Abstract
Muller’s ratchet predicts fitness losses in small populations of asexual organisms because of the irreversible accumulation of deleterious mutations and genetic drift. This effect should be enhanced if population bottlenecks intervene and fixation of mutations is not compensated by recombination. To study whether Muller’s ratchet could operate in a retrovirus, 10 biological clones were derived from a human immunodeficiency virus type 1 (HIV-1) field isolate by MT-4 plaque assay. Each clone was subjected to 15 plaque-to-plaque passages. Surprisingly, genetic deterioration of viral clones was very drastic, and only 4 of the 10 initial clones were able to produce viable progeny after the serial plaque transfers. Two of the initial clones stopped forming plaques at passage 7, two others stopped at passage 13, and only four of the remaining six clones yielded infectious virus. Of these four, three displayed important fitness losses. Thus, despite virions carrying two copies of genomic RNA and the system displaying frequent recombination, HIV-1 manifested a drastic fitness loss as a result of an accentuation of Muller’s ratchet effect.
Genetic variation provides the background on which evolution acts in living organisms. RNA viruses display extreme genetic variation (13, 15, 25, 47) which, although it is an energetically inefficient process, offers clear adaptive advantages. Variation in RNA viruses is the result of an error-prone replication machinery and of the lack of repair and proofreading mechanisms (6, 12–14, 47). Variant genomes are continuously arising in any replicating viral population, forming a complex swarm of related genomes termed quasispecies (18) which are subjected to positive and negative selective forces, as well as to other nonselective mechanisms such as genetic drift or random sampling events like genetic bottlenecks.
Retroviruses display unique features in their replication cycles that have implications for genome variation. One is the need for reverse retrotranscription of the genomic viral RNA, an error-prone process (3). Another is the presence of two copies of the genomic RNA per virus particle, in what has been termed pseudodiploidy (27). The process of retrotranscription, with the requirement of enzyme strand transfers, provides the mechanistic basis for a high recombination potential. However, an important source of genetic variation in human immunodeficiency virus type 1 (HIV-1) results from point mutations, which have been identified in the emergence of mutants resistant to retroviral inhibitors (30, 34, 43), or in the nonsyncytial-to-syncytial switch, which occurs in the course of infection (20, 29) and is associated with changes in coreceptor usage (42).
RNA viruses are increasingly recognized as useful models to test concepts and theoretical predictions of molecular evolution. Some advantages of RNA viruses as model systems include their short duplication times, small genomes, and high rates of genetic variation and the possibility of using large population sizes. Among the principles of population biology which have been addressed with RNA viruses are the Red Queen hypothesis and the competitive exclusion principle, which were tested by using vesicular stomatitis virus (VSV) (5, 12). The first principle describes fitness gains in two variants when they are cocultured in the same environment, although one overgrows the other. The more general competitive exclusion principle states that when two organisms coexist in the same environment with limited resources, one will eventually overgrow the other (reviewed in reference 12). Another prediction of classical population genetics that found ample confirmation with RNA viruses is the operation of Muller’s ratchet (33). In its initial formulation it recognized the tendency of populations of asexual organisms to lose fitness due to the accumulation of deleterious mutations which could not be compensated by sex or recombination (33). The Muller’s ratchet effect was experimentally documented for the first time with bacteriophage φ6 by using an experimental design that involved serial plaque-to-plaque transfers (4) and then with VSV (17), foot-and-mouth disease virus (FMDV) (19), and bacteria (1). The plaque transfers, which represent severe bottleneck events, produced in all of these cases decreases in average fitness, some of which were very intense.
HIV-1 is currently one of the most thoroughly studied human pathogens. Although many in vivo studies have been performed on HIV-1 evolution (reviewed in references 7 and 44), studies on the in vitro evolution of HIV-1 in cell culture are scarce. HIV-1 is an attractive model to analyze the possible operation of Muller’s ratchet for theoretical and practical reasons. The HIV-1 virion contains two copies of the genome, and the virus displays high recombination rates (9, 11, 26, 28, 40) which, from a theoretical point of view, could potentially counteract the debilitating effects of Muller’s ratchet on virus fitness (4, 6, 17, 32, 33, 35, 36). Also, HIV-1 infection has a diverse natural history, with different routes of infection (by sexual transmission, by vertical transmission, or by infected blood or blood products), and these routes of infection may often involve different infecting doses (small in sexual transmission, larger in drug user transmission, and probably even larger in transfusion-associated cases) resulting, in some cases, in bottlenecks. In addition, infected individuals show different patterns of disease progression. In this background, it is interesting to study how genetic bottlenecks may affect fitness values of HIV-1, a problem that has not been approached experimentally with retroviruses.
In the present study, fitness evolution was analyzed by using 10 biological clones derived from an HIV-1 isolate. Each HIV-1 clone was subjected to 15 serial plaque-to-plaque transfers, taking advantage of a plaque assay in MT-4 cells (22). The results document a high fitness heterogeneity among the initial clones and strong fitness losses in the viruses subjected to repeated bottleneck passages, to the point that 6 of the 10 initial clones did not survive through 15 bottleneck transfers.
MATERIALS AND METHODS
Cells, viruses, and biological cloning.
