Abstract
A 46-kDa receptor, coxsackievirus-adenovirus (Ad) receptor (CAR), mediates cell attachment of a number of different Ad serotypes; however, not all Ad serotypes utilize this receptor for infection. Moreover, the precise amino acid sequences in the Ad fiber protein that mediate cell attachment have yet to be identified. We investigated the interaction of subgroup D Ads with human ocular cells. Ad serotype 37 (Ad37), a virus associated with epidemic keratoconjunctivitis, but not a closely related virus serotype, Ad19p, exhibited preferential binding to and infection of human conjunctival cells. A single amino acid substitution in the Ad19p fiber distal domain (knob), Glu240 to Lys, conferred binding to conjunctival cells, while the reverse substitution in the Ad37 fiber abrogated cell binding. These findings provide new information on the fiber sequences that regulate Ad host cell tropism.
Human adenoviruses (Ads) are associated with a significant number of gastrointestinal and respiratory infections in young children and are also a leading cause of viral conjunctivitis. Several subgroup D viruses, including Ad serotype 8 (Ad8), Ad19p, and Ad37, have been frequently isolated from patients during outbreaks of epidemic keratoconjunctivitis (EKC) (5, 14). EKC can be distinguished from viral conjunctivitis in that it involves the cornea. Persistent Ad infection in EKC can threaten long-term visual function. At present, there is little information on the viral and cellular factors that govern the tropism of certain Ad serotypes for specific ocular cell types.
The Ad fiber protein is an oligomeric molecule comprised of three identical polypeptide subunits of 30 to 65 kDa (12). The fiber contains a conserved N-terminal tail that mediates interaction with the Ad penton base protein, a variable-length elongated (shaft) domain, and a C-terminal knob that mediates high-affinity interaction with cell receptors (10). Recent studies (11) have indicated that multiple Ad serotypes from different subgroups recognize a 46-kDa cell receptor that also serves as the receptor for the subgroup B coxsackieviruses (designated CAR [coxsackievirus-Ad receptor]) (4, 13). However, not all Ads use CAR to bind to host cells. The identity of cell receptors for many other Ads remain to be determined.
The three-dimensional structure of the Ad5 fiber N-terminal knob domain has been determined (17). A comparison of the fiber knob sequences of different Ad serotypes together with the crystal structure of the protein has suggested that the sites for CAR binding interaction are located on residues lining the walls and central depression of the fiber knob (4, 13). However, this has not been formally demonstrated. Moreover, the locations of receptor binding sites for other Ad serotypes on the fiber protein have not been identified.
The amino acid sequences of the Ad19p and Ad37 fiber proteins have been reported (1). Interestingly, only two differences (amino acid residues K/E240 and N/D340) were noted in the knob regions of these two related viruses. In the studies presented here, we sought to determine whether either of these amino acid sequences played a role in Ad interaction with cell receptors.
MATERIALS AND METHODS
Cell lines, viruses, and plaque assays.
Human epithelial cells A549, 293 (Ad5-transformed embryonic kidney cells), and Chang C conjunctival cells (American Type Culture Collection [ATCC]) were maintained in complete Dulbecco modified Eagle medium containing 10% fetal bovine serum. Ad37 (ATCC) was propagated in Chang C cells, while Ad2 and Ad19p (ATCC) were produced in A549 cells. High-titered virus culture supernates were added to 90 to 95% confluent cell monolayers in 162-cm2 tissue culture flasks. When more than 60% of the cells showed evidence of cytopathic effect the cells were detached by using 10 mM EDTA, resuspended in 0.5 to 1.0 ml of complete Dulbecco modified Eagle medium, and then subjected to three cycles of freezing-thawing to release viral particles from infected cells. Virions were subsequently purified by banding on 16 to 40% continuous CsCl density gradients as previously described (6) and then dialyzed against 10 mM Tris-buffered saline (pH 8.1) containing 10% glycerol. Viral protein content was measured by the Bio-Rad protein assay. The number of viral particles was calculated based on the known molecular weight of Ad2 virions (1 μg = 4 × 109 particles).
