Abstract
Alternative splicing is an important mechanism that enhances protein functional diversity. To date, our understanding of alternative splicing variants has been based on mRNA transcript data, but due to the difficulty in predicting protein structures, protein tertiary structures have been largely unexplored. However, with the release of AlphaFold, which predicts three-dimensional models of proteins, this challenge is rapidly being overcome. Here, we present a dataset of 315 predicted structures of abnormal isoforms in 18 uveal melanoma patients based on second- and third-generation transcriptome-sequencing data. This information comprises a high-quality set of structural data on recurrent aberrant isoforms that can be used in multiple types of studies, from those aimed at revealing potential therapeutic targets to those aimed at recognizing of cancer neoantigens at the atomic level.
Subject terms: Computational biology and bioinformatics, Diseases
Background & Summary
Alternative splicing (AS) can influence transcriptome and proteome diversity, as evidence shows that approximately 95% of genes with multiple exons produce multiple isoforms1,2. Therefore, it is not surprising that the gene isoforms play important roles in many biological processes, such as processes related to development, pluripotency and apoptosis3–5. Aberrant isoforms have been implicated in multiple human tumors, including uveal melanoma (UM), showing extensive changes via alternative splicing and the expression of critical gene isoforms6–8. Specific splicing isoforms are important for the initiation, metastasis and drug resistance of cancer, and some AS events have been shown to be significantly related to patient survival9–11. Although the role of a few splicing isoforms in cancer has been studied, 3D protein structure prediction on a scale that covers the transcriptome and can be used for evaluating biological functionality remains unexplored.
The suitability of short-read mRNA sequencing (short-read RNA-seq) in the discovery of AS events is limited because of the mapping uncertainty of short read lengths or assembly problems12. Long-read mRNA sequencing (long-read RNA-seq) shows advantages over short-read RNA-seq in isoform detection because long reads directly cover the entire transcript without the need of reconstruction, which is needed for short reads13–15. However, because of the high sequencing error rate (~15%) of raw long-read RNA-seq data, it is still challenging to determine the precise splicing sites with only long-read RNA-seq data16. Hybrid sequencing, combining long-read RNA-seq reads with high quality short-read RNA-seq reads and taking advantage of both platforms, improves the identification of AS events and gene isoforms17,18. Although great efforts have been made to study the alternative splicing mechanisms and functions of different isoforms, our knowledge of the 3D structure of splicing isoforms is very limited19,20. This lack of information means that for a large majority of spliced isoforms, no documented structures have been deposited in the Protein Data Bank (PDB), causing a large knowledge gap; hence, accurate prediction of protein structure is one of the most challenging goals in biology21,22. As structures carry vital information about how different isoforms with a certain degree of sequence homology perform different functions, it is necessary to investigate the 3D structure of abnormal isoforms to explore their functions23. The most recent achievement in related technology, AlphaFold, a deep-learning-based approach, has been proven to be highly successful in predicting the 3D structures of proteins based on their amino acid sequences24. This is a significant advance that might have a profound impact on the study of protein dysfunction and the discovery of new polypeptides with potential medical applications25.
In this study, we provide an information resource based on the predicted structures of 315 novel isoforms obtained by long-read RNA-seq and short-read RNA-seq of transcriptome data from 18 UM patients. To better understand the structural differences of abnormal isoforms and the potential effects, we compared the structural differences between 295 abnormal-gene-encoded isoforms and their normal gene-encoded protein counterparts. We also identified 13 potential AS-derived neoantigens in 10 abnormal isoforms with altered amino acid sequences. These data constitute particularly valuable information on aberrant isoform structures that intersects with that on abnormal isoforms in other datasets, which can be used for an investigation into the roles of these isoforms in multiple cancer types. This study also offers new insights into the structure-based prediction of neoantigens and potential drug targets.
Methods
Patient samples
A total of 20 patients with primary UM who visited Shanghai Ninth People’s Hospital between 2018 and 2021 were selected for sampling. The detail information of the 20 UM patients is listed in Table 1. The process of sample collection adhered to the tenets of the Declaration of Helsinki and was approved by the Ethics Committee of Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine (SH9H-2021-T82-1). All patient samples were donated freely, with written informed consent and with the full cooperation of each patient. We have confirmed that in our ethics statement, patients consent to the disclosure of genomic data. The critical exclusion criteria include previous treatment of chemotherapy or radiotherapy. Sterilized instruments collected the samples, and immediately underwent snap-frozen in liquid nitrogen and stored at a temperature below −80 °C.
