Abstract
Epstein-Barr virus nuclear antigen 2 (EBNA2) is essential for B-cell immortalization by EBV, most probably by its ability to transactivate a number of cellular and viral genes. EBNA2-responsive elements (EBNA2REs) have been identified in several EBNA2-regulated viral promoters, each of them carrying at least one RBP-Jκ recognition site. RBP-Jκ recruits EBNA2 to the EBNA2RE and, once complexed to EBNA2, is converted from a repressor into an activator. An activated form of the cellular receptor Notch also interacts with RBP-Jκ, providing a link between EBNA2 and Notch signalling. To determine whether activated Notch is able to transactivate EBNA2-responsive viral promoters, we performed cotransfection experiments with activated mouse Notch1 (mNotch1-IC) and luciferase constructs of the BamHI C, LMP1, and LMP2A promoters. We present here evidence that mNotch1-IC transactivates viral promoters known to be regulated by EBNA2. As shown for EBNA2, mutations or deletions of the RBP-Jκ sites diminish or eliminate mNotch1-IC-mediated transactivation of the promoters, pointing to an essential role for Notch–RBP-Jκ interaction. In addition to RBP-Jκ, other cellular factors may bind within the EBNA2REs of viral promoters. While some factors appear to play an important role in both EBNA2- and mNotch1-IC-mediated transactivation, others are only important for the activity of either EBNA2 or mNotch1-IC. We could observe specific mNotch1-IC-responsive regions, thereby throwing more light upon which cofactors interact with EBNA2 and mNotch1-IC, thus enabling them to become functionally transactivators in vivo.
Epstein-Barr Virus (EBV), a lymphotrophic human gammaherpesvirus, infects primary resting B cells and induces unlimited proliferation, a process called immortalization or transformation. In the immortalized cell lines, only a subset of viral genes is expressed that codes for six EBV nuclear antigens (EBNA1, -2, -3A, -3B, -3C, and -LP) and three latent membrane protein antigens (LMP1, -2A, and -2B).
Together with EBNA-LP, EBNA2 is the first viral gene expressed after EBV infection and is essential for both the initiation and maintenance of immortalization (9, 23, 31). EBNA2 has been shown to act as a transcriptional activator. It modulates the transcription of different cellular genes, including the B-cell activation markers CD21 and CD23 (7, 10, 65) and the proto-oncogene c-fgr (33). In addition, it transactivates the viral promoters of the latent membrane proteins LMP1, LMP2A, and LMP2B (14, 60, 70, 71) and the BamHI C promoter, which drives transcription of the six EBNAs. Therefore, the principal role of EBNA2 in B-cell transformation seems to be in transcriptional regulation.
In the promoters of several EBNA2-regulated genes, EBNA2-responsive elements (EBNA2REs) have been identified (29, 38, 62, 67, 72). All EBNA2REs have a large (70 to 80 bp) complex structure and can confer EBNA2 responsiveness on heterologous promoters. EBNA2 does not bind to DNA directly but is recruited to EBNA2REs by the cellular protein RBP-Jκ binding to the conserved core sequence CGTGGGAA. RBP-Jκ is present in all EBNA2-responsive regions known so far and recruits EBNA2 to those elements (21, 25, 63, 73). In all EBNA2REs, the RBP-Jκ binding site is essential but is not sufficient to mediate EBNA2 responsiveness, even if it occurs in tandem as in the LMP2A promoter. Other binding sites have been observed in the EBNA2REs, suggesting that additional cellular proteins are likely to participate in the transactivation process. One of them was identified as Spi1, also called PU.1, a member of the Ets family of transcription factors, which plays a pivotal role in regulating transcription of the LMP1 promoter (30, 37). Spi1 is active in myeloid and B cells.
RBP-Jκ (also designated as KBF2 or CBF1) was originally purified and characterized by Matsunami et al. (43) and Hamaguchi et al. (22). The protein is ubiquitously expressed and highly conserved through evolution. The RBP-Jκ homologue in Drosophila spp. is the neurogenic protein Suppressor of Hairless [Su(H)]. In insects, as well as in mammals, Su(H)/RBP-Jκ acts downstream of Notch (4, 39).
In vertebrates, the Notch signal transduction pathway has an essential function during embryogenesis and is involved in differentiation processes of neuronal precursors, myoblasts, and Malpighian tubules (2, 8, 34, 50). Some data suggest that Notch signalling is also involved in the renewal and differentiation of hematopoietic cells. Notch is expressed in CD34-positive hematopoietic stem cells (47). It influences the choice between CD4 and CD8, as well as the choice between the alpha-beta versus the gamma-delta T-cell lineage (54, 69). An important role of Notch in the T-cell system is also indicated by the fact that constitutive Notch activation is a characteristic feature of a subset of T-cell leukemias and lymphomas in humans, cats, and mice (13, 20, 55). Furthermore, an inhibitory effect on the granulocyte differentiation has been observed (5, 40, 48). Notch appears to be expressed in B cells, but so far there are no studies of the role of Notch signalling in B-cell differentiation.
In mammalian cells, RBP-Jκ is localized in the nucleus bound to RBP-Jκ binding sites, where it usually acts as a transcriptional repressor (12, 27, 64). Activation of the transmembrane receptor Notch by its ligand delta or jagged leads to proteolytic cleavage of Notch, followed by the translocation of the intracellular part of Notch (Notch-IC or activated Notch) to the nucleus, where it transactivates genes previously repressed by RBP-Jκ (35, 56, 59, 61). Thus, EBNA2 can be regarded as a functional homologue of Notch-IC.
To get further insight into this functional homology between EBNA2 and Notch-IC in B cells, we studied whether Notch-IC is able to transactivate the known viral EBNA2-responsive promoters. It has already been demonstrated that both EBNA2 and an activated mouse Notch1 transactivate promoter reporter gene constructs carrying a multimerized RBP-Jκ binding site (27, 41, 58). However, nothing is known about the Notch responsiveness of EBNA2-regulated promoters. We compared EBNA2 and an activated mouse Notch1 concerning their transactivation of the viral EBNA2REs to determine whether the EBNA2REs can be upregulated by Notch-IC. We wanted to see whether the same cis-acting sequences are necessary for EBNA2 and Notch-IC responsiveness and also whether it was possible to characterize specific Notch-responsive regions.
MATERIALS AND METHODS
Cell lines and culture conditions.
Raji is a Burkitt’s lymphoma cell line (52), and HeLa is a cervical carcinoma epithelial cell line (19). BL41-P3HR1 was obtained after infection of the EBV-negative Burkitt’s lymphoma cell line BL41 with the P3HR1 virus strain (7). HH514 is a single cell clone of P3HR1 (53). The cell line BL41-P3HR1-5E was obtained after stable transfection of BL41-P3HR1 cells with a plasmid encoding an EBNA2-estrogen receptor fusion protein (ER-EBNA2) (32).
The cell lines were grown in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM glutamine, 1 mM sodium pyruvate, penicillin (100 U/ml), and streptomycin (100 μg/ml). β-Estradiol was added to the cell culture medium for BL41-P3HR1-5E cells at a final concentration of 1 μM. HeLa cells were grown in RPMI 1640 medium and rotated at 30 rpm and at 37°C to avoid adherence. All other cell lines were incubated at 37°C in an atmosphere of 5% CO2 and diluted 1:3 with fresh medium twice a week.
Plasmids.
The mNotch1-IC expression vector pSG5 mNotch1-IC (27) was kindly provided by T. Henkel. The EBNA2 expression plasmid pGa986-20 was generated by ligation of a 5.1-kb HindIII (blunt-end)/BglII fragment from pU294-6 (72) with a 4.1-kb EcoRI (blunt-end)/BglII fragment from pSG5 (Stratagene). The luciferase reporter construct pGa981-16, carrying a multimerized RBP-Jκ binding site, has been described by Minoguchi et al. (49). Most of the LMP1 promoter luciferase constructs have been described by Laux et al. (38): LMPLUC0, LMPLUC9, E/βg, 80/βg, BA18, BA20, and pGa50-7. pHoe58-3 was generated by site-specific mutagenesis of BA20 by using Vent DNA polymerase according to the method described by Byrappa et al. (6). The following primers were chosen to delete three central bases of the second RBP-Jκ motif (positions 169823 to 169825 according to Baer et al. [3]): RBP2s, AGC GGC AGT GTA ATC TGC AC, and RBP2as, ACA ACA CTA CGC ATC CCC CC.