The infectivity of Ad37 was determined by plaquing the virus on Chang C cells. Various dilutions of virus in a total volume of 2 ml were added to cell monolayers in six-well tissue culture plates and incubated for 2 h at 37°C. The cells were then washed with serum-free medium and overlaid with 3 ml of 0.5% agarose (SeaKem; FMC) in medium. The cells were observed daily for the appearance of plaques, and quantitation was performed on day 7 postinfection. Infections of A549 and 293 cells by Ad37 and Ad19p were performed in identically. Infectivity of Ad2 was determined by plaquing on 293 cells.
Recombinant wild-type and mutant Ad fiber proteins.
Recombinant Ad2 fiber protein was produced in insect cells using baculovirus and purified by anion-exchange chromatography as previously described (16). Recombinant Ad19p and Ad37 fiber proteins containing an N-terminal polyhistidine sequence were produced in bacteria by using the pET expression system (Novagen). The highly similar Ad19p and Ad37 fiber DNAs (GenBank accession no. X94485 and X94484, respectively) were PCR amplified from viral genomic DNA by using 5′ and 3′ primers with the sequences CAATCTAGATCAAACAGGCTCCGGGTGGAA and GCAACTCGAGTCATTCTTGGGCAATATAGG, respectively (the underlined sequences correspond to XbaI [5′ primer] and XhoI [3′ primer] endonuclease restriction sites). PCRs were performed at 94°C (denaturation), 55°C (annealing), and 72°C (extension; 30 cycles), using PCR polymerase (Qiagen). The amplified DNA fragments were digested with XbaI and XhoI and then subcloned into the bacterial expression vector pEM1 (NheI/XhoI cloning sites). Following transformation of Escherichia coli BL21(DE3), individual colonies were selected for fiber expression by induction with 1 mM isopropyl-β-d-thiogalactopyranoside for 1 h at 37°C. Colonies displaying high fiber expression as assessed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis analysis were then used for large-scale fiber production. The recombinant fiber proteins were isolated from bacterial lysates by using Ni+ chelating columns as recommended by the manufacturer (Qiagen).
To construct single-site mutations in the wild-type Ad37 fiber, additional PCRs were performed as follows. Using pEM1/Ad37 DNA as a template, we carried out two separate PCRs: one with the wild-type 3′ primer (see above) and a primer for the mutant sequence 5′-TTTTTATTTCTGGATTTGTCTT (the mutated sequence is underlined); and a second reaction with the 5′ wild-type primer and a mutant primer (ACAAATCCAGAAATAAAAAGTT-3′). An equimolar amount of each of the gel-purified PCR fragments was then mixed and subjected to a second round of PCR amplification using the 5′ and 3′ wild-type fiber primers to generate the entire fiber DNA sequence containing the mutations. The PCR products were then digested with XhoI/XbaI and cloned into pEM1 DNA as described above. A mutant Ad19p fiber expression plasmid was similarly prepared with the primers ACAAATCCAAAAATAAAAAGTT-3′ and TTTTTATTTTTGGATTTGTCTT-5′ and with pEM1/Ad19p DNA as a template. The DNA sequence of each of the fiber construct was subjected to automated sequencer analysis and confirmed to be correct.
Purified wild-type and mutant fiber proteins were analyzed on 8% polyacrylamide gels under denaturing (2% SDS, boiled) or native (no SDS, nonboiled) conditions.
Ad fiber and virus binding assays.
