Abstract
The interaction of the chemokine stromal cell-derived factor 1 (SDF-1) with its receptor CXCR4 is vital for cell trafficking during development, is capable of inhibiting human immunodeficiency virus type 1 (HIV-1) utilization of CXCR4 as a coreceptor, and has been implicated in delaying disease progression to AIDS in vivo. Because of the importance of this chemokine-chemokine receptor pair to both development and disease, we investigated the molecular basis of the interaction between CXCR4 and its ligands SDF-1 and HIV-1 envelope. Using CXCR4 chimeras and mutants, we determined that SDF-1 requires the CXCR4 amino terminus for binding and activates downstream signaling pathways by interacting with the second extracellular loop of CXCR4. SDF-1-mediated activation of CXCR4 required the Asp-Arg-Tyr motif in the second intracellular loop of CXCR4, was pertussis toxin sensitive, and did not require the distal C-terminal tail of CXCR4. Several CXCR4 mutants that were not capable of binding SDF-1 or signaling still supported HIV-1 infection, indicating that the ability of CXCR4 to function as a coreceptor is independent of its ability to signal. Direct binding studies using the X4 gp120s HXB, BH8, and MN demonstrated the ability of HIV-1 gp120 to bind directly and specifically to the chemokine receptor CXCR4 in a CD4-dependent manner, using a conformationally complex structure on CXCR4. Several CXCR4 variants that did not support binding of soluble gp120 could still function as viral coreceptors, indicating that detectable binding of monomeric gp120 is not always predictive of coreceptor function.
Chemokines are a soluble peptide family that modulate the immune response by virtue of their chemoattractive and signaling properties (see reference 51 for a review). Chemokines are divided into two major classes, CC and CXC, based on the spacing of their two highly conserved Cys residues. Stromal cell-derived factor 1 (SDF-1) is an 8-kDa CXC chemokine originally isolated from a bone marrow stromal cell line (60) that activates a wide variety of primary cells and cell lines (2, 9, 48). The importance of this chemokine in immunomodulation, organogenesis, and hematopoiesis has been highlighted by the characterization of SDF-1 and CXCR4 knockout mice (47, 59, 69). Both exhibit significant developmental abnormalities, indicating that chemokines can play a critical role during development in addition to their well-characterized role in the mature immune response.
The importance of SDF-1 to human disease has also been highlighted by the discovery that a naturally occurring polymorphism in the SDF-1 gene is correlated with slower progression to AIDS in human immunodeficiency virus (HIV)-infected individuals (66). While the mechanism behind this observation has yet to be fully explained, the only known receptor for SDF-1, CXCR4 (8, 48), is the major HIV type 1 (HIV-1) coreceptor used by X4 strains of the virus (also referred to as T-tropic or syncytium-inducing strains) (5, 27). Interaction between the viral envelope (Env) protein and a coreceptor such as CXCR4 triggers conformational changes in Env that lead to membrane fusion and entry of the viral genome into the host cell cytoplasm. SDF-1, like other coreceptor ligands, can block HIV-1 from utilizing CXCR4 and entering a cell (8, 48). Since the emergence of X4 strains of HIV-1 in vivo is correlated with a rapid decline in CD4+ T cells in infected individuals (42), the availability of CXCR4 to X4 strains of HIV-1 in vivo is likely to be a major factor determining the protective effect of the SDF-1 mutation.
Despite its protective effects, the ability of SDF-1 to block HIV-1 coreceptor utilization is variable, often weak, and largely dependent on the Env protein of HIV-1 that mediates the fusion process (62). Previous studies have shown that the extracellular loops (ECLs) of CXCR4, particularly the first and second ECLs (ECL1 and ECL2), are important for coreceptor activity, but the results also suggest that Env-CXCR4 interactions can vary depending on the virus strain studied (10, 40, 50). The identification of small-molecule antagonists of CXCR4 and readily selected strains of HIV-1 that can resist inhibitor challenges highlights the flexibility of Env and the need to understand the interaction of ligands with CXCR4 to design more effective antiretroviral agents (20, 21, 38, 46, 49, 56). Recent advances in detecting direct Env interactions with CCR5 have enhanced our understanding of the role of the chemokine receptors in fusion (37, 41, 52, 61, 67, 68), but direct interactions of X4 Envs with CXCR4 have been difficult to study (4, 34, 39, 43).
To better understand the basis for SDF-1-mediated disease protection, SDF-1-induced signaling, and CXCR4 coreceptor function, we analyzed the interactions between SDF-1, HIV-1 Env, and CXCR4. We identified a principal SDF-1 binding determinant on the CXCR4 amino terminus and a distinct region on ECL2 of CXCR4 that mediates activation of the receptor by SDF-1. Our data are consistent with models proposed by Crump et al. and Heveker et al. in which the RFFESH motif of SDF-1 (amino acids 12 to 17) mediates binding to the amino terminus of CXCR4, while the first two amino acids of SDF-1 (Lys-Pro) mediate activation of CXCR4 by interacting with ECL2 (16, 36). HIV-1 fusion required regions of CXCR4 that overlapped the binding and activation regions used by SDF-1, but the ability of CXCR4 to signal was clearly distinct from its ability to function as a coreceptor, similar to CCR5. Binding of the gp120 subunit of X4 Envs to CXCR4 was dependent on a conformationally complex structure on CXCR4. However, several mutants of CXCR4 that exhibited no detectable binding of X4 gp120s could still function as fusion coreceptors, suggesting that binding of monomeric gp120 to CXCR4 does not necessarily predict coreceptor activity.
MATERIALS AND METHODS
CXCR4 chimeras and mutants.
The CXCR4 chimeras used in this study and the pT4 plasmid encoding human CD4 have been described previously (40). Chimeras were produced by joining CXCR2 and CXCR4 clones in the pcDNA3 vector and are named based on the parental receptor from which the extracellular domains are derived. For example, 2444 contains the amino terminus of CXCR2 and the first, second, and third ECLs of CXCR4. In brief, chimeras were joined at the following CXCR4 residues: 2444b (Gly-64), 4442 (Ile-243), 2442 (Cys-28, Ile-243), 2244 (Asp-133), and 2242 (Asp-133, Ile-243). 4222 and 2444 were joined reciprocally at the common Cys in the amino terminus of CXCR4 (Cys-28) and CXCR2 (Cys-39). Junctions are depicted graphically in Fig. 7. CXCR4Δtail truncates the C terminus of CXCR4 to residue 316 and mutates Thr-311 and Ser-312 to Ala to eliminate all Ser and Thr residues in the carboxy terminus. Construction of the CXCR4 point mutants used in this study are described elsewhere (65).
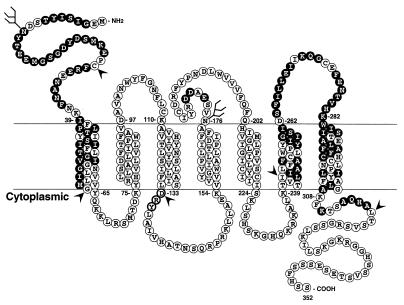
FIG. 7.
Coreceptor utilization overlaps, but is distinct from, SDF-1 binding and activation sites. The primary amino acid sequence of CXCR4 is shown, with shaded residues indicating regions substituted in CXCR4-CXCR2 chimeras or in CXCR4 mutants that are required for SDF-1 binding or activation. Residues within these regions that are not shaded are conserved between CXCR4 and CXCR2. The DRY motif in the second intracellular loop that is required for signaling is highlighted with darker circles. Arrowheads indicate CXCR4 residue junctions at which chimeras or truncation mutants were constructed.
Cells.
The human astroglioma cell line U87-MG (ATCC HTB-14) was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. The quail fibrosarcoma cell line QT6 and the human kidney cell line 293T were provided by Paul Bates (University of Pennsylvania). COS-SH cells are one subclone of the COS cell lineage and were obtained from Mike Malim (University of Pennsylvania). All cells were maintained in DMEM (Dulbecco’s modified Eagle medium, high glucose) supplemented with 10% fetal bovine serum, 2 mM glutamine, and 2 mM penicillin-streptomycin.
Ca2+ mobilization assays.
