, which may allow it to wrap multiple features of nucleosome surfaces, shielding DNA from damage. The region of Dsup spanning the HMGN-like nucleosome binding domain is negatively charged and could facilitate interactions with positively charged histone tails. Meanwhile, the Dsup C-terminus is enriched in positively charged amino acids, which would facilitate ionic interactions with negatively charged DNA15. These interactions with histones and DNA are likely to cooperatively recruit Dsup to chromatin, and further promote non-specific coating of multiple surfaces. We observe a functional redundancy in the various Dsup interactions with chromatin in vivo, since individually disrupting the HMGN-like domain (Dsup 3R/3E) or adjacent C-terminal region (Dsup HMGN ΔC), that respectively compromised interactions with the nucleosome acidic patch / histone tails or DNA, still improved yeast survival on exposure to oxidative damage (Fig. 7c). Functional redundancy in the multiple interactions Dsup makes with chromatin may facilitate its recruitment even if certain surfaces are blocked by other chromatin / DNA binding proteins, potentiating its ability to coat the genome.
The observation that Dsup binding to the nucleosome was largely agnostic to most histone PTMs in vitro (Fig. 5d,e) is consistent with our finding that Dsup covers the entire in vivo genome rather than being enriched / excluded from certain regions with their particular histone modifications (Fig. 4b). Importantly, Ramazzottius varieomatus histones are highly conserved with human histones (as used in the dCypher assay) (Suppl. Table 1E), while yeast and human histones are even more similar. As a result, we consider that the observed interactions between Dsup and human or yeast histones are relevant for how the protein helps to protect tardigrades from irradiation.
In initial testing we expressed Dsup from a range of yeast promoters of various strengths, but only the very strong TDH3 promoter enabled protection from oxidative damage (not shown and Fig. 1b). Of note, this Dsup expression level was equivalent to that of histone H2B (Fig. 1a), suggesting Dsup may be in sufficient abundance for at least two molecules per yeast nucleosome. Given that highly expressed Dsup protects the genome from oxidative DNA damage (Fig. 1c), is bound to chromatin genome-wide (Fig. 4a,b), and redundantly interacts with multiple nucleosome surfaces (Fig. 5), it is likely that Dsup non-specifically coats the in vivo genome to physically protect from oxidative damage, as was proposed from the previous in vitro studies18. It may be relevant to note that when we yeast-codon optimized the tardigrade Dsup protein in an attempt to promote still higher expression levels, the resulting yeast were inviable, suggesting that too much Dsup is deleterious. In agreement, expression of codon-optimized Dsup in cultured rat neurons, had detrimental effects42.
There is precedent for proteins binding to the genome to provide protection from irradiation and H2O2. Previous studies have demonstrated that chromatin compaction protects DNA from free radical-mediated damage caused by ionizing radiation or iron43–45. These findings are consistent in vivo and in vitro, suggesting a direct protection of DNA from damage rather than a particular feature of the cellular environment. Compacted chromatin also provides protection from ROS damage after direct incubation with H2O246. Additionally, the deletion of proteins involved in chromatin assembly and disassembly, including the remodelers ISWI, Chd1, and INO80, renders chromatin more sensitive to DNA damage47.
It is intriguing that Dsup expression protected yeast from oxidative damage, but not from MMS, bleomycin, or UV: indeed, it actually increased sensitivity to these agents (Fig. 1b). Future studies should examine whether there is delayed repair of the DNA lesions generated by these genotoxins, potentially due to Dsup hindering access of the repair machinery. We note, however, that the growth rate of Dsup yeast was not reduced (Fig. 2d), indicating they are fully capable of transcriptional regulation, DNA replication and mitosis - other events one could imagine might also be prone to complications from the genome being coated with Dsup protein - but that did not appear to be the case.
These findings provide precedent for the development of organisms that can survive and live longer in the face of oxidative damage, potentially expanding the range of applications for developing therapeutic interventions by biotechnology, and furthering efforts towards human resistance to extraterrestrial effects.