Table 1.
The detail information of UM patients.
| Patient | Age (yrs) | Gender (male/female) | Ethnicity | Largest basal diameter*thickness (mm) |
|---|---|---|---|---|
| Case 1 | 30–35 | male | Chinese | 18.55*8.54 |
| Case 2 | 30–35 | male | Chinese | 19.53*10.67 |
| Case 3 | 45–50 | male | Chinese | 12.98*7.78 |
| Case 4 | 45–50 | female | Chinese | 14.07*6.59 |
| Case 5 | 45–50 | male | Chinese | 18.28*9.53 |
| Case 6 | 45–50 | male | Chinese | 11.65*9.19 |
| Case 7 | 45–50 | male | Chinese | 10.58*8.66 |
| Case 8 | 45–50 | female | Chinese | 12.43*11.25 |
| Case 9 | 50–55 | male | Chinese | 11.95*8.12 |
| Case 10 | 50–55 | male | Chinese | 13.22*14.42 |
| Case 11 | 50–55 | female | Chinese | 16.64*9.83 |
| Case 12 | 50–55 | female | Chinese | 18.9*13.4 |
| Case 13 | 55–60 | female | Chinese | 15.06*10.07 |
| Case 14 | 55–60 | male | Chinese | 14.83*6.63 |
| Case 15 | 65–70 | female | Chinese | 14.7*8.83 |
| Case 16 | 65–70 | male | Chinese | 10.5*5.02 |
| Case 17 | 70–75 | male | Chinese | 15.23*7.79 |
| Case 18 | 70–75 | male | Chinese | 10.01*11.13 |
| Case 19 | 70–75 | male | Chinese | 15.95*5.25 |
| Case 20 | 75–80 | male | Chinese | 12.25*5.15 |
Illumina sequencing
Total RNA was isolated using Trizol Reagent (Invitrogen Life Technologies), then the concentration, quality and integrity of RNA were determined by NanoDrop spectrophotometer (Thermo Scientific). Three micrograms of RNA were used as input material for the RNA sample preparations. Sequencing libraries were generated which was then sequenced on Illumina NovaSeq. 6000 platform by Shanghai Personal Biotechnology Cp. Ltd.
Nanopore sequencing
Total RNA was isolated using the Trizol Reagent (Invitrogen Life Technologies), and the concentration, quality and integrity were determined by NanoDrop spectrophotometer (Thermo Scientific). A total of 1ug RNA was prepared for cDNA libraries using the cDNA-PCR Sequencing Kit (SQK-PCS109) according to the instructions of Nanopore Technologies (ONT). Defined PCR adaptors were directly added to both ends of the first-strand cDNA by reverse transcriptase. After 14-cycle of PCR by LongAmp Tag (NEB), the PCR products were subjected to ONT adaptor ligation using T4 DNA ligase (NEB). Agencourt XP beads were used for DNA purification according to ONT protocol. The final cDNA libraries were added to FLO-MIN109 flow cells and were sequenced on the PromethION platform. GUPPY (version 3.2.6) was used for basecalling to convert the fast5 format data to fastq format.
