Abstract
Most chronic liver diseases progress to liver fibrosis, which, when left untreated, can lead to cirrhosis and hepatocellular carcinoma. MicroRNA (miRNA)-targeted therapeutics have become attractive approaches to treat diseases. In this study, we investigated the therapeutic effect of miR-155 inhibition in the bile duct ligation (BDL) mouse model of liver fibrosis and evaluated the role of miR-155 in chronic liver fibrosis using miR-155-deficient (miR-155 knockout [KO]) mice. We found increased hepatic miR-155 expression in patients with cirrhosis and in the BDL- and CCl4-induced mouse models of liver fibrosis. Liver fibrosis was significantly reduced in miR-155 KO mice after CCl4 administration or BDL. To assess the therapeutic potential of miR-155 inhibition, we administered an rAAV8-anti-miR-155 tough decoy in vivo that significantly reduced liver damage and fibrosis in BDL. BDL-induced protein levels of transforming growth factor β (TGF-β), p-SMAD2/3, and p-STAT3 were attenuated in anti-miR-155-treated compared with control mice. Hepatic stellate cells from miR-155 KO mice showed attenuation in activation and mesenchymal marker expression. In vitro, miR-155 gain- and loss-of-function studies revealed that miR-155 regulates activation of stellate cells partly via STAT3 signaling. Our study suggests that miR-155 is the key regulator of liver fibrosis and might be a potential therapeutic target to attenuate fibrosis progression.
Keywords: MT: Non-coding RNAs, anti-miR-155 tough decoy, SMAD2/3, STAT3, CCl4, BDL, fibrosis, stellate cells, TGFbeta, Caspase3
Graphical abstract

Szabo and colleagues found an imperative role of miR-155 in liver fibrosis. Most chronic liver diseases progress to fibrosis, which can lead to cirrhosis. Therapeutic inhibition or deficiency of miR-155 caused reduction in liver fibrosis in preclinical mouse models. miR-155-targeted therapy might be an effective approach to treat liver fibrosis.
Introduction
The burden of chronic liver disease is increasing globally. Liver fibrosis is the final common pathway in progression of almost all chronic liver diseases. Nonalcoholic fatty liver disease (NAFLD), nonalcoholic steatohepatitis (NASH), hepatitis B or C infection, alcohol abuse, autoimmune diseases, and cholestatic diseases are the major contributing factors to fibrosis.1,2 If left untreated, persistent and excessive liver damage leads to inflammation, fibrosis, cirrhosis, and, ultimately, hepatocellular carcinoma (HCC).3,4 Initially, liver fibrosis is a healing response to injury; however, when prolonged injury progresses to advanced fibrosis and cirrhosis, it becomes irreversible, causes impaired liver function, and leads to high morbidity and mortality.3 Independent of etiology, hepatic fibrosis is an important predictor of prognosis in chronic liver disease.2,3 Despite this major clinical need, development of therapeutic approaches to attenuate liver fibrosis has been challenging, and to date there is no treatment for liver fibrosis.5
Fibrogenesis is a complex mechanism involving different cell types and cellular and extracellular signaling. This is a dynamic, complex process that involves recruitment and activation of different cell types, such as inflammatory cells, hepatic stellate cells (HSCs), and other extracellular matrix (ECM)-producing cells, including portal fibroblasts, hepatocytes, cholangiocytes, and bone marrow-derived cells.4,5,6 During liver injury, hepatocyte damage prompts inflammatory signaling and aberrant activation of inflammatory pathways, resulting in increased synthesis and altered deposition of ECM components as well as impaired regeneration and wound healing responses.4,5,6
HSCs are the main liver cells involved in fibrogenesis, and irrespective of etiology, activation of HSCs is the common pathway leading to fibrosis.7,8 Various factors are known to stimulate and inhibit activation of HSCs, and recently, microRNAs (miRNAs) have emerged as new regulators of HSCs.9,10 miRNAs are small non-coding endogenous RNAs that are involved in fine-tuning of cellular pathways. miRNAs regulate expression of genes by either mRNA degradation or translational repression in normal physiological and disease milieus.11 In recent years, miRNA-based therapeutics have gained attention; various preclinical studies have been carried out to test their efficacy in vivo, and several miRNA-based therapeutics have been advanced into the clinical phase.12,13
Various miRNAs, such as the miR-29 family, miR-34, and miR-21, have been shown to regulate the liver fibrogenesis pathway.9,14 miR-155 is a multifactorial miRNA that regulates various cellular processes, such as inflammation, metabolism, oncogenesis, and metastasis; is uniquely expressed in non-parenchymal and parenchymal cells; and plays a key role in various diseases.11,15 Increased miR-155 expression has been reported in fibrotic disorders of the kidneys,16 lungs,17 and liver.11 miR-155 regulates various transcription factors involved in metabolism and fibrogenesis, such as peroxisome proliferator-activated receptor gamma (PPARγ), liver X receptor alpha (LXRα), CCAAT/enhancer-binding protein alpha (C/EBPα), C/EBPβ, and silent mating-type information regulation 2 homolog (SIRT1).11,18,19,20
Because of the pleiotropic effects of miR-155, we hypothesized that miR-155 targets various genes to regulate the fibrogenesis pathway, which will have a synergistic effect on the liver fibrosis phenotype. In this study, we investigated the therapeutic potential of miR-155 inhibition in a well-established mouse model of liver fibrosis, bile duct ligation (BDL). Also, the role of miR-155 in advanced liver fibrosis was evaluated using the miR-155 knockout (KO) mouse model system after CCl4 treatment. Here, we demonstrated that either deletion of miR-155 (miR-155 KO mice) or in vivo inhibition of miR-155 (rAAV8-miR-155 tough decoy [TuD]) results in attenuation of CCl4- or BDL-induced chronic liver fibrosis. The results also revealed that miR-155 regulates activation of HSCs.
Results
Attenuation of fibrosis in miR-155 KO mice after chronic liver injury
miR-155 is a multifunctional miRNA and plays important roles in various cellular pathways.15 Here, we found an increase in hepatic miR-155 levels in patients with alcohol-associated cirrhosis and in Hepatitis C Virus (HCV) infection-associated cirrhosis (Figure 1A). Therefore, to elucidate the precise role of miR-155 in advanced liver fibrosis, we used well-established mouse models of liver injury: CCl4 and BDL. CCl4 causes hepatocyte damage, necrosis, inflammation, fibrosis, and cirrhosis, whereas BDL is the most widely used experimental model to explore special aspects of occlusive cholestasis.21 BDL stimulates proliferation of biliary epithelial cells and oval cells, and continuous cholestasis leads to chronic inflammation, which damages bile duct cells and liver cells and causes fibrosis.21 A significant increase of miR-155 expression was observed in the livers of wild-type (WT) mice after 9 weeks of CCl4 treatment compared oil-treated control mice (Figure 1B). miR-155 levels were undetected in the livers of miR-155 KO mice. CCl4 treatment caused induction of bridging fibrosis in WT, mice as indicated by Sirius red-stained collagen fibers, whereas miR-155 KO mice exhibited attenuation of bridging fibrosis (Figure 1C). Consistent with this finding, we found increases in hydroxyproline content (Figure 1D) and procollagen 1α transcripts (Figure 1E) in CCl4-treated WT mice, and a significant attenuation of these parameters was observed in miR-155 KO mice. CCl4-treated WT mice displayed an increase in alpha smooth muscle actin (αSMA) protein levels that was significantly attenuated in miR-155 KO CCl4 mice (Figure 1F).