The LMP2A promoter luciferase construct (pTP1Luc/-45) has been described by Zimber-Strobl et al. (72). For cloning of mutated or deleted forms of the 81-bp EBNA2RE in front of the LMP2A minimal promoter, we have inserted a polylinker containing a SnaBI site and a NcoI site in the StuI site of pTP1Luc/-45 (pGa59/19). pGa59/19 was digested with SnaBI and NcoI and ligated with oligonucleotides containing SnaBI and NcoI sites at their ends, respectively. To obtain the BamHI C promoter (Cp) luciferase constructs pHoe19-7 and pHoe25-7, a KpnI/BamHI fragment of p633 (23) containing the BamHI C promoter region was cloned into a pBluescript vector (pBluescript II KS(+); Stratagene), which was cut with these enzymes. This subclone was digested with Sau3A and StuI or with Sau3A and Ecl136. The 744-bp (Sau3A/StuI) and 251-bp (Sau3A/Ecl136) fragments were isolated and cloned into the BglII/StuI sites of pBLLUC5 (38), resulting in the plasmids pHoe19-7 and pHoe25-7, respectively.
For cloning of the EBNA2RE of Cp and mutated constructs in front of the β-globin minimal promoter and the reporter gene luciferase, pGa59/19 was digested with SnaBI and NcoI and ligated with the corresponding oligonucleotides containing the corresponding restriction sites at their ends. These constructs again were digested with SnaBI and AatII; the resulting 180-bp fragments contained the oligonucleotides. pGa50-7, the β-globin minimal promoter construct, was digested with StuI and AatII and ligated with the 180-bp fragment, yielding pHoe230-1 (OCpWT), pHoe230-2 (OCpMuta), and pHoe 230-3 (OCpMutb).
The mNotch1-IC expression plasmid pLS710-6 for constitutive expression of mNotch1-IC in stable cell lines was generated by ligation of the 2.8-kb SalI fragment of pSG5-mNotch1-IC with the vector pHEBOPL (51), which was linearized by SalI.
All plasmids generated by PCR or after cloning of an oligonucleotide were verified by DNA sequencing.
Oligonucleotides.
The following oligonucleotides used for the LMP2A promoter luciferase constructs have been described previously: O54 (70), O40, O80, MutC and MutH (45), and RBPMut (73). The position of the oligonucleotide O59 according to the EBV genomic sequence described by Baer et al. (3) is in the region from 166236 to 166289.
The oligonucleotides used for the BamHI C promoter luciferase constructs are in the following positions according to the Cp enhancer sequence (−429 to −254) published by Jin and Speck (29): OCpWT, −391 to −320; OCpMuta, OCpWT carrying mutations at positions −373 to −371; and OCpMutb, OCpWT carrying mutations at positions −357 to −353.
Transfection of cells.
Electroporation was carried out as described previously (72) with slight modifications. Cells were electroporated in RPMI 1640 without fetal calf serum at room temperature. Then, 500 μl of fetal calf serum was added to the cells immediately after electroporation. After 10 min cells were resuspended in 10 ml of prewarmed RPMI 1640 containing 10% fetal calf serum and standard supplements.
For establishment of the stable cell line BL41-P3HR1 mNotch1-IC, the transfected cells were kept in medium with 200 μg of hygromycin per ml.
Luciferase assays.
Cells were harvested and lysed as described previously (45). First, 10 μl of each probe was mixed with 150 μl of test buffer (25 mM glycylglycine [pH 7.8], 5 mM ATP, 15 mM MgSO4) on a 96-well plate. Then, after the addition of 100 μl of 11 mM luciferin in 0.5 M Tris-HCl (pH 7.8) to each reaction, the bioluminescence in relative light units was measured with a Micro-Lumat LB 96 P (Berthold, Wildbach, Germany).
Protein immunoblots.
For Western blot analysis cellular extracts were prepared by sonification in H8 lysis buffer (20 mM Tris, pH 7.0; 2 mM EGTA; 2 mM EDTA; 6 mM β-mercaptoethanol; 50 mM NaF; 100 mM NaCl; 1% sodium dodecyl sulfate [SDS]). The protein concentration was determined, and equal amounts of protein were separated on a Laemmli 10% polyacrylamide–SDS gel. Proteins were transferred onto nitrocellulose filters (Amersham Hybond ECL), and protein expression was analyzed with the anti-FLAG monoclonal antibody M2 (Eastman Kodak). Immunoreactive proteins were detected by peroxidase-coupled secondary antibodies and enhanced chemiluminescence (ECL System; Amersham).
Nuclear extract preparation and EMSA.
Nuclear extracts were prepared by a modification of the method of Dignam et al. (11) as described by Zimber-Strobl et al. (72). Binding reactions for electrophoretic mobility shift assay (EMSA) were carried out as described previously (72). In supershift experiments, either 2 μl of tissue culture supernatant containing anti-EBNA2 monoclonal antibody R3 (rat immunoglobulin G2a [36]) or 2 μl of anti-FLAG monoclonal antibody M2 was added to the reaction mixture.
Methylation interference analysis.
The methylation interference assay is based on a G reaction of the Maxam-Gilbert sequencing reaction (44) and was performed as described by Meitinger et al. (45).
Radioactively labelled probes.
The oligonucleotides O40 and O54 were synthesized with 5′-protruding ends, which were filled in with Klenow polymerase in the presence of [32P]dCTP (3,000 Ci/mmol) and unlabeled dATP, dGTP, and TTP.
RESULTS
Effects of different amounts of mNotch1-IC and EBNA2 upon the transactivation of a RBP-Jκ multimer construct.
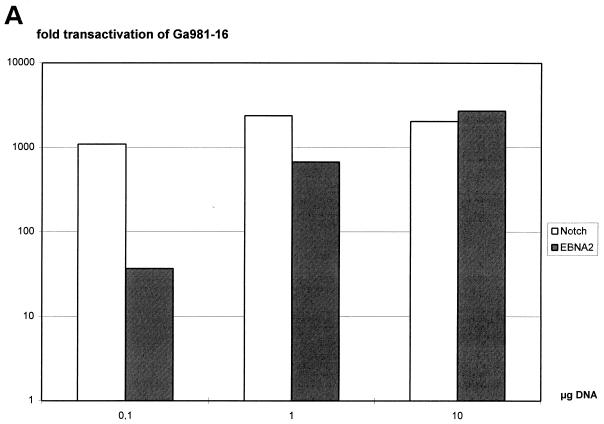
It has already been shown that both mNotch1-IC and EBNA2 can strongly transactivate a luciferase reporter construct containing a hexamer of the two RBP-Jκ binding sites of the LMP2A promoter in front of the β-globin minimal promoter pGa981-16 (58). Since transactivation of this construct depends on the RBP-Jκ binding site only, it was used to standardize the transactivation capacity of EBNA2 and mNotch1-IC. Portions (0.1, 1, or 10 μg) of the EBNA2 and mNotch1-IC expression plasmids were transiently cotransfected with the promoter reporter gene construct pGa981-16 in the EBNA2-negative cell line BL41-P3HR1. The total amount of expression plasmid was adjusted to 10 μg with the vector pSG5. Transactivation rates were determined by standardizing the luciferase activities to the values obtained after transfection of 10 μg of pSG5. In Fig. 1A, the fold transactivation after cotransfection of different amounts of mNotch1-IC and EBNA2 expression plasmids are shown. While cotransfection of 0.1, 1, or 10 μg of the mNotch1-IC expression plasmid resulted in comparable transactivation rates, transactivation values significantly increased after cotransfection of rising amounts of the EBNA2 expression plasmid. Since transfection of 10 μg of the mNotch1-IC and EBNA2 expression plasmids resulted in approximately the same transactivating activity, this concentration was used to study mNotch1-IC responsiveness of the EBNA2-regulated viral promoters.
FIG. 1.
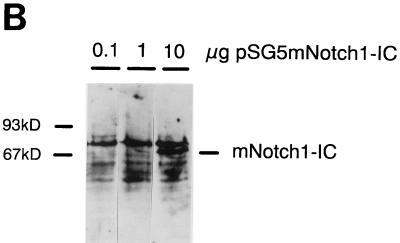
Luciferase activity of the RBP-Jκ multimer construct upon titration of mNotch1-IC and EBNA2 expression plasmids. (A) Twenty micrograms of the reporter construct pGa981-16 was cotransfected with 0.1, 1, or 10 μg of the expression plasmids pSG5 mNotch1-IC (black boxes) or pGa986-20 (white boxes) into the EBNA2-negative cell line BL41-P3HR1. The amount of expression plasmid was adjusted to 10 μg with the vector pSG5. The fold transactivation was standardized to the value obtained from cotransfection with 10 μg of the vector control pSG5. Results are averages from two independent experiments. (B) At 24 h after the transfection of 0.1, 1, or 10 μg of pSG5 mNotch1-IC in BL41-P3HR1 cells, the cells were harvested. Equal amounts of protein were loaded onto the gel, and expression of mNotch1-IC was examined by Western blotting with the anti-FLAG antibody. Available antibodies failed to detect EBNA2 at this concentration.
Expression levels of mNotch1-IC in transient-transfection assays.