Hybrid-sequencing strategy
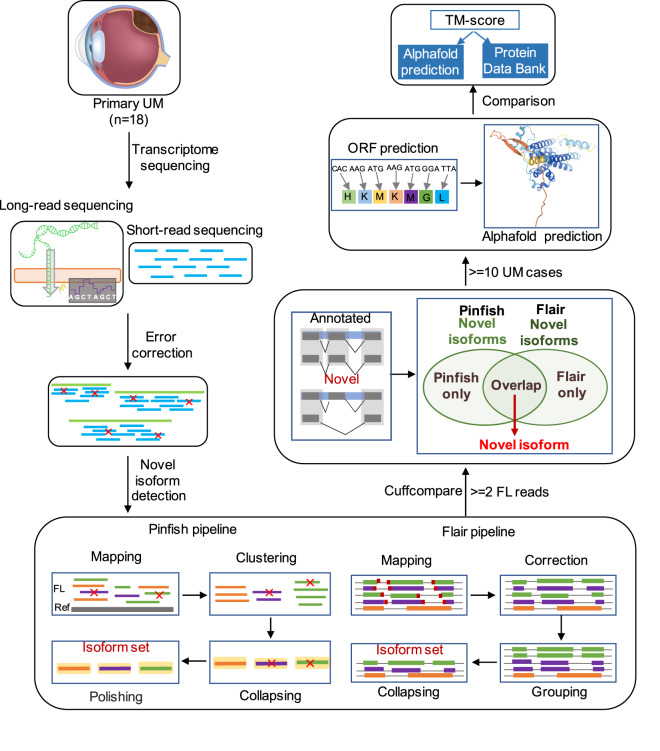
Hybrid error correction, a simple and cost-effective approach involved with high quality short-read RNA-seq data, was used to improve the quality of long reads (Fig. 1). Here we used LoRDEC to correct the errors of full-length (FL) sequences17. LoRDEC is a new and efficient hybrid correction algorithm based on De Bruijn Graphs (DBG) of short reads. Achieving a comparable accuracy, LoRDEC runs six times faster and requires 93% less memory than PacBioToCA and LSC. LoRDEC first reads the short reads, builds their DBG of order k and then corrects each long read one after the other independently.
Fig. 1.
The overall workflow of this study.
Pinfish pipeline
The corrected FL reads for each sample were aligned to hg38 using minimap2 with the command ‘minimap2 -ax splice’26. Spliced_bam2gff was used to convert sorted BAM files with spliced alignments (from minimap2) into GFF2 format. With sorted GFF2 file as input, based on the median of exon boundaries from all transcripts in the cluster, cluster_gff clusters reads with similar exon/intron structures into a rough consensus set of clusters. Then, by mapping all reads to the median length of read within each cluster generated by cluster_gff, polish_clusters creates an error corrected read and polishes it using racon27. Finally, taking polished and consistent transcripts as input, collapse_partials filters transcripts which are likely caused by 5′ end degradation and collapses input transcripts into a polished and collapsed transcripts set of each UM case (Fig. 1).
Flair pipeline
Flair_align aligns FL reads of each sample to hg38 using minimap2 and converts the SAM output to BED format. Flair_correct corrects mis-aligned splice sites with genome annotations and short-read splice junctions generated by STAR28. Finally, flair_collapse defines high-confidence transcript sets from corrected long reads14 (Fig. 1).
Novel isoform detection
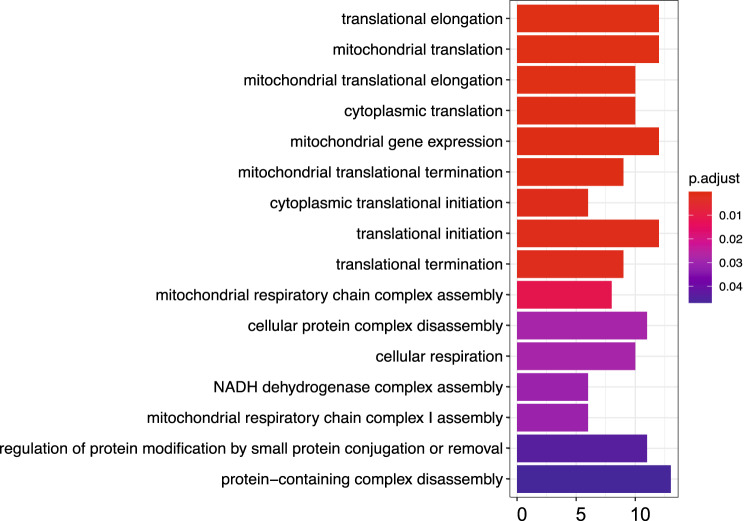
We filtered out transcripts supported by less than two FL reads. With the cuffcompare tool in the Cufflinks package29, we compared the high-confidence isoforms output by flair and pinfish pipelines with the “RefSeq” gene annotation file, respectively. Cuffcompare explores the structure of each isoform, and matches reference transcripts that agrees on the coordinates and orders of all their exons, as well as strand. Isoforms set were further classified into eight groups based on their exon structures (splicing junctions) after the cuffcompare process. Isoform labeled by “ = ” and “j” tags in the output “.tracking” file was considered as an annotated and unannotated (novel) isoform, respectively. We got a median of 8,989 annotated and a median of 9,150 novel isoform candidates based on pinfish pipeline (Table 2), with a median of 11,117 annotated and a median of 12,366 novel isoform candidates from flair pipeline (Table 2). Finally, we defined novel isoforms as those were identified as novel isoform candidates in both flair and pinfish pipelines of each case. In order to further define high-confidence and recurrent novel isoform set for further analysis, we only remained 315 novel isoforms which have been identified in more than 10 UM cases (Fig. 1). We then performed GO enrichment analysis and found regulation of translation initiation, elongation and termination related pathways were significantly enriched in these isoform related genes (Fig. 2).