MATERIALS AND METHODS
Yeast strains, primers, and plasmids
The pRS306-PTDH3-Dsup plasmid was created by Gibson cloning (NEB Gibson Assembly® Cloning Kit) as follows. Plasmid pRS30648 was digested with SacI and BglII. The TDH3 promoter was PCR amplified (primer sequences in Suppl. Table 1A) from yeast genomic DNA with forward primer pTDH3_SacI_F (giving homology to Sac1 digested end of pRS306), and reverse primer pTDH3_R (giving homology to 5’ end of Dsup gene). The tarigrade (Ramazzottius varieornatus) Dsup gene (aa1–445; encoding protein accession P0DOW4, Suppl. Table 1E) including an N-terminal 6xHis and C-terminal FLAG tag was amplified from plasmid pET21b-nHis6-Rvar-DSUP-cFLAG (kind gift from James Kadonaga18), with primers Rvar_Dsup_F and Rvar_Dsup_R, respectively giving homology to the 3’ end of the TDH3 promoter and the 5’ end of the ADH1 terminator. The ADH1 terminator was amplified from yeast genomic DNA using primers tADH1_F and tADH1_BglII_R, respectively giving homology to the 3’ end of the Dsup gene and the BglII digested end of pRS306. Gibson cloning of amplified DNA fragments was carried out following kit directions.
Plasmid pRS306-PTDH3-Dsup was digested with MfeI and integrated into yeast strain BY4741 49 at site of the endogenous TDH3 promoter to make Dsup strain, RGY002 (pTDH3–6His_Dsup_FLAG: full list of yeast strains and their phenotypes in Suppl. Table 1B). Further mutations of the Dsup gene, including deletion of the C-terminus to derive Dsup ΔC (Δ359–445; RAY136), addition of an NLS (PKKKRKVPKKKRKV) to derive Dsup ΔC+NLS (RAY228), glutamic acid substitution of three arginines in the HMGN-like sequence (R363E/R364E/R367E) to derive Dsup 3R/3E (RAY153), and insertion of a stop codon (at Dsup codon 2) to derive Empty vector strain (RAY149), were made after integration using CRISPR-Cas9 mediated genome editing50. Dsup HMGN ΔC+NLS (Δ374–460, RAY274) was derived from Dsup ΔC+NLS by reintroduction of the HMGN consensus sequence region (aa360–373). Primer sequences used to generate guide RNAs and HDR template DNA are in Suppl. Table 1A.
Plasmid p415TEF cyto roGFP2-Grx1-NLS was made from p415TEF cyto roGFP2-Grx1 (kind gift from Tobias Dick; Addgene plasmid # 65004)31 by traditional cloning. First, the roGFP2-Grx1 sequence was PCR-amplified from plasmid p415TEF cyto roGFP2-Grx1 using primers that added a 2xNLS sequence (PKKKRKVPKKKRKV) to the Grx1 C-terminus. The resulting PCR product was digested with BamHI and HindIII and ligated to similarly digested plasmid p415TEF cyto roGFP2-Grx1.
Yeast culture and handling was performed using standard methods. Growth of strains expressing Dsup was in SC-ura media (unless otherwise indicated). All strains were isogenic to BY474149 (Suppl. Table 1B).
Immunoblot analysis
~107 exponentially growing yeast cells (ODλ600 0.8–1.0) were collected by centrifugation, washed once with water, and flash frozen in liquid nitrogen before being resuspended in 100 μL modified Laemmli buffer 51 and boiled for five minutes. Proteins were resolved by 10% SDS-PAGE, membrane transferred, and immunoblotted with antibodies to FLAG (Sigma F1804, 1:1,000) and GAPDH (Sigma A9521, 1:10,000).
Growth curve analysis
Yeast were grown to saturation overnight in YPD at 30°C and diluted to ODλ600 0.1–0.2. Growth measurements (ODλ600) of cultures grown from three independent colonies were taken every 30 minutes and plotted over time. Growth curves were fitted with an exponential regression using Microsoft Excel, and doubling times calculated as the slope of the curve during exponential phase. Doubling times of independent growth curves were compared using a student’s t-test.