The HIV-1 parental population, isolate s61 (41), was obtained from a 4-year-old child and was isolated by standard coculture procedures. Ten biological clones were obtained from this viral population by using an MT-4 plaque assay as previously described (22, 41). In this method, the plaques produced do not result in lysis of the monolayer; rather, there is a formation of plaques (visible by the naked eye or by microscopy) due to the accumulation of cells that support viral replication following infection of a single cell (22). Plaques appear 7 to 10 days after infection. Viruses from randomly chosen, well-isolated plaques are the origin of viral populations designated A1 to K1. In subsequent plaque transfers, viral clones are designated with the same letter followed by a number that indicates the total number of plaque isolation passages (for example, B6 is clone B1 after five serial plaque-to-plaque transfers). Virus from each plaque was resuspended in 300 μl of culture medium, diluted 1:10, and used to infect MT-4 monolayers. The number of plaques was quantified in each passage for each virus. The plaque assay on MT-4 cells was used for titration of all viral stocks used in the study. Titers were expressed as PFU per milliliter. When a viral population did not yield plaques on MT-4 cells, its infectivity was also tested in cultures of MT-2 cells (22) or in peripheral blood mononuclear cells.
HTA.
DNA extraction was carried out by using the Instagene purification matrix (Bio-Rad) according to the manufacturer’s instructions. The heteroduplex tracking assay (HTA) (10) was carried out with a cDNA copy of a 650-bp fragment corresponding to the V1-V2 region of the env gene. To amplify this DNA, a nested PCR was carried out with primers 91ECU (5′CTTAGGCATCTCCTATGGC3′; positions 5535 to 5555 [numbering is as in reference 39]) and 92ECD (5′GGAGCAGCAGGAAGCAC3′; complementary to positions 7370 to 7388) and nested primers 99ECU (5′AGAGCAGAAGACAGTGGC3′; positions 5783 to 5801) and 52EV1D (5′TAATGTATGGGAATTGGCTCAA3′; complementary to positions 6429 to 6451). Amplifications were carried out for 35 cycles of 94°C for 1 min, 55°C for 55 s, and 72°C for 1 min, with an extension cycle of 10 min at 72°C.
In order to determine the proportion of each molecular species during competition passages, amplified cDNA of clone J1 was labeled with [α-32P]dCTP (3,000 Ci/mmol) in a PCR amplification. About 10,000 cpm of this radioactive PCR probe was mixed with 50 to 100 ng of unlabeled second-round PCR product from the competing virus in annealing buffer (0.1 M NaCl, 10 mM Tris HCl [pH 7.8], and 2 mM EDTA). The DNA mixtures were denatured at 94°C for 2 min and then quickly cooled (10). Heteroduplexes were resolved in denaturing 8% polyacrylamide–15% urea gels in TBE (88 mM Tris-borate, 89 mM boric acid, and 2 mM EDTA). Autoradiograms were obtained by exposing gels on a Fuji 2000 instrument for 2 h. The quantification of the ratio of the J1 cDNA (homoduplex) to the cDNA of the competing virus (heteroduplex) was determined by densitometry with the help of the PCBAS program.
Detection of mixed viral populations by HTA.
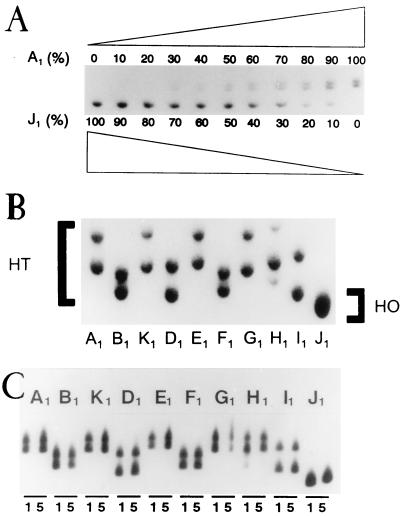
To test the reliability of the HTA for the detection and quantification of viral populations, cultures with different proportions of clones A1 and J1 were grown and proviral DNA was subjected to HTA analysis (Fig. 1A). The assay was able to detect the presence of a homoduplex amounting to about 10% of the total DNA and of a heteroduplex amounting to about 30% (Fig. 1A). DNA from each of the biological clones analyzed in this study displayed a heteroduplex (A1 to I1) with a distinct mobility when J1 was used as a probe (Fig. 1B). Each clone showed a distinct gel mobility reflecting its specific nucleotide sequence. For clone H1, two DNAs with different mobilities were detected (Fig. 1B), but the origin of this heterogeneity was not investigated. Each clone maintained its characteristic HTA pattern during five passages in cell culture under the same conditions used for the competition assays (Fig. 1C). Also, independent experiments with the same DNA preparations produced identical HTA patterns (data not shown).
FIG. 1.
HTA as a tool for quantification of mixed viral populations. (A) Quantification of two viruses present in different proportions. Mixed infections with clones J1 and A1 were carried out with decreasing (from 100 to 0%) and increasing (from 0 to 100%) proportions of the two viruses. Proviral DNA obtained from the infections was subjected to HTA with, as a labeled probe, cDNA of the V1-V2 region of the env gene, as described in Materials and Methods. (B) The same HTA analysis with DNAs of all initial clones, A1 to J1, using cDNA of clone J1 as a probe. All clones formed heteroduplexes (HT), whereas J1 formed a homoduplex (HO). The H1 population was probably a mixture of two clones. (C) Stability of the HTA pattern of each initial clone after five passages in cell culture. HTA patterns of amplified proviral DNAs from the first (lanes 1) and fifth (lanes 5) passages are shown.
Fitness assay.
