Abstract
The present study is an attempt to establish a fast, highly reproducible transformation with a simplified regeneration system in soybean targeting the apical meristem. The modified half-seed explants from soybean cultivar (cv.) JS335 were subjected to different time intervals of sonication (0, 1, 10, 20, and 30 min) and vacuum infiltration (0, 1, 10, 20, and 30 min) in the presence of Agrobacterium tumefaciens strain EHA105 harbouring pCAMBIA1301. The explants were then co-cultivated and subjected to a modified plant regeneration process that involves only two steps (1) primary shoot regeneration, and (2) in vitro rooting of primary shoot. The rooted plantlets were hardened and maintained in the greenhouse until maturity. Sonication treatment of 10 min, followed by plant regeneration using a modified method, recorded the highest transformation efficiency of 26.3% compared to other time duration tested. Furthermore, 10 min of vacuum infiltration alone resulted in even higher transformation efficiency after regeneration, reaching 28.0%. Interestingly, coupling sonication and vacuum infiltration for 10 min respectively produced the highest transformation efficiency after regeneration of 38.0%. The putative transformants showed gus expression in mature leaves, trifoliate leaves, flowers, and pods. The presence of hpt II was also confirmed in putative transformants, with an amplicon size of 500 bp. Quantitative real-time PCR confirmed the existence of hpt II as one to two copies in the soybean genome of T0 plants. Furthermore, the segregation pattern was observed in the T1 generation soybean plants which were confirmed using PCR for hpt II. The optimized protocol when tested with other Indian soybean cultivars showed an enhanced transformation efficiency ranging from 19.3% (cv. MAUS47) to 36.5% (cv. CO1). This optimized protocol could provide a reliable platform to overcome the challenges that are associated with the genetic engineering of soybean.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-023-03715-8.
Keywords: Soybean, Modified half-seed explant, Regeneration, Sonication, Vacuum infiltration
Introduction
Soybean (Glycine max (L.) Merrill), an economically valuable crop, is largely used for consumption and industrial applications (Widholm et al. 2010). The global population growth and the consistent demand for soy products are leading to a continuous increase in the production and demand for soybeans. Consequently, significant efforts have been dedicated to improving the regeneration system and the effectiveness of transforming soybeans. These efforts show great potential for developing superior soybean varieties with desired characteristics. Up until now, soybean regeneration has been achieved through somatic embryogenesis, direct organogenesis, and indirect organogenesis. However, poor regeneration has been a major obstacle in the indirect organogenesis method for soybeans. Most of the research conducted on soybeans has focused on somatic embryogenesis or direct organogenesis. Regarding soybean transformation, various intrinsic factors such as Agrobacterium strains, types of explants, composition of culture media, duration of co-cultivation, and plant selection markers have been extensively investigated to enhance the efficiency of the transformation process. Moreover, extrinsic factors like physical wounding of explants, sonication, and vacuum infiltration have been optimized to achieve higher transformation efficiency in soybeans.
Despite various studies aimed at improving soybean transformation efficiency using Agrobacterium infection, the success rate has been very low due to genotype dependency and low regeneration of transformants (Kumari et al. 2016; Liu et al. 2004). Moreover, poor shoot elongation and long regeneration duration are other important limiting factors for the effective regeneration of transformants (Ma and Wu 2008). Thus, there is an urgent need to look for alternative ways to develop transformed soybean to meet the global demand. In this regard, the current study aims to develop a fast, reliable, and efficient soybean transformation system incorporating sonication and vacuum infiltration thereby targeting the apical meristem of modified half-seed explants. Moreover, the highlight of the present study is the hassle-free and fast regeneration of transformed plants from infected half-seed explants using a simplified regeneration method that involves just two steps (1) primary shoot regeneration, and (2) in vitro rooting of primary shoot. The optimized protocol has also been tested with 10 cultivars to check its efficiency.
Materials and methods
Indian soybean cultivars (cv.) JS335, PUSA 9712, CO1, TAMS-38, JS71-05, JS93-05, NRC7, MAUS47, PK416, and Punjab 1 were procured from ICAR-Indian Institute of Soybean Research, Indore, Madhya Pradesh, India, and the cultivars were grown and maintained at the research garden, Department of Biotechnology, Bharathiar University, Coimbatore, Tamil Nadu, India. The optimization was carried out using the soybean cultivar (cv.) JS335 (Fig. 1a).
Fig. 1.