We performed Western blots to determine protein levels of mNotch1-IC in transient-transfection assays. First, 0.1, 1, or 10 μg of the vector pSG5 mNotch1-IC was transfected into BL41-P3HR1 cells, and protein extracts were prepared after 24 h. By Western blotting with the anti-FLAG monoclonal antibody M2, mNotch1-IC expression could be detected after transfection of 10 μg of the expression vector, whereas the signal was barely visible with smaller amounts. This indicates that protein levels correlate with the amount of the transfected expression plasmid. Not surprisingly, the biological activity of mNotch1-IC can be detected at a much lower concentration than the protein. The same is true for EBNA2, which is virtually undetectable by Western blotting with the available antibody R3 at the concentrations used to measure its activity.
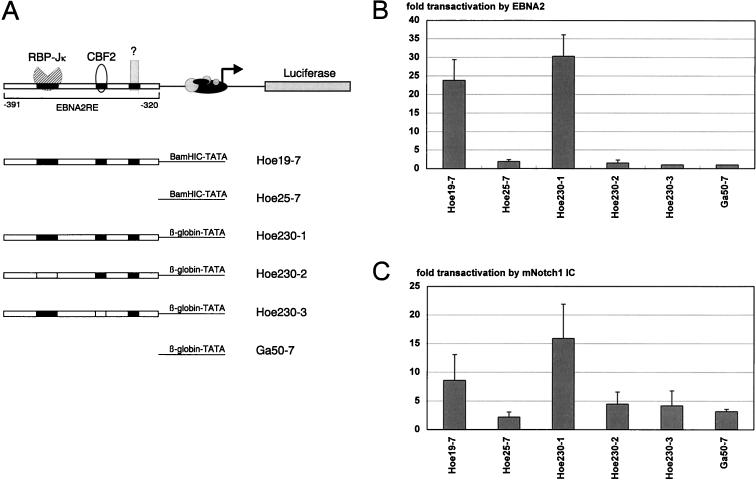
Activation of the BamHI C promoter by mNotch1-IC and EBNA2.
For the BamHI C promoter it was shown that an enhancer located upstream is transactivated by EBNA2 (60). To elucidate whether activated mNotch1 can also upregulate this promoter, we constructed plasmids containing the BamHI C promoter with and without the enhancer element that were linked to the luciferase reporter gene (luc) (Fig. 2A). Transient transfections of the reporter plasmids without an additional expression vector, with the control vector pSG5, or with the expression plasmids pSG5 mNotch1-IC and pSG5 EBNA2 (pGa986-20) were carried out in EBNA2-negative BL41-P3HR1 cells. The transactivation of mNotch1-IC or EBNA2 was standardized to the pSG5 values. Mean values of four representative experiments are shown in Fig. 2B and C. Cotransfection of the mNotch1-IC expression vector with pHoe19-7, which contains the EBNA2-responsive enhancer in front of the BamHI C promoter, induced an 8.6-fold increase in luciferase expression, whereas the construct without the enhancer element (pHoe25-7) mediated a 2.2-fold induction. Similar results were obtained with the EBNA2 expression vector with a 23.8-fold transactivation of pHoe19-7 and a 1.9-fold transactivation of pHoe25-7. This indicates that the EBNA2-responsive enhancer can mediate transcriptional activation by constitutively active mNotch1 to the BamHI C promoter. The degree of transactivation with mNotch1-IC is about two- to threefold lower than that obtained with EBNA2.
FIG. 2.
The EBNA2RE of the BamHI C promoter can confer mNotch1-IC and EBNA2 responsiveness on either the BamHI C or the β-globin minimal promoter. (A) Schematic representation of the BamHI C promoter luciferase constructs used in the cotransfection assays. In the upper part of the illustration the essential regions of the EBNA2RE of the BamHI C promoter are shown. The two regions interact with RBP-Jκ and CBF2, respectively. In the lower part, the EBNA2REs and mutated sequences of the essential regions are indicated by solid and open boxes, respectively. (B and C) Portions (10 μg) of the expression plasmids pSG5 mNotch1-IC (B) or pGa986-20 (C) were cotransfected with 20 μg of the reporter construct into the EBNA2-negative cell line BL41-P3HR1. The fold transactivation was standardized to the value from cotransfection with 10 μg of the vector control pSG5. The mean values and the standard deviations of four independent experiments are shown.
Effects of specific elements of the BamHI C promoter EBNA2RE upon mNotch1-IC- and EBNA2-mediated transactivation.
Three elements within the EBNA2-responsive enhancer have been described as important for EBNA2-dependent activity (29), with two of them being more critical than the third. The sequence of the promoter distal element 1 corresponds to the RBP-Jκ recognition sequence, whereas the second element is described as the CBF2 binding site (17). To study the effect of these motifs in mNotch1-IC-induced transactivation, we constructed plasmids containing the enhancer (−391 to −320, according to the sequence published by Jin and Speck [29]), with the two elements either unmutated (pHoe230-1) or with only one of them mutated (pHoe230-2 and pHoe230-3). They were linked to the luciferase gene under the control of the β-globin minimal promoter. A plasmid consisting only of the β-globin minimal promoter in front of the luciferase gene (pGa50-7) was used as a negative control (Fig. 2A). Transient-cotransfection assays were performed with these constructs as described above, and the mean results of four experiments are shown in Fig. 2B and C. The mNotch1-IC expression vector resulted in a 15.9-fold transactivation of the unmutated enhancer construct, whereas the EBNA2 expression vector induced a 30.3-fold luciferase expression. Transfection of the β-globin minimal promoter construct led to a 3.2-fold induction by mNotch1-IC, which was not observed for EBNA2.
Mutations in either the RBP-Jκ or the CBF2 binding site reduced the ability of mNotch1-IC and EBNA2 to transactivate to the level observed with the minimal promoter construct. Regarding EBNA2, these results correspond to those published by Jin and Speck (29) and Fuentes-Pananá and Ling (17). We conclude that the RBP-Jκ and CBF2 binding sites are important for mNotch1-IC as well as for EBNA2 responsiveness.
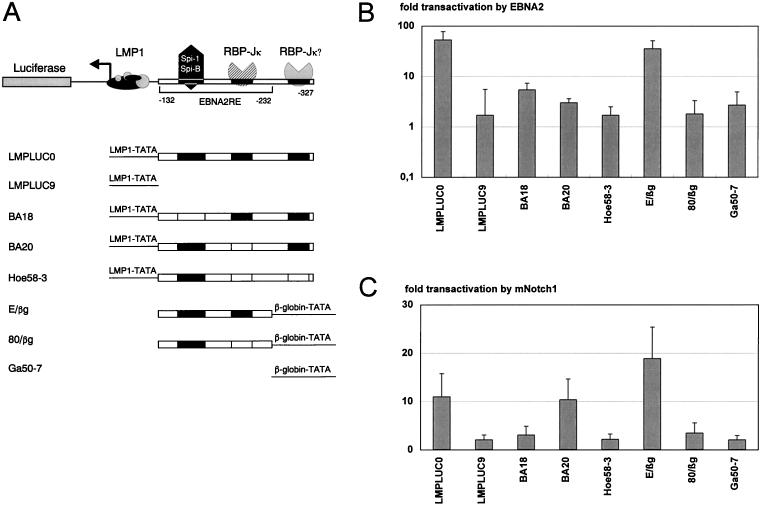
Response of the LMP1 promoter to constitutively active mNotch1 and EBNA2.
For the LMP1 promoter, an 80-bp element (EBNA2RE) located between positions −152 and −232, relative to the RNA start site, has been shown to be sufficient for transactivation by EBNA2 (38). Within the EBNA2RE, two sequence elements harboring Spi1 (−157 to −174) and RBP-Jκ (−217 to −225) binding sites, respectively, have been shown to be essential to mediate an EBNA2 response. A second potential RBP-Jκ recognition site is located 65 bp upstream of the first and outside the previously described 80-bp EBNA2RE (Fig. 3A), but this has not yet been functionally analyzed. To study the response of the LMP1 promoter to mNotch1-IC, transient-transfection assays were performed as described above with the luciferase reporter plasmids LMPLUC0 containing the whole LMP1-promoter region and LMPLUC9 lacking the EBNA2RE (38). Cotransfection of pSG5 mNotch1-IC with LMPLUC0 induced an 11-fold luciferase expression, whereas LMPLUC9 mediated a 2.1-fold transactivation by mNotch1-IC (Fig. 3B).
FIG. 3.