Response to ligand was determined in transiently transfected COS-SH cells. For transfection, cells were split at 106 cells/10-cm-diameter plate 24 h prior to transfection. Plasmids encoding chemokine receptors were mixed (10 μg) with DEAE cocktail (5.5 ml of DMEM, 55 μl of l-glutamine [100×], 55 μl of amphotericin B [Fungizone; Sigma] [100×], 55 μl of penicillin-streptomycin [100×], 55 μl of nutridoma [Boehringer Mannheim Biochemicals], 55 μl of DEAE [Pharmacia], 16.5 μl of chloroquine) and shaken vigorously. Following a 15-min incubation, the DNA-DEAE suspension was added to COS-SH cells which had been washed twice with incomplete DMEM. DNA-DEAE was incubated at 37°C for 2.5 h. Cells were shocked in 10% dimethyl sulfoxide for 2 min, washed twice with incomplete DMEM, and then placed in complete medium. Following expression for 16 to 20 h, cells were trypsinized and replated in dishes to grow for an additional 24 h. Cells were loaded with 5 μM Fura-2/AM (Molecular Probes) in the dark at 37°C for 1 h. Cells were removed from plates by incubation in phosphate-buffered saline (PBS) without Ca2+ or Mg2+ and were resuspended in Dulbecco’s PBS containing Ca2+ and Mg2+ (BioWhittaker). Ca2+ mobilization was measured in an Aminco-Bowman Luminescence Spectrometer in a constantly stirring cuvette and in a volume of 1.5 ml. Excitation of cells was monitored at 340 and 380 nm, and the Ca2+ concentration was calculated as previously described (33), using an assumed Kd of 224. SDF-1α, interleukin-8 (IL-8), and GROα (Peprotech) were used at a final concentration of 62.5 nM (500 ng/ml) and had no background activity on COS-SH cells in this assay. Thrombin receptor agonist peptide (TAP; referred to elsewhere as the PAR-1 agonist peptide) was used at a final concentration of 27 μM and consists of the amino acid residues SFLLRN. Pertussis toxin was used at a final concentration of 100 ng/ml and was incubated with cells 8 to 16 h before use of cells for Ca2+ mobilization, flow cytometry, binding, or infection.
Flow cytometry.
In preparation for flow cytometry (fluorescence-activated cell sorting [FACS]), cells were removed from the plate with 5 mM EDTA in PBS, centrifuged, resuspended in staining buffer (PBS with 0.1% bovine serum albumin) supplemented with 25% normal rat serum and 25% normal rabbit serum, and placed on ice. Cells were stained with primary monoclonal antibodies (MAbs), washed with staining buffer, and then stained with goat anti-mouse antibody conjugated to either fluorescein isothiocyanate or phycoerythrin fluorochrome (Biosource, Camarillo, Calif.). Fluorescence was monitored on a FACScan instrument with a 15-mW 488-nm blue argon laser (Becton Dickinson, San Jose, Calif.), and data from 10,000 cells were analyzed with CellQuest version 3.0.1 software (Becton Dickinson).
Binding assays.
For chemokine binding assays, 5 × 105 293T cells transiently transfected by CaPO4 with 4 μg of DNA were resuspended in 75 μl of binding buffer (50 mM HEPES [pH 7.4], 150 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 5% bovine serum albumin). Subsequently, 0.1 nM 125I-SDF-1α (specific activity, 2,200 Ci/mmol; NEN-Dupont) was added to cells in 25 μl of binding buffer for a total volume of 100 μl. Cells were incubated at room temperature for 1 h. Unbound radioactivity was removed by filtering cells through Whatman GF/C filters presoaked in 0.3% polyethyleneimine (Sigma) and washing them two times with 4 ml of wash buffer (50 mM HEPES [pH 7.4], 500 mM NaCl, 5 mM MgCl2, 1 mM CaCl2). Filters were counted in a Wallac 1470 Wizard gamma counter.
Env binding assays were performed similarly to SDF-1 binding assays except that binding buffer did not include NaCl. The inclusion of NaCl in Env binding assays eliminated detectable Env binding, while inclusion of NaCl in SDF-1 binding assays was required for specific binding to CXCR4. BH8 and HXB gp120s were produced by using vaccinia virus as previously described (23) and was >90% pure, as demonstrated by Coomassie blue staining. MN gp120, produced via baculovirus by ImmunoDiagnostics, was obtained through the NIH AIDS Reagent Repository. Five to 20 μg of each protein was iodinated by using Iodogen (Pierce) to specific activities of 5.7 μCi/μg (HXB), 1.7 μCi/μg (BH8), and 3.4 μCi/μg (MN).
Infection studies.
Viral stocks were prepared as previously described (11, 15) by transfecting 293T cells by CaPO4 with plasmids encoding the HXB2 or NL4-3 env and the NL4-3 luciferase virus backbone (pNL-Luc-E−R−). The resulting supernatant was stored at −80°C. For infection, U87-MG cells were plated in 24-well plates and transfected with the desired plasmids (1.5 to 2 μg of each). Medium was changed after 4 h, and cells were allowed to express overnight. Cells were infected the next day with 100 μl of viral supernatant in a total volume of 500 μl in the presence of 8 μg of DEAE-dextran per ml. Cells were lysed at 3 days postinfection by resuspension in 150 μl of 0.5% Triton X-100–PBS, and 50 μl of the resulting lysate was assayed for luciferase activity in a Wallac Microbeta scintillation and luminescence counter, using a luciferase assay kit from Promega. All values were within the linear range of luciferase detection.
RESULTS
CXCR4 domains required for SDF-1-induced signaling.
To understand how the chemokine SDF-1 and its cognate receptor CXCR4 interact, we tested a panel of previously described CXCR4-CXCR2 chimeras and mutants (40) for the ability to bind and signal in response to SDF-1. CXCR2 (30% identical to CXCR4) signals upon binding the chemokines IL-8 and GROα (1) but does not bind or respond to SDF-1 and does not serve as a coreceptor for HIV-1 (19). We used a Ca2+ mobilization assay to determine which chimeras could signal in response to SDF-1, IL-8, or GROα. COS-SH cells were transiently transfected with the indicated chimeras, loaded with the Ca2+-sensitive fluorescent dye Fura-2/AM, and assayed for Ca2+ mobilization following addition of the indicated chemokine. Untransfected cells did not signal in response to SDF-1, IL-8, or GROα but did respond appropriately to these chemokines when the cognate receptor was expressed (Fig. 1). The concentration of SDF-1 used in this assay, 500 ng/ml (62.5 nM), has previously been shown to stimulate CXCR4 to near-maximal levels (8, 48). The effects of chemokine receptor surface expression levels are accounted for below.
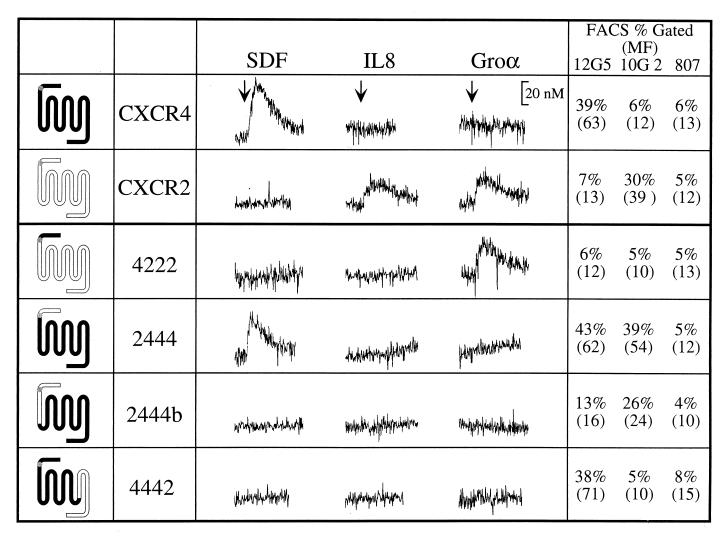
FIG. 1.