Transformation of soybean cv. JS335 using Agrobacterium tumefaciens strain EHA105 harboring pCAMBIA1301 and regeneration from modified half-seed explants. a Soybean seeds of cv. JS335; b one-day-old imbibed seeds; c half- seed explants prepared from the imbibed seeds (red arrow indicates the presence of plumule and blue arrow indicates the removal of radicle); d, e primary shoot regenerated on MS medium containing 2.2 µM BAP and 3 mg l−1 hygromycin B after 15 days and 30 days of culture respectively; f, g in vitro rooting of primary shoots on MS medium containing 4.9 µM IBA and 2 mg l−1 hygromycin B after 15 days and 30 days of culture respectively; h putatively transformed soybean plantlet; i transformed soybean plant grown in containment room
To begin the experiment, the seeds of soybean cv. JS335 were subjected to surface sterilization and imbibed in sterile water for a period of 24 h (Fig. 1b). Following imbibition, the seed coat was removed, and the cotyledons were separated. Only the cotyledon containing the embryonic axis was utilized for the study. Additionally, the radicle of the embryo, which was attached to the cotyledon, was carefully dissected to obtain the modified half-seed explant (Fig. 1c). For primary shoot regeneration, explants were inoculated on MS medium supplemented with different concentrations of 6-Benzylaminopurine (BAP) (0–8.8 μM) and cultured for 30 days. The explants were sub-cultured into a fresh medium with respective hormonal concentrations at 15 days intervals. For rooting of primary shoots, MS medium supplemented with different concentrations of Indole-3 butyric acid (IBA) (0–9.8 μM) was used and the culture was incubated for 30 days. In-order to select the primary shoot after transformation, minimum inhibitory concentration (MIC) was determined in modified half-seed explants by inoculating in regeneration medium (MS + 2.2 μM BAP; pH 5.7) with different concentrations of hygromycin B (0–5 mg l−1) and incubating for 30 days. In addition, the explants were sub-cultured at 15 days intervals. Subsequently, the established primary shoots were transferred to a rooting medium (MS + 4.9 μM IBA; pH 5.7) with different concentrations of hygromycin B (0 to 3 mg l−1) and incubated for 30 days for selection at the rooting stage. All the cultures were maintained at 25 ± 2 °C under a 16/8-h photoperiod.
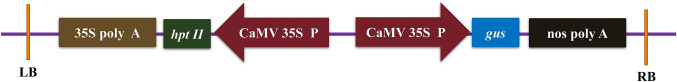
Agrobacterium tumefaciens strain EHA105 harbouring pCAMBIA1301 was used for transformation (Fig. 2). The T-DNA region of the binary vector contains hygromycin phosphotransferase II (hpt II) as the plant selection marker and gus as a reporter gene. The vector backbone carries the neomycin phosphotransferase II (npt II) for bacterial selection. Agrobacterium culture was prepared by inoculating a single colony into 30 ml LB broth containing antibiotics such as kanamycin (50 mg l−1) and rifampicin (25 mg l−1). The culture was incubated at 28 °C for 16 h at 180 rpm. The bacterial culture was centrifuged at 6000 rpm for 15 min, and the pellet was suspended in a liquid MS medium. Additionally, a filter-sterilized solution of 200 μM acetosyringone was added to the bacterial suspension, which was then incubated for 1 h at 28 °C at 180 rpm. The absorbance of bacterial suspension was adjusted to 1.0 at OD600 prior to infection.
Fig. 2.
Linear map of the plasmid (T-DNA) pCAMBIA1301 present within the Agrobacterium tumefaciens strain EHA105 used for the transformation experiments. The T- DNA region of pCAMBIA1301 showing the assembly of hpt II expression cassette (CaMV 35S P: hpt II: 35S poly A), and gus expression cassette (CaMV 35S P: gus: nos poly A). CaMV 35S P: Cauliflower mosaic virus 35S promoter, hpt II: hygromycin phosphotransferase II, 35S poly A: Cauliflower mosaic virus 35S poly A terminator. gus: β-glucuronidase, nos poly A: nopaline synthase poly A terminator, LB left border, RB right border