Table 2.
Statistics of annotated and novel isoforms among 18 UM cases.
| Case | Annotated | Novel candidates | Novel | ||
|---|---|---|---|---|---|
| Pinfish | Flair | Pinfish | Flair | ||
| Case 2 | 7118 | 8622 | 3074 | 4905 | 1020 |
| Case 3 | 10004 | 12655 | 15696 | 20561 | 3956 |
| Case 4 | 11702 | 14581 | 15639 | 20352 | 4500 |
| Case 5 | 5678 | 7818 | 4074 | 6866 | 1141 |
| Case 6 | 9599 | 11606 | 10321 | 13437 | 3601 |
| Case 7 | 11428 | 13740 | 15518 | 17572 | 4335 |
| Case 9 | 7523 | 9571 | 11110 | 11451 | 2788 |
| Case 10 | 6724 | 8477 | 3237 | 5270 | 958 |
| Case 11 | 8901 | 11153 | 9144 | 13214 | 3041 |
| Case 12 | 10893 | 13317 | 12302 | 16035 | 3718 |
| Case 13 | 4031 | 6114 | 6591 | 11919 | 1552 |
| Case 14 | 9185 | 11081 | 9157 | 11289 | 2565 |
| Case 15 | 11062 | 13508 | 14755 | 23147 | 4235 |
| Case 16 | 2599 | 4298 | 4602 | 6847 | 1010 |
| Case 17 | 9077 | 12108 | 8052 | 12814 | 2522 |
| Case 18 | 12400 | 14773 | 15879 | 21276 | 4934 |
| Case 19 | 8444 | 10813 | 7038 | 10729 | 2687 |
| Case 20 | 3041 | 4873 | 7208 | 10365 | 1726 |
Fig. 2.
Gene ontology enrichment of genes with novel isoforms.
Alphafold structure prediction
For novel isoforms, we first extracted corresponding DNA sequences from the human reference genome (hg38) using “samtools faidx” based on the coordinates and orders of all their exons, as well as the strand30. The tool of ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/) was subsequently employed to search for open reading frames (ORFs)31. The 3D structure was predicted using AlphaFold-Multimer version 2.2.0 using Shanghai Jiao Tong University’s supercomputing resources (more specifically, one NVIDIA Volta V100 GPUs with 32GB graphics processing unit (GPU) memory)24. The version and parameters of AlphaFold-Multimer databases used were outlined as below:
“python run_alphafold.py
--use_gpu_relax
--data_dir = $DIR
--uniref90_database_path = $DIR/uniref90/uniref90.fasta
--mgnify_database_path = $DIR/mgnify/mgy_clusters_2018_12.fa
--bfd_database_path = $DIR/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt
--uniclust30_database_path = $DIR/uniclust30/uniclust30_2020_06/UniRef30_2020_06
--pdb_seqres_database_path = $DIR/pdb_seqres/pdb_seqres.txt
--template_mmcif_dir = $DIR/pdb_mmcif/mmcif_files
--obsolete_pdbs_path = $DIR/pdb_mmcif/obsolete.dat
