Abstract
The major immediate-early (MIE) gene of human cytomegalovirus (HCMV) encodes several MIE proteins (MIEPs) produced as a result of alternative splicing and polyadenylation of the primary transcript. Previously we demonstrated that the HCMV MIEPs expressed from the entire MIE gene could rescue the temperature-sensitive (ts) transcriptional defect in the ts13 cell line. This defect is caused by a ts mutation in TAFII250, the 250-kDa TATA binding protein-associated factor (TAF). These and other data suggested that the MIEPs perform a TAF-like function in complex with the basal transcription factor TFIID. In addition to the transcriptional defect, the ts mutation in ts13 cells results in a defect in cell cycle progression which ultimately leads to apoptosis. Since all of these defects can be rescued by wild-type TAFII250, we asked whether the MIEPs could rescue the cell cycle defect and/or affect the progression to apoptosis. We have found that the MIEPs, expressed from the entire MIE gene, do not rescue the cell cycle block in ts13 cells grown at the nonpermissive temperature. However, despite the maintenance of the cell cycle block, the ts13 cells which express the MIEPs are resistant to apoptosis. MIEP mutants, which have previously been shown to be defective in rescuing the ts transcriptional defect, maintained the ability to inhibit apoptosis. Hence, the MIEP functions which affect transcription appear to be separable from the functions which inhibit apoptosis. We discuss these data in the light of the HCMV life cycle and the possibility that the MIEPs promote cellular transformation by a “hit-and-run” mechanism.
The major immediate-early (MIE) gene (Fig. 1) of human cytomegalovirus (HCMV) encodes several MIE proteins (MIEPs) produced as a result of alternative splicing and polyadenylation of the primary transcript (46, 72, 74). Two of these proteins appear in greater abundance in the lytic infection and have been extensively examined for their ability to affect RNA polymerase II transcription in a cell- and promoter-specific manner (1–4, 7–11, 13–15, 19, 20–22, 24, 25, 27–30, 34–36, 38, 40–47, 50–55, 58, 61, 62, 68, 70, 71, 73, 77–79, 82–84). They are IEP72 (72 kDa; also called IE1, IE1491aa, or ppUL123) and IEP86 (86 kDa; also called IE2, IE2579aa, or ppUL122a). The MIEPs function in transcription by participating in direct protein-protein interactions with cellular promoter binding factors (9, 18, 21, 24, 32, 33, 38, 41, 62, 68, 84). In addition, the MIEPs directly contact cell cycle-regulatory proteins to abrogate some of their functions (17, 20, 44, 53, 69).
FIG. 1.
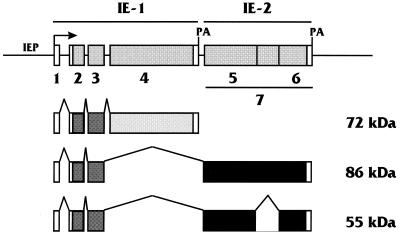
Diagram of the MIE gene of HCMV showing alternatively spliced and polyadenylated (PA) transcripts produced from its primary transcript. The transcripts encoding the 72-, 86-, and 55-kDa MIEPs (IEP72, IEP86, and IEP55) are shown. The various exons are numbered; open reading frames (ORFs) within exons are shaded. The IE1 region is composed of exons 1 to 4, and the IE2 region consists of exon 7. Note that IEP55 differs from IEP86 by having an extra splice, resulting in the removal of an IEP86 unique region in exon 7. The IE2-derived ORFs flanking the UR are labeled as exons 5 and 6, specific to IEP55. In some diagrams of the MIE region the exon we have called 7 is numbered as exon 5.
We have previously suggested that IEP72 and IEP86 perform a function similar to that of the TATA binding protein-associated factors (TAFs) as components of the basal transcription factor TFIID (40). One of the pieces of supporting evidence for this came from the use of the temperature-sensitive (ts) BHK-21 cell line, ts13. The mutation causing the ts phenotype is in TAFII250 (26), the largest TAF in TFIID. When ts13 cells are cultured at the nonpermissive temperature the ts mutation in TAFII250 results in promoter- and activator-specific effects on cellular gene transcription and defects in cell cycle progression leading, ultimately, to apoptosis (23, 49, 57, 64, 66, 75, 80, 81). Expression of wild-type (WT) human TAFII250 (hTAFII250) rescues these transcriptional and cell cycle defects, and the cells do not become apoptotic (23, 49, 57, 64, 66, 75, 80, 81). In support of a TAF-like function of the MIEPs we demonstrated that expression of the MIEPs from the entire MIE gene (Fig. 1) rescued the ts transcriptional defect in TAFII250 (40). In addition, specific mutants, previously shown to be defective in transcriptional activation, failed to rescue the ts defect (40).
In the present studies we asked whether the MIEPs could also rescue the cell cycle defect and/or affect the progression to apoptosis. Various studies have addressed the function of the MIEPs in cell cycle and growth control. The MIEPs upregulate p53 but inhibit its ability to activate transcription (48, 69). Similarly, the MIEPs can inhibit the ability of Rb family members to repress E2F activity (20). Both IEP72 and IEP86 can bind to p105/Rb and inhibit its ability to induce the flat-cell phenotype in Saos-2 cells (17). The ability of the MIEPs to interact directly with both p53 and p105/Rb suggests that the MIEPs may function similarly to proteins of the small DNA tumor viruses, which abrogate the G1-to-S-phase checkpoint controls. However, results are conflicting concerning the significance of these interactions in the ability of the MIEPs to stimulate the cell cycle (5, 17, 53, 67). It has been reported that human cytomegalovirus (HCMV) infection of permissive cells can cause G1 arrest and a block to S-phase entry in infected cells (6, 16, 39). Regarding apoptosis, the MIEPs have been shown to block E1A and tumor necrosis factor alpha-induced apoptosis in HeLa cells (85).