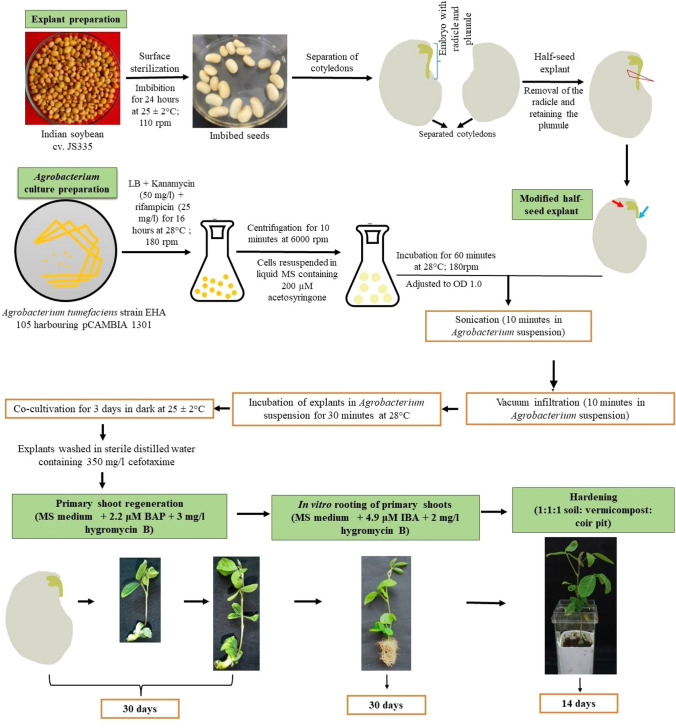
For genetic transformation, the modified half-seed explants were inoculated into 30 ml Agrobacterium suspension and sonicated for different durations (0, 1, 10, 20, and 30 min). Similarly, the explants were subjected to vacuum infiltration for different time intervals (0, 1, 10, 20, and 30 min) in 30 ml Agrobacterium suspension. Finally, the explants were subjected to combined treatments of sonication (10 min) and vacuum infiltration (10 min) in the presence of Agrobacterium. After different treatments, the explants were then incubated in fresh Agrobacterium suspension (30 ml) at 28 °C for 30 min. After 30 min, the explants were blot-dried and placed in a co-cultivation medium (MS + 200 μM acetosyringone; pH 5.7) and cultured in complete darkness at 25 ± 2 °C for 3 days. Subsequently, the explants were then thoroughly washed with sterile distilled water containing 350 mg l−1 cefotaxime and cultured in regeneration medium (MS + 2.2 μM BAP + 3 mg l−1 hygromycin B; pH 5.7) for 30 days for selection of primary shoots. The excised primary shoots were then cultured in a rooting medium (MS medium + 4.9 μM IBA + 2 mg l−1 hygromycin B; pH 5.7) for 30 days. The rooted plantlets were carefully removed from the medium, washed with sterile distilled water, hardened for 2 weeks in paper cups and later transferred to earthen pots. The complete methodology of modified regeneration protocol along with transformation procedure in soybean is presented in Fig. 3.
Fig. 3.
Schematic representation of soybean transformation and regeneration using modified half-seed explants. Red arrow indicates the presence of plumule, and blue arrow indicates the removal of radicle from the half-seed explant
The survived plants (T0) were randomly selected and subjected to histochemical GUS assay (Jefferson et al. 1987). Additionally, survived plants (T0) were analysed for the hpt II integration by polymerase chain reaction (PCR) using hpt II specific primers. The forward primer for hpt II was: 5ʹ ATGAAAAAGCCTGAACT 3ʹ and the reverse primer for hpt II was: 5ʹ TCCATCACAGTTTGCC 3ʹ. The copy number of hpt II integration was analysed using the method adapted from Li et al. (2017) in randomly selected PCR-positive hpt II plants. To study the stability and segregation pattern of the transgene, T1 seeds obtained from the T0 plants (Gm JS335-7, Gm JS335-8, and Gm JS335-9 showing single copy) were cultured on MS medium containing 25 mg l−1 hygromycin for 15 days. Later, PCR analysis was conducted to check for the presence of the transgene (hpt II) in the T1 generation. The optimized protocol was applied to other cultivars of the Indian soybean (PUSA 9712, CO1, TAMS-38, JS71-05, JS93-05, NRC7, MAUS47, PK416, and Punjab 1) to test its efficacy. In both tissue culture and MIC experiments, a total of 100 explants or 100 primary shoots were utilized for the respective treatments. These experiments were repeated twice to ensure accuracy and reliability. As for the transformation experiments, 100 explants were employed for the respective treatments, and the experiments were repeated three times. The resulting data were presented as mean values with the standard error (SE). Statistical analysis was performed using SPSS software version 20, specifically employing Duncan’s multiple range test (DMRT) to determine significant differences at a significance level of P < 0.05. For segregation ratio analysis, the SE and Chi-square analysis were used (Gomez and Gomez 1984; Hada et al.2018). Significance was determined for values with a P < 0.05.
Results and discussion
We have conducted studies on various parameters to improve transformation after regeneration efficiency in Indian soybean cultivars to address the challenges associated with soybean transformation, including low regeneration rates and the absence of cultivar-independent protocols.
