Abstract
INTRODUCTION:
LOAD is a complex neurodegenerative disease characterized by multiple progressive stages, glucose metabolic dysregulation, AD pathology, and inexorable cognitive decline. Discovery of metabolic profiles unique to sex, APOE-genotype and stage of disease progression could provide critical insights for personalized LOAD medicine.
METHODS:
Sex- and APOE-specific metabolic networks were constructed based on changes in 127 metabolites of 656 serum samples from the ADNI cohort.
RESULTS:
Application of advanced analytical platform identified metabolic drivers and signatures clustered with sex and/or APOEɛ4, establishing patient-specific biomarkers predictive of disease state that significantly associated with cognitive function. Presence of the APOEɛ4 shifts metabolic signatures to a phosphatidylcholine-focused profile overriding sex-specific differences in serum metabolites of AD patients.
DISCUSSION:
These findings provide an initial but critical step in developing a diagnostic platform for personalized medicine by integrating metabolomic profiling and cognitive assessments to identify targeted precision therapeutics for AD patient subgroups through computational network modeling.
Keywords: Late-onset Alzheimer’s disease, computational systems biology, metabolic network, metabolomics, ADNI, APOE ε4, sex-specific metabolic changes, metabolic biomarkers, precision medicine
1. BACKGROUND
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder without a cure. Recent clinical trial failures targeting β-amyloid (Aβ) or hyperphosphorylated tau protein (p-tau) underscore the importance of understanding disease-driving mechanisms. The primary risk factors of late-onset AD (LOAD), the predominant form, include age, female sex, and the presence of the APOEε4 allele [1, 2]. LOAD is a multifactorial disorder with perturbations in glucose and insulin signaling, energy and lipid homeostasis, mitochondrial function, oxidative stress, inflammation, and neurotransmission [3, 4]. Recent progress in dissecting sex-specific mechanisms of AD has become possible through the implementation of systems-level approaches and availability of clinically characterized samples including ADNI cohort tissue and biofluids samples from AD patients and cognitively normal individuals enabling large-scale multi-omics studies. Metabolomics is the newest omics that measures thousands of metabolites reflecting alterations in genetic, transcriptomic, proteomic profiles, and influences from the environment [5, 6]. A large number of studies using metabolomcis and lipidomics platforms has provided new biochemical insights about disease mechanisms, early changes in disease and provided support that peripheral metabolic changes inform about central changes and ATN markers of disease [7–20].
We recently conducted stratified linear regression analyses of serum metabolites from 1517 ADNI participants to determine the association of metabolic signatures with disease diagnosis (Dx) and A-T-N biomarkers (CSF Aβ1–42 (A); CSF p-tau (T); FDG-PET (N)) [14] . Changes in metabolites associated with the Dx or A-T-N were influenced by sex and APOE and related to altered energy homeostasis [14]. We applied a recently developed computational predictive network model [21–24] to construct sex- and APOE-specific metabolic networks of CN and LOAD patients from the ADNI cohort in respect to sex and genotype and to clinical diagnosis cognitive parameters. We confirmed previous findings, demonstrated that metabolic panels associate with cognitive assessment, and identified metabolic drivers of LOAD. These findings further support the application of blood-based metabolomics as a precision medicine tool for disease stage profiling, prognosis, and identification of novel therapeutic targets.
2. METHODS
2.1. Participants
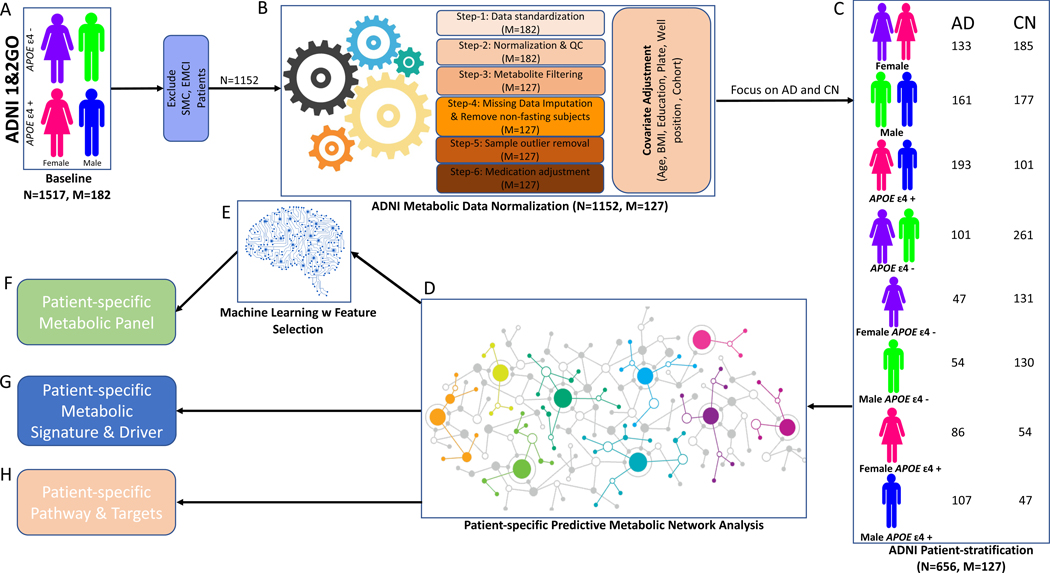
Figure 1 and Table 1 summarize information on ADNI participants utilized in this study.
Figure 1. Analytical pipeline utilized in the study.
The analytical pipeline included 1152 samples from ADNI cohort. Patients with self-memory complain (SMC) and early mild cognitive impairment (EMCI) were removed leaving 1152 samples from AD, late mild cognitive impairment (LMCI) and CN (A). Data were normalized; the residuals were obtained after covariate adjustment (B). 362 CN and 294 AD samples were stratified into eight groups based on sex and APOE genotype (C). A predictive network model was built (D) to derive patient-specific metabolic signatures and drivers of progression from CN to AD in each group (G). The DE analysis identified significant changes in metabolites. Metabolic biomarker panels (F) were derived using machine learning models (E). Patient-specific pathways were identified based on metabolic signatures and drivers (H). N, the number of participants; M, the number of metabolites; ADNI, AD Neuroimaging Initiative; ADNI 1&2GO, phase 1 and phase 2/GO of ADNI; QC, quality control; AD, Alzheimer disease; CN, cognitive normal; BMI, body mass index.
Table 1.
Characteristics of the 1152 ADNI subjects in this study
| Demographics | CN | LMCI | AD |
|---|---|---|---|
| Sample size | 362 | 496 | 294 |
| Sex (M/F) | 177/185 | 307/189 | 161/133 |
| Age(yr.) | 74.61(+/−5.66) | 74.11(+/−7.57) | 74.71(+/−7.85) |
| BMI (kg/m2) | 27.04(+/−4.51) | 26.49(+/−4.32) | 25.87(+/−4.71) |
| Education(yr.) | 16.20(+/−2.79) | 15.86(+/−2.91) | 15.19(+/−2.99) |
| APOE ɛ4 +/− | 101/261 | 265/231 | 193/101 |
| Clinic Assessment | |||
| ADAS-Cog total score | 5.99(+/−3.04) | 11.56(+/−4.50) | 19.34(+/−6.75) |
| Memory function (ADNI_MEM) | 0.95(+/−0.53) | −0.05(+/−0.57) | −0.73(+/−0.52) |
| Executive function (ADNI_EF) | 0.74(+/−0.70) | 0.014(+/−0.78) | −0.82(+/−0.84) |
| CSF Pathology | |||
| CSF p181-Tau | 25.54(+/−14.80) | 35.36(+/−17.36) | 41.64(+/−19.63) |
| CSF Aβ1–42 | 207.67(+/−54.47) | 163.48(+/−53.57) | 143.64(+/−41.86) |
Table 1. Characteristics of the 1152 ADNI participants included in this study
Metabolomics datasets from the Biocrates p180 platform used in the current analyses for the ADNI-1 and ADNI-GO/2 cohorts are available via the Accelerating Medicines Partnership-Alzheimer’s Disease (AMP-AD) Knowledge Portal and can be accessed at http://dx.doi.org/10.7303/syn5592519 (ADNI-1) and http://dx.doi.org/10.7303/syn9705278 (ADNI GO-2). The full complement of clinical and demographic data for the ADNI cohorts are hosted on the LONI data sharing platform and can be requested at http://adni.loni.usc.edu/data-samples/access-data/. Abbreviations: ADAS-Cog, Alzheimers Disease Assessment Scale – Cognitive Subscale; BMI, Body Mass Index; CN, cognitively normal; LMCI, Late Mild Cognitive Impairment; AD, Alzheimer’s Disease; yr., years. APOEε4−/+: non-carriers and carriers of the APOE ε4 allele, ADAS-Cog: Alzheimer’s Disease Assessment Scale-Cognitive Subscale, CSF Aβ1–42: Cerebrospinal fluid Amyloid beta 1–42 protein. CSF p181-Tau: Cerebrospinal fluid phosphorylated Tau protein at threonine 181 (p181tau).