For cell binding experiments, purified recombinant fiber proteins or viral particles were labeled with 125I by using Iodogen (Pierce). Briefly, 500-μg aliquots of virus or recombinant fiber proteins were incubated for 20 min at 22°C in an Iodogen-coated tube containing 1 mCi of Na125I. Iodinated proteins were separated from free 125I by passage through a PD10 column (Pharmacia). Generally, fiber proteins were labeled to a specific activity of 1 × 107 to 2 × 107 cpm/μg, while viral particles were labeled to a specific activity 5 × 106 to 8 × 106 cpm/μg. Binding of radiolabeled virus particles or of fiber protein was then assayed by incubating 106 Chang C, A549, or 293 cells in suspension with 106 cpm of the labeled virus proteins at 4°C for 2 h. Nonspecific binding was determined by incubating cells and labeled proteins in the presence of a 100-fold excess of unlabeled virus or fiber protein. After the cells were washed four times in ice-cold phosphate-buffered saline, specific binding was calculated by subtracted the nonspecific binding from the total counts per minute bound. For competition experiments, Chang C cells were preincubated with a 100-fold excess of unlabeled recombinant Ad2 or Ad37 fiber protein for 1 h at 4°C prior to addition of 106 cpm of 125I-labeled Ad37 virus particles.
Scatchard analysis of Ad37 fiber binding was performed as follows. After being detached by using 10 mM EDTA, Chang C cells were washed several times in serum-free medium to remove residual EDTA and resuspended to 2 × 107/ml in medium. Cell aliquots of 100 μl were then incubated with various amounts of 125I-labeled Ad37 fiber in a total volume of 200 μl for 2 h at 4°C with constant agitation. The cells were then washed four times with PBS, and the cell pellets were counted in a gamma counter. Nonspecific binding (approximately 10% of the total) was determined by incubating the cells in a 100-fold excess of unlabeled Ad37 fiber protein. The data were then analyzed by Scatchard analyses using the EBDA-LIGAND PC software program (Biosoft, Ferguson, Mo.).
Sequence analysis and molecular modeling of Ad fiber proteins.
Sequences of the fiber knob domains of Ad37 and Ad19p were aligned with those of Ad5, Ad2, and Ad7 by using the computer-based software program AMPS (2, 3, 7). The crystal structure of the Ad5 fiber knob (17) was used a template for Ad37 and Ad19p modeling. Sequence alignments suggested an insertion and a deletion in the Ad5 template sequence with respect to the Ad37 and Ad19p target sequences. Therefore, residue changes, insertions, and deletions in Ad39 and Ad19p, according to the sequence alignment, were modeled onto the Ad5 structure by using the graphics program O (7).
RESULTS
Comparison of Ad2 and Ad37 interactions with conjunctival and epithelial cells.
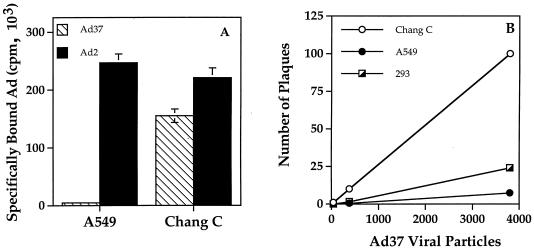
To better understand the molecular basis of Ad host cell tropism, we evaluated the binding of 125I-labeled Ad37 to human epithelial or conjunctival cells. As it is not practical to examine virus interactions with primary human conjunctival cells due to the relatively low abundance of these cells in human tissue and their limited proliferative capacity, we used the immortalized Chang C human conjunctival cell line. Ad37 bound at 30-fold higher levels to Chang C conjunctival cells than to A549 lung epithelial cells (Fig. 1A). Ad2, a serotype belonging to subgroup C, showed similar levels of binding to both A549 and Chang C cells. Conjunctival cells were also more susceptible to Ad37 infection than A549 or 293 human embryonic kidney cells (Fig. 1B). These studies indicated that Ad37, a virus associated with EKC, displays preferential binding to and infection of human conjunctival cells.
FIG. 1.
Comparison of Ad37 and Ad2 interactions with different cell types. (A) Specific binding of 125I-labeled Ad37 and Ad2 to A549 or Chang C cells. Nonspecific binding, which was subtracted from the total counts per minute bound, was determined by incubation in the presence of a 100-fold excess of unlabeled virus particles. (B) Susceptibility of A549, 293, and Chang C cells to Ad37 infection, determined by plaque assay.
Role of the Ad37 fiber protein in virus attachment.