The EBNA2RE of the LMP1 promoter can confer mNotch1-IC and EBNA2 responsiveness on either the LMP1 or the β-globin minimal promoter. (A) Schematic representation of the LMP1 promoter luciferase constructs used in the cotransfection assays. In the upper part of the figure the essential regions of the LMP1 promoter EBNA2RE are shown. One sequence element interacts with RBP-Jκ; the other interacts with Spi1. A potential second RBP-Jκ site located beyond the EBNA2RE is indicated. In the lower part the EBNA2REs and mutated sequences of the essential regions are indicated by solid and open boxes, respectively. (B and C) Portions (10 μg) of the expression plasmids pSG5 mNotch1-IC (B) or pGa986-20 (C) were cotransfected with 20 μg of the reporter construct into the EBNA2-negative cell line BL41-P3HR1. The fold transactivation was standardized to the value from cotransfection with the vector control pSG5. The mean values and the standard deviations of four independent experiments are shown.
EBNA2, on the other hand, exhibited a 53.4-fold transactivation of LMPLUC0 and no induction of LMPLUC9.
The Spi1 and RBP-Jκ binding sites play an important role for mNotch1-IC- as well as EBNA2-mediated transactivation.
To study the role of the Spi1 and RBP-Jκ recognition sites upon mNotch1-IC-mediated transactivation, we used reporter plasmids with either a mutated Spi1 binding site (BA18), a deleted first RBP-Jκ binding site (BA20), or with a mutated first and potential second RBP-Jκ binding site (pHoe58-3). Mutation of the Spi1 binding site dramatically decreased inducibility by activated mNotch1 as well as by EBNA2 (Fig. 3B and C). This indicates that the binding of Spi1/SpiB plays a critical role in the response of the LMP1 promoter by both EBNA2 and activated mNotch1. Deletion of the first RBP-Jκ binding site (BA20) significantly reduced transactivation by EBNA2 but not by mNotch1-IC. To test whether the potential second RBP-Jκ binding site is able to mediate response to mNotch1-IC, we introduced a second mutation that also destroyed the second RBP-Jκ site (pHoe58-3). As a consequence, mNotch1-IC-mediated transactivation decreased to 2.2-fold. This indicates that mNotch1-IC can functionally interact with this second RBP-Jκ binding site, whereas EBNA2 cannot.
To elucidate whether mNotch1-IC can use the first RBP-Jκ site within the EBNA2RE as well, we tested the reporter gene plasmid E/βg, containing the 80-bp EBNA2RE in front of the β-globin minimal promoter and the luciferase gene. This construct could be transactivated about 35-fold by EBNA2 (Fig. 3C) and 19-fold by mNotch1-IC (Fig. 3B), providing evidence that mNotch1-IC can interact with the first RBP-Jκ binding site as well. Mutation of this RBP-Jκ site in plasmid 80/βg dramatically decreased transactivation by mNotch1-IC as well as by EBNA2. The construct pGa50-7 was used as a negative control.
These results demonstrate that the Spi1 recognition site is essential for both mNotch1-IC- and EBNA2-mediated transactivation, whereas the first RBP-Jκ site can be compensated by the presence of the potential second one, resulting in complete mNotch1-IC responsiveness.
Activation of the LMP2A promoter by mNotch1-IC and EBNA2.
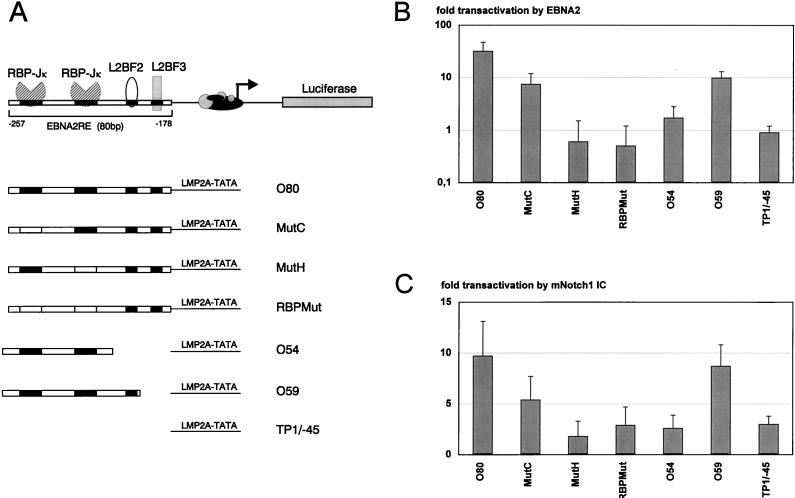
To investigate whether mNotch1-IC is able to transactivate the LMP2A promoter, we performed transient-transfection assays in BL41-P3HR1 cells as described above. The reporter plasmid pTP1Luc/O80, containing the wild-type EBNA2RE of the LMP2A promoter in front of the LMP2A minimal promoter and the luciferase gene (Fig. 4A), was cotransfected with mNotch1-IC and EBNA2 expression vectors, respectively. A 9.7-fold transactivation by mNotch1-IC (Fig. 4B) was observed, compared to a 31.8-fold induction by EBNA2 (Fig. 4C). pTP1Luc/-45 was used as negative control.
FIG. 4.
The EBNA2RE of the LMP2A promoter can confer mNotch1-IC and EBNA2 responsiveness on the LMP2A minimal promoter. (A) Schematic representation of the LMP2A promoter luciferase constructs used in the cotransfection assays. In the upper part of the figure the essential regions of the EBNA2RE of the LMP2A promoter are shown. Two regions interact with RBP-Jκ, whereas the two others are designated as L2BF2 and L2BF3. In the lower part of the figure the EBNA2REs and mutated sequences of the essential regions are indicated by solid and open boxes, respectively. (B and C) Portions (10 μg) of the expression plasmids pSG5 mNotch1-IC (B) or pGa986-20 (C) were cotransfected with 20 μg of the reporter construct, respectively, into the EBNA2-negative cell line BL41-P3HR1. The fold transactivation was standardized to the value from cotransfection with the vector control pSG5. The mean values and the standard deviations of four independent experiments are shown.
Effects of specific elements of the LMP2A EBNA2RE upon mNotch1-IC- and EBNA2-mediated transactivation.
To elucidate the importance of the two RBP-Jκ sites, we used the constructs pTP1Luc/MutC, pTP1Luc/MutH, and pTP1Luc/RBPMut, in which either the first, the second, or both sites were mutated, respectively. Mutation of the first RBP-Jκ binding site led to a 50% decrease of mNotch1-IC-mediated transactivation, whereas mutation within the second or both motifs resulted in a nearly complete abolishment of transactivation by mNotch1-IC. These results are very similar to those described for EBNA2 (Fig. 4B and Meitinger et al. [45]).
We have shown previously that the two 11-bp RBP-Jκ binding sites are essential but not sufficient for transactivation by EBNA2 (45). To determine whether the two RBP-Jκ sites would be sufficient for activation by mNotch1-IC, the construct containing the two RBP-Jκ sites in front of the LMP2A minimal promoter was transfected into BL41-P3HR1 cells together with mNotch1-IC. As shown in Fig. 4B and C, the 54-bp element (positions −262 to −209 corresponding to the LMP2A transcription start site) is neither sufficient for EBNA2- nor for mNotch1-IC-mediated transactivation. The addition of 5 bp at the promoter proximal site restored activation by EBNA2 partially (8.7-fold) and by mNotch1-IC almost completely (9.8-fold).
These results revealed that (i) mNotch1-IC can transactivate the EBNA2RE of the LMP2A promoter, although to a lesser extent than EBNA2 (approximately 30% of EBNA2); (ii) the two RBP-Jκ sites are essential for mNotch1-IC- as well as EBNA2-mediated transactivation, whereby the second site is more important than the first; (iii) the sequences between positions −209 and −198 are essential for both mNotch1-IC- and EBNA2-mediated transactivation; and (iv) the sequences between −198 and −178 further increase transactivation by EBNA2 but not by mNotch1-IC.
mNotch1-IC can interact with the EBNA2RE of the LMP2A promoter.
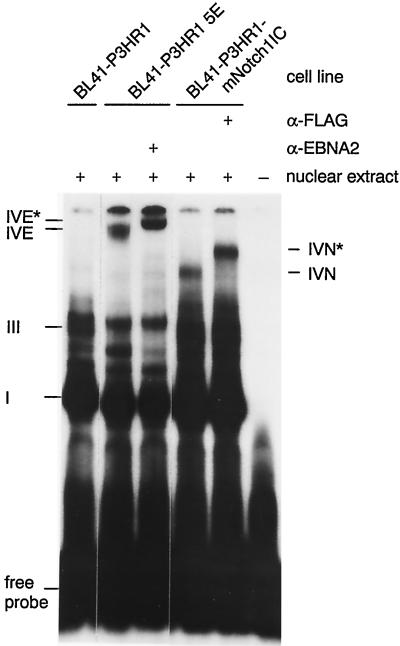
We have shown previously that EBNA2 interacts with RBP-Jκ, which binds to a duplicated 11-bp motif within the EBNA2RE of the LMP2A promoter (45). To analyze whether similar complexes are formed between RBP-Jκ within the EBNA2RE and mNotch1-IC, we performed EMSAs with the 54-bp oligonucleotide as a radioactively labelled probe and nuclear extracts of BL41-P3HR1 cells either untransfected or stably transfected with mNotch1-IC containing a FLAG epitope. Nuclear extracts of the cell line BL41-P3HR1-5E containing ER-EBNA2 (32) were used as a control. The result of the EMSA is shown in Fig. 5. With BL41-P3HR1 extracts, complexes I and III could be detected, reflecting occupation of one or both RBP-Jκ binding sites with a cellular factor (45). Nuclear extracts of the cell line BL41-P3HR1-5E revealed the EBNA2-containing complex IVE, which could be supershifted by the monoclonal rat anti-EBNA2 antibody R3.
