Abstract
The adeno-associated virus (AAV) Rep78 and Rep68 proteins are required for site-specific integration of the AAV genome into the AAVS1 locus (19q13.3-qter) as well as for viral DNA replication. Rep78 and Rep68 bind to the GAGC motif on the inverted terminal repeat (ITR) and cut at the trs (terminal resolution site). A similar reaction is believed to occur in AAVS1 harboring an analogous GAGC motif and a trs homolog, followed by integration of the AAV genome. To elucidate the functional domains of Rep proteins at the amino acid level, we performed charged-to-alanine scanning mutagenesis of the N terminus (residues 1 to 240) of Rep78, where DNA binding and nicking domains are thought to exist. Mutants were analyzed for their abilities to bind the GAGC motif, nick at the trs homolog, and integrate an ITR-containing plasmid into AAVS1 by electrophoretic mobility shift assay, trs endonuclease assay, and PCR-based integration assay. We identified the residues responsible for DNA binding: R107A, K136A, and R138A mutations completely abolished the binding activity. The H90A or H92A mutant, carrying a mutation in a putative metal binding site, lost nicking activity while retaining binding activity. Mutations affecting DNA binding or trs nicking also impaired the site-specific integration, except for E66A and E239A. These results provide important information on the structure-function relationship of Rep proteins. We also describe an aberrant nicking of Rep78. We found that Rep78 cuts predominantly at the trs homolog not only between the T residues (GGT/TGG), but also between the G and T residues (GG/TTGG), which may be influenced by the sequence surrounding the GAGC motif.
Adeno-associated virus (AAV) type 2 (referred to here as AAV) is a nonpathogenic parvovirus with a linear, single-stranded DNA genome of 4.7 kb with positive or negative polarity (reviewed in references 6, 28, and 42). Both ends of the genome show a unique T-shaped hairpin configuration, termed an inverted terminal repeat (ITR), which is required in cis for viral DNA replication and packaging. Between the ITRs are two open reading frames corresponding to rep and cap. The rep gene encodes four overlapping nonstructural proteins (Rep78, Rep68, Rep52, and Rep40), while the cap gene codes for structural proteins (VP1, VP2, and VP3). On the genome are three promoters, p5, p19, and p40, designated according to their map positions. The unspliced and spliced transcripts from the p5 promoter encode Rep78 and Rep68, respectively, while Rep52 and Rep40 are translated from unspliced and spliced transcripts from the p19 promoter. Biochemical characterization has revealed that the larger Rep proteins, Rep78 and Rep68, possess site-specific, strand-specific endonuclease activity and ATP-dependent helicase activity (22). Rep78 and Rep68 bind the ITR (23) via the Rep binding sites (RBS), consisting of tandem repeats of GAGC tetramer (10, 40), and nick at the terminal resolution site (trs) (22). The site-specific nicking is followed by unwinding of the terminal hairpin, resulting in AAV DNA replication. Thus, Rep78 and Rep68 resemble a prokaryotic initiator protein in rolling-circle replication that introduces a strand-specific, site-specific cut into the DNA replicon (2, 25, 62). Besides functioning as key proteins in viral DNA replication, Rep78 and Rep68 also transregulate AAV promoters as well as heterologous promoters (5, 20, 34, 35, 67).
AAV integrates into the host chromosomal DNA (9, 29), preferentially at the AAVS1 locus on chromosome 19 (19q13.3-qter) (30–32, 51). The AAVS1 locus harbors a sequence element analogous to the GAGC repeats and the trs (65). Linden et al. defined a 33-bp sequence as the minimum required for site-specific integration (36); this sequence contains both the GAGC motif and the trs homolog. Electrophoretic mobility shift assays (EMSA) demonstrated that the large Rep proteins bind to the analogous GAGC motif (10, 65) and mediate the complex formation between the AAV terminal hairpin and AAVS1 sequence (65). An in vitro study showed that the large Rep proteins nick at the trs homolog and initiate asymmetric DNA replication (59). Therefore, a similar reaction observed on the hairpin DNA during replication of the AAV genome is believed to occur in AAVS1, leading to the integration of the AAV genome (36, 37).
Rep78 and Rep68 consist of 621 and 536 amino acid residues, respectively, and their functions are essentially the same, although some differences have been noted (43, 44, 63). To elucidate the functional domains of multifunctional Rep proteins, a number of mutational analyses have been performed (39, 47, 60, 61, 64, 65, 68, 69). A deletion mutagenesis study by McCarty et al. showed that amino acid residues 134 to 242 and 415 to 490 are at least essential for the specific binding to AAV hairpin DNA (39). Yang and Trempe reported that residues 25 to 62, 88 to 113, 125 to 256, and 346 to 400 were necessary for binding to AAV hairpin DNA as determined by insertion and deletion mutagenesis (69). A deletion analysis by Owens et al. showed that the N-terminal portion mediated binding to the GAGC motif and that the central region of the Rep proteins was important for stabilizing the protein-DNA complex (47). Weitzman et al. reported that removal of the N-terminal 29 amino acids abolished the binding activity and that the C-terminally truncated Rep protein (residues 1 through 448) retained an affinity with AAV hairpin DNA (64). Moreover, several mutational studies, including those mentioned above, were conducted by introducing single amino acid mutations at residues that were highly conserved among the parvoviruses. McCarty et al. reported that W242L and P415H mutant Rep proteins lost ITR binding activity and that E379K and E379Q caused low binding activity (39). Walker et al. noted that Y156F, Y224F, Y307F, Y311F, E379A, K391A, I393A, K404A, I417A, T419A, and D429E mutants showed weak or no binding activities to AAV hairpin DNA (39, 60, 61). These reports also described mutations affecting trs endonuclease activity. Weitzman et al. reported that C-terminally truncated Rep (residues 1 to 476) retained trs nicking activity, while Rep containing residues 1 to 466 lost this activity (64). Walker et al. found that mutations in the helicase motif of the Rep protein (residues 330 to 422) abrogated helicase and trs endonuclease activities (61). Moreover, by systematically substituting phenylalanine for tyrosine residues, they revealed that tyrosine at position 152 was a candidate for the active-site tyrosine residue linking to the 5′ end of nicked DNA (60). These mutational analyses of the Rep protein assigned the DNA binding domain and the domain for trs endonuclease roughly to at least the N-terminal half. Other mutational analyses of Rep proteins focused on regulation of AAV promoters (33, 48) and heterologous promoters (20), replication of herpes simplex virus (27), and oncogene-mediated transformation (68).
The comprehensive identification of amino acid residues that are responsible for the function of the Rep protein is important for dissecting the functional domains of pleiotropic proteins. However, no systematic study has been conducted to determine the functional domain at the amino acid level in more detail, especially in the N-terminal region. Because charged amino acids (arginine, lysine, histidine, aspartic acid, and glutamic acid) are more likely to be located on the surface of the protein (14, 26) and are also capable of forming ion pairs and hydrogen bonds, they may play a role in enzyme catalysis and in the recognition of interacting proteins. Based on these hypotheses, the strategy of substituting alanine for charged amino acids systematically, i.e., charged-to-alanine scanning mutagenesis (3, 17), has been employed to identify the functional regions of proteins. To determine which amino acid residues of Rep protein were important for DNA binding as well as for trs nicking, we substituted alanine for all charged amino acids in the N-terminal half of the Rep78 protein, where DNA binding and nicking domains are thought to exist. We analyzed a set of mutant Rep proteins for binding to the GAGC motif, trs endonuclease activity, and site-specific integration into AAVS1.
Rep78 and Rep68 have been shown to introduce a site-specific, strand-specific nick predominantly between the thymidine residues at the trs (AGT/TGG) (22) and trs homolog (GGT/TGG) (59). However, Batchu and Hermonat reported that the nicking site is not always restricted to the trs (4). We also describe an aberrant nicking of Rep78 protein. By using the AAVS1 sequence as a substrate, we found that Rep78 preferentially cuts not only between the T residues (GGT/TGG) but also between the G and T residues (GG/TTGG). The nick site for Rep protein may be influenced by the sequence surrounding the minimal element required for the site-specific and strand-specific nick, i.e., the RBS and the trs homolog.
MATERIALS AND METHODS
Site-directed mutagenesis.