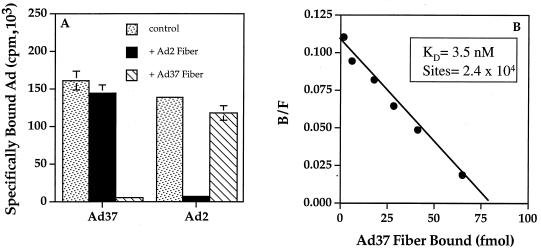
Previous studies (15) demonstrated that the Ad2 fiber protein mediates high-affinity interaction of the virus with host cells whereas cell integrins promote the subsequent step of internalization/penetration. To determine whether the Ad37 fiber protein was also responsible for attachment of Ad37 to cells, we examined the binding of 125I-labeled Ad37 to conjunctival cells in the presence or absence of soluble recombinant fiber proteins derived from different Ad serotypes. The specific binding of Ad37 to conjunctival cells was competed by soluble recombinant Ad37 fiber but not by the fiber protein of Ad2, a subgroup C virus (Fig. 2A). In parallel studies, soluble Ad2 fiber was capable of blocking attachment of Ad2 virions to cells but did not alter Ad37 virus attachment. These studies indicate that the Ad37 fiber protein is responsible for virus attachment and that the receptor for Ad37 is distinct from that of Ad2. In further studies, we examined by Scatchard analysis binding of the Ad37 fiber protein to its cellular receptor. The Ad37 fiber saturation binding data revealed that the Ad37 fiber protein exhibits high affinity for conjunctival cells (KKd = 3.5 nM) and that there are 2 × 104 fiber binding sites per cell (Fig. 2B).
FIG. 2.
Quantitative binding analysis of Ad37 interaction with conjunctival cells. (A) Binding of 125I-labeled Ad37 or Ad2 virions to Chang C cells assayed in the presence or absence (control) of a 100-fold excess of unlabeled Ad37 or Ad2 fiber proteins; (B) Scatchard analysis of recombinant Ad37 fiber binding, performed as described in Materials and Methods. The number of receptor binding sites for Ad37 fiber protein was calculated by dividing the value extrapolated on the x axis (78.5 fmol, equivalent to 4.7 × 1010 molecules of fiber), based on a molecular mass of 132 kDa for the trimeric fiber protein), by the number of cells (2 × 106). B, bound; F, free.
Different group D Ads exhibit distinct binding and infection properties.
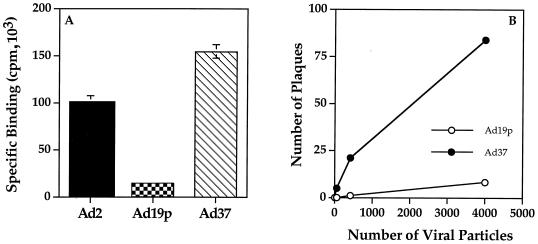
As other subgroup D Ads are also associated with ocular infections, we examined the interaction of one of them Ad19p, with conjunctival cells. Somewhat surprisingly, we observed only low levels of Ad19p binding to conjunctival cells compared to Ad37 (Fig. 3A). The low level of Ad19p binding to these cells also correlated with a substantial reduction in infectivity compared to Ad37 (Fig. 3B). These findings indicated that Ad37 exhibits a selective tropism for Chang C conjunctival cells and raised the possibility that differences in sequences of the fiber proteins of Ad37 and Ad19p could account for this.
FIG. 3.
Relative binding efficiency and infection of Chang C conjunctival cells by different Ad serotypes. (A) Binding of radiolabeled Ad2, Ad19p, and Ad37 to Chang C cells, measured as described in Materials and Methods; (B) infection of Chang C cells with various amounts of Ad19p and Ad37 virus particles, assayed by plaque formation.
