SUMMARY
The human UBR5 is a single polypeptide chain HECT-type E3 ubiquitin ligase essential for embryonic development in mammals. Dysregulated UBR5 functions like an oncoprotein to promote cancer growth and metastasis. Here we report that UBR5 assembles into a dimer and tetramer. Our cryo-EM structures reveal that two crescent-shaped UBR5 monomers assemble head-to-tail to form the dimer, and two dimers bind face-to-face to form the cage-like tetramer with all four catalytic HECT domains facing the central cavity. Importantly, the N-terminal region of one subunit and the HECT of the other form an “intermolecular jaw” in the dimer. We show the jaw-lining residues are important for function, suggesting that the intermolecular jaw functions to recruit ubiquitin-loaded E2 to UBR5. Further work is needed to understand how oligomerization regulates the UBR5 ligase activity. This work provides a framework for structure-based anticancer drug development and contributes to a growing appreciation of E3 ligase diversity.
INTRODUCTION
Ubiquitination is a common post-translational modification (PTM) that involves the attachment of ubiquitin (Ub) to its protein substrates1. It occurs in eukaryotes and regulates a variety of biological processes such as signal transduction, apoptosis, immune response, and DNA repair1,2. The Ubiquitin Proteasome System (UPS) is responsible for a large portion of regulated proteostasis in eukaryotes 3. In this system, a cascade of three enzymes (the Ub-activating enzyme E1, Ub-conjugating E2, and Ub ligase E3) act sequentially to ubiquitylate target proteins, with the E3 predominantly determining the substrate specificity 4,5. Most E3s can be classified into four families: The “Really Interesting New Gene” (RING) family, the “Ring-Between-Ring” (RBR) family, the U-box family, and the “Homology to E6AP C-Terminus” (HECT) family6. The RING family members are often formed by multi-protein complexes and mediate direct transfer of Ub from an Ub-loaded E2 to a substrate without forming an E3–Ub intermediate 7,8. The U-box family members contain a conserved ~70 amino acids U-box motif which is similar to the RING motif but lacks the zinc chelating residues. The U-box family E3 ligase transfers Ub in a similar manner to the RING family9. In contrast, the RBR and HECT family members are usually single polypeptide chains and catalyze a two-step reaction in which Ub is first transferred from Ub–E2 to the E3 to form a Ub–E3 covalent intermediate, followed by a second transfer of the Ub to a substrate 4,10–13. Humans encode 600-700 E3 ligases, accounting for ~5% of the genome 14–16. Perhaps due to their giant size and often flexible architecture, only a dozen or so full-length E3 ligase structures have been reported 17,18, including the yeast Ubr1 19, the human and parasite HUWE1 4,13, the yeast GID3 complex 20,21, the chicken Fanconi anemia core complex 22, the mouse AAA-E3 ligase RNF213 23, the human ASB9 24, the human BRCA1-BARD E3 ligase 25,26, and two human E3-E3 super-assemblies, SCF–ARIH1 and CUL5–ARIH2 27,28. The majority of E3 ligase structures were determined by cryo-EM, which has become a method of choice to solve the structures of many large and partially flexible protein complexes 29.
Humans encode seven N-end rule pathway E3 ligases, UBR1-7, four of which (UBR1, UBR2, UBR4 and UBR5) contain an N-degron recognition UBR Box 30. UBR5 is unique among them with a HECT domain and therefore is the only one that belongs to the HECT family 31. UBR5 is also known as EDD or EDD1 (E3 ligase identified by Differential Display) and HYD (hyperplastic discs) 32,33. UBR5 assembles K11-K48 linked branched Ub chains34, and has a wide range of substrates, including β-catenin, TopBP1, TERT, CDK9, ATMIN, PEPCK1, CAPZA1, CDC73 , which are implicated in multiple cellular processes, such as DNA damage, metabolism, transcription, apoptosis, and immunoregulation 35–41, and UBR5 has been implicated in disassembly of mitotic checkpoint complexes 42. Hence, UBR5 is a key regulator of cell signaling relevant to broad areas of cancer biology 34,43. In addition, UBR5 is highly expressed in a substantial proportion of breast, ovarian, prostate, gastric, and pancreatic cancers 44–48. Recently, UBR5 E3 pathway was shown to play a key role in the aggression of breast and ovarian cancers by enabling the cancer cells to survive and resist immune attack and standard treatments 44,46, UBR5 destabilizes the tumor suppressor CDC73 to promote aggressive breast cancer 49. The tumor-promoting effect of UBR5 has been largely attributed to its ubiquitin ligase activity, highlighting the potential for targeting UBR5 for anticancer drug development and the importance of determining UBR5’s structure and molecular mechanism 46.
UBR5 contains 2799 amino acids and has an estimated mass of 309 kDa (Figure 1A). The HECT domain is at the C-terminus with a larger N-lobe that interacts with E2s and a C-lobe containing the catalytic Cys2768 50. In addition to HECT, UBR5 also contains a Ub association domain (UBA) that interacts with Ub 51,52, two predicted small β-barrel domains (SBB1 and SBB2), a 70-residue zinc finger UBR-Box that recognizes the N-degron 31,53,54, and an MLLE (the term comes from a signature motif, MLLEKITG) domain that mediates protein-protein interactions 55. Structures of the C-lobe and MLLE have been reported 50,55. Additionally, previous studies have determined the crystal structures of the isolated HECT domains of several HECT E3 ligases 56–58. However, the structure of the full-length UBR5 has been lacking. We have determined the cryo-EM structure of the human full-length UBR5 at up to 2.66 Å resolution and revealed that UBR5 assembles into dimers and tetramers in solution. The high-resolution structures contribute to the ubiquitin field, shed light on the UBR5 catalyzed transthiolation reaction mechanism, and provide a platform for developing UBR5 inhibitors for the potential treatment of cancers. Future studies are required to understand whether the UBR5 dimer and tetramer possess distinct functions or substrates.
Figure 1. Cryo-EM of the UBR5 dimer and tetramer.
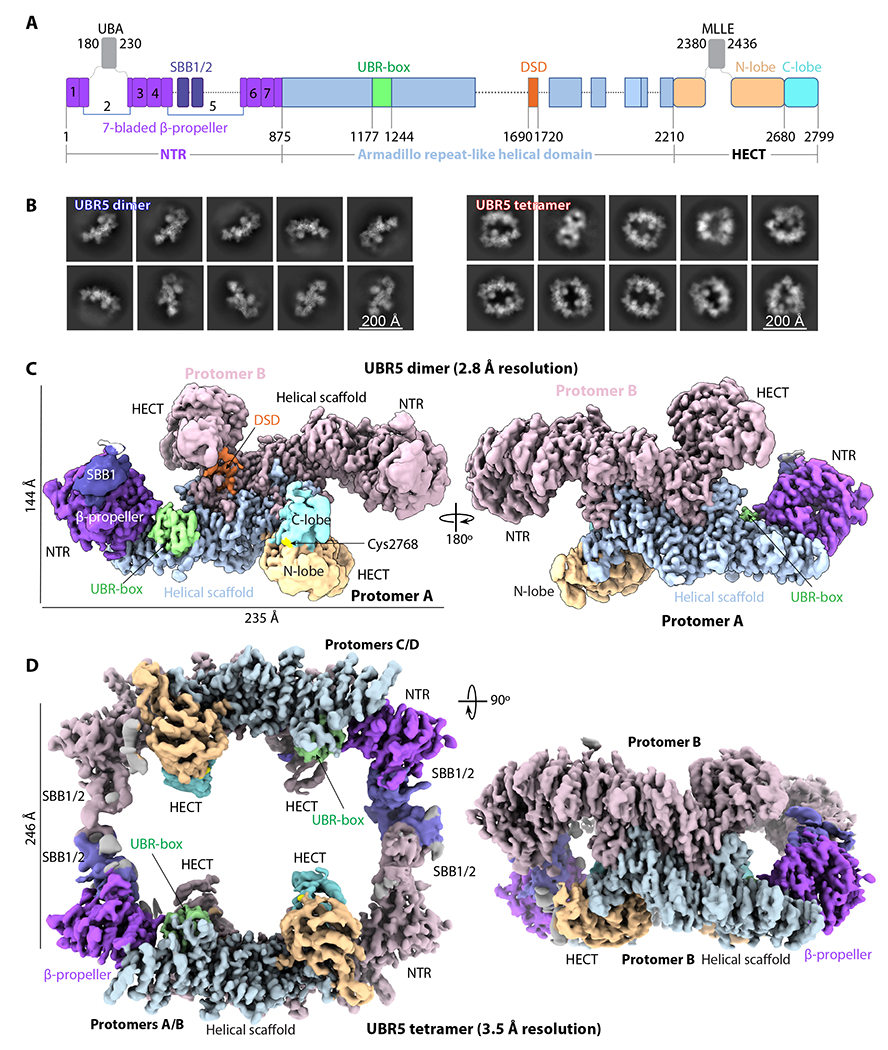
(A) Domain architecture of the human E3 ligase UBR5. Unresolved regions in the EM map are shown as dash lines. The disordered UBA and MLLE domains are shown as gray squares. (B) Selected 2D class averages of the dimer (left) and tetramer (right). (C-D) Cryo-EM 3D maps of the dimer (C) and tetramer (D). Domains are colored as in (A).
RESULTS
UBR5 assembles into a dimer and a tetramer as revealed by cryo-EM
We recombinantly expressed human UBR5 with an N-terminal FLAG tag in insect cells and purified the protein via affinity and size exclusion chromatography. Purified UBR5 eluted from the gel filtration column in two partially overlapping broad peaks at volumes that were consist with a dimer (620 kDa) and a tetramer (1.2 MDa) (Figure S1A–B). PEPCK1 was previously shown to be a UBR5 substrate in vivo 59,60. We demonstrated that the purified UBR5 composed of mixture of dimer and tetramer was enzymatically active as it ubiquitylated PEPCK1 in vitro in the presence of Ub, E1, and E2D2, while the catalytically inactive UBR5 mutant protein (C2768S) was unable to do so (Figure S1C). Cryo-EM and 2D image classification of the putative dimer elution peak contained mostly dimers and the putative tetramer elution peak contained mostly tetramers, although dimer and tetramer particles were found in each sample (Figure S1D and 1B). We derived a cryo-EM 3D map of the dimer at an average resolution of 2.8 Å in C1 symmetry with an overall dimension of 108 Å × 144 Å × 235 Å (Figure 1C, Figures S2–S3, Table S1). Resolution increased to 2.66 Å when 2-fold symmetry was applied. However, the two ends of the dimer map, corresponding to the N-terminal region (NTR) and the HECT region, are of lower resolution. Therefore, we performed focused refinement on the two regions separately, and improved the map quality in these regions, leading to a high-resolution C2 symmetric composite EM map of the dimer (Figures S2 and S4). Next, we combined an untilted and a 30° tilted dataset to obtain a composite EM map of the UBR5 tetramer at 3.5 Å average resolution (Figures S5–S6). The 3D map was determined in the C1 symmetry, because application of the C2 or D2 symmetry led to a loss of densities in the HECT and NTR, indicative of mobility in these regions. The tetramer is assembled by interaction between the NTR regions of the two dimers, leading to more density but not increased resolution of the junction region. The tetramer structure has an overall dimension of 221 Å × 238 Å × 126 Å, with the four catalytic HECT domains all facing inside the large central cavity (Figure 1D).