FIG. 5.
Analysis of DNA-protein interactions within the EBNA2RE 5′ domain of the LMP2A promoter. An EMSA is shown with nuclear extracts of BL41-P3HR1 cells and of BL41-P3HR1 cells stably transfected with either EBNA2 or mNotch1-IC, which were incubated with the 32P-labelled 54-bp oligonucleotide O54 consisting of positions −209 to −262 relative to the LMP2A RNA start site. To show that EBNA2 or mNotch1-IC is a component of the complex IVE/IVN, the monoclonal anti-EBNA2 antibody R3 or the anti-FLAG monoclonal antibody M2 was added, respectively. Complexes were separated on a 4% polyacrylamide gel. The positions of complexes I, III, IVE, IVN, IVE*, and IVN* are indicated.
With nuclear extracts of BL41-P3HR1 cells stably transfected with mNotch1-IC, an additional complex designated IVN was detected; this complex could be supershifted with the anti-FLAG monoclonal antibody M2. This indicates that mNotch1-IC is able to interact with the LMP2A promoter in gel shift analysis.
DNA-protein interactions within the EBNA2RE of the LMP2A promoter downstream of the second RBP-Jκ binding site.
The transfection experiments indicated that sequences between positions −209 and −198 are essential for both EBNA2- and mNotch1-IC-mediated transactivation, whereas the promoter proximal sequences between positions −197 and −178 increase transactivation by EBNA2 but not by mNotch1-IC.
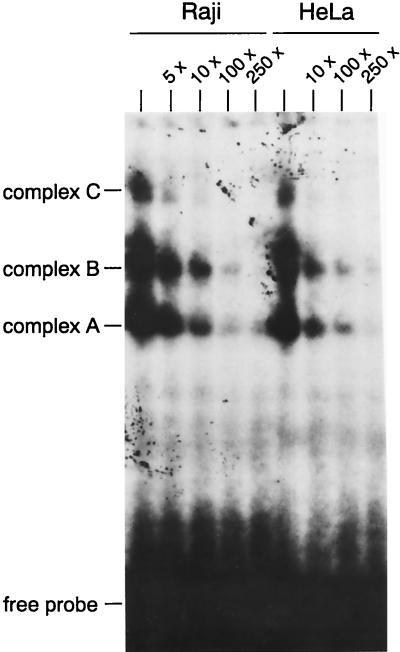
To look for cellular proteins which might be responsible for these effects, we performed a gel retardation assay with the radioactively labeled O40 (45). This oligonucleotide consists of the 3′ region of the EBNA2RE without the two RBP-Jκ binding sites (positions −217 to −178). The two RBP-Jκ sites were excluded from the oligonucleotide probe because protein complexes containing RBP-Jκ are so dominant that other interactions may not be detectable by EMSA. To show possible differences between lymphoid and nonlymphoid cells, we incubated the probe with nuclear extracts of Raji and HeLa cells (Fig. 6). Both extracts formed three complexes (A, B, and C) with oligonucleotide O40; all of them can be competed by the addition of increasing amounts of unlabelled competitor DNA (5× to 250×). The DNA-protein interactions within the 3′ region of the EBNA2RE are not lymphoid specific and are much weaker than those formed by RBP-Jκ and the two RBP-Jκ binding sites (data not shown).
FIG. 6.
Analysis of DNA-protein interactions within the EBNA2RE 3′ domain of the LMP2A promoter. An EMSA is shown with oligonucleotide O40 consisting of positions −217 to −178 relative to the LMP2A RNA start site as a radioactively labelled probe. The probe was incubated with Raji and HeLa nuclear extracts. For competition, a 5- to 250-fold molar excess of unlabelled oligonucleotide was added to the gel shift reactions. Complexes were separated on a 4% polyacrylamide gel. The positions of complexes A, B, and C are shown.
DNA regions protected by the complexes A, B, and C.
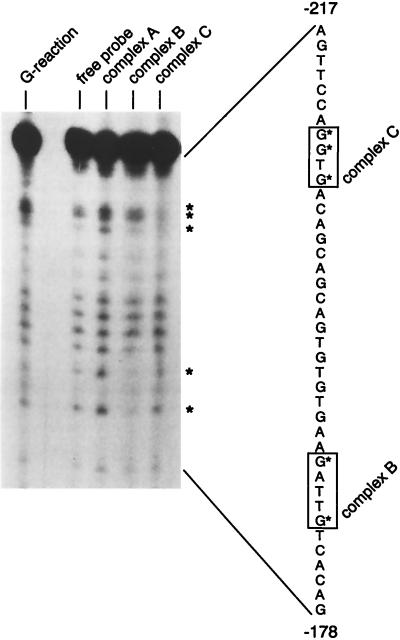
To identify the nucleotides involved in the complexes A, B, and C, we performed a methylation interference experiment with the oligonucleotide O40 and the nuclear extract of HeLa cells. Complexes A, B, and C and the free oligonucleotide were recovered after gel retardation. No protected guanines could be detected in complex A. In complex B the guanines at positions −188 and −184 were weakened. The analysis of complex C identified a protected region of three guanines located between positions −210 to −207 (Fig. 7).
FIG. 7.
Methylation interference analysis of the EBNA2RE 3′ region. The radioactively labelled and randomly methylated oligonucleotide O40 consisting of positions −217 to −178 of the 3′ region was incubated with HeLa nuclear extracts. The protected guanines are shown for complexes A, B, and C in comparison with the free probe. As a control reaction, a Maxam-Gilbert G reaction is shown on the lefthand side. On the righthand side the sequence of the 3′ region from positions −217 to −178 is shown. Protected sequences are marked with asterisks. Minimal extensions of the protection are indicated by boxes.
These experiments provide evidence that, apart from RBP-Jκ, there are at least two additional sites in the promoter proximal region of the EBNA2RE interacting with DNA binding proteins, which we designate L2BF2 and L2BF3. L2BF2 interacting with sequences between −210 and −207 seems to be essential for EBNA2- as well as for mNotch1-IC-mediated transactivation of the LMP2A promoter. L2BF3 binding to the promoter proximal region (positions −198 to −178, including complex B) appears to contribute to EBNA2- but not to mNotch1-IC-mediated transactivation of the LMP2A promoter.
DISCUSSION
EBNA2 is known to be essential in EBV-induced B-cell immortalization, most likely because of its ability to transactivate several cellular and viral genes. In most of these promoters EBNA2-responsive elements could be identified, all of them carrying at least one RBP-Jκ recognition site (29, 38, 72). EBNA2 is tethered to the EBNA2-responsive promoter elements by interaction with the cellular repressor protein RBP-Jκ (21, 25, 63, 73), which is known to be a component of the Notch receptor signal transduction pathway and to directly interact with activated Notch (28, 35). Therefore, we sought to determine whether EBNA2 can be regarded as a viral homologue of Notch. We have already shown that the luciferase reporter construct pGa981-16, containing a hexamer of the two RBP-Jκ binding sites of the LMP2A promoter, is strongly transactivated by mNotch1-IC as well as EBNA2 (58). We used this construct to standardize the transactivation capacity of mNotch1-IC and EBNA2. Transfection of 10 μg of the mNotch1-IC and EBNA2 expression plasmids, respectively, resulted in similar transactivation rates of the RBP-Jκ multimer construct. Therefore, this concentration was used to analyze the mNotch1-IC responsiveness of the EBNA2-regulated viral promoters. The ability to transactivate the RBP-Jκ multimer construct rather than the protein expression level was used for standardization, because the biological activity can be measured very sensitively by using the same reporter plasmid for mNotch1-IC and EBNA2. On the other hand, it is critical to compare the expression levels of mNotch1-IC and EBNA2 by Western blotting, since different antibodies are used which cannot be compared in their affinities.