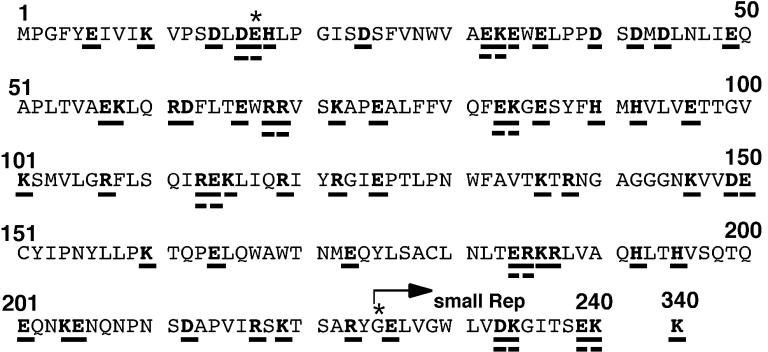
pCMVR78, expressing Rep78 alone under the control of the cytomegalovirus promoter, was described before (58). The T3 promoter was located upstream of the open reading frame of Rep78. This construct harbors a substitution of glycine for methionine at position 225 so as not to translate small Rep proteins, which had proved to exhibit normal AAV duplex replicative form replication (7) and to retain DNA binding and trs endonuclease activities, as described elsewhere (47). pCMVR78 was a derivative of pIM45 containing all of the AAV genome except the ITR sequence (38). Sequencing of our pIM45 revealed that the G at nucleotide (nt) 50 of the Rep78 and -68 open reading frame was mutated to A (compared to the published sequence data for the AAV genome [57]), resulting in a missense mutation of glycine at position 17 to glutamic acid. Therefore, pCMVR78 expressing Rep78 with G17E and M225G mutations (compared to authentic Rep78) was used as a template for the mutagenesis (Fig. 1). In this paper, the G17E M225G mutant is called wild type for convenience. All charged amino acids (R, K, H, D, and E) in the N-terminal half of Rep78 (amino acid residues 1 through 240) were replaced with alanine (Fig. 1) by using the Transformer site-directed mutagenesis kit (Clontech) according to the manufacturer’s instructions. Because all of the residues for mutation were changed to alanine, mutant Rep proteins were designated by the wild-type amino acids for simplicity. For example, E6 represents an alanine substitution for glutamic acid at position 6. When charged amino acids existed in a cluster, two charged residues were changed to alanine simultaneously (DE16, EK32, EK57, RD61, RR68, EK83, RE113, ER184, KR186, KE204, DK233, and EK239). DE16 indicates that two residues, D and E at positions 16 and 17, respectively, were mutated to alanine simultaneously. If these double mutations affected the function of Rep proteins, an individual residue was mutated to alanine independently for further analyses; e.g., in the case of DE16, D16 and E17 were also constructed. We also constructed the K340H mutant, in which the nucleoside triphosphate (NTP)-binding lysine at position 340 was changed to histidine as reported by Chejanovsky and Carter (8). All of the mutations were verified by sequencing both strands.
FIG. 1.
Charged-to-alanine scanning mutagenesis of the N-terminal region of the Rep78 protein. All of the charged amino acids, i.e., arginine (R), lysine (K), histidine (H), aspartic acid (D), and glutamic acid (E), in the N-terminal half (residues 1 to 240) were mutated to alanine. Residues replaced are underlined. Because all of the residues for mutation were changed to alanine, for simplicity, mutant Rep proteins were designated by the wild-type amino acids. Most mutant Rep proteins have a single alanine substitution. However, two charged residues were simultaneously changed to alanine where charged residues existed in a cluster (DE16, EK32, EK57, RD61, RR68, EK83, RE113, ER184, KR186, KE204, DK233, and EK239). When these double mutations affected the function of Rep protein, both residues were mutated to alanine independently for further analyses. For example, in the case of DE16, D16A and E17A were also constructed. The K340H mutant, in which the NTP-binding lysine at position 340 was changed to histidine as described before (8), was included in our assay. ∗, all of the mutants and the “wild-type” Rep78 harbor two mutations compared to authentic Rep78: M225G so as not to synthesize small Rep proteins and G17E. The translation start site of the small Rep proteins is indicated by an arrow.
Western blotting.
Two micrograms of plasmid DNA was transfected into 2 × 105 293 cells/well in six-well plates by a calcium phosphate precipitation method. Twenty-four hours later, cells were rinsed with ice-cold phosphate-buffered saline and lysed in a lysis buffer (10 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% Nonidet P-40) supplemented with 1 mM phenylmethylsulfonyl fluoride and 500 U of aprotinin per ml. Five micrograms of lysate was subjected to 7.5% polyacrylamide gel electrophoresis. Separated proteins were transferred to polyvinylidene difluoride membranes (Millipore). The membranes were incubated with anti-Rep antibody 76.3 (Progen, Heidelberg, Germany), which recognizes only the unspliced-form Rep proteins, Rep78 and Rep52, at a dilution of 1:50 in TBST (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.05% Tween 20) for 1 h at room temperature and then were incubated with anti-mouse immunoglobulin G labeled with horseradish peroxidase for 30 min at room temperature in TBST. Chemiluminescent signals were detected by using the ECL system (Amersham). The N-terminal region (residues 1 to 240) in which mutations were introduced in the present study was not believed to include the epitope recognized by anti-Rep 76.3.
EMSA.
Rep78 and mutant Rep78 proteins were synthesized in vitro by using the TnT T3 Coupled Reticulocyte Lysate System (Promega) as instructed by the manufacturer. To monitor the synthesis of Rep proteins, l-[35S]methionine and l-[35S]cysteine (redivue Pro-mix l-[35S] in vitro cell labeling mix; Amersham) were added to reaction mixtures. Three microliters of product was resolved by sodium dodecyl sulfate (SDS)–7.5% polyacrylamide gel electrophoresis, dried, and analyzed on an imaging analyzer (BAS-1500; Fuji, Tokyo, Japan). EMSA was performed as described previously (24). Briefly, 3 μl of in vitro-synthesized Rep78 or mutant Rep protein in the absence of radiolabeled amino acids was incubated for 15 min at 30°C with 20,000 cpm of 5′-end-labeled RBS oligonucleotide probe (equimolar amounts of 5′-CGGCGCTCGCTCGCTCGCTGGGCG and 5′-CGCCCAGCGAGCGAGCGAGCGCCG, containing the GAGC repeats (underlined) in AAVS1, annealed to each other) in a 10-μl solution of 10 mM HEPES-KOH (pH 7.9), 50 mM KCl, 0.1 mM EDTA, 0.05% bovine serum albumin (BSA), 10% glycerol, and 1 μg of sheared calf thymus DNA. In some experiments, before addition of the probe and sheared calf thymus DNA, Rep protein was incubated with 0.1 μl of anti-Rep antibody 76.3 for 30 min on ice. In competition experiments, a 20- or 100-fold molar excess of unlabeled RBS oligonucleotide was included in the reaction mixture. Reaction products were separated on 4% nondenaturing polyacrylamide gels, dried, and then analyzed on a BAS-1500 imaging analyzer.
Assay for trs endonuclease activity.
The site-specific nicking activity was assayed as reported by Im and Muzyczka (22). 32P-5′-end-labeled AAV hairpin DNA was prepared as described previously (46). We also used a sequence element derived from AAVS1 as a substrate. pRVK (a kind gift from K. I. Berns) containing the EcoRI-KpnI fragment of AAVS1 was double digested with StyI and PvuII. The resulting 200-bp fragment harboring a minimum element required for site-specific integration was blunt ended and subcloned into the HincII site of pBluescript II (Stratagene), which still retained the StyI site (pS1). Similarly, the 109-bp SmaI fragment derived from pRVK, the same as the P1 fragment described by Weitzman et al. (65), was cloned into the HincII-SmaI sites of pBluescript II (pS2). pS1 and pS2 were digested with StyI or SmaI, respectively, end labeled with [γ-32P]ATP (Amersham) and polynucleotide kinase, and then cut with XhoI. The resulting 198- or 113-bp fragment was used for the assay. Three microliters of in vitro-translated Rep78 or mutant Rep protein was mixed with 15,000 cpm of the substrate in a 10-μl solution containing 25 mM HEPES-KOH (pH 7.5), 10 mM MgCl2, 1 mM dithiothreitol, 2% glycerol, 0.1 μg of BSA, and 0.4 mM ATP and then incubated for 1 h at 37°C. After addition of 3× loading buffer (0.5% SDS, 50 mM EDTA, 40% [vol/vol] glycerol, 0.1% [wt/vol] xylene cyanol, 0.1% [wt/vol] bromophenol blue), samples were boiled for 5 min and then applied to 6 or 8% nondenaturing polyacrylamide gels. When nicking products were observed, we concluded that mutant Rep proteins retained trs endonuclease activity even if the amounts of the products were small. To determine the lengths of released fragments produced by Rep proteins, the reaction mixtures were extracted with phenol-chloroform, ethanol precipitated, and then suspended in Tris-EDTA (TE). The product derived from the StyI-XhoI fragment was resolved on a 6% denaturing sequencing gel along with sequencing ladders. To prepare the DNA sequence ladders, the sequencing reaction was performed with a Taq cycle sequencing kit (Takara, Tokyo, Japan) with some modifications: the reaction mixture contained dGTP instead of 7-deaza dGTP, 10% dimethyl sulfoxide, the 32P-5′-end-labeled 27-nt oligonucleotide 5′-CTTGGGGCGGTGGGGGGCCAGCGGCAG (nt 311 to 337; numbering of nucleotides is based on the sequence data for AAVS1 reported by Kotin et al. [30]), and pS1 as a template. A chemical sequencing reaction was also performed to produce DNA sequence ladders with the StyI-XhoI or SmaI-XhoI fragment for the nicking assay.
PCR-based assay for site-specific integration at the AAVS1 locus.