The UBR5 dimer structure
The high-quality EM map of the UBR5 dimer allowed us to build an atomic model for most regions of UBR5, except for the UBA, SBB2, and MLLE domains that are flexibly linked to the core of the dimer complex (Figure 2A, Figure S7, Video S1). Two UBR5 subunits assemble head-to-tail via their respective helical scaffold to form an extended dimer interface of 7936 Å2. The crystal structures of isolated UBA and MLLE domains are known 51,55. UBA is flexible in the UBR5 dimer because it is tethered to the NTR by a preceding 100-residue linker and a following 120-residue linker. Contrary to a previous bioinformatic analysis55, MLLE does not precede the HECT, instead it is inserted into the N-lobe of the HECT domain and is flexibly connected by a 63-residue-long linker on each side. Strikingly, the NTR of one subunit is close to the HECT domain of the other subunit, forming a gap that we speculate could bind an E2–Ub. For this reason, we tentatively term the gap an “intermolecular E2–Ub jaw”. Individual UBR5 molecules adopt a crescent shape with a bridging helical scaffold (residues 876-2209) interrupted by the well-ordered E3 substrate N-terminal recognition UBR-box (1177-1244). The NTR (1-875) and the C-terminal HECT domain (2210-2799) straddles the two ends of the rigid helical scaffold platform, enclosing a space for E3 substrate binding.
Fig. 2. The UBR5 dimer structure.
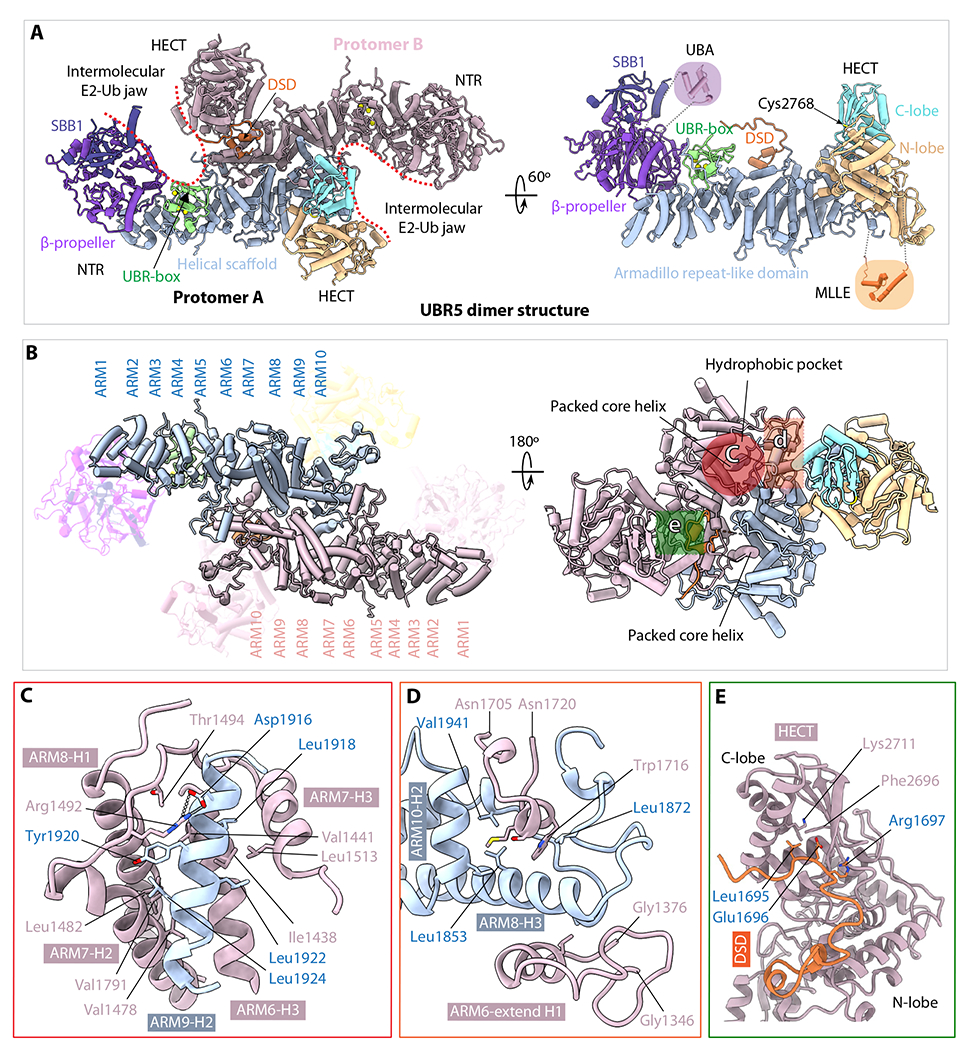
(A) The dimer structure in cartoon viewed from top along the 2-fold symmetry axis (left) and a monomer in a side view (right). Th structures are colored as in Figure 1A. The crystal structures of the UBA (PDB ID 2QHO) and MLLE (PDB ID 3NTW) are shown in shadowed cartoons for illustrative purpose only; they are invisible in the EM map. (B) Top and bottom views of the middle Armadillo-like helical scaffold that primarily mediates UBR5 dimerization. The three major interacting regions are marked by three colored shapes in the right panel. (C) Close-up view of the hydrophobic interface region marked by the red circle in (B). Residues involved in dimerization such as the salt bridge between Arg1492 and Asp1916 are shown as sticks. (D) Close-up view of the interface region marked by the orange square in (B). This region contains both hydrophobic and H-bonding interactions. (E) Close-up view of the region marked by green square in (B), which involves the domain-swapped dimerization (DSD) motif.
The helical scaffold is composed of 10 Armadillo-like (ARM1-10) repeats (Figure 2B). The second half of the helical scaffold, ARM5-10, is near the 2-fold symmetry axis and is primarily responsible for the dimer interface. The two symmetry-related ARM5-10 motifs interact with each other hydrophobically. Specifically, the ARM9 interface helix 2 (ARM9-H2) of one subunit inserts into an extended hydrophobic pocket formed by ARM6-8 of the partner subunit; this interface is further stabilized by a salt bridge between Arg1492 of one subunit and Asp1916 of the other (Figure 2B–C). The peripheral dimer interface is also largely hydrophobic and is formed by the ARM8 H3 helix inserting into a space in the partner subunit between the ARM6 extended H1 helix and an extended loop (1705-1720) (Figure 2D). Interestingly, the dimer interface is further stabilized by a domain-swapped dimerization loop (DSD loop, aa 1690-1700) interacting with both N- and C-lobes of the HECT domain (Figure 2E). The presence of the DSD loop in the catalytic domain likely has functions beyond dimerization, because E3 ligase activity involves the relative movement of the N- and the C-lobes, as described below.
The N-terminal region (NTR)
The NTR of a UBR5 dimer is located above ARM1-4 that are not involved in the helical scaffold dimerization (Figure 3A–B). The NTR is composed of SBB1 and β-propeller with SBB1 sitting above the β-propeller. The SBB1 contains five β-strands with an SH3-like fold. The SH3 fold is typically involved in protein-protein interaction or nucleic acid binding 61 or in substrate degron recognition 17. The UBR5 SBB1 lines the intermolecular E2-Ub jaw and could be involved in substrate binding. The β-propeller domain is seven-bladed with a central pore, with each blade containing four β-strands (Figure 3B). Blade 1 is composed of the β-strands from both ends of the NTR (Figure 1A and S7) and contains an extra α-helix that connects the β-propeller domain with the middle helical scaffold. Blades 2 to 7 are arranged clockwise to complete the β-propeller fold with blade 1. Blade 2 features two extended loops that link the unresolved, mobile UBA domain. SBB1-2 are inserted into blade 5. Finally, blade 7 interacts with the UBR-box (Figure 3A). β-propeller is also often involved in protein-protein interaction 62, and several multi-protein E3s including KLHDC1-3, KLHDC10, and Gid11 recruit either C-degrons or N-degrons to the central pore of their respective β-propellers 17. However, the central pore of the UBR5 β-propeller is partially covered by SBB1 on top. Because the C-terminal catalytic HECT and MLLE directly interact with UBR5 E3 substrates 40,59,63, it will be interesting to investigate if the NTR (SBB1/2 and β-propeller) may also contribute to substrate binding or interact with other protein partners.
Figure 3. The NTR and UBR-box structures.