Chromatin fractionation analysis
~4×108 exponentially growing yeast cells (ODλ600 0.8–1.0) were collected by centrifugation, washed once with ice cold 10% glycerol, and flash frozen in liquid nitrogen. After thawing on ice, the cell pellet was washed (100 mM Tris pH 9.4, 10 mM DTT), resuspended in the same buffer, and rested on ice for 10 minutes. Cells were pelleted by centrifugation, washed in spheroplasting buffer (10 mM HEPES, 1.2 M Sorbitol, 0.5 mM PMSF), resuspended in spheroplasting buffer containing 56 μg/mL Zymolyase 100T (US Biological), and incubated at 30°C with rotation for 1 hour. Spheroplasts were collected by gentle centrifugation, washed once in spheroplasting buffer, and once in wash buffer (1 M sorbitol, 20 mM Tris pH 7.5, 20 mM KCL, 2 mM EDTA, 0.5 mM PMSF, 0.1 μM spermine, 0.25 μM spermidine, Calbiochem Protease Inhibitor Cocktail Set IV (1:100)). Cells were gently resuspended and lysed in 250 μL Lysis Buffer (wash buffer with 400 mM sorbitol) for 10 minutes on ice.
Half of the volume after lysis (Input fraction) was boiled for 5 min in 5x Laemmli buffer, while the other half was pelleted at 14,000 x g for 15 minutes (chromatin fraction). The supernatant was collected (non-chromatin fraction) and boiled for 5 minutes in 5x Laemmli buffer, and the pellet (chromatin fraction) resuspended in 1x Laemmli buffer and boiled for 5 minutes. 7.5% of the total volume of each sample was resolved by 12.5% SDS-PAGE, membrane transferred, and immunoblotted with anti-FLAG (Sigma F1804, 1:1,000) to detect Dsup. Successful fractionation was confirmed with anti-H2A (Abcam ab18255, 1:5,000) as a chromatin bound protein, and anti-GAPDH (Sigma A9521, 1:20,000) as a non-chromatin bound protein.
Immunofluorescence analysis
Yeast indirect immunofluorescence was carried out following published methods 52. 2.5 OD of early-mid log phase cells (ODλ600 0.5–0.6) were crosslinked in 4% formaldehyde for 20 mins at room temperature, then spheroplasted in 500 μg/mL Zymolyase 100T (US Biological) for 30 minutes at 30°C with rotation. Spheroplasted cells were applied to a 10-chamber poly-lysine coated microscope slide and permeabilized by a six minute incubation in methanol at −20°C, immediately followed by a 30 second incubation in acetone at −20°C. After blocking in 5% BSA, slides were incubated with primary antibodies to H2A (Abcam ab18255, 1:1.000), GAPDH (Sigma A9521, 1:5,000), or FLAG (Sigma F1804, 1:1,000). Incubation with Alexa Fluor® 594 or 488 secondary antibodies (BioLegend) followed, and coverslips were mounted using ProLong™ Gold Antifade Mountant with DAPI (Invitrogen). Images were taken using an Olympus BX63 Fluorescence Microscope with a DP80 Camera and 60X objective.
Acute and chronic damage sensitivity analysis
To measure resistance to acute hydrogen peroxide (H2O2) exposure, cells were grown in liquid YPD media until mid-log, harvested by centrifugation, and resuspended to 0.6 OD in fresh media containing H2O2 (0, 4, 6, or 8 mM). After 90 minutes growth (30°C, with shaking), cultures were diluted and spread on SC-ura agar plates. After two days at 30°C, colonies were counted and averaged across three technical replicates. Three experiments were performed from separate starting colonies, and statistical analysis performed using a student’s t-test.
The response to chronic H2O2 exposure was examined using a serial dilution assay. Cells were grown in liquid culture until mid-log (ODλ600 0.5–1.0), harvested by centrifugation, and resuspended in sterile water to ODλ600 1.0. Five-fold serial dilutions were made in a 96-well plate, and yeast spotted using a sterile 6x8-prong inoculating manifold onto YPD agar plates containing indicated concentrations of H2O2. Similar methods were used to evaluate sensitivity to methyl methanesulfonate (at the indicated concentrations in YPD) and Zeocin (at the indicated concentrations in YPD). For ultraviolet light sensitivity, yeast serial dilutions were onto YPD plates and exposed to UV (at the doses (J/cm2) indicated in figure legends) using a crosslinker [Stratalinker]. Plates were incubated for 3 days at 30°C.