Two micrograms of pCMVR78, mutant Rep expression plasmid, or blank vector was transfected into 2 × 105 293 cells/well in six-well plates along with 2 μg of pW1, harboring a lacZ expression cassette flanked by ITRs, by a standard calcium phosphate precipitation method. Twenty-four hours later, cells were rinsed with ice-cold phosphate-buffered saline and lysed in 200 μl of a solution of 50 mM Tris-HCl (pH 7.6), 100 mM NaCl, 20 mM EDTA, and 1% SDS. The samples were mixed with 20 μg of proteinase K, incubated for 6 h at 55°C, extracted with an equal volume of phenol-chloroform, ethanol precipitated, and then suspended in 200 μl of TE. The PCR to detect site-specific integration was carried out as reported previously (58) with minor modifications. One microliter of isolated genomic DNA was subjected to a thermal cycling reaction in a 20-μl reaction mixture containing 1× thermophilic DNA polymerase buffer [10 mM KCl, 20 mM Tris-HCl (pH 8.8), 10 mM (NH4)2SO4, 4 mM MgSO4, 0.1% Triton X-100 (New England Biolabs {NEB})], 1 μM 5′-CGGCCTCAGTGAGCGAGCGAGC and 5′-CGGGGAGGATCCGCTCAGAGGACA, and 2 U of Deep Vent Exo(−) DNA polymerase (NEB). The cycling conditions were as follows: 99°C for 1 min, followed by 35 cycles of 99°C for 10 s and 72°C for 4 min. Ten microliters of the PCR mixture was transferred to a hybridization membrane (Hybond-N+; Amersham) by using a dot blot apparatus and hybridized with an AAVS1 probe. The probe was a random-primed 32P-labeled PCR fragment generated in a reaction with 20 ng of pRVK as a template and primers 5′-ACTTTGAGCTCTACTGGCTTC and 5′-GGAGGATCCGCTCAGAGG. The membranes were then analyzed on a BAS-1500 imaging analyzer. The assay was repeated at least four times.
RESULTS
Expression of Rep78 and mutant Rep proteins.
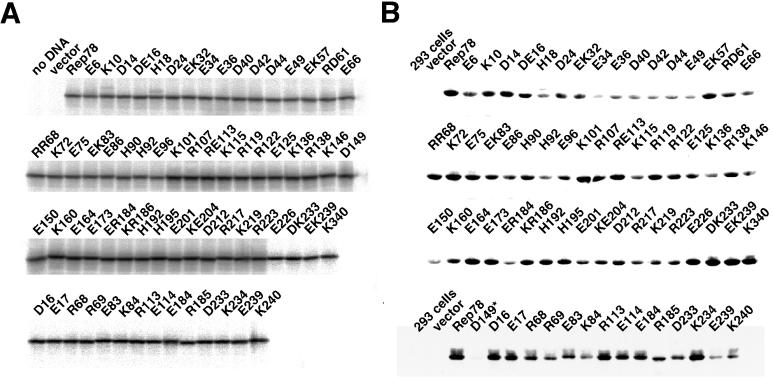
In vitro transcription-translation with a mutant Rep expression plasmid performed in the presence of [35S]methionine-cysteine produced a major single band of about 75 kDa (Fig. 2A). The intensities of the bands determined by densitometric scanning were not significantly different from one another, indicating that the amount of in vitro-synthesized Rep protein in each reaction was essentially the same. Expression of mutant Rep proteins in 293 cells was also detected by immunoblotting (Fig. 2B). However, some mutant Rep proteins (e.g., E34, E149, and E150) were expressed poorly, a result reproducibly observed. In particular, the D149 mutant (asterisk in Fig. 2B) showed a faint band that could be detected only by longer exposure. Repeated Western blotting with another anti-Rep antibody, 294.4 (19) (a kind gift of J. A. Kleinschmidt), which recognized all wild-type Rep proteins (Rep78, -68, -52, and -40) could not detect a stronger signal (data not shown). Extensive sequencing of the expression cassette of the D149 mutant revealed no mutations except for the intended change, suggesting that the D149 mutant Rep protein is subject to proteolytic degradation in 293 cells.
FIG. 2.
(A) Wild-type and mutant Rep78 proteins synthesized in vitro. Mutant Rep proteins were produced by using a coupled in vitro transcription-translation system in the presence of [35S]methionine-cysteine. Two microliters of reaction mixture was resolved on an SDS–7.5% polyacrylamide gel. Unprogrammed lysate (no DNA) and lysate programmed with vector (vector) were also loaded. A major single band of about 75 kDa was detected in each lane. The intensities of the bands were not significantly different from one another as revealed by densitometry, indicating that the amount of in vitro-synthesized Rep protein in each reaction was essentially the same. (B) 293 cells were transfected with plasmids expressing mutant Rep proteins under the control of the cytomegalovirus promoter. One day after transfection, cells were harvested, and 5 μg of lysate was electrophoresed on SDS–7.5% polyacrylamide gels. Proteins were transferred onto polyvinylidene difluoride membranes, and Rep proteins were detected with anti-Rep antibody 76.3. Unlike in vitro-synthesized mutant Rep proteins, several mutant Rep proteins (e.g., E34, D149, and E150) were expressed reproducibly at lower levels. ∗, prolonged exposure revealed a band corresponding to Rep protein.
EMSA.
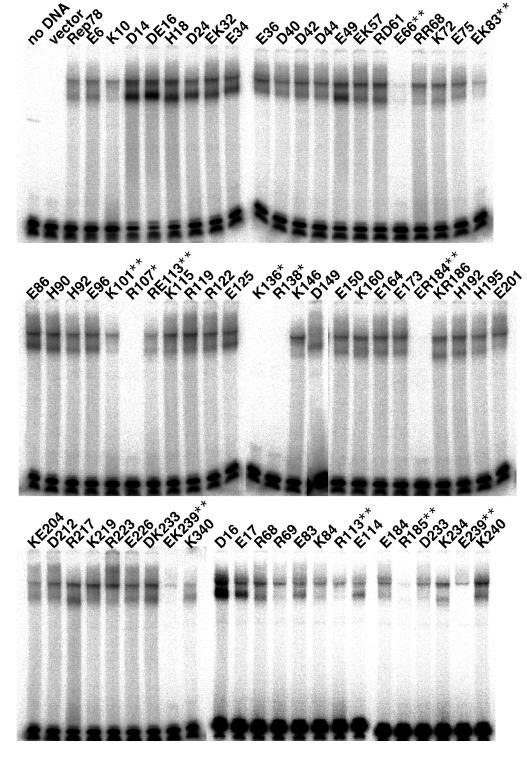
One of our main goals is to determine which amino acid residues are important for the specific binding to the GAGC repeats. Figure 3 shows multiple shifted bands (bracket) when in vitro-translated Rep78 was included in a reaction (lane 2) and that these bands became competed out gradually by increasing the amount of cold competitor (lanes 3 and 4). The multiple shifted bands in EMSA were considered to be produced by oligomerization of Rep78, as described elsewhere (18, 54). The higher-mobility EMSA product (arrowhead) was completely depleted when anti-Rep antibody 76.3 was added in the reaction (lanes 2 and 5), whereas other slower bands (asterisks) are not likely to be supershifted. The interaction of the monoclonal antibody 76.3 with Rep protein may be affected by the multimeric state of the Rep protein. The epitope of Rep protein reacting with the antibody 76.3 is in the central region of large Rep proteins (see Materials and Methods). This region is reported to be essential for the homo-oligomerization of Rep protein (54). Thus, multimerization may hinder the epitope from reacting with the antibody, and/or self-association itself may cause the conformational change of Rep protein, leading to failure to interact with the antibody. Figure 4 shows EMSA of mutant Rep proteins. When the R107, K136, or R138 mutant was used, no bands could be detected reproducibly even after prolonged exposure (asterisks in Fig. 4), suggesting that amino acids R107, K136, and R138 were important for the specific binding to the GAGC motif. The E66, EK83, RE113, ER184, and EK239 mutants showed diminished binding activity to the probe (double asterisks in Fig. 4). EK83, RE113, ER184, and EK239 were double mutants. To determine which residue(s) was involved in GAGC binding, the single-substitution mutants E83, K84, R113, E114, E184, R185, E239, and K240 were constructed and were examined as described above. As shown in the lower panel of Fig. 4, binding activity was not significantly different for E83 and K84, although EK83 consistently showed reduced binding activity to the RBS probe. The double mutation EK83 might disturb the tertiary conformation of the Rep protein, which was not apparent when a single mutation (E83 or K84) was introduced. R113 showed a lower binding activity than E114. In a comparison of band densities between the E184 and R185 mutants, R185 clearly had a lower binding activity. In the case of E239 and K240, E239 lost binding activity predominantly. Thus, the impaired binding to the RBS probe observed in double mutants RE113, ER184, and EK239 was attributed mainly to the R113, R185, and E239 mutations, respectively.
FIG. 3.