2.2. Metabolomics data acquisition, normalization, and covariate adjustment
Metabolomics data normalization followed a six-step procedure [11] (Figure 1B, Online Method-1). Final 127 metabolites are listed in Table S1, and the final 362 CN and 294 AD samples were stratified into eight groups based on sex and APOE genotype (Table S2).
2.3. Predictive network Modeling
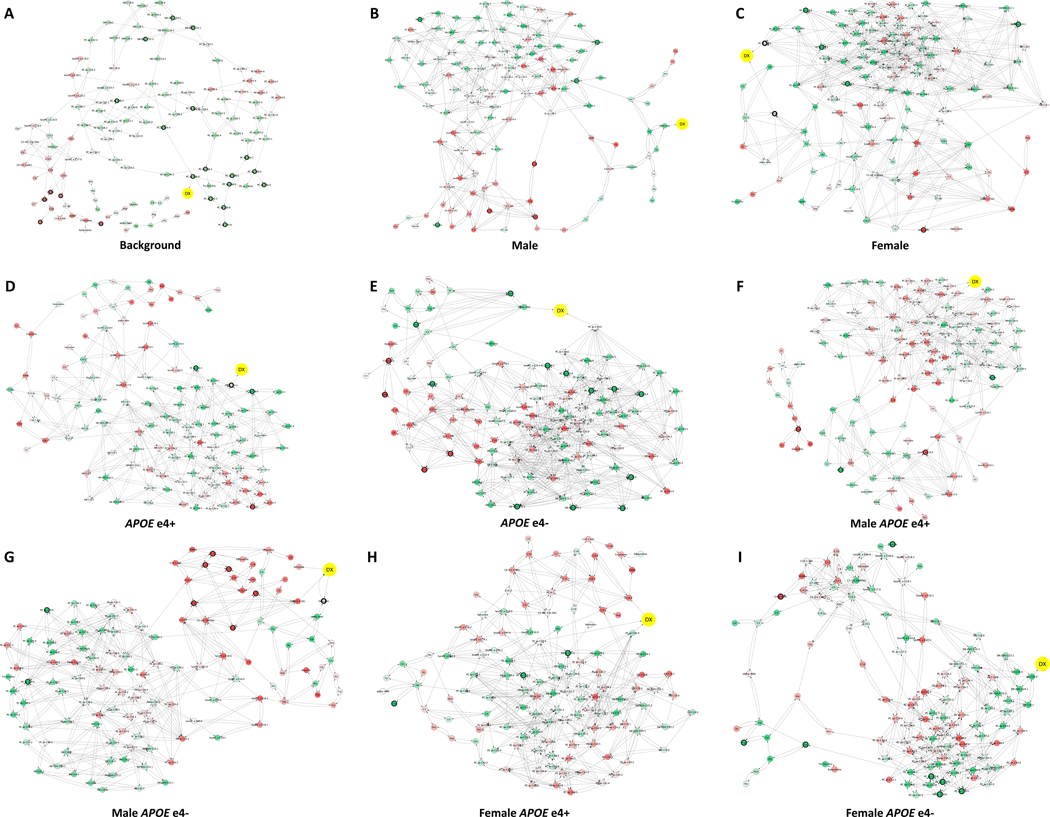
For each patient group, metabolites of AD and CN subjects were integrated with Dx into the predictive network modeling pipeline [22–25]. The network model consists of metabolites and Dx as nodes and causal interactions between them (Figure 2, Online Method-2).
Figure 2. Sex- and APOE-specific consensus predictive metabolic network.
To build consensus causal predictive metabolic network, we subsampled 100 datasets and constructed 100 metabolic networks per patient group. The 95% confidence interval is calculated per edge. The consensus network models were used to identify the upstream metabolites and pathways associated with AD in background with all 656 AD and CN samples (A), males (B), females (C), APOEɛ4+ (D), APOEɛ4- (E), male APOEɛ4+ (F), male APOEɛ4- (G), female APOEɛ4+ (H), female APOEɛ4- (I). Dx, disease diagnosis. Red color indicates metabolites metabolite level is increased in AD comparing to CN; green color indicates metabolite level is decreased in AD comparing to CN. Significant DE metabolites are indicated with black circles.
2.4. Empirical non-parametric bootstrap and consensus network analysis
For each patient group, the 95% confidence interval (CI) of each edge was evaluated with the empirical bootstrap method (Online Method-3). To derive the patient-specific consensus metabolic networks, we included top 10% of edges with 95% confidence per patient group (Table S3). The metabolic signature was extracted as the 3-step upstream subnetwork of Dx in the patient-specific network (Table S4).
2.5. Evaluation of heterogeneity of key drivers
The heterogeneity of key drivers was evaluated by calculating the significance of robustness and confidence of patient-specific key driver in each patient group (Table S5, Online Method-4).
2.6. Differential expression (DE) analysis
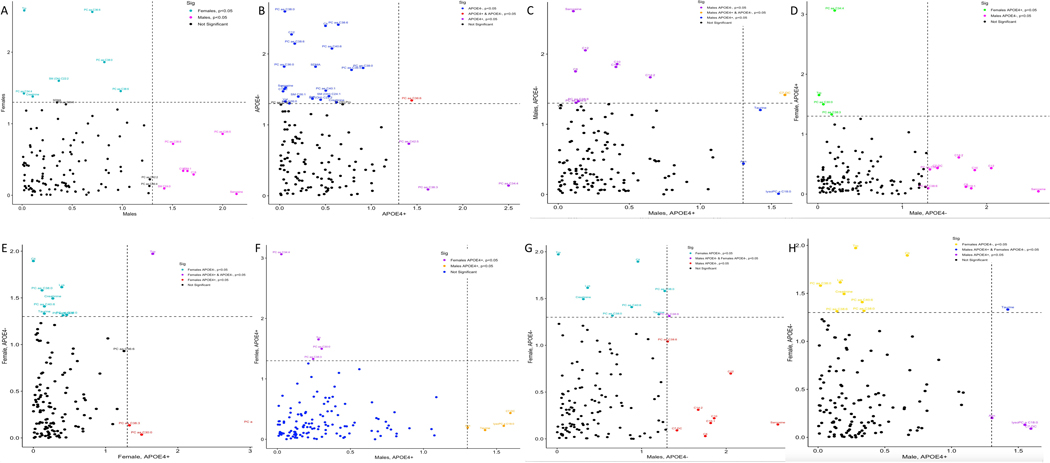
Following covariate adjustment, metabolites were subjected to t-test using Limma R package [26] between AD and CN samples in each group (Figure 4, S1, Table S6).
Figure 4. Sex- and APOE-specific metabolic differential expression analysis.
The significant P-value<0.05 of differentially produced metabolites are compared between patient groups to illustrate the specificity and commonality of AD-associated metabolic signatures to sex and APOE genotype. (A) Male vs Female; (B) APOEɛ4+ vs APOEɛ4-; (C) Male APOEɛ4+ vs Male APOEɛ4-; (D) Male APOEɛ4- vs Female APOEɛ4+; (E) Female APOEɛ4+ vs Female APOEɛ4-; (F) Male APOEɛ4+ vs Female APOEɛ4+; (G) Male APOEɛ4- vs Female APOEɛ4+; (H) Male APOEɛ4+ vs Female APOEɛ4-.