For elucidating the potential functional similarity of activated mouse Notch1 and EBNA2, we compared their transactivation activity upon the viral BamHI C, the LMP1, and the LMP2A promoters. mNotch1-IC was shown to transactivate each of the three viral promoters to ca. 10-fold, indicating an interaction between mNotch1-IC and the EBNA2REs. We also observed a two- to threefold mNotch1-IC-mediated transactivation upon the three promoter constructs lacking the whole EBNA2RE. This may indicate a slight transcriptional induction through the basal transcription complex by mNotch1-IC which is not observed with EBNA2.
The EBNA2-mediated transactivation of the BamHI C, the LMP1, and the LMP2A promoters is three- to fivefold higher than the upregulation observed by mNotch1-IC. This observation can be explained by the set of additional proteins binding within the EBNA2REs of the viral promoters. It is also possible that in BL41-P3HR1 cells there are B-cell-specific proteins interacting with EBNA2 but not with mNotch1-IC. Therefore, it would be interesting to study the effects of mNotch1-IC in non-B cells.
Constructs with different mutations in the EBNA2REs were used to get further insight as to which cofactors are necessary for mNotch1-IC or EBNA2 responsiveness.
The BamHI C promoter controls the expression of the entire family of EBNA genes in latently infected lymphoblastoid cells and is activated by EBNA2 through an upstream enhancer element containing RBP-Jκ and CBF2 binding sites (17). For the EBNA2-mediated upregulation, it has been shown that mutations of the RBP-Jκ or the CBF2 binding site dramatically reduce the level of transactivation. Both binding sites are equally important for the upregulation of the promoter by mNotch1-IC as well as by EBNA2. This suggests that mNotch1-IC and EBNA2 transactivate the BamHI C promoter by a similar mechanism.
Within the LMP1 promoter an EBNA2RE could be identified, located between positions −152 and −232, relative to the RNA start site, and containing RBP-Jκ and Spi1/SpiB binding sites (30, 37, 62). Previously, we have shown that the Spi1 and the RBP-Jκ binding sites are both essential for EBNA2-mediated transactivation of the LMP1 promoter (38). Here we provide evidence that mNotch1-IC is able to upregulate the LMP1 promoter in transient-transfection assays. It was striking that the Spi1 binding site is crucial for EBNA2 as well as for mNotch1-IC responsiveness. Until now, a direct interaction of the native EBNA2 protein with Spi1 could not be demonstrated. Probably, Spi1 is an auxilliary factor that binds within the EBNA2RE, contributing to EBNA2- and mNotch1-IC-mediated transactivation. It has been hypothesized that the expression pattern of Spi1 determines that EBNA2 can transactivate the LMP1 promoter only in B cells and not in T and epithelial cells (15, 66). The importance of the Spi1 binding site for mNotch1-IC-mediated upregulation of the LMP1 promoter supports the notion that Notch signalling physiologically plays a role in B cells.
Surprisingly, the RBP-Jκ binding site described within the EBNA2RE was not essential for mNotch1-IC responsiveness in the context of the whole promoter. This apparent paradox was solved by the finding that there is a second potential RBP-Jκ binding site in the LMP1 promoter (core sequence TGTGGGAA) which is located outside of the previously defined EBNA2RE, 65 bp upstream of the RBP-Jκ site characterized so far. This second RBP-Jκ binding site is essential for LMP1 promoter transactivation by mNotch1-IC. After mutation of both RBP-Jκ binding sites, we could no longer observe the transactivation by mNotch1-IC. Since mNotch1-IC as well as EBNA2 were able to transactivate a construct consisting only of the EBNA2RE without the potential second RBP-Jκ site and since mutation of the first site in this context completely abolished transactivation by EBNA2 as well as by mNotch1-IC, we conclude that mNotch1-IC, in contrast to EBNA2, can use both RBP-Jκ binding sites in combination with the Spi1 site. This points to a significance of the size and/or conformational differences between the mNotch1-IC and EBNA2 binding domains.
Johannsen et al. (30) showed that four other factors bind to the EBNA2RE of the LMP1 promoter. It would be interesting to elucidate the role of these cellular factors in Notch-mediated transactivation.
Concerning the LMP2A promoter, it has been demonstrated that the whole 80-bp EBNA2RE is crucial for complete EBNA2 transactivation, whereby binding of RBP-Jκ and EBNA2 to the two RBP-Jκ sites is essential but not sufficient (45, 72). The promoter proximal RBP-Jκ binding site has a higher affinity for the cellular repressor protein and is more important for EBNA2-mediated transactivation. Previous deletion analysis of the LMP2A promoter provided evidence that beyond the two RBP-Jκ sites there are at least two other sites (L2BF2 and L2BF3) in the promoter proximal region of the EBNA2RE which play an important role in the EBNA2 responsiveness of the LMP2A promoter. mNotch1-IC transactivates the wild-type EBNA2RE of the LMP2A promoter ca. 10-fold. Mutational analysis revealed that mNotch1-IC behaves similarly to EBNA2: (i) the RBP-Jκ interaction is essential but not sufficient, and transactivation requires the interaction with at least one additional factor; and (ii) extension of the O54 fragment carrying the two RBP-Jκ sites by 5 bp at the promoter proximal site resulted in a ca. 10-fold transactivation by EBNA2 and mNotch1-IC. This suggests that both EBNA2 and mNotch1-IC cooperate with L2BF2. In contrast to EBNA2, L2BF3 had no further effect for Notch responsiveness.
The reporter experiments are all performed in the EBNA2-negative cell line BL41-P3HR1. In Fig. 3B and C it is shown that after transient transfection of EBNA2 or mNotch1-IC, LMP1 is upregulated. Therefore, it cannot be completely excluded that the overall results are a combination of EBNA2/mNotch1-IC and LMP1 effects. However, Cordier et al. (10) have shown that LMP1 expression is not detectable in cell clones of BL41-P3HR1 cells stably expressing ENBA2. In addition, the cell line BL41-P3HR1 mNotch1-IC does not express LMP1 (58a). Since the transient-transfection experiments are performed in the same cell line, we suppose that the described promoter inductions are exclusively EBNA2/mNotch1-IC effects.
Furthermore, we analyzed DNA-protein interactions with the EBNA2RE of the LMP2A promoter and mNotch1-IC. In gel retardation assays we could demonstrate that mNotch1-IC interacts with the two RBP-Jκ sites of the LMP2A promoter.
So far nothing is known about specific DNA-protein interactions in the promoter proximal part of the LMP2A promoter. Therefore, we investigated potential cellular factors binding to the region that is important for the EBNA2- and mNotch1-IC-mediated transactivation. Using only the promoter proximal part of the EBNA2RE (positions −217 to −178, relative to the RNA start site) as radioactively labelled probe in a gel retardation assay, we could show three specific DNA-protein interactions (complexes A, B, and C) with Raji and HeLa nuclear extracts, indicating specific DNA-protein interactions in the promoter proximal part. These DNA-binding proteins are not B cell specific. The intensity of the complexes is weak and therefore cannot be detected when using the complete EBNA2RE as radioactively labelled probe because of the strong intensity of the RBP-Jκ complexes (data not shown). This can be explained either by a weak DNA-protein interaction or by a low concentration of the interacting proteins in the nuclear extracts. With methylation interference experiments we studied which DNA sequences in the complexes A, B, and C are in contact with the DNA-binding proteins. The analysis of complex A did not reveal contacting guanines. This complex is either unspecific or the binding protein does not directly contact guanines. Analysis of complex B showed a weakening of those two bands representing the guanines in positions −188 and −184. Investigation of complex C revealed that three almost successive guanines at positions −210, −209, and −207 are involved in the interaction. The guanines are within the region which together with the RBP-Jκ binding sites is essential to mediate a 10-fold transactivation by EBNA2. These data suggest that EBNA2 and mNotch1-IC use at least two identical factors, RBP-Jκ and L2BF2, to transactivate the LMP2A promoter. L2BF3, binding at positions −188 to −184, might cooperate with EBNA2 but not with mNotch1-IC. Since both cellular factors responsible for EBNA2- and mNotch1-IC-mediated transactivation of the LMP2A promoter are non-B cell specific, it is possible that LMP2A transcription in EBNA2-negative cells such as Hodgkins lymphoma cells and nasopharyngeal carcinoma cells is induced by Notch signalling. In addition, Notch signalling could be responsible for LMP2A expression in EBV-infected CD19+ CD23− CD80− resting B cells in vivo (46).
Here we have shown that the intracellular part of mNotch1 mediates transactivation of viral EBNA2-responsive promoters. In this context the interaction with RBP-Jκ plays a crucial role for both EBNA2 and mNotch1-IC. Furthermore, both proteins need the interaction with other cellular factors: CBF2 for the BamHI C promoter, Spi1 for the LMP1 promoter, and L2BF2 for the LMP2A promoter. The transactivating effects of EBNA2 are three- to fivefold higher than the mNotch1-IC effects. Because mNotch1-IC is as efficient as EBNA2 in transactivating the promoter containing the multimerized RBP-Jκ site pGa981-16, we conclude that the higher activity of EBNA2 for the viral promoters is due to more efficient interaction with other factors required for cooperation. Apparently, the EBNA2REs have been optimized for mediating an EBNA2 response, and mNotch1-IC cannot interact with every one of the factors involved. It would be interesting to see whether the inverse situation could be observed in the HES promoter, i.e., whether there are factors interacting only with Notch and not with EBNA2.