SDF-1 activation requires the proximal amino terminus and the third ECL of CXCR4. Transiently transfected COS-SH cells were stimulated with the indicated chemokine and assayed for mobilization of Ca2+. All cells were subsequently stimulated with TAP to ensure cell integrity (data not shown). Experiments were repeated at least three times. The names and general structures of chimeric constructs are indicated on the left. The percentage of cells scored as receptor positive (% Gated) and the mean fluorescence of staining (MF; indicated in parentheses) as measured by flow cytometry (FACS) of parallel sets of cells are indicated on the right. MAb 12G5 recognizes the first and second ECLs of CXCR4, 10G2 recognizes the distal amino terminus of CXCR2, and 807 is an isotype-matched (IgG2a) control MAb. Chimera 4222 is not capable of being recognized by any of the antibodies used here but has previously been shown to be expressed on the cell surface (40).
Our results indicate that while the distal amino terminus (the first 27 residues up to the conserved Cys) of CXCR4 was neither necessary (chimera 2444) nor sufficient (4222) for activation by SDF-1, the proximal amino terminus (carboxy terminal to the conserved Cys) near the transmembrane region (2444b) was required for SDF-1 activation. Chimera 4442 did not respond to SDF-1, suggesting that the third ECL of CXCR4 may also play an important role in CXCR4 activation. However, we cannot rule out the possibility that the failure of 4442 to signal is due to indirect effects of ECL3 (and adjoining transmembrane domains) substitution on the molecule’s overall conformation. Several additional chimeras were constructed in order to identify the contributions of other regions of CXCR4, such as ECL1 and ECL2, but these mutants (4244, 4424, 2224, 2442b, and 4422) were not expressed on the cell surface.
SDF-1 requires residues in ECL2 and second intracellular loop of CXCR4 for signaling.
To identify specific residues of CXCR4 that contribute to SDF-1-induced signaling, we used site-directed mutants of CXCR4 (65). We focused on ECL2 because the second ECLs of both CXCR4 and CCR5 make major contributions to HIV-1 coreceptor activity (7, 10, 38, 40) and, in the case of CCR5, to chemokine binding specificity (53). Since SDF-1 and the V3 loop of X4 Envs (implicated in coreceptor interaction [13]) are highly basic, our mutants focused on negatively charged residues within this domain. CXCR4-QAAN changes a conspicuous stretch of negatively charged amino acids, Glu-Ala-Asp-Asp (EADD), in ECL2 to the residues Gln-Ala-Ala-Asn (QAAN). When tested in Ca2+ mobilization assays (Fig. 2A), mutant CXCR4-QAAN failed to signal, highlighting the role of ECL2 residues in SDF-1-mediated signal transduction. Another mutation of an acidic residue in ECL2, D193K (Asp 193 changed to Lys), had no effect on CXCR4 signaling.
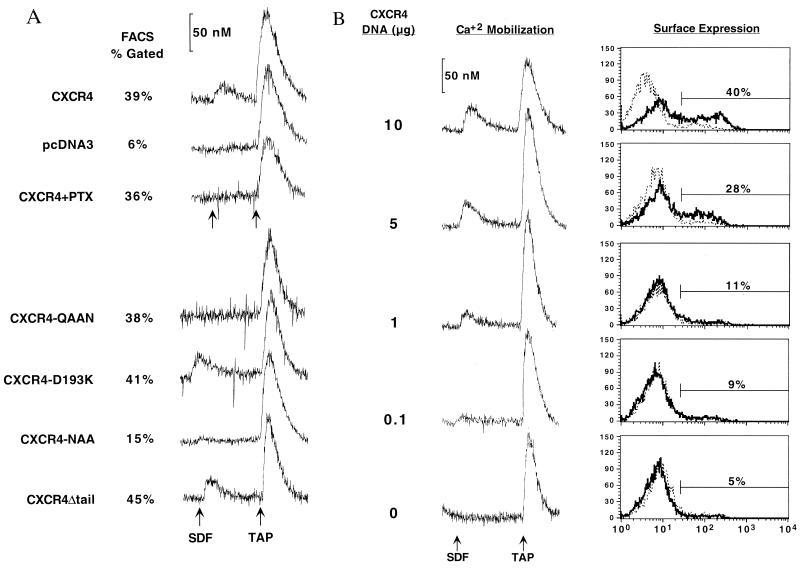
FIG. 2.
SDF-1 activation requires residues in the second extracellular and second intracellular loops of CXCR4, is pertussis toxin sensitive, and does not require the distal C terminus of CXCR4. (A) Transiently transfected COS-SH cells were stimulated with SDF-1 and then with the positive control TAP and assayed for mobilization of Ca2+. Experiments were repeated two to three times. The percent gated cells as measured by flow cytometry of parallel sets of cells is indicated. PTX indicates the addition of pertussis toxin 16 h prior to assay, and pcDNA3 indicates that cells were transfected with control vector DNA that does not express any chemokine receptor. (B) Ca2+ mobilization assay sensitivity. COS-SH cells were transfected with diminishing amounts of CXCR4 plasmid DNA, as indicated, keeping total DNA constant at 10 μg by using plasmid pcDNA3. Parallel sets of cells were tested for Ca2+ mobilization in response to SDF-1 and were tested for surface expression of CXCR4 by flow cytometry using MAb 12G5 (black tracing) and control MAb 807 (dotted tracing). The percentage of cells staining positive for 12G5 within the gate indicated is given on the right. Additional transfected CXCR4 DNA (20 μg) did not significantly increase the percent gated population or Ca2+ mobilization response (data not shown).
Important cytoplasmic residues of CXCR4 that contributed to SDF-1-mediated signal transduction were also identified. The Asp-Arg-Tyr motif (DRY box) is highly conserved among G-protein-coupled receptors, and its mutation in well-studied receptors such as rhodopsin, the α- and β-adrenergic receptors (28–30, 64), and CCR5 (3, 7, 22, 26, 32) eliminates signaling. Mutation of this motif in CXCR4 to Asn-Ala-Ala (NAA) largely eliminated the ability of the CXCR4-NAA mutant to signal (Fig. 2A). We note, however, that CXCR4-NAA may retain at least partial G-protein-coupling capability, as an extremely small Ca2+ mobilization signal was consistently noted. Truncation of the Ser-Thr-rich region of the distal C terminus that contains potential sites of receptor phosphorylation had no effect on the ability of CXCR4 to signal (CXCR4Δtail).
Surface expression and detection limitation of Ca2+ mobilization.
Because adequate cell surface expression of chemokine receptors is a prerequisite for detectable receptor activity, Ca2+ mobilization assays were performed in conjunction with flow cytometry (FACS) on parallel sets of cells (Fig. 1 and 2A). For FACS analysis we used MAbs 12G5, which recognizes a conformation-dependent epitope composed of the first and second ECLs of CXCR4 (10, 25, 40), 10G2, which recognizes a linear epitope on the CXCR2 amino terminus (14), and 807, which is an isotype-matched (immunoglobulin G2a [IgG2a]) control antibody. Surface staining of COS-SH cells confirmed the expression of chimeras such as 4442 that failed to respond to SDF-1 (Fig. 1). Chimera 2444b was detected on the surface by 12G5 at levels below wild-type but significantly above background levels. The ability of 10G2 to detect the linear amino-terminal epitope of this particular chimera may more accurately reflect its surface expression levels since the construction of this chimera may partially disturb the conformational epitope recognized by 12G5. Chimera 4222 could not be detected by FACS since the epitopes for 12G5 and 10G2 are not present on it, but its ability to signal in response to GROα indicates that it was expressed at functional levels, and expression of this chimera has been confirmed previously by using other antibodies (40).
Due to the reduced expression levels of some chimeras, we addressed the sensitivity of our Ca2+ mobilization assay by transfecting limiting dilutions of CXCR4 into COS-SH cells followed by both Ca2+ mobilization and CXCR4 surface expression measurements in parallel sets of cells (Fig. 2B). Our results indicated that detection of Ca2+ mobilization was at least as sensitive as the ability to detect CXCR4 on the surface of these cells by FACS with 12G5. Thus, mutants of CXCR4 that were expressed on the surface of cells at reduced levels, such as 2444b, can be assayed for Ca2+ mobilization with confidence. We conclude that the inability of 2444b, 4442, CXCR4-QAAN, and CXCR4-NAA to produce a measurable Ca2+ mobilization response was due not to detection limitations but to their inability to transduce a signal in response to SDF-1.
SDF-1 requires the amino terminus of CXCR4 for binding.