2.7. Machine learning model and feature selection
To derive biomarker panel for each patient group, we employed a two-step machine learning procedure consisting of quantifying the feature importance in the first step followed by training elasticnet and XGBoost models to select a subset of features from input features (Online Method-5). To evaluate the prediction accuracy of every patient-specific panel, we performed 5-fold cross-validation in each group and repeated 100 times. The prediction performance was evaluated by calculating the averaged area under the curve (AUC).
2.8. Biomarker association with clinical features
For each biomarker panel, we extracted principal components that explained >90% of the variance in data. The response variables (clinical cognitive test scores) were regressed on these principal components, and ANOVA with F-statistics were used to calculate the fitness of regression. Multiple testing was adjusted by calculating the FDR value and significance reported based on FDR<0.05 (Table S8).
3. RESULTS
3.1. Sex- and APOE-specific consensus metabolic networks identify distinct metabolic signatures and drivers of LOAD
The analytical pipeline utilized in the study is presented in Figure 1. Consensus networks provide metabolic signatures defined as subnetworks containing metabolites within 3-steps upstream of Dx node. Metabolic key drivers are the immediate (1-step) metabolite(s) upstream of Dx node in each network (Figure 2, Table S4). To investigate the common metabolic signature of LOAD, we first built a background network by using 656 AD and CN samples without patient stratification (Figure 2A). This network identified changes in six phosphatidylcholines (PCs) with PC aa C36:6 as an immediate upstream driver regardless of sex or APOE genotype (Table S4A). The DE analysis confirmed significant changes in thirteen PCs, three sphingomyelins (SMs), four acylcarnitines and citrulline (Table S6A).
To reveal sex-specific differences, we built consensus metabolic networks using residuals of 161 AD and 177 CN males and 133 AD and 185 CN females. The male consensus network (Figure 2B) identified changes in amino acids valine, isoleucine, lysine and tryptophan mediated by alpha-amino adipic acid (alpha-AAA) (Table S4B). The DE analysis confirmed increased levels of three acylcarnitines and a decrease in sarcosine, two PCs and one sphingomyelin (SM) (Table S6B). Levels of valine, a metabolite directly connected to alpha-AAA, were decreased by more than 7-fold (P = 0.13, Table S6B). The female consensus network (Figure 2C) was dominated by reduced levels of four PCs, one SM and tryptophan, and an increase in creatinine (Table S6C). These data suggest that AD was mainly associated with changes in amino acids in males, and PCs and tryptophan in females.
To define APOE-specific metabolic signatures, we built consensus networks using residuals of 193 AD and 101 CN APOEɛ4+ and 101 AD and 261 CN APOEɛ4-. The APOEɛ4+ consensus network (Figure 2D) revealed a homogeneous signature of six PCs mediated by PC aa C34:4 (Table S4D). The DE analysis confirmed significant changes in four PCs (Table S6D). The APOEɛ4- consensus network (Figure 2E) identified a mixed signature of valine, isoleucine, alpha-AAA, tryptophan, creatinine, lysine, proline, two acylcarnitines, and 26 PCs mediated by PC aa C38:0 and alpha-AAA (Table S4E). The DE analysis confirmed a significant decrease in nine PCs, four SMs, sarcosine, lysine, and valine, and an increase in creatinine and citrulline, three acylcarnitines and a pro-inflammatory agent symmetric dimethylarginine (Table S6E). These results demonstrate that APOE ɛ4 allele specifically affects the metabolism of PCs.
Next, we built male APOEε4+, male APOEε4-, female APOEε4+, and female APOEε4- consensus networks by using 107/47, 54/130, 86/54 and 47/131 AD/CN residuals, respectively (Figure 2F–2I). The male APOEɛ4+ network (Figure 2F) identified a homogeneous signature of 22 PCs and five SMs (Table S4F). The DE analysis revealed significant decrease in taurine and carnitine C7-DC, and an increase in asparagine and lyso-PC a C18:0. While not statistically significant, levels of alanine and lysine decreased by more than 8- and 3-fold, respectively, whereas levels of glycine, threonine, ornithine, and glutamate increased by more than 20-, 5-, 3- and 3-fold, respectively. The male APOEɛ4- consensus network (Figure 2G) identified changes in ten acylcarnitines, five amino acids and three lyso-PCs (Table S4G). A decrease in sarcosine and two PCs and an increase in six acylcarnitines were significant in LOAD compared to CN. While not significant, levels of branched chain amino acids (BCAA) valine and isoleucine and amino acids lysine, glutamate, isoleucine, and arginine were decreased while levels of citrulline, glycine, creatinine, alanine and taurine were increased from 2 to 13-fold (Table S4G).
The female APOEɛ4+ consensus network (Figure 2H) identified a PC-dominant signature (Table S4H). DE analysis revealed significantly decreased PCs and essential amino acid L-tryptophan. Levels of alanine and lysine decreased by more than 18- and 6-fold, respectively, whereas glycine, proline, and arginine increased by more than 6-, 4- and 4-fold, respectively, though not statistically significant. In female APOEɛ4- carriers, the consensus network discovered a mixed signature of eight SMs and three PCs (Figure 2I, Table S4I). The DE analysis revealed significantly decreased four PCs and amino acids L-tryptophan, taurine, lysine, as well as significantly increased citrulline and creatine. Summary for each group is presented in Table 2.