Fitness determination was performed by growth competition experiments as previously described (24). Briefly, the assay consists of the coinfection of cultures with known amounts of the virus to be tested together with a reference clone. These cocultures are performed at different initial proportions, and the cultures are allowed to compete for a number of infections, after which the ratio of the two viruses is quantified by a phenotypic assay (4, 17, 19) or by a genotypic assay, as was done in this work (Fig. 1B). The proportion of the competing variant with respect to the reference strain (Rn) is divided by its proportion in the initial mixture (Ro), and this value (Rn/Ro) is plotted versus the competition passage to derive the fitness vector (24). In all cases, three competition passages were carried out by infecting MT-4 cells with mixtures of the clonal population to be tested and J1 at initial ratios of 1:9, 1:1, and 9:1. For each competition passage, 5 × 104 MT-4 cells were infected with 5 × 10−3 PFU of the mixture of viruses (multiplicity of infection of 0.1 PFU per cell). Virus was recovered from the culture supernatant when a cytopathic effect was evident (about 5 to 7 days postinfection). Fresh MT-4 cells were then infected with 50 μl of this supernatant diluted 1:10. Similar results were obtained for competitions carried out with the different initial ratios. Unless stated otherwise, the results presented are those obtained with the 1:1 initial ratio. The proportion of each clone in the competition passages was determined by using the HTA and divided by the proportion of the two clones in the starting competition mixtures. Ratios were used to calculate the fitness vectors as previously described (24). The slope of the fitness vector represents the fitness value of the corresponding viral clone; by this procedure, the fitness value of the reference clone J1 is zero.
RESULTS
Serial plaque-to-plaque transfers of biological HIV-1 clones.
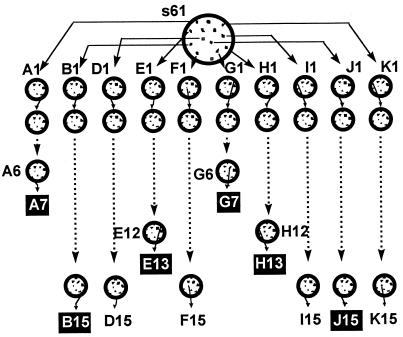
To study whether Muller’s ratchet can operate in HIV-1, viruses from 10 biological clones of isolate s61, termed A1, B1, D1, E1, F1, G1, H1, I1, J1, and K1, were obtained as described in Materials and Methods. Each viral clone was subjected to repeated plaque-to-plaque transfers in MT-4 cells (Fig. 2). Unexpectedly, 15 serial plaque-to-plaque passages could be completed for only six clones: B1, D1, F1, I1, J1, and K1. Plaque formation was not observed with clones A1 and G1 at passage 8 and with clones E1 and H1 at passage 14. Repeated plating of these viruses failed to produce visible plaques. In the case of clone H1, very small plaques were observed at transfer 13, following 12 to 14 days of plaque development, which represents a 7-day delay relative to normal plaque formation. Further plating of this population did not yield any plaques. This loss was preceded by a decreasing number of plaques in previous transfers. There were significant differences in the virus titers obtained among the different clones and within a clone during the plaque transfers (data not shown). Although titers varied in a rather irregular fashion, there was a trend towards decreasing titers with increasing plaque transfers. Visible plaques were obtained from populations D15, F15, K15, and I15, but no infectious virus was recovered from clones B15 and J15. These viruses were unable to grow not only in MT-4 cells (Fig. 2) but also in other cell lines, such as MT-2 or peripheral blood mononuclear cells (data not shown). These results highlight the frequent loss of infectivity of HIV-1 clones upon serial bottleneck passages.
FIG. 2.
Schematic representation of the derivation of HIV-1 clones and of serial plaque-to-plaque transfers. Procedures for isolation of virus from individual plaques of the HIV-1 isolate s61 on MT-4 cells are detailed in Materials and Methods. Viruses from randomly chosen individual plaques were diluted and plated again (circles and arrows). Filled boxes indicate the viral populations from which infectious virus could not be rescued upon subsequent plating or infection in liquid medium in different cell lines (as described in Materials and Methods). The 15 intended serial plaque transfers could be completed only for clones B1, D1, F1, I1, J1, and K1, although infectious virus could be rescued only from populations D15, F15, I15, and K15.
Fitness decrease upon plaque-to-plaque transfers of HIV-1.