In the soybean direct organogenesis system, the explants will be initially subjected to multiple shoot induction. Then attempts will be made to elongate the shoots, and after elongation, the shoots will be cultured for in vitro rooting. In addition, this process takes approximately 3 or more months to obtain an in vitro rooted plantlet that will be ready for hardening. In order to achieve regeneration using a direct organogenesis system, the radicle, and plumule of the half-seed explants have to be excised and need to be placed in cytokinin containing medium to trigger the meristematic cells to produce multiple shoots. However, in this method, most of the shoots will fail to elongate affecting the regeneration response (Ether et al. 2013). To overcome this limitation with half-seed explants, we have modified the explant preparation in a way that we removed only the radicle and left the plumule intact to obtain the modified half-seed explants. The presence of plumule in the modified half-seed explants directed the regeneration system towards primary shoot development followed by subsequent in vitro rooting diverting it from the conventional direct organogenesis process that includes multiple shoot induction, shoot elongation, and rooting. Moreover, this diversion also bypassed the shoot elongation step which was critical in affecting the regeneration efficiency. In the present investigation, the primary shoots developed and elongated in the same BAP medium avoiding the necessity of a separate shoot elongation process. Also, using this method, we were able to produce rooted plantlets that are ready for hardening within 60 days (30 days for primary shoot regeneration and 30 days for rooting) which was comparatively less than similar reports on soybean. The aforesaid advantages of using this modified half-seed method also highly favoured efficient regeneration in transformation experiments.
In the present study, modified half-seed explants produced the highest response in inducing the primary shoot regeneration (93.0%) in MS medium supplemented with 2.2 µM BAP (Supplementary Table 1). In addition, the maximum in vitro rooting of primary shoots (75.5%) was observed in the MS medium supplemented with 4.9 µM IBA (Supplementary Table 2). Our findings were similar to those of Arun et al. (2015) and Chen et al. (2018), where the aforementioned concentrations of BAP and IBA showed the best response in inducing shoots and roots in soybean. The MIC of hygromycin B in primary shoot regeneration was found to be 3 mg l−1 and the MIC of hygromycin B in in vitro rooting of primary shoots was 2 mg l−1. The use of hygromycin B as a potent plant selection marker in soybean was established by Olhoft et al. (2006).
In this study, the transformed modified half-seed explants were successfully regenerated using an optimized regeneration method. Among the different treatments applied, the modified half-seed explants that underwent a 10-min sonication treatment exhibited the highest number of primary shoots that survived (54.6), along with a substantial number of rooted shoots (44.3) and plants that survived after a 2-week hardening period (26.3). The transformation efficiency for this treatment was calculated to be 26.3% (Table 1). However, it is worth noting that the transformation efficiency decreased when the sonication time was reduced to 1 min or increased to 20 and 30 min, as indicated in Table 1. The highly active and rapidly dividing meristematic cells that were used for genetic transformation are present in the primary shoot. Sonication creates micro-wounds through which Agrobacterium could easily reach the meristematic cells and enhance the transformation efficiency (Trick and Finer 1997). Our study is consistent with the findings of Hada et al. (2018), where the optimum sonication time was found to be 10 min, and increasing the sonication time decreased the transformation efficiency from 36.2% to 12.1%. Guo et al. (2015) also claimed that sonication for 2 s improved transformation efficiency in soybean from 2.5 to 5.7%. Vacuum infiltration has been well validated as an efficient method to improve the rate of transformation by creating negative atmospheric pressure, enabling easy passage for Agrobacterium to target the meristematic cells (Subramanyam et al. 2013). Among different time durations for vacuum infiltration tested (0, 1, 10, 20, and 30 min), a treatment duration of 10 min showed the maximum number of survived primary shoots (51.6), number of rooted shoots (40.6), number of plants that survived after 2 weeks of hardening (28.0), with the transformation efficiency of 28.0% (Table 1). Furthermore, it was observed that extending the vacuum infiltration time beyond 10 min had a negative impact on the transformation efficiency. This decrease in efficiency can be attributed to the injury caused to the explants due to excessive vacuum. Similarly, reducing the vacuum infiltration time to 1 min resulted in a decreased transformation efficiency of 11.6%. In the case of the treatment involving sonication for 10 min combined with vacuum infiltration for 10 min, the number of primary shoots that survived in the selection medium was recorded as 54.6 (Fig. 1d and e). Additionally, the number of rooted shoots was 46.6 (Fig. 1f and g). Following the hardening process, a total of 38.0 plants successfully survived and acclimatized (Fig. 1h and i). Importantly, the transformation efficiency significantly improved to 38.0%, which is considerably higher than the efficiency observed in the explants treated with sonication or vacuum infiltration alone (Table 1). These findings align with the previous studies conducted on soybeans by Arun et al. (2015) and Hada et al. (2018), which also suggested that combining sonication and vacuum infiltration can enhance the transformation efficiency, as demonstrated in the present study.