Replicative lifespan analysis
Cells were grown overnight to early-mid (ODλ600 0.2–0.6) and diluted to OD 0.1 in freshly-filtered YPD. This innoculum was added to an iBiochips automated dissection chip to achieve single cell loading as per manufacturer’s instructions. Light microscopy images of cells were acquired every 20 minutes over four days using an Evos FL Auto two-cell imaging microscope and associated software (ImageJ). At least 50 cells were counted per condition, with survival curves calculated on Graphpad Prism 9, and statistical analysis performed with a log-rank test.
Chronological lifespan analysis
Chronological lifespan was measured according to published methods53. Data is presented as average and standard deviation across three independent cultures, each of which is an average of two technical replicates.
Redox analysis
Cells expressing nuclear roGFP (p415TEF roGFP2-Grx1-NLS) were grown to mid-log (ODλ600 0.6–0.8). Cells were diluted in SC-ura media to ODλ600 0.6 in 5 mL flow cytometry tubes. Fluorescence at 405 nm and 488 nm was measured on a flow cytometer (BD Biosciences BD® LSR II) immediately before direct addition of H2O2 (2 mM or 10 mM). Subsequent fluorescence measurements were taken every 20 minutes over 80 minutes.
The mean of the 405/488 nm values for each timepoint was calculated using FlowJo, with the value at time 0 normalized to 1 for each strain. Data is presented as the mean and standard deviation of three independent cultures and compared using a student’s t-test.
ELISA for 8-OHdG
30 mL yeast cultures were grown at 30°C in shaking flasks until ODλ600 0.6. Cells were harvested by centrifugation, and half of each culture resuspended in either 15 mL of fresh SC-ura media or that containing 10 mM H2O2. After two hours growth at 30°C, cells were again harvested by centrifugation and genomic DNA isolated (Thermo Scientific Yeast DNA extraction kit). Genomic DNA was resuspended in 50 μL of nuclease-free water and stored overnight at 4°C.
DNA concentrations were measured using a NanoDrop spectrometer, diluted in water to 2 mg/mL, boiled for five minutes at 95°C, then immediately placed on ice for 10 minutes (to denature double-stranded DNA). 50 μg of DNA (25 μL) were sequentially incubated with Nuclease P1 (NEB: 1 unit for 2 hours at 37°C in provided buffer) and alkaline phosphatase (NEB Quick CIP: 10 units for 1 hour at 37°C in provided buffer supplemented with 100 mM Tris pH 8). Samples were incubated to denature enzymes (10 minutes at 95°C), then spun at 6000 x g for 5 minutes. DNA concentrations were measured on a NanoDrop spectrometer to ensure even loading onto the ELISA plate.
ELISA to 8-hydroxy 2 deoxyguanosine was performed as per kit instructions (Abcam ab201734). 15 μg of DNA was loaded into each of three triplicate wells for each sample (with three independent cultures measured for each condition). Absorbance at 450 nm was measured using a plate reader.
CUT&RUN analysis
Nuclei from yeast cells expressing Dsup alleles (Suppl. Table 1B) were purified according to published methods54 with slight modifications. Yeast were grown in 500 mL of SC-ura media to ODλ600 0.6–0.8. Cells were spheroplasted using 500 μL of 2 mg/mL Zymolyase 100T (37°C for ~ 30 mins; until a 50 μL aliquot mixed with 1 mL of 10% SDS had an ODλ600 ~10% of the starting value). Remainder of the nuclei isolation was performed as previously54, and 1 mL aliquots containing 5 × 107 nuclei were slow-frozen in an isopropanol chamber at −80°C overnight.
For CUT&RUN nuclei were rapidly thawed (2–3 minutes at 37°C), and 100 μL of suspension (5 × 106 nuclei) used per reaction with the CUTANA™ ChIC/CUT&RUN Kit (version 3.2; EpiCypher). After immunotethering (to Rabbit IgG (EpiCypher), SNAP-Certified™ anti-H3K4me3 (EpiCypher), or anti-FLAG (DYKDDDDK Tag; ThermoFisher MA1-91878): Suppl. Table 1C) MNase digestion was performed for two hours at 4°C, and DNA eluted in 12 μL final volume.