The failure of a receptor to signal in response to SDF-1 can be attributed either to its inability to bind SDF-1 or to its inability to be activated by a bound SDF-1 molecule. To distinguish between these possibilities, we analyzed the ability of the same panel of chimeras and mutants to bind iodinated SDF-1. To maximize sensitivity, we used transiently transfected 293T cells, which are capable of high levels of transient expression. The low levels of endogenous CXCR4 (estimated to be <200 copies per cell [63]) on 293T cells did not interfere with our analyses. Similar results were also obtained with transiently transfected QT6 cells, a quail cell line that expresses no known chemokine receptors (data not shown). COS-SH cells exhibited high background binding under the conditions used and thus were unsuitable for this analysis. Using limiting dilutions of transfected CXCR4 DNA, we found that SDF-1 binding could be detected even when CXCR4 expression levels were nearly undetectable as measured by FACS analysis with 12G5 (data not shown).
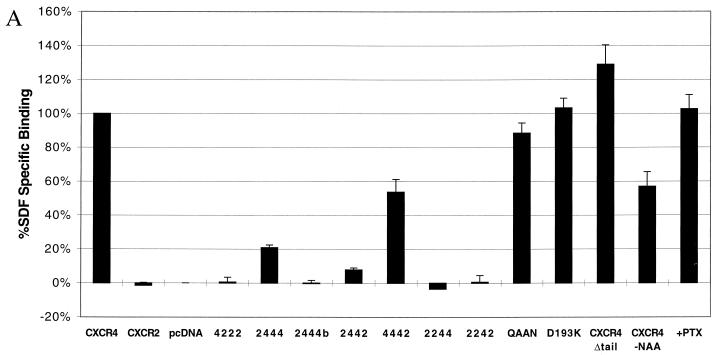
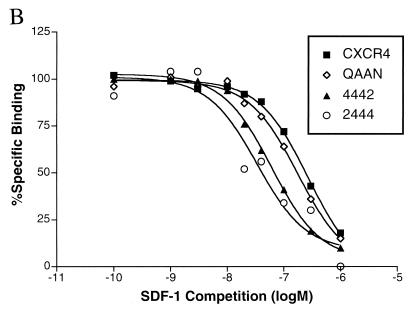
Binding assays performed with CXCR4 mutants and chimeras (Fig. 3A) demonstrated a dependence on the amino terminus of CXCR4. Chimera 2444 exhibited only minimal binding of SDF-1, while chimera 2444b was unable to bind SDF-1. These results suggest that the amino terminus of CXCR4, particularly the region after Cys-28, is critical for SDF-1 binding. A chimeric receptor identical to 2444 but using the distal amino terminus of CCR5 instead of CXCR2 produced binding and signaling results identical to that of chimera 2444, thus confirming the role of the distal amino terminus in SDF-1 binding (data not shown). Most notably, CXCR4-QAAN was capable of binding SDF-1 despite its failure to signal, suggesting that these residues in ECL2 are critical for signal transduction mediated by SDF-1. Homologous competition assays (Fig. 3B) indicated that our conclusions are not based on widely varying affinity differences. Calculated Ki values, as derived by the method of Swillens (45, 58), for CXCR4, QAAN, 4442, and 2444 were 85, 68, 37, and 38 nM, respectively.
FIG. 3.
(A) SDF-1 binding requires the proximal amino terminus of CXCR4. 293T cells transiently transfected with the indicated constructs were tested for binding of iodinated SDF-1. Data shown represent the mean and standard error of experiments repeated two to four times. Values for cells transfected with pcDNA3 were considered background and were subtracted from all measurements. Typical values of total bound radioactivity for transfected cells were 20,000 cpm for CXCR4 and 3,000 cpm for pcDNA3. All chimeric constructs were also tested for binding of iodinated GROα, but despite robust binding to CXCR2, iodinated GROα was incapable of binding any of these chimeras above a minimal 10% specific binding (data not shown). (B) Affinity of SDF-1 for CXCR4 variants. A total of 2 × 105 transiently transfected 293T cells were used for competition binding of iodinated SDF-1 with unlabeled SDF-1. Results are the average of two independent experiments, and values are normalized to binding levels without competition (100%) and with maximum competition (0%). Maximum plateau levels before normalization are represented in panel A. Results were analyzed by nonlinear regression using GraphPad Prism version 2.0 (45).
HIV-1 coreceptor utilization of CXCR4 is independent of the ability of CXCR4 to signal or to bind SDF-1.
We have previously used a subset of the mutants presented here to map the coreceptor utilization of CXCR4 by HIV-1 in a cell-cell fusion assay (40). Here we extended this analysis by using a virus infection assay and by correlating our results with the regions of the receptor required for SDF-1 binding and signaling and gp120 binding (below). The ability of our chimeras and mutants to support viral entry was assessed in an assay using recombinant virions that express luciferase after integration and that can be pseudotyped with a desired Env (11, 15). For this assay we used transiently transfected human U87-MG cells because of their ability to support viral expression and their high transfection efficiency. Limiting dilutions of transfected CXCR4 DNA demonstrated that coreceptor activity could be detected with this assay even when coreceptor levels were undetectable by FACS (data not shown).
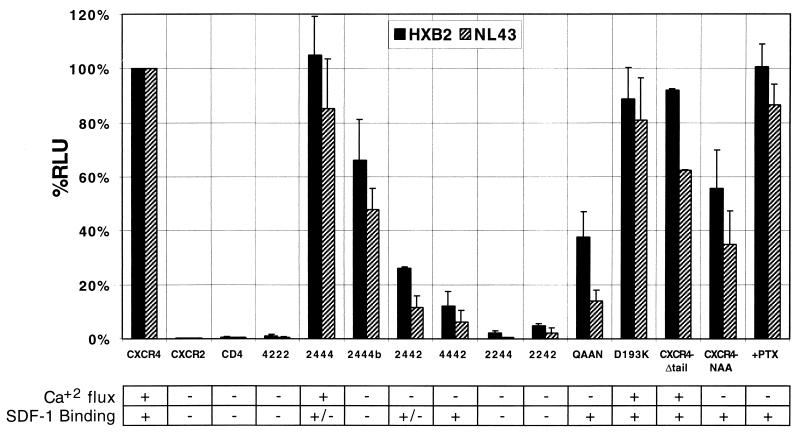
The distal amino terminus was not required for viral entry, since replacement of the distal amino terminus (2444) did not affect the coreceptor activity of CXCR4 (Fig. 4). Further substitution of the amino terminus (2444b) diminished the coreceptor’s ability to support HIV-1 infection, but the reduced surface expression levels of 2444b may account for this minimal decrease. ECL1 appeared to make a major contribution to coreceptor activity, since replacement of this region (2244) eliminated coreceptor activity, but the lower surface expression levels of this mutant (<10% of the wild-type level [data not shown]) may account for this result. However, chimera 2244 does support cell-cell fusion with other Envs (40). Residues in ECL2 (CXCR4-QAAN) were extremely important for coreceptor function, as replacement of these few residues diminished the ability of CXCR4 to support HIV-1 entry. Finally, residues in ECL3 also contributed to coreceptor activity, since chimera 4442 supported entry less efficiently than wild-type CXCR4 (Fig. 4). Thus, residues in all four extracellular regions of CXCR4 appear to contribute to coreceptor activity, in agreement with previous analyses of CXCR4 chimeras and mutants by cell-cell fusion (10, 40, 50).
FIG. 4.
HIV-1 Env utilizes multiple regions of the coreceptor CXCR4 for viral fusion. HIV-luciferase reporter viruses pseudotyped with the X4 Envs of HXB2 and NL4-3 were used to infect U87-MG cells transiently transfected with the constructs indicated. All cells were transfected with pT4, and the vector control (CD4) was cotransfected with plasmid vector alone (pcDNA3) instead of vector expressing a chemokine receptor. Pertussis toxin (PTX) was added 8 to 16 h prior to infection of cells expressing CXCR4 and either removed at the time of infection or maintained in culture during infection, with identical results. The results of SDF-1 binding (Fig. 3) and Ca2+ mobilization data (Fig. 1 and 2) are summarized below (+, near wild-type activity; +/−, <50% of wild-type activity; −, no significant activity detectable). Chimeras 2442 and 2242 did not respond to SDF-1 by Ca2+ mobilization but have been shown to be on the cell surface by FACS at near wild-type levels (data not shown). Chimera 2244 is expressed on the surface, but at <10% of the wild-type level (data not shown). Data shown are the average and standard error of independent experiments repeated at least three times. RLU, relative light units.