Table 1.
Influence of sonication and vacuum infiltration on the transformation efficiency of soybean cv. JS335 (modified half-seed explant) infected and co-cultivated with Agrobacterium tumefaciens strain EHA105 harboring pCAMBIA1301
| Sonication (minutes) | Vacuum infiltration (minutes) | Number of explants infected | Number of primary shoot surviveda | Number of rooted shootsb | Number of plants survived after hardeningc | Number of hpt II PCR positive plantsd | Transformation efficiency (%)e |
|---|---|---|---|---|---|---|---|
| 0 | 0 | 100 | 15.3 ± 0.8j | 9.6 ± 1.4j | 6.6 ± 0.6j | 6.6 ± 0.6j | 6.6 ± 0.6j |
| 1 | – | 100 | 35.6 ± 1.3g | 20.6 ± 0.8g | 12.3 ± 1.4f | 12.3 ± 1.4f | 12.3 ± 1.4f |
| 10 | – | 100 | 54.6 ± 0.8b | 44.3 ± 1.4b | 26.3 ± 0.6c | 26.3 ± 0.6c | 26.3 ± 0.6c |
| 20 | – | 100 | 42.3 ± 1.4e | 31.6 ± 1.4e | 19.3 ± 1.4e | 19.3 ± 1.4e | 19.3 ± 1.4e |
| 30 | – | 100 | 27.3 ± 0.8h | 18.3 ± 0.8h | 9.3 ± 0.8i | 9.3 ± 0.8i | 9.3 ± 0.8i |
| – | 1 | 100 | 39.0 ± 1.1f | 28.3 ± 0.8f | 11.6 ± 0.6g | 11.6 ± 0.6g | 11.6 ± 0.6g |
| – | 10 | 100 | 51.6 ± 0.3c | 40.6 ± 0.3c | 28.0 ± 1.4b | 28.0 ± 1.4b | 28.0 ± 1.4b |
| – | 20 | 100 | 44.3 ± 1.4d | 35.3 ± 0.3d | 21.3 ± 0.6d | 21.3 ± 0.6d | 21.3 ± 0.6d |
| – | 30 | 100 | 21.3 ± 0.8i | 17.0 ± 0.6i | 10.3 ± 0.8h | 10.3 ± 0.8 h | 10.3 ± 0.8 h |
| 10 | 10 | 100 | 54.6 ± 1.1a | 46.6 ± 0.3a | 38.0 ± 0.5a | 38.0 ± 0.5a | 38.0 ± 0.5a |
Mean values of three independent experiments (±) with standard errors (n = 100 × 3). Values with the different letters within columns are significantly different according to Duncan’s multiple range test (DMRT) at a 5% level
aTotal number of primary shoots survived on regeneration medium (MS + 2.2 μM BAP + 3 mg l−1 hygromycin B) after 30 days of culture
bTotal number of primary shoots responded for the root development after 30 days of culture on rooting medium (MS + 4.9 μM IBA + 2 mg l−1 hygromycin B)
cTotal number of putatively transformed plants that survived in the greenhouse after hardening
dTotal number of putatively transformed plants showing the presence of hpt II
eTransformation efficiency = number of hpt II PCR positive plants/total number of infected explants × 100
The superscript letter f, g, h, i, j shows that these values are significantly different according to DMRT
From the histochemical GUS assay, it was observed that the mature leaf (Fig. 4a), trifoliate leaves (Fig. 4c), flower (Fig. 4e), and pod (Fig. 4g) from putative transformants developed an intense blue colour and tested positive for gus expression. The mature leaf (Fig. 4b), trifoliate leaves (Fig. 4d), flower (Fig. 4f), and pod (Fig. 4h) from non-transformed plants did not show the gus expression. In the present study, the transformation efficiency was calculated based on the presence of the hpt II in transformed plants. The T0 plants that survived after hardening were subjected to this analysis. The amplicon size of 500 bp (Fig. 4i, Lane 4–8) indicated the presence of hpt II in transformed plants. pCAMBIA1301 plasmid served as the positive control (Fig. 4i, Lane 2) whereas non- transformed plants did not show any amplification for hpt II (Fig. 4i, Lane 3). Overall maximum transformation efficiency of 38.0% was achieved when modified half-seed explants were subjected to sonication (10 min) and vacuum infiltration (10 min).
Fig. 4.