5 ng of DNA was used to prepare sequencing libraries with the Ultra II DNA Library Prep Kit (NEB #E7645L). Libraries were sequenced on an Illumina NextSeq 2000 platform, obtaining an average of ~1.1 million paired-end reads per reaction (Suppl. Table 2). Paired-end fastq files were aligned to the sacCer3 reference genome using Bowtie2. Duplicate (SAMtools) and multi-aligned (Picard) reads were filtered, and the resulting unique reads for comparable reactions normalized by an E. coli scaling factor (1/ % E. coli Reads) (bedtools), and further normalized to RPKM bigwig files (DeepTools). Integrative Genomics Viewer (IGV) was utilized for the visualization of peaks from bigwig files. All sequencing data has been deposited in the NCBI Gene Expression Omnibus (GEO) with accession number GSE237436.
PTM-defined nucleosomes
All mononucleosomes (EpiCypher; Suppl. Table 1D) were created from fully-defined (PTM or mutant) octamers wrapped by 5’ biotinylated 147×601 DNA (Suppl. Table 1E) unless stated otherwise, with modifications confirmed by mass-spectrometry and immunoblotting (if an antibody was available)33,55. Histone tail truncations were by direct expression of the indicated histone prior to octamer assembly (H3.1 NΔ2, H3.1 NΔ32, H3.3 NΔ32, or H4 NΔ15), or trypsin digestion of assembled unmodified nucleosome (tail-less).
dCypher binding assays
dCypher assays on the Alpha (Amplified luminescence proximity homogeneous assay) platform to examine the interaction of WT or mutant 6His-Dsup-FLAG (kind gift from James Kadonaga)18: the Queries [Suppl. Table 1E]) with free DNA (147×601 Widom sequence) or fully defined nucleosomes (the Targets: Suppl. Table 1D) were performed as previously33,55 with minor modifications.
In 384-well plates, 5 μL Dsup queries were serially titrated (in duplicate) against a fixed concentration of target (10 nM biotinylated nucleosome or 2.5 nM free DNA (147x601)). After incubation (30 minutes), interactions were detected with addition of a 10 μL mix of AlphaScreen streptavidin Donor (Revvity, 6760002) and nickel-chelate Acceptor beads (Revvity, AL108M). Following a final incubation (60 minutes), Alpha counts were measured using a PerkinElmer 2104 EnVision plate reader (680 nm laser excitation, 570 nm emission filter ± 50 nm bandwidth). Experiments were performed to assess [Query : Target] binding over a range of assay conditions (20 mM Tris pH 7.5, 0.01% BSA, 0.01% NP-40, 1 mM DTT with additives as noted), including the impact of ionic strength (50 – 250 mM NaCl) and competitor salmon sperm DNA (salDNA; 0 – 20 μg/mL). All incubations were performed at room temperature in subdued lighting. Binding curves were plotted in GraphPad Prism 9.0 using 4-parameter logistic nonlinear regression.
Binding curves [Query : Target] were generated using a non-linear 4PL curve fit in Prism 9.0 (GraphPad) to yield EC50rel values33,55 (Suppl. Table 3). Where necessary, values beyond the Alpha hook point (indicating bead saturation / competition with unbound Query) were excluded and top signal constrained to average max signal for Target. In cases where signal never reached plateau, those were constrained to the average max signal within the assay (relative to unmodified nucleosome). In remaining cases, when a targets maximal signal never achieved half of max signal relative to unmodified nucleosome, an EC50rel was deemed not determinable (ND).
ACKNOWLEDGEMENTS
We are indebted to Drs. Jim Kadonaga, George Kassavetis, Grisel Cruz-Becerra, and Carolina Chavez for discussing pre-published work and generously sharing plasmids and purified proteins. We are grateful to Andres Mansisidor for help generating plasmids and yeast strains, and the Weill Cornell Flow Cytometry Core for technical support. EpiCypher is supported by National Institutes of Health (NIH) grants R44 HG010640, R44 GM117683, R44 GM136172 and R44 CA212733. JKT is supported by NIH grants R35 GM139816 and R01 CA95641. RRA was supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the NIH under award number T32 GM007739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program.