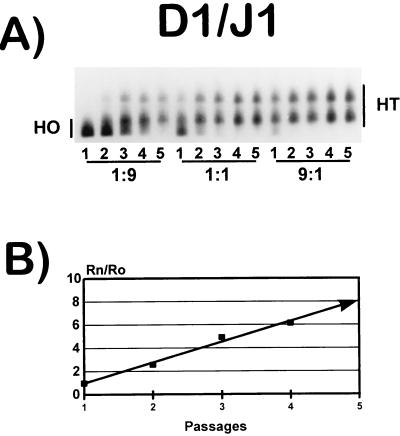
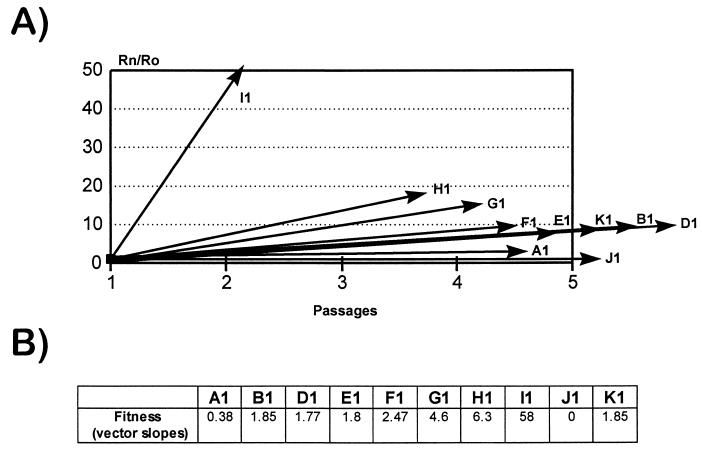
The determination of relative fitness values was carried out by growth competition experiments with mixed infections of each viral population to be tested and clone J1, which was used as a reference (24). As an example, results of an experiment corresponding to the fitness determination for clone D1 are shown in Fig. 3. The HTA patterns obtained for five serial competition passages between D1 and J1, mixed at the three initial proportions of 1:9, 1:1, and 9:1 are shown (Fig. 3A), as is the fitness vector derived from the competition series with the 1:1 initial proportion (Fig. 3B). The fitness vectors and fitness values for all of the starting populations A1 to K1 are displayed in Fig. 4A and B, respectively. The fitness values ranged from 0.38 (clone A1) to 58 (clone I1) relative to that for J1.
FIG. 3.
Fitness vector determination for HIV-1 clone D1. The fitness value was obtained from competition passages between clones D1 and J1 as described in Materials and Methods. A total of five competition passages were carried out with, as starting viruses, mixtures of D1 and J1 at ratios of 1:9, 1:1, and 9:1. (A) Result of the HTA assay. HT (heteroduplex) represents the proportion of D1 virus; HO (homoduplex) represents the proportion of J1. (B) From the 1:1 initial mixture, the proportions of D1 and J1 DNAs (measured and quantified by densitometry of the autoradiogram) (Rn) were obtained for the five competition passages. This proportion was compared to the ratio of the two viruses in the initial mixtures (Ro). The value in each passage (solid squares) was used to derive the fitness vector for D1 (24). Details of all procedures involved are given in Materials and Methods.
FIG. 4.
Fitness vectors and corresponding fitness values of the initial clones. (A) Fitness vectors of all of the initial clones, determined as indicated in Materials and Methods and in the legend to Fig. 3. In all lineages, the 1:1 competition cultures were used in the calculation of the vectors, except for clones I1 and H1. In these viruses the vectors were drawn with 1:9 competition cultures because of the very high fitness value of clone I1, which completely suppressed the J1 population, and because of the presence of two populations in clone H1 (Fig. 1B). Vectors were drawn with data from at least three competing passages. (B) Slopes of fitness vectors (fitness values) for the initial HIV-1 clones. Clone J1 was used as a reference, and consequently its slope is zero.
Fitness values were obtained for each of the clonal populations that yielded infectious virus after the 15 serial plaque transfers: populations D15, F15, I15, and K15 (Fig. 5). Fitness losses were observed in all cases except for clone F1, which experienced a 27% fitness gain at transfer 15 (Table 1). Clone I1 displayed a 99% fitness loss, suggesting that this clone may also be evolving towards extinction. Therefore, serial plaque-to-plaque transfers of HIV-1 clones led either to loss of virus infectivity or to strong fitness decreases in 9 of 10 clones tested.
FIG. 5.
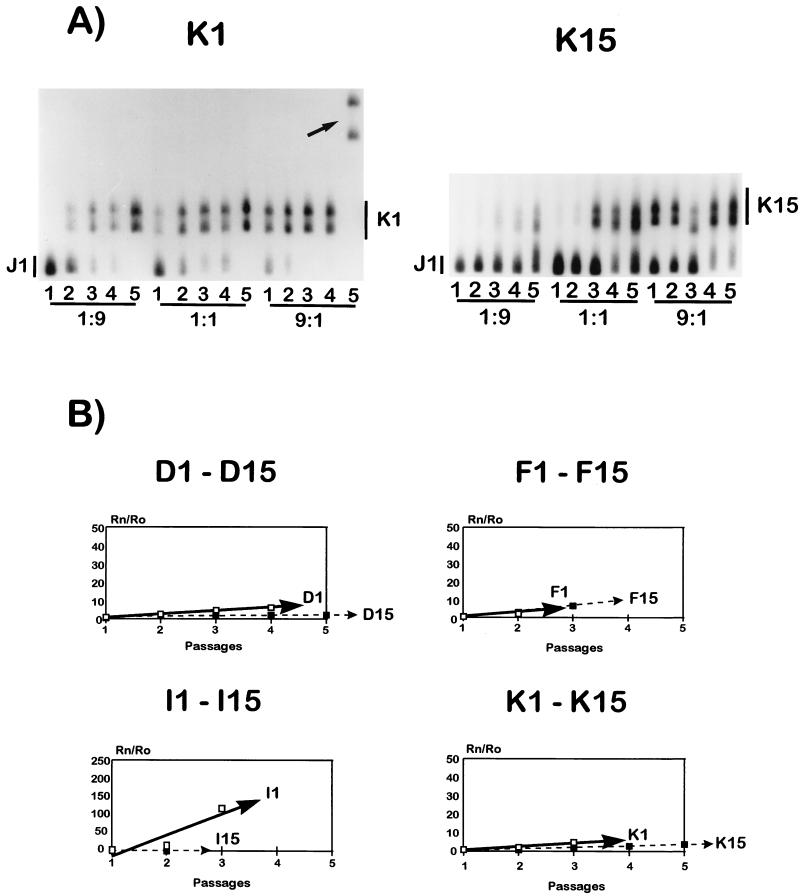
Fitness alterations produced in the viral populations after 15 serial plaque-to-plaque transfers. (A) HTA patterns obtained from the competitions at the 1:9, 1:1, and 9:1 proportions with cDNAs from the initial clone K1 and the final clone K15, using J1 cDNA as a probe. The arrow points to a new variant genome with a different HTA mobility arising at passage 5 of the competition series carried out at a 1:9 ratio of K1 and J1 in competition culture. (B) Comparison of the vectors in lineages for D1 to D15, F1 to F15, I1 to I15, and K1 to K15. The continuous lines represent initial fitness vectors, and the dashed lines represent final fitness vectors. For I1 to I15, a different scale has been used due to the wider range of values plotted. All of the vectors corresponding to final populations (transfers 15) are below the corresponding values for the initial clones, except for clone F1 (see text). Procedures are described in Materials and Methods, and fitness values are summarized in Table 1.