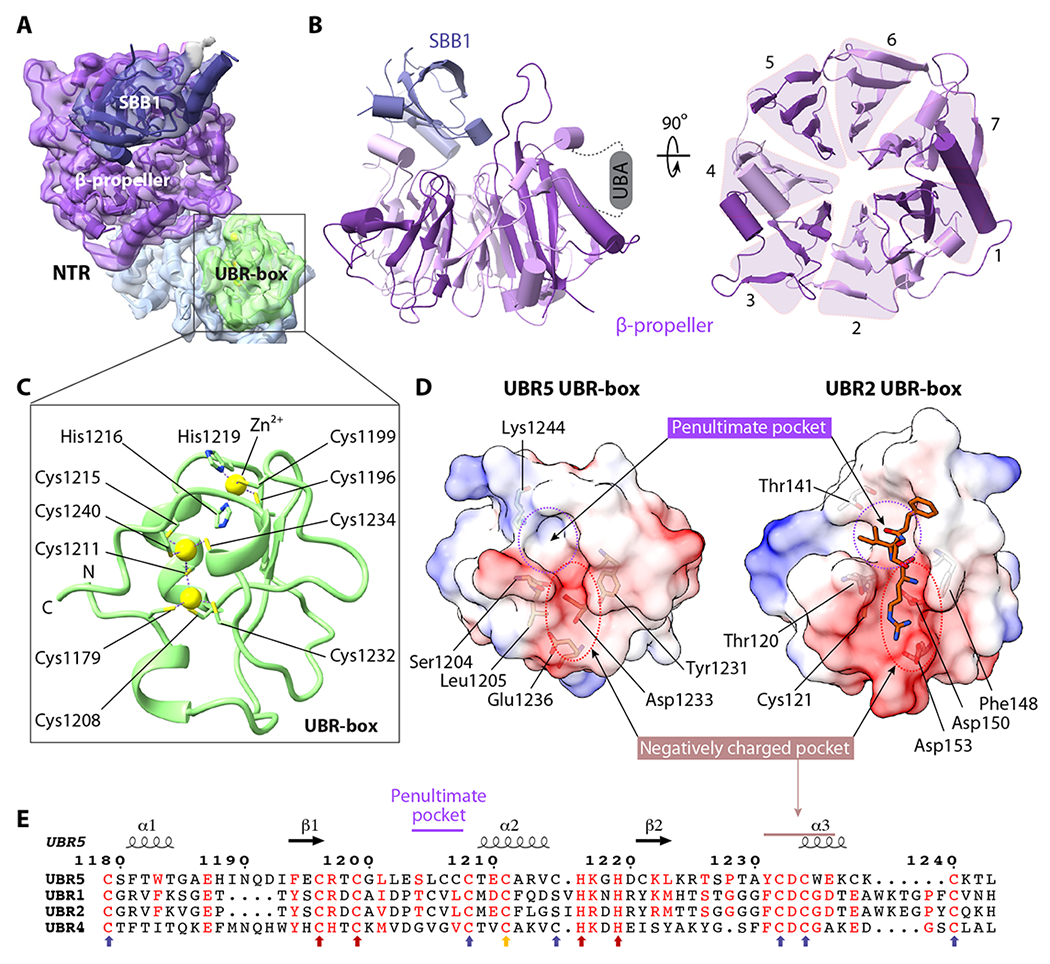
(A) Focus-refined EM map of the NTR and UBR-box in transparent surface view superimposed with atomic model in cartoons and colored as in Figure 1B. (B) Left: Side view of the β-propeller and the small β-barrel 1 (SBB1) in the NTR. Right: Top view of the seven-blades β-propeller. (C) Structure of the UBR-box with two zinc fingers coordinating three zinc ions (yellow spheres). The coordinating cysteine and histidine residues are in sticks. (D) Electrostatic surface views of the UBR5 UBR-box (left) and the UBR2 UBR-box bound to an N-degron peptide shown in orange sticks (right, PDB ID 3NY3). The two N-degron binding subsites are marked by dashed red and purple circles, respectively. (E) Sequence alignment of the UBR boxes of human UBR5, UBR1, UBR2, and UBR4. The conserved Cys and His residues coordinating the first Zn2+ are indicated by red arrows. The six Cys that coordinate the remaining two Zn2+ are indicated by blue arrows. Cys1211 participates in coordination of two zincs and is indicated by a yellow arrow.
The UBR-box contains a conserved N-degron recognition pocket
So far, all known structures of the UBR-box are from the RING E3 ligases 19,53,54,64. However, UBR5 is a HECT-family ligase. The UBR5 UBR-box coordinates three zinc ions with two zinc fingers having little regular secondary structure (Figure 3C), which is similar to other UBR-box structures. The first zinc finger is a typical Cys2His2 type, with Cys1196, Cys1199, His1216, and His1219 coordinating the top zinc ion. The second zinc finger contains the middle and the bottom zinc ions. The middle zinc is coordinated by Cys1179, Cys1208, Cys1232, and Cys1211, and the bottom zinc is coordinated by Cys1211, Cys1215, Cys1234, and Cys1240. Therefore, Cys1211 participates in the coordination of two zinc ions. This Zn2/Cys7 coordination differs from the well-known binuclear zinc clusters, such as the Zn2/Cys6 pattern observed in the yeast Gal4 protein or the Zn2/Cys6-His pattern observed in the RING UBR boxes 31 (Figure S8).
The UBR5 UBR-box contains a negatively charged pocket for recognizing the N-terminal residue and a hydrophobic pocket for the second residue (Figure 3D), consistent with the knowledge that UBR5 targets type 1 N-degrons in which the first amino acid is positively charged Arg, Lys, or His 17,31. UBR-box in UBR5 aligns well with UBR-box structures (Figure S9), with only two amino acids in the peptide bound UBR2 UBR-box structure larger than those of UBR5 UBR-box (Figure 3D). The negatively charged pocket is well conserved among E3s with the N-degron related UBR box (Figure 3E). The penultimate hydrophobic pocket is also conserved, but to a lesser extent. These observations suggest that UBR5 likely uses the UBR box to recognize the substrates. But the substrate specificity requires further study.
The UBR5 tetramer is assembled via SBB2-SBB2 interaction between two dimers
The 3D EM map of the UBR5 tetramer readily fits two UBR5 dimers that fuse into a cage like structure (Figure 4A). Thus, the tetrameric architecture places the four UBR-boxes and four HECT domains all facing the inside of the large central chamber. This organization suggests that the UBR5 tetramer may encapsulate a substrate and add multiple Ub to the substrate simultaneously, like the GID E3 chelator 17. The primary interfaces between the two dimers are clearly located in the SBB2 region, which is not resolved in the UBR5 dimer structure. We used the AlphaFold-Multimer to predict the SBB1-2 structure and the dimerization interface 65. The predicted interface between two SBB1-2 structures is of high confidence and consistent among the top five predicted structures (Figure S10). Importantly, the predicted SBB1-2 dimer structure fits well with the EM density at the dimer-dimer interface (Figure 4B). Therefore, the intermolecular SBB2-SBB2 interaction likely contributes to the tetramer assembly. The interface is stabilized by hydrophobic interactions involving Pro701, Leu705, and Leu710, as well as H-bonds between Glu715 and Tyr677 and between Asp674 and the backbone nitrogen atom of Leu710 (Figure 4C). The tetramer interface is ~800 Å2 per junction, much smaller than the dimer interface, suggesting that the tetramer is less stable and may be converted to dimers, but the dimer is likely a stable functional unit.
Figure 4. The UBR5 tetramer structure.
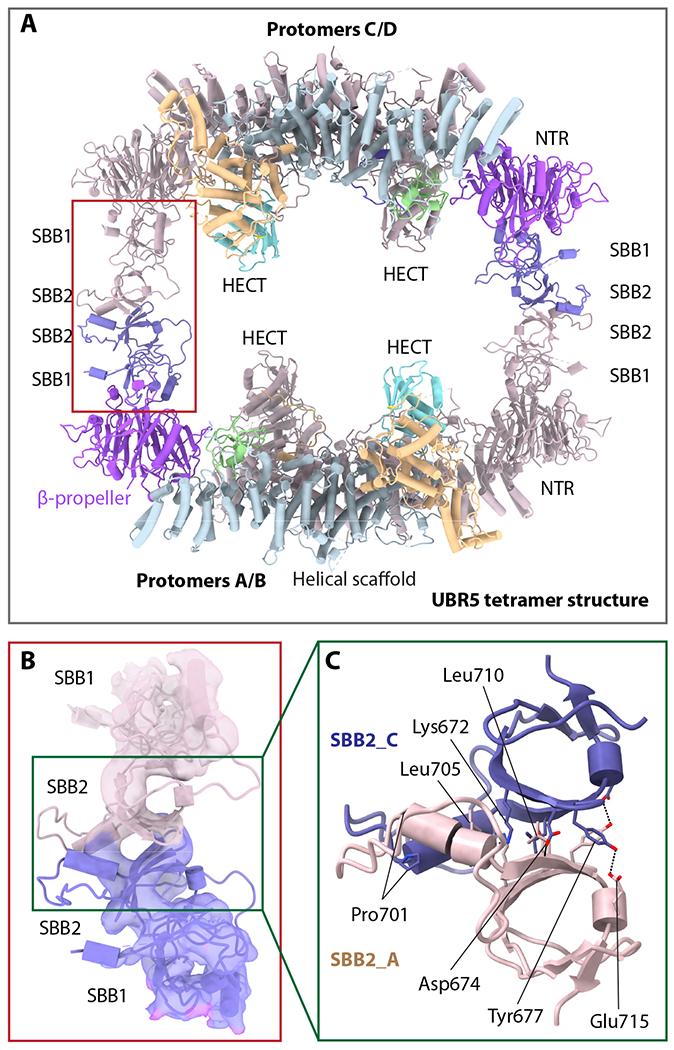
(A) Atomic model of the tetramer UBR5 in cartoon view. Two UBR5 chains (A and C) are colored as in Fig. 1c, and the two remaining chains in salmon. The red rectangle marks the tetramerization interface between two dimers that is mediated by the SBB2-SBB2 interaction. (B) Close-up view of the red rectangle region in panel a showing the EM density of SBB1/2 of protomers A and C in transparent surface superimposed with atomic model in cartoons. (C) Close-up view of the interface in the green box in panel b showing the SBB2 residues involved in tetramerization as predicted by AlphaFold-multimer.
The UBR5 dimer undergoes large conformational changes around the E2–Ub jaw
The N-lobe and C-lobe of the HECT domain are connected by a flexible linker. In the truncated homologous HECT domains, these two lobes were previously shown to arrange either in an L or an inverted T conformations based on the orientation of the C-lobe with respect to N-lobe 56–58. The UBR5 HECT is in the L-conformation (Figure 5A), the Ub–E2 bound NEDD4L HECT is in the inverted-T configuration with the C-lobe sitting in the middle of the long axis of the N-lobe (Figure 5B). When this structure is superimposed with UBR5 HECT, the E2-linked Ub is in the backside of UBR5 C-lobe (Figure 5C). The UBR5 C-lobe needs to rotate ~130° to align with the C-lobe of NEDD4L HECT that interacted with the Ub–E2. We asked if the UBR5 dimer indeed undergoes large conformational changes as suggested above. We performed a 3-dimensional variability analysis (3DVA) 66. We found that the apo UBR5 dimer in solution fluctuates among a series of conformations (Video S2), with the NTR and HECT undergoing the most significant motions, including the lateral expansion of the NTR and an up to 55° rotation of the HECT domain (Figure 5D). These intrinsic motions are likely related to and perhaps enable the above-described conformational changes based on structural comparison (Figure 5C). When the structure of the Ub–E2 bound NEDD4L HECT is superimposed with the UBR5 HECT in the UBR5 dimer, the Ub–E2 fits well inside the E2–Ub jaw (Figure 5E), indicating that the UBR5 intermolecular jaw is most likely involved in the Ub transferring from E2–Ub to UBR5 (Figure 5F).