The experiments presented below show that MIEP expression does not rescue the cell cycle block. However, despite the maintenance of the block, the expression of the MIEPs in ts13 cells inhibited the progression to apoptosis. MIEP mutants which failed to rescue the ts transcriptional defect maintained the ability to inhibit apoptosis, suggesting that the MIEP functions which mediate transcription are separable from the functions which affect apoptosis. These findings are discussed in light of the HCMV life cycle and data (67) suggesting that the MIEPs may promote cellular transformation by a “hit-and-run” mechanism.
MATERIALS AND METHODS
Plasmids.
All plasmids were propagated in Escherichia coli DH5α or HB101. Plasmid DNA for transfections was prepared as previously described (40). Plasmids pRL43A and pSVHwt contain the genomic HCMV MIE region (Fig. 1) and express HCMV IE regions IE1 and IE2 from the MIEP (37, 40, 52, 73). Plasmids which produced mutant IE proteins included pSVHm2, pSVHm13, pSVHm14, pSVHm16, and pSVHm19 (73). Plasmid pc-hCD4, expressing the human CD4 (hCD4) cDNA, was constructed by digesting p-ts-CD4 (a gift of Michael Malim) with EcoRI and blunting it with Klenow polymerase. The hCD4 insert was then liberated with a BamHI digest. This was cloned into pcDNA3 (Invitrogen), which had been digested with HindIII, blunted with Klenow polymerase, and then digested with BamHI. Plasmid pCMV-βgal was a gift of Michael Malim.
Cell culture and lipofection.
The cell line BHK-21 ts13 was propagated and maintained as previously described (40). Cells (2 × 105) were plated and lipofected by procedures supplied by the manufacturer (Gibco/BRL) with 1 μg of total plasmid per plate. The lipofection efficiency was 15 to 30%, as determined by fluorescence-activated cell sorter (FACS) analysis or by transfection of pCMV-βgal followed by LacZ staining.
FACS analysis of ts13 cells.
Cultures of ts13 cells were lipofected with 1 μg each of pXJ40E/B, pCMV-hTAFII250, and pRL43a. pc-hCD4 (1.5 μg) was colipofected to allow sorting of lipofected cells. The lipofections, done in triplicate, were incubated at either 32 or 39°C for 24 to 28 h. A previously described whole-cell isolation protocol was used (60), with modifications, to allow simultaneous detection of cell surface and nuclear staining. Specifically, cells were scraped into 1 ml of staining medium (1× phosphate-buffered saline [PBS]–1% bovine serum albumin [BSA]). The cells (106) were resuspended in 50 μl of a 1:5 dilution of fluorescein isothiocyanate (FITC)-conjugated anti-hCD4 antibody (Sigma). Nonstained controls received only 50 μl of staining medium. All tubes were incubated for 30 min on ice, washed with staining medium, and then resuspended in 0.5 ml of ice-cold PBS. The cells were fixed by the addition of 0.5 ml of ice-cold 0.5% paraformaldehyde for 1 h at 4°C. For permeabilization, the cells were pelleted by centrifugation, resuspended in 1 ml of 0.2% Tween 20 in PBS (prewarmed to 37°C), and incubated for 15 min at 37°C. One milliliter of PBS containing 0.2% BSA and 0.1% sodium azide was added to each sample with brief mixing followed by centrifugation to pellet the cells. For DNA labeling, the cells were resuspended in PBS containing 0.2% BSA, 0.1% sodium azide, 10 μg of propidium iodide (Sigma)/ml, and 50 μg of RNase (Sigma)/ml. These were mixed well and incubated for 18 to 24 h at 4°C.
The cells were analyzed with a Becton-Dickinson FACScan running CellQuest software for data collection. Only CD4-positive cells were gated for analysis of DNA content. Cell cycle analysis was performed with the program ModFit (Verity). The data shown is representative of five separate lipofections.
TUNEL for detection of apoptosis.
ts13 cells were grown on sterilized glass coverslips in 35-mm-diameter plates. The cells on individual coverslips were lipofected with 1 μg of one of the following plasmids: XJ40E/B, pCMV-hTAFII250, pc53-wt, pSVHwt, or one of the pSVH mutant plasmids described above. The cells were analyzed at 24 to 28 h postlipofection. The coverslips were washed on the plates three times with 1 ml of PBS, pH 7.4, and then fixed in fresh 4% paraformaldehyde in PBS, pH 7.4, for 30 min at room temperature. The coverslips were washed three more times with 1 ml of PBS, pH 7.4, and then permeabilized in PBS, pH 7.4, containing 0.1% Triton X-100–0.1% sodium citrate for 10 min at 4°C and washed twice with 1 ml of PBS, pH 7.4.
Terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) reactions were performed with the fluorescein cell death detection kit (Boehringer Mannheim) with a modified protocol. Specifically, the cells were labeled on the coverslips with 50 μl of reaction mixture containing terminal deoxynucleotidyltransferase and a nucleotide mix containing FITC-conjugated dUTP. The reaction mixtures were incubated at 37°C for 60 min. The coverslips were washed three times with PBS, pH 7.4, and then blocked by incubation in 1 ml of blocking buffer (PBS, pH 7.4, containing 1.0% Triton X-100, 0.5% Tween 20, and 0.1% BSA) for 30 min at room temperature. After removal of the blocking buffer, the coverslips were incubated with appropriate primary antibodies in 250 μl of blocking buffer for 1 h at room temperature. These antibodies included AB810 (anti-IEP72 and -IEP86) at 1:200, anti-p53 (Ab-2) at 1:100, and anti-hTAFII250 at 1:100 diluted in blocking buffer. Following incubation, the coverslips were washed three times in PBS, pH 7.4, and then incubated in 250 μl of tetramethyl rhodamine isothiocyanate (TRITC)-conjugated anti-F(ab′)2 fragments for 30 min at room temperature. The anti-F(ab′)2 secondary antibody was diluted 1:1,000 for Ab810, 1:500 for anti-p53, and 1:250 for anti-hTAFII250. The coverslips were washed extensively in PBS, pH 7.4, dried well, and then mounted on microscope slides with Permount (Fisher). The coverslips were sealed to the slides. At least three fields of each coverslip were photographed for visualization of FITC and TRITC staining, and representative fields are presented.
RESULTS
HCMV MIEPs do not rescue the cell cycle block in ts13 cells.
In our previous analysis of ts13 cells, we found that the HCMV MIEPs expressed from a plasmid containing the entire genomic MIE region (Fig. 1) rescued the transcriptional defect in TAFII250 (40). Under these conditions all of the possible MIEPs can be made from the variably spliced and polyadenylated primary transcript (i.e., IEP86, IEP72, and possibly others). In addition, the ratio of the MIEPs produced is determined, at least in part, by RNA processing. Individual MIEPs (IEP72 and IEP86) encoded by cDNA plasmid constructions were also tested in these transfection experiments (40). IEP86 alone could only modestly rescue the transcriptional defect, and this occurred only at very low plasmid input concentrations. IEP72 alone had no effect at any plasmid input concentration, and coexpression of each protein in cotransfection experiments showed no additional effects. Given the lack of effect of the individual proteins, we chose to do the subsequent experiments with plasmid pRL43a, which contains the entire MIE genomic region (Fig. 1).
The ts mutation in TAFII250 produces a defect in cell cycle progression indicated by a block in late G1. This block can be rescued by transfection of a plasmid encoding hTAFII250 (49, 64, 66). In order to determine whether the MIEPs would similarly rescue this cell cycle defect, we measured the total DNA content of ts13 cells transfected with various plasmids: pRL43a, described above; pCMVhTAFII250, which expresses hTAFII250; or a control plasmid encoding no eucaryotic protein. Each transfection also contained a plasmid which encoded the hCD4 molecule. The expression of CD4 allowed the specific sorting and analysis of only the transfected cells (see Materials and Methods).
Figure 2 shows a summary of five experiments performed in triplicate. When the control plasmid-transfected cells were incubated for 24 h at the nonpermissive temperature (39°C), the percentage of cells in the G0/G1 population increased relative to similarly transfected cells grown at the permissive temperature (32°C). This correlated with a decrease in the percentage of cells in S phase at the nonpermissive temperature and is in agreement with previously published reports (63, 76). When cells were transfected with the WT hTAFII250-expressing plasmid, the percentage of cells in G0/G1 and S returned to the levels seen at the permissive temperature, agreeing with previous data showing that expression of WT hTAFII250 rescues the ts cell cycle defect in ts13 cells (49). Transfection of the genomic MIE region gave results similar to those of the control transfected cells, indicating that the expression of the MIEPs failed to rescue the ts cell cycle defect. The conditions used in these experiments were the same as those used to demonstrate that the MIEPs can rescue the ts transcriptional defect. In addition, MIEP expression was verified in each experiment; specific data pertaining to the expression of the MIEPs in ts13 cells has been previously presented (40).
FIG. 2.
Effect of MIEP expression on the block to cell cycle progression in ts13 cells. Cultures of ts13 cells were transfected with pXJ40E/B (control), pRL43a (encoding the MIEPs), or pCMV-hTAFII250 (encoding hTAFII250). In addition, the cells were cotransfected with plasmid pc-hCD4, which expresses CD4. The transfected cells were then subjected to FACS analysis for DNA content; only CD4-positive cells were gated so that the transfected cells would be preferentially analyzed (see Materials and Methods). Cell cycle modeling and quantitation were performed with ModFit software. The bars show the percentage of cells in the various phases of the cell cycle. The data show representative results from an experiment where samples were tested in triplicate. Similar results were obtained in five separate experiments. The first (solid) bar in each phase indicates control plasmid-transfected cells at 32°C, the second (lightly shaded) bar indicates control plasmid-transfected cells at 39°C, the third (darkly shaded) bar indicates MIEP-expressing cells at 39°C, and the fourth (open) bar indicates hTAFII250-expressing cells at 39°C. The error bars indicate standard deviations.
These data indicate that the HCMV MIEPs do not rescue the cell cycle defect in ts13 cells when tested under conditions where they are known to rescue the transcriptional defect. In fact, repeated experiments indicated that expression of the MIEPs consistently resulted in even fewer cells entering S phase at the nonpermissive temperature. Hence, the MIEPs may augment, or duplicate, the mechanisms blocking the cell cycle in ts13 cells at the nonpermissive temperature.
Apoptosis in ts13 cells at 39°C is blocked by the HCMV MIEPs.
The ts mutation in ts13 cells and in tsBN462 cells (containing a similar ts mutation in TAFII250) induce apoptosis when the cells are grown at the nonpermissive temperature (64). Introduction of WT hTAFII250 into these cells has been shown to eliminate apoptosis (64). Therefore, we asked whether the MIEPs could block apoptosis in ts13 cells at the nonpermissive temperature despite the fact that they did not activate the cell cycle.