Figure 1.
miR-155 KO mice exhibit attenuation of CCl4- or BDL-induced liver fibrosis
Hepatic miR-155 levels were increased in patients with alcohol-associated or chronic HCV infection-associated cirrhosis or control individuals (n = 10–12/group), as determined by TaqMan miRNA qRT-PCR assay. RNU48 was used to normalize Cq values (A). C57BL/6 wild-type (WT) or miR-155 KO male mice received either corn oil (n = 6) or CCl4 (n = 9) for 9 weeks or underwent either sham (n = 4) or bile duct ligation (BDL; n = 8) for 2 weeks. Hepatic miR-155 expression was analyzed by qRT-PCR, and snoRNA-202 was used as an internal control (B and G). miR-155 KO mice showed a decrease in liver fibrosis markers. Representative Sirius red-stained (collagen fibers) liver sections are shown (C and H), and the ImageJ-quantified stained area is presented (H) (n = 8). Total collagen amount was determined from liver by measuring hydroxyproline content biochemically (D) (n = 6–8) and pro-collagen 1α expression by qRT-PCR (E). αSMA mRNA (I) and protein levels (F and J) were evaluated by qRT-PCR and western blot analysis, respectively. Western blot analysis of vimentin expression is shown in (K). Densitometry units are presented as fold change compared with either oil or sham WT mice after normalization with GAPDH (F and J–K, n = 6–8). 18S mRNA was used to normalize the Cq values. Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 compared with oil-treated or sham WT mice.
Next, we measured expression of miR-155 in the BDL model and found a significant increase in miR-155 levels in WT mice 14 days after BDL compared with sham mice (Figure 1G), and no changes in its levels were observed 2 days after BDL. To elucidate the role of miR-155 in this model, WT or miR-155 KO mice underwent either BDL or sham treatment for 14 days. WT mice displayed an increase in liver fibrosis, as indicated by Sirius red-stained collagen fibers, and in contrast, miR-155 KO mice showed significantly reduced fibrosis after BDL (Figure 1H). While expression of αSMA was increased at the mRNA (Figure 1I) and protein (Figure 1J) levels in WT BDL mice compared with sham controls, its expression was significantly attenuated in miR-155 KO BDL mice (Figures 1I and 1J). Vimentin, a mesenchymal marker, was increased in WT mice after BDL, whereas miR-155 KO BDL mice displayed a significantly decrease in its expression (Figure 1K). These results suggest a regulatory role of miR-155 in promoting liver fibrosis irrespective of etiology.
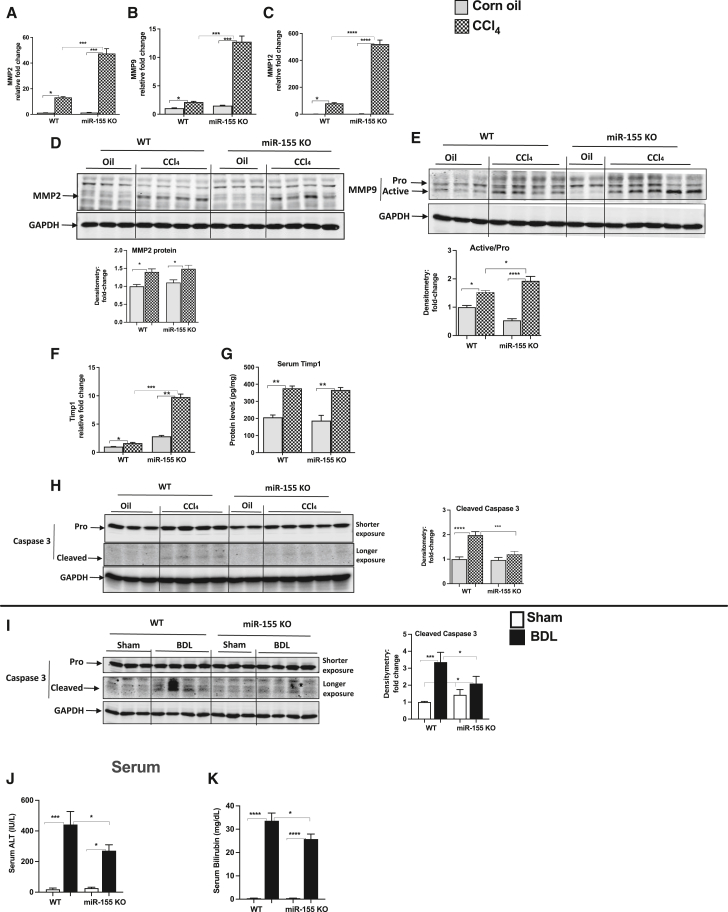
Increase in matrix metalloproteinases and attenuation of cell death markers in miR-155 KO mice
Matrix metalloproteinases (MMPs) play a central role in fibrosis;22 therefore, we determined their expression and found increases in MMP2, MMP9, and MMP12 transcripts in WT CCl4-treated mice, and this increase in MMPs was further augmented in miR-155 KO CCl4 mice (Figures 2A–2C). MMP2 protein expression was increased to a similar extent in both genotypes (CCl4-treated WT or miR-155 KO mice) (Figure 2D). A further significant increase in active MMP9 (active versus pro form) was observed in miR-155 KO mice compared with WT mice after CCl4 treatment (Figure 2E). Because we found increased transcription of MMPs, next we checked the levels of tissue inhibitors of MMPs (TIMP1) and found enhanced TIMP1 mRNA transcripts in miR-155 KO mice compared with WT mice after CCl4 treatment (Figure 2F). At the protein level, a similar increase in serum TIMP1 levels was found between the genotypes (Figure 2G).
Figure 2.
Increase in MMPs and attenuation of cell death markers in miR-155 KO mice
Expression of MMP2 (A), MMP9 (B), and MMP12 (C) was determined by qRT-PCR from the liver tissues. MMP2 (D) and MMP9 (E) protein levels were evaluated by western blot analysis. mRNA (F) and serum (G) levels of TIMP1 were determined by qRT-PCR and ELISA, respectively. A decrease in cell death markers was found in miR-155 KO mice. Caspase-3 protein levels were determined by western blot analysis (H and I). Densitometry units are presented as fold change compared with either WT oil-treated (H) or sham (I) mice after normalization with GAPDH (n = 6–8). Serum ALT (J) and bilirubin (K) levels were evaluated using a biochemical assay. Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 compared with oil-treated or sham WT mice.
CCl4 treatment as well as BDL induce hepatocyte damage; therefore, we evaluated the expression of caspase-3 because it is involved in cell death.23 In CCl4- or BDL-treated mice, cleaved caspase-3 levels were increased in WT mice, whereas miR-155 KO mice displayed either no increase in CCl4-induced cleaved caspase-3 levels (Figure 2H), or significantly attenuated levels were found in BDL-induced fibrosis (Figure 2I). Further, BDL in WT mice resulted in robustly increased serum alanine aminotransferase (ALT) levels that were significantly attenuated in miR-155 KO BDL mice (Figure 2J). In obstructive cholestasis, secretion of bile into the bile duct and intestine is impaired, which causes accumulation of bile acids and bilirubin into the circulation.24 Therefore, we checked bilirubin levels and found an increase in bilirubin levels in the serum of WT mice after BDL compared with sham controls, and miR-155 deficiency attenuated its increase after BDL (Figure 2K).