Further experiments are in progress to compare the behavior of mNotch1-IC and EBNA2 in stably transfected cell lines and to investigate the influence of the chromatin structure upon Notch-induced effects. In this context we want to answer the question whether mNotch1-IC can maintain B-cell immortalization.
ACKNOWLEDGMENTS
We thank Elisabeth Kremmer for providing the monoclonal antibody anti-EBNA2 R3 and Thomas Henkel for the expression vector pSG5 mNotch1-IC.
This work was supported by Die Deutsche Forschungsgemeinschaft (Str 461/1-1; Forschergruppe, Multiprotein-Komplexe in der Genexpression, and Sonderforschungsbereich 217), the EU (Molekulare Pathogenese menschlicher Tumorvirusinfektionen), and Fonds der Chemischen Industrie.
REFERENCES
- 1.Abbot S D, Rowe M, Cadwallader K, Gordon J, Ricksten A, Rymo L, Rickinson A B. Epstein-Barr virus nuclear antigen 2 induces expression of the virus-coded latent membrane protein. J Virol. 1990;64:2126–2134. doi: 10.1128/jvi.64.5.2126-2134.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Notch signalling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 3.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, Tufnell P S, Barell B G. DNA sequence and expression of the B 95-8 Epstein-Barr virus genome. Nature (London) 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 4.Bailey A M, Posakony J W. Suppressor of Hairless directly activates transcription of Enhancer of split Complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 5.Bigas A, Martin D I, Milner L A. Notch1 and Notch2 inhibit myeloid differentiation in response to different cytokines. Mol Cell Biol. 1998;18:2324–2333. doi: 10.1128/mcb.18.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrappa S, Gavin D K, Gupta K C. A highly efficient procedure for site-specific mutagenesis of full-length plasmids using Vent DNA polymerase. Genome Res. 1995;5:404–407. doi: 10.1101/gr.5.4.404. [DOI] [PubMed] [Google Scholar]
- 7.Calender A, Billaud M, Aubry J P, Banchereau J, Vuillaume M, Lenoir G M. Epstein-Barr virus (EBV) induces expression of B-cell activation markers on in vitro infection of EBV-negative B-lymphoma cells. Proc Natl Acad Sci USA. 1987;84:8060–8064. doi: 10.1073/pnas.84.22.8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature (London) 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- 9.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordier M, Calender A, Billaud M, Zimber U, Rousselet G, Pavlish O, Banchereau J, Tursz T, Bornkamm G, Lenoir G M. Stable transfection of Epstein-Barr virus (EBV) nuclear antigen 2 in lymphoma cells containing the EBV P3HR1 genome induces expression of B-cell activation molecules CD21 and CD23. J Virol. 1990;64:1002–1013. doi: 10.1128/jvi.64.3.1002-1013.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dignam J D, Leboritz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dou S, Zeng X, Cortes P, Erdjument-Bromage H, Tempst P, Honjo T, Vales L D. The recombination signal sequence-binding protein RBP-2N functions as a transcriptional repressor. Mol Cell Biol. 1994;14:3310–3319. doi: 10.1128/mcb.14.5.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. TAN-1, the human homolog of the Drosophila Notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 14.Fahraeus R, Jansson A, Richsten A, Sjoeblom A, Rymo L. Epstein-Barr virus-encoded nuclear antigen 2 activates the viral latent membrane protein promoter by modulating the activity of a negative regulatory element. Proc Natl Acad Sci USA. 1990;87:7390–7394. doi: 10.1073/pnas.87.19.7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fahraeus R, Jansson A, Sjoeblom A, Nilsson T, Klein G, Rymo L. Cell phenotype-dependent control of Epstein-Barr virus latent membrane protein 1 gene regulatory sequences. Virology. 1993;195:71–80. doi: 10.1006/viro.1993.1347. [DOI] [PubMed] [Google Scholar]
- 16.Fortini M E, Artavanis-Tsakonas S. The suppressor of hairless protein participates in Notch receptor signalling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 17.Fuentes-Pananá E M, Ling P D. Characterization of the CBF2 binding site within the Epstein-Barr virus latency C promoter and its role in modulating EBNA2-mediated transactivation. J Virol. 1998;72:693–700. doi: 10.1128/jvi.72.1.693-700.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galson D L, Harsold J O, Bishop T R, Schalling M, D’Andrea A D, Jones C, Auron P E, Housman D E. Mouse β-globin DNA-binding protein B1 is identical to a proto-oncogene, the transcription factor Spi-1/PU.1, and is restricted in expression to hematopoietic cells and the testis. Mol Cell Biol. 1993;13:2929–2941. doi: 10.1128/mcb.13.5.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gey G, Coffman W, Kubicek M. Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Cancer Res. 1952;12:264–271. [Google Scholar]
- 20.Girard L, Hanna Z, Beaulieu N, Hoemann C D, Simar C, Kozak C A, Jolicoeur P. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 1996;10:1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- 21.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the Jκ recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamaguchi Y, Matsunami N, Yamamoto Y, Honjo T. Purification and characterization of a protein that binds to the recombination signal sequence of the immunoglobulin J kappa segment. Nucleic Acids Res. 1989;17:9015–9026. doi: 10.1093/nar/17.22.9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature (London) 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 24.Henderson S, Rowe M, Gregory C, Croom-Carter D, Wang F, Longnecker R, Kieff E, Rickinson A B. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B-cells from programmed cell death. Cell. 1991;65:1107–1115. doi: 10.1016/0092-8674(91)90007-l. [DOI] [PubMed] [Google Scholar]
- 25.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 26.Hsieh J J, Hayward S D. Masking of the CBF-1/RBP-Jκ transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- 27.Hsieh J J, Henkel T, Salmon P, Robey E, Peterson M G, Hayward S D. Truncated mammalian Notch1 activates CBF1/RBP-Jκ-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarriault S, Brou C, Logeat F, Schroeter E M, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature (London) 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 29.Jin X W, Speck S H. Identification of critical cis elements involved in mediating Epstein-Barr virus nuclear antigen 2-dependent activity of an enhancer located upstream of the viral BamHI C promoter. J Virol. 1992;66:2846–2852. doi: 10.1128/jvi.66.5.2846-2852.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johannsen E, Koh E, Mosialos G, Tong X, Kieff E, Grossman S R. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J Virol. 1995;69:253–262. doi: 10.1128/jvi.69.1.253-262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kempkes B, Spitkovsky D, Jansen-Duerr P, Ellwart J W, Kremmer E, Delecluse H J, Rottenberger C, Bornkamm G W, Hammerschmidt W. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 1995;14:88–96. doi: 10.1002/j.1460-2075.1995.tb06978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kempkes B, Zimber-Strobl U, Eissner G, Pawlita M, Falk M, Hammerschmidt W, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 (EBNA2)-estrogen receptor fusion proteins complement the EBNA2-deficient Epstein-Barr virus strain P3HR1 in transformation of primary B cells but suppress growth of human B cell lymphoma lines. J Gen Virol. 1996;77:227–237. doi: 10.1099/0022-1317-77-2-227. [DOI] [PubMed] [Google Scholar]
- 33.Knutson J C. The level of c-fgr RNA is increased by EBNA-2, an Epstein-Barr virus gene required for B-cell immortalization. J Virol. 1990;64:2530–2536. doi: 10.1128/jvi.64.6.2530-2536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kopan R, Nye J S, Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994;120:2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 35.Kopan R, Schroeter E H, Weintraub H, Nye J S. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci USA. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kremmer E, Kranz B R, Hille A, Klein K, Eulitz M, Hoffmann-Fezer G, Feiden W, Herrmann K, Delecluse H J, Delsoi G, Bornkamm G W, Mueller-Lantzsch N, Graesser F A. Rat monoclonal antibodies differentiating between the Epstein-Barr virus nuclear antigen 2A (EBNA2A) and 2B (EBNA2B) Virology. 1995;208:336–342. doi: 10.1006/viro.1995.1157. [DOI] [PubMed] [Google Scholar]