Coverslips containing ts13 cells grown at 32°C were transfected with pSVHwt, which encodes the WT MIEP genomic region, and incubated at 39°C. Twenty-four to 28 h posttransfection, the cells were fixed and (i) subjected to the TUNEL assay for immunofluorescence detection (with FITC) of apoptosis and (ii) treated with anti-IE72 and -IE86 antibody for immunofluorescence detection of the IE proteins with TRITC-conjugated secondary F(ab′)2 fragments. Control experiments (not shown) with mock-transfected cells indicated no immunofluorescence due to cross-reactivity of the anti-IE72 and -IE86 antibody with cellular proteins.
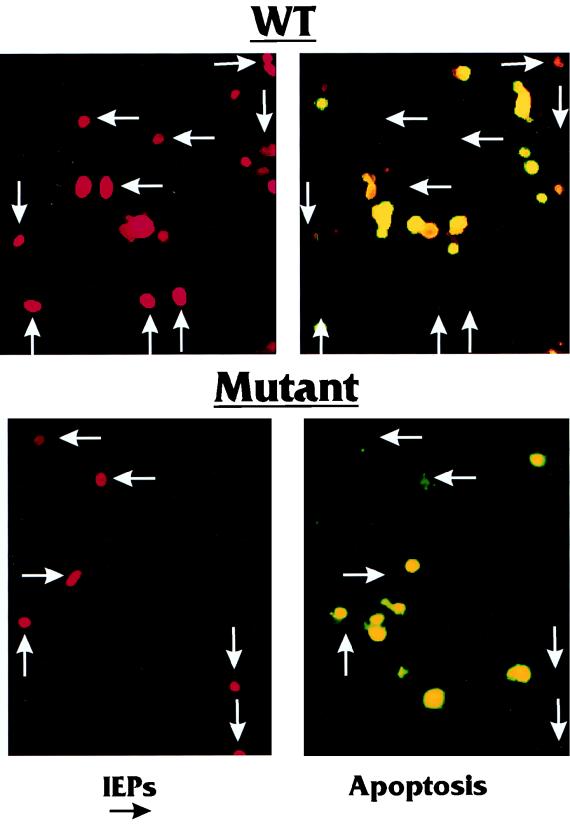
Figure 3 (upper two panels) shows a representative field of cells expressing the WT MIEPs. The right panel shows the results of the TUNEL assay for apoptosis (yellow nuclei). As can be seen, many cells in the field were positive for apoptosis; a similar assay with cells grown at 32°C indicated that very few cells were apoptotic (not shown). The left panel shows the immunofluorescence of cells in the same field which are producing the MIEPs (red nuclei). Comparison of the two panels shows that cells which produce the MIEPs are rarely apoptotic, and vice versa.
FIG. 3.
Effect of MIEP expression on apoptosis in ts13 cells grown at the nonpermissive temperature. Coverslips containing ts13 cells grown at 32°C were transfected with either pSVHwt (upper panels), which encodes the WT MIEPs, or pSVHm13 (lower panels), which encodes MIEPs which are defective in transcriptional activation (40). The cultures were incubated at 39°C for 24 to 28 h posttransfection and then fixed. The fixed coverslips were tested for (i) immunofluorescence detection of the MIEPs (left panel in each set) and (ii) immunofluorescence detection of apoptosis (by TUNEL assay; right panel in each set) (see Materials and Methods for details). Identical fields of cells are shown for MIEP detection (red) and apoptosis (yellow). For comparison, the arrows indicate the positions of MIEP-expressing cells in the fields.
In previous studies (40) we characterized a number of MIEP mutants which had reduced abilities to rescue the transcriptional defect in ts13 cells. The steady-state amounts of IEP86 and IEP72 produced by these mutants in ts13 cells is equivalent to that produced by WT MIEP-expressing plasmids (40). One of the mutants, m13 (with a mutation in amino acid 359 of IEP86 [73]), was virtually unable to rescue the transcriptional defect. Transfection of the m13 MIEP-expressing plasmid had no greater effect than transfection of a control plasmid (40).
As shown in the lower two panels of Fig. 3, we examined the ability of the mutant m13 MIEPs to block apoptosis. Comparison of the panels again shows that cells expressing the m13 MIEPs are rarely apoptotic and vice versa. Hence, the MIEP function(s) needed to block apoptosis is separate from the functions needed to rescue the transcriptional defect in ts13 cells at 39°C. In addition, we tested m2 (with a mutation in amino acid 59, which is common to both IEP72 and IEP86 [73]), another mutant severely affected in transcriptional activation, and found that it also maintained the ability to block apoptosis (data not shown).
In control experiments (not shown) we confirmed that ts13 cells expressing hTAFII250 at the nonpermissive temperature were not apoptotic, which agrees with previous results (64). It can be argued that the results we see could occur if the subpopulation of transfectable cells were somehow resistant to apoptosis. However, this does not seem to be the case. Previous experiments (64) have suggested that overexpression of p53 only partially rescues the apoptotic induction of tsBN462 cells at 39°C (64). In control experiments (not shown) where WT p53 was overexpressed we detected only partial rescue of apoptosis; i.e., many TUNEL-positive cells also showed p53 immunofluorescent staining due to overexpression of p53.