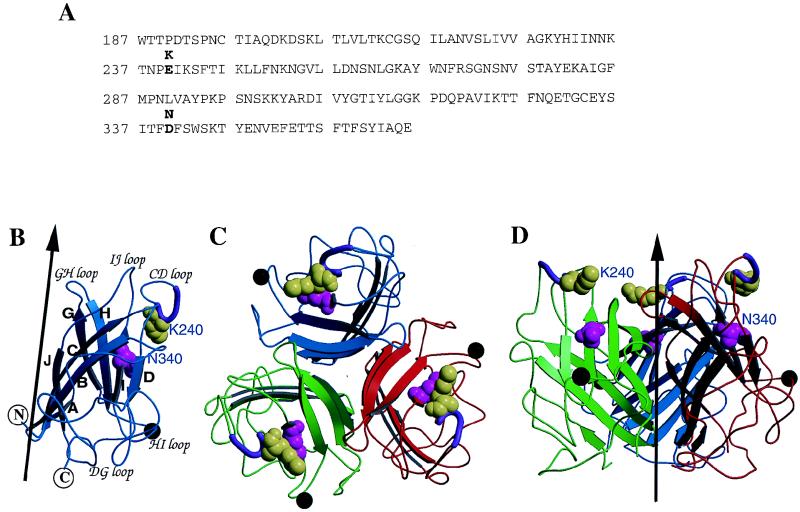
As an initial assessment of the role of individual amino acid residues in fiber binding, we performed molecular modeling of the Ad37 and Ad19p fiber domains based on the crystal structure of the Ad5 fiber (17). The sequences of the Ad37 and Ad5 fiber domains are similar (53% identity overall). There are only two amino acid residue differences in the fiber distal domain (knob) (1): at position 240, where Ad37 fiber contains lysine while Ad19p has glutamic acid (Fig. 4A); and at position 340, where Ad37 contains asparagine while Ad19p contains aspartic acid. Modeling of the Ad37 fiber knob required an insertion of four residues between amino acids 451 and 452 of the CD loop, while a deletion of six residues was carried out on the HI loop between residues 539 and 546 in the template of the Ad5 fiber crystal structure (Fig. 4B). The final model suggested that the two amino acid changes in the fiber knob are located at the apex of the exposed CD loop (K240E) or within the individual folding domains of the knob monomers (N340D). The three-dimensional model also indicates that residue K240E is more solvent exposed than residue N340D. These studies suggested that K240 rather than N340 could play the major role in receptor recognition on conjunctival cells.
FIG. 4.
Sequences and modeling of the Ad37 and Ad19p fiber knob domains. (A) Amino acid sequence of the Ad19p fiber knob domain (5), with the different residues (K240 and N340) present in Ad37 indicated in bold. (B) Ribbon diagram of the modeled Ad19p fiber knob domain. Shown are an insertion of four residues in the CD loop between residues 451 and 452 (purple tube) and a deletion of six residues on the HI loop, between residues 539 and 546 (black sphere). The two residues that are different between Ad37 and Ad19p are K240E and N340D, shown in yellow-green and magenta space-filling models, respectively. The black arrow represents the trimeric fiber axis. Secondary structural elements are labeled according to the Ad5 fiber structure (12). (C) Ribbon representation of the trimeric knob of Ad37 viewing down the fiber axis. Each fiber domain is indicated with a different color. (D) Side view of the trimeric knob. The fiber axis is represented by the dark arrow. The side chain of K240E is highly solvent accessible (60% exposed) compared to that of N340D, which is only 20% exposed.
Identity of the amino acid residue that regulates Ad37 fiber binding.