Table 2.
Summarize of Sex- and APOE-specific metabolic signature, key drivers and DE metabolites in each patient group.
| A. Overall.DE | Overall.KD | Overall Signature |
|---|---|---|
| C12↑ | PC aa C36:6 | PC aa C36:6 |
| C18↑ | PC ae C38:0 | |
| C18:1↑ | PC aa C34:4 | |
| C18:2↑ | PC ae C40:6 | |
| Citruline↑ | PC aa C30:0 | |
| PC aa C34:4↓ | PC aa C38:4 | |
| PC aa C36:0↓ | ||
| PC aa C36:5↓ | ||
| PC aa C36:6↓ | ||
| PC aa C38:0↓ | ||
| PC aa C38:3↓ | ||
| PC aa C38:6↓ | ||
| PC aa C40:6↓ | ||
| PC aa C42:6↓ | ||
| PC ae C36:5↓ | ||
| PC ae C38:0↓ | ||
| PC ae C38:6↓ | ||
| PC ae C40:1↓ | ||
| SM (OH) C22:1↓ | ||
| SM (OH) C22:2↓ | ||
| SM (OH) C24:1↓ |
| B. Male.DE | Male.KD | Male.Signature |
|---|---|---|
| C18↑ | alpha-AAA | alpha-AAA |
| C18:1↑ | Val | |
| C18:2↑ | Ile | |
| PC ae C36:5↓ | Lys | |
| PC ae C38:6↓ | Trp | |
| Sarcosine↓ | ||
| SM C24:0↓ |
| C. Female.DE | Female.KD | Female.Signature |
|---|---|---|
| Creatinine↑ | PC aa C36:6 | PC aa C36:6 |
| PC aa C34:4↓ | Trp | Trp |
| PC aa C36:6↓ | PC aa C30:0 | |
| PC aa C38:6↓ | PC ae C38:0 | |
| PC ae C38:0↓ | PC ae C40:6 | |
| SM (OH) C22:2↓ | PC aa C34:4 | |
| Trp↓ | PC aa C36:5 | |
| PC aa C38:6 | ||
| Tyr | ||
| C3 | ||
| Val | ||
| PC ae C30:0 | ||
| PC aa C32:1 | ||
| PC aa C32:0 | ||
| PC aa C38:0 | ||
| PC aa C40:6 | ||
| PC ae C40:1 | ||
| PC ae C42:3 | ||
| PC ae C42:2 | ||
| PC aa C42:1 | ||
| PC ae C40:5 | ||
| PC ae C38:6 | ||
| PC aa C34:3 | ||
| PC aa C40:4 | ||
| PC aa C38:4 | ||
| PC aa C38:5 | ||
| PC ae C40:4 | ||
| Ala | ||
| Asn | ||
| C4 | ||
| alpha-AAA | ||
| C0 | ||
| Ile | ||
| Lys |
| D. Male.APOE e4+.DE | Male.APOE e4+.KD | Male.APOE e4+.Signature |
|---|---|---|
| Asn↑ | PC ae C36:3 | PC ae C36:3 |
| C7-DC↓ | PC aa C40:2 | PC aa C40:2 |
| lysoPC a C18:0↑ | PC ae C34:3 | |
| Taurine↓ | PC ae C34:2 | |
| PC ae C34:1 | ||
| SM C16:0 | ||
| PC aa C42:2 | ||
| PC aa C40:3 | ||
| PC ae C32:1 | ||
| PC ae C30:0 | ||
| PC ae C36:4 | ||
| PC ae C36:2 | ||
| PC ae C38:3 | ||
| PC aa C32:0 | ||
| PC ae C34:0 | ||
| PC ae C36:1 | ||
| PC aa C36:1 | ||
| SM C24:1 | ||
| SM C24:0 | ||
| PC ae C32:2 | ||
| PC ae C40:3 | ||
| SM C16:1 | ||
| SM (OH) C14:1 | ||
| PC aa C42:1 | ||
| PC ae C42:3 | ||
| PC ae C42:1 | ||
| PC ae C42:2 |
| E. Female.APOE e4+.DE | Female.APOE e4+.KD | Female.APOE e4+.Signature |
|---|---|---|
| PC aa C30:0↓ | PC aa C34:4 | PC aa C34:4 |
| PC aa C34:4↓ | PC ae C36:4 | PC ae C36:4 |
| PC aa C38:3↓ | PC aa C30:0 | |
| Trp↓ | PC aa C34:3 | |
| PC aa C36:6 | ||
| PC aa C40:4 | ||
| PC aa C38:5 | ||
| PC ae C34:3 | ||
| PC ae C36:5 | ||
| PC ae C36:3 | ||
| PC ae C38:5 | ||
| PC ae C38:4 | ||
| PC ae C30:0 | ||
| PC aa C32:1 | ||
| PC aa C32:0 | ||
| PC ae C34:0 | ||
| PC ae C38:0 | ||
| PC aa C40:6 | ||
| PC aa C36:5 | ||
| PC aa C38:6 | ||
| PC aa C38:3 | ||
| PC ae C42:1 | ||
| PC ae C40:4 | ||
| PC aa C40:5 | ||
| PC aa C38:4 | ||
| PC ae C40:1 | ||
| PC aa C42:5 | ||
| PC ae C38:6 | ||
| PC ae C32:2 | ||
| PC ae C32:1 | ||
| PC ae C34:2 | ||
| SM C16:0 | ||
| PC ae C34:1 | ||
| PC aa C40:3 | ||
| PC ae C36:2 | ||
| PC ae C40:5 | ||
| PC ae C44:5 | ||
| SM (OH) C16:1 | ||
| C18 |
| F. Male.APOE e4-.DE | Male.APOE e4-.KD | Male.APOE e4-.Signature |
|---|---|---|
| C10↑ | C6 (C4:1-DC) | C6 (C4:1-DC) |
| C12↑ | Sarcosine | Sarcosine |
| C14:2↑ | alpha-AAA | |
| C16:1↑ | C8 | |
| C7-DC↑ | C10 | |
| C8↑ | C16:1 | |
| PC aa C38:6↓ | C10:2 | |
| PC ae C38:6↓ | C5-DC (C6-OH) | |
| Sarcosine↓ | C12 | |
| C14:1 | ||
| C18:1 | ||
| lysoPC a C16:0 | ||
| lysoPC a C18:0 | ||
| C9 | ||
| lysoPC a C17:0 | ||
| Glu | ||
| Val | ||
| Ile |
| G. Female.APOE e4-.DE | Female.APOE e4-.KD | Female.APOE e4-.Signature |
|---|---|---|
| Citrulline↑ | SM C26:0 | SM C26:0 |
| Creatinine↑ | SM (OH) C22:1 | |
| Lysine↓ | SM C26:1 | |
| PC aa C38:0↓ | SM (OH) C24:1 | |
| PC aa C38:6↓ | SM C24:0 | |
| PC aa C40:6↓ | SM (OH) C22:2 | |
| PC ae C38:0↓ | SM C24:1 | |
| Taurine↓ | PC ae C40:3 | |
| Trp↓ | PC ae C40:2 | |
| SM (OH) C16:1 | ||
| PC ae C44:5 |
3.3. Heterogeneity in patient group-specific metabolic key drivers
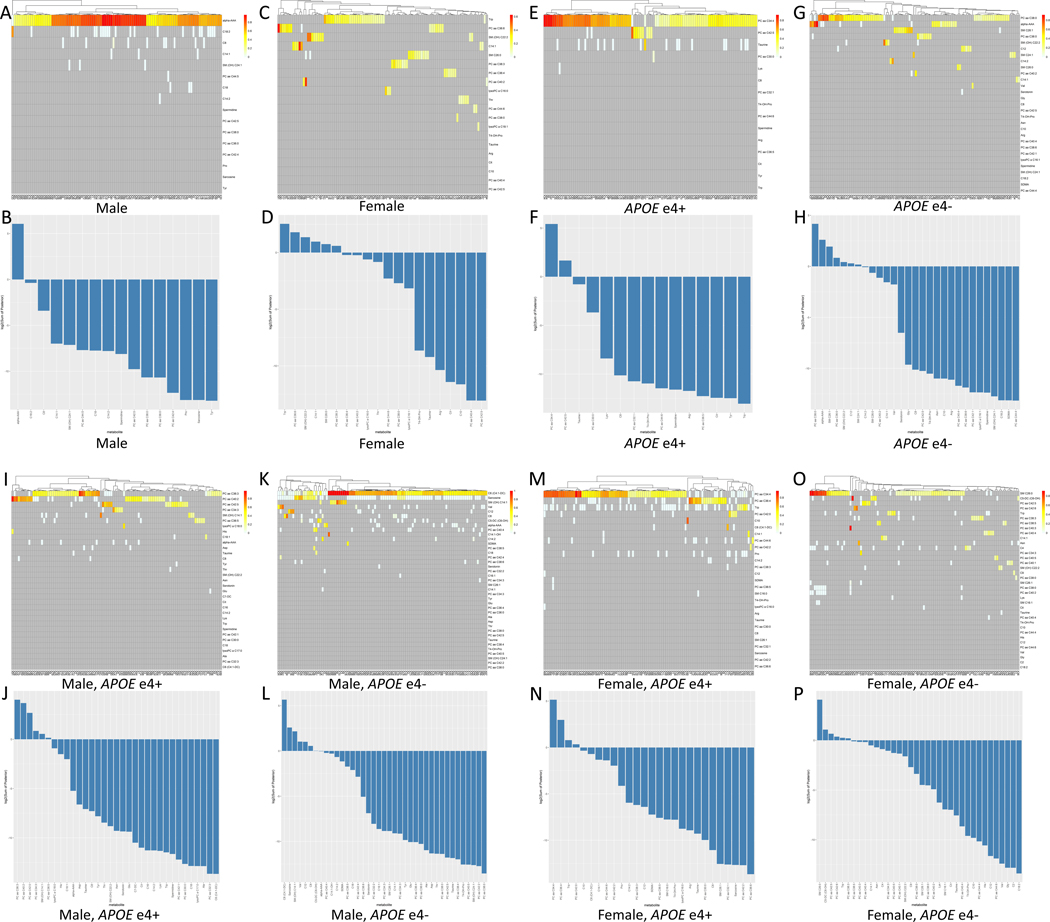
The consensus network analysis revealed distinct patterns of homogeneity/heterogeneity in the metabolic signatures within each patient group. While this network captures the most robust signal relative to all individuals in each patient group, it doesn’t address the endogenous metabolic heterogeneity associated with different subpopulations within the same group. To further investigate inherent metabolic heterogeneity, we calculated the significance (FDR adjusted) of robustness and confidence for every key driver in each group (Table S5). Robustness of metabolic drivers was determined based on their connection to Dx with positive 95% CI, significant robustness, and confidence. Despite multiple potential metabolic drivers identified in each subpopulation group, the following metabolites robustly connected to Dx: males: alpha-AAA; females: PC aa C36:6 and tryptophan; APOEɛ4+: PC aa C34:4; APOEɛ4-: PC aa C38:0, alpha-AAA, and serotonin; male APOEɛ4+: PC ae C36:3 and PC aa C40:2; male APOEɛ4-: C6 and sarcosine; female APOEɛ4+: PC aa C34:4, PC ae C36:4 and L-tryptophan; and female APOEɛ4-: SM C26:0 (Figure 3, Table 2, Table S5).