rAAV-mediated in vivo inhibition of miR-155 results in reduction of BDL-induced fibrosis
Because we found a reduction in liver fibrosis in miR-155 KO mice after CCl4 treatment or BDL, we hypothesized that direct inhibition of miR-155 in vivo will attenuate fibrosis. To inhibit miR-155 in vivo, we developed an recombinant adeno-associated virus (rAAV8) vector system expressing either anti-miR-155 TuD or rAAV8-scramble (control) (Figures S1A and S1B). AAV-mediated delivery of anti-miR-TuD is an effective way to inhibit miRNA function in vivo.25 WT mice were treated with either rAAV8-scramble (rAAV control) or rAAV8-anti-miR-155-TuD (rAAV miR-155 TuD) 4 days prior to BDL (Figure 3A). We found a significant increase in primary (pri) miR-155 levels in rAAV control-BDL mice after 14 days of treatment, and this increase was prevented in rAAV miR-155 TuD-treated mice (Figure 3B). BDL-induced mature miR-155 expression was also attenuated in anti-miR-155 TuD-treated mice compared with control mice (Figure 3C). Gaussia activity was checked for transgene expression (Figure S1C). Consistent with miR-155 KO findings, we found a reduction in liver fibrosis after inhibition of miR-155 in vivo. Anti-miR-155 TuD-treated mice displayed attenuation of BDL-induced Sirius red-stained collagen fibers compared with control mice (Figure 3D). Further, mRNA (Figure 3E) and protein levels (Figure 3F) of αSMA were increased in BDL-rAAV control mice, and this increase in αSMA expression was significantly attenuated in rAAV miR-155 TuD-treated mice after BDL (Figures 3E and 3F). Vimentin expression was also increased in BDL-rAAV control mice, while its levels were significantly reduced in BDL-rAAV miR-155 TuD-treated mice (Figure 3F). Next, we evaluated liver/hepatocyte damage markers, such as ALT (serum) and caspase-3 levels (liver), and found significant attenuation of BDL-induced ALT (Figure 3G), and a BDL-induced increase in cleaved caspase-3 was prevented (Figure 3H) in rAAV miR-155 TuD mice compared with rAAV control mice.
Figure 3.
Attenuation of BDL-induced liver fibrosis in mice treated with rAAV8-miR-155 TuD
WT male mice received the control or miR-155 TuD rAAV8 vector, followed by sham or BDL treatment (n = 4–7) for indicated times (A). Primary (B) and mature miR-155 (C) expression was quantified by TaqMan qRT-PCR assay. GAPDH (pri) or snoRNA-202 (mature) was used to normalize Cq values. In vivo inhibition of miR-155 caused attenuation of BDL-induced liver fibrosis. Representative Sirius red-stained (collagen fibers) liver sections are shown, and the ImageJ-quantified stained area is presented (D). αSMA mRNA levels were quantified, and 18S mRNA was used to normalize the Cq (E). αSMA and vimentin protein levels were evaluated by western blot analysis (F). Serum ALT levels were assessed using a biochemical assay (G). Protein levels of caspase-3 (H) were determined by western blot analysis. Densitometry units are presented as fold change compared with respective controls after normalization with GAPDH (F and H, n = 4–7). Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 compared with control vector sham-treated mice.
miR-155 inhibition decreases activation of fibrotic genes in BDL-induced fibrosis
Transforming growth factor β (TGF-β) is a master profibrogenic cytokine known to induce fibrosis by activation of quiescent HSCs through canonical (SMAD) or non-canonical (STAT3, c-Jun N-terminal kinases [JNK], extracellular signal-regulated kinase [ERK], etc.) pathways,26 and miR-155 has been shown to target these genes.20 Therefore, we evaluated the effect of miR-155 inhibition and found that BDL caused an increase in serum TGF-β levels in control mice, which were significantly decreased in rAAV miR-155 TuD mice (Figure 4A). A similar observation was made in miR-155 KO mice, where WT mice showed a significant increase in serum TGF-β levels, which was prevented in KO mice after BDL (Figure 4B). Next, we checked SMAD expression and found a BDL-induced SMAD2/3 expression in rAAV control BDL mice, whereas this increase in SMADs was prevented in rAAV miR-155 TuD mice (Figure 4C). Because TGF-β induces phosphorylation of SMADs,26 we next checked activation of SMAD2/3. BDL caused a significant increase in phosphorylation of SMAD2/3 in rAAV control mice, which was either prevented (p-SMAD2) or attenuated (p-SMAD3) in rAAV miR-155 TuD mice (Figure 4D). STAT3 signaling plays an essential role in liver fibrosis, and an increase in STAT3 expression has been reported in humans and rodent models of fibrosis.27 miR-155 regulates STAT3 signaling;28 therefore, we assessed hepatic STAT3 levels and found an increase in phospho-STAT3 levels in rAAV control BDL mice (Figure 4E). This increase in phospho-STAT3 was significantly attenuated in rAAV miR-155 TuD mice (Figure 4E). Similarly the BDL-induced increase in phospho-STAT3 was significantly decreased in miR-155 KO mice compared with WT-BDL mice (Figure 4F). Altogether, these findings indicate that inhibition of miR-155 using an rAAV miR-155 TuD vector system can attenuate progression of liver fibrosis in a mouse model via the TGF-β-SMAD2/3 and STAT3 pathways. These results also mirror the findings observed in miR-155 KO BDL or CCl4-treated mice and confirm an essential regulatory role of miR-155 in liver fibrosis.
Figure 4.
miR-155 inhibition decreases activation of fibrotic genes in BDL-induced fibrosis
WT male mice received the control or miR-155 TuD rAAV8 vector followed by sham or BDL treatment as indicated above. An increase in TGF-β protein levels was prevented after miR-155 inhibition (A) or deletion (B), as determined by ELISA from the serum samples. miR-155 inhibition caused a reduction in BDL-induced SMAD2/3 (total and phosphorylated) and activation of STAT3. The protein levels of SMAD2/3 (C), phospho-SMAD2/3 (D), and STAT3 (total and phosphorylated, E and F) were determined by western blot analysis. Densitometry units are presented as fold change compared with the respective controls after normalization with either GAPDH (C and D) or total STAT3 (E and F). Data are presented as mean ± SEM (n = 4–7). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 compared with control vector sham or sham WT mice.
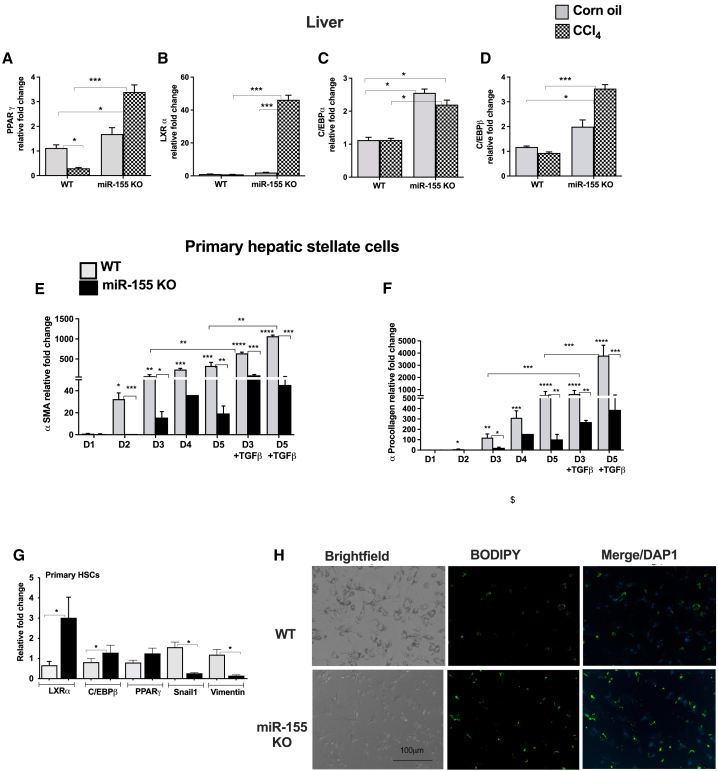
Primary HSCs of miR-155 KO mice display decreased markers of activation
Adipogenic transcription factors are central in driving fibrogenesis.7 Because we found attenuation of collagen fibers and HSC activation markers (αSMA and vimentin) in miR-155 KO mice, we sought to evaluate the expression of transcription factors involved in fibrotic events, such as PPARγ, LXRα, C/EBPα, and C/EBPβ (miR-155 target genes). We found that PPARγ transcripts were significantly decreased in WT CCl4 mice, whereas miR-155 KO CCl4-treated mice displayed a significant increase in its expression (Figure 5A). Compared with control mice, a significant increase in LXRα (Figure 5B), C/EBPα (Figure 5C), and C/EBPβ (Figure 5D) transcripts was found in miR-155 KO mice after CCl4 treatment (Figures 5B–5D). Even oil-treated miR-155 KO mice displayed an increase in baseline expression of C/EBPα and C/EBPβ (Figures 5C and 5D).