EMSA of wild-type Rep78 synthesized in vitro. Each lane contained 20,000 cpm of 32P-labeled RBS oligonucleotide probe. Lane 1 contained unprogrammed lysate. When lysate programmed with pCMVR78 was incubated with the probe, multiple shifted bands (bracket) were observed (lane 2). Shifted bands disappeared gradually as the amount of cold competitor added increased (lanes 3 and 4). The higher-mobility band (arrowhead) is completely depleted when anti-Rep antibody 76.3 is included in the reaction, whereas other slower bands (∗) are not likely to be supershifted (compare lane 5 to lane 2).
FIG. 4.
EMSA of mutant Rep78 synthesized in vitro. Three microliters of in vitro-synthesized Rep78 or mutant Rep protein in the absence of radiolabeled amino acids was incubated for 15 min at 30°C with 20,000 cpm of 5′-end-labeled RBS oligonucleotide probe in a 10-μl solution of 10 mM HEPES-KOH (pH 7.9), 50 mM KCl, 0.1 mM EDTA, 0.05% BSA, 10% glycerol, and 1 μg of sheared calf thymus DNA. Reaction products were separated on 4% nondenaturing polyacrylamide gels, dried, and then analyzed on a BAS-1500 imaging analyzer. Unprogrammed lysate (no DNA) and lysate programmed with vector (vector) were also included as negative controls. ∗, complete loss of binding activity; ∗∗, consistently lower binding activity compared to the wild type.
Rep78 does not necessarily nick between the T residues.
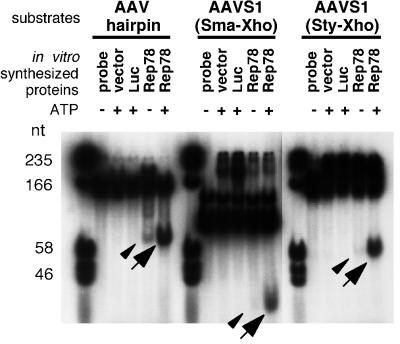
First, to confirm that our assay system for trs endonuclease activity worked well, we incubated end-labeled hairpin DNA or AAVS1 fragments with in vitro-synthesized Rep78. Figure 5 demonstrates that Rep78 cleaved substrates in the presence of ATP (arrows), while luciferase, used as a negative control, failed to release a nicking product. Liberated fragments of the same size, although in much smaller amounts, were also detected in the presence of Rep78 even when ATP was not added (arrowheads). The site-specific, strand-specific nicking activity is ATP dependent (22). Thus, the nicking activity observed without ATP may be due to the presence of remaining NTP which was included in the transcription-translation system we used to synthesize RNA. To determine which strand was cut, the nicking reaction was performed by using templates with one 5′ end labeled with [γ-32P]ATP. We could detect the liberated fragments only when the strand harboring the GGTTGG element was end labeled (data not shown), suggesting that in vitro-translated Rep78 protein cut the strand with the trs homolog, as reported previously (59).
FIG. 5.
trs endonuclease activity of in vitro-synthesized Rep78 protein. Three different substrates were used. γ-32P-end-labeled AAV hairpin DNA was prepared as described previously with minor modification (see Materials and Methods). The SmaI- and StyI-XhoI fragments (113 and 198 bp, respectively) derived from AAVS1 harbored a minimum sequence element required for site-specific integration (Fig. 6A and B). Three microliters of in vitro-translated Rep78 protein was mixed with 15,000 cpm of probe in a 10-μl solution containing 25 mM HEPES-KOH (pH 7.5), 10 mM MgCl2, 1 mM dithiothreitol, 2% glycerol, and 0.1 μg of BSA, in the presence or absence of 0.4 mM ATP, and then incubated for 1 h at 37°C. Instead of Rep78, lysate programmed with blank vector (vector) or with luciferase DNA (Luc) was also used. After addition of 3× loading buffer (0.5% SDS, 50 mM EDTA, 40% [vol/vol] glycerol, 0.1% [wt/vol] xylene cyanol, 0.1% [wt/vol] bromophenol blue), samples were boiled for 5 min and then applied to 8% nondenaturing polyacrylamide gels. Arrows indicate the nicking products. Note the products that were present even when ATP was not included (arrowheads). However, when the AAVS1 fragment (SmaIXhoI or StyI-XhoI) was incubated with Rep78 in the absence of ATP, the amount of released fragment was small.
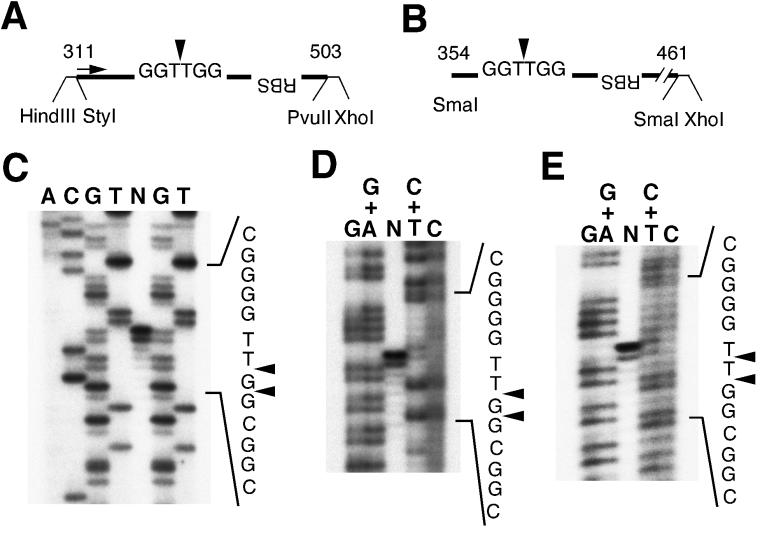
To determine the site of cutting by Rep78, the products were resolved on a 6% denaturing sequence gel along with DNA sequence ladders (Fig. 6). The nicking products derived from the StyI-XhoI fragment were run with sequencing ladders prepared by a chain termination method with Taq polymerase (Fig. 6C) or by a chemical modification procedure (Fig. 6D). Figure 6C shows that the main cutting sites were GG/TTGG and G/GTTGG, not GGT/TGG. Figure 6D shows a comparison of the nicking products with DNA sequence ladders produced by a chemical reaction. Considering that there is about a 1-base difference between ladders generated by primer extension and by chemical modification, the main nick site is deduced to be between the G and T residues. Another product derived from the SmaI-XhoI fragment was resolved along with chemical sequencing ladders (Fig. 6E). The major nicking site was GGT/TGG, which was different from the result obtained by using the StyI-XhoI fragment.
FIG. 6.
Lengths of nicking fragments produced by in vitro-synthesized Rep78. (A) The StyI-XhoI (PvuII) fragment was used for the nicking reaction. The major nicking site of Rep protein reported elsewhere is shown (arrowhead). The sequencing primer (nt 311 to 337) to produce DNA ladders is indicated by an arrow. (B) The other substrate for the trs nicking reaction. (C) The nicking products derived from the StyI-XhoI fragment (lane N) were resolved on a 6% denaturing sequencing gel along with sequencing ladders. The sequencing ladders were prepared by using the Takara Taq cycle sequencing kit with some modifications: the reaction mixture contained dGTP instead of 7-deaza dGTP, 10% dimethyl sulfoxide, and 32P-5′-end-labeled 27-nt oligonucleotide (see panel A). Arrowheads indicate the cutting sites. (D) The nicking products derived from the StyI-XhoI fragment (lane N) were also separated on a 6% gel with sequence ladders produced by a chemical reaction. The deduced cut sites are indicated by arrowheads. There is about a 1-base difference between ladders produced by primer extension and by chemical reaction. (E) The nicking products derived from the SmaI-XhoI fragment were resolved on a 6% denaturing sequence gel with chemically prepared sequence ladders. Arrowheads indicates the deduced nicking sites.
trs endonuclease activities of mutant Rep proteins.
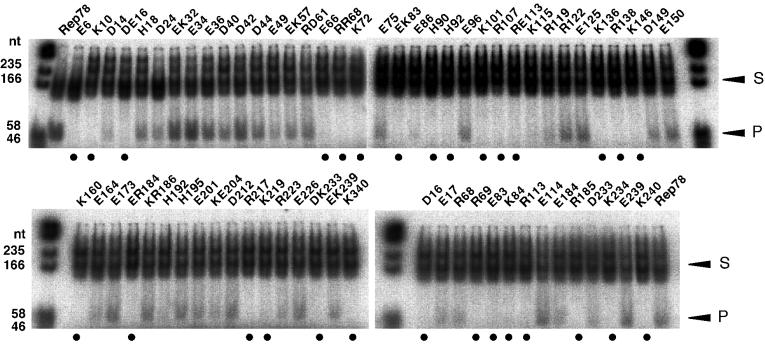
By using the 32P-end-labeled StyI-XhoI fragment as a substrate (Fig. 6A), the trs endonuclease activities of mutant Rep proteins were assayed (Fig. 7). The E6, K10, DE16, E66, RR68, K72, EK83, H90, H92, K101, R107, RE113, K136, R138, K146, K160, ER184, R217, K219, DK233, and K340H mutants were negative for nicking activity (closed circles). As the double mutants, DE16, RR68, EK83, RE113, ER184, and DK233 lost nicking activity, D16, E17, R68, R69, E83, K84, R113, E114, E184, R185, D233, and K234 mutants were further examined. Among them, D16, R69, E83, K84, R113, R185, K234, and K240 were revealed to be defective for the site-specific, strand-specific cutting reaction. The mutants, R107, K136, and R138, that failed to bind the GAGC motif were also defective for the trs nicking activity. Moreover, E66, K101, R113, and R185, showing decreased binding activity in EMSA, also lost nicking activity. The observation that mutant Rep proteins showing no or reduced binding to the GAGC motif were negative for trs endonuclease activity is consistent with the conclusion by McCarty et al. that the specific binding to the GAGC motif is a prerequisite for nicking at the trs (39), although there is one exception, E239, which had decreased binding activity but still retained trs nicking activity.