Figure 3. Sex- and APOE-specific metabolic heterogeneity.
In each group, the confidence and robustness of metabolic drivers were shown in the heatmap where the X-axis represents 100 different networks and Y-axis represents candidate key drivers in 100 networks. Each row in the heatmap represents a vector of 100 posterior probability values of the edge from a key driver to Dx derived from 100 networks, and the bar plot is ranked based on the log2 of the sum of the 100 posterior values per key driver in male (A,B), female (C,D), APOEɛ4+ (E,F), APOEɛ4- (G,H), male APOEɛ4+ (I,J), male APOEɛ4- (K,L), female APOEɛ4+ (M,N), female APOEɛ4- (O,P).
3.4. Metabolic network cross-validation using sex- and APOE-specific biomarker panel
To validate metabolic networks and key drivers, we trained an ensemble of machine learning models to select a subset of metabolites based on the network model and drivers with changes significantly associated with the disease state in each patient group. The prediction performance was evaluated with averaged AUC by cross-validation in the ADNI data (Figure 5). In each patient group, we trained different models and compared AUCs of each model with eight sets of input features. Set 1: all 127 metabolites; Set 2: significant DE metabolites; Set 3: network-derived metabolites; Set 4: combination of all 127 metabolites plus age, education, BMI; Set 5: significant DE metabolites plus demographics; Set 6: network-derived metabolites plus age, education length, BMI; Set 7: network-derived metabolites plus significant DE metabolites; Set 8: network-derived metabolites plus significant DE metabolites and age, education, BMI. The network-derived metabolites were extracted from the neighbor (within 3-step undirected) subnetwork of the Dx node in respectful networks.
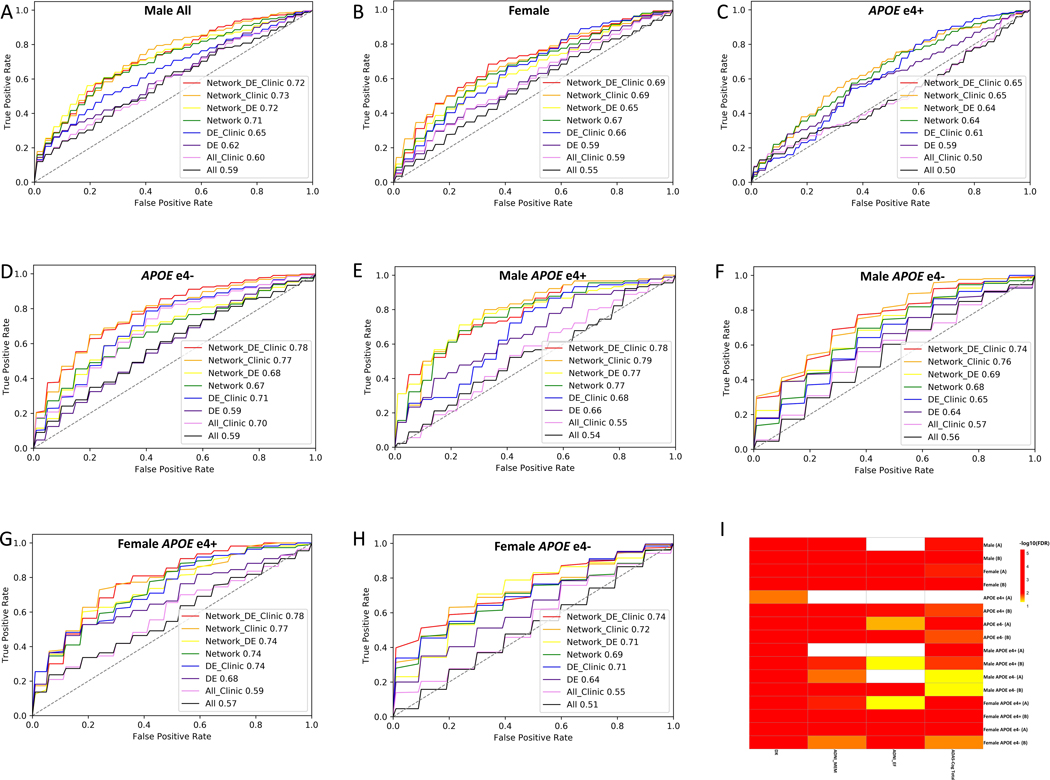
Figure 5. Biomarker panel and cross-validation accuracy for AD diagnosis.
The prediction performance of diagnostic biomarker panels derived from different sets of features are compared in each patient group, The number in the figure represents the averaged cross-validation AUC with 8 feature sets respectively in male (A), female (B), APOEɛ4+ (C), APOEɛ4- (D), male APOEɛ4+ (E), male APOEɛ4- (F), female APOEɛ4+ (G), male APOEɛ4- (H); All, all 127 metabolites in the data; All Clinic, all 127 metabolites in the data combines with age, BMI and/or education; DE: significant DE metabolites; DE Clinic: significant DE metabolites combined with age, BMI and/or education; Network: biomarkers derived from metabolic network; Network_DE: biomarkers derived from the combination of significant DE and metabolic network; Network_DE_Clinic: biomarkers derived from combination of significant DE metabolites, metabolic network and age, BMI, and/or education. Network_Clinic: biomarkers derived from metabolic network and age, BMI and/or education; (I) Two selected optimal biomarker panel association with clinical assessment and cognitive decline: The association of the two selected biomarker panel with and without Age, BMI and Education in each patient group (A: biomarker pane derived from metabolic network; B: biomarker pane derived from combination metabolic network plus age, BMI and/or education) with diagnosis (Dx) and clinical assessments (ADAS-Cog Total Score, memory function (ADNI_MEM) and executive function (ADNI_EF)).
We found that the prediction accuracy (AUC) of the network-derived metabolites (Set 3, Figure 5 green line) robustly and significantly outperformed those predicted by using all metabolites in the data (Set 1, Figure 5 black line) and only significant (P<0.05) DE metabolites derived from the linear regression model (Set 2, Figure 5 purple line) across all patient groups. Adding patient demographics to Sets 1 and 3 greatly improved individual prediction accuracy. However, the same pattern was observed in their relative accuracy, i.e., the AUC produced by the network-derived metabolite with patient demographics (Set 6, Figure 5 orange line) consistently outperformed the accuracy predicted by adding demographics to either all metabolites in the data (Set 4, Figure 5 pink line) or only significant (P<0.05) DE metabolites (Set 5, Figure 5 blue line) across all patient groups. When significant DE metabolites were added to the combination of network-derived features with patient demographics (Set 8, Figure 5 red line), a marginal improvement in AUC was observed in all APOEɛ4-, female APOɛ4+ and female APOEɛ4- groups, with a slight decrease in AUC was observed in males, male APOEɛ4+, male APOEɛ4-, suggesting that significant DE metabolites derived from linear regression added no further power in prediction of Dx given the network structure and patient demographics. Data suggest that the metabolic network-derived features with or without age, BMI, and education significantly improved the prediction accuracy compared to the other feature sets, DE metabolites and all 127 metabolites in the data, across all patient groups. These data indicate that the predictive network model utilized in this study is more sensitive than traditional regression method in detecting weaker relations of metabolic changes with disease state in sex- and APOE-specific patient groups. The best performing feature set with or without demographics, respectively, was selected for each group as the biomarker panel (Table S7).