GUS analysis and molecular confirmation of putative transformants regenerated from modified half-seed explants infected with Agrobacterium tumefaciens strain EHA105 harboring pCAMBIA1301. a Expression of gus in mature leaf from putatively transformed plants; b mature leaf from non-transformed plant; c expression of gus in trifoliate leaves from putatively transformed plants; d trifoliate leaves from non-transformed plant; e expression of gus in flower from putatively transformed plants; f flower from non-transformed plant; g expression of gus in pod from putatively transformed plants; h pod from non-transformed plant; i molecular confirmation for the presence of hpt II in putatively transformed soybean plants. Lane 1: DNA ladder (1 Kb); lane 2: pCAMBIA1301 plasmid (positive control); lane 3: soybean genomic DNA from non-transformed plants (negative control); lanes 4–8: genomic DNA from putatively transformed soybean plants with expected amplicon (500 bp) of hpt II
In this present study, the copy number of hpt II in T0 plants was determined by quantitative real-time PCR using Actin as the reference gene. The results revealed that the copy number of hpt II ranged between one and two. The T0 transgenic soybean lines GmJS335-2 and GmJS335-3 had two copies, while lines GmJS335-1, GmJS335-4, GmJS335-5, GmJS335-6, GmJS335-7, GmJS335-8, and GmJS335-9 had one copy of hpt II (Supplementary Table 3). The quantitative real-time PCR is replacing the traditional method of detecting the copy number of the foreign gene via the southern blot technique due to various advantages such as accuracy, lower cost, higher stability, and ease of operation. This technique has been successfully employed in several crops, including cotton (Yang 2012), wheat (Gadaleta et al. 2011), rice (Wei et al. 2011), maize (Yuan et al. 2010), tomato (Wang et al. 2011), and soybean (You-wen et al. 2012). Additionally, the segregation pattern observed in the T1 generation demonstrated Mendelian inheritance with a ratio of 3:1 in one plant (GmJS335 8) at a significance level of 0.05% (Supplementary Table 4). The optimized protocol has been further applied to evaluate the transformation efficiency in other cultivars of soybean. In the present investigation, cv. JS335 was found to be having the highest transformation efficiency of 38.0% followed by the cv. CO1 (36.5%) and cv. TAMS-38 (33.6%). Among the different cultivars examined, cv. MAUS47 displayed the lowest transformation frequency, recorded at 19.3% (Table 2). Overall, the method developed in this study proved to be fast and highly efficient in obtaining transgenic lines within a relatively short duration of 60 days to obtain rooted plantlets. In comparison, previous studies conducted by Arun et al. (2015), Hada et al. (2018), and Wang et al. (2022) demonstrated soybean transformation systems utilizing conventional direct organogenesis, which required longer regeneration times of 123 days, 104.5 days, and 97 days, respectively. Therefore, this simplified transformation and modified regeneration protocol can be utilized effectively for developing transgenic soybean varieties with desired traits.
Table 2.
Influence of sonication and vacuum infiltration on the transformation efficiency of different soybean cultivars infected and co-cultivated with Agrobacterium tumefaciens strain EHA105 harboring pCAMBIA1301
| Cultivar | Number of explants infected | Number of primary shoot surviveda | Number of rooted shootsb | Number of plants survived after hardeningc | Number of hpt II PCR positive plantsd | Transformation efficiency (%)e |
|---|---|---|---|---|---|---|
| JS335 | 100 | 54.6 ± 1.1a | 46.6 ± 0.3a | 38.0 ± 0.5a | 38.0 ± 0.5a | 38.0 ± 0.5a |
| PUSA 9712 | 100 | 47.3 ± 1.6e | 33.9 ± 0.3g | 27.3 ± 1.1g | 27.3 ± 1.1g | 27.3 ± 1.1g |
| CO1 | 100 | 52.4 ± 1.4c | 41.6 ± 0.8d | 36.5 ± 0.6b | 36.5 ± 0.6b | 36.5 ± 0.6b |
| TAMS-38 | 100 | 51.6 ± 0.3d | 43.5 ± 1.6b | 33.6 ± 0.3c | 33.6 ± 0.3c | 33.6 ± 0.3c |
| JS71-05 | 100 | 44.6 ± 0.6 g | 36.6 ± 0.3f | 28.3 ± 1.2f | 28.3 ± 1.2f | 28.3 ± 1.2f |
| JS93-05 | 100 | 53.5 ± 1.2b | 42.3 ± 0.9c | 31.3 ± 0.3d | 31.3 ± 0.3d | 31.3 ± 0.3d |
| NRC7 | 100 | 45.3 ± 0.6f | 37.6 ± 1.2e | 29.6 ± 1.4e | 29.6 ± 1.4e | 29.6 ± 1.4e |