DISCUSSION
The ability of the HCMV MIEPs, particularly IEP72 and IEP86, to promiscuously activate transcription has been documented in many reports (1–4, 7–11, 13–15, 19–22, 24, 25, 27–30, 34–36, 38, 40–47, 50–55, 58, 61, 62, 68, 70, 71, 73, 77–79, 82–84). In large part, transcriptional activation appears to involve interactions of these proteins with both non-basal-transcription factors (e.g., Tef-1, Sp1, AP-1 family members, CREB, and Egr-1) and basal-transcription factors, such as TBP and TAFs (9, 21, 24, 32, 33, 38, 40, 41, 62, 68, 83). IEP72 and IEP86 can interact simultaneously with hTAFII130 through regions required for transcriptional activation (40). These interactions may account for the synergy of transcriptional activation seen when both proteins are expressed in a cell. These and other data have led us to propose that the MIEPs can perform a TAF-like function (40). Both IEP72 and 86 can interact with multiple components of TFIID in vitro. In addition, they copurify with TFIID and coimmunoprecipitate with purified TFIID from nuclear extracts from infected cells (40). Finally, expression of the MIEPs can rescue the ts transcriptional defect in TAFII250 in the tsBHK-21 cell line ts13 when grown at the nonpermissive temperature (40).
In addition to the transcriptional defect caused by the ts mutation in TAFII250, ts13 cells grown at the nonpermissive temperature are defective in G1 progression and S-phase entry and eventually become apoptotic (23, 49, 63–66, 76). Each of these ts conditions, the transcriptional defect, the cell cycle defect, and the resultant apoptosis, can be rescued by the expression of a WT form of TAFII250 (23, 26, 40, 49, 56, 57, 64, 65, 75, 80, 81).
Viral proteins have been shown to overcome these ts defects in ts13 cells grown at the nonpermissive temperature. As mentioned previously, the MIEPs can rescue the ts transcriptional defect (40). This ability is shared by simian virus 40 large T antigen (12). Additionally, large T antigen can rescue the cell cycle defect (42a), and this appears to sustain the cells in such a way that apoptosis does not occur. Simian virus 40 large T antigen and the MIEPs are similar in that they are promiscuous transcriptional activators, apparently targeting similar components of the transcription complex. However, large T antigen is known for its obvious activation of the cell cycle, whereas HCMV infection, although shown to upregulate the expression of some markers of normal cellular proliferation, appears to induce a late G1 block (5, 6, 16, 17, 31, 39, 58). Taken together, these facts seem to show that HCMV infection upregulates specific cellular replicative functions while blocking S-phase entry. Thus, the question to be asked was whether the MIEPs could rescue the specific cell cycle defect in ts13 cells at the nonpermissive temperature and/or inhibit the apoptosis which results under these conditions.
In the present studies we have found that expression of the MIEPs in ts13 cells at the nonpermissive temperature did not increase the level of cells in S phase as measured by propidium iodide staining and FACS analysis. In fact, numerous analyses indicated that the MIEPs slightly enhanced the G1 block caused by the ts13 ts mutation. This agrees with previous data indicating a G1 block by the MIEPs in primary foreskin fibroblasts (39) and with data indicating the inability of the MIEPs to overcome G1 arrest in Rb-transfected Saos-2 cells (17) and in fibroblasts treated with actinomycin D (5). Thus, the inability to rescue the ts cell cycle defect in ts13 cells is to be expected and is supportive of a model, in normal human cells, where a function of the MIEPs is to establish and/or maintain a cell cycle block that inhibits the G1-to-S transition.
A consequence of blocking the cell cycle in ts13 cells at the nonpermissive temperature is the onset of apoptosis. Our data show that in this system the MIEPs effectively block the onset of apoptosis. Additionally, this function of the MIEPs is separate from the function which mediates rescue of the transcriptional defect in ts13 cells. Specifically, mutations in the MIEPs which eliminate the transcriptional rescue do not affect the ability of the MIEPs to block apoptosis.
How might our observations relate to the life cycle of HCMV? Clearly the ts13 cell line is not permissive to HCMV infection. However, we feel that our data provide insight into the functions of the MIEPs in permissive cells, since there is no comparably mutated human cell line. Many studies suggest that specific cellular genes must be induced for viral replication and that this apparently occurs under conditions where the cells will become blocked in G1, a point in the cell cycle where the cellular milieu is apparently preferred for optimal viral replication (5, 16, 17, 31, 39, 58). The transcriptional activation functions, and/or the TAF-like functions, of the MIEPs can apparently accomplish the induction of cellular gene expression which will initiate the cell cycle block at the appropriate point. However, an HCMV lytic cycle is very long, even under the most permissive conditions; for example, detectable progeny virions only begin to appear after 72 h postinfection. It would be deleterious for the success of the viral infection if the cells began to die due to the long and stressful effects of the viral infection and the cell cycle block. The data reported here and the data of others (85) suggest that the MIEPs contain a function, separable from the transcriptional activation function, which blocks apoptosis. This antiapoptosis function would maintain cells in the population long after they would normally have been eliminated by apoptosis.
What might be the consequences of the inhibition of apoptosis if the MIEPs were expressed in cells where the progression of the lytic infection may be incomplete or blocked? We would propose that damaged cells, which should be removed from the population, would be maintained. Shen et al. (67) have shown that the MIEPs can cooperate with adenovirus E1a to generate transformed foci with primary baby rat kidney cells. Their data suggest that the MIEPs are mutagenic and the resultant mutations in cellular genes predispose some of the cells to transformation in cooperation with E1a. Interestingly, MIEP gene expression appears to be transient, as viral DNA is not present in clonal cell lines derived from the transformed foci. Therefore, it has been proposed that the MIEP-induced mutagenesis contributes to oncogenesis through a hit-and-run mechanism. Our data may extend this proposal by suggesting that the MIEP may be not only mutagenic but also able to maintain the cells, through inhibition of apoptosis, in such a way that deleterious DNA damage can accumulate. Hence a putative “transient immortalization” state induced by the MIEP may be a causative effect leading to unexpected pathology, such as cancer and atherosclerosus, which have long been associated, but never definitively correlated, with HCMV.