3.5. Blood-based metabolic biomarker panels associate with cognitive decline
To evaluate the association of selected biomarker panels with clinical cognitive assessment (diagnosis, ADAS-Cog score, memory, and executive function), we calculated the eigen expression to recapitulate the primary variance component (the first principal component) for each panel and fitted a linear regression model between the eigen expression of the 1st principal component and cognitive measures. Significance of association is shown in Figure 5I (Table S8). Network-derived biomarker panels for each patient group with or without demographics were all significantly associated with the Dx. Of 16 panels (Table S7, two selected panels per group times 8 patient groups), 13 and 15 panels were significantly associated with memory (ADNI_MEM) and the overall cognition (ADAS-Cog Total Score), respectively. Of 16 panels, 10 were significantly associated with the executive function composite score (ADNI_EF). Two biomarker panels approached statistical significance with the executive function composite score: network-derived features with demographics for male APOEɛ4+ (FDR=0.0667) and network-derived features without demographic for female APOEɛ4+ (FDR=0.0678). Two biomarker panels approached statistical significance with overall cognition score: network-derived features with (FDR=0.0655) and without (FDR=0.0667) demographics. These results indicate that patient-specific metabolic networks and network-derived biomarker panels are associated with clinical cognitive assessment.
4. Discussion
Using advanced computational method, we demonstrated that LOAD is associated with metabolomic profiles defined by sex and APOE genotype. Based on patient group-specific network models, we identified key drivers, differentially produced metabolites and metabolic signatures of the disease. Unstratified analyses identified changes in lipid homeostasis, with carnitines, PCs and SMs as most affected metabolites that differentiated AD from CN. Further stratification by sex revealed that metabolic changes in AD males were associated with amino acids while lipids remained predominantly affected in females. Stratification by sex and APOE did not affect lipid-dominant metabolic signatures in females while in APOEɛ4+ males, metabolic drivers and signatures changed from amino acids, especially BCAAs, to lipids comparable to APOEɛ4+ females. In APOEɛ4- males and females, metabolic changes were more diverse compared to APOEɛ4+ and included lipids and amino acids. The identified metabolic alterations are consistent with previous reports where changes in blood levels of PCs, SMs, acylcarnitines, ceramides, and amino acids differentiated MCI and AD from CN [11, 14, 18, 19, 27–30].
In addition to replicating sex-specific differences in serum metabolites associated with AD [14], our analyses also generated multiple novel and important findings by identifying metabolic signatures and key drivers in patient groups stratified by the intersection of sex and APOE genotype, which has not been previously reported. We demonstrate that i) previously observed amino acid-centered metabolic signatures and drivers in AD males are true for males without APOEɛ4; (ii) metabolic signatures and drivers for APOEɛ4+ males shifted from amino acids to lipids (PCs and SMs) similar to changes observed in APOEɛ4+ females. This important finding highlights the ability of APOEɛ4 genotype to signifinatly influence metabolic changes overriding sex-specific differences observed in serum metabolites in AD males and females. (iii) The metabolic shift is more subtle between APOEɛ4+ and APOEɛ4- females, which is confined to different lipid species, i.e., the shift from a SM-dominant metabolic signatures and drivers in APOEɛ4- to a PC-dominant signatures and drivers in APOEɛ4+. Overall, our novel findings indicate that APOEɛ4 genotype drives metabolic signature to a phosphatidylcholine-focused profile regardless of the patient sex.
The replicative validity and novelty of our findings emphasizes the importance of these metabolic pathways for AD. We identified changes in numerous individual lipids, including PCs and lysoPCs, as key drivers or components of metabolic signatures associated with cognition. PCs and SMs are integral constituents of the plasma membrane. The reduced PC levels observed in AD may reflect abnormal membrane functions including synaptic transmission and processing of the amyloid precursor protein contributing to Aβ production [31]. Furthermore, alterations in PCs may contribute to increased inflammation, one of the underlying mechanisms of LOAD [32, 33]. The panel of PCs and carnitines predicted the conversion from CN to AD/aMCI with sensitivity and specificity of 90% [27, 34] yielding improvements to previous reports where stratification was not used [35–37]. L-Carnitine and acylcarnitines play an essential role in energy metabolism transporting activated long-chain fatty acids into mitochondria for β-oxidation. They also mediate the metabolism of BCAAs, neuromodulation, antioxidant and anti-apoptotic functions in the brain [38, 39]. Consistent with our findings, changes in multiple carnitines (e.g., C12, C12:1, C14:1 and C8) contributed to discriminating AD from CN [40–42]. A recent study with the same metabolomics data conducted in both ante-mortem blood and post-mortem brain samples in two community-based longitudinal aging and dementia cohorts reported that decanoylcarnitine C10, pimelylcarnitine C7-DC, and tetradecadienylcarnitine C14:2 significantly predicted a lower AD risk after a 4.5-year follow-up, independent of age, sex, and education [34]. However, the most important changes in carnitines and amino acids detected in AD patients associate with sex-specific dysregulation of energy metabolism [2, 43].
Altered glucose uptake in the brain detected using FDG-PET occurs decades before onset of AD symptoms, suggesting that metabolic deficits are an upstream event specific to LOAD [43, 44]. Thus, changes in carnitines, fatty acids and amino acids, BCAA in particular, may indicate differential compensatory mechanisms for alternative energy substrates in AD males and females [45, 46]. High levels of carnitines may indicate a buildup of fatty acids, suggesting increased energy demands coupled with impaired energy production via mitochondrial β-oxidation [46]. Male-specific metabolic signatures identified herein included alpha-AAA and BCAA valine and isoleucine. BCAAs are important energy carrying molecules associated with cognitive decline and brain atrophy in AD [47]. Changes in their levels could indicate a switch to increased energy consumption via degradation of amino acids. The biogenic amine alpha-AAA is a degradation product of lysine and is involved in mechanisms of neurotransmission [48, 49]. Higher levels of serum alpha-AAA are associated with decreased cognitive function [5]. Consistent with previous observations, we detected positive associations of AD cognitive function with multiple amino acids, including tryptophan, citrulline, sarcosine, aspartic acid, and taurine [5, 11, 14, 18–20].
Our study has several limitations. First, the AbsoluteIDQ-p180 system is a targeted metabolomic platform with limited set of metabolites, including amino acids (21), biogenic amines (21), hexose (1), acylcarnitines (40), lysophosphatidylcholines (14), phosphatidylcholines (76), and sphingolipids (15). Utilization of this platform enabled a direct comparison of the results reported herein, generated using advanced analytical computational analysis, to previously generated findings using the same platform[14]. Our novel computational systems biology approach enabled findings that strongly support utility of the targeted metabolomic biomarker translational approach for individualized medicine. Future large-scale metabolomics analyses could provide greater detail that support the metabolic pathways reported herein while also identifying additional pathways. From a translational perspective the replication of affected pathways using the targeted metabolomic platform coupled with a novel systems biology computational approach enabled identification of sex, APOE genotype and AD stage specific phenotypes. These findings provide the foundation for personalized therapeutic interventions and simultaneously a biomarker strategy to determine target engagement and therapeutic efficacy. Second, the metabolic data are inherently susceptible to environmental influences and personal factors. Such variability is further amplified in stratified analyses like ours, where although starting with thousands of patients, stratification reduces group size resulting in smaller detection power. Thus, most of significant DE didn’t survive multiple-testing correction. Therefore, potentially important findings could be missed by using conventional analytical methods such as DE and linear regression. We addressed this problem by utilizing a more sensitive network model than conventional methods. Our network model exploited the conditional independence derived from the robust covariance structure to overcome the relatively small effect-size in metabolic data due to random noise and small detection power due to reduced number of patients by stratification, which is not well handled by linear regression and correlation-based methods[14]. Herein, we demonstrated that our network approach is more robust and sensitive for detecting true associations over conventional methods. This may explain why previous studies have not discovered that APOEɛ4 status overrides sex-specific difference in serum AD metabolites though a similar effect was observed in a recent study with humanized APOE mice [30]. Although our network approach to some extent can mathematically alleviate the issue of low detection power, the number of subjects in each group was relatively small and studies in larger longitudinal cohorts are warranted to confirm these results. Our Alzheimer Disease Metabolomics Consortium (ADMC) is conducting comprehensive metabolic profiling across metabolomic platforms to provide broad biochemical coverage of the metabolome to map metabolic failures across trajectory of disease.