FIG. 7.
trs endonuclease activities of mutant Rep proteins synthesized in vitro. The 32P-5′-end-labeled StyI-XhoI fragment derived from AAVS1 was used as a template (S). Reaction mixtures were the same as described in the legend to Fig. 5. Substrates and nicking products (P) were separated on 8% nondenaturing gels. When nicking products were observed, we concluded that mutant Rep proteins retained trs endonuclease activity even if the amounts of the products were small. Nicking products below a background level are indicated by closed circles.
Site-specific integration into AAVS1.
Examining mutant Rep proteins for their ability to mediate site-specific integration also provided us with important information on Rep proteins. To test whether mutant Rep proteins could introduce the lacZ cassette flanked by ITRs into AAVS1, the PCR-based assay for detecting site-specific integration was conducted. One primer in the PCR corresponded to the A region of the ITR, and the second was specific for AAVS1. The PCR products were blotted on membranes, and signals were detected by using an AAVS1 probe. In a previous study we demonstrated that strong signals were observed when Rep78 or Rep68 was supplied in trans (58). Representative data from at least four independent experiments are shown in Fig. 8. Mutant Rep proteins that failed to show nicking activity were also defective in the ability to integrate the ITR plasmid into AAVS1 (e.g., E6 and K10 [see Fig. 9]), except for E66, which showed weak binding and was negative for endonuclease activity and positive for ability to introduce the ITR plasmid into AAVS1. The mutant Rep proteins analyzed here were classified into four groups, with two exceptions (E66 and E239). One group is characterized by there being no effect of the mutations on the functions of Rep protein (e.g., E17 and D24); the second is characterized by having all of the functions tested here affected by the mutations (K10, R69, K101, R107, K136, R138, and R185 mutants). These mutants are possibly defective because of defective binding to the GAGC motif, which is required for the full activity of Rep protein. The D14, H18, E36, D42, EK57, RD61, K115, E150, E164, and R223 mutants are in the third group, which is characterized by weak or no activity for introduction of site-specific integration while binding and nicking activities are intact. The E6, D16, K72, E83, H90, H92, K146, K160, R217, K219, K234, K240, and K340H mutants belong to the fourth group, which is characterized by being binding positive, nicking negative, and integration negative. The loss of ability to mediate site-specific integration may be partly due to insufficient nicking activity. It should be noted that the site-specific integration by Rep protein requires the trs homolog element which is a target for the nicking by Rep proteins as well as the GAGC motif (37). As expected, the K340H mutant, having a mutation of the NTP-binding lysine residue, retained binding activity but lost both nicking activity and the ability to integrate site specifically, which is consistent with a previous report (47).
FIG. 8.
Ability of mutant Rep proteins to introduce ITR plasmid into AAVS1. Two micrograms of pCMVR78, mutant Rep expression plasmids, or blank vector was transfected into 2 × 105 293 cells/well in six-well plates along with 2 μg of pW1, harboring a lacZ expression cassette flanked by ITRs, by a standard calcium phosphate precipitation method. Twenty-four hours later, total cellular DNA was isolated and suspended finally in 200 μl of TE. PCR to detect site-specific integration was carried out as reported previously with minor modifications: 1 μl of isolated genomic DNA was subjected to a thermal cycling reaction in a 20-μl reaction mixture containing 1× thermophilic DNA polymerase buffer [10 mM KCl, 20 mM Tris-HCl (pH 8.8), 10 mM (NH4)2SO4, 4 mM MgSO4, 0.1% Triton X-100 (NEB)], 1 μM 5′-CGGCCTCAGTGAGCGAGCGAGC and 5′-CGGGGAGGATCCGCTCAGAGGACA, and 2 U of Deep Vent Exo(−) DNA polymerase (NEB). The cycling conditions were 99°C for 1 min followed by 35 cycles of 99°C for 10 s and 72°C for 4 min. Ten microliters of the PCR mixture was transferred to a hybridization membrane (Hybond-N+; Amersham) by using a dot blot apparatus and hybridized with a 32P-labeled AAVS1 probe. The membranes were then analyzed on a BAS-1500 imaging analyzer. The assay was repeated at least four times. p, positive control for hybridization.
FIG. 9.
Summary of mutant Rep78 proteins and their functions. The abilities of mutant Rep78 proteins to bind to the GAGC motif, nick at the trs, and mediate integration into AAVS1 were compared to those of wild-type Rep78. Open circles indicate that the activities are comparable to those of the wild type. Shaded circles indicate reduced activities. Closed circles indicate that activities are below background levels. EMSA was performed in triplicate, and the assay to detect site-specific integration was repeated at least four times. When nicking products were observed in the trs endonuclease assay, we concluded that the mutant Rep proteins retained nicking activity even if the amounts of the products were small. If data obtained by using single-substitution mutants are available, results for double mutants are omitted.
As shown in Fig. 2B, some mutant Rep proteins (E34, D149, and D150) were poorly expressed in 293 cells. Nevertheless, these mutants produced intense signals in the PCR-based assay for site-specific integration (Fig. 8). Thus, the site-specific integration at AAVS1 occurs even when the expression level of Rep protein is low.
DISCUSSION
The large Rep proteins, Rep78 and Rep68, are key proteins in the replication of the AAV genome and in site-specific integration. Rep78 and Rep68 bind the GAGC motif on the terminal hairpin, an event followed by a site-specific, strand-specific nick at the trs. AAV integrates preferentially into the AAVS1 locus on chromosome 19. Recent studies have revealed the mechanism underlying the site-specific integration (6, 12). In AAVS1, Rep proteins are believed to bind the analogous GAGC repeats (10, 59) and then nick at the trs homolog (59), similar to the phenomenon that occurs on the terminal hairpin. The binding of Rep proteins to the GAGC repeats is thought to be the first event in the site-specific integration as well as in AAV DNA replication. Determination of the amino acid residues that are responsible for the binding will provide us with important information on Rep proteins, especially on the structure-function relationship. To identify and dissect the functional domains of multifunctional Rep proteins, many mutational analyses have been performed by several groups (39, 47, 60, 61, 64, 65, 68, 69). However, no systematic study had been conducted to determine the functional domains of Rep proteins at the amino acid level. In the present study, we performed charged-to-alanine scanning mutagenesis of the N-terminal half of the Rep78 protein, where DNA binding and nicking domains are thought to exist. Mutant Rep78 proteins were analyzed for their abilities to bind the GAGC motif, nick at the trs homolog, and integrate an ITR plasmid into AAVS1. We chose a charged-to-alanine scanning strategy to demonstrate the important residues for the function of Rep protein because (i) charged amino acids are more likely to be located on the surface of the proteins and (ii) they are also capable of forming ion pairs and hydrogen bonds, and therefore they may be involved in enzyme catalysis and in the recognition of interacting proteins. In addition, a mutation to alanine is less likely to severely disrupt the tertiary conformation of the molecule. As expected, alanine scanning clearly showed the critical residues for the function of Rep protein: we found that the R69A mutant showed lower binding activity and was negative for both trs endonuclease and site-specific integration, whereas mutation of the adjacent residue R68 did not have any effect on the functions of Rep protein tested (Fig. 9). The same was true for E184A and R185A.
We identified the amino acid residues that are responsible for the specific binding to the GAGC motif: R107, K136, and R138 mutations completely abolished the binding activity. Moreover, K10, E66, R69, K101, R113, R185, and E239 mutants had lower binding activities than the wild type. The residues involved in the specific binding are possibly in contact with the GAGC motif. Most of these disruptive residues affecting DNA binding are basic. Thus, it is probable that there is electrostatic complementarity between an electropositive binding epitope and an electronegative phosphate backbone of the RBS. An insertion and deletion mutational analysis by Yang and Trempe showed that residues 25 to 62, 88 to 113, and 125 to 256 are important for DNA binding in the N-terminal portion of the Rep protein (69). Another deletion mutational study by McCarty et al. showed that residues 134 to 242 were essential for DNA binding in the N-terminal region (38). The present study is not directly comparable to the previous mutational analyses because the mutations introduced are different. However, all of the amino acid residues, R107, K136, and R138, that are revealed to be essential for binding in the present study are within the domains described before. All of the mutations affecting the specific binding to the probe in EMSA caused loss of not only trs endonuclease activity but also the ability to integrate site specifically (Fig. 9), except for E66 and E239. These results support the idea that the binding of Rep proteins to the GAGC motif is a prerequisite for the site-specific and strand-specific nicking activity (39).