Our infection results also demonstrated that signaling and coreceptor function are independent activities of CXCR4. The CXCR4 mutant CXCR4-NAA, which largely failed to signal, supported HIV-1 entry. Consistent with a previous report (35), treatment of cells with pertussis toxin eliminated detectable signal transduction by CXCR4 (Fig. 2A) but did not eliminate viral entry, integration, or long terminal repeat expression, all of which are required for the final detection of luciferase in this assay. Several CXCR4 mutants that were incapable of binding SDF-1 (2444b) or that did not signal in response to any chemokine ligand (2442, 4442, and CXCR4-QAAN) still supported HIV-1 virus entry, providing further evidence that SDF-1 binding and CXCR4 activation are independent of CXCR4 coreceptor function.
Direct binding of X4 Envs to CXCR4.
Direct binding of HIV-1 Envs to chemokine receptors has been demonstrated for both CXCR4 (4, 34, 39, 43) and CCR5 (37, 41, 52, 61, 67, 68). However, since chemokine receptors do not normally serve as the primary binding receptors for HIV-1, it is not clear what type of contact between Env and the coreceptors is necessary for Env-mediated fusion. Coreceptor mutants that dissociate Env binding from triggering the conformational changes that lead to fusion will be valuable in dissecting the functional domains of CXCR4 and defining their role in virus-membrane fusion.
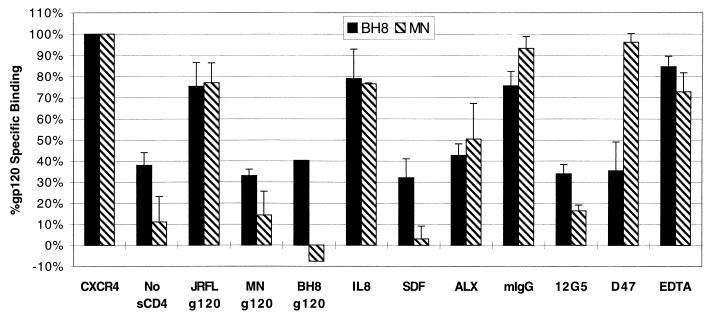
To address the relationship between the ability of CXCR4 to support Env-mediated fusion and gp120 binding, we adapted the SDF-1 binding assay to detect direct binding of X4 Envs to cells expressing CXCR4 or mutant receptors. We used iodinated gp120s from the X4 HIV-1 strains HXB, BH8, and MN (6, 12). Soluble CD4 (sCD4) was included in all assays except where noted. As shown in Fig. 5, binding of gp120 to cells expressing CXCR4 was observed only in the presence of sCD4, consistent with the conformational changes induced by CD4 that are believed to expose the chemokine receptor binding site on gp120 (52, 54, 55, 61, 67). In addition, binding was observed only when cells expressed CXCR4; we detected no binding to cells expressing CXCR2 or CCR5 (Fig. 6). Binding of the iodinated gp120s to CXCR4-positive cells was inhibited by unlabeled BH8 and MN gp120s but not by the R5 JRFL gp120 (Fig. 5). CXCR4-gp120 binding was also inhibited by SDF-1, ALX40-4C (a CXCR4 antagonist [21]), and a MAb directed against CXCR4 (12G5). Binding was not inhibited by IL-8 or a control mouse MAb (mIgG). In addition, a MAb (D47) specifically directed against the V3 loop of BH8 prevented BH8, but not MN, binding to CXCR4-expressing cells (Fig. 5). Since calcium ions are required for Env-mediated fusion in a post-CD4 binding step (18), we conducted Env binding assays in a modified binding buffer containing no divalent cations and including 10 mM EDTA. These conditions had no effect on gp120 binding, indicating that the requirement of divalent cations for HIV fusion is not at the level of coreceptor binding.
FIG. 5.
Binding of X4 gp120s directly to CXCR4. The X4 gp120s from BH8 and MN were iodinated and used for binding to 4 × 105 transfected 293T cells expressing CXCR4. All conditions contained 100 nM sCD4 except where indicated. Values represent the average and standard error of two to three independent experiments. To best represent the signal-to-noise levels achieved in this assay, only background binding to filters alone was subtracted from values. Raw values of binding and background binding to cells not expressing CXCR4 are presented in Fig. 6. BH8 exhibited high background binding in the presence of cells regardless of blocking or transfection conditions, and thus the minimal binding of BH8 in the presence of cells was 30% of total binding. Blocking agents and concentrations were as follows: JRFL gp120 (R5), MN gp120 (X4), and BH8 gp120 (X4) Envs (250 to 500 nM); 12G5 (anti-CXCR4), D47 (BH8-specific anti-V3 loop), and mIgG (pooled mouse IgG) MAbs (10 μg/ml); IL-8 (CXCR2 ligand) and SDF-1α (CXCR4 ligand) chemokines (100 nM); EDTA (10 mM); and ALX40-4C (anti-CXCR4 antagonist) (5 to 10 μM).
FIG. 6.
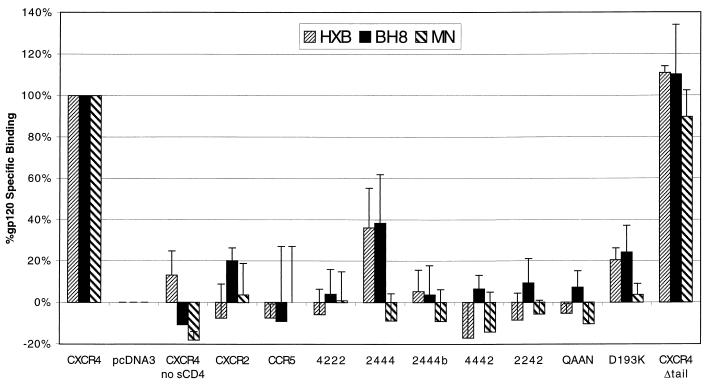
Multiple regions of CXCR4 are required for detectable binding of X4 HIV-1 gp120s. Radiolabeled HXB, BH8, and MN gp120 proteins were used for binding to transiently transfected 293T cells as for Fig. 5. Cells were transfected with the constructs indicated, and background values of binding to cells transfected with pcDNA3 vector alone were subtracted from all measurements. Values represent the average and range of two independent experiments. Constructs have been tested two to four times. Representative raw values for binding to cells containing CXCR4, cells transfected with pcDNA3, and binding to the filter alone were 3,300, 1,600, and 800 cpm for HXB (42,000 cpm added), 2,600, 1,500, and 900 cpm for BH8 (100,000 cpm added), and 7,500, 3,200, and 2,700 cpm for MN (80,000 cpm added). HXB and BH8 are nearly identical clones of the X4 HIV-1 strain IIIB that were prepared and tested completely independently but yielded nearly identical results. For measurement of steady-state kinetics, the proportion of radioligand bound (2 to 9%) is within the optimal range for linear detection of radioligand binding (<10%). Values for binding to membrane-bound CD4 were two- to threefold higher than values for binding to CXCR4 in the presence of sCD4 (data not shown). Radiolabeled JRFL gp120 control exhibited no significant binding to CXCR4 despite robust binding to CCR5 (data not shown).
To address the role that gp120 binding plays in coreceptor function of CXCR4, we screened the panel of CXCR4 chimeras and mutants to determine their ability to bind iodinated gp120 (Fig. 6). Our results indicate that detectable binding of X4 Envs to CXCR4 requires nearly all extracellular regions of CXCR4. Even relatively minor changes to CXCR4, such as D193K, QAAN, and 2444, significantly diminished gp120 binding. This result is consistent with our finding that nearly all regions of CXCR4 contribute to coreceptor function but is surprising since most of these mutants supported HIV-1 infection at some level (Fig. 4). The one mutant that fully supported X4 Env binding, CXCR4Δtail, is expressed at slightly higher levels than wild-type CXCR4 and was capable of binding gp120 accordingly. Thus, detectable binding of monomeric gp120 to CXCR4 does not necessarily correlate with the ability of a coreceptor to support virus infection.