ACKNOWLEDGMENTS
We thank Mike Malim, Wafik el-Deiry, and Bert Vogelstein for plasmids and cell lines. We give special thanks to Jonni Moore, Hank Pletcher, and Seong-Joo Jeong for help in FACS analysis and Diana Palmeri and Bruce Friedman for helpful discussions regarding immunofluorescence. We also thank all the members of the Alwine laboratory for helpful discussion and support.
D.M.L. was partially supported by a predoctoral fellowship from the Southeastern Pennsylvania Affiliate of the American Heart Association. This work was supported by Public Health Service grant CA28379 awarded to J.C.A. by the National Cancer Institute. Cheers to all.
REFERENCES
- 1.Arlt H, Lang D, Gebert S, Stamminger T. Identification of binding sites for the 86-kilodalton IE2 protein of human cytomegalovirus within an IE2-responsive viral early promoter. J Virol. 1994;68:4117–4125. doi: 10.1128/jvi.68.7.4117-4125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baracchini E, Glezer E, Fish K, Stenberg R M, Nelson J A, Ghazal P. An isoform variant of the cytomegalovirus immediate-early auto repressor functions as a transcriptional activator. Virology. 1992;188:518–529. doi: 10.1016/0042-6822(92)90506-k. [DOI] [PubMed] [Google Scholar]
- 3.Barry P A, Pratt-Lowe E, Peterlin B M, Luciw P A. Cytomegalovirus activates transcription directed by the long terminal repeat of human immunodeficiency virus type 1. J Virol. 1990;64:2932–2940. doi: 10.1128/jvi.64.6.2932-2940.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biegalke B J, Geballe A P. Sequence requirements for activation of the HIV-1 LTR by human cytomegalovirus. Virology. 1991;183:381–385. doi: 10.1016/0042-6822(91)90151-z. [DOI] [PubMed] [Google Scholar]
- 5.Bonin L R, McDougall J K. Human cytomegalovirus IE2 86-kilodalton protein binds p53 but does not abrogate G1 checkpoint function. J Virol. 1997;71:5861–5870. doi: 10.1128/jvi.71.8.5861-5870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bresnahan W, Boldogh I, Thompson E A, Albrecht T. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology. 1996;224:150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- 7.Burns L J, Waring J F, Reuter J J, Stinski M F, Ginder G D. Only the HLA class I gene minimal promoter elements are required for transactivation by human cytomegalovirus. Blood. 1993;81:1558–1566. [PubMed] [Google Scholar]
- 8.Caswell R, Bryant L, Sinclair J. Human cytomegalovirus immediate-early 2 (IE2) protein can transactivate the human hsp70 promoter by alleviation of Dr1-mediated repression. J Virol. 1996;70:4028–4037. doi: 10.1128/jvi.70.6.4028-4037.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caswell R C, Hagemeier C, Ghiou C J, Hayward G, Kouzarides T, Sinclair J. The human cytomegalovirus 86k immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J Gen Virol. 1993;74:2691–2698. doi: 10.1099/0022-1317-74-12-2691. [DOI] [PubMed] [Google Scholar]
- 10.Cherrington J M, Mocarski E S. Human cytomegalovirus ie1 transactivates the promoter-enhancer via an 18-base-pair repeat element. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colberg-Poley A M, Santomenna L D, Harlow P P, Benfield P A, Tenney D J. Human cytomegalovirus US3 and UL36-38 immediate-early proteins regulate gene expression. J Virol. 1992;66:95–105. doi: 10.1128/jvi.66.1.95-105.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damania B, Alwine J C. TAF-like function of SV40 large T antigen. Genes Dev. 1996;10:1369–1381. doi: 10.1101/gad.10.11.1369. [DOI] [PubMed] [Google Scholar]
- 13.Davis M G, Kenney S C, Kamine J, Pagano J S, Huang E-S. Immediate-early gene region of human cytomegalovirus trans-activates the promoter of human immunodeficiency virus. Proc Natl Acad Sci USA. 1987;84:8642–8646. doi: 10.1073/pnas.84.23.8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Depto A S, Stenberg R M. Functional analysis of the true late human cytomegalovirus pp28 upstream promoter: cis-acting elements and viral trans-acting proteins necessary for promoter activation. J Virol. 1992;66:3241–3246. doi: 10.1128/jvi.66.5.3241-3246.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Depto A S, Stenberg R M. Regulated expression of the human cytomegalovirus pp65 gene: octamer sequence in the promoter is required for activation by viral gene products. J Virol. 1989;63:1232–1238. doi: 10.1128/jvi.63.3.1232-1238.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dittmer D, Mocarski E S. Human cytomegalovirus infection inhibits G1/S transition. J Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortunato E A, Sommer M H, Yoder K, Spector D H. Identification of domains within the human cytomegalovirus major immediate-early 86-kilodalton protein and the retinoblastoma protein required for physical and functional interaction with each other. J Virol. 1997;71:8176–8185. doi: 10.1128/jvi.71.11.8176-8185.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furnari B A, Poma E, Kowalik T F, Huong S-M, Huang E-S. Human cytomegalovirus immediate-early gene 2 protein interacts with itself and with several novel cellular proteins. J Virol. 1993;67:4981–4991. doi: 10.1128/jvi.67.8.4981-4991.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghazal P, Young J, Giulietti E, DeMattei C, Garcia J, Gaynor R, Stenberg R M, Nelson J A. A discrete cis element in the human immunodeficiency virus long terminal repeat mediates synergistic trans activation by cytomegalovirus immediate-early proteins. J Virol. 1991;65:6735–6742. doi: 10.1128/jvi.65.12.6735-6742.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagemeier C, Caswell R, Hayhurst G, Sinclair J, Kouzarides T. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 1994;13:2897–2903. doi: 10.1002/j.1460-2075.1994.tb06584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagemeier C, Walker S, Caswell R, Kouzarides T, Sinclair J. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J Virol. 1992;66:4452–4456. doi: 10.1128/jvi.66.7.4452-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagemeier C, Walker S M, Sissons P J G, Sinclair J H. The 72K IE1 and 80K IE2 proteins of human cytomegalovirus independently trans-activate the c-fos, c-myc and hsp70 promoters via basal promoter elements. J Gen Virol. 1992;73:2385–2393. doi: 10.1099/0022-1317-73-9-2385. [DOI] [PubMed] [Google Scholar]