| MAUS47 | 100 | 37.6 ± 0.3j | 26.9 ± 0.3j | 19.3 ± 0.6j | 19.3 ± 0.6j | 19.3 ± 0.6j |
| PK416 | 100 | 43.3 ± 1.2h | 32.3 ± 0.9h | 26.0 ± 0.9h | 26.0 ± 0.9h | 26.0 ± 0.9h |
| Punjab 1 | 100 | 41.5 ± 0.3i | 29.0 ± 1.4i | 20.6 ± 1.4i | 20.6 ± 1.4i | 20.6 ± 1.4i |
Mean values of three independent experiments (±) with standard errors (n = 100 × 3). Values with the different letters within columns are significantly different according to Duncan’s multiple range test (DMRT) at a 5% level
aTotal number of primary shoots survived on regeneration medium (MS + 2.2 μM BAP + 3 mg l−1 hygromycin B) after 30 days of culture
bTotal number of primary shoots responded for the root development after 30 days of culture on rooting medium (MS + 4.9 μM IBA + 2 mg l−1 hygromycin B)
cTotal number of putatively transformed plants that survived in the greenhouse after hardening
dTotal number of putatively transformed plants showing the presence of hpt II
eTransformation efficiency = number of hpt II PCR positive plants/total number of infected explants × 100
The superscript letter f, g, h, i, j shows that these values are significantly different according to DMRT
Conclusion
In this study, we have successfully developed a simple regeneration system from modified half-seed explants, consisting of two steps: primary shoot regeneration and rooting. This system has been effectively adapted to regenerate transgenic plants from modified half-seed explants infected with Agrobacterium, and it offers the advantage of a shorter regeneration period. Additionally, the incorporation of sonication and vacuum infiltration techniques has significantly enhanced the transformation efficiency in our study. Moreover, this transformation and regeneration system has demonstrated its efficacy across various soybean cultivars, indicating its wide applicability. We believe that this simple protocol holds great potential for commercial trait improvement in diverse soybean varieties.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are thankful to the University Grants Commission: Basic Startup Research Grant (No.F.30-410/2018 (BSR); dt.: 29.06.2018), New Delhi, Government of India, for financial assistance provided to Muthukrishnan Arun, and also thanks to ICAR -Indian Institute of soybean research, Indore, Madhya Pradesh, India for providing soybean seeds to carry out the research work.
Author contributions
MA and KS: Conception and idea. KS: performed the experimental work with the assistance of MA. NV and KS wrote the manuscript with the assistance of MA. KS and NV: preparation of tables and figures. CA: helped in performing experimental analysis. PG and CA: helped in the critical reviewing of the manuscript. Finally, all the authors approved for the manuscript.
Data availability
Data generated in this work is included in the manuscript and in the supplementary material. This will be made available on request.
Declarations
Conflict of interest
There are no conflicts of interest to declare.
Ethical approval for involving human participants and/or animals
Not applicable, since this article does not contain any studies with human participants or animals performed by any of the authors.
References
- Arun M, Subramanyam K, Mariashibu TS, Theboral J, Shivanandhan G, Manickavasagam M, Ganapathi A. Application of sonication in combination with vacuum infiltration enhances the Agrobacterium-mediated genetic transformation in indian soybean cultivars. Appl Biochem Biotechnol. 2015;175:2266–2287. doi: 10.1007/s12010-014-1360-x. [DOI] [PubMed] [Google Scholar]
- Chen L, Cai Y, Liu X, Yao W, Guo C, Sun S, Wu C, Jiang B, Han T, Hou W. Improvement of soybean Agrobacterium-mediated transformation efficiency by adding glutamine and asparagine into the culture media. Int J Mol Sci. 2018;19:1–17. doi: 10.3390/ijms19103039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ether YB, Jadhav PV, Moharil MP, Dudhare MS, Kale P, Nandanwar RS, Mane SS, Dani R. Epigenesis through in vitro regeneration in soybean amenable to genetic transformation. VEGETOS. 2013;26(2):245–254. doi: 10.5958/j.2229-4473.26.2.081. [DOI] [Google Scholar]