Figure 5. UBR5 undergoes large conformational changes around the E2–Ub jaw.
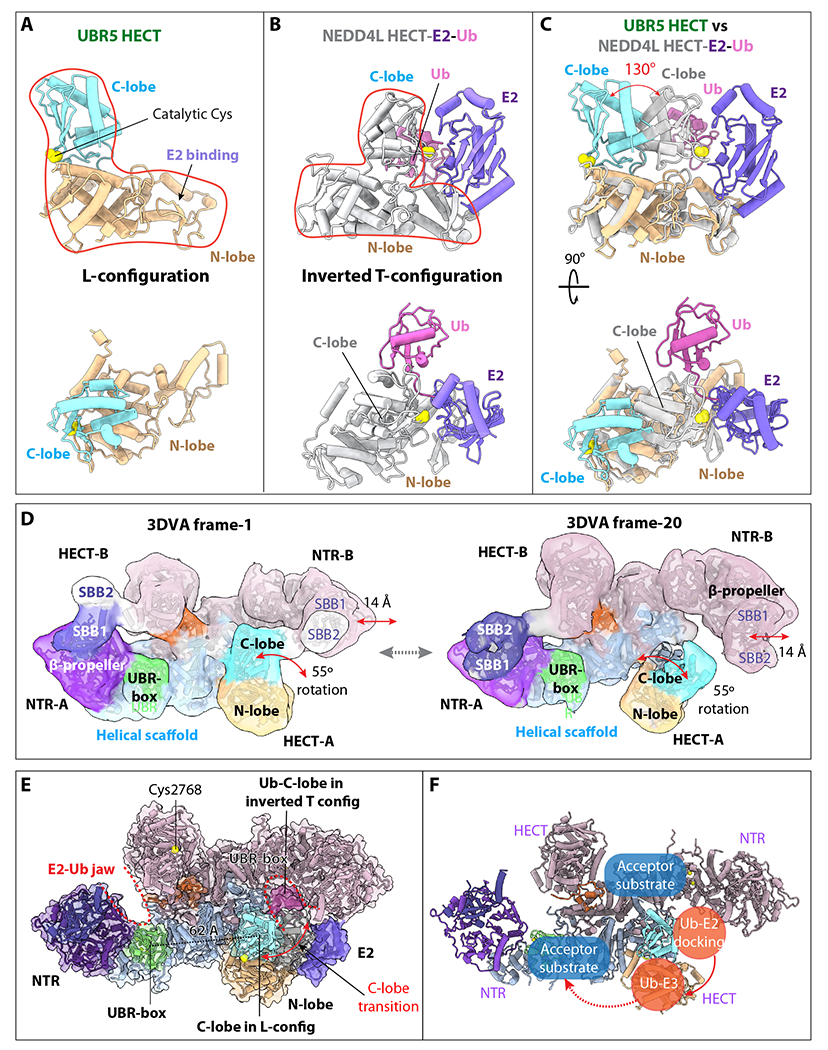
(A) UBR5 HECT domain in the L-conformation in two orthogonal views. (B) The NEDD4L-HECT-E2-Ub structure (PDB ID 3JWO) in the same view. (C) Superimposition of the UBR5-HECT (this study) and NEDD4L-HECT-E2-Ub (PDB ID 3JWO) structures. The UBR5 HECT N-lobe is poised to bind E2, but the C-lobe needs to rotate 130° to reach the C-lobe position of the NEDD4L-HECT for transthiolation reaction. (D) The two most distinct conformations of the UBR5 dimer as determined by 3DVA, showing a 14 Å lateral movement of the NTR and a 55° rotation of the HECT. SBB2 above SBB1 was observed in this lower resolution variability analysis, but missing in the 2.8 Å 3D map, indicating its high mobility in the dimer. (E) Ub-E2 docked in the right intermolecular jaw of the UBR5 dimer. The distance between the C-lobe and UBR-box is 62 Å. (F) Possible substrate ubiquitylation pathway. The curved red arrow indicates that E3 Ub transthiolation reaction occurs in the intermolecular jaw. The dashed red arrow indicates the Ub transfer route for substrate ubiquitylation.
Residues lining the E2–Ub jaw are important for UBR5 ligase function
Motions of the jaw-forming NTR and HECT point to their function in the initial Ub-loaded E2 recruitment for transthiolation as well as in the following E2 release and Ub-linked HECT reorientation for substrate ubiquitylation. To gain insights into this process, we first identified jaw-lining residues that potentially interact with Ub or E2. Superimposition of our UBR5 HECT with published Ub bound or Ub–E2 bound HECT structures showed that UBR5 HECT N-lobe likely binds E2 (UbcH5B) via hydrophobic interactions involving Met2575 and Tyr2576, and UBR5 C-lobe binds Ub via hydrophobic interactions involving Phe2732, Leu2789, and Ala2790 (Figure 6A). The published UBR5 UBA-Ub complex structure has revealed the hydrophobic interactions involving Ub Leu8 and UBR5 Val196 and Leu224 51. Therefore, we made five single mutations lining the “jaw”, V196K, L224K, Y2576A, F2732A, and A2790W, plus the active site mutation C2768S, purified the mutant UBR5 proteins, and performed the E2 discharge assay. Because the dimer and tetramer peaks partially overlapped, the UBR5 samples were a mixture of both. In this assay, we labeled Ub with a fluorescent dye, loaded Ub by E1 onto a E2 (E2D2), then added purified WT and mutant UBR5 proteins, and monitored the transthiolation reaction in which Ub was transferred from E2 to UBR5. We found the WT UBR5 had robust Ub discharge activity from E2 and the catalytically inactive C2768S mutant protein had no activity (Figure 6B–D). The A2790W and F2732A substitutions at the catalytic C-lobe interface with Ub had little activity. Single mutation Y2576A in the E2-interacting HECT N-lobe reduced but did not abolish the UBR5 activity. The UBA domain is inserted in the NTR β-propeller. We found that mutating the two Ub-interacting residues (V196K and L224K) reduced the activity. This result is consistent with the previous finding that substitution of V196 and L224 disrupted interaction between UBA and Ub 51. We further generated an NTR-deletion UBR5 (Δ1-875) and found that the truncation compromised but did not abolish the activity. These results support our proposal that the intermolecular jaw is important for UBR5 function. Further study is required to find out how these mutations affect the Ub transfer to substrate proteins.
Figure 6. Ub transfer assay from E2-Ub to UBR5 by WT and mutations around the E2–Ub jaw.
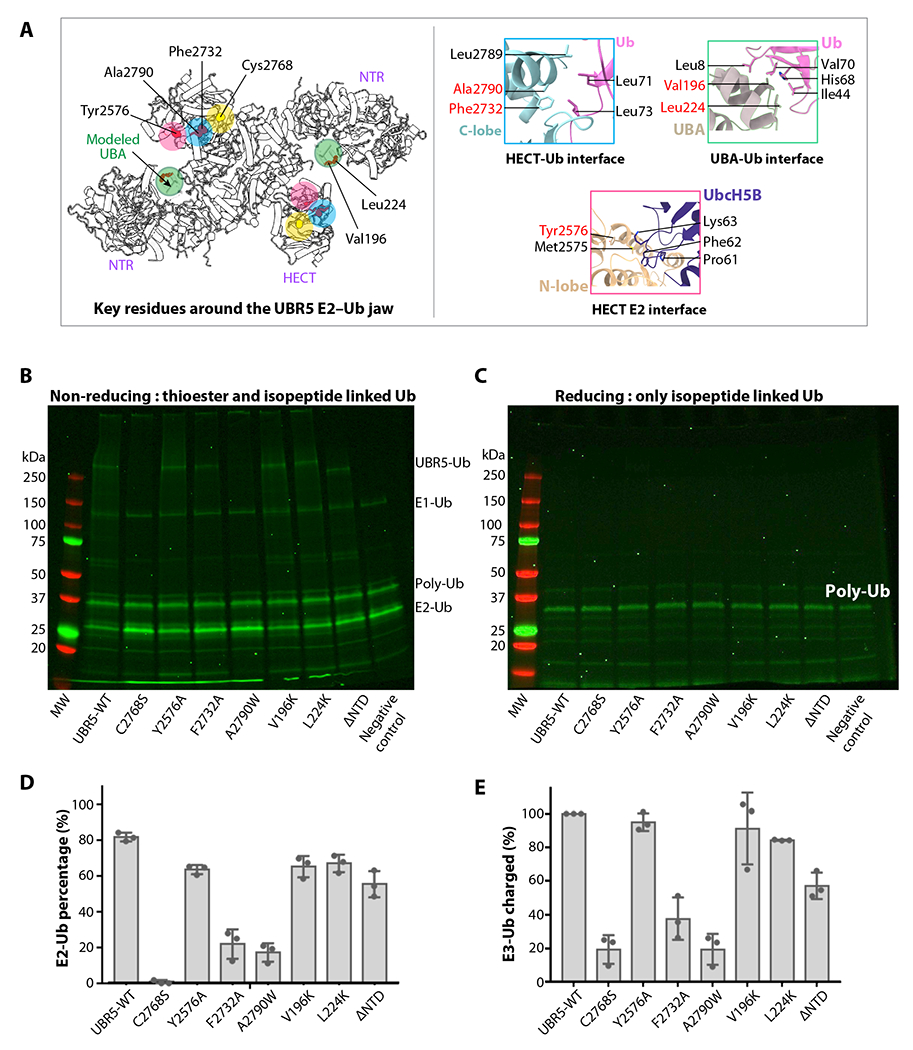
(A) Key residues involved in E2 and Ub binding are displayed as red spheres, and their locations are highlighted by colored circles. The catalytic cysteine is in yellow. The right panels show the catalytic pocket and three predicted interfaces between HECT C-lobe and Ub, between HECT N-lobe and Ub, and between UBA and Ub, based on alignment with the isolated HECT–Ub structures shown in Figure 5A. The UBA location is based on the published isolated UBR5 UBA–Ub complex structure (PDB ID 2QHO). (B-C) In-gel fluorescence of the E2 discharge assay by purified WT and seven mutant UBR5 proteins under non-reducing (B) and reducing agent (5mM β-mercaptoethanol, C). (D) Quantification of the E2-Ub bands. (E) Quantification of the ubiquitylated WT and mutant UBR5 proteins. In panels d-e, ΔNTR refers to UBR5 truncating N-terminal residues 1-875. All values represent means ± SD obtained from three independent experiments.