--uniprot_database_path = $DIR/uniprot/uniprot.fasta
--model_preset = multimer
--max_template_date = 2022-1-1
--db_preset = full_dbs
--output_dir = output
--fasta_paths = input.fasta”
TM-score calculation for structural comparison
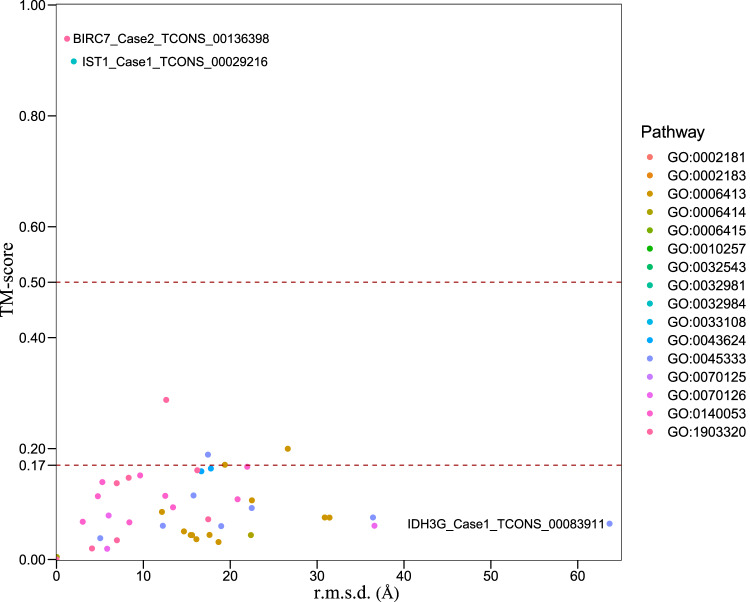
Typical structure files were downloaded from Uniprot (https://www.uniprot.org) according to the priority order of EM, NMR, X-ray and alphafold predicted sources of structures. We used TM-score (https://zhanggroup.org/TM-score) to compare the predictive results with typical structures32. Protein pairs with a TM-score > 0.5 are mostly in the same fold while those with a TM-score < 0.5 are mainly not in the same fold, and some of them with a TM-score < 0.17 just have random structural similarity33. We then check the distribution of comparison scores of novel isoforms based on the gene ontology enrichment results above (Fig. 3). As only 32 pairs (32/295) of both ratios of aligned length to novel protein and aligned length to canonical protein are bigger than 0.9, which suggested that most of novel isoforms have random or low structures similarities with their canonical proteins. TM-score results of each paired comparison were deposited at figshare34.
Fig. 3.
Structure comparison of novel isoforms with typical proteins.
Neoantigens prediction
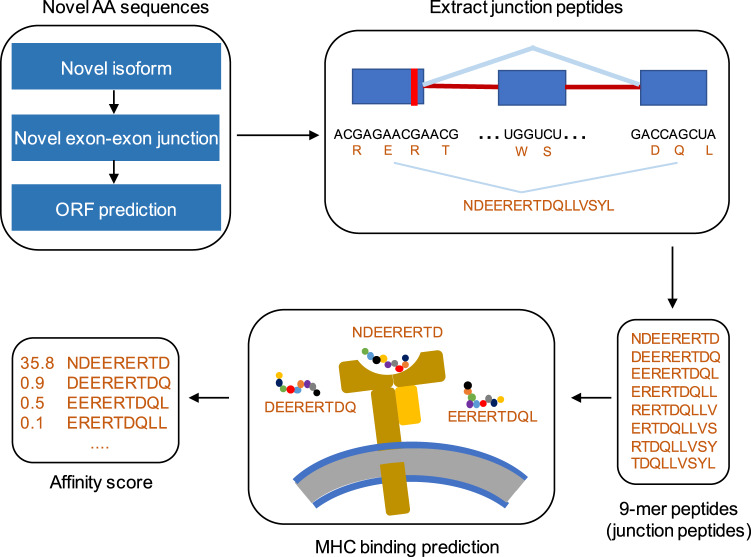
For predicting AS-derived neoantigens, we first used a custom script to extract the unannotated splicing site information from all novel isoforms and defined such splicing junctions as neojunctions. Based on neojunction loci, we obtained all polypeptides generated by a neojunction. We used a custom script to extract all possible 9-amino acid sequences from these polypeptides. NetMHCPan was used to perform MHC-I binding affinity prediction. NetMHCpan methods inform if a sequence is a strong MHC binder if the % Rank is below the specified threshold (0.5%), and define the peptide as a weak binder if the % Rank is above the threshold of the strong binders but below the specified threshold (2%)35. Peptides with strong binding or weak binding affinity were defined as neoantigens (Fig. 4). Finally, we got 13 potential AS-derived neoantigens from 10 highly recurrent abnormal isoforms (present in at least 10 UM cases) due to changes in amino acid sequences (Table 3).