TABLE 1.
Fitness values of viable clones initially and at passage 15
| Clonea | Fitness valueb at passage:
|
Fitness variation (%)c | |
|---|---|---|---|
| 1 | 15 | ||
| A1 | 0.38 | —d | |
| B1 | 1.85 | — | |
| D1 | 1.77 | 0.26 | −85 |
| E1 | 1.8 | — | |
| F1 | 2.47 | 3.15 | +27 |
| G1 | 4.6 | — | |
| H1 | 6.3 | — | |
| I1 | 58 | 0.8 | −99 |
| J1 | 0 | — | |
| K1 | 1.85 | 0.68 | −63 |
The origin of the HIV-1 clones is detailed in Materials and Methods.
Fitness values were calculated from the corresponding fitness vectors as described in Materials and Methods and from Fig. 4 and 5.
Variation determined by comparing fitness values of transfer 15 clones to the corresponding initial values. +, fitness gain; −, fitness loss.
—, no fitness values could be determined because no virus able to produce plaques was obtained either at passage 15 or at a previous passage (see Fig. 2 and Results).
DISCUSSION
Severe Muller’s ratchet effect in HIV-1.
The results reported here with HIV-1 extend previous observations of the deleterious effects of repeated population bottlenecks on the fitness of bacteriophage φ6 (4), VSV (17), and FMDV (19) to a retrovirus. Despite important biological differences among the four viral systems, in all cases decreases in average fitness have been observed, providing clear support for the accentuation of Muller’s ratchet effect when viral populations are subjected to repeated bottleneck passages (1, 4, 6, 12, 17, 32, 33). Despite the fact that HIV-1 has two genomic RNA molecules in each particle and shows high rates of molecular recombination (9, 11, 28, 40), only 4 of 10 clones maintained plaque-forming potential over 15 plaque transfers. The fitness loss for HIV-1 was more severe than the reductions observed with other RNA viruses when a similar experimental design was used (Table 2).
TABLE 2.
Number of clones unable to form plaques after serial bottleneck passages of different RNA virusesa
| Virus | No. of clones unable to form plaques/total no. of clones analyzed | No. of bottleneck passages performed |
|---|---|---|
| φ6 | 0/20 | 40 |
| VSV | 0/16 | 20 |
| FMDV | 1/20 | 30 |
| HIV | 6/10 | 15 |
To determine the relative fitness in HIV-1 populations, we have applied a genetic method (HTA) instead of a phenotypic assay as previously used for other RNA viruses (19, 24). This genetic technique offers the advantage that it can be used with any virus in which genetic differences from a reference virus are detectable. However, the use of a genetic method does not impose any selective pressure against variants, present or emerging in the quasispecies, during competition experiments as is the case with phenotypic selection (4, 17, 19). In the present study, new variants were detected in the course of some of the competition passages, like the one found in the fifth passage of the competition culture between K1 and J1 carried out at a proportion of 1:9 (arrow in Fig. 5A). Additional variant genomes were also seen in the competitions between B1 and J1 and between A1 and J1 carried out at a ratio of 1:9 between the two competing viruses. However, a repetition of these two competition experiments with the same starting inocula did not produce variant heteroduplexes (results not shown). These observations suggest that such variants arose as a result of stochastic genetic variation events in the course of each competition. Viral populations showing these variant genomes were excluded from the calculation of fitness values. It could be argued that recombinants that could arise in the course of the serial competitive infections involved in the fitness assays could modify our measurements of relative fitness values. This is highly unlikely, because it implies that a recombinant between the reference J1 clone (with the lowest fitness among the clones studied) and the clone to be tested (harboring deleterious mutations) would be more fit than the parental J1 clone itself. Moreover, values obtained for homologous recombination with viral vectors and reporter genes in retroviruses are from 2 × 10−5 to 2 × 10−4 recombinations/nucleotide/cycle (26, 45, 51). These rates indicate that viable recombinant viruses are very unlikely to rise in the micromethod applied in the competition experiments because of the small viral inoculum (5 × 103 PFU), the low number of replication cycles (around seven cycles in each passage), and the small population size produced at the end of each passage (for example, around 3.5 × 104 PFU in first passage and 2.4 × 104 PFU in the second passage).