In summary, we provide a compelling systems biology analytical platform for metabolomics data analysis. We identified sex- and APOE-specific metabolic signatures associated with clinical diagnosis and cognitive assessment and key metabolic drivers that could be evaluated as therapeutic targets with a potential to shift the trajectory of the disease. The metabolic signatures and key drivers demonstrated clear metabolic differences in sex and APOE genotype and highlighted the potential of APOEɛ4 genotype overriding sex-difference in human serum metabolic associated with AD. In addition, we identified serum metabolic panels significantly associated with clinical diagnosis and cognitive assessment in each patient subgroup. This is the first study to establish patient-specific serum metabolic biomarkers predictive of disease diagnosis that significantly associated with clinical cognitive assessment for individual groups of patients stratified by sex and APOE genotype (Figure 5I, Table S9). Based on the biomarker panel of network-derived metabolites and demographic features, we identified that education attainment and BMI are two most common biomarkers shared by 5 out of 8 patient groups, followed by tryptophan (4 out of 8), a set of PCs (PC aa C42:6, PC ae C36:5, PC ae C40:2, PC ae C42:5, PC ae C36:0) and age (3 out of 8). Interestingly, we identified valine, creatinine, lysine, C16, SM C26:1, lysoPC a C16:1, lysoPC a C18:0, lysoPC a C18:2, lysoPC a C20:3, PC aa C32:1, PC ae C38:5, PC ae C42:3 as unique markers for males; alpha-AAA and sarcosine as specific markers for APOEɛ4- males; C14:1-OH, PC aa C40:3, PC ae C30:0, SM (OH) C22:1 and taurine as unique markers for APOEɛ4+ males; PC aa C34:4 as a specific marker for APOEɛ4- females; and PC aa C38:0 as a specific marker for APOEɛ4+ females.
Our study provides an initial but critical step towards developing personalized and precision medicine for AD and an operational strategy to achieve that goal, which integrates clinical cognitive assessment, metabolomic profiling, and computational network model to identify targeted therapeutic strategies for subsets of patients.
Supplementary Material
Table S1. Summary of 127 metabolites used in this study.
Table S2. Sample summary of sex- and APOE-specific patient groups.
Table S4. Summary of sex- and APOE-specific metabolic signature and key drivers derived from the network in each patient group.
Table S5. Robustness and confidence of key drivers in each patient group.
Table S3. Summary of sex- and APOE-specific metabolic network in each patient group.
Table S7. Summary of metabolic biomarker panel in each patient group.
Table S8. Summary of biomarker metabolites association with Dx, clinical cognition assessment. The values are FDR values.
Table S6. Summary of sex- and APOE-specific DE signatures.
Table S9. Summary of common and specific biomarkers across 8 patient groups.
Acknowledgement
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12–2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
This research was supported by grants from the National Institutes of Health and partnership with Alzheimer’s Disease Metabolomics Consortium (ADMC) [grant numbers RF1AG057457 and R56AG062620 to R.C., RF1AG059093 to R.B., R.C. and R.K., R01AG057931 to R.B. and R.C, RF1AG55549, R01NS107265 and R01AG062135 to E.T.]. The results published here are in whole or in part based on data obtained from the AD Knowledge Portal ( https://adknowledgeportal.org ). Metabolomics data is provided by the Alzheimer’s Disease Metabolomics Consortium (ADMC) and funded wholly or in part by the following grants and supplements thereto: NIA R01AG046171, RF1AG051550, RF1AG057452, R01AG059093, RF1AG058942, U01AG061359, U19AG063744 and FNIH: #DAOU16AMPA awarded to Dr. Kaddurah-Daouk at Duke University in partnership with a large number of academic institutions. As such, the investigators within the ADMC, not listed specifically in this publication’s author’s list, provided data along with its pre-processing and prepared it for analysis, but did not participate in analysis or writing of this manuscript. A complete listing of ADMC investigators can be found at: https://sites.duke.edu/adnimetab/team/.
We thank Drs. Matthias Arnold (MA) and Gabi Kasternmuller (GK) from Institute of Bioinformatics and Systems Biology, Helmholtz Zentrum München for their discussions. Additionally, MA, R.K-D, and GK are supported by NIA grants RF1 AG058942 and R01 AG057452. MA and GK are also supported by funding from Qatar National Research Fund NPRP8-061-3-011. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
R.C. is the founder of INTelico Therapeutics LLC and co-founder of PATH Biotech LLC. RKD in an inventor on a series of patents on use of metabolomics for the diagnosis and treatment of CNS and ther diseases and holds founding equity in Metabolon Inc., Chymia LLC and PsyProtix. This study was not supported by any of above companies.
List of Abbreviations
- APOE
apolipoprotein E
- ADNI
Alzheimer’s Disease Neuroimaging Initiative
- ADMC
Alzheimer Disease Metabolomics Consortium
- LOAD
Late-onset Alzheimer’s disease
- APOEε4
ε4 allele of the Apolipoprotein E (APOE)
- AD
Alzheimer’s Disease
- Aβ
amyloid beta
- CN
cognitively normal
- CSF
cerebrospinal fluid
- MCI
mild cognitive impairment
- Aβ1–42
amyloid beta peptide 1–42
- p-tau
hyperphosphorylated-tau protein
- FDG-PET
fluorodeoxyglucose-positron emission tomography
- Dx
disease diagnosis
- BMI
body mass index
- BN
Bayesian network
- CI
confidence interval
- XGBoost
eXtreme Gradient Boosting
- AUC
area under curve
- ANOVA
analysis of variance
- FDR
false discovery rate
- PC
phosphatidylcholine
- DE
Differential Expression
- SM
sphingomyelin
- alpha-AAA
alpha-amino adipic acid
- APOEɛ4-
APOEε4 negative/non-carrier
- APOEɛ4+
APOEε4 positive/carrier
- C7-DC
Pimelylcarnitine
- lyso-PC
Lysophosphatidylcholine
- BCAA
branched chain amino acid
- ADAS-Cog
The Alzheimer’s Disease Assessment Scale–Cognitive Subscale
- ADNI_MEM
Alzheimer’s Disease Neuroimaging Initiative Memory Test Score
- ADNI_EF
Alzheimer’s Disease Neuroimaging Initiative Executive Function Test Score
- aMCI
amnestic mild cognitive impairment
Footnotes
Competing interest statement
The other authors declare no competing interests.