Fig. 4.
Overview of the neojunction-derived putative neoantigens detection.
Table 3.
Sequences, binding scores and levels of neoantigens.
| Isoform ID | Peptide | Score | Bind Level |
|---|---|---|---|
| Case20_TCONS_00011217 | CQVDGLIFL | 0.65303 | WB |
| Case20_TCONS_00011217 | SLHCQVDGL | 0.4821 | WB |
| Case20_TCONS_00118164 | WLIHKTTKL | 0.59746 | WB |
| Case20_TCONS_00118487 | SQADKFLSL | 0.51318 | WB |
| Case14_TCONS_00003591 | GLFFSHAGV | 0.68942 | SB |
| Case14_TCONS_00032920 | AILEIGAGV | 0.71785 | SB |
| Case14_TCONS_00047054 | CVLHELFHL | 0.5366 | WB |
| Case14_TCONS_00047054 | RILCVLHEL | 0.70888 | SB |
| Case14_TCONS_00065906 | AFWDWSVEA | 0.48128 | WB |
| Case14_TCONS_00066759 | KLSHPMVAI | 0.63195 | WB |
| Case14_TCONS_00069531 | FMRLPLISV | 0.64744 | WB |
| Case14_TCONS_00069531 | RLPLISVAL | 0.54154 | WB |
| Case5_TCONS_00029306 | LLAQLGFPL | 0.78058 | SB |
Data Records
The datasets presented here have been stored at GEO under GSE20646436. Our cohort includes a total of 20 cases, of which 20 cases have short-read RNA-seq data and 18 cases have long-read RNA-seq data. This study is based on the 18 cases with both short-read and long-read RNA-seq data. AlphaFold-Multimer predicted structures files are accessible at figshare37. Each folder, whose name consists of gene name and isoform ID, corresponds to each isoform structure files (for example, “AAMDC_TCONS_00022447”, which means isoform “TCONS_00022447” of gene AAMDC). Each folder contains multiple format text files which represent the predicted structures information. Among all predicted structures files, “ranked_0.pdb” file has the highest confidence.
Technical Validation
Sequencing quality of long-read RNA-seq data
Pychopper package (https://github.com/nanoporetech/pychopper) was used to identify, orient and rescue FL cDNA reads. The number of total long reads ranged from 4,788,440 to 14,048,314 in which the FL reads were between 4,112,595 and 12,828,344 (Table 4). We observed an average of 7.05 million FL reads (accounting for 87.49% of total long reads) with an average read quality of 10.78 (ranging from 9.9 to 12.8), confirming the high confidence in the quality of sequencing data (Table 4).
Table 4.
Sequencing statistics of 18 UM cases.
| Case | Data (Gb) | Total reads | FL reads | Mean read length | Mean read quality |
|---|---|---|---|---|---|
| Case 2 | 2.8 | 6549553 | 5623406 | 1094 | 12.8 |
| Case 3 | 8.5 | 8102441 | 6632235 | 1046 | 10.1 |
| Case 4 | 7.2 | 7820006 | 6670112 | 918 | 10 |
| Case 5 | 3.4 | 4788440 | 4215364 | 691 | 10.6 |
| Case 6 | 6.3 | 10669535 | 9770237 | 589 | 10.5 |
| Case 7 | 7.8 | 8210582 | 7042750 | 953 | 10 |
| Case 9 | 4.8 | 7326024 | 6503192 | 646 | 10.6 |
| Case 10 | 3.1 | 4798635 | 4112595 | 1069 | 12.7 |
| Case 11 | 6.1 | 8495028 | 7596197 | 706 | 10.9 |
| Case 12 | 7.1 | 7675607 | 6652587 | 932 | 10.8 |
| Case 13 | 5.3 | 8593514 | 7783746 | 606 | 10.7 |
| Case 14 | 4.9 | 4843576 | 4127856 | 994 | 9.9 |
| Case 15 | 7.1 | 7745207 | 6780170 | 863 | 12.8 |
| Case 16 | 6 | 13066897 | 11847478 | 448 | 10.2 |
| Case 17 | 5.3 | 7943859 | 7142291 | 652 | 10.6 |
| Case 18 | 9.2 | 8144175 | 6649344 | 1130 | 10 |
| Case 19 | 3.7 | 5510264 | 4937671 | 670 | 10.6 |
| Case 20 | 6.1 | 14048314 | 12828344 | 423 | 10.2 |
Acknowledgements
This work was supported by National Key R&D Program of China (2021YFC2701103 to J.S.) and General Program of National Natural Science Foundation of China (No.81972667 to J.S.) and Shanghai Municipal Education Commission-Two Hundred Talent (No.20191817 to J.S.).