There are several mechanisms that either alone or in combination could contribute to greater fitness losses induced by serial genetic bottlenecks in HIV-1 than in other RNA viruses previously examined (4, 6, 17, 19, 35, 36) (Table 2). One possible mechanism stems from the plating system of HIV-1 on MT-4 cells, which does not result in the formation of lytic plaques. Rather, despite some cell killing, sites of virus production appear as localized clusters of virus-producing cells (22). Also, titers obtained from HIV-1 plaques are low, with values around 103 to 104 total PFU per plaque, compared with titers in the range of 109 to 1010 PFU per plaque for VSV or φ6 and around 105 PFU per plaque for FMDV. Therefore, the number of replication rounds and the possibility of competition among components of HIV-1 quasispecies during plaque development could be more restricted for HIV-1 than for other viruses. The lower chance of local quasispecies optimization within each plaque would result in cumulative lower fitness values after repeated plaque-to-plaque passages. Another possibility is that the mutational input in the case of HIV-1 may be larger than that for nonretroviral riboviruses. Such a difference does not seem to be supported by comparative analyses of mutation rates and frequencies (14, 16, 31, 37, 47). However, a number of environmental modifications (including biases in nucleotide substrate concentrations) can affect the retroviral mutation rates (2, 46), and it cannot be excluded that such an effect could operate during HIV-1 multiplication in MT-4 cells. Yet another explanation could lie in the origin of the clones used in this study: they were obtained from a natural HIV-1 isolate which was passaged only once in MT-4 cells before biological cloning and thus had limited adaptation to the cell culture system. This possibility seems unlikely, since no difference in Muller’s ratchet effect was detected when FMDV populations with different degrees of adaptation to the BHK-21 cell line, on which this effect was tested, were compared (19).
A rapid debilitating effect as a result of a limited number of bottleneck events could have implications for the evolution of the pathogenic potential of HIV-1 in vivo. Although the number of infectious particles that are transmitted and initiate viral multiplication is not known, it cannot be excluded that one or a limited number of HIV-1 particles may initiate an infection (48, 52). In this case, fitness differences between clones due to deleterious mutations may influence the outcome of the infectious process. Also, the implementation of hard triple- or multidrug therapy produces drastic and sustained reductions in the sizes of viral populations, which may contribute to fitness losses of the surviving HIV-1 (21, 23).
Fitness variations among components of the mutant spectra.
Viral quasispecies are important reservoirs of genetically and phenotypically relevant variants (reviewed in references 15 and 25). Examples are the preexistence in many natural quasispecies of HIV-1 mutants resistant to antiretrovial inhibitors (30, 34, 43) and the presence of phenotypic variants within viral populations (12–15). In the present analysis, biological clones derived from a field HIV-1 isolate displayed large fitness differences (Fig. 4 and Table 1) with regard to replication in MT-4 cells. The observed clonal variations in relative fitness values (Fig. 4 and Table 1) range from no difference between clones B1 and K1 to 1.04-fold between clones D1 and B1 to 153-fold between clones A1 and I1. However, even small fitness differences can become quantitatively important with regard to the dominance of viral subpopulations, given the large number of replication rounds during the natural infection with HIV-1 (8). Obviously, fitness values determined in MT-4 cells may not parallel fitness in lymphocytes or other natural host cells of HIV-1. It seems unlikely, however, that fitness heterogeneity is restricted to MT-4 cells.
The urgent need for the development of an effective anti-AIDS vaccine is reinforced by the explosive expansion of the AIDS epidemic in developing countries (38). Although other approaches are also possible, current concepts of RNA virus population heterogeneity and dynamics suggest that an immune response against a large number of B-cell and T-cell epitopes can be achieved with attenuated virus (50). The dynamics of viral quasispecies suggest that a broad, multivalent immune response may diminish the chances of selecting escape mutants (14). Obviously, safety concerns must always be considered in the development of live retrovirus vaccines, and even more so since the detection of in vivo recombination of attenuated simian immunodeficiency virus strains in rhesus monkeys (49). Experiments to investigate the nature of the genetic lesions that mediate the severe fitness losses observed in HIV-1 are now in progress. Knowledge of such genetic alterations may help in the design of highly debilitated strains of HIV-1.
ACKNOWLEDGMENTS
We thank A. del Pozo and G. Gimenez for the photographic work. C. Escarmis is acknowledged for helpful discussions and advice. MT-2 and MT-4 cells were kindly provided by D. Richman (University of California, San Diego).
Work at Centro Nacional de Biología Fundamental was supported by grants from FIS (97-0117 and 98-0054/02), and work at Centro de Biología Molecular “Severo Ochoa” was supported by grants from DGES (PM97-0060-CO2-01), FIS (98/0054-01), and Fundación Ramón Areces.