DISCUSSION
Our cryo-EM analysis has shown that a UBR5 monomer adopts a crescent shape comprising ten ARM repeats forming the central helical scaffold, an NTR containing multiple potential protein-protein interaction modules, and a HECT domain that ubiquitylates target substrates 57. Unexpectedly, we found that UBR5 oligomerizes into a dimer and a tetramer. While UBR5 dimerization is mediated by the central helical scaffold, leaving both NTR and HECT domains unconstrained by the dimerization interface, UBR5 tetramerization is mediated by intermolecular SBB2-SBB2 interaction between two UBR5 dimers. Because the SBB2 interface is small relative to the dimer surface, the tetramer is likely less stable than the dimer. It is possible that the tetramer interface may open up to either become a dimer or recruit more dimers to assemble a larger cage, as found in the multiprotein GID E3 complex 17. In fact, the high-resolution cryo-EM structure of UBR5 is enabling us to develop potent UBR5 inhibitors.
Previous studies have suggested that Ub transfer from E2 to E3 HECT occurs when the HECT is in the inverted T conformation and Ub transfer from E3 HECT to substate occurs when the HECT is in the L-conformation 57,67,68. Our 3DVA analysis of UBR5 dimer shows the C-lobe of HECT undergoes larger scale rotation in the absence of a E2–Ub (Figure 4A). We suggest such intrinsic dynamism may account for the UBR5’s ability to transition from the L to inverted T conformation during catalysis. Our structural and functional studies suggest that the first step of the Ub transthiolation reaction between the Ub–E2 and UBR5 occurs inside the intermolecular jaw formed between the NTR of one subunit and the HECT of the partner subunit in the inverted T conformation. Upon accepting Ub from E2 to form the Ub-UBR5 intermediate, the catalytic C-lobe likely reverts to the observed L conformation. The next step of ubiquitylation reaction occurs between the E3-bound Ub and a protein substrate. The mechanism of the second Ub transfer is complicated by UBR5 oligomerization. In the case of the UBR5 dimer, the E3-bound Ub is likely transferred to a substrate bound to the same UBR5 subunit (Figure 5E).
The E3 activity can be regulated by multiple factors, including post-translational modifications, intermolecular and intramolecular interactions, and accessory proteins or adaptors 69–73. In UBR5, the enzyme’s ability to form dimer and tetramer is notable. Of particular interest is the domain-swapped dimerization (DSD) motif projecting from one subunit into the HECT domain of the partner subunit. This motif wedges between the N-lobe and C-lobe at the back of the partner HECT domain. Because the C-lobe is expected to undergo large conformational changes during catalysis, the DSD motif likely regulates the UBR5 activity. In the UBR5 tetramer, the DSD motif could extend into the E2–Ub jaw to potentially influence the tetramer interface, thereby regulating the tetramer assembly (Figure 1D). Further studies are required to determine how oligomerization affects the UBR5 activity, and whether there are distinct biological functions or substrates between the dimer and the tetramer.
STAR★Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents and resources may be directed to, and will be fulfilled by the Lead Contact Huilin Li (Huilin.Li@vai.org)
Material availability
All expression plasmids used in this study will be made available on request. This study did not generate new unique reagents.
Data and code availability
The cryo-EM 3D maps of the human UBR5 have been deposited at the Electron Microscopy Data Bank database with accession code EMD-27201 (dimer in C1 symmetry) EMD-27822 (dimer in C2 symmetry), and EMD-28646 (tetramer in C1 symmetry). The corresponding atomic models were deposited at the RCSB Protein Data Bank database with accession codes 8D4X,8E0Q, and 8EWI. These data are publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
E2D2 and PEPCK1 were expressed in BL21(DE3) E. coli cells grown in LB Broth media. Recombinant UBR5 WT and UBR5 mutants’ proteins for cryo-EM and biochemical assay were expressed in Sf9 insect cells.
METHOD DETAILS
Bacterial strain and plasmids
pET28-mE1 was a gift from Jorge Eduardo Azevedo (Addgene plasmid # 32534; http://n2t.net/addgene:32534; RRID: Addgene 32534). The human UBR5 sequence codon-optimized for insect cell expression with an N-terminal Flag tag was synthesized and sequence confirmed by GenScript. The human ubiquitin sequence with an inserted N-terminal cysteine residue was synthesized by Eurofins. Human His6-E2D2 was a gift from Dr. Efrat Finkin-Groner at Tri-Institutional Therapeutics Discovery Institute. The PEPCK1 expression plasmid was a gift from Dr. Lewis Cantley at Harvard Medical School.
The human UBR5 sequence with an N-terminal Flag tag was cloned into pFastBac with a polyhedrin promoter. The UBR5 mutants were generated by PCR based mutagenesis. The NTR deletion of UBR5 (876-2799 aa) was cloned into the pFastBac vector using the same restriction sites as the UBR5 wild type. The mouse E1 and human E2D2 were cloned into the pET22b vectors. Ub with an added N-terminal cysteine residue was cloned into a pET28a vector to make Ub with an N-terminal 6xHis-tag and a thrombin cleavage site. The human PEPCK1 was cloned into the pET28a vector with an N-terminal His tag. These plasmids were individually transformed into E. coli BL21 (DE3) cells (Thermo Fisher). All plasmid sequences were confirmed by DNA sequencing.
Protein expression and purification
UBR5 and UBR5 mutants were expressed in Sf9 insect cells (Novagen) using the Bac-to-Bac baculovirus expression system (ThermoFisher). Cell cultures were grown in ESF 921 serum-free medium (Expression Systems) to a density of 2.5 × 106 cells/mL and then infected with the UBR5 baculovirus (3%-5% v/v) for 48-60 h post-transfection. We found the use of a higher UBR5 baculovirus infection ratio and the longer time after transfection leads to a higher population of UBR5 tetramer. The cells from 1.6 L culture were collected, flash frozen, and stored at −80 °C for further usage. Cells were lysed by sonication in lysis buffer (25 mM HEPES, PH7.5, 200 mM NaCl, 10% glycerol, 1 tablet EDTA-free protease inhibitors cocktail), centrifugated in a Ti-45 rotor at 40,000x rpm for 1 h, and the supernatant was incubated with 0.5 mL FLAG-antibody-coated beads at 4 °C for 2-3 h. Beads were washed with 50 mL lysis buffer, and the proteins were eluted with 5 mL of lysis buffer with 0.2 mg/mL 3 × FLAG peptides. The proteins were concentrated using centrifugal concentrators (Amicon, 100 kDa) and polished by size exclusion chromatography (SEC, Superose 6 Increase, GE Healthcare) in buffer containing 25 mM HEPES, PH 7.5, 100 mM NaCl, 0.5 mM TCEP. Fraction peaks corresponding to the dimer and tetramer were pooled separately for cryo-EM analysis.
All recombinant strains were grown in 2 L LB medium at 37°C. When the cell density reached an OD600 value of 0.8, 0.2 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was added, and the culture was continued for 12 h at 16°C. We then collected the cells, resuspended the cells in buffer A (20 mM HEPES pH 7.6, 150 mM NaCl, and 10% glycerol), and lysed the cells with a homogenizer (SPX Corporation). The lysate was centrifuged at 20,000x g for 40 min, and the supernatant was collected and loaded into a 5-ml Ni-NTA column (Cytiva). The proteins were eluted using buffer A plus 300 mM imidazole. The E1, E2, and PEPCK1 proteins were further purified by a Superdex 200 column in buffer A. The N-terminal 6xHis-tag on Ub was cleaved by incubating with thrombin at 4°C overnight. The tag cleaved Ub was further purified by size exclusion chromatography through a Superdex 75 column (GE Healthcare) in buffer A.
Cryo-EM sample preparation and data collection
The holey carbon grids (Quantifoil Au R2/1, 300 gold mesh) were glow discharged in the Ar/O2 mixture for 30 s using a Gatan 950 Solarus plasma cleaning system with a power of 15 W. Aliquots of 3 μl of purified UBR5 solution at a concentration of 0.7 mg/ml were placed on the freshly treated EM grids. The grids were blotted with qualitative cellulose filter paper (TED PELLA, INC) for 3 s after each sample application with the blotting force set to 3 and flash-frozen in liquid ethane using an FEI Vitrobot Mark IV. Temperature and relative humidity were maintained at 6°C and 100%, respectively. We prepared cryo-EM grids for both the dimer and tetramer elution peaks then loaded the grids into an FEI Titan Krios electron microscope operated at a high tension of 300 kV. Cryo-electron micrographs were collected automatically with the SerialEM program 81 at a nominal magnification of 105,000 X in a K3 summit direct electron detector (Gatan) in multi-hole mode, with the objective lens defocus being varied between −1.0 to −2.0 μm. The energy filter was not used during data collection. The K3 camera was operated in the super-resolution counting mode. During a 1.5-s exposure time, a total of 75 frames were recorded with a total dose of 65 e−/Å2. The calibrated physical pixel size was 0.828 Å at the specimen level for all digital micrographs. All datasets were collected with the sample stage tilt angle set to 0°, except for one dataset of the UBR5 tetramer that was recorded with a tilt angle of 30° to alleviate the preferred orientation problem of the tetramer.