DISCUSSION
To define the interaction of the chemokine receptor CXCR4 with its ligands, we used a panel of CXCR4 mutants to distinguish between SDF-1 binding, SDF-1-mediated CXCR4 activation, HIV-1 gp120 binding, and HIV-1 coreceptor activity of CXCR4. The regions identified in this study that contribute to SDF-1 binding and activation are summarized graphically in Fig. 7. The amino-terminal region of CXCR4 constituted an important SDF-1 binding domain. Replacement of the first 27 residues of CXCR4 (up to the first Cys residue) with the corresponding region from CXCR2 decreased SDF-1 binding, while replacement of the entire amino-terminal domain completely abrogated SDF-1 binding. Whether SDF-1 interacts directly with this region or whether these mutations affect overall CXCR4 structure is not known, but it is important to note that chimera 2444b supported efficient HIV-1 infection and MAb 12G5 binding, two conformationally sensitive interactions. In contrast to the N terminus, alteration of the second and third ECLs of CXCR4 had little effect on SDF-1 binding.
While the amino-terminal domain of CXCR4 was critical for ligand binding, residues in ECL2 comprising an acidic EADD sequence were critical for receptor activation. Thus, CXCR4-QAAN bound SDF-1 as well as wild-type CXCR4 but failed to signal. Residues in the third ECL of CXCR4 may also contribute to signaling, as demonstrated by the undetectable signaling response of 4442, but we cannot exclude residues in the adjacent transmembrane domains of ECL3 from influencing these results. We also found that the conserved DRY motif in the second intracellular loop of CXCR4 was important for signaling, consistent with previous characterization of this motif in other chemokine receptors and G-protein-coupled receptors (22, 26, 28–30, 32, 64).
Receptor mutants that failed to bind detectable levels of chemokine also failed to signal in response to ligand binding with two exceptions: chimera 2444 signaled in response to SDF-1, and chimera 4222 signaled in response to GROα. While we have not quantified 50% effective concentrations for these chimeras to determine if their activation is quantitatively comparable to that of the wild type, we note that similar effects are well documented in the literature and have been observed with other chemokine receptors. For example, multiple CXCR2 (1) and CCR2 (53) chimeras that exhibit only minimal detectable binding nonetheless signal robustly in response to cognate chemokine ligands, suggesting that detection of high-affinity binding is not absolutely required for signal transduction.
Our results are consistent with a previously proposed two-site model of chemokine-chemokine receptor interaction in which the amino terminus of the chemokine receptor plays a major role in the initial binding of the chemokine, while interaction of the chemokine with the loops of the receptor transmits an activation signal (1, 17, 31, 44, 57). The recent determination of the nuclear magnetic resonance structure of SDF-1 and the accompanying analysis of SDF-1 mutants (16) and of SDF-1-derived peptides (36) provides a model for the interaction of SDF-1 and CXCR4 that complements our current work. Crump et al. showed that SDF-1 binds to CXCR4 by using the RFFESH motif at amino acids 12 to 17 of SDF-1 and subsequently mediates activation of CXCR4 with the first two amino acids of SDF-1 (Lys-Pro) (16). Heveker et al. used a peptide-based strategy to reach very similar conclusions about the functional structures of SDF-1 (36). These two complementary studies of SDF-1 suggest that the two amino-terminal residues of SDF-1 are absolutely critical for signaling, that additional residues in the amino terminus distal to the CXC motif (residues 3 to 8) also contribute to signaling, and that residues proximal to the CXC motif that are focused near positions 12 to 14 (RFF) are critical for SDF-1 binding.
By analogy to other chemokine receptors such as CXCR2, both Crump et al. and Heveker et al. speculate that the primary binding event of SDF-1 occurs at the amino terminus of CXCR4 and that the activation of the receptor occurs through a pocket formed by the loops of CXCR4 (16, 36). In conjunction with these SDF-1 mapping data, our data suggest a model in which the binding of SDF-1 to CXCR4 involves SDF-1 residues R12, F13, and F14 binding directly to the CXCR4 amino terminus, with the proximal amino terminus of CXCR4 playing an especially critical role. The cumulative data also suggest that activation of CXCR4 occurs, at least in part, by contact of SDF-1 residues K1 and P2 with ECL2 of CXCR4. Additional biophysical evidence to confirm this model of SDF-1–CXCR4 interaction is clearly required.
Previous studies have demonstrated that signaling by the chemokine receptor CCR5 is not required for coreceptor function (3, 7, 22, 26, 32), but with the exception of a study that included pertussis toxin in an infection (35), we are not aware of similar studies that eliminate the ability of other coreceptors to signal. We eliminated CXCR4 signaling by altering a predicted G-protein-coupling motif (CXCR4-NAA), by chemical uncoupling of G-protein interaction (pertussis toxin), and by creating mutants that are unable to mediate SDF-1-signal transduction (2444b, 2442, 4442, and CXCR4-QAAN). Nevertheless, most of these modifications did not eliminate coreceptor function. Our analysis has thus separated the abilities of CXCR4 to bind SDF-1, to signal in response to SDF-1, and to act as a coreceptor for HIV-1.
Using virus infection assays, we found that HIV-1 Env utilized a conformationally complex structure involving each of the major extracellular regions of CXCR4 for coreceptor function, in agreement with our previous results using a cell-cell fusion assay (40). The contribution of many regions of CXCR4 to coreceptor function implies that a highly conformational structure created by all extracellular regions of CXCR4 interacts with Env. We addressed the possibility that the failure of some coreceptor mutants to support viral fusion is due to their inability to bind Env. The ability to divide coreceptor function into two discrete steps, Env binding and Env triggering, would help identify important chemokine receptor structures that mediate Env conformational changes and would increase our understanding of the fusion mechanism of HIV. By adapting the conditions of chemokine binding, we established a reliable and specific binding assay for detecting X4 Env binding to CXCR4. While this assay is not as robust as similar assays using R5 Envs, multiple controls, including an Env-specific MAb, Env proteins of different coreceptor tropisms, a CXCR4-specific MAb, and CXCR4 antagonists and agonists, demonstrated the specificity of this assay.
We found that monomeric gp120 binding to CXCR4 did not correlate with the ability of CXCR4 to support Env-mediated fusion. Several CXCR4 mutants and chimeras that efficiently supported virus infection were either diminished in the capacity to bind gp120 or completely unable to do so. We have obtained similar results for R5 gp120 binding to CCR5, in which even small perturbations of the CCR5 protein can completely disrupt detectable gp120 binding without strongly affecting coreceptor activity (reference 24 and our unpublished results). Since CD4 serves as the primary receptor for HIV-1 Env, a strong interaction of gp120 with CXCR4 may not be required for coreceptor function. Alternatively, oligomeric Env may interact more strongly with CXCR4 than the monomeric gp120 molecules used in this study. In addition, the interaction of Env with CXCR4 may be followed rapidly by conformational changes in Env that lead to membrane fusion, making even a low-affinity interaction essentially irreversible in the context of virus infection. The dissociation of coreceptor binding of Env and coreceptor fusion activity is a step toward understanding the molecular basis of how the chemokine receptors function as fusion coreceptors.
ACKNOWLEDGMENTS
We thank Jane Sung, Sarah Baik, Joe Rucker, Rolf Windh, Dave Manning, Thue Schwartz, Joe Hesselgesser, and Richard Horuk for technical advice and support. MAbs 10G2 and 807 were graciously supplied by Caroline Hebert and Jin Kim (Genentech) and by Francisco Gonzalez-Scarano (University of Pennsylvania), respectively. A number of reagents used in these experiments were provided by the NIH AIDS Research Reference and Reagent Program.
This work was supported by NIH grants AI-35383 and AI-40880 to Robert W. Doms and Howard Hughes Medical Institute predoctoral fellowships to Joanne F. Berson and Benjamin J. Doranz.