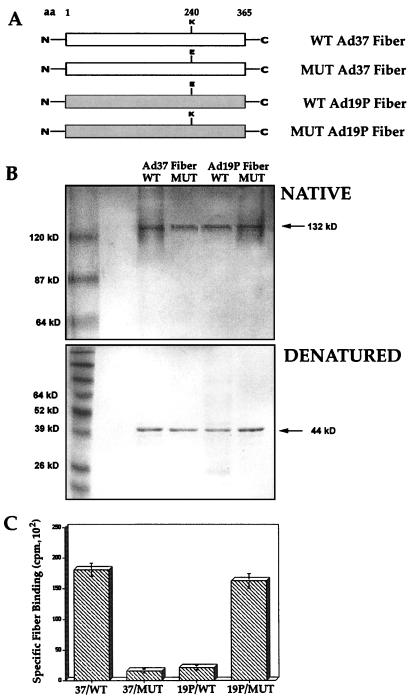
To investigate whether the K240E sequence in the Ad fiber knob played a role in binding to conjunctival cells, we generated recombinant forms of wild-type and site-specific mutants of the Ad37 (K to E) and Ad19p (E to K) fiber proteins in bacteria (Fig. 5A). The wild-type and mutant fiber proteins were expressed in their native trimeric form (132 kDa) as assessed by electrophoresis on a non-SDS (native) polyacrylamide gel (Fig. 5B). The purified proteins were assayed for the ability to bind to conjunctival cells. Recombinant wild-type Ad37 but not the wild-type Ad19p fiber exhibited significant binding to conjunctival cells (Fig. 5C). A single amino acid substitution in the Ad37 fiber protein (K240 to E) abrogated binding. In contrast, the reverse substitution in the Ad19p fiber (E240 to K) restored binding to conjunctival cells (Fig. 5C). The E-to-K mutant Ad19p fiber was also capable of competing the binding of 125I-labeled Ad37 to cells (data not shown). In parallel studies, substitution of the amino acids at position 340 in the two different fiber proteins did not affect binding (data not shown). Together, these studies indicate that K240 plays a major role in conferring virus binding of Ad37 to conjunctival cells.
FIG. 5.
Structures and binding properties of Ad37 and Ad19p fiber proteins containing a single amino acid (aa) substitution in the knob domain. (A) Schematic representation of the recombinant Ad37 and Ad19p wild-type (WT) and mutant (MUT) proteins; (B) Analysis of recombinant fiber proteins by electrophoresis on polyacrylamide gels under native (no SDS, nonboiled) or denaturing (2% SDS, boiled) conditions; (C) binding of wild-type or mutant 125I-labeled fiber proteins to Chang C cells, analyzed as described in Materials and Methods.
DISCUSSION
We undertook this study to gain further insight into the molecular basis by which human Ad recognizes cellular receptors. Previous studies demonstrated that a number of different Ad serotypes, representing distinct subgroups, adhere to cells via interaction with a 46-kDa protein (CAR). CAR is a member of the immunoglobulin superfamily whose normal cell function is unknown. The fact that not all Ad serotypes, including several members of subgroup B, do not use CAR for binding (11) indicates that other, as yet unidentified cell receptors play a role in infection by several other Ads. To gain insight into the identity of some of these receptors, we examined host cell interactions of several subgroup D viruses associated with EKC in the anticipation that they might display selective tropism for certain ocular cell types. Interestingly Ad37 but not Ad19p showed selective binding and infection of Chang C conjunctival cells, and this was shown to be mediated by the fiber protein (Fig. 1 and 3). As is true for multiple Ad serotypes (9), Ad37 also interacts with αv integrins, the receptors used for internalization and penetration (8).
Since there are only two amino acid differences in the fiber distal domain between these highly related Ad serotypes (1), and one of these residues, K240E, is predicted to lie at the apex of the exposed CD loop on the fiber knob (Fig. 4), we examined the role of the lysine246 residue in fiber binding. Consistent with the model predictions, substitution of the lysine to glutamic acid at position 240 abrogated Ad37 fiber binding whereas the reverse substitution in Ad19p conferred the ability of this protein to bind human conjunctival cells (Fig. 5C). These studies confirm that a single amino residue in an exposed loop of the fiber knob promote virus interaction with its cellular receptor. It is likely that other amino acid sequences in the Ad37 fiber knob also contribute to virus attachment; however, it appears that K240 plays a pivotal role in this event.