To elucidate the amino acid residues involved in binding to the GAGC repeats clearly, further mutational studies will be needed. Mutations should be introduced into the regions adjacent to the charged residues that are responsible for the specific binding. Moreover, a comprehensive mutational study of regions other than the N-terminal half should be performed, because DNA binding domains exist in other regions of Rep proteins, as reported previously (39, 47, 60, 61, 64, 69). Furthermore, a three-dimensional structure solved by X-ray crystallography or nuclear magnetic resonance spectroscopy will help us identify the amino acid residues that interact with DNA. We agree that the binding domain for the GAGC motif is mainly in the N-terminal halves of the large Rep proteins and that the other regions help stabilize the DNA-protein complex (47). The stabilization may be contributed to by multimerization of the Rep molecules and/or by nonspecific DNA binding. It is noteworthy that Rep52, which lacks the N-terminal region we analyzed, has a helicase activity (53), which should bind DNA. Moreover, Ryan et al. suggested that Rep protein also binds DNA other than GAGC motif (50), although it remains to be elucidated whether DNA binding not via the RBS is specific or not. In addition, simian virus 40 large T antigen has been reported not only to bind a specific DNA motif but also to have nonspecific DNA binding activity (52). Several nondisruptive mutations have been reported to affect specific binding to the GAGC motif, such as W242L (39), I393A, and I417A (61). As hydrophobic amino acid residues such as W and I are not likely to contact DNA directly, we believe that W242, I393, and I417 are in the core of the Rep molecule and that the mutations mentioned above cause disruption of the tertiary conformation, leading to reduced binding activity.
The domain of trs endonuclease activity is thought to be in the N-terminal and the central regions of the Rep protein. A few mutational analyses dissected DNA binding and nicking domains (60, 61, 64), although these were not examined extensively. Focusing on the N-terminal region, only Y121F and Y152F mutants were binding positive and nicking negative, as shown by Walker et al. (60). Our study with charged-to-alanine scanning mutagenesis identified the amino acid residues that were important for nicking activity but not essential for DNA binding (Fig. 9). Mutations affecting trs endonuclease activity existed over the entire N-terminal half (Fig. 7), which contrasts with the relatively small number of charged residues important for GAGC binding (Fig. 4).
Based on homologies with metalloenzymes, prokaryotic initiator proteins in rolling-circle DNA replication have a conserved motif, called the two-His motif, that might be involved in the metal ion coordination required for the activities of the replication proteins (21). This motif consists of HuHuuu, where u is any hydrophobic residue. Rep78 and Rep68 also possess this motif, HMHVLV (residues 90 to 95) and require Mg2+ for their functions, including trs endonuclease and helicase activities (11, 22, 66). Interestingly, both H90 and H92 mutants lost trs endonuclease activity while retaining GAGC binding activity (Fig. 4, 7, and 9). Thus, HMHVLV (residues 90 to 95) may bind Mg2+. A metal binding site composed of H-x-H should be in a β-sheet as shown by modeling experiments (1). The prediction of the secondary structure of the Rep protein by the PHDsec (EMBL) or the Chou-Fasman algorithm showed that the two-His motif in the Rep protein is indeed in a β-sheet. To confirm this, the three-dimensional structure of the Rep protein should be solved. A mutational analysis of a two-His motif in the NS1 protein, which is homologous to Rep78 and Rep68, of minute virus of mice (a member of the parvoviruses) by Nüesch et al. revealed that the histidine residues were essential for nicking activity (45). Besides a two-His motif, prokaryotic initiator proteins possess another conserved motif, uxxYux(x)K, where u is hydrophobic and x is any residue, encompassing tyrosine residue forming the covalent link with nicked DNA (21). By homology with initiator proteins, Y at position 156 is predicted to be a linking tyrosine and to be followed by lysine at position 160, with a 3-amino-acid spacer (IPNYLLPK). The mutant with an alanine substitution for the lysine at position 160 was negative for trs endonuclease activity while retaining DNA binding activity. However, Walker et al. concluded that the tyrosine residue at position 152 was a candidate for the linking tyrosine, based on their observation that a Y152F mutant lost trs endonuclease activity while DNA binding and helicase activity were not apparently affected (60), on the assumption that that the tyrosine residue linking to DNA is associated only with nicking at the trs and not with unwinding of DNA or with the specific binding to DNA. On the other hand, they found that the Y156F mutant lost nicking and helicase activities, with reduced DNA binding activity.
The large Rep proteins play a critical role in the site-specific integration of the AAV genome into AAVS1. Therefore, we analyzed whether mutant Rep proteins were capable of mediating the site-specific integration into the AAVS1 locus by using a sensitive method to detect site-specific integration of plasmid DNA containing ITR sequences (Fig. 8 and 9). Mutant Rep proteins with charged-to-alanine mutations were classified into four groups: (i) no change in functions, (ii) all of the functions tested affected, (iii) binding positive and both nicking and integration negative, and (iv) both binding and nicking positive and integration negative. However, there are two exceptions. The E66 mutant, showing weak binding, was negative for endonuclease activity and positive for the ability to introduce the ITR plasmid into AAVS1. There is a possible explanation for this discrepancy. In vitro studies, such as EMSA and trs endonuclease assay, may not be sensitive enough. In cells, mutant Rep proteins with diminished binding activity and with weak trs nicking activity may be able to achieve site-specific integration. Unknown factors might compensate for the dysfunction of mutant Rep proteins. Recently, Rep protein has been shown to associate with a nuclear protein, high-mobility-group chromosomal protein 1, which promotes the formation of Rep protein-DNA complexes and stimulates the activity of Rep protein in the nicking reaction (15). Another exception is the E239 mutant, which shows a weak binding, intact trs endonuclease activity, and low site specificity. The explanation is that a reduced binding activity is not enough to deprive the mutant Rep protein of trs endonuclease activity, and this mutation may also affect other functions of Rep protein that are required for site-specific integration. Targeted integration of the AAV genome to AAVS1 is a complex phenomenon that requires host factors (e.g., DNA polymerase). The mechanism underlying this unique feature has not completely elucidated. The present study is not able to demonstrate why the mutants in the fourth group (binding positive, nicking positive, and integration negative), such as the D14, H18, E36, D42, EK57, RD61, K115, E150, E164, and R223 mutants (Fig. 9), are defective for site-specific integration. It will be interesting to determine whether these mutants still retain helicase activity, one of the well-characterized activities of Rep protein (22). Moreover, the set of mutant Rep proteins that we constructed would serve as a useful tool for characterizing the unknown aspects of Rep protein in site-specific integration, such as association with an undetermined host factor(s).
Sequences of AAVs other than type 2 have been reported, i.e., those of AAV type 3 (AAV3) (41), AAV4 (13), AAV3B, and AAV6 (49). All of the amino acid residues at which mutations affected the functions of Rep protein in this study were well conserved or showed conservative change except for one residue; in the present study, the E66 mutant showed reduced binding and no trs nicking activities. Only AAV6 has Q at position 66; AAV3, AAV3B, and AAV4 have E at that position. Moreover, as the RBS sequences on ITRs are conserved well among these AAVs (AAV2 and -6, GAGC GAGC GAGC GCGC; AAV3 and -3B, GAGC GAGC GAGT GCGC; AAV4, GAGT GAGT GAGC GAGC), it is noteworthy that the amino acid residues responsible for the specific binding to the GAGC motif are also conserved well among different types of AAV. These comparisons demonstrated that the residues at which substitution with alanine affected the function of Rep proteins were probably important for other types of AAV as well.