Author contributions
Z.Z. and C.L. performed the experiments, analyzed the results and wrote the manuscript. X.L., Q.L., J.L., L.Z. and X.S. provided research materials and project consultation. J.S. designed the experiments, analyzed the results and wrote the manuscript.
Code availability
Here we list the details of the software used for data analysis. Pychopper (https://github.com/epi2me-labs/pychopper), version 2, was used to identify, orient and trim FL Nanopore cDNA reads. LoRDEC, version 1.4.1, was used for correcting errors of long-read RNA-seq data based on short-read RNA-seq data. Pinfish (https://github.com/nanoporetech/pinfish), version 0.1.0, which is a collection of tools helping to make sense of long-read RNA-seq data. Flair (https://github.com/BrooksLabUCSC/flair), version 1.5.0, was used for isoform definition with long-read RNA-seq data. Cuffcompare, version 2.2.1, was used to identify novel isoform based on gene annotation information. Samtools, version 1.9, was used to extract sequence according to the coordinates of novel isoforms. ORFfinder, version 0.4.3, was used to predict ORFs based on nucleotide sequences. AlphaFold Multimer (an extension of AlphaFold2, version 2.2.0) was used to predict the 3D structures of novel isoforms. Then, we also use some in-house scripts to filter and prepare the input and output files, which have been deposited in github (https://github.com/ZhangNestor/magic).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhe Zhang, Chen Li.
Contributor Information
Zhe Zhang, Email: zhangzistiger@gmail.com.
Xinhua (James) Lin, Email: james@sjtu.edu.cn.
Jianfeng Shen, Email: jfshen@shsmu.edu.cn.
References
- 1.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 2.Jiang W, Chen L. Alternative splicing: Human disease and quantitative analysis from high-throughput sequencing. Comput Struct Biotechnol J. 2021;19:183–195. doi: 10.1016/j.csbj.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen K, Dai X, Wu J. Alternative splicing: An important mechanism in stem cell biology. World J Stem Cells. 2015;7:1–10. doi: 10.4252/wjsc.v7.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnal SC, Lopez-Oreja I, Valcarcel J. Roles and mechanisms of alternative splicing in cancer - implications for care. Nat Rev Clin Oncol. 2020;17:457–474. doi: 10.1038/s41571-020-0350-x. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Qian J, Gu C, Yang Y. Alternative splicing and cancer: a systematic review. Signal Transduct Target Ther. 2021;6:78. doi: 10.1038/s41392-021-00486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kahles A, et al. Comprehensive Analysis of Alternative Splicing Across Tumors from 8,705 Patients. Cancer Cell. 2018;34:211–224 e216. doi: 10.1016/j.ccell.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alsafadi S, et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat Commun. 2016;7:10615. doi: 10.1038/ncomms10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Group PTC, et al. Genomic basis for RNA alterations in cancer. Nature. 2020;578:129–136. doi: 10.1038/s41586-020-1970-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Climente-Gonzalez H, Porta-Pardo E, Godzik A, Eyras E. The Functional Impact of Alternative Splicing in Cancer. Cell Rep. 2017;20:2215–2226. doi: 10.1016/j.celrep.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 11.Stanley RF, Abdel-Wahab O. Dysregulation and therapeutic targeting of RNA splicing in cancer. Nat Cancer. 2022;3:536–546. doi: 10.1038/s43018-022-00384-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steijger T, et al. Assessment of transcript reconstruction methods for RNA-seq. Nat Methods. 2013;10:1177–1184. doi: 10.1038/nmeth.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolisetty MT, Rajadinakaran G, Graveley BR. Determining exon connectivity in complex mRNAs by nanopore sequencing. Genome Biol. 2015;16:204. doi: 10.1186/s13059-015-0777-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang AD, et al. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat Commun. 2020;11:1438. doi: 10.1038/s41467-020-15171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aw JGA, et al. Determination of isoform-specific RNA structure with nanopore long reads. Nat Biotechnol. 2021;39:336–346. doi: 10.1038/s41587-020-0712-z. [DOI] [PubMed] [Google Scholar]