REFERENCES
- 1.Ahuja S K, Lee J C, Murphy P M. CXC chemokines bind to unique sets of selectivity determinants that can function independently and are broadly distributed on multiple domains of human interleukin-8 receptor B. J Biol Chem. 1996;271:225–232. doi: 10.1074/jbc.271.1.225. [DOI] [PubMed] [Google Scholar]
- 2.Aiuti A, Webb I J, Bleul C, Springer T, Gutierrez-Ramos J C. The chemokine SDF-1 is a chemoattractant for human CD34+ hematopoietic cells and provides a new mechanism to explain the mobilization of CD34+ progenitors to peripheral blood. J Exp Med. 1997;185:111–120. doi: 10.1084/jem.185.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alkhatib G, Locati M, Kennedy P E, Murphy P M, Berger E A. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: independence from G protein signaling and importance of coreceptor downmodulation. Virology. 1997;234:340–348. doi: 10.1006/viro.1997.8673. [DOI] [PubMed] [Google Scholar]
- 4.Bandres J C, Wang Q F, O’Leary J, Baleaux F, Amara A, Hoxie J A, Zolla-Pazner S, Gorny M K. Human immunodeficiency virus (HIV) envelope binds to CXCR4 independently of CD4, and binding can be enhanced by interaction with soluble CD4 or by HIV envelope deglycosylation. J Virol. 1998;72:2500–2504. doi: 10.1128/jvi.72.3.2500-2504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger E A, Doms R W, Fenyö E M, Korber B T M, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 6.Berson J F, Long D, Doranz B J, Rucker J, Jirik F R, Doms R W. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S G, Caron M G, Cullen B R. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 9.Bleul C C, Fuhlbrigge R C, Casasnovas J M, Aiuti A, Springer T A. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1) J Exp Med. 1996;184:1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brelot A, Heveker N, Pleskoff O, Sol N, Alizon M. Role of the first and third extracellular domains of CXCR-4 in human immunodeficiency virus coreceptor activity. J Virol. 1997;71:4744–4751. doi: 10.1128/jvi.71.6.4744-4751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen B K, Saksela K, Andino R, Baltimore D. Distinct modes of human immunodeficiency virus type 1 proviral latency revealed by superinfection of nonproductively infected cell lines with recombinant luciferase-encoding viruses. J Virol. 1994;68:654–660. doi: 10.1128/jvi.68.2.654-660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choe H, Farzan M, Konkel M, Martin K, Sun Y, Marcon L, Cayabyab M, Berman M, Dorf M E, Gerard N, Gerard C, Sodroski J. The orphan seven-transmembrane receptor Apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J Virol. 1998;72:6113–6118. doi: 10.1128/jvi.72.7.6113-6118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 14.Chuntharapai A, Lee J, Hebert C Z, Kim K J. Monoclonal antibodies detect different distribution patterns of IL-8 receptor A and IL-8 receptor B on human peripheral blood leukocytes. J Immunol. 1994;153:5682–5688. [PubMed] [Google Scholar]
- 15.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 16.Crump M P, Gong J H, Loetscher P, Rajarathnam K, Amara A, Arenzana-Seisdedos F, Virelizier J L, Baggiolini M, Sykes B D, Clark-Lewis I. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeMartino J A, Riper G V, Siciliano S J, Molineaux C J, Konteatis Z D, Rosen H, Springer M S. The amino terminus of the human C5a receptor is required for high affinity C5a binding and for receptor activation by C5a but not C5a analogs. J Biol Chem. 1994;269:14446–14450. [PubMed] [Google Scholar]
- 18.Dimitrov D S, Broder C C, Berger E A, Blumenthal R. Calcium ions are required for cell fusion mediated by the CD4-human immunodeficiency virus type 1 envelope glycoprotein interaction. J Virol. 1993;67:1647–1652. doi: 10.1128/jvi.67.3.1647-1652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doms R W, Moore J P. HIV coreceptor use: a molecular window into viral tropism. In: Korber B, Foley B, Leitner T, Myers G, Hahn B, McCutchan F, Mellors J, Kuiken C, editors. Human retroviruses and AIDS 1997. Los Alamos, N. Mex: Los Alamos National Laboratory, Theoretical Biology and Biophysics, part III; 1998. pp. 1–12. [Google Scholar]
- 20.Donzella G A, Schols D, Lin S W, Esté J A, Nagashima K A, Maddon P J, Allaway G P, Sakmar T P, Henson G, De Clerq E, Moore J P. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4:72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 21.Doranz B J, Grovit-Ferbas K, Sharron M P, Mao S, Goetz M B, Daar E S, Doms R W, O’Brien W A. A small-molecule inhibitor directed against the chemokine receptor CXCR4 prevents its use as an HIV-1 coreceptor. J Exp Med. 1997;186:1395–1400. doi: 10.1084/jem.186.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doranz B J, Lu Z, Rucker J, Zhang T, Sharron M, Cen Y, Wang Z, Guo H, Du J, Accavitti M A, Doms R W, Peiper S C. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Earl P L, Broder C C, Long D, Lee S A, Peterson J, Chakrabarti S, Doms R W, Moss B. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J Virol. 1994;68:3015–3026. doi: 10.1128/jvi.68.5.3015-3026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edinger, A. L., C. Blanpain, M. Parmentier, and R. W. Doms. Functional dissection of CCR5 coreceptor function through the use of CD4-independent SIV strains. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 25.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 26.Farzan M, Choe H, Martin K A, Sun Y, Sidelko M, Mackay C R, Gerard N P, Sodroski J, Gerard C. HIV-1 entry and macrophage inflammatory protein-1β-mediated signaling are independent functions of the chemokine receptor CCR5. J Biol Chem. 1997;272:6854–6857. doi: 10.1074/jbc.272.11.6854. [DOI] [PubMed] [Google Scholar]
- 27.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 28.Franke R R, König B, Sakmar T P, Khorana H G, Hofmann K P. Rhodopsin mutants that bind but fail to activate transducin. Science. 1990;250:123–125. doi: 10.1126/science.2218504. [DOI] [PubMed] [Google Scholar]
- 29.Franke R R, Sakmar T P, Graham R M, Khorana H G. Structure and function in rhodopsin: studies of the interaction between the rhodopsin cytoplasmic domain and transducin. J Biol Chem. 1992;267:14767–14774. [PubMed] [Google Scholar]
- 30.Fraser C M, Chung F-Z, Wang C-D, Venter J C. Site-directed mutagenesis of human beta-adrenergic receptors: substitution of aspartic acid-130 by asparagine produces a receptor with high-affinity agonist binding that is uncoupled from adenylate cyclase. Biochemistry. 1988;85:5478–5482. doi: 10.1073/pnas.85.15.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gayle R B, Sleath P R, Srinivason S, Birks C W, Weerawarna K S, Cerretti D P, Kozlosky C J, Nelson N, Bos T V, Beckmann M P. Importance of the amino terminus of the interleukin-8 receptor in ligand interactions. J Biol Chem. 1993;268:7283–7289. [PubMed] [Google Scholar]
- 32.Gosling J, Monteclaro F S, Atchison R E, Arai H, Tsou C, Goldsmith M A, Charo I F. Molecular uncoupling of C-C chemokine receptor 5-induced chemotaxis and signal transduction from HIV-1 coreceptor activity. Proc Natl Acad Sci USA. 1997;94:5061–5066. doi: 10.1073/pnas.94.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grynkiewicz G, Poenie M, Tsien R Y. A new generation of Ca+2 indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 34.Hesselgesser J, Halks-Miller M, DelVecchio V, Peiper S C, Hoxie J, Kolson D L, Taub D, Horuk R. CD4-independent association between HIV-1 gp120 and CXCR4: functional chemokine receptors are expressed in human neurons. Curr Biol. 1997;7:112–121. doi: 10.1016/s0960-9822(06)00055-8. [DOI] [PubMed] [Google Scholar]