Image processing
For the untilted dataset, a total of 13,090 raw movie micrographs were collected and motion-corrected using the program MotionCorr 2.0 75. Then the micrographs were imported into cryoSPARC (version 3.2.0) 74 to perform the patch-based contrast transfer function (CTF) estimation and correction. A total of 12,471 micrographs with CTF signals extending to 4.1 Å were retained in further processing. We first used the blob particle picking (70-300 Å diameter) to generate 2D templates for subsequent automatic particle picking. In total, 4,262,102 particles were automatically picked and extracted. Two rounds of 2D classifications were then performed on particle images with 4x binning and particles in the classes with clear structural features were selected. A total of 1,931,379 particles was used to calculate five starting 3D models. One major class was chosen to perform a new round of 3D classification to remove bad particles. Two major 3D classes were combined to perform further 3D refinement and post-processing, resulting in the 2.80-Å 3D density map with 844,403 particles in C1 symmetry. Application of 2-fold symmetry during 3D reconstruction and refinement led to the final 3D map at an overall resolution of 2.66 Å. The map resolution was estimated by the gold-standard Fourier shell correlation at a correlation cut-off value of 0.143 82. We further performed focused refinement on the NTR and the HECT region separately. The focus refined NTR and HECT maps had an average resolution at 2.94 Å and 3.01 Å, respectively. Finally, we combined the focus-refined maps with the original C2 symmetric map to generate a composite EM map for the UBR5 dimer.
The NTR and the C-terminal HECT region were of lower resolution, indicating that these regions are more dynamics. We next used the 3D variability analysis (3DVA) program 66 to investigate the conformational heterogeneity in the UBR5 dimer. The final dataset with 844,403 particle images were down sampled by a factor of 4 and subjected to non-uniform refinement in C1 symmetry to generate an appropriate mask. 3DVA was carried out to solve three principal components with the filter resolution set to 8 Å; and other parameters used their default values. The distribution of reaction coordinates across particles in the principal components were smooth, confirming the compositional homogeneity of the UBR5 dataset and the conformational flexibility of the NTR and HECT domain.
The 30° tilted dataset of the tetramer peak sample contained a total of 17,373 raw movie micrographs. The micrographs were motion-corrected using the program MotionCorr 2.0 75 and combined with the untitled micrographs imported into cryoSPARC (version 3.2.0) 74 to perform the patch-based contrast transfer function (CTF) estimation and correction. A total of 28,570 micrographs with CTF signals extending to 4.0 Å were retained for further process; these included 12,453 untiled micrographs and 16,117 tiled micrographs. We also generated 2D templates for the tetramer particles from the untilted dataset and used template picking (290 Å diameter) to select an initial dataset of 1,793,574 tetramer particles. Several rounds of 2D classifications were then performed on 4x binned particle images. Particles in the 2D classes with clear structural features were retained, resulting in a selected dataset of 1,027,335 particles, which was used to calculate three starting 3D models. We performed heterogenous refinement on the particle images belonging to the best start model and obtained 3 additional maps; particles belonging to the two good maps were then combined (401,468 particles) for further refinement and post-processing, resulting in the 3.5-Å 3D density map in C1 symmetry. Application of 2-fold symmetry or 4-fold symmetry during 3D reconstruction and refinement led to a high-resolution 3D map but with reduced densities in certain regions. The resolution of the C1 map was estimated by the gold-standard Fourier shell correlation at a correlation cut-off value of 0.143 82. We further performed focused refinements on two UBR5 dimers and the dimer-dimer interface regions separately. The focus refined UBR5 dimer maps and the interface region map had an average resolution of 3.4 Å. We finally combined these partial maps with the original C1 map to generate a composite 3D map for the UBR5 tetramer.
Model building, refinement, and validation
We first used the AlphaFold2 server installed in a local workstation to obtain a predicted UBR5 atomic model 65. The high-confidence regions, including the N-terminal small β-barrel 1 (SBB1) and β-propeller, the UBR box, the helical repeat, and the C-terminal HECT domain, were docked individually into the cryo-EM maps using UCSF Chimera 79. We then manually adjusted each domain and built missing residues in our high-resolution cryo-EM map in Coot 76. We also referenced to the high-resolution (2.66 Å) 2-fold symmetric composite 3D map to build several regions with weaker densities. The completed model was subjected to several iterations of real-space refinement in PHENIX 77 and manual adjustment in Coot 76. For modeling the UBR5 tetramer map, two dimer models were readily fitted into the composite EM map. But the dimer-dimer interface region had a lower resolution that was insufficient for manual model building. We used the AlphaFold-Multimer server to predict a dimer structure of the SBB1-SBB2 sequence. All predicted dimer structures had a high confidence value over 95% with a consistent SBB2-SBB2 interface. The predicted SBB2-SBB2 complex structure was fitted well in the dimer-dimer interface region of the UBR5 tetramer map. Finally, all atomic models were validated using MolProbity 83. The original EM maps in both C1 and C2 symmetry and the maps sharpened by deepEMhancer were used to refine the atomic models 80. The statistics of the model refinement are shown in Supplementary Table 1. Structural figures were prepared in UCSF ChimeraX 78.
In vitro E2 discharge Assay for UBR5
Purified Ub was first loaded onto the purified E2. Purified Cys-Ub was modified with Dylight800 maleimide at pH 7.4 by mixing them at a molar ratio of 1:1.5 and incubating the mixture for 1 h, then 5 mM DTT was added to quench the modification reaction. Excess Dylight800 was removed by a Superdex75 column. The fluorescently labeled Ub was incubated with 0.5 μM recombinant mouse E1 and 8 μM recombinant human E2D2 in 50 mM HEPES pH 7.5, 150 mM NaCl and 3 mM Mg2+-ATP for 30 min at room temperature to produce the Ub charged E2 (Ub–E2D2). The reaction was quenched with 20 mM EDTA. Next, the Ub–E2D2 at the final concentration of 0.5 μM was incubated with 0.75 μM purified wild-type or mutant UBR5 proteins for 3 min at room temperature. Each reaction was in 20 μl volume and the reaction buffer contained 50 mM HEPES pH 7.5 and 150 mM NaCl. The reactions were quenched with 4x SDS sample loading buffer with or without a reducing agent (5 mM β-mercaptoethanol) and run on the 4-12% SDS-PAGE gels. The gels were scanned by fluorescent imaging in a ChemiDoc MP imager (Bio-Rad). The intensities of the Ub–E2D2 bands were quantified by ImageJ and normalized to the Ub–E2D2 only control.
In vitro ubiquitylation assay
Our in vitro ubiquitylation assay solution contained 0.2 μM purified E1, 2 μM purified E2D2, 50 μM ubiquitin (R&D Systems), either 0.5 μM wildtype UBR5 or catalytically inactive mutant UBR5(C2768A), and 45 μM His-tagged PEPCK1 in 40 μl reaction buffer (50 mM HEPES pH7.5, 150 mM NaCl, 10 mM MgCl2, 10 mM AMPPNP). AMPPNP was used instead of ATP because AMPPNP supports ubiquitination 84 but prevents degradation of ubiquitinated proteins by the ATP-dependent 26S proteasome. The reaction system was incubated with 20 μl Ni-NTA agarose beads for 120 min at 37°C. The reaction time was chosen based on a previous in vitro substrate modification by a E3 ligase 4. Then, the beads were washed two times by incubating with the wash buffer (50 mM HEPES pH7.5, 150 mM NaCl, 50 mM imidazole). Finally, the ubiquitylated PEPCK1 was eluted with 500 mM imidazole in 50 mM HEPES pH7.5, 150 mM NaCl. The eluent was mixed with the reducing 2X SDS loading buffer, analyzed by SDS-PAGE, and subsequently immunoblotted with an FK2 antibody (Cayman Chemical, Cat # 14220).
QUANTIFICATION AND STATISTICAL ANALYSIS
Comparisons between groups were made by unpaired two-tailed Student’s t test. Statistical analyses were performed using GraphPad Prism (GraphPad Software Inc.). Data are expressed as the mean ± SD
Supplementary Material
Video S1. The overall structure of UBR5 in a dimeric form, showing the model-to-map fitting quality and structural details, related to Figures 1 and 2.