Figure 5.
miR-155 KO HSCs display decreased markers of activation
An increase in expression of adipogenic genes was found in miR-155 KO mice after CCl4 treatment. The expression of PPARγ (A), LXRα (B), C/EBPα (C), and C/EBPβ (D) was measured from the livers by qRT-PCR (A, n = 6–8). HSCs isolated from miR-155 KO mice showed a decrease in activation markers and increase in fat droplets. HSCs from WT or miR-155 KO chow-fed mice were cultured in vitro for the indicated times, and expression of αSMA (E), procollagen 1α (F), and LXRα, C/EBPβ, PPARγ, snail1, and vimentin (G) was determined by qRT-PCR (n = pool of 12–15 mice). 18S mRNA was used to normalize the Cq values. BODIPY staining of HSCs was performed after 5 days of in vitro culture (H). Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 compared with control WT mice (A–D) or WT cells (E–H).
Next, we evaluated the potential role of miR-155 in HSCs using an in vitro culture model system. WT HSCs showed a sustained robust and time-dependent increase in αSMA (Figure 5E) and pro-collagen 1α (Figure 5F) mRNA transcripts over the course of activation in culture (days 2–5), and a further significant increase in αSMA and pro-collagen 1α levels was observed after TGF-β treatment. In contrast, a significant attenuation of αSMA and pro-collagen 1α levels was found in miR-155 KO stellate cells with or without TGF-β treatment (Figures 5E and 5F).
Quiescent HSCs highly express putative adipogenic transcription factors,7 and we found that miR-155 KO stellate cells exhibited increases in LXRα, C/EBPβ, and PPARγ transcripts compared with WT stellate cells (Figure 5G). The mesenchymal markers, snail1 and vimentin transcripts, were significantly decreased in miR-155 KO stellate cells (Figure 5G). Consistent with these findings, we found that miR-155 KO stellate cells contained more lipid droplets (BODIPY stained) compared with WT stellate cells even after 5 days in culture (Figure 5H). All of these findings suggest attenuated HSCs activation in the absence of miR-155 and indicate a role of miR-155 in HSC activation.
miR-155 regulates activation of HSCs
Next, we evaluated expression of miR-155 in activated HSCs and found an increase in miR-155 levels in WT cells after 5 days of in vitro culture (Figure 6A). To elucidate the role of miR-155 in regulation of HSC activation, we carried out in vitro gain- and loss-of-function mechanistic studies using LX-2 cells, a human HSC cell line. miR-155 overexpression using a miR-155 mimic induced increases in miR-155 expression with or without TGF-β treatment in LX-2 cells (Figure 6B). miR-155 overexpression in TGF-β-naive cells resulted in a significant increase in the mRNA (Figure 6C) and protein levels (Figure 6D) of αSMA compared with negative control-treated cells. A further significant increase in αSMA expression (mRNA and protein) was found after TGF-β treatment in miR-155-overexpressing cells compared with negative control cells (Figures 6C and 6D). In contrast, miR-155 inhibition using a miRNA-155-specific inhibitor (Figure 6E) resulted in a significant decrease in TGF-β-induced αSMA mRNA (Figure 6F) and protein expression (Figure 6G) compared with negative control-treated cells. Activation of STAT3 in HSCs is associated with fibrogenesis,29 and we found attenuated STAT3 signaling after miR-155 inhibition or deletion in vivo (Figure 4). Consistent with the in vivo findings, we found that in vitro overexpression of miR-155 promoted (Figure 6H) whereas inhibition of miR-155 inhibited (Figure 6I) phosphorylation of STAT3 in LX-2 cells with or without TGF-β treatment. Further, to evaluate the role of STAT3 in miR-155-mediated HSC activation, a small inhibitor of STAT3, Stattic, was used.30 Stattic is known to inhibit dimerization, activation, and nuclear translocation of STAT3.31 Our results indicated that the miR-155 inhibitor or Stattic alone decreases phosphorylation of STAT3 in naive or TGF-β-treated cells (Figure 6J). Cells treated with the miR-155 inhibitor and Stattic showed a further decrease in activation of STAT3. No significant changes in total STAT3 were found (Figure 6J). The miR-155 inhibitor or Stattic caused significant inhibition of TGF-β-induced αSMA expression (Figure 6J). Collagen 1 levels were significantly decreased in naive or TGF-β-treated cells after miR-155 inhibitor or Sttatic treatment (Figure 6K). No further decrease in αSMA and collagen 1 protein levels was found in cells treated with the miR-155 inhibitor and Sttatic compared with Sttatic-only-treated cells (Figures 6J and 6K). Altogether, these findings suggest a role of miR-155 in regulating HSCs activation, partly via STAT3.
Figure 6.
miR-155 regulates activation of HSCs
miR-155 expression was increased in WT- HSCs after in vitro culture. miR-155 levels were quantified using qRT-PCR (n = 6) (A). miR-155 overexpression or inhibition in LX-2 cells caused an increase (C and D) or attenuation (F and G) of αSMA levels, respectively. miR-155 (B and E) and αSMA mRNA (C and F) levels were quantified by qRT-PCR. Sttatic, a small inhibitor of STAT3, was used to inhibit activation of STAT3 (J). The protein levels of αSMA (D, G, and J), STAT3 and phospho-STAT3 (H–J), and collagen 1 (K) were determined by western blot analysis. Densitometry units are presented as fold change compared with respective control cells after normalization with GAPDH or STAT3 (n = 3–4). Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 compared with respective control cells.
Discussion
Advanced liver fibrosis and cirrhosis (the end stage of fibrosis) are major causes of morbidity and mortality worldwide, and liver transplantation is the only effective therapy.4,6 Fibrosis is a healing response to liver injury caused by different etiologies of chronic liver injury. Because liver fibrogenesis is a complex phenomenon with dynamic interactions between cellular and molecular processes, defining common mechanisms of liver fibrosis is a prerequisite for innovation of new therapeutic targets. miRNA-based therapeutics are an emerging area of research, and various miRNA therapies are currently undergoing clinical trials.
In this study, we demonstrated the therapeutic potential of miR-155 inhibition in liver fibrosis and identified its role in HSC activation. miR-155 is highly expressed in immune cells and upregulated in various cancers and chronic diseases.15 Here, we found upregulated miR-155 expression in human livers with cirrhosis as well as in mice after CCl4- or BDL-induced liver fibrosis. miR-155 KO mice exhibited attenuation of CCl4 -induced chronic liver fibrosis. This was further confirmed in BDL-induced liver fibrosis, where we found attenuation of the fibrotic phenotype in miR-155 KO mice and after therapeutic inhibition of miR-155 with anti-miR-155 TuD.
During liver injury, damaged hepatocytes, inflammatory factors, and cytokines trigger the various signaling pathways involved in hepatic fibrogenesis, which causes activation of αSMA and increased collagen synthesis in HSCs.4,22 Our results revealed decreases in BDL- or CCl4-induced cell death (ALT and cleaved caspase-3 levels) and attenuation of αSMA expression in miR-155 KO mice. Previously, we reported diminished diet-induced (NAFLD)19 or alcohol-induced (ALD; alcohol associatde liver disease)11 liver injury in miR-155 KO mice, suggesting a role of miR-155 in liver injury irrespective of etiology.