References
- 1.Jansen IE, et al. , Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet, 2019. 51(3): p. 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo L, et al. , Sex Differences in Alzheimer’s Disease: Insights From the Multiomics Landscape. Biol Psychiatry, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Talwar P, et al. , Dissecting Complex and Multifactorial Nature of Alzheimer’s Disease Pathogenesis: a Clinical, Genomic, and Systems Biology Perspective. Mol Neurobiol, 2016. 53(7): p. 4833–64. [DOI] [PubMed] [Google Scholar]
- 4.Zhang B, et al. , Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell, 2013. 153(3): p. 707–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkins JM and Trushina E, Application of Metabolomics in Alzheimer’s Disease. Front Neurol, 2017. 8: p. 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niedzwiecki MM, et al. , High-resolution metabolomic profiling of Alzheimer’s disease in plasma. Ann Clin Transl Neurol, 2020. 7(1): p. 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trushina E, et al. , Identification of altered metabolic pathways in plasma and CSF in mild cognitive impairment and Alzheimer’s disease using metabolomics. PLoS One, 2013. 8(5): p. e63644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han X, et al. , Metabolomics in early Alzheimer’s disease: identification of altered plasma sphingolipidome using shotgun lipidomics. PLoS One, 2011. 6(7): p. e21643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaddurah-Daouk R, et al. , Metabolomic changes in autopsy-confirmed Alzheimer’s disease. Alzheimers Dement, 2011. 7(3): p. 309–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaddurah-Daouk R, et al. , Alterations in metabolic pathways and networks in Alzheimer’s disease. Transl Psychiatry, 2013. 3: p. e244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toledo JB, et al. , Metabolic network failures in Alzheimer’s disease: A biochemical road map. Alzheimers Dement, 2017. 13(9): p. 965–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nho K, et al. , Altered bile acid profile in mild cognitive impairment and Alzheimer’s disease: Relationship to neuroimaging and CSF biomarkers. Alzheimers Dement, 2019. 15(2): p. 232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nho K, et al. , Association of Altered Liver Enzymes With Alzheimer Disease Diagnosis, Cognition, Neuroimaging Measures, and Cerebrospinal Fluid Biomarkers. JAMA Netw Open, 2019. 2(7): p. e197978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold M, et al. , Sex and APOE epsilon4 genotype modify the Alzheimer’s disease serum metabolome. Nat Commun, 2020. 11(1): p. 1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernath MM, et al. , Serum triglycerides in Alzheimer disease: Relation to neuroimaging and CSF biomarkers. Neurology, 2020. 94(20): p. e2088–e2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huynh K, et al. , Concordant peripheral lipidome signatures in two large clinical studies of Alzheimer’s disease. Nat Commun, 2020. 11(1): p. 5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baloni P, et al. , Metabolic Network Analysis Reveals Altered Bile Acid Synthesis and Metabolism in Alzheimer’s Disease. Cell Rep Med, 2020. 1(8): p. 100138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nho K, et al. , Serum metabolites associated with brain amyloid beta deposition, cognition and dementia progression. Brain Commun, 2021. 3(3): p. fcab139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horgusluoglu E, et al. , Integrative metabolomics-genomics approach reveals key metabolic pathways and regulators of Alzheimer’s disease. Alzheimers Dement, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Batra R, et al. , The landscape of metabolic brain alterations in Alzheimer’s disease. bioRxiv, 2021: p. 2021.11.15.468698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang R and Schadt E, Patent US2019019864. SYSTEMS AND METHODS FOR PREDICTIVE NETWORK MODELING FOR COMPUTATIONAL SYSTEMS, BIOLOGY AND DRUG TARGET DISCOVERY 2019. [Google Scholar]
- 22.Petyuk VA, et al. , The human brainome: network analysis identifies HSPA2 as a novel Alzheimer’s disease target. Brain, 2018. 141(9): p. 2721–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carcamo-Orive I, et al. , Analysis of Transcriptional Variability in a Large Human iPSC Library Reveals Genetic and Non-genetic Determinants of Heterogeneity. Cell Stem Cell, 2017. 20(4): p. 518–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carcamo-Orive I, et al. , Predictive network modeling in human induced pluripotent stem cells identifies key driver genes for insulin responsiveness. PLoS Comput Biol, 2020. 16(12): p. e1008491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruti Rajan Patel KZ, Marc Y.R. Henrion, Beckmann Noam D., Moein Sara, Alamprese Melissa L., Allen Mariet, Wang Xue, Chan Gail, Pertel Thomas, Nejad Parham, Reddy Joseph S., Carrasquillo Minerva M., Bennett David A, Ertekin-Taner Nilüfer, De Jager Philip L., Schadt# Eric E., Bradshaw# Elizabeth M., Chang# Rui, Single Cell-type Integrative Network Modeling Identified Novel Microglial-specific Targets for the Phagosome in Alzheimer’s disease. bioRxiv, 2020. [Google Scholar]
- 26.Ritchie ME, et al. , limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res, 2015. 43(7): p. e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mapstone M, et al. , Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med, 2014. 20(4): p. 415–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klavins K, et al. , The ratio of phosphatidylcholines to lysophosphatidylcholines in plasma differentiates healthy controls from patients with Alzheimer’s disease and mild cognitive impairment. Alzheimers Dement (Amst), 2015. 1(3): p. 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oeckl P and Otto M, A Review on MS-Based Blood Biomarkers for Alzheimer’s Disease. Neurol Ther, 2019. 8(Suppl 2): p. 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shang Y, et al. , Evidence in support of chromosomal sex influencing plasma based metabolome vs APOE genotype influencing brain metabolome profile in humanized APOE male and female mice. PLoS One, 2020. 15(1): p. e0225392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kosicek M and Hecimovic S, Phospholipids and Alzheimer’s disease: alterations, mechanisms and potential biomarkers. Int J Mol Sci, 2013. 14(1): p. 1310–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Che H, et al. , Comparative study of the effects of phosphatidylcholine rich in DHA and EPA on Alzheimer’s disease and the possible mechanisms in CHO-APP/PS1 cells and SAMP8 mice. Food Funct, 2018. 9(1): p. 643–654. [DOI] [PubMed] [Google Scholar]
- 33.Hur JY, et al. , The innate immunity protein IFITM3 modulates gamma-secretase in Alzheimer’s disease. Nature, 2020. 586(7831): p. 735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huo Z, et al. , Brain and blood metabolome for Alzheimer’s dementia: findings from a targeted metabolomics analysis. Neurobiol Aging, 2020. 86: p. 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casanova R, et al. , Blood metabolite markers of preclinical Alzheimer’s disease in two longitudinally followed cohorts of older individuals. Alzheimers Dement, 2016. 12(7): p. 815–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li D, et al. , Plasma phospholipids and prevalence of mild cognitive impairment and/or dementia in the ARIC Neurocognitive Study (ARIC-NCS). Alzheimers Dement (Amst), 2016. 3: p. 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li D, et al. , Prospective associations of plasma phospholipids and mild cognitive impairment/dementia among African Americans in the ARIC Neurocognitive Study. Alzheimers Dement (Amst), 2017. 6: p. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones LL, McDonald DA, and Borum PR, Acylcarnitines: role in brain. Prog Lipid Res, 2010. 49(1): p. 61–75. [DOI] [PubMed] [Google Scholar]
- 39.Mihalik SJ, et al. , Increased levels of plasma acylcarnitines in obesity and type 2 diabetes and identification of a marker of glucolipotoxicity. Obesity (Silver Spring), 2010. 18(9): p. 1695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciavardelli D, et al. , Medium-chain plasma acylcarnitines, ketone levels, cognition, and gray matter volumes in healthy elderly, mildly cognitively impaired, or Alzheimer’s disease subjects. Neurobiol Aging, 2016. 43: p. 1–12. [DOI] [PubMed] [Google Scholar]
- 41.Cristofano A, et al. , Serum Levels of Acyl-Carnitines along the Continuum from Normal to Alzheimer’s Dementia. PLoS One, 2016. 11(5): p. e0155694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzalez-Dominguez R, et al. , Metabolomic profiling of serum in the progression of Alzheimer’s disease by capillary electrophoresis-mass spectrometry. Electrophoresis, 2014. 35(23): p. 3321–30. [DOI] [PubMed] [Google Scholar]
- 43.Cunnane SC, et al. , Brain energy rescue: an emerging therapeutic concept for neurodegenerative disorders of ageing. Nat Rev Drug Discov, 2020. 19(9): p. 609–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mosconi L, Glucose metabolism in normal aging and Alzheimer’s disease: Methodological and physiological considerations for PET studies. Clin Transl Imaging, 2013. 1(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lynch CJ and Adams SH, Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol, 2014. 10(12): p. 723–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma S and Black SM, Carnitine Homeostasis, Mitochondrial Function, and Cardiovascular Disease. Drug Discov Today Dis Mech, 2009. 6(1–4): p. e31–e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tynkkynen J, et al. , Association of branched-chain amino acids and other circulating metabolites with risk of incident dementia and Alzheimer’s disease: A prospective study in eight cohorts. Alzheimers Dement, 2018. 14(6): p. 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang YE, Lysine metabolism in the rat brain: the pipecolic acid-forming pathway. J Neurochem, 1978. 30(2): p. 347–54. [DOI] [PubMed] [Google Scholar]
- 49.Guidetti P and Schwarcz R, Determination of alpha-aminoadipic acid in brain, peripheral tissues, and body fluids using GC/MS with negative chemical ionization. Brain Res Mol Brain Res, 2003. 118(1–2): p. 132–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of 127 metabolites used in this study.
Table S2. Sample summary of sex- and APOE-specific patient groups.
Table S4. Summary of sex- and APOE-specific metabolic signature and key drivers derived from the network in each patient group.
Table S5. Robustness and confidence of key drivers in each patient group.
Table S3. Summary of sex- and APOE-specific metabolic network in each patient group.
Table S7. Summary of metabolic biomarker panel in each patient group.
Table S8. Summary of biomarker metabolites association with Dx, clinical cognition assessment. The values are FDR values.
Table S6. Summary of sex- and APOE-specific DE signatures.
Table S9. Summary of common and specific biomarkers across 8 patient groups.