- Gadaleta A, Giancaspro A, Cardone M, Blanco A. Real-time PCR for the detection of precise transgene copy number in durum wheat. Cell Mol Biol Lett. 2011 doi: 10.2478/s11658-011-0029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez KA, Gomez AA. Statistical procedures for agricultural research. John wiley & sons; 1984. [Google Scholar]
- Guo B, Guo Y, Wang J, Zhang L, Jin L, Hong H, Chang R, Qiu L. Co-treatment with surfactant and sonication significantly improves Agrobacterium-mediated resistant bud formation and transient expression efficiency in soybean. J Integr Agric. 2015;14:1242–1250. doi: 10.1016/S2095-3119(14)60907-2. [DOI] [Google Scholar]
- Hada A, Krishnan V, Mohamed Jaabir MS, Kumari A, Jolly M, Praveen S, Sachdev A. Improved Agrobacterium tumefaciens-mediated transformation of soybean [Glycine max (L.) Merr.] following optimization of culture conditions and mechanical techniques. Vitr Cell Dev Biol Plant. 2018;54:672–688. doi: 10.1007/s11627-018-9944-8. [DOI] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S, Krishnan V, Dahuja A, Vinutha T, Jolly M, Sachdev A. A rapid method for optimization of Agrobacterium- mediated transformation of Indian soybean. Indian J Biochem Biophys. 2016;53:218–226. [Google Scholar]
- Li S, Cong Y, Liu Y, Wang T, Shuai Q, Chen N, Gai J, Li Y. Optimization of Agrobacterium-mediated transformation in soybean. Front Plant Sci. 2017;8:1–15. doi: 10.3389/fpls.2017.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H-K, Yang C, Wei Z-M. Efficient Agrobacterium tumefaciens-mediated transformation of soybeans using an embryonic tip regeneration system. Planta. 2004;219:1042–1049. doi: 10.1007/s00425-004-1310-x. [DOI] [PubMed] [Google Scholar]
- Ma X, Wu T. Rapid and efficient regeneration in soybean [Glycine max (L.) Merrill] from whole cotyledonary node explants. Acta Physiol Plant. 2008;30:209–216. doi: 10.1007/s11738-007-0109-3. [DOI] [Google Scholar]
- Olhoft PM, Donovan CM, Somers DA. Soybean (Glycine max) transformation using mature cotyledonary node explants. Methods Mol Biol. 2006;343:385–396. doi: 10.1385/1-59745-130-4:385. [DOI] [PubMed] [Google Scholar]
- Subramanyam K, Rajesh M, Jaganath B, Vasuki A, Theboral J, Elayaraja D, Karthik S, Manickavasagam M, Ganapathi A. Assessment of factors influencing the Agrobacterium-mediated in planta seed transformation of brinjal (Solanum melongena L.) Appl Biochem Biotechnol. 2013;171:450–468. doi: 10.1007/s12010-013-0359-z. [DOI] [PubMed] [Google Scholar]
- Trick H, Finer J. SAAT: sonication-assisted Agrobacterium-mediated transformation. Transgenic Res. 1997;6:329–336. doi: 10.1023/A:1018470930944. [DOI] [Google Scholar]
- Wang W-W, Zhu C-Q, Liu X-H, Chen K-S, Xu C-J. Techniques for rapid preparation of tomato leaf DNA and its application in real-time quantitative PCR-based transgene detection. Hered. 2011;33:1017–1022. doi: 10.3724/SP.J.1005.2011.01017. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li Z, Chen X, Gu Y, Zhang L, Qiu L. An efficient soybean transformation protocol for use with elite lines. Plant Cell Tissue Organ Cult. 2022;151:457–466. doi: 10.1007/s11240-022-02312-6. [DOI] [Google Scholar]
- Wei J, Chen S, Zeng L, Yang J, Liu X, Zhu X. Quantitative expression analysis of rice bacterial blight resistant candidate genes of Xa7 by real-time fluorescent quantitative PCR. Mol Plant Breed. 2011;9:9–16. [Google Scholar]
- Widholm JM, Finer JJ, Vodkin LO, Trick HN, LaFayette P, Li JPW. Soybean. I. Berlin: Springer; 2010. In genetic modification of plants; pp. 473–498. [Google Scholar]
- Yang X. Analysis of the copy number of exogenous genes in transgenic cotton using real-time quantitative PCR and the 2-△△CT method. Afr J Biotechnol. 2012 doi: 10.5897/AJB11.4117. [DOI] [Google Scholar]
- You-wen Q, Xue-jun G, Bang-ruo Q, Lu L, Zhen Z. Establishment of TaqMan real-time quantitative PCR assay for Foreign gene copy numbers in transgenic soybean. J Northeast Agric Univ (english Ed) 2012;19:48–52. doi: 10.1016/s1006-8104(13)60050-1. [DOI] [Google Scholar]
- Yuan L, Sun H-W, Yang C-L, Shang Y-F, Lu X-B, Zhao L. Analysis of junction sequence in the transgenic maize MON88017 and the methods of qualitative PCR detection. ACTA Agron Sin. 2010;36:361–364. doi: 10.3724/SP.J.1006.2010.00361. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated in this work is included in the manuscript and in the supplementary material. This will be made available on request.