ECM degradation and remodeling are controlled by a fine balance between MMPs and TIMPs, a crucial step in resolution of liver fibrosis. Depending on the cell injury milieu, MMPs either promote or resolve fibrosis.32 In the CCl4 mouse model, MMP2-deficient mice have been found to exhibit augmentation of liver fibrosis and TIMP1.22 Also, MMP2 has been shown to inhibit type I collagen synthesis in activated HSCs.32 It has been demonstrated that Kupffer cell (KC)-derived MMP9 plays a key regulatory role in matrix remodeling and architectural repair during fibrosis regression, and increased MMP9 expression is associated with faster resolution of fibrosis.33
Consistent with these reports, we found an increase in the active form of MMP9 protein in miR-155 KO mice upon induction of advanced fibrosis with CCl4. This was associated with decreases in procollagen 1α levels, hydroxyproline content, and Sirius red-stained collagen fibers in miR-155 KO mice. During fibrosis resolution, elevation of the ratio of MMP9/TIMP1 has been reported to promote fibrosis resolution.33 In accordance with this study, we found increased TIMP1 expression in miR-155 KO CCl4-treated mice. miR-155 has been shown to regulate MMP2 and MMP9 levels either directly or indirectly.34 MMP9 is highly expressed in KCs,33 and previously we have demonstrated a role of miR-155 in KCs/macrophages.35 It is plausible that enhanced MMP9 expression in miR-155 KO mice may promote fibrosis resolution and could be one of the factors of the observed phenotype in our study.
miRNA-based therapeutics have advantages over existing RNA-based therapies because miRNAs are naturally occurring molecules and act by targeting multiple genes in one pathway, causing a wider specific response.36 Currently, various miRNAs delivery methods are being used, such as lipid nanoparticles, polymers, viral vector systems, and exosomes. rAAV-mediated miRNA delivery is a simple and efficient way to explore the potential of miRNA-based therapeutics in mammals.25 Our approach to inhibit miRNA-155 function in the liver using the AAV8-anti-miR-155 TuD vector was effective to ameliorate BDL-induced liver fibrosis. Markers of liver injury, such as ALT and cleaved caspase-3, and mesenchymal markers, such as vimentin, αSMA, and Sirius red-stained collagen fibers, were reduced in anti-miR-155 TuD mice. Indeed, these findings mirrored the phenotype of miR-155 KO mice (BDL or CCl4), suggesting an imperative role of miR-155 in liver fibrosis.
TGF-β/SMAD signaling plays an essential role in fibrosis. Smad2/3 are the intracellular mediators of the TGF-β signal transduction pathway, and activation of SMAD2/3 signaling promotes fibrosis by regulating expression of various profibrotic genes.3 Our results revealed attenuation of Smad2/3 expression (total and phosphorylated) after therapeutic inhibition of miR-155 in vivo, which was associated with reduced fibrosis.
HSCs play a pivotal role in development of liver fibrosis. During fibrogenesis, HSCs are activated and differentiated into myofibroblasts, resulting in increased amounts of ECM.7 Adipogenic transcription factors, such as PPARγ, LXRα, C/EBPα, and C/EBPβ, are highly expressed in quiescent HSCs.7 Transcriptional silencing of the PPARγ gene is required for conversion of HSCs into myofibroblasts, and we have demonstrated previously that miR-155 directly regulates the PPARγ gene.11 A decrease in hepatic PPARγ levels has been reported in fibrotic livers, and PPAR agonists have been shown to improve liver fibrosis.37 Consistent with these studies, we found a reduction in PPARγ levels in WT mice, whereas miR-155 KO mice exhibited a significant increase in its expression after CCl4 treatment. Also, an increase in LXRα, C/EBPα, and C/EBPβ transcripts was observed in miR-155 KO mice. Because quiescent HSCs express high levels of adipogenic factors, and miR-155 has been shown to regulate them, we hypothesized that miR-155 regulates activation of HSCs.
Our results revealed that miR-155 KO stellate cells are less fibrogenic because expression of αSMA and procollagen 1α was decreased in culture-activated miR-155 KO cells with or without TGF-β treatment. Also, an increase in adipogenic transcription factors and decrease in mesenchymal markers (vimentin and snail1) were detected in culture-activated miR-155 KO stellate cells. Consistent with these findings, an increase in the number of quiescent HSCs was found in culture-activated miR-155 KO stellate cells. Collectively, our findings suggest that miR-155 KO stellate cells are less activated, suggesting a role of miR-155 in HSC activation.
The role of miR-155 in HSCs was further elucidated using LX-2 cells, a human HSC cell line, and our in vitro results suggest that miR-155 regulates activation of stellate cells because overexpression of miR-155 resulted in an increase in αSMA expression with or without TGF-β treatment. Consistent with in vivo findings, where absence of miR-155 (KO mice, CCl4 or BDL) or miR-155 inhibition in vivo (anti-miR-155 TuD) caused a reduction in αSMA levels, in vitro inhibition of miR-155 in LX-2 cells resulted in a decrease in αSMA expression after TGF-β treatment. Our in vitro findings suggest that miR-155 regulates STAT3 signaling because overexpression of miR-155 in LX-2 cells induced STAT3 phosphorylation and vice versa. Further, Stattic-mediated inhibition of STAT3 phosphorylation caused attenuation of TGF-β-induced activation of LX-2 cells. These findings corroborated previous studies, where activation of STAT3 in HSCs has been shown to be associated with fibrogenesis.29,38 Our results further indicate a role of miR-155 in HSC activation via its ability to modulate various genes of fibrogenesis.
The STAT3 signaling pathway integrates several profibrotic signals and is a key mediator of liver fibrosis, and miR-155 has been shown to activate STAT3 signaling.27,39 STAT3 overexpression has been found in liver fibrosis, and its activation can have an anti- or pro-inflammatory effect because of its model-dependent and cell-specific functions.27,29,38 Pharmacological inhibition of STAT3 has been shown to attenuate development of liver fibrosis, suggesting that it promotes fibrogenesis.27,30,38 In accordance with this, we found that therapeutic inhibition of miR-155 (anti-miR-155 TuD mice) or deficiency of miR-155 (KO mice) caused attenuation of the BDL-induced increase in STAT3 phosphorylation and reduction in fibrosis.
Besides HSCs, KCs are core mediators in the pathogenesis of chronic liver disease, and we have shown previously that macrophage miR-155 contributes to liver inflammation,35 and increased miR-155 in KCs and hepatocytes is associated with steatohepatitis.11 The anti-miR-155 TuD vector is under control of the U6 promoter, a strong ubiquitous promoter that should express in all/or most of the cells.25 Considering the complexity of the in vivo system, it is likely that the fibrotic phenotype we observed in the absence of miR-155 (KO mice) or its inhibition (anti-miR-155 TuD) is a result of its synergistic effects on different liver cell types (such as KCs, HSCs, and hepatocytes), and on various genes (such as STAT3, TGF-β/SMAD signaling, PPARγ, LXRα, C/EBPα, C/EBPβ, MMPs, and αSMA) directly or indirectly. The role of miR-155 in liver fibrosis seems to be independent of etiology because we have shown previously that miR-155 KO mice exhibit attenuation of early (alcohol)11 or mid-stage (diet; NASH20 or NAFLD19) liver fibrosis, underscoring the hierarchy of miR-155 regulation. These important criteria should be taken into consideration for miR-155-based therapeutics.
Overall, our findings highlight miR-155 as an important regulator of liver fibrosis via its ability to modulate multiple targets/cell types of the fibrogenesis pathway. In conclusion, our results establish a role of miR-155 in activation of HSCs and in promoting liver fibrosis. Therefore, therapeutic inhibition of miR-155 might be an effective approach to ameliorate liver fibrosis.
