Abstract
Neottiabifidus, a new mycoheterotrophic orchid, found in Maolan National Nature Reserve in Guizhou Province, China, is described and illustrated here. The new species is close to N.nidus-avis, N.kiusiana and N.papilligera but differs in having a finely pubescent rachis with fewer flowers, a finely pubescent pedicel, and a fishtail-shaped lip that is deeply bilobed to the middle of the lip, with the lobes diverging at an acute angle (45°) to each other and mesochile with many papillae. Additionally, N.bifidus is well supported as a new species by molecular phylogenetic results based on ITS and chloroplast genome. The chloroplast genome of the novelty, which contains an LSC region of 33,819 bp, SSC region of 5,312 bp and IRs of 46,762 bp was assembled and annotated. A key to mycoheterotrophic Neottia species in China is also provided.
Key words: Neottiabifidus , new species, Orchidaceae, saprophytic orchid
Introduction
The genus Neottia Guett. comprises 81 accepted species, including 63 autotrophic species and 18 mycoheterotrophic species (https://powo.science.kew.org, Mu et al. 2017; Chen and Jin 2021), distributed widely in north temperate areas with a few species extending into alpine regions in the mountains of tropical Asia (Govaerts et al. 2019; Chen and Jin 2021). East Asia is one of the diversity centers for this genus with more than 70% of Neottia species occurring in this region (So and Lee 2020). Formerly, Neottia was divided into Listera and Neottia (Bentham 1881; Pfitzer 1887; Schlechter 1926; Brieger et al. 1974; Dressler 1981; Rasmussen 1982) by the distinct morphological differences possessed by autotrophic plants (Listera) with two opposite leaves (sometimes three or more) in the middle of the stem, while mycoheterotrophic plants are achlorophyllous and possess densely fleshy bird's nest like roots. In 2003, Govaerts cited another genus in Tribe Neottieae Lindl., Holopogon, as a synonym of Neottia (Govaerts 2003).
There are 52 species and one variation of Neottia in China, amongst which 14 species are mycoheterotrophic (https://powo.science.kew.org, Mu et al. 2017; Chen and Jin 2021). During our fieldwork in the Maolan National Nature Reserve, Libo County, Guizhou Province, China in 2021, an unknown species of mycoheterotrophic Neottia was found in the evergreen broad-leaved forest. Based on morphological characters and molecular evidence, it was considered as a new species of Neottia and is described below.
Materials and methods
Morphological characteristics of the new species were observed, measured and photographed, based on living plants in Maolan National Nature Reserve, Guizhou. The studied specimens are deposited at The National Orchid Conservation Center of China and the Orchid Conservation & Research Center of Shenzhen. The general morphology was derived from fresh specimens and photographs were taken with a DSLR camera. To investigate the systematic position of the new species, the plastid genome and the nuclear ribosomal internal transcribed spacers (nrITS) marker were used in molecular phylogenetic analysis. Total genomic DNA was extracted from fresh flowers and stems (voucher specimens J.B.Chen 00599) using a plant genomic DNA kit and then sent to Novogene (Beijing, China) for the library (350 bp) preparation for genome skimming sequencing. Paired-end (150 bp) sequencing was conducted on the Illumina Hiseq 6000 platform (San Diego, CA), producing approximately 8 Gb reads. The plastid genome was assembled using GetOrganelle (Jin et al. 2020) with the chloroplast genome of Neottiacamtschatea (L.) Rchb. F.(NC_030707) and Neottialisteroides Lindl. (NC_030713) as the reference sequences. After assembly, the obtained scaffolds and contigs were annotated by Geneious Prime (Biomatters Ltd., Auckland, New Zealand) (Kearse et al. 2012) and Plastid Genome Annotator (Qu et al. 2019). The annotated complete chloroplast genome was deposited in GenBank with accession number OP279442. nrITS were also sequenced for the new species in this study. The PCR reactions and Sanger Sequencing were performed by Sangon Biotech (Shanghai, China). The primers used in this study are presented in Table 1. In total, 70 species (incl. 29 species of Neottia) from seven genera were used for molecular phylogenetic analyses (Table 2). The nrITS dataset consists of six genera and 66 species and the plastid genome dataset consists of five genera and 27 species, respectively. Five species of Cionisaccus, Ophrys and Serapias were selected as outgroup taxa based on Li et al. (2016). All plastid genomes were aligned by MAFFT 7.3 (ffT-NS-i × 1000 strategy) after removing one inverted repeat (IR) region of each sample (Katoh and Standley 2013). Poorly-aligned regions were removed by trimAl 1.2 with default settings before phylogenetic analyses (Capella-Gutiérrez et al. 2009). Maximum Likelihood (ML) analyses were conducted in IQTREE 1.6 using the SH-aLRT test and ultrafast bootstrap (UFBoot) feature (–alrt 1000 –bb 1000 –nt AUTO) (Nguyen et al. 2015; Hoang et al. 2018).
Table 1.
Primers used in this study.
| Primer | Sequence (5’to3’) | Origin |
|---|---|---|
| ITS-17SE | ACGAATTCATGGTCCGGTGAAGTGTTCG | Sun et al. 1994 |
| ITS-26SE | TAGAATTCCCCGGTTCGCTCGCCGTTAC | Sun et al. 1994 |
Table 2.
GenBank accession numbers for sequence data, a dash (-) indicates missing data and an asterisk (*) denotes sequences obtained in this study.
| Species | nrITS | cp |
|---|---|---|
| Aphyllorchiscaudata | FJ454866 | - |
| Aphyllorchisgollanii | MZ463253 | - |
| Aphyllorchismontana | FJ454867 | - |
| Aphyllorchispallida | MZ463252 | - |
| Cephalantherabijiangensis | MZ463242 | - |
| Cephalantheradamasonium | AY146446 | NC_041179 |
| Cephalantheraepipactoides | KY512499 | - |
| Cephalantheraerecta | MZ463245 | - |
| Cephalantheraexigua | FJ454868 | - |
| Cephalantherafalcata | AB856493 | - |
| Cephalantherafalcatavar.flava | MZ463241 | - |
| Cephalantherahumilis | MZ463240 | NC_030706 |
| Cephalantheralongibracteata | MK306540 | NC_041180 |
| Cephalantheralongifolia | AY146447 | NC_030704 |
| Cephalantherananchuanica | JN706696 | - |
| Cephalantherananlingensis | KT338669 | - |
| Cephalantherarubra | AY146445 | NC_041181 |
| Epipactisalbensis | AY154384 | NC_041182 |
| Epipactisatrorubens | JN847403 | - |
| Epipactisduriensis | AY351377 | - |
| Epipactisfageticola | AY351382 | - |
| Epipactisflava | FJ454869 | - |
| Epipactishelleborine | MZ463247 | MK608776 |
| Epipactisleptochila | FJ454870 | - |
| Epipactislusitanica | AY351381 | - |
| Epipactismairei | MZ463250 | NC_030705 |
| Epipactismicrophylla | FR750399 | MH590352 |
| Epipactismuelleri | FJ454871 | - |
| Epipactispalustris | AY146448 | NC_041187 |
| Epipactispapillosa | MZ463248 | - |
| Epipactispurpurata | JN847416 | MH590354 |
| Epipactisroyleana | MZ463249 | - |
| Epipactisthunbergii | MK306477 | NC_046817 |
| Epipactisveratrifolia | KF727435 | NC_030708 |
| Epipactisvoethii | FR750400 | - |
| Neottiaacuminata | KT338755 | - |
| Neottiaalternifolia | MZ463268 | - |
| Neottiabicallosa | MZ463271 | - |
| Neottiabifidus | OP265395* | OP279442* |
| Neottiabifolia | MG216639 | - |
| Neottiaborealis | MG216431 | - |
| Neottiabrevicaulis | MZ463258 | - |
| Neottiacamtschatea | KJ023677 | NC_030707 |
| Neottiacordata | KJ023678 | NC_041189 |
| Neottiasuzukii | MH321188 | NC_041447 |
| Neottiadivaricata | MZ463257 | - |
| Neottiafugongensis | MZ463256 | NC_030711 |
| Neottiahybrid sp. | MZ463255 | - |
| Neottiajaponica | KT338756 | NC_041446 |
| Neottiakaroana | MZ463270 | - |
| Neottiakiusiana | KT338757 | MN537563 |
| Neottialisteroides | MZ463262 | NC_030713 |
| Neottiameifongensis | MZ463267 | - |
| Neottiamucronata | MZ463261 | - |
| Neottianidus-avis | AY351383 | JF325876 |
| Neottianujiangensis | MZ463254 | - |
| Neottiaovata | - | NC_030712 |
| Neottiapapilligera | KT338758 | - |
| Neottiapinetorum | KT338759 | KU551269 |
| Neottiapuberula | MH808061 | - |
| Neottiasmallii | AF521058 | - |
| Neottiasmithiana | MZ463263 | - |
| Neottiawardii | MZ463260 | - |
| Neottiawuyishanensis | MZ409849 | - |
| Cionisaccusprocera | - | MW589517 |
| Ophrysapifera | AY699976 | - |
| Ophrysfusca subsp. | - | AP018716 |
| Ophrysinsectifera | AY699950 | - |
| Ophryssphegodes | - | AP018717 |
| Serapiascordigera | AY364884 | - |
Results
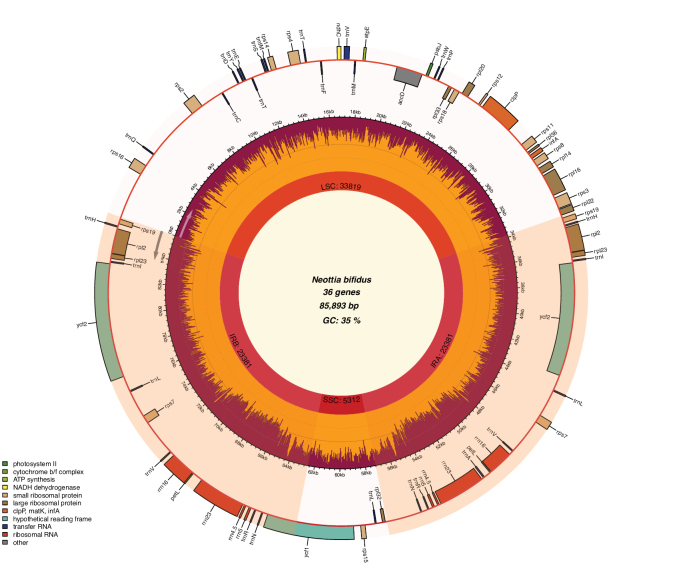
The whole chloroplast genome of N.bifidus showed a typical quadripartite structure containing a pair of inverted repeats (IRs) separated by a large single-copy (LSC) region and a small single-copy (SSC) region (Fig. 1). The complete plastid genome sequence of N.bifidus was 85,893 bp in length containing an LSC region of 33,819 bp, SSC region of 5,312 bp and IRs of 46,762 bp. The chloroplast genome contained 72 genes, including 36 protein-coding genes, 28 tRNA genes and eight rRNA genes (Table 3). The overall GC content is 35%.
Figure 1.
Chloroplast genome map of N.bifidus.
Table 3.
Genes present in the chloroplast genome of Neottiabifidus.
| Group of genes | Gene |
|---|---|
| Photosystem I | - |
| Photosystem II | psbJ |
| Cytochrome b/f complex | petL* |
| ATP synthase | atpE |
| NADH dehydrogenase | ndhC |
| Rubis CO large subunit gene | - |
| RNA polymerase | - |
| Small ribosomal proteins | rps2, rps3, rps4, rps7*, rps8, rps11, rps12, rps14, rps15, rps16, rps18, rps19* |
| Large ribosomal proteins | rpl2*, rpl14, rpl16, rpl20, rpl22, rpl23*, rpl32, rpl33, rpl36 |
| tRNA | trnA-UGC, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnH-GUG*, trnI-CAU*, trnL-CAA*, trnL-UAG, trnM-CAU, trnN-GUU*, trnP-UGG, trnQ-UUG, trnR-ACG*, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC*, trnV-UAC, trnW-CCA, trnY-GUA |
| rRNA | rrn4.5*, rrn5*, rrn16*, rrn23* |
| Translational initiation factor | infA |
| Subunits of Acetyl-CoA-carboxylase | accD |
| Protease | clpP |
| Conserved open reading frames | ycf1, ycf2* |
Note: * means duplicated gene in IRs.
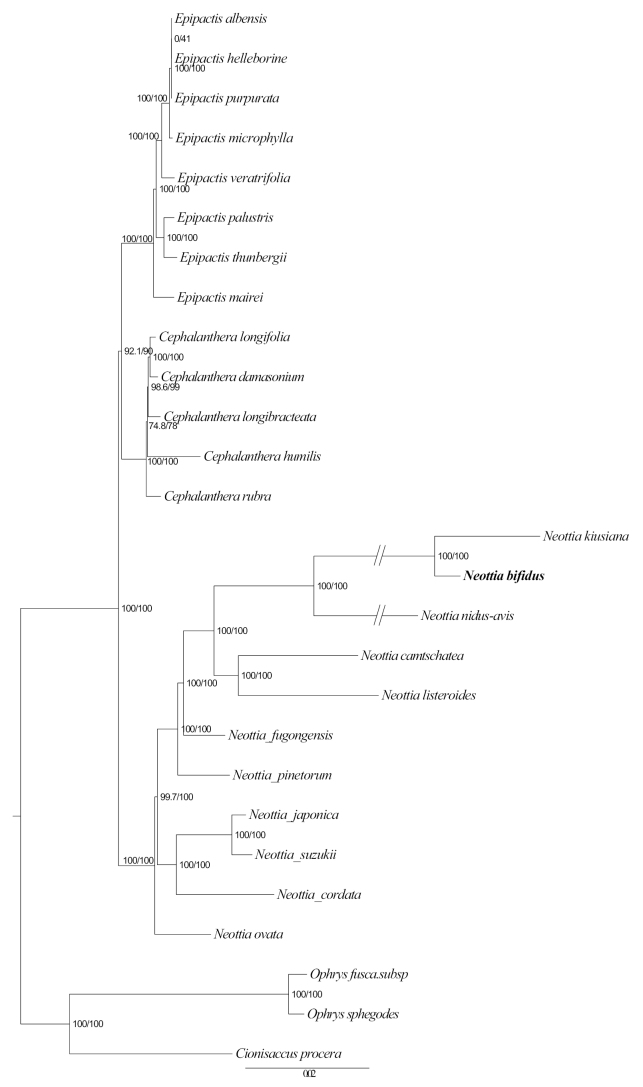
The phylogenetic analyses indicated that this unknown species is far from other autotrophic species, but has a better clustering relationship with leafless holomycotrophic species in Neottia. The phylogenetic tree, based on the plastid genome, indicated that it is close to N.kiusiana T.Hashim. & S.Hatus. (KT338757) with high support (SH-aLRT 100%, UfBoot 100%) and then sister to N.nidus-avis (L.) Rich. (JF325876) also with strong support (SH-aLRT 100%, UfBoot 100%) (Fig. 2). The phylogenetic tree, based on nrITS, showed that the new species is sister to N.kiusiana and N.papilligera Schltr. with high support (SH-aLRT 100%, UfBoot 100%) (Fig. 3).
Figure 2.
Phylogram of Neottieae, based on the plastid genome. The numbers near the nodes are the values of SH-aLRT test (left) and the ultrafast bootstrap (right).
Figure 3.
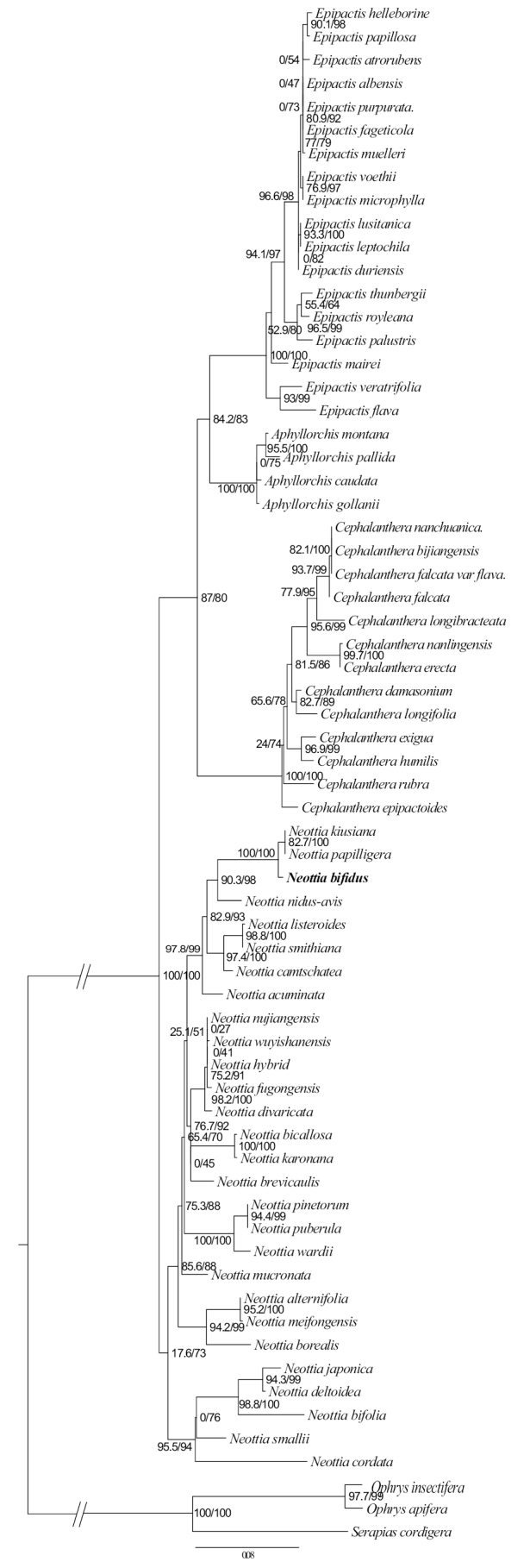
Phylogram of Neottieae, based on nrITS. The numbers near the nodes are the values of SH-aLRT test (left) and the ultrafast bootstraps (right).
Taxonomy
. Neottia bifidus
M.N.Wang sp. nov.
0410D15E-A8CF-5619-96E6-8BE5ED9F437F
urn:lsid:ipni.org:names:77324361-1
Figs 4 , 5 Chinese name: 鱼尾鸟巢兰
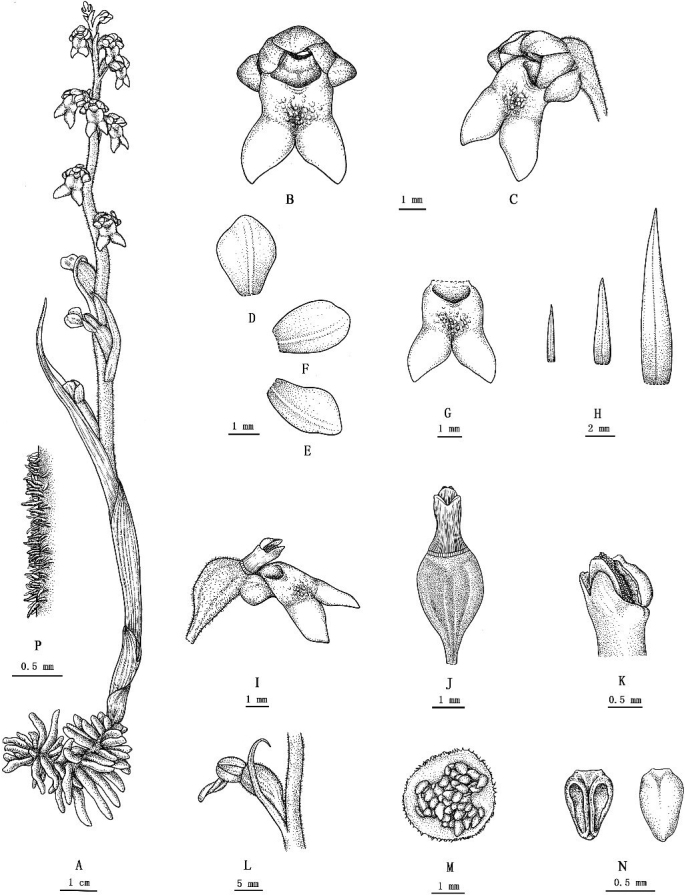
Figure 4.
Neottiabifidus M.N.Wang, sp. nov. A whole plant B flower (front view) C flower (side view) D dorsal sepal E lateral sepal F petal G lip (front view) H bracts I ovary, column and lip (side view) J ovary and column (ventral view) K column L fruit with bract M fruit (cross section) N anther cap P hairy on rachis.
Figure 5.
Neottiabifidus M.N.Wang, sp. nov. Photographed by M. N. Wang & W. H. Rao. A habit B whole plant and hairy on rachis C inflorescence D flower (front view) E ovary and flower (side view) and hairy on ovary F dorsal sepal G, H lateral sepals I, J petals K lip (front view, back view and side view) L bract M ovary and column N column O fruit with bract P fruit (cross section) Q Anther cap.
Type.
China. Guizhou Province, Qiannan Buyi and Miao Autonomous Prefecture, Libo County, the Maolan National Nature Reserve, 825 m elev., 23 April 2021, J.B.Chen 00599 (holotype: NOCC).
Diagnosis.
Neottiabifidus is morphologically similar to N.nidus-avis, N.kiusiana and N.papilligera but differs in having a finely pubescent rachis, with fewer flowers; finely pubescent pedicel; and fish-tail-shaped lip, deeply 2-lobed to the centre of mid-lip, lobes diverging at an acute angle (45°) to one another, mesochile with many papilloses (Table 4).
Table 4.
Morphological comparison of Neottiabifidus and similar species.
| Morphological characters | N.bifidus | N.kiusiana (Yukawa et al. 2009) | N.papilligera (Chen et al. 2009) | N.nidus-avis (Jersáková et al. 2022) |
|---|---|---|---|---|
| Plant height | 15–19 cm | 6–21 cm | 27–30 cm | 15–60 cm |
| Rachis | Rachis densely pubscent, laxly and irregularly 9–15-flowered. | Rachis sparsely glandular hairy, with 10–28 flowers. | Rachis glabrous or pubescent, with much more than 20 flowers. | Rachis glabrous, with much more than 20 flowers. |
| Pedicel | Pubscent | Glabrous | Glabrous | Glabrous |
| Lip | Lip 2-lobed to the centre of mid-lip; hypochile without purple dots; mesochile with many papilloses; epichile 2-lobed, lobes triangular, fish-tail-shaped, diverging at an acute angle (45°) to one another. | Lip 2-lobed (not up to the centre of mid-lip); hypochile purple-dotted adaxially; epichile 2-lobed, lobes transversely oblique-rectangular, rectangular or oblong, diverging at an acute angle (45°) to one another. | Lip apex deeply 2-lobed; lobes narrowly oblong, usually twisted, diverging at an obtuse angle (120°–170°) to one another. | Lip apex deeply 2-lobed, diverging at an obtuse angle (120°–170°) to one another. |
Terrestrial herbs, leafless, holomycotrophic, 10–19 cm tall. Rhizome short, with many stout, fleshy fascicled roots. Stem erect, terete, leafless, pubscent, with 2–3 sheaths at base; sheaths tubular, 2–3 cm, membranous, glabrous, with 4–7 dark brown veins, upper ones much longer than lower ones; rachis 7–13 cm, pubscent, laxly and irregularly 9–15-flowered; floral bracts membranous, glabrous, narrowly lanceolate, ovate-lanceolate, obtuse to subacute, 0.7–2.1 cm long, lowermost ones much longer than flowers, 1.1–1.3 × 2.6–3 cm, gradually diminishing in upper ones which are shorter than ovaries. Flowers resupinate, pale brown; pedicel and ovary 0.6–1.5 cm long, pubescent. Sepals membranous, ovate to obovate, pale brown, nearly equal in size; dorsal sepal cucullate, 2.3–2.4 × 1.6–1.8 mm, apex obtuse, glabrous; lateral sepals cucullate, strongly cupped, 2.4–2.5 × 1.4–1.5 mm, apex obtuse, glabrous. Petals membranous, ovate to obovate, pale brown, nearly equal in size to dorsal sepal. Lip spreading downwards, subrectangular, 3.8–5 mm long, small and semi-transparent at early anthesis, becoming larger and yellowish-brown at late anthesis, apex deeply 2-lobed to the center of mid-lip; hypochile rectangular, concave at base; mesochile with many papilloses; epichile 2-lobed, lobes extending outwards, triangular, fish-tail-shaped, 2.3–2.5 × 1.5–1.6 mm, diverging at an acute angle (45°) to one another, apex obtuse, margins of apices and inner sides repand or erose. Column cylindrical, 2.8–3 mm long; anther inclined towards rostellum, elliptic, ca. 0.7 mm; stigma ca. 0.9 mm, lamellate, 2-lobed; rostellum shorter than anther. Capsule elliptic, with persistent sepals and petals, 1–1.5 cm long.
Etymology.
The species epithet refers to the fish-tail-shaped lip of the new species.
Distribution and habitat.
Neottiabifidus is currently known only from the type locality in Libo, Guizhou, China. It grows in humus-rich soil under broad-leaved forests at elevations of 700–900 m and is found growing with Miliusasinensis Finet & Gagnep. (Annonaceae), Platycaryastrobilacea Siebold & Zucc (Juglandaceae), Micheliamartini (H. Lév.) Finet & Gagnep. ex H. Lév. (Magnoliaceae), Mallotusphilippensis (Lamarck) Müll. Arg. (Euphorbiaceae), Symplocosadenophylla Wall. (Symplocaceae), Chimonobambusaangustifolia C. D. Chu & C. S. Chao (Poaceae), Murrayaexotica L. (Rutaceae), Gomphandratetrandra (Wall.) Sleumer (Stemonuraceae), Diospyrosmollis Griff. (Ebenaceae), Strobilantheshongii Y. F. Deng & F. L. Chen (Acanthaceae), etc.
Phenology.
Flowering and fruiting from Apr–May.
Conservation status.
During our fieldwork, only one population with less than 10 individuals was discovered in Maolan National Nature Reserves (213 km2). Most individuals were found growing along the roadside and are easily disturbed by human activities. According to the guidelines for using the IUCN Red List Categories and Criteria (IUCN 2022), the new species should be temporarily assigned as ‘Critically Endangered’ by its limited populations, localities and vulnerable habitats.
Note.
Neottiabifidus is morphologically - related to three species, namely, N.nidus-avis, N.kiusiana and N.papilligera, but it is readily distinguished from them, based on morphological characters given in Table 4.
Key to mycoheterotrophic species of Neottia in China
| 1 | Stigma terminal; rostellum absent | 2 |
| – | Stigma lateral or rarely subterminal; rostellum present, usually above concave stigma | 4 |
| 2 | Flowers purplish-red | Neottiagaudissartii (Holopogongaudissartii) |
| – | Flowers green | 3 |
| 3 | Flowers actinomorphic, lip very similar to the petals | N.pekinensis (Holopogonpekinensis) |
| – | Flowers zygomorphic, lip bilobed at the apex, utterly different from the petals | N.smithiana (Holopogonsmithianus) |
| 4 | Lip entire; column (excluding anther and rostellum) less than 0.5 mm | 5 |
| – | Lip bilobed at apex; column (excluding anther and rostellum) 1.5–4 mm | 6 |
| 5 | Floral rachis glabrous; flowers resupinate | N.acuminata |
| – | Floral rachis villous; flowers not resupinate | N.taibaishanensis |
| 6 | Lip distinctly concave at base | 7 |
| – | Lip not concave at base | 9 |
| 7 | Apical lobes of lip parallel or diverging at an acute angle to one another | N.bifidus |
| – | Apical lobes of lip diverging at an obtuse angle to one another | 8 |
| 8 | Apical lobes of lip 2.5–3 mm; sinus of lip without a short tooth between lobes | N.papilligera |
| – | Apical lobes of lip less than 1 mm; sinus of lip with a short tooth between lobes | N.brevilabris |
| 9 | Lip with a pair of triangular auricles at base | N.tenii |
| – | Lip without a pair of auricles at base | 10 |
| 10 | Lip obovate, 6–10 mm wide | N.megalochila |
| – | Lip narrowly obovate-oblong or cuneate, 1.5–4 mm wide | 11 |
| 11 | Lip narrowly obovate-oblong, 6–9 × 3–4 mm | N.listeroides |
| – | Lip cuneate, 10–12 × 1.5–2 mm | N.camtschatea |
Supplementary Material
Citation
Wang M-N, Wu X-Y, Tan C-J, Yu P, Rao W-H, Chen J-S, Li J, Chen J-B (2023) Neottia bifidus (Orchidaceae, Epidendroideae, Neottieae), a new mycoheterotrophic species from Guizhou, China. PhytoKeys 229: 215–227. https://doi.org/10.3897/phytokeys.229.103107
Funding Statement
National Natural Science Foundation of China (Grant No. 32001245)
Contributor Information
Jian Li, Email: lij@cnocc.cn.
Jian-Bing Chen, Email: conservation@cnocc.cn.
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
This work was financially supported by the National Natural Science Foundation of China (Grant No. 32001245) and the Science, Technology and Innovation Commission of Shenzhen Municipality (Grant No. KCXFZ20211020164200001).
Author contributions
Data curation: MNW, XYW. Funding acquisition: MNW. Investigation: MNW,WHR, JL, CJT, PY. Methodology:MNW, XYW. Project administration: JL. Software: XYW. Supervision: JL, JBC. Visualization: JSC. Writing-original draft: MNW. Writing-review and editing: MNW
Author ORCIDs
Mei-Na Wang https://orcid.org/0000-0001-5998-6028
Xin-Yi Wu https://orcid.org/0000-0002-8623-1867
Cheng-Jiang Tan https://orcid.org/0009-0008-4558-4907
Ping Yu https://orcid.org/0009-0003-3319-7145
Wen-Hui Rao https://orcid.org/0000-0002-2177-6700
Jie-Shan Chen https://orcid.org/0009-0000-8859-6053
Jian Li https://orcid.org/0000-0002-0096-6257
Jian-Bing Chen https://orcid.org/0000-0001-9085-0663
Data availability
All of the data that support the findings of this study are available in the main text.
References
- Bentham G. (1881) Notes on Orchideae. Journal of the Linnean Society of London, Botany 18: 281–360. 10.1111/j.1095-8339.1881.tb01258.x [DOI] [Google Scholar]
- Brieger FG. (1974) Unterfamilie: Neottioideae. In: Schlechter R. (Ed.) Die Orchideen; ihre Beschreibung, Kultur und Ziichtung.3. Auflage. Brieger FG, Paul Parey, Berlin, 284–358.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. (2009) trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15): 1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BH, Jin XH. (2021) Neottiawuyishanensis (Orchidaceae: Neottieae), a new species from Fujian, China. Plant Diversity 43(5): 426–431. 10.1016/j.pld.2021.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SC, Gale SW, Cribb PJ. (2009) Neottia Guett. In: Wu ZY, Raven PH, Hong DY (Eds) Flora of China (Vol. 25). Science Press, Beijing & Missouri Botanical Garden Press, St. Louis, 187.
- Dressler RL. (1981) The Orchids: Natural History and Classification. Harvard University Press, Cambridge, 332 pp. [Google Scholar]
- Govaerts R. (2003) World Checklist of Monocotyledons Database in ACCESS: 1-71827. The Board of Trustees of the Royal Botanic Gardens, Kew.
- Govaerts R, Bernet P, Kratochvil K, Gerlach G, Carr G, Alrich P, Pridgeon AM, Pfahl J, Campacci MA, Holland Baptista D, Tigges H, Shaw J, Cribb PJ, George A, Kreuz K, Wood JJ. (2019) World Checklist of Orchidaceae. Facilitated by the Royal Botanic Gardens, Kew. http://wcsp.science.kew.org/ [Accessed 2 October 2019]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. (2018) UfBoot2: Improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35(2): 518–522. 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUCN (2022) Guidelines for Using the IUCN Red List Categories and Criteria. Version15.1. https://www.iucnredlist.org/resources/redlistguidelines [Accessed October 2022]
- Jersáková J, Minasiewicz J, Selosse MA. (2022) Biological flora of Britain and Ireland: Neottianidus-avis. Journal of Ecology 110(9): 2246–2263. 10.1111/1365-2745.13953 [DOI] [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, de Pamphilis CW, Yi TS, Li DZ. (2020) GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biology 21(1): 241. 10.1186/s13059-020-02154-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013) MAffT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Molecular Biology and Evolution 30(4): 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12): 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MH, Zhang GQ, Lan SR, Liu ZJ. (2016) A molecular phylogeny of Chinese orchids. Journal of Systematics and Evolution 54(4): 349–362. 10.1111/jse.12187 [DOI] [Google Scholar]
- Mu XY, Liu B, Zhu YX, Tong L, Lin Q, Zhang ZX. (2017) Holopogonpekinensis (Orchidaceae), a new heteromycotrophic species from Northern China. Phytotaxa 326(2): 151–155. 10.11646/phytotaxa.326.2.7 [DOI] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32(1): 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfitzer EHH. (1887) Entwurf einer natürlichen Anordnung der Orchideen. Winter’s Universitätsbuchhandlung, Heidelberg, 108 pp. 10.5962/bhl.title.166408 [DOI] [Google Scholar]
- Qu XJ, Moore MJ, Li DZ, Yi TS. (2019) PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15(1): 50. 10.1186/s13007-019-0435-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen FN. (1982) The gynostemium of the neottioid orchids. Opera Botanica 65: 1–96. [Google Scholar]
- Schlechter R. (1926) Das System der Orchidaceen. Notizblatt des Botanischen Gartens und Museums zu BerlinDahlem 9(88): 563–591. 10.2307/3994326 [DOI] [Google Scholar]
- So J, Lee N. (2020) Phylogenetic analysis of Neottiajaponica (Orchidaceae) based on ITS and matK regions. Korean Journal of Plant Taxonomy 50(4): 385–394. 10.11110/kjpt.2020.50.4.385 [DOI] [Google Scholar]
- Sun Y, Skinner DZ, Liang GH, Hulbert SH. (1994) Phylogenetic analysis of Sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theoretical and Applied Genetics 89(1): 26–32. 10.1007/BF00226978 [DOI] [PubMed] [Google Scholar]
- Yukawa T, Yagame T, Lee NS. (2009) A Taxonomic reappraisal of Neottia (Orchidaceae) from east Asia. Bulletin of the National Museum of Nature and Science, Series B (Botany)(Tokyo), Japan 35: 57–62. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data that support the findings of this study are available in the main text.