- 35.Hesselgesser J, Liang M, Hoxie J, Greenberg M, Brass L F, Orsini M J, Taub D, Horuk R. Identification and characterization of the CXCR4 chemokine receptor in human T cell lines: ligand binding, biological activity, and HIV-1 infectivity. J Immunol. 1998;160:877–883. [PubMed] [Google Scholar]
- 36.Heveker N, Montes M, Germeroth L, Amara A, Trautmann A, Alizon M, Schneider-Mergener J. Dissociation of the signalling and antiviral properties of SDF-1-derived small peptides. Curr Biol. 1998;8:369–376. doi: 10.1016/s0960-9822(98)70155-1. [DOI] [PubMed] [Google Scholar]
- 37.Hill C M, Deng H, Unutmaz D, Kewalramani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Labrosse B, Brelot A, Heveker N, Sol N, Schols D, De Clercq E, Alizon M. Determinants for sensitivity of human immunodeficiency virus coreceptor CXCR4 to the bicyclam AMD3100. J Virol. 1998;72:6381–6388. doi: 10.1128/jvi.72.8.6381-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Evidence for cell-surface association between fusin and the CD4-gp120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 40.Lu Z, Berson J F, Chen Y, Turner J D, Zhang T, Sharron M, Jenks M H, Wang Z, Kim J, Rucker J, Hoxie J A, Peiper S C, Doms R W. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc Natl Acad Sci USA. 1997;94:6426–6431. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin K A, Wyatt R, Farzan M, Choe H, Marcon L, Desjardins E, Robinson J, Sodroski J, Gerard C, Gerard N P. CD4-independent binding of SIV gp120 to rhesus CCR5. Science. 1997;278:1470–1473. doi: 10.1126/science.278.5342.1470. [DOI] [PubMed] [Google Scholar]
- 42.Miedema F, Meyaard L, Koot M, Klein M R, Roos M T L, Groenink M, Fouchier R A M, Van’t Wout A B, Tersmette M, Schellekens P T A, Schuitemaker H. Changing virus-host interactions in the course of HIV-1 infection. Immunol Rev. 1994;140:35–72. doi: 10.1111/j.1600-065x.1994.tb00864.x. [DOI] [PubMed] [Google Scholar]
- 43.Missé D, Cerutti M, Schmidt I, Jansen A, Devauchelle G, Jansen F, Veas F. Dissociation of the CD4 and CXCR4 binding properties of human immunodeficiency virus type 1 gp120 by deletion of the first putative alpha-helical conserved structure. J Virol. 1998;72:7280–7288. doi: 10.1128/jvi.72.9.7280-7288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monteclaro F S, Charo I F. The amino-terminal extracellular domain of the MCP-1 receptor, but not the RANTES/MIP-1α receptor, confers chemokine selectivity. J Biol Chem. 1996;271:19084–19092. doi: 10.1074/jbc.271.32.19084. [DOI] [PubMed] [Google Scholar]
- 45.Motulsky H J. GraphPad Prism version 2.0. San Diego, Calif: GraphPad Software, Inc.; 1995. [Google Scholar]
- 46.Murakami T, Nakajima T, Koyanagi Y, Tachibana K, Fujii N, Tamamura H, Yoshida N, Waki M, Matsumoto A, Yoshie O, Kishimoto T, Yamamoto N, Nagasawa T. A small molecule CXCR4 inhibitor that blocks T cell line-tropic HIV-1 infection. J Exp Med. 1997;186:1389–1393. doi: 10.1084/jem.186.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- 48.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 49.O’Brien W A, Sumner-Smith M, Mao S, Sadeghi S, Zhao J, Chen I Y. Anti-human immunodeficiency virus type 1 activity of an oligocationic compound mediated via gp120 V3 interactions. J Virol. 1996;70:2825–2831. doi: 10.1128/jvi.70.5.2825-2831.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Picard L, Wilkinson D A, McKnight A, Gray P W, Hoxie J A, Clapham P R, Weiss R A. Role of the amino-terminal domain of CXCR-4 in human immunodeficiency virus type 1 entry. Virology. 1997;231:105–111. doi: 10.1006/viro.1997.8506. [DOI] [PubMed] [Google Scholar]
- 51.Premack B A, Schall T J. Chemokine receptors: gateways to inflammation and infection. Nat Med. 1996;2:1174–1178. doi: 10.1038/nm1196-1174. [DOI] [PubMed] [Google Scholar]
- 52.Rizzuto C D, Wyatt R, Hernández-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 53.Samson M, LaRosa G, Libert F, Paindavoine P, Detheux M, Vassart G, Parmentier M. The second extracellular loop of CCR5 is the major determinant of ligand specificity. J Biol Chem. 1997;272:24934–24941. doi: 10.1074/jbc.272.40.24934. [DOI] [PubMed] [Google Scholar]
- 54.Sattentau Q J, Moore J P. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sattentau Q J, Moore J P, Vignaux F, Traincard F, Poignard P. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67:7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schols D, Struyf S, Van Damme J, Esté J A, Henson G, De Clerq E. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J Exp Med. 1997;186:1383–1388. doi: 10.1084/jem.186.8.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siciliano S J, Rollins T E, DeMartino J, Konteatis Z, Malkowitz L, VanRiper G, Bondy S, Rosen H, Springer M S. Two-site binding of C5a by its receptor: an alternative binding paradigm for G protein-coupled receptors. Proc Natl Acad Sci USA. 1994;91:1214–1218. doi: 10.1073/pnas.91.4.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swillens S. Interpretation of binding curves obtained with high receptor concentrations: practical aid for computer analysis. Mol Pharmacol. 1995;47:1197–1203. [PubMed] [Google Scholar]
- 59.Tachibana K, Hirota S, Iizasa H, Yoshida H, Kawabata K, Kataoka Y, Kitamura Y, Matsushima K, Yoshida N, Nishikawa S-I, Kishimoto T, Nagasawa T. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature. 1998;393:591–594. doi: 10.1038/31261. [DOI] [PubMed] [Google Scholar]
- 60.Tashiro K, Tada H, Heilker R, Shirozu M, Nakano T, Honjo T. Signal sequence trap: a cloning strategy for secreted proteins and type 1 membrane proteins. Science. 1993;261:600–603. doi: 10.1126/science.8342023. [DOI] [PubMed] [Google Scholar]
- 61.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 62.Trkola A, Paxton W A, Monard S P, Hoxie J A, Siani M A, Thompson D A, Wu L, Mackay C R, Horuk R, Moore J P. Genetic subtype-independent inhibition of human immunodeficiency virus type 1 replication by CC and CXC chemokines. J Virol. 1998;72:396–404. doi: 10.1128/jvi.72.1.396-404.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ueda H, Siani M A, Gong W, Thompson D A, Brown G G, Wang J M. Chemically synthesized SDF-1α analogue, N33A, is a potent chemotactic agent for CXCR4/Fusin/LESTR-expressing human leukocytes. J Biol Chem. 1997;272:24966–24970. doi: 10.1074/jbc.272.40.24966. [DOI] [PubMed] [Google Scholar]
- 64.Wang C-D, Buck M A, Fraser C M. Site-directed mutagenesis of alpha2A-adrenergic receptors: identification of amino acids involved in ligand binding and receptor activation by agonists. Mol Pharmacol. 1991;40:168–179. [PubMed] [Google Scholar]
- 65.Wang Z X, Berson J F, Zhang T Y, Cen Y H, Sun Y, Sharron M, Lu Z H, Peiper S C. CXCR4 sequences involved in coreceptor determination of human immunodeficiency virus-1 tropism: unmasking of activity with M-tropic env glycoproteins. J Biol Chem. 1998;273:15007–15015. doi: 10.1074/jbc.273.24.15007. [DOI] [PubMed] [Google Scholar]
- 66.Winkler C, Modi W, Smith M W, Nelson G W, Wu X, Carrington M, Dean M, Honjo T, Tashiro K, Yabe D, Buchbinder S, Vittinghoff E, Goedert J J, O’Brien T R, Jacobson L P, Detels R, Donfield S, Willoughby A, Gomperts E, Vlahov D, Phair J, O’Brien S J ALIVE Study; Hemophilia Growth and Development Study (HGDS); Multicenter AIDS Cohort Study (MACS); Multicenter Hemophilia Cohort Study (MHCS); San Francisco City Cohort (SFCC) Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 67.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 68.Wu L, LaRosa G, Kassam N, Gordon C J, Heath H, Ruffing N, Chen H, Humblias J, Samson M, Parmentier M, Moore J P, Mackay C R. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J Exp Med. 1997;186:1373–1381. doi: 10.1084/jem.186.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zou Y R, Kottmann A H, Huroda M, Taniuchi I, Littman D R. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]