Materials and methods
Mice and liver injury models
Eight- to twelve-week-old male C57Bl/6 WT or miR-155-deficient (KO, total body) mice were used to carry out in vivo CCl4 studies. The study was approved by the University of Massachusetts Medical School (UMMS; Worcester, MA, USA) and Beth Israel Deaconess Medical Center (BIDMC; Boston, MA, USA) institutional animal use and care committees. The breeding pairs of miR-155 KO and C56BL/6 WT control mice were obtained from the Jackson Laboratory (Bar Harbor, ME, USA). The colonies were maintained in the animal facility of UMMS and BIDMC as described previously.11 To induce chronic liver fibrosis, WT or miR-155 male KO mice (n = 6–9) were treated either with corn oil (vehicle control) or CCl4 (0.6 mL/kg intraperitoneally [i.p.]) twice weekly for 9 weeks. Mice were sacrificed 72 h after the last CCl4 injection. For BDL, 10- to 14-week-old male mice (WT or miR-155 KO, n = 4–8) were used, and the surgical procedure was performed as described previously.24 Briefly, mice were anesthetized with isoflurane, placed on the operating pad, and shaved. Shaved skin was disinfected with 70% ethanol, a midline incision was made in the upper abdomen, and the common bile duct was identified, isolated, and ligated with two surgical knots. After irrigating the abdomen with 0.9% NaCl, the abdomen and peritoneum were closed with a running suture. Sham treatment was performed similarly but without ligation of the bile duct. Animals were monitored during recovery, and to reduce the depressant effects of surgery on food and water consumption, they were treated with the systemic analgesic agent buprenorphine (1.2 mg/kg).
At the conclusion of the experiment, blood was collected by cheek bleeding, and mice were sacrificed. Liver tissues were harvested and snap frozen for proteins and in RNAlater (QIAGEN, Germany) for RNA extraction. All samples were stored at −80°C until further analysis. For AAV-mediated inhibition of miR-155 in vivo, 10- to 12-week-old male mice (WT, n = 4–7) were used, and 6 × 1011 viral particles of rAAV8 containing either anti-miR-155 TuD or rAAV8-scrambled (control) were administered intravenously 4 days prior to initiation of BDL surgery (Figures S1A and S1B).25 Mice were sacrificed 14 days after surgery.
Histopathology
A portion of the liver tissue was fixed in 10% phosphate-buffered formalin, embedded in paraffin blocks, and processed for histological analysis. Sections were stained with Sirius red using standard protocols. The slides were observed under light microscopy at 4× magnification. Quantification of Sirius red staining was performed using ImageJ software.40
Human samples
Human liver samples were obtained from the National Institutes of Health Liver Tissue Cell Distribution System (Minneapolis, MN, USA). Liver samples were from patients with alcohol-associated cirrhosis (n = 10), HCV patients with cirrhosis (n = 12), and respective controls.
Cell culture and transfection
LX-2 cells were cultured in DMEM low-glucose medium containing 2% fetal bovine serum (FBS) and 1% antibiotics as described previously.41 LX-2 cells were kindly gifted by Dr. Scot Friedman (Mount Sinai School of Medicine, New York, NY, USA). For transfection studies, cells were transfected either with miRNA mimic negative control #1 (negative control) or miR-155 mimic at 6 pmol or miRNA inhibitor negative control #1 (negative control) or miR-155-inhibitor at 30 pmol for 24 h using Lipofectamine RNAi Max reagent (Thermo Fisher Scientific). The concentrations of the miRNA mimic and inhibitors were used according to the manufacturer’s recommendations (Thermo Fisher Scientific). 24 h after transfection, cells were treated with 10 ng/mL TGF-β for 48 h or left untreated. At the end of treatments, the medium was removed, and cells were washed with 1× PBS twice and lysed with Qiazol for total RNA or whole-cell protein extraction. For STAT3 inhibition, 24 h after transfection with the miR-155 inhibitor or negative control (30 pmol), LX-2 cells were treated with 10 μM Stattic (a small-molecule inhibitor of STAT3 activation)30 with or without TGF-β for 48 h or left untreated, and cells were lysed and used for western blot analysis.
HSC isolation
HSCs were isolated from WT and miR-155 KO chow-fed mice by in situ liver perfusion using 0.075% collagenase as described previously.42 Briefly, after cannulation of the portal vein, livers were perfused in situ with 1× EGTA perfusion buffer for 5 min at 37°C at a flow rate of 5.5 mL/min and perfused with digestion buffer I (0.075% collagenase in GBSS) for ∼10 min. The perfused liver was removed, placed into a sterile Petri dish in digestion buffer II (0.009% collagenase in GBSS), and further disintegrated with sterile scissors. The suspension was transferred to a 50-mL conical tube containing digestion buffer II and incubated at 37°C in a rotating incubator (150–200 rpm) for 20–30 min. After incubation, the suspension was filtered using a 70-μm cell strainer (BD Biosciences), and the filtrate was diluted with GBSS to 50 mL and centrifuged at 500 rpm for 5 min at room temperature. The supernatant was transferred to a new tube and centrifuged, and this step was repeated twice to remove hepatocytes. The last centrifugation step was performed at 2,000 rpm for 10 min at 4°C. The supernatant was discarded, the cell pellet was suspended in GBSS and transferred to a new 15-mL tube, and then centrifugation was performed at 1,600 rpm for 10 min at 4°C. HSCs were purified via differential centrifugation using OptiPrep density gradients (Sigma, USA). The supernatant was removed carefully, and the cell pellet was suspended in 15% OptiPrep and mixed gently (layer 1). After this, 11.5% OptiPrep (layer 2) was added very slowly to layer 1 so that the two layers did not mix. On the top of layer 2, GBSS washing buffer was added (layer 3), and the tubes were centrifuged at 3,000 rpm for 17 min at 4°C. The cell fraction at the interface of layer 2 and layer 3 was gently transferred to a new 15-mL tube. The cell suspension was diluted in GBSS washing buffer and centrifuged at 1,600 rpm for 10 min at 4°C, and this step was repeated twice. After the second wash, the cell pellet was suspended in culture medium in a laminar hood and plated at a density of 5 × 104 cells/well onto 6-well plates in RPMI 1640 medium containing 10% FBS, 10% horse serum (Gibco, 26050-088), and 1% antibiotics for indicated times. The cells were harvested on day 1, day 3, and day 5 for total RNA isolation. On day 3 and day 5, cells were either treated with 5 ng/mL TGF-β or left untreated.
BODIPY staining
For staining of neutral lipids, primary HSCs were isolated from WT and miR-155 KO chow diet-fed mice and cultured for 5 days as described above. On day 5, cells were washed with 1× PBS twice and incubated with BODIPY 493/503 reagent (Thermo Fisher Scientific, catalog number D3922) at a concentration of 1 μM for 20 min. Cells were stained with DAPI for nuclei (1: 10,000) and observed under a microscope at 20× magnification (Evon).
Enzyme-linked immunosorbent assay (ELISA)
Protein levels of TIMP1 and TGF-β were measured from serum samples using ELISA according to the manufacturer’s instructions (R&D Systems).
Biochemical analysis
ALT levels were measured from serum samples using a kinetic method (TECO Diagnostics, catalog number A560-240) according to the manufacturer’s instructions. Bilirubin levels were determined from serum using the Bilirubin Colorimetric Assay Kit (Sigma, catalog number MAK126) according to the manufacturer’s instructions.
Hydroxyproline assay
The collagen content was evaluated by quantification of the hydroxyproline amount from the liver tissues as described previously.11 Briefly, liver tissues were homogenized in water, and 12 N HCl was added to hydrolyze the samples in a glass tube at 120°C for 3 h, followed by measurement according to manufacturer’s instructions (BioVision, K555-100). Hydroxyproline levels were calculated against standard curves of 4-hydroxy-L-proline and expressed as micrograms of hydroxyproline per gram of liver tissue.