The results of this study indicate that a specific cell receptor may facilitate Ad37 attachment to conjunctival cells. Our preliminary studies indicate that the cell receptor is a protein, since treatment of cells with several different proteases abrogates Ad37 binding. Interestingly, other subgroup D viruses such as Ad8 may use the same receptor since Ad8 partially competes Ad37 binding to conjunctival cells (data not shown). Further characterization of this receptor should provide further insights into the earliest events in Ad infection of cells and may allow the development of therapeutic approaches to restrict ocular infections by subgroup D Ads.
ACKNOWLEDGMENTS
This work was supported by NIH grants HL54352 and EY11431.
We express our appreciation to Catalina Hope and Joan Gausepohl for preparation of the manuscript.
Footnotes
Manuscript no. 11946-IMM of The Scripps Research Institute.
REFERENCES
- 1.Arnberg N, Mei Y-F, Wadell G. Fiber genes of adenoviruses with tropism for the eye and the genital tract. Virology. 1997;227:239–244. doi: 10.1006/viro.1996.8269. [DOI] [PubMed] [Google Scholar]
- 2.Barton G J. Protein multiple sequence alignment and flexible pattern. Methods Enzymol. 1990;183:403–428. doi: 10.1016/0076-6879(90)83027-7. [DOI] [PubMed] [Google Scholar]
- 3.Barton G J, Sternberg M J. A strategy for the rapid multiple alignment of protein sequences. J Mol Biol. 1987;198:327–337. doi: 10.1016/0022-2836(87)90316-0. [DOI] [PubMed] [Google Scholar]
- 4.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 5.Curtis S, Wilkinson G W G, Westmoreland D. An outbreak of epidemic keratoconjunctivitis caused by adenovirus type 37. J Med Microbiol. 1998;47:91–94. doi: 10.1099/00222615-47-1-91. [DOI] [PubMed] [Google Scholar]
- 6.Everitt E, Meador S A, Levine A S. Synthesis and processing of the precursor to the major core protein of adenovirus type 2. J Virol. 1977;21:199–214. doi: 10.1128/jvi.21.1.199-214.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 8.Mathias P, Galleno M, Nemerow G R. Interactions of soluble recombinant integrin avβ5 with human adenoviruses. J Virol. 1998;72:8669–8675. doi: 10.1128/jvi.72.11.8669-8675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathias P, Wickham T J, Moore M, Nemerow G. Multiple adenovirus serotypes use αv integrins for infection. J Virol. 1994;68:6811–6814. doi: 10.1128/jvi.68.10.6811-6814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Philipson L, Lonberg-Holm K, Pettersson U. Virus-receptor interaction in an adenovirus system. J Virol. 1968;2:1064–1075. doi: 10.1128/jvi.2.10.1064-1075.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roelvink P W, Lizonova A, Lee J G M, Li Y, Bergelson J M, Finberg R W, Brough D E, Kovesdi I, Wickham T J. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, E, and F. J Virol. 1998;72:7909–7915. doi: 10.1128/jvi.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruigrok R W H, Barge A, Albiges-Rizo C, Dayan S. Structure of adenovirus fibre. II. Morphology of single fibres. J Mol Biol. 1990;215:589–596. doi: 10.1016/S0022-2836(05)80170-6. [DOI] [PubMed] [Google Scholar]
- 13.Tomko R P, Xu R, Philipson L. HCAR and MAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viswalingam N D. Adenovirus keratoconjunctivitis: an enigma. Eye. 1993;7(Pt. 3 Suppl.):5–7. [PubMed] [Google Scholar]
- 15.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Integrins αvβ3 and αvβ5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 16.Wickham T J, Nemerow G R. Optimization of growth methods and recombinant protein production in BTI-Tn-5B1-4 insect cells using the baculovirus expression system. Biotechnol Prog. 1993;9:25–30. doi: 10.1021/bp00019a004. [DOI] [PubMed] [Google Scholar]
- 17.Xia D, Henry L J, Gerard R D, Deisenhofer J. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 Å resolution. Structure. 1994;2:1259–1270. doi: 10.1016/s0969-2126(94)00126-x. [DOI] [PubMed] [Google Scholar]