We also demonstrated that in vitro-synthesized Rep78 protein nicked preferentially not always between the thymidine residues. We compared the lengths of the nicking products derived from two different AAVS1 substrates with DNA sequencing ladders and concluded that the major nicking site fluctuates between GGT/TGG and GG/TTGG. The nicking product derived from the SmaI-XhoI fragment, which is essentially the same as the P1 fragment described before (65), was released by cutting between the T residues (GGT/TGG). On the other hand, the Rep78 protein liberated a nicking product from the longer StyI-XhoI substrate by cutting between the G and T residues (GG/TTGG), which was confirmed by comparing the nick product to sequence ladders produced by both chain termination and chemical modification methods. We used another substrate for the nicking reaction, which was prepared by digesting pS1 with HindIII and XhoI (Fig. 6A). Again, the main nick site was GG/TTGG (data not shown). We also performed a trs nicking reaction by using 61-nt synthetic oligonucleotides carrying the RBS and the trs homolog, as reported by Urcelay et al. (59). Our result is consistent with their conclusion, showing that the cut site was between the T residues (data not shown). Im and Muzyczka reported that Rep68 isolated from HeLa cells infected with AAV and adenovirus type 2 nicked at the trs on the AAV hairpin between the T residues (22). Kotin and coworkers reported that bacterially expressed Rep78 as a fusion protein with maltose-binding protein cut at the trs and the trs homolog between the T residues (10, 59). These results are inconsistent with our present findings that the main nick site for in vitro-translated Rep78 fluctuates between GGT/TGG and GG/TTGG. This may be due to the different assay systems; we used in vitro-synthesized Rep78 protein for the trs endonuclease assay, but previous studies on the nicking site used HeLa cell-isolated or bacterially expressed Rep protein. Moreover, our reaction mixture contained a lysate for in vitro transcription and translation, which may cause the aberrant nicking. Interestingly, Ryan et al. reported that Rep protein binds a sequence motif other than the RBS within the ITR (50). The second motif they identified that interacts with Rep protein is CCTTTGG, which is quite similar to the 5′ end of our StyI-XhoI substrate, CTTGG. In addition, they suggested other binding sites for Rep protein within the AAV hairpin. Thus, the nicking by Rep protein may be influenced by the sequence surrounding the minimal element required for the site-specific and strand-specific nicking, i.e., the RBS and the trs homolog. We are currently trying to determine the sequence element affecting the nick site by deletion analysis of the StyI-XhoI substrate. Moreover, the ends of the AAV genome are reported to be heterogeneous (16), and Snyder et al. described nicking products that were heterogeneous in size and that it was possible for Rep protein to nick at other sites (56). Although they also demonstrated that a tyrosine residue in Rep68 was linked to a thymidine residue at the trs (55), they did not exclude the possibility that Rep proteins attached covalently to other residues. Batchu and Hermonat also reported that Rep78 produced in Escherichia coli as a fusion protein with maltose-binding protein showed multiple nicking sites besides a nick at the trs (4). Thus, under certain conditions, large Rep proteins may nick at different sites.
ACKNOWLEDGMENTS
We are deeply grateful to K. I. Berns for providing pRVK, to J. A. Kleinschmidt for anti-Rep antibody 294.4, and to Y. Iwaki and T. Inaba for valuable advice.
This work was supported in part by grants from the Ministry of Health and Welfare of Japan and Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Arnold F H, Haymore B L. Engineered metal-binding proteins: purification to protein folding. Science. 1991;252:1796–1797. doi: 10.1126/science.1648261. [DOI] [PubMed] [Google Scholar]
- 2.Baas P D, Jansz H S. Single-stranded DNA phage origins. Curr Top Microbiol Immunol. 1988;136:31–70. doi: 10.1007/978-3-642-73115-0_3. [DOI] [PubMed] [Google Scholar]
- 3.Bass S H, Mulkerrin M G, Wells J A. A systematic mutational analysis of hormone-binding determinants in the human growth hormone receptor. Proc Natl Acad Sci USA. 1991;88:4498–4502. doi: 10.1073/pnas.88.10.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batchu R B, Hermonat P L. Dissociation of conventional DNA binding and endonuclease activities by an adeno-associated virus Rep78 mutant. Biochem Biophys Res Commun. 1995;210:717–725. doi: 10.1006/bbrc.1995.1718. [DOI] [PubMed] [Google Scholar]
- 5.Beaton A, Palumbo P, Berns K I. Expression from the adeno-associated virus p5 and p19 promoters is negatively regulated in trans by the Rep protein. J Virol. 1989;63:4450–4454. doi: 10.1128/jvi.63.10.4450-4454.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berns K I, Giraud C. Biology of adeno-associated virus. Curr Top Microbiol Immunol. 1996;218:1–23. doi: 10.1007/978-3-642-80207-2_1. [DOI] [PubMed] [Google Scholar]
- 7.Chejanovsky N, Carter B J. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology. 1989;173:120–128. doi: 10.1016/0042-6822(89)90227-4. [DOI] [PubMed] [Google Scholar]
- 8.Chejanovsky N, Carter B J. Mutation of a consensus purine nucleotide binding site in the adeno-associated virus rep gene generates a dominant negative phenotype for DNA replication. J Virol. 1990;64:1764–1770. doi: 10.1128/jvi.64.4.1764-1770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung A K, Hoggan M D, Hauswirth W W, Berns K I. Integration of the adeno-associated virus genome into cellular DNA in latently infected human Detroit 6 cells. J Virol. 1980;33:739–748. doi: 10.1128/jvi.33.2.739-748.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiorini J A, Weitzman M D, Owens R A, Urcelay E, Safer B, Kotin R M. Biologically active Rep proteins of adeno-associated virus type 2 produced as fusion proteins in Escherichia coli. J Virol. 1994;68:797–804. doi: 10.1128/jvi.68.2.797-804.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiorini J A, Wiener S M, Owens R A, Kyöstiö S R, Kotin R M, Safer B. Sequence requirements for stable binding and function of Rep68 on the adeno-associated virus type 2 inverted terminal repeats. J Virol. 1994;68:7448–7457. doi: 10.1128/jvi.68.11.7448-7457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiorini J A, Wiener S M, Yang L, Smith R H, Safer B, Kilcoin N P, Liu Y, Urcelay E, Kotin R M. The roles of AAV Rep proteins in gene expression and targeted integration. Curr Top Microbiol Immunol. 1996;218:25–33. doi: 10.1007/978-3-642-80207-2_2. [DOI] [PubMed] [Google Scholar]
- 13.Chiorini J A, Yang L, Liu Y, Safer B, Kotin R M. Cloning of adeno-associated virus type 4 (AAV4) and generation of recombinant AAV4 particles. J Virol. 1997;71:6823–6833. doi: 10.1128/jvi.71.9.6823-6833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chothia C. The nature of the accessible and buried surfaces in proteins. J Mol Biol. 1976;105:1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- 15.Costello E, Saudan P, Winocour E, Pizer L, Beard P. High mobility group chromosomal protein 1 binds to the adeno-associated virus replication protein (Rep) and promotes Rep-mediated site-specific cleavage of DNA, ATPase activity and transcriptional repression. EMBO J. 1997;16:5943–5954. doi: 10.1093/emboj/16.19.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fife K H, Berns K I, Murray K. Structure and nucleotide sequence of the terminal regions of adeno-associated virus DNA. Virology. 1977;78:475–477. doi: 10.1016/0042-6822(77)90124-6. [DOI] [PubMed] [Google Scholar]
- 17.Gibbs C S, Zoller M J. Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J Biol Chem. 1991;266:8923–8931. [PubMed] [Google Scholar]
- 18.Hermonat P L, Batchu R B. The adeno-associated virus Rep78 major regulatory protein forms multimeric complexes and the domain for this activity is contained within the carboxy-half of the molecule. FEBS Lett. 1997;401:180–184. doi: 10.1016/s0014-5793(96)01469-x. [DOI] [PubMed] [Google Scholar]
- 19.Hölscher C, Hörer M, Kleinschmidt J A, Zentgraf H, Bürkle A, Heilbronn R. Cell lines inducibly expressing the adeno-associated virus (AAV) rep gene: requirements for productive replication of rep-negative AAV mutants. J Virol. 1994;68:7169–7177. doi: 10.1128/jvi.68.11.7169-7177.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hörer M, Weger S, Butz K, Hoppe-Seyler F, Geisen C, Kleinschmidt J A. Mutational analysis of adeno-associated virus Rep protein-mediated inhibition of heterologous and homologous promoters. J Virol. 1995;69:5485–5496. doi: 10.1128/jvi.69.9.5485-5496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ilyina T V, Koonin E V. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Im D-S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 23.Im D-S, Muzyczka N. Factors that bind to adeno-associated virus terminal repeats. J Virol. 1989;63:3095–3104. doi: 10.1128/jvi.63.7.3095-3104.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inaba T, Shapiro L H, Funabiki T, Sinclair A E, Jones B G, Ashmun R A, Look A T. DNA-binding specificity and trans-activating potential of the leukemia-associated E2A-hepatic leukemia factor fusion protein. Mol Cell Biol. 1994;14:3403–3413. doi: 10.1128/mcb.14.5.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inamoto S, Yoshioka Y, Ohtsubo E. Site- and strand-specific nicking in vitro at oriT by the traY-traI endonuclease of plasmid R100. J Biol Chem. 1991;266:10086–10092. [PubMed] [Google Scholar]
- 26.Janin J. Surface and inside volumes in globular proteins. Nature. 1979;277:491–492. doi: 10.1038/277491a0. [DOI] [PubMed] [Google Scholar]