Video S2. The 3DVA analysis result of the entire apo UBR5 dimer, showing the most dramatic changes occur in the N-terminal region and the C-terminal catalytic HECT domain of UBR5, related to Figure 5.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
|
| ||
| E. coli DH5alpha competent cells | ThermoFisher | Cat# 18265017 |
| E. coli BL21(DE3) competent cells | ThermoFisher | Cat# EC0114 |
| E. coli DH10Bac competent cells | ThermoFisher | Cat# 10361012 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Protease inhibitor | MilliporeSigma | Cat# 11873580001 |
| DyLight™ 800 Maleimide | Thermo Fisher | Cat # 46621 |
| Plasmid Mega Kit | Qiagen | Cat # 12181 |
|
| ||
| Deposited data | ||
|
| ||
| UBR5 structure model (C1 symmetry in dimer) | This study | PDB: 8D4X |
| UBR5 structure model (C2 symmetry in dimer) | This study | PDB: 8E0Q |
| UBR5 structure model (C1 symmetry in tetramer) | This study | PDB: 8EWI |
| UBR5 structure map (C1 symmetry in dimer) | This study | EMDB: EMD-27201 |
| UBR5 structure model (C2 symmetry in dimer) | This study | EMDB: EMD-27822 |
| UBR5 structure model (C1 symmetry in tetramer) | This study | EMDB: EMD-28646 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Sf9 insect cells | Invitrogen | Cat# 11496015 |
|
| ||
| Oligonucleotides | ||
|
| ||
| GCGCGGAATTCATGGACTACAAGGACGACGACGACAAGATGCAGCACATCCTGAGGTGCGACTACG | Eurofins Scientific | UBR5(876-2799)_forward |
| CGAAAGCGGCCGCTTATTACACGAAACCGAAGTTCTTGGTCT | Eurofins Scientific | UBR5(876-2799)_reverse |
| CCGCGCGGCAGCCATATGTGTCAGATCTTCGTGAAAACCC | Eurofins Scientific | Cys-Ub_forward |
| GTGGTGGTGGTGCTCGAGTCAACCACCTCTCAGACGCAGG | Eurofins Scientific | Cys-Ub_reverse |
|
| ||
| Recombinant DNA | ||
|
| ||
| pFastBac_UBR5 | This study | N/A |
| pET28a_Cys-ub | This study | N/A |
| pET28a-PEPCK1 | This study | N/A |
|
| ||
| Software and algorithms | ||
| cryoSPARC v3.2.0 | Punjani et al., 201774 | https://cryosparc.com/ |
| MotionCor2 | Zheng et al., 201775 | https://emcore.ucsf.edu/ucsf-software |
| Coot v0.9.4 | Emsley et al., 200476 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Phenix v1.20 | Adams et al., 201077 | https://phenix-online.org/documentation/reference/autobuild_gui.html |
| ChimeraX | Goddard et al.,201878 | https://www.cgl.ucsf.edu/chimerax/docs/user/index.html |
| Chimera | Pettersen et al., 200479 | http://www.cgl.ucsf.edu/chimera/ |
| DeepEMhancer | Sanchez-Garcia et al., 202180 | https://github.com/rsanchezgarc/deepEMhancer |
| AlphaFold2 | Jumper et al., 202165 | https://github.com/deepmind/alphafold |
|
| ||
| Others | ||
| Quantifoil Au R2/1 | SPI Supplies | Cat# 4330G-FA |
| easiGlow glow discharger | PELCO | Model# 91000 |
| Vitrobot | Thermo Fisher | N/A |
| Superose 6 Increase10/300 GL | Sigma-Aldrich | Cat# GE29-0915-96 |
ACKNOWLEDGEMENTS
Cryo-EM datasets were collected at the David Van Andel Advanced Cryo-Electron Microscopy Suite in Van Andel Institute. We thank G. Zhao and X. Meng for facilitating data collection. We thank L. Cantley at Harvard Medical School for the gift of PEPCK1 expression plasmid. This work was supported by the US Department of Defense grant W81XWH2110261 (to X.M.), the US National Institutes of Health grant GM131754 (to H.L.), and the Van Andel Institute (to H.L.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Finley D, and Chau V (1991). Ubiquitination. Annu Rev Cell Biol 7, 25–69. 10.1146/annurev.cb.07.110191.000325. [DOI] [PubMed] [Google Scholar]
- 2.Laney JD, and Hochstrasser M (1999). Substrate targeting in the ubiquitin system. Cell 97, 427–430. 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 3.Morreale FE, and Walden H (2016). Types of Ubiquitin Ligases. Cell 165, 248–248 e241. 10.1016/j.cell.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Grabarczyk DB, Petrova OA, Deszcz L, Kurzbauer R, Murphy P, Ahel J, Vogel A, Gogova R, Faas V, Kordic D, et al. (2021). HUWE1 employs a giant substrate-binding ring to feed and regulate its HECT E3 domain. Nat Chem Biol 17, 1084–1092. 10.1038/s41589-021-00831-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weissman AM (2001). Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2, 169–178. 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 6.Yang Q, Zhao J, Chen D, and Wang Y (2021). E3 ubiquitin ligases: styles, structures and functions. Mol Biomed 2, 23. 10.1186/s43556-021-00043-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petroski MD, and Deshaies RJ (2005). Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6, 9–20. 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 8.Duda DM, Scott DC, Calabrese MF, Zimmerman ES, Zheng N, and Schulman BA (2011). Structural regulation of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol 21, 257–264. 10.1016/j.sbi.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cyr DM, Hohfeld J, and Patterson C (2002). Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci 27, 368–375. 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 10.Lechtenberg BC, Rajput A, Sanishvili R, Dobaczewska MK, Ware CF, Mace PD, and Riedl SJ (2016). Structure of a HOIP/E2~ubiquitin complex reveals RBR E3 ligase mechanism and regulation. Nature 529, 546–550. 10.1038/nature16511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walden H, and Rittinger K (2018). RBR ligase-mediated ubiquitin transfer: a tale with many twists and turns. Nat Struct Mol Biol 25, 440–445. 10.1038/s41594-018-0063-3. [DOI] [PubMed] [Google Scholar]
- 12.Rotin D, and Kumar S (2009). Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 10, 398–409. 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 13.Hunkeler M, Jin CY, Ma MW, Monda JK, Overwijn D, Bennett EJ, and Fischer ES (2021). Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition. Mol Cell 81, 3468–3480 e3467. 10.1016/j.molcel.2021.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, and Joazeiro CA (2008). Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS One 3, e1487. 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berndsen CE, and Wolberger C (2014). New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol 21, 301–307. 10.1038/nsmb.2780 [DOI] [PubMed] [Google Scholar]
- 16.George AJ, Hoffiz YC, Charles AJ, Zhu Y, and Mabb AM (2018). A Comprehensive Atlas of E3 Ubiquitin Ligase Mutations in Neurological Disorders. Front Genet 9, 29. 10.3389/fgene.2018.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherpa D, Chrustowicz J, and Schulman BA (2022). How the ends signal the end: Regulation by E3 ubiquitin ligases recognizing protein termini. Mol Cell 82, 1424–1438. 10.1016/j.molcel.2022.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horn-Ghetko D, and Schulman BA (2022). New classes of E3 ligases illuminated by chemical probes. Curr Opin Struct Biol 73, 102341. 10.1016/j.sbi.2022.102341. [DOI] [PubMed] [Google Scholar]
- 19.Pan M, Zheng Q, Wang T, Liang L, Mao J, Zuo C, Ding R, Ai H, Xie Y, Si D, et al. (2021). Structural insights into Ubr1-mediated N-degron polyubiquitination. Nature 600, 334–338. 10.1038/s41586-021-04097-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiao S, Langlois CR, Chrustowicz J, Sherpa D, Karayel O, Hansen FM, Beier V, von Gronau S, Bollschweiler D, Schafer T, et al. (2020). Interconversion between Anticipatory and Active GID E3 Ubiquitin Ligase Conformations via Metabolically Driven Substrate Receptor Assembly. Mol Cell 77, 150–163 e159. 10.1016/j.molcel.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Sherpa D, Chrustowicz J, Qiao S, Langlois CR, Hehl LA, Gottemukkala KV, Hansen FM, Karayel O, von Gronau S, Prabu JR, et al. (2021). GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme. Mol Cell 81, 2445–2459 e2413. 10.1016/j.molcel.2021.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shakeel S, Rajendra E, Alcon P, O’Reilly F, Chorev DS, Maslen S, Degliesposti G, Russo CJ, He S, Hill CH, et al. (2019). Structure of the Fanconi anaemia monoubiquitin ligase complex. Nature 575, 234–237. 10.1038/s41586-019-1703-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahel J, Lehner A, Vogel A, Schleiffer A, Meinhart A, Haselbach D, and Clausen T (2020). Moyamoya disease factor RNF213 is a giant E3 ligase with a dynein-like core and a distinct ubiquitin-transfer mechanism. Elife 9. 10.7554/eLife.56185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lumpkin RJ, Baker RW, Leschziner AE, and Komives EA (2020). Structure and dynamics of the ASB9 CUL-RING E3 Ligase. Nat Commun 11, 2866. 10.1038/s41467-020-16499-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Q, Botuyan MV, Zhao D, Cui G, Mer E, and Mer G (2021). Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation. Nature 596, 438–443. 10.1038/s41586-021-03716-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witus SR, Burrell AL, Farrell DP, Kang J, Wang M, Hansen JM, Pravat A, Tuttle LM, Stewart MD, Brzovic PS, et al. (2021). BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1. Nat Struct Mol Biol 28, 268–277. 10.1038/s41594-020-00556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horn-Ghetko D, Krist DT, Prabu JR, Baek K, Mulder MPC, Klugel M, Scott DC, Ovaa H, Kleiger G, and Schulman BA (2021). Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly. Nature 590, 671–676. 10.1038/s41586-021-03197-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kostrhon S, Prabu JR, Baek K, Horn-Ghetko D, von Gronau S, Klugel M, Basquin J, Alpi AF, and Schulman BA (2021). CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation. Nat Chem Biol 17, 1075–1083. 10.1038/s41589-021-00858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai X-C, McMullan G, and Scheres SH (2015). How cryo-EM is revolutionizing structural biology. Trends Biochem Sci 40, 49–57. 10.1016/j.tibs.2014.10.005 [DOI] [PubMed] [Google Scholar]
- 30.Kim JG, Shin HC, Seo T, Nawale L, Han G, Kim BY, Kim SJ, and Cha-Molstad H (2021). Signaling Pathways Regulated by UBR Box-Containing E3 Ligases. Int J Mol Sci 22. 10.3390/ijms22158323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sriram SM, and Kwon YT (2010). The molecular principles of N-end rule recognition. Nat Struct Mol Biol 17, 1164–1165. 10.1038/nsmb1010-1164. [DOI] [PubMed] [Google Scholar]
- 32.Callaghan MJ, Russell AJ, Woollatt E, Sutherland GR, Sutherland RL, and Watts CK (1998). Identification of a human HECT family protein with homology to the Drosophila tumor suppressor gene hyperplastic discs. Oncogene 17, 3479–3491. 10.1038/sj.onc.1202249. [DOI] [PubMed] [Google Scholar]
- 33.Mansfield E, Hersperger E, Biggs J, and Shearn A (1994). Genetic and molecular analysis of hyperplastic discs, a gene whose product is required for regulation of cell proliferation in Drosophila melanogaster imaginal discs and germ cells. Dev Biol 165, 507–526. 10.1006/dbio.1994.1271 [DOI] [PubMed] [Google Scholar]