REFERENCES
- 1.Anderson D I, Hughes D. Muller’s ratchet decreases fitness of a DNA-based microbe. Proc Natl Acad Sci USA. 1996;93:906–907. doi: 10.1073/pnas.93.2.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Back N K T, Nijhuis M, Keulen W, Boucher C A B, Oude B B, Essink A B P, van Kullenburg A, van Gennip A H, Berkout B. Reduced replication of 3TC-resistant variants in primary cells due to a processivity defect of reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 3.Bebeneck K, Kunkel T. The fidelity of retrovial reverse transcriptases. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 85–162. [Google Scholar]
- 4.Chao L. Fitness of RNA virus decreased by Muller’s ratchet. Nature. 1990;348:454–455. doi: 10.1038/348454a0. [DOI] [PubMed] [Google Scholar]
- 5.Clarke D K, Duarte E A, Elena S F, Moya A, Domingo E, Holland J. The Red Queen reigns in the kingdom of RNA viruses. Proc Natl Acad Sci USA. 1994;91:4821–4824. doi: 10.1073/pnas.91.11.4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke D K, Duarte E A, Moya A, Elena S F, Domingo E, Holland J. Genetic bottlenecks and population passages cause profound fitness differences in RNA viruses. Proc Natl Acad Sci USA. 1993;67:222–228. doi: 10.1128/jvi.67.1.222-228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 8.Coffin J M. Plasma viral load, CD4+ cell counts, and HIV-1 production by cells. Science. 1996;271:670–671. doi: 10.1126/science.271.5249.671. [DOI] [PubMed] [Google Scholar]
- 9.Cornelissen M, Kampinga G, Zorgdrager F, Goudsmit J. Human immunodeficiency virus type 1 subtypes defined by env show high frequency of recombinant gag genes. J Virol. 1996;70:6777–6780. doi: 10.1128/jvi.70.11.8209-8212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rübsamen-Walgmann H, Mullins J I. Genetic relationship determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 11.Diaz R S, Sabino E C, Mayer A, Mosley J W, Busch M P, Group T T S S. Dual human immunodeficiency virus type 1 infection and recombination in a dually exposed transfusion recipient. J Virol. 1995;69:3273–3281. doi: 10.1128/jvi.69.6.3273-3281.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domingo E, Escarmis C, Sevilla N, Moya A, Elena S F, Quer J, Novella I S, Holland J J. Basic concepts in RNA virus evolution. FASEB J. 1996;10:859–863. doi: 10.1096/fasebj.10.8.8666162. [DOI] [PubMed] [Google Scholar]
- 13.Domingo E, Holland J J. High error rates, population equilibrium and evolution of RNA replication systems. In: Domingo E, Holland J J, Ahlquist P, editors. RNA genetics. III. Variability of RNA genomes. Boca Raton, Fla: CRC Press; 1988. pp. 3–36. [Google Scholar]
- 14.Domingo E, Holland J J. Mutations rates and rapid evolution of RNA viruses. In: Morse S S, editor. The evolutionary biology of viruses. New York, N.Y: Raven Press; 1994. pp. 161–184. [Google Scholar]
- 15.Domingo E, Martínez-Salas E, Sobrino F, de la Torre J C, Portela A, Ortin J, López-Galíndez C, Perez-Breña P, Villanueva N, Nájera R, VandePol S, Steinhauer D, DePolo N, Holland J J. The quasispecies (extremely heterogenous) nature of viral RNA genome populations: biological relevance—a review. Gene. 1985;40:1–8. doi: 10.1016/0378-1119(85)90017-4. [DOI] [PubMed] [Google Scholar]
- 16.Drake J. Rates of spontaneous mutation among RNA viruses. Proc Natl Acad Sci USA. 1993;90:4171–4175. doi: 10.1073/pnas.90.9.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duarte E, Clarke D, Moya A, Domingo E, Holland J J. Rapid fitness losses in mammalian RNA virus due to Muller’s ratchet. Proc Natl Acad Sci USA. 1992;89:6015–6019. doi: 10.1073/pnas.89.13.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elgen M. Self-organization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- 19.Escarmís C, Dávila M, Charpentier N, Bracho A, Moya A, Domingo E. Genetic lesions associated with Muller’s ratchet in an RNA virus. J Mol Biol. 1996;264:255–267. doi: 10.1006/jmbi.1996.0639. [DOI] [PubMed] [Google Scholar]
- 20.Fouchier R A M, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schultemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goudsmit J, de Ronde A, de Roolj E, de Boer R. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J Virol. 1997;71:4479–4484. doi: 10.1128/jvi.71.6.4479-4484.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harada S, Koyanagi Y, Yamamoto N. Infection of HTLV-III/LAV in HTLV-I-carrying cells MT-2 and MT-4 and application in a plaque assay. Science. 1985;229:563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]
- 23.Harrigan P R, Bloor S, Larder B A. Relative replicative fitness of zidovudine-resistant human immunodeficiency virus type 1 isolates in vitro. J Virol. 1998;72:3773–3778. doi: 10.1128/jvi.72.5.3773-3778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holland J J, de la Torre J C, Clarke D K, Duarte E. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J Virol. 1991;65:2960. doi: 10.1128/jvi.65.6.2960-2967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holland J J, de la Torre J C, Steinhauer D A. RNA virus populations as quasispecies. Curr Top Microbiol Immunol. 1992;176:1–20. doi: 10.1007/978-3-642-77011-1_1. [DOI] [PubMed] [Google Scholar]
- 26.Hu W S, Temin H M. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 27.Hu W S, Temin H M. Genetic consequences of packing two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci USA. 1990;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kampinga G A, Simonon A, Van de Perre P, Karlta E, Msellati P, Goudsmit J. Primary infections with HIV-1 of women and their offspring in Rwanda. Findings of heterogeneity at seroconversion, coinfection, and recombinations of HIV-1 subtypes A and C. Virology. 1997;227:63–76. doi: 10.1006/viro.1996.8318. [DOI] [PubMed] [Google Scholar]