Western blot analysis
Whole-cell lysates were extracted from the frozen liver tissues or cells as described previously,20 and an equal amount of protein was used for western blotting. The denatured samples were separated in polyacrylamide gel, transferred onto a nitrocellulose membrane, and probed with specific primary antibodies, followed by horseradish peroxidase (HRP)-labeled secondary antibodies. Primary antibodies for α smooth muscle actin (Abcam, MA, USA, catalog number ab15734), MMP2 (Abcam, catalog number 92536), MMP9 (Abcam, catalog number 38898), caspase-3 (Cell Signaling Technology, MA, USA), vimentin (CST, catalog number 5741S), STAT3 (CST, catalog number 12640), phospho-STAT3 (CST, catalog number 9145), SMAD2/3 (CST, catalog number 8685), p-SMAD2/3 (CST, catalog number 8828), collagen 1 (Abcam, catalog number ab138492), and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Proteintech, catalog number 60004-1-Ig) were used.
RNA extraction and quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the livers or cells using the Direct-zol RNA MiniPrep Kit with on-column DNA digestion (Zymo Research, CA, USA) as described previously.11 For mRNA analysis, cDNA was transcribed from total RNA using iScript Reverse Transcription Supermix for qRT-PCR (Bio-Rad, Hercules, CA, USA). Quantitative real-time PCR was performed using iCycler (Bio-Rad). The mean of the quantification cycle (Cq) of all samples was normalized to 18S mRNA expression, and relative expression was calculated by the ΔΔCt method. Primer sequences were the same as we have described previously.11 For miRNA quantification, cDNA synthesis and qRT-PCR were performed from total RNA using TaqMan miRNA assays (Applied Biosystems, USA). Cq of the target miRNA was normalized either with small nucleolar RNA (snoRNA)-202 (mouse) or RNU48 (human) Cq. For detection of pri-miR-155, cDNA was generated using iScript Reverse Transcription Supermix for qRT-PCR (Bio-Rad), and TaqMan primers and probes specific for mmu-pri-miR-155-FAM and mouse GAPDH-fluorescein (FAM) (control) were used (Applied Biosystems, USA). The qPCR, western blot, and in-vitro experiments were performed with independent replicates.
Statistical analysis
Statistical significance was determined using either Mann-Whitney test or one-way ANOVA. Data are shown as mean ± SEM and considered statistically significant at p < 0.05. GraphPad Prism software v.7 or v.8 was used for analysis.
Acknowledgments
The authors thank Drs. Dora Lippai, Timea Cask, Joseph Cerasuolo, Kevin Pelland, and the late Karen Kodys for assistance with animal work. Research reported in this publication was supported by the National Institute on Alcohol Abuse and Alcoholism of the National Institutes of Health under award R01AA0207440 (to G.S.).
Author contributions
S.B., conceptualization, methodology, investigation, validation, formal analysis, visualization, and writing; Y.Z., conceptualization, methodology, investigation, validation, formal analysis, visualization, and writing – review & editing; P.T.N., D.C., A.Z., and Y.W., investigation, validation, and visualization ; J.X., methodology and investigation; G.G., methodology and resources; G.S., conceptualization, resources, funding acquisition, writing – review & editing, and supervision.
Declaration of interests
G.S. is the Editor-in-Chief of Hepatology Communication. G.S. reports being a consultant for Durect Corp., Generon/Evive, Terra Firma, Quest, Pandion Therapeutics, Surrozen, Takeda, Novartis, Merck, and Pfizer. G.S. has received grants from NIAAA and NIA and has stock options with Glympse Bio, Satellite Bio, and Zomagen. She holds intellectual property rights with Up to Date.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2023.07.012.
Supplemental information
Data and code availability
Data supporting the findings of this study are available upon request.
References
- 1.Kisseleva T., Brenner D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat. Rev. Gastroenterol. Hepatol. 2021;18:151–166. doi: 10.1038/s41575-020-00372-7. [DOI] [PubMed] [Google Scholar]
- 2.Lackner C., Tiniakos D. Fibrosis and alcohol-related liver disease. J. Hepatol. 2019;70:294–304. doi: 10.1016/j.jhep.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Walton K.L., Johnson K.E., Harrison C.A. Targeting TGF-β Mediated SMAD Signaling for the Prevention of Fibrosis. Front. Pharmacol. 2017;8:461. doi: 10.3389/fphar.2017.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuda M., Seki E. Hepatic Stellate Cell-Macrophage Crosstalk in Liver Fibrosis and Carcinogenesis. Semin. Liver Dis. 2020;40:307–320. doi: 10.1055/s-0040-1708876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman S.L. Hepatic Fibrosis: Emerging Therapies. Dig. Dis. 2015;33:504–507. doi: 10.1159/000374098. [DOI] [PubMed] [Google Scholar]
- 6.Atta H., El-Rehany M., Hammam O., Abdel-Ghany H., Ramzy M., Roderfeld M., Roeb E., Al-Hendy A., Raheim S.A., Allam H., Marey H. Mutant MMP-9 and HGF gene transfer enhance resolution of CCl4-induced liver fibrosis in rats: role of ASH1 and EZH2 methyltransferases repression. PLoS One. 2014;9:e112384. doi: 10.1371/journal.pone.0112384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.She H., Xiong S., Hazra S., Tsukamoto H. Adipogenic transcriptional regulation of hepatic stellate cells. J. Biol. Chem. 2005;280:4959–4967. doi: 10.1074/jbc.M410078200. [DOI] [PubMed] [Google Scholar]
- 8.Trivedi P., Wang S., Friedman S.L. The Power of Plasticity-Metabolic Regulation of Hepatic Stellate Cells. Cell Metabol. 2021;33:242–257. doi: 10.1016/j.cmet.2020.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ezhilarasan D. MicroRNA interplay between hepatic stellate cell quiescence and activation. Eur. J. Pharmacol. 2020;885:173507. doi: 10.1016/j.ejphar.2020.173507. [DOI] [PubMed] [Google Scholar]
- 10.Sabater L., Locatelli L., Oakley F., Hardy T., French J., Robinson S.M., Sen G., Mann D.A., Mann J. RNA sequencing reveals changes in the microRNAome of transdifferentiating hepatic stellate cells that are conserved between human and rat. Sci. Rep. 2020;10:21708. doi: 10.1038/s41598-020-78776-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bala S., Csak T., Saha B., Zatsiorsky J., Kodys K., Catalano D., Satishchandran A., Szabo G. The pro-inflammatory effects of miR-155 promote liver fibrosis and alcohol-induced steatohepatitis. J. Hepatol. 2016;64:1378–1387. doi: 10.1016/j.jhep.2016.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 13.Hanna J., Hossain G.S., Kocerha J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019;10:478. doi: 10.3389/fgene.2019.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roderburg C., Urban G.-W., Bettermann K., Vucur M., Zimmermann H., Schmidt S., Janssen J., Koppe C., Knolle P., Castoldi M., et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 15.Mahesh G., Biswas R. MicroRNA-155: A Master Regulator of Inflammation. J. Interferon Cytokine Res. 2019;39:321–330. doi: 10.1089/jir.2018.0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W., Li X., Tang Y., Chen C., Jing R., Liu T. miR-155-5p Implicates in the Pathogenesis of Renal Fibrosis via Targeting SOCS1 and SOCS6. Oxid. Med. Cell. Longev. 2020;2020:6263921. doi: 10.1155/2020/6263921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christmann R.B., Wooten A., Sampaio-Barros P., Borges C.L., Carvalho C.R.R., Kairalla R.A., Feghali-Bostwick C., Ziemek J., Mei Y., Goummih S., et al. miR-155 in the progression of lung fibrosis in systemic sclerosis. Arthritis Res. Ther. 2016;18:155. doi: 10.1186/s13075-016-1054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller A.M., Gilchrist D.S., Nijjar J., Araldi E., Ramirez C.M., Lavery C.A., Fernández-Hernando C., McInnes I.B., Kurowska-stolarska M. MiR-155 has a protective role in the development of non-alcoholic hepatosteatosis in mice. PLoS One. 2013;8:e72324. doi: 10.1371/journal.pone.0072324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bala S., Ganz M., Babuta M., Zhuang Y., Csak T., Calenda C.D., Szabo G. Steatosis, inflammasome upregulation, and fibrosis are attenuated in miR-155 deficient mice in a high fat-cholesterol-sugar diet-induced model of NASH. Lab. Invest. 2021;101:1540–1549. doi: 10.1038/s41374-021-00626-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Csak T., Bala S., Lippai D., Kodys K., Catalano D., Iracheta-Vellve A., Szabo G. MicroRNA-155 Deficiency Attenuates Liver Steatosis and Fibrosis without Reducing Inflammation in a Mouse Model of Steatohepatitis. PLoS One. 2015;10:e0129251. doi: 10.1371/journal.pone.0129251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bao Y.-L., Wang L., Pan H.-T., Zhang T.-R., Chen Y.-H., Xu S.-J., Mao X.L., Li S.W. Animal and Organoid Models of Liver Fibrosis. Front. Physiol. 2021;12:666138. doi: 10.3389/fphys.2021.666138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onozuka I., Kakinuma S., Kamiya A., Miyoshi M., Sakamoto N., Kiyohashi K., Watanabe T., Funaoka Y., Ueyama M., Nakagawa M., et al. Cholestatic liver fibrosis and toxin-induced fibrosis are exacerbated in matrix metalloproteinase-2 deficient mice. Biochem. Biophys. Res. Commun. 2011;406:134–140. doi: 10.1016/j.bbrc.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 23.Thapaliya S., Wree A., Povero D., Inzaugarat M.E., Berk M., Dixon L., Papouchado B.G., Feldstein A.E. Caspase 3 inactivation protects against hepatic cell death and ameliorates fibrogenesis in a diet-induced NASH model. Dig. Dis. Sci. 2014;59:1197–1206. doi: 10.1007/s10620-014-3167-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuang Y., Xu H.C., Shinde P.V., Warfsmann J., Vasilevska J., Sundaram B., Behnke K., Huang J., Hoell J.I., Borkhardt A., et al. Fragile X mental retardation protein protects against tumour necrosis factor-mediated cell death and liver injury. Gut. 2020;69:133–145. doi: 10.1136/gutjnl-2019-318215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie J., Ameres S.L., Friedline R., Hung J.-H., Zhang Y., Xie Q., Zhong L., Su Q., He R., Li M., et al. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat. Methods. 2012;9:403–409. doi: 10.1038/nmeth.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabregat I., Caballero-Díaz D. Transforming Growth Factor-β-Induced Cell Plasticity in Liver Fibrosis and Hepatocarcinogenesis. Front. Oncol. 2018;8:357. doi: 10.3389/fonc.2018.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Öztürk Akcora B., Vassilios Gabriël A., Ortiz-Perez A., Bansal R. Pharmacological inhibition of STAT3 pathway ameliorates acute liver injury in vivo via inactivation of inflammatory macrophages and hepatic stellate cells. FASEB Bioadv. 2020;2:77–89. doi: 10.1096/fba.2019-00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen R., Wang Y., Wang C.-X., Yin M., Liu H.-L., Chen J.-P., Han J.-Q., Wang W.-B. MiRNA-155 mediates TAM resistance by modulating SOCS6-STAT3 signalling pathway in breast cancer. Am. J. Transl. Res. 2015;7:2115–2126. [PMC free article] [PubMed] [Google Scholar]
- 29.Nunez Lopez O., Bohanon F.J., Wang X., Ye N., Corsello T., Rojas-Khalil Y., Chen H., Chen H., Zhou J., Radhakrishnan R.S. STAT3 Inhibition Suppresses Hepatic Stellate Cell Fibrogenesis: HJC0123, a Potential Therapeutic Agent for Liver Fibrosis. RSC Adv. 2016;6:100652–100663. doi: 10.1039/C6RA17459K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakraborty D., Šumová B., Mallano T., Chen C.-W., Distler A., Bergmann C., Ludolph I., Horch R.E., Gelse K., Ramming A., et al. Activation of STAT3 integrates common profibrotic pathways to promote fibroblast activation and tissue fibrosis. Nat. Commun. 2017;8:1130. doi: 10.1038/s41467-017-01236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schust J., Sperl B., Hollis A., Mayer T.U., Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem. Biol. 2006;13:1235–1242. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 32.Radbill B.D., Gupta R., Ramirez M.C.M., DiFeo A., Martignetti J.A., Alvarez C.E., Friedman S.L., Narla G., Vrabie R., Bowles R., et al. Loss of matrix metalloproteinase-2 amplifies murine toxin-induced liver fibrosis by upregulating collagen I expression. Dig. Dis. Sci. 2011;56:406–416. doi: 10.1007/s10620-010-1296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng M., Ding J., Wang M., Zhang J., Zhu X., Guan W. Kupffer-derived matrix metalloproteinase-9 contributes to liver fibrosis resolution. Int. J. Biol. Sci. 2018;14:1033–1040. doi: 10.7150/ijbs.25589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang L., Zheng Z., Zhou Q., Bai X., Fan L., Yang C., Su L., Hu D. miR-155 promotes cutaneous wound healing through enhanced keratinocytes migration by MMP-2. J. Mol. Histol. 2017;48:147–155. doi: 10.1007/s10735-017-9713-8. [DOI] [PubMed] [Google Scholar]
- 35.Bala S., Marcos M., Kodys K., Csak T., Catalano D., Mandrekar P., Szabo G. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor {alpha} (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease. J. Biol. Chem. 2011;286:1436–1444. doi: 10.1074/jbc.M110.145870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winkle M., El-Daly S.M., Fabbri M., Calin G.A. Noncoding RNA therapeutics - challenges and potential solutions. Nat. Rev. Drug Discov. 2021;20:629–651. doi: 10.1038/s41573-021-00219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boyer-Diaz Z., Aristu-Zabalza P., Andrés-Rozas M., Robert C., Ortega-Ribera M., Fernández-Iglesias A., Broqua P., Junien J.-L., Wettstein G., Bosch J., Gracia-Sancho J. Pan-PPAR agonist lanifibranor improves portal hypertension and hepatic fibrosis in experimental advanced chronic liver disease. J. Hepatol. 2021;74:1188–1199. doi: 10.1016/j.jhep.2020.11.045. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z., Li J., Xiao W., Long J., Zhang H. The STAT3 inhibitor S3I-201 suppresses fibrogenesis and angiogenesis in liver fibrosis. Lab. Invest. 2018;98:1600–1613. doi: 10.1038/s41374-018-0127-3. [DOI] [PubMed] [Google Scholar]
- 39.Zhao R., Dong R., Yang Y., Wang Y., Ma J., Wang J., Li H., Zheng S. MicroRNA-155 modulates bile duct inflammation by targeting the suppressor of cytokine signaling 1 in biliary atresia. Pediatr. Res. 2017;82:1007–1016. doi: 10.1038/pr.2017.87. [DOI] [PubMed] [Google Scholar]
- 40.Crowe A.R., Yue W. Semi-quantitative Determination of Protein Expression using Immunohistochemistry Staining and Analysis: An Integrated Protocol. Bio. Protoc. 2019;9:e3465. doi: 10.21769/BioProtoc.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu L., Hui A.Y., Albanis E., Arthur M.J., O’Byrne S.M., Blaner W.S., Mukherjee P., Friedman S.L., Eng F.J. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park J.-K., Ki M.-R., Lee H.-R., Hong I.-H., Ji A.-R., Ishigami A., Park S.-I., Kim J.-M., Chung H.-Y., Yoo S.-E., Jeong K.S. Vitamin C deficiency attenuates liver fibrosis by way of up-regulated peroxisome proliferator-activated receptor-gamma expression in senescence marker protein 30 knockout mice. Hepatology. 2010;51:1766–1777. doi: 10.1002/hep.23499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available upon request.