- 37.Laux G, Adam B, Strobl L J, Moreau-Gachelin F. The Spi1/PU.1 and Spi-B Ets family transcription factors and the recombination signal binding protein RBP-Jκ interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 1994;13:5624–5632. doi: 10.1002/j.1460-2075.1994.tb06900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laux G, Dugrillon C, Eckert B, Adam B, Zimber-Strobl U, Bornkamm G W. Identification and characterization of an Epstein-Barr virus nuclear antigen 2-responsive element in the bidirectional promoter region of latent membrane protein and terminal protein 2 genes. J Virol. 1994;68:6947–6958. doi: 10.1128/jvi.68.11.6947-6958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lecourtois M, Schweisguth F. The neurogenic Suppressor of Hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split Complex genes triggered by Notch signaling. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- 40.Li L, Milner L A, Deng Y, Iwatat M, Banta A, Graf L, Marcovina S, Friedman C, Trask B J, Hood L, Torok-Storb B. The human homolog of rat Jagged1 expressed by marrow stroma inhibits differentiation of 32D cells through interaction with Notch1. Immunity. 1998;8:43–55. doi: 10.1016/s1074-7613(00)80457-4. [DOI] [PubMed] [Google Scholar]
- 41.Lu F M, Lux S E. Constitutively active human Notch1 binds to the transcription factor CBF1 and stimulates transcription through a promoter containing a CBF1-responsive element. Proc Natl Acad Sci USA. 1996;93:5663–5667. doi: 10.1073/pnas.93.11.5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 43.Matsunami N, Hamaguchi Y, Yamamoto Y, Kuze K, Kangawa K, Matsuo H, Kawaichi M, Honjo T. A protein binding to the J kappa recombination sequence of immunoglobulin genes contains a sequence related to the integrase motif. Nature (London) 1989;342:934–937. doi: 10.1038/342934a0. [DOI] [PubMed] [Google Scholar]
- 44.Maxam A, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 45.Meitinger C, Strobl L J, Marschall G, Bornkamm G W, Zimber-Strobl U. Crucial sequences within the Epstein-Barr virus TP1 promoter for EBNA2-mediated transactivation and interaction of EBNA2 with its responsive element. J Virol. 1994;68:7497–7506. doi: 10.1128/jvi.68.11.7497-7506.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyashita E M, Yang B, Lam K M, Crawford D H, Thorley-Lawson D A. A novel form of Epstein-Barr virus latency in normal B-cells in vivo. Cell. 1995;80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- 47.Milner L A, Kopan R, Martin D I, Bernstein I D. A human homologue of the Drosophila developmental gene, Notch, is expressed in CD34+ hematopoietic precursors. Blood. 1994;83:2057–2062. [PubMed] [Google Scholar]
- 48.Milner L A, Bigas A, Kopan R, Brashem-Stein C, Bernstein I D, Martin D I K. Inhibition of granulocytic differentiation by mNotch1. Proc Natl Acad Sci USA. 1996;93:13014–13019. doi: 10.1073/pnas.93.23.13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Minoguchi S, Taniguchi Y, Kato H, Okazaki T, Strobl L J, Zimber-Strobl U, Bornkamm G W, Honjo T. RBP-L, a transcription factor related to RBP-Jκ. Mol Cell Biol. 1997;17:2679–2687. doi: 10.1128/mcb.17.5.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nye J S, Kopan R, Axel R. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development. 1994;120:2421–2430. doi: 10.1242/dev.120.9.2421. [DOI] [PubMed] [Google Scholar]
- 51.Polack A, Strobl L J, Feederle R, Schweizer M, Koch E, Eick D, Wiegand H, Bornkamm G W. The intron enhancer of the immunoglobulin kappa gene activates c-myc but does not induce the Burkitt-specific promoter shift. Oncogene. 1991;6:2033–2040. [PubMed] [Google Scholar]
- 52.Pulvertaft R J V. A study of malignant tumours in Nigeria by short term tissue culture. J Clin Pathol. 1965;18:261–273. doi: 10.1136/jcp.18.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rabson M, Gradoville L, Heston L, Miller G. Non-immortalizing P3J-HR-1 Epstein-Barr virus: a deletion mutant of its transforming parent, Jijoye. J Virol. 1982;44:834–844. doi: 10.1128/jvi.44.3.834-844.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robey E, Chang D, Itano A, Cado D, Alexander H, Lans D, Weinmaster G, Salmon P. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell. 1996;87:483–492. doi: 10.1016/s0092-8674(00)81368-9. [DOI] [PubMed] [Google Scholar]
- 55.Rohn J L, Lauring A S, Linenberger M L, Overbaugh J. Transduction of Notch2 in feline leukemia virus-induced thymic lymphoma. J Virol. 1996;70:8071–8080. doi: 10.1128/jvi.70.11.8071-8080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakai T, Taniguchi Y, Tamura K, Minoguchi S, Fukuhara T, Strobl L J, Zimber-Strobl U, Bornkamm G W, Honjo T. Functional replacement of the intracellular region of the Notch1 receptor by Epstein-Barr virus nuclear antigen 2. J Virol. 1998;72:6034–6039. doi: 10.1128/jvi.72.7.6034-6039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schroeter E H, Kisslinger J A, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature (London) 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 58.Strobl L J, Hoefelmayr H, Stein C, Marschall G, Brielmeier M, Laux G, Bornkamm G W, Zimber-Strobl U. Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-Jκ. Immunobiology. 1997;198:299–306. doi: 10.1016/s0171-2985(97)80050-2. [DOI] [PubMed] [Google Scholar]
- 58a.Strobl, L. J., et al. Unpublished data.
- 59.Struhl G, Adachi A. Nuclear access and action of Notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 60.Sung N S, Kenney S, Gutsch D, Pagano J S. EBNA-2 transactivates a lymphoid-specific enhancer in the BamHI C promoter of Epstein-Barr virus. J Virol. 1991;65:2164–2169. doi: 10.1128/jvi.65.5.2164-2169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcriptional factor RBP-Jκ/Su(H) Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 62.Tsang S F, Wang F, Izumi K M, Kieff E. Delineation of the cis-acting element mediating EBNA-2 transactivation of latent infection membrane protein expression. J Virol. 1991;65:6765–6771. doi: 10.1128/jvi.65.12.6765-6771.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waltzer L, Logeat F, Brou C, Israel A, Sergeant A, Manet E. The human Jκ recombination signal sequence binding protein (RBP-Jκ) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 1994;13:5633–5638. doi: 10.1002/j.1460-2075.1994.tb06901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waltzer L, Bourillot P Y, Sergeant A, Manet E. RBP-J kappa repression activity is mediated by a co-repressor and antagonized by the Epstein-Barr virus transcription factor EBNA2. Nucleic Acids Res. 1995;23:4939–4945. doi: 10.1093/nar/23.24.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang F, Gregory C D, Rowe M, Rickinson A B, Wang D, Birkenbach M, Kikutani H, Kishimoto T, Kieff E. Epstein-Barr virus nuclear antigen 2 specifically induces expression of the B-cell activation antigen CD23. Proc Natl Acad Sci USA. 1987;84:3452–3456. doi: 10.1073/pnas.84.10.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang F, Tsong S, Kurilla M G, Cohen J I, Kieff E. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP. J Virol. 1990;64:3407–3416. doi: 10.1128/jvi.64.7.3407-3416.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang F, Kikutani, Tsang S F, Kishimoto T, Kieff E. Epstein-Barr virus nuclear protein 2 transactivates a cis-acting CD23 DNA element. J Virol. 1991;65:4101–4106. doi: 10.1128/jvi.65.8.4101-4106.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Warren S P, Aster J C, Scott M L, Hasserjian R P, Soffer B, Sklar J, Baltimore D. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Washburn T, Schweighoffer E, Gridley T, Chang D, Fowlkes B J, Cado D, Robey E. Notch activity influences the alphabeta versus gammadelta T cell lineage decision. Cell. 1997;88:833–843. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- 70.Woisetschlaeger, M., C. N. Yandava, L. A. Furmanski, J. L. Strominger, and S. H. Speck. Promoter switching in Epstein-Barr virus during the initial stages of infection of B lymphocytes. Proc. Natl. Acad. Sci. USA 87:1725–1729. [DOI] [PMC free article] [PubMed]
- 71.Zimber-Strobl U, Suentzenich K O, Laux G, Eick D, Cordier M, Calender A, Billaud M, Lenoir G M, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 activates transcription of the terminal protein 1 gene. J Virol. 1991;65:415–423. doi: 10.1128/jvi.65.1.415-423.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zimber-Strobl U, Kremmer E, Graesser F, Marschall G, Laux G, Bornkamm G W. The Epstein-Barr virus nuclear antigen 2 interacts with an EBNA2 responsive cis-element of the terminal protein 1 gene promoter. EMBO J. 1993;12:167–175. doi: 10.1002/j.1460-2075.1993.tb05642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zimber-Strobl U, Strobl L J, Meitinger C, Hinrichs R, Sakai T, Furukuwa T, Honjo T, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-Jκ, the homologue of Drosophila Suppressor of Hairless. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]