- 16.Watson M, Warr A. Errors in long-read assemblies can critically affect protein prediction. Nat Biotechnol. 2019;37:124–126. doi: 10.1038/s41587-018-0004-z. [DOI] [PubMed] [Google Scholar]
- 17.Salmela L, Rivals E. LoRDEC: accurate and efficient long read error correction. Bioinformatics. 2014;30:3506–3514. doi: 10.1093/bioinformatics/btu538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang JR, Holt J, McMillan L, Jones CD. FMLRC: Hybrid long read error correction using an FM-index. BMC Bioinformatics. 2018;19:50. doi: 10.1186/s12859-018-2051-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varadi M, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sommer, M. J. et al. Structure-guided isoform identification for the human transcriptome. Elife11, 10.7554/eLife.82556 (2022). [DOI] [PMC free article] [PubMed]
- 21.UniProt C. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armstrong DR, et al. PDBe: improved findability of macromolecular structure data in the PDB. Nucleic Acids Res. 2020;48:D335–D343. doi: 10.1093/nar/gkz990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhlman B, Bradley P. Advances in protein structure prediction and design. Nat Rev Mol Cell Biol. 2019;20:681–697. doi: 10.1038/s41580-019-0163-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jumper J, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinheiro F, Santos J, Ventura S. AlphaFold and the amyloid landscape. J Mol Biol. 2021;433:167059. doi: 10.1016/j.jmb.2021.167059. [DOI] [PubMed] [Google Scholar]
- 26.Li H. New strategies to improve minimap2 alignment accuracy. Bioinformatics. 2021;37:4572–4574. doi: 10.1093/bioinformatics/btab705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaser R, Sovic I, Nagarajan N, Sikic M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017;27:737–746. doi: 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rombel IT, Sykes KF, Rayner S, Johnston SA. ORF-FINDER: a vector for high-throughput gene identification. Gene. 2002;282:33–41. doi: 10.1016/s0378-1119(01)00819-8. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 33.Xu J, Zhang Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics. 2010;26:889–895. doi: 10.1093/bioinformatics/btq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang N. 2023. TM_scores.xlsx. Figshare. [DOI]
- 35.Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z, Shen JF. 2022. GEO. //identifiers.org/geo/GSE206464
- 37.Zhang N. 2023. Alphafold structure files of novel isoforms. Figshare. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
Data Availability Statement
Here we list the details of the software used for data analysis. Pychopper (https://github.com/epi2me-labs/pychopper), version 2, was used to identify, orient and trim FL Nanopore cDNA reads. LoRDEC, version 1.4.1, was used for correcting errors of long-read RNA-seq data based on short-read RNA-seq data. Pinfish (https://github.com/nanoporetech/pinfish), version 0.1.0, which is a collection of tools helping to make sense of long-read RNA-seq data. Flair (https://github.com/BrooksLabUCSC/flair), version 1.5.0, was used for isoform definition with long-read RNA-seq data. Cuffcompare, version 2.2.1, was used to identify novel isoform based on gene annotation information. Samtools, version 1.9, was used to extract sequence according to the coordinates of novel isoforms. ORFfinder, version 0.4.3, was used to predict ORFs based on nucleotide sequences. AlphaFold Multimer (an extension of AlphaFold2, version 2.2.0) was used to predict the 3D structures of novel isoforms. Then, we also use some in-house scripts to filter and prepare the input and output files, which have been deposited in github (https://github.com/ZhangNestor/magic).