- 27.Kleinschmidt J A, Möhler M, Weindler F W, Heilbronn R. Sequence elements of the adeno-associated virus rep gene required for suppression of herpes-simplex-virus-induced DNA amplification. Virology. 1995;206:254–262. doi: 10.1016/s0042-6822(95)80040-9. [DOI] [PubMed] [Google Scholar]
- 28.Kotin R M. Prospects for the use of adeno-associated virus as a vector for human gene therapy. Hum Gene Ther. 1994;5:793–801. doi: 10.1089/hum.1994.5.7-793. [DOI] [PubMed] [Google Scholar]
- 29.Kotin R M, Berns K I. Organization of adeno-associated virus DNA in latently infected Detroit 6 cells. Virology. 1989;170:460–467. doi: 10.1016/0042-6822(89)90437-6. [DOI] [PubMed] [Google Scholar]
- 30.Kotin R M, Linden R M, Berns K I. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 1992;11:5071–5078. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kotin R M, Menninger J C, Ward D C, Berns K I. Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13-qter. Genomics. 1991;10:831–834. doi: 10.1016/0888-7543(91)90470-y. [DOI] [PubMed] [Google Scholar]
- 32.Kotin R M, Siniscalco M, Samulski R J, Zhu X D, Hunter L, Laughlin C A, McLaughlin S, Muzyczka N, Rocchi M, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kyöstiö S R, Owens R A, Weitzman M D, Antoni B A, Chejanovsky N, Carter B J. Analysis of adeno-associated virus (AAV) wild-type and mutant Rep proteins for their abilities to negatively regulate AAV p5 and p19 mRNA levels. J Virol. 1994;68:2947–2957. doi: 10.1128/jvi.68.5.2947-2957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Labow M A, Graf L H, Jr, Berns K I. Adeno-associated virus gene expression inhibits cellular transformation by heterologous genes. Mol Cell Biol. 1987;7:1320–1325. doi: 10.1128/mcb.7.4.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Labow M A, Hermonat P L, Berns K I. Positive and negative autoregulation of the adeno-associated virus type 2 genome. J Virol. 1986;60:251–258. doi: 10.1128/jvi.60.1.251-258.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Linden R M, Ward P, Giraud C, Winocour E, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1996;93:11288–11294. doi: 10.1073/pnas.93.21.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linden R M, Winocour E, Berns K I. The recombination signals for adeno-associated virus site-specific integration. Proc Natl Acad Sci USA. 1996;93:7966–7972. doi: 10.1073/pnas.93.15.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCarty D M, Christensen M, Muzyczka N. Sequences required for coordinate induction of adeno-associated virus p19 and p40 promoters by Rep protein. J Virol. 1991;65:2936–2945. doi: 10.1128/jvi.65.6.2936-2945.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCarty D M, Ni T H, Muzyczka N. Analysis of mutations in adeno-associated virus Rep protein in vivo and in vitro. J Virol. 1992;66:4050–4057. doi: 10.1128/jvi.66.7.4050-4057.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCarty D M, Pereira D J, Zolotukhin I, Zhou X, Ryan J H, Muzyczka N. Identification of linear DNA sequences that specifically bind the adeno-associated virus Rep protein. J Virol. 1994;68:4988–4997. doi: 10.1128/jvi.68.8.4988-4997.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muramatsu S, Mizukami H, Young N S, Brown K E. Nucleotide sequencing and generation of an infectious clone of adeno-associated virus 3. Virology. 1996;221:208–217. doi: 10.1006/viro.1996.0367. [DOI] [PubMed] [Google Scholar]
- 42.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 43.Ni T H, McDonald W F, Zolotukhin I, Melendy T, Waga S, Stillman B, Muzyczka N. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J Virol. 1998;72:2777–2787. doi: 10.1128/jvi.72.4.2777-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ni T H, Zhou X, McCarty D M, Zolotukhin I, Muzyczka N. In vitro replication of adeno-associated virus DNA. J Virol. 1994;68:1128–1138. doi: 10.1128/jvi.68.2.1128-1138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nüesch J P, Cotmore S F, Tattersall P. Sequence motifs in the replicator protein of parvovirus MVM essential for nicking and covalent attachment to the viral origin: identification of the linking tyrosine. Virology. 1995;209:122–135. doi: 10.1006/viro.1995.1236. [DOI] [PubMed] [Google Scholar]
- 46.Owens R A, Trempe J P, Chejanovsky N, Carter B J. Adeno-associated virus rep proteins produced in insect and mammalian expression systems: wild-type and dominant-negative mutant proteins bind to the viral replication origin. Virology. 1991;184:14–22. doi: 10.1016/0042-6822(91)90817-u. [DOI] [PubMed] [Google Scholar]
- 47.Owens R A, Weitzman M D, Kyöstiö S R, Carter B J. Identification of a DNA-binding domain in the amino terminus of adeno-associated virus Rep proteins. J Virol. 1993;67:997–1005. doi: 10.1128/jvi.67.2.997-1005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pereira D J, McCarty D M, Muzyczka N. The adeno-associated virus (AAV) Rep protein acts as both a repressor and an activator to regulate AAV transcription during a productive infection. J Virol. 1997;71:1079–1088. doi: 10.1128/jvi.71.2.1079-1088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rutledge E A, Halbert C L, Russell D W. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J Virol. 1998;72:309–319. doi: 10.1128/jvi.72.1.309-319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryan J H, Zolotukhin S, Muzyczka N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J Virol. 1996;70:1542–1553. doi: 10.1128/jvi.70.3.1542-1553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samulski R J, Zhu X, Xiao X, Brook J D, Housman D E, Epstein N, Hunter L A. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simmons D T, Wun Kim K, Young W. Identification of simian virus 40 T-antigen residues important for specific and nonspecific binding to DNA and for helicase activity. J Virol. 1990;64:4858–4865. doi: 10.1128/jvi.64.10.4858-4865.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith R H, Kotin R M. The Rep52 gene product of adeno-associated virus is a DNA helicase with 3′-to-5′ polarity. J Virol. 1998;72:4874–4881. doi: 10.1128/jvi.72.6.4874-4881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith R H, Spano A J, Kotin R M. The Rep78 gene product of adeno-associated virus (AAV) self-associates to form a hexameric complex in the presence of AAV ori sequences. J Virol. 1997;71:4461–4471. doi: 10.1128/jvi.71.6.4461-4471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snyder R O, Im D-S, Muzyczka N. Evidence for covalent attachment of the adeno-associated virus (AAV) Rep protein to the ends of the AAV genome. J Virol. 1990;64:6204–6213. doi: 10.1128/jvi.64.12.6204-6213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Snyder R O, Samulski R J, Muzyczka N. In vitro resolution of covalently joined AAV chromosome ends. Cell. 1990;60:105–113. doi: 10.1016/0092-8674(90)90720-y. [DOI] [PubMed] [Google Scholar]
- 57.Srivastava A, Lusby E W, Berns K I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Surosky R T, Urabe M, Godwin S G, McQuiston S A, Kurtzman G J, Ozawa K, Natsoulis G. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J Virol. 1997;71:7951–7959. doi: 10.1128/jvi.71.10.7951-7959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Urcelay E, Ward P, Wiener S M, Safer B, Kotin R M. Asymmetric replication in vitro from a human sequence element is dependent on adeno-associated virus Rep protein. J Virol. 1995;69:2038–2046. doi: 10.1128/jvi.69.4.2038-2046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walker S L, Wonderling R S, Owens R A. Mutational analysis of the adeno-associated virus Rep68 protein: identification of critical residues necessary for site-specific endonuclease activity. J Virol. 1997;71:2722–2730. doi: 10.1128/jvi.71.4.2722-2730.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walker S L, Wonderling R S, Owens R A. Mutational analysis of the adeno-associated virus type 2 Rep68 protein helicase motifs. J Virol. 1997;71:6996–7004. doi: 10.1128/jvi.71.9.6996-7004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang P Z, Projan S J, Henriquez V, Novick R P. Origin recognition specificity in pT181 plasmids is determined by a functionally asymmetric palindromic DNA element. EMBO J. 1993;12:45–52. doi: 10.1002/j.1460-2075.1993.tb05630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weger S, Wistuba A, Grimm D, Kleinschmidt J A. Control of adeno-associated virus type 2 cap gene expression: relative influence of helper virus, terminal repeats, and Rep proteins. J Virol. 1997;71:8437–8447. doi: 10.1128/jvi.71.11.8437-8447.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weitzman M D, Kyöstiö S R, Carter B J, Owens R A. Interaction of wild-type and mutant adeno-associated virus (AAV) Rep proteins on AAV hairpin DNA. J Virol. 1996;70:2440–2448. doi: 10.1128/jvi.70.4.2440-2448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weitzman M D, Kyöstiö S R, Kotin R M, Owens R A. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wonderling R S, Kyöstiö S R, Owens R A. A maltose-binding protein/adeno-associated virus Rep68 fusion protein has DNA-RNA helicase and ATPase activities. J Virol. 1995;69:3542–3548. doi: 10.1128/jvi.69.6.3542-3548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wonderling R S, Owens R A. The Rep68 protein of adeno-associated virus type 2 stimulates expression of the platelet-derived growth factor B c-sis proto-oncogene. J Virol. 1996;70:4783–4786. doi: 10.1128/jvi.70.7.4783-4786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang Q, Kadam A, Trempe J P. Mutational analysis of the adeno-associated virus rep gene. J Virol. 1992;66:6058–6069. doi: 10.1128/jvi.66.10.6058-6069.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang Q, Trempe J P. Analysis of the terminal repeat binding abilities of mutant adeno-associated virus replication proteins. J Virol. 1993;67:4442–4447. doi: 10.1128/jvi.67.7.4442-4447.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]