- 34.Yau RG, Doerner K, Castellanos ER, Haakonsen DL, Werner A, Wang N, Yang XW, Martinez-Martin N, Matsumoto ML, Dixit VM, and Rape M (2017). Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control. Cell 171, 918–933 e920. 10.1016/j.cell.2017.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cojocaru M, Bouchard A, Cloutier P, Cooper JJ, Varzavand K, Price DH, and Coulombe B (2011). Transcription factor IIS cooperates with the E3 ligase UBR5 to ubiquitinate the CDK9 subunit of the positive transcription elongation factor B. J Biol Chem 286, 5012–5022. 10.1074/jbc.M110.176628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hay-Koren A, Caspi M, Zilberberg A, and Rosin-Arbesfeld R (2011). The EDD E3 ubiquitin ligase ubiquitinates and up-regulates beta-catenin. Mol Biol Cell 22, 399–411. 10.1091/mbc.E10-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Honda Y, Tojo M, Matsuzaki K, Anan T, Matsumoto M, Ando M, Saya H, and Nakao M (2002). Cooperation of HECT-domain ubiquitin ligase hHYD and DNA topoisomerase II-binding protein for DNA damage response. J Biol Chem 277, 3599–3605. 10.1074/jbc.M104347200. [DOI] [PubMed] [Google Scholar]
- 38.Rutz S, Kayagaki N, Phung QT, Eidenschenk C, Noubade R, Wang X, Lesch J, Lu R, Newton K, and Huang OW (2015). Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells. Nature 518, 417–421. Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Singh S, Jung HY, Yang G, Jun S, Sastry KJ, and Park JI (2013). HIV-1 Vpr protein inhibits telomerase activity via the EDD-DDB1-VPRBP E3 ligase complex. J Biol Chem 288, 15474–15480. 10.1074/jbc.M112.416735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida M, Yoshida K, Kozlov G, Lim NS, De Crescenzo G, Pang Z, Berlanga JJ, Kahvejian A, Gehring K, and Wing SS (2006). Poly (A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J 25, 1934–1944. 10.1038/sj.emboj.7601079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang T, Cronshaw J, Kanu N, Snijders AP, and Behrens A (2014). UBR5-mediated ubiquitination of ATMIN is required for ionizing radiation-induced ATM signaling and function. Proc Natl Acad Sci U S A 111, 12091–12096. 10.1073/pnas.1400230111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaisari S, Miniowitz-Shemtov S, Sitry-Shevah D, Shomer P, Kozlov G, Gehring K, and Hershko A (2022). Role of ubiquitin-protein ligase UBR5 in the disassembly of mitotic checkpoint complexes. Proc Natl Acad Sci U S A 119. 10.1073/pnas.2121478119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shearer RF, Iconomou M, Watts CK, and Saunders DN (2015). Functional Roles of the E3 Ubiquitin Ligase UBR5 in Cancer. Mol Cancer Res 13, 1523–1532. 10.1158/1541-7786.MCR-15-0383. [DOI] [PubMed] [Google Scholar]
- 44.Liao L, Song M, Li X, Tang L, Zhang T, Zhang L, Pan Y, Chouchane L, and Ma X (2017). E3 Ubiquitin Ligase UBR5 Drives the Growth and Metastasis of Triple-Negative Breast Cancer. Cancer Res 77, 2090–2101. 10.1158/0008-5472.CAN-16-2409. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y, Zhao J, Mao Y, Lin G, Li F, and Jiang Z (2020). UBR5 over-expression contributes to poor prognosis and tamoxifen resistance of ERa+ breast cancer by stabilizing beta-catenin. Breast Cancer Res Treat 184, 699–710. 10.1007/s10549-020-05899-6. [DOI] [PubMed] [Google Scholar]
- 46.Song M, Yeku OO, Rafiq S, Purdon T, Dong X, Zhu L, Zhang T, Wang H, Yu Z, and Mai J (2020). Tumor derived UBR5 promotes ovarian cancer growth and metastasis through inducing immunosuppressive macrophages. Nat Commun 11, 1–16. 10.1038/s41467-020-20140-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding F, Zhu X, Song X, Yuan P, Ren L, Chai C, Zhou W, and Li X (2020). UBR5 oncogene as an indicator of poor prognosis in gastric cancer. Exp Ther Med 20, 1-1. 10.3892/etm.2020.9135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Zhang W, Gao J, Du M, Li H, Li M, Cong H, Fang Y, Liang Y, and Zhao D (2021). E3 Ubiquitin Ligase UBR5 Promotes the Metastasis of Pancreatic Cancer via Destabilizing F-Actin Capping Protein CAPZA1. Front Oncol 11, 677. 10.3389/fonc.2021.634167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiang G, Wang S, Chen L, Song M, Song X, Wang H, Zhou P, Ma X, and Yu J (2022). UBR5 targets tumor suppressor CDC73 proteolytically to promote aggressive breast cancer. Cell Death Dis 13, 451. 10.1038/s41419-022-04914-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matta-Camacho E, Kozlov G, Menade M, and Gehring K (2012). Structure of the HECT C-lobe of the UBR5 E3 ubiquitin ligase. Acta Crystallogr Sect F Struct Biol Cryst Commun 68, 1158–1163. 10.1107/S1744309112036937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kozlov G, Nguyen L, Lin T, De Crescenzo G, Park M, and Gehring K (2007). Structural basis of ubiquitin recognition by the ubiquitin-associated (UBA) domain of the ubiquitin ligase EDD. J Biol Chem 282, 35787–35795. 10.1074/jbc.M705655200. [DOI] [PubMed] [Google Scholar]
- 52.Ohtake F, Tsuchiya H, Saeki Y, and Tanaka K (2018). K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc Natl Acad Sci U S A 115, E1401–E1408. 10.1073/pnas.1716673115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi WS, Jeong BC, Joo YJ, Lee MR, Kim J, Eck MJ, and Song HK (2010). Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases. Nat Struct Mol Biol 17, 1175–1181. 10.1038/nsmb.1907. [DOI] [PubMed] [Google Scholar]
- 54.Matta-Camacho E, Kozlov G, Li FF, and Gehring K (2010). Structural basis of substrate recognition and specificity in the N-end rule pathway. Nat Struct Mol Biol 17, 1182–1187. 10.1038/nsmb.1894. [DOI] [PubMed] [Google Scholar]
- 55.Munoz-Escobar J, Matta-Camacho E, Kozlov G, and Gehring K (2015). The MLLE domain of the ubiquitin ligase UBR5 binds to its catalytic domain to regulate substrate binding. J Biol Chem 290, 22841–22850. 10.1074/jbc.M115.672246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh S, Ng J, Nayak D, and Sivaraman J (2019). Structural insights into a HECT-type E3 ligase AREL1 and its ubiquitination activities in vitro. J Biol Chem 294, 19934–19949. 10.1074/jbc.RA119.010327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lorenz S (2018). Structural mechanisms of HECT-type ubiquitin ligases. Biol Chem 399, 127–145. 10.1515/hsz-2017-0184. [DOI] [PubMed] [Google Scholar]
- 58.Buetow L, and Huang DT (2016). Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat rev Mol cell biol 17, 626–642. 10.1038/nrm.2016.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen Q, Qiu Z, Wu W, Zheng J, and Jia Z (2018). Characterization of interaction and ubiquitination of phosphoenolpyruvate carboxykinase by E3 ligase UBR5. Biol Open 7. 10.1242/bio.037366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, Xiong Y, Guan KL, and Zhao S (2011). Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell 43, 33–44. 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Youkharibache P, Veretnik S, Li Q, Stanek KA, Mura C, and Bourne PE (2019). The Small beta-Barrel Domain: A Survey-Based Structural Analysis. Structure 27, 6–26. 10.1016/j.str.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 62.Chen CK, Chan NL, and Wang AH (2011). The many blades of the beta-propeller proteins: conserved but versatile. Trends Biochem Sci 36, 553–561. 10.1016/j.tibs.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 63.Jiang H, He X, Feng D, Zhu X, and Zheng Y (2015). RanGTP aids anaphase entry through Ubr5-mediated protein turnover. J Cell Biol 211, 7–18. 10.1083/jcb.201503122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Munoz-Escobar J, Matta-Camacho E, Cho C, Kozlov G, and Gehring K (2017). Bound Waters Mediate Binding of Diverse Substrates to a Ubiquitin Ligase. Structure 25, 719–729 e713. 10.1016/j.str.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 65.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Punjani A, and Fleet DJ (2021). 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J Struct Biol 213, 107702. 10.1016/j.jsb.2021.107702. [DOI] [PubMed] [Google Scholar]
- 67.Kamadurai HB, Qiu Y, Deng A, Harrison JS, Macdonald C, Actis M, Rodrigues P, Miller DJ, Souphron J, Lewis SM, et al. (2013). Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. Elife 2, e00828. 10.7554/eLife.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kamadurai HB, Souphron J, Scott DC, Duda DM, Miller DJ, Stringer D, Piper RC, and Schulman BA (2009). Insights into ubiquitin transfer cascades from a structure of a UbcH5B∼ ubiquitin-HECTNEDD4L complex. Mol cell 36, 1095–1102. 10.1016/j.molcel.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shah SS, and Kumar S (2021). Adaptors as the regulators of HECT ubiquitin ligases. Cell Death Differ 28, 455–472. 10.1038/s41418-020-00707-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sander B, Xu W, Eilers M, Popov N, and Lorenz S (2017). A conformational switch regulates the ubiquitin ligase HUWE1. Elife 6, e21036. 10.7554/eLife.21036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiesner S, Ogunjimi AA, Wang H-R, Rotin D, Sicheri F, Wrana JL, and Forman-Kay JD (2007). Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 130, 651–662. 10.1016/j.cell.2007.06.050 [DOI] [PubMed] [Google Scholar]
- 72.Gallagher E, Gao M, Liu Y-C, and Karin M (2006). Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc Natl Acad Sci U S A 103, 1717–1722. 10.1073/pnas.0510664103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ogunjimi AA, Briant DJ, Pece-Barbara N, Le Roy C, Di Guglielmo GM, Kavsak P, Rasmussen RK, Seet BT, Sicheri F, and Wrana JL (2005). Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol Cell 19, 297–308. 10.1016/j.molcel.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 74.Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 75.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 77.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, and Ferrin TE (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27, 14–25. 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 80.Sanchez-Garcia R, Gomez-Blanco J, Cuervo A, Carazo JM, Sorzano COS, and Vargas J (2021). DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun Biol 4, 874. 10.1038/s42003-021-02399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mastronarde DN (2018). Advanced data acquisition from electron microscopes with SerialEM. Microscopy and Microanalysis 24, 864–865. [Google Scholar]
- 82.Scheres SH, and Chen S (2012). Prevention of overfitting in cryo-EM structure determination. Nat Methods 9, 853–854. 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, et al. (2018). MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci 27, 293–315. 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, Hershko A, Pagano M, and Draetta GF (2003). Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature 426, 87–91. 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. The overall structure of UBR5 in a dimeric form, showing the model-to-map fitting quality and structural details, related to Figures 1 and 2.
Video S2. The 3DVA analysis result of the entire apo UBR5 dimer, showing the most dramatic changes occur in the N-terminal region and the C-terminal catalytic HECT domain of UBR5, related to Figure 5.
Data Availability Statement
The cryo-EM 3D maps of the human UBR5 have been deposited at the Electron Microscopy Data Bank database with accession code EMD-27201 (dimer in C1 symmetry) EMD-27822 (dimer in C2 symmetry), and EMD-28646 (tetramer in C1 symmetry). The corresponding atomic models were deposited at the RCSB Protein Data Bank database with accession codes 8D4X,8E0Q, and 8EWI. These data are publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