- 23.Hayashida T, Sekiguchi T, Noguchi E, Sunamoto H, Ohba T, Nishimoto T. The CCG1/TAFII250 gene is mutated in thermosensitive G1 mutants of the BHK21 cell line derived from golden hamster. Gene. 1994;141:267–270. doi: 10.1016/0378-1119(94)90583-5. [DOI] [PubMed] [Google Scholar]
- 24.Hayhurst G P, Bryant L A, Caswell R C, Walker S M, Sinclair J H. CCAAT box-dependent activation of the TATA-less human DNA polymerase α promoter by the human cytomegalovirus 72-kilodalton major immediate-early protein. J Virol. 1995;69:182–188. doi: 10.1128/jvi.69.1.182-188.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hermiston T W, Malone C L, Witte P R, Stinski M F. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J Virol. 1987;61:3214–3221. doi: 10.1128/jvi.61.10.3214-3221.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hisatake K, Hasegawa S, Takada R, Nakatani Y, Horikoshi M, Roeder R G. The p250 subunit of native TATA box-binding factor TFIID is the cell-cycle regulatory protein CCG1. Nature. 1993;362:179–181. doi: 10.1038/362179a0. [DOI] [PubMed] [Google Scholar]
- 27.Huang L, Malone C L, Stinski M F. A human cytomegalovirus early promoter with upstream negative and positive cis-acting elements: IE2 negates the effect of the negative element, and NF-Y binds to the positive element. J Virol. 1994;68:2108–2117. doi: 10.1128/jvi.68.4.2108-2117.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunninghake G W, Monks B G, Geist L J, Monick M M, Monroy M A, Stinski M F, Webb A C, Dayer J-M, Auron P E, Fenton M J. The functional importance of a cap site-proximal region of the human prointerleukin 1B gene is defined by viral protein trans-activation. Mol Cell Biol. 1992;12:3439–3448. doi: 10.1128/mcb.12.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iskendarian A C, Huang L, Reilly A, Stenberg R M, Anders D G. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J Virol. 1996;70:383–392. doi: 10.1128/jvi.70.1.383-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwamoto G K, Monick M M, Clark B D, Auron P E, Stinski M F, Hunninghake G W. Modulation of interleukin 1 beta gene expression by the immediate early genes of human cytomegalovirus. J Clin Investig. 1990;85:1853–1857. doi: 10.1172/JCI114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jault F M, Jault J-M, Ruchti F, Fortunato E A, Clark C, Corbeil J, Richman D D, Spector D H. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J Virol. 1995;69:6697–6704. doi: 10.1128/jvi.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jupp R, Hoffmann S, Depto A, Stenberg R M, Ghazal P, Nelson J A. Direct interaction of the human cytomegalovirus IE86 protein with the cis repression signal does not preclude TBP from binding to the TATA box. J Virol. 1993;67:5595–5604. doi: 10.1128/jvi.67.9.5595-5604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jupp R, Hoffmann S, Stenberg R M, Nelson J A, Ghazal P. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J Virol. 1993;67:7539–7546. doi: 10.1128/jvi.67.12.7539-7546.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerry J A, Priddy M A, Jervey T Y, Kohler C P, Staley T L, Vanson C D, Jones T R, Iskendarian A C, Anders D G, Stenberg R M. Multiple regulatory events influence human cytomegalovirus DNA polymerase (UL54) expression during viral infection. J Virol. 1996;70:373–382. doi: 10.1128/jvi.70.1.373-382.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerry J A, Priddy M A, Stenberg R M. Identification of sequence elements in the human cytomegalovirus DNA polymerase gene promoter required for activation by viral gene products. J Virol. 1994;68:4167–4176. doi: 10.1128/jvi.68.7.4167-4176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klucher K M, Sommer M, Kadonaga J T, Spector D H. In vivo and in vitro analysis of transcriptional activation mediated by the human cytomegalovirus major immediate-early proteins. Mol Cell Biol. 1993;13:1238–1250. doi: 10.1128/mcb.13.2.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LaFemina R L, Pizzorno M C, Mosca J D, Hayward G S. Expression of the acidic nuclear immediate-early protein (IE1) of human cytomegalovirus in stable cell lines and its preferential association with metaphase chromosomes. Virology. 1989;172:583–600. doi: 10.1016/0042-6822(89)90201-8. [DOI] [PubMed] [Google Scholar]
- 38.Lang D, Gebert S, Arlt H, Stamminger T. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J Virol. 1995;69:6030–6037. doi: 10.1128/jvi.69.10.6030-6037.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu M, Shenk T. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J Virol. 1996;70:8850–8857. doi: 10.1128/jvi.70.12.8850-8857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lukac D M, Harel N Y, Tanese N, Alwine J C. TAF-like functions of human cytomegalovirus immediate early proteins. J Virol. 1997;71:7227–7239. doi: 10.1128/jvi.71.10.7227-7239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukac D M, Manuppello J R, Alwine J C. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J Virol. 1994;68:5184–5193. doi: 10.1128/jvi.68.8.5184-5193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luu P, Flores O. Binding of SP1 to the immediate-early protein-responsive element of the human cytomegalovirus DNA polymerase promoter. J Virol. 1997;71:6683–6691. doi: 10.1128/jvi.71.9.6683-6691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42a.Maguire, T. G., and J. C. Alwine. Unpublished data.