Footnotes
COMPETING INTERESTS
EpiCypher is a commercial developer and supplier of reagents (e.g., fully PTM-defined nucleosomes (dNucs) and SNAP-CUTANA® K-MetStat nucleosomes) and platforms (e.g., dCypher and CUTANA CUT&RUN) used in this study. LK, ARH, RW, MWC, MRM and MCK are currently employed by (and own shares in) EpiCypher. RJE and HEW were previously employed by (and own shares in) EpiCypher. MRM and MCK are board members of EpiCypher.
REFERENCES
- 1.Jonsson K. I., Holm I. & Tassidis H. Cell Biology of the Tardigrades: Current Knowledge and Perspectives. Results Probl Cell Differ 68, 231–249, doi: 10.1007/978-3-030-23459-1_10 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Horikawa D. D. et al. Establishment of a rearing system of the extremotolerant tardigrade Ramazzottius varieornatus: a new model animal for astrobiology. Astrobiology 8, 549–556, doi: 10.1089/ast.2007.0139 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Mole R. H. The LD50 for uniform low LET irradiation of man. Br J Radiol 57, 355–369, doi: 10.1259/0007-1285-57-677-355 (1984). [DOI] [PubMed] [Google Scholar]
- 4.Hashimoto T. et al. Extremotolerant tardigrade genome and improved radiotolerance of human cultured cells by tardigrade-unique protein. Nat Commun 7, 12808, doi: 10.1038/ncomms12808 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirke J., Jin X. L. & Zhang X. H. Expression of a Tardigrade Dsup Gene Enhances Genome Protection in Plants. Mol Biotechnol 62, 563–571, doi: 10.1007/s12033-020-00273-9 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Vignard J., Mirey G. & Salles B. Ionizing-radiation induced DNA double-strand breaks: a direct and indirect lighting up. Radiother Oncol 108, 362–369, doi: 10.1016/j.radonc.2013.06.013 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Ricci C. et al. The Tardigrade Damage Suppressor Protein Modulates Transcription Factor and DNA Repair Genes in Human Cells Treated with Hydroxyl Radicals and UV-C. Biology (Basel) 10, doi: 10.3390/biology10100970 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poetsch A. R. The genomics of oxidative DNA damage, repair, and resulting mutagenesis. Comput Struct Biotechnol J 18, 207–219, doi: 10.1016/j.csbj.2019.12.013 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finkel T., Serrano M. & Blasco M. A. The common biology of cancer and ageing. Nature 448, 767–774, doi: 10.1038/nature05985 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Kennedy S. R., Loeb L. A. & Herr A. J. Somatic mutations in aging, cancer and neurodegeneration. Mech Ageing Dev 133, 118–126, doi: 10.1016/j.mad.2011.10.009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lodato M. A. et al. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 359, 555–559, doi: 10.1126/science.aao4426 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tubbs A. & Nussenzweig A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 168, 644–656, doi: 10.1016/j.cell.2017.01.002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crow J. F. The origins, patterns and implications of human spontaneous mutation. Nat Rev Genet 1, 40–47, doi: 10.1038/35049558 (2000). [DOI] [PubMed] [Google Scholar]
- 14.van den Boogaard W. M. C., Komninos D. S. J. & Vermeij W. P. Chemotherapy Side-Effects: Not All DNA Damage Is Equal. Cancers (Basel) 14, doi: 10.3390/cancers14030627 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minguez-Toral M., Cuevas-Zuviria B., Garrido-Arandia M. & Pacios L. F. A computational structural study on the DNA-protecting role of the tardigrade-unique Dsup protein. Sci Rep 10, 13424, doi: 10.1038/s41598-020-70431-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vuzman D., Azia A. & Levy Y. Searching DNA via a "Monkey Bar" mechanism: the significance of disordered tails. J Mol Biol 396, 674–684, doi: 10.1016/j.jmb.2009.11.056 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Lobley A., Swindells M. B., Orengo C. A. & Jones D. T. Inferring function using patterns of native disorder in proteins. PLoS Comput Biol 3, e162, doi: 10.1371/journal.pcbi.0030162 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chavez C., Cruz-Becerra G., Fei J., Kassavetis G. A. & Kadonaga J. T. The tardigrade damage suppressor protein binds to nucleosomes and protects DNA from hydroxyl radicals. Elife 8, doi: 10.7554/eLife.47682 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cubenas-Potts C. & Corces V. G. Architectural proteins, transcription, and the three-dimensional organization of the genome. FEBS Lett 589, 2923–2930, doi: 10.1016/j.febslet.2015.05.025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McBryant S. J., Adams V. H. & Hansen J. C. Chromatin architectural proteins. Chromosome Res 14, 39–51, doi: 10.1007/s10577-006-1025-x (2006). [DOI] [PubMed] [Google Scholar]
- 21.Nanduri R., Furusawa T. & Bustin M. Biological Functions of HMGN Chromosomal Proteins. Int J Mol Sci 21, doi: 10.3390/ijms21020449 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuroda S., Otaka S. & Fujisawa Y. Fermentable and nonfermentable carbon sources sustain constitutive levels of expression of yeast triosephosphate dehydrogenase 3 gene from distinct promoter elements. J Biol Chem 269, 6153–6162 (1994). [PubMed] [Google Scholar]
- 23.Dizdaroglu M., Jaruga P., Birincioglu M. & Rodriguez H. Free radical-induced damage to DNA: mechanisms and measurement. Free Radic Biol Med 32, 1102–1115, doi: 10.1016/s0891-5849(02)00826-2 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Stadtman E. R. Protein oxidation and aging. Science 257, 1220–1224, doi: 10.1126/science.1355616 (1992). [DOI] [PubMed] [Google Scholar]
- 25.Kirkwood T. B. & Kowald A. The free-radical theory of ageing--older, wiser and still alive: modelling positional effects of the primary targets of ROS reveals new support. Bioessays 34, 692–700, doi: 10.1002/bies.201200014 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Barker M. G., Brimage L. J. & Smart K. A. Effect of Cu,Zn superoxide dismutase disruption mutation on replicative senescence in Saccharomyces cerevisiae. FEMS Microbiol Lett 177, 199–204, doi: 10.1111/j.1574-6968.1999.tb13732.x (1999). [DOI] [PubMed] [Google Scholar]
- 27.Ueda T., Catez F., Gerlitz G. & Bustin M. Delineation of the protein module that anchors HMGN proteins to nucleosomes in the chromatin of living cells. Mol Cell Biol 28, 2872–2883, doi: 10.1128/MCB.02181-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashimoto T. & Kunieda T. DNA Protection Protein, a Novel Mechanism of Radiation Tolerance: Lessons from Tardigrades. Life (Basel) 7, doi: 10.3390/life7020026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adam S. A., Lobl T. J., Mitchell M. A. & Gerace L. Identification of specific binding proteins for a nuclear location sequence. Nature 337, 276–279, doi: 10.1038/337276a0 (1989). [DOI] [PubMed] [Google Scholar]
- 30.Jamieson D. J. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14, 1511–1527, doi: (1998). [DOI] [PubMed] [Google Scholar]
- 31.Morgan B. et al. Multiple glutathione disulfide removal pathways mediate cytosolic redox homeostasis. Nat Chem Biol 9, 119–125, doi: 10.1038/nchembio.1142 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Skene P. J. & Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6, doi: 10.7554/eLife.21856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marunde M. R., Popova I. K., Weinzapfel E. N. & Keogh M. C. The dCypher Approach to Interrogate Chromatin Reader Activity Against Posttranslational Modification-Defined Histone Peptides and Nucleosomes. Methods Mol Biol 2458, 231–255, doi: 10.1007/978-1-0716-2140-0_13 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Dilworth D. et al. A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization. Nat Chem Biol 18, 56–63, doi: 10.1038/s41589-021-00898-0 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spangler C. J. et al. Structural basis of paralog-specific KDM2A/B nucleosome recognition. Nat Chem Biol 19, 624–632, doi: 10.1038/s41589-023-01256-y (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skrajna A. et al. Comprehensive nucleosome interactome screen establishes fundamental principles of nucleosome binding. Nucleic Acids Res 48, 9415–9432, doi: 10.1093/nar/gkaa544 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson M. & Woodbury N. W. Thermodynamics of specific and nonspecific DNA binding by two DNA-binding domains conjugated to fluorescent probes. Biophys J 81, 1793–1804, doi: 10.1016/S0006-3495(01)75830-4 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kugler J. E., Deng T. & Bustin M. The HMGN family of chromatin-binding proteins: dynamic modulators of epigenetic processes. Biochim Biophys Acta 1819, 652–656, doi: 10.1016/j.bbagrm.2012.01.013 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato H. et al. Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proc Natl Acad Sci U S A 108, 12283–12288, doi: 10.1073/pnas.1105848108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith E. & Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell 40, 689–701, doi: 10.1016/j.molcel.2010.11.031 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghoneim M., Fuchs H. A. & Musselman C. A. Histone Tail Conformations: A Fuzzy Affair with DNA. Trends Biochem Sci 46, 564–578, doi: 10.1016/j.tibs.2020.12.012 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Escarcega R. D. et al. The Tardigrade damage suppressor protein Dsup promotes DNA damage in neurons. Mol Cell Neurosci 125, 103826, doi: 10.1016/j.mcn.2023.103826 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Enright H. U., Miller W. J. & Hebbel R. P. Nucleosomal histone protein protects DNA from iron-mediated damage. Nucleic Acids Res 20, 3341–3346, doi: 10.1093/nar/20.13.3341 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brambilla F. et al. Nucleosomes effectively shield DNA from radiation damage in living cells. Nucleic Acids Res 48, 8993–9006, doi: 10.1093/nar/gkaa613 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takata H. et al. Chromatin compaction protects genomic DNA from radiation damage. PLoS One 8, e75622, doi: 10.1371/journal.pone.0075622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ljungman M. & Hanawalt P. C. Efficient protection against oxidative DNA damage in chromatin. Mol Carcinog 5, 264–269, doi: 10.1002/mc.2940050406 (1992). [DOI] [PubMed] [Google Scholar]
- 47.Singh A. K. & Mueller-Planitz F. Nucleosome Positioning and Spacing: From Mechanism to Function. J Mol Biol 433, 166847, doi: 10.1016/j.jmb.2021.166847 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Sikorski R. S. & Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27, doi: 10.1093/genetics/122.1.19 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brachmann C. B. et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132, doi: (1998). [DOI] [PubMed] [Google Scholar]
- 50.Aguilar R. R., Shen Z. J. & Tyler J. K. A Simple, Improved Method for Scarless Genome Editing of Budding Yeast Using CRISPR-Cas9. Methods Protoc 5, doi: 10.3390/mps5050079 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horvath A. & Riezman H. Rapid protein extraction from Saccharomyces cerevisiae. Yeast 10, 1305–1310, doi: 10.1002/yea.320101007 (1994). [DOI] [PubMed] [Google Scholar]
- 52.Dunham M. J., Dunham M. J., Gartenberg M. R. & Brown G. W. Methods in yeast genetics and genomics : a Cold Spring Harbor Laboratory course manual / Maitreya J. Dunham, University of Washington, Marc R. Gartenberg, Robert Wood Johnson Medical School, Rutgers, The State University of New Jersey, Grant W. Brown, University of Toronto. 2015 edition / edn, (Cold Spring Harbor Laboratory Press, 2015). [Google Scholar]
- 53.Postnikoff S. D. & Harkness T. A. Replicative and chronological life-span assays. Methods Mol Biol 1163, 223–227, doi: 10.1007/978-1-4939-0799-1_17 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Brahma S. & Henikoff S. CUT&RUN Profiling of the Budding Yeast Epigenome. Methods Mol Biol 2477, 129–147, doi: 10.1007/978-1-0716-2257-5_9 (2022). [DOI] [PubMed] [Google Scholar]
- 55.Morgan M. A. J. et al. A trivalent nucleosome interaction by PHIP/BRWD2 is disrupted in neurodevelopmental disorders and cancer. Genes Dev 35, 1642–1656, doi: 10.1101/gad.348766.121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]