- 29.Kulken C L, de Jong J J, Baan E, Keulen W, Tersmette M, Goudsmit J. Evolution of the V3 envelope domain in proviral sequences and isolates of human immunodeficiency virus type 1 during transitions of the viral biological phenotype. Proc Natl Acad Sci USA. 1992;66:4622–4627. doi: 10.1128/jvi.66.7.4622-4627.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lech W J, Wang G, Yang Y L, Chee Y, Dorman K, McCrae D, Lazzeroni L C, Erickson J W, Sinshelmer J S, Kaplan A H. In vivo sequence diversity of the protease of human immunodeficiency virus type 1: presence of protease inhibitor-resistant variants in untreated subjects. J Virol. 1996;70:2038–2043. doi: 10.1128/jvi.70.3.2038-2043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mansky L M, Temin H M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maynard-Smith J. The evolution of sex. Cambridge, United Kingdom: Cambridge University Press; 1976. [Google Scholar]
- 33.Muller H. The relation of recombination to mutational advance. Mutat Res. 1984;1:2–29. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 34.Nájera I, Holguín A, Quiñones-Mateu M E, Muñoz-Fernández M A, Nájera R, López-Galíndez C, Domingo E. pol gene quasispecies of human immunodeficiency virus: mutations associated with drug resistance in virus from patients undergoing no drug therapy. J Virol. 1995;69:23–31. doi: 10.1128/jvi.69.1.23-31.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novella I S, Elena S F, Moya A, Domingo E, Holland J J. Repeated transfer of small RNA virus populations leading to balanced fitness with infrequent stochastic drift. Mol Gen Genet. 1996;252:733–738. doi: 10.1007/BF02173980. [DOI] [PubMed] [Google Scholar]
- 36.Novella I S, Elena S F, Moya A, Domingo E, Holland J J. Size of genetic bottlenecks leading to virus fitness loss determined by mean initial population fitness. Proc Natl Acad Sci USA. 1995;69:2869–2872. doi: 10.1128/jvi.69.5.2869-2872.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Preston B P, Dougherty J P. Mechanisms of retroviral mutation. Trends Microbiol. 1996;4:16–21. doi: 10.1016/0966-842x(96)81500-9. [DOI] [PubMed] [Google Scholar]
- 38.Quinn T C. Population migration and the spread of types 1 and 2 human immunodeficiency viruses. Proc Natl Acad Sci USA. 1994;91:2407–2414. doi: 10.1073/pnas.91.7.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whichorn E, Baumelsteir K, Ivanoff L, Petteway S R J, Pearson M L, Lautenbergen J A, Papas T S, Chrayeb J, Chang N T, Gallo R C, Wong-Staal F. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 40.Robertson D L, Sharp P M, McCutchan F E, Hahn B H. Recombination in HIV-1. Nature. 1995;374:124–127. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 41.Sánchez-Palomino S, Rojas J M, Martínez M A, Fenyö E M, Nájera R, Domingo E, López-Galindez C. Dilute passage promotes expression of genetic and phenotypic variants of human immunodeficiency virus type 1 in cell culture. J Virol. 1993;67:2938–2943. doi: 10.1128/jvi.67.5.2938-2943.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scarlatti G, Tresoldi E, Björndal A, Fredriksson R, Colognesl C, Deng H K, Malnatl M S, Plebani A, Siccardi A G, Littman D R, Fenyö E M, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 43.Schinazi R, Larder B, Mellors J. Mutations in retroviral genes associated with drug resistance. Int Antiviral News. 1997;5:129–142. [Google Scholar]
- 44.Seillier-Molsiwitsch F, Margolin B H, Swanstrom R. Genetic variability of the human immunodeficiency virus: statistical and biological issues. Annu Rev Genet. 1994;28:559–596. doi: 10.1146/annurev.ge.28.120194.003015. [DOI] [PubMed] [Google Scholar]
- 45.Temin H M. Retrovirus variation and reverse transcription: abnormal strand transfer results in retrovirus genetic variation. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vartanian J-P, Meyerhans A, Äsjö B, Wain-Hobson S. Selection, recombination, and G→A hypermutation of human immunodeficiency virus type 1 genomes. J Virol. 1991;65:1779–1788. doi: 10.1128/jvi.65.4.1779-1788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wain-Hobson S. Running the gamut of retroviral variation. Trends Microbiol. 1996;4:135–141. doi: 10.1016/0966-842x(96)10023-8. [DOI] [PubMed] [Google Scholar]
- 48.Wolinsky S M, Wike G M, Korber B T M, Hutto C, Parks W P, Rosenblum L L, Kunstman K J, Furtado M R, Muñoz J L. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science. 1992;255:1134–1137. doi: 10.1126/science.1546316. [DOI] [PubMed] [Google Scholar]
- 49.Wooley D P, Smith R A, Czajak S, Desrosiers R. Direct demonstration of retroviral recombination in a rhesus monkey. J Virol. 1997;71:9650–9653. doi: 10.1128/jvi.71.12.9650-9653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wyand M S, Manson K H, Lackner A A, Desrosiers R C. Resistance of neonatal monkeys to live attenuated vaccine strains of simian immunodeficiency virus. Nat Med. 1997;3:32–35. doi: 10.1038/nm0197-32. [DOI] [PubMed] [Google Scholar]
- 51.Zhang J, Temin H. Rate and mechanism of nonhomologous recombination during a single cycle of retroviral replication. Science. 1993;259:234–238. doi: 10.1126/science.8421784. [DOI] [PubMed] [Google Scholar]
- 52.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]