- 43.Malone C L, Vesole D H, Stinski M F. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J Virol. 1990;64:1498–1506. doi: 10.1128/jvi.64.4.1498-1506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Margolis M J, Pajovic S, Wong E L, Wade M, Jupp R, Nelson J A, Azizkhan J C. Interaction of the 72-kilodalton human cytomegalovirus IE1 gene product with E2F1 coincides with E2F-dependent activation of dihydrofolate reductase transcription. J Virol. 1995;69:7759–7767. doi: 10.1128/jvi.69.12.7759-7767.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michelson S, Alcami J, Kim S-J, Danielpour D, Bachelerie F, Picard L, Bessia C, Paya C, Virelizier J-L. Human cytomegalovirus infection induces transcription and secretion of transforming growth factor 1. J Virol. 1994;68:5730–5737. doi: 10.1128/jvi.68.9.5730-5737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mocarski E S. Cytomegaloviruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 47.Monick M M, Geist L J, Stinski M F, Hunninghake G W. The immediate early genes of human cytomegalovirus upregulate expression of the cellular genes myc and fos. Am J Respir Cell Mol Biol. 1992;7:251–256. doi: 10.1165/ajrcmb/7.3.251. [DOI] [PubMed] [Google Scholar]
- 48.Muganda P, Mendoza O, Hernandez J, Qian Q. Human cytomegalovirus elevates levels of the cellular protein p53 in infected fibroblasts. J Virol. 1994;68:8028–8034. doi: 10.1128/jvi.68.12.8028-8034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noguchi E, Sekiguchi T, Nohiro Y, Hayashida T, Hirose E, Hayashi N, Nishimoto T. Minimal essential region of CCG1/TAFII250 required for complementing the ts cell cycle mutants, tsBN462 and ts13 cells, of hamster BHK21 cells. Somatic Cell Genet. 1994;20:505–513. doi: 10.1007/BF02255841. [DOI] [PubMed] [Google Scholar]
- 50.Paya C V, Virelizier J-L, Michelson S. Modulation of T-cell activation through protein kinase C- or A-dependent signalling pathways synergistically increases human immunodeficiency virus long terminal repeat induction by cytomegalovirus immediate-early proteins. J Virol. 1991;65:5477–5484. doi: 10.1128/jvi.65.10.5477-5484.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pizzorno M C, Mullen M-A, Chang Y-N, Hayward G S. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J Virol. 1991;65:3839–3852. doi: 10.1128/jvi.65.7.3839-3852.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pizzorno M C, O’Hare P, Sha L, LaFemina R L, Hayward G S. trans-activation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J Virol. 1988;62:1167–1179. doi: 10.1128/jvi.62.4.1167-1179.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poma E E, Kowalik T F, Zhu L, Sinclair J H, Huang E-S. The human cytomegalovirus IE1-72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J Virol. 1996;70:7867–7877. doi: 10.1128/jvi.70.11.7867-7877.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rando R F, Srinivasan A, Feingold J, Gonczol E, Plotkin S. Characterization of multiple molecular interactions between human cytomegalovirus (HCMV) and human immunodeficiency virus type 1 (HIV-1) Virology. 1990;176:87–97. doi: 10.1016/0042-6822(90)90233-h. [DOI] [PubMed] [Google Scholar]
- 55.Reifel-Miller A E, Lee C-H. Detection of an IE responsive element(s) in the BamHI J fragment of human cytomegalovirus AD169. Virology. 1990;177:496–504. doi: 10.1016/0042-6822(90)90514-r. [DOI] [PubMed] [Google Scholar]
- 56.Ruppert S, Tjian R. Human TAFII250 interacts with RAP74: implications for RNA polymerase II initiation. Genes Dev. 1995;9:2747–2755. doi: 10.1101/gad.9.22.2747. [DOI] [PubMed] [Google Scholar]
- 57.Ruppert S, Wang E H, Tjian R. Cloning and expression of human TAFII250: a TBP-associated factor implicated in cell-cycle regulation. Nature (London) 1993;362:175–179. doi: 10.1038/362175a0. [DOI] [PubMed] [Google Scholar]
- 58.Salvant B S, Fortunato E A, Spector D H. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J Virol. 1998;72:3729–3741. doi: 10.1128/jvi.72.5.3729-3741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sambucetti L C, Cherrington J M, Wilkinson G W G, Mocarski E S. NF-κB activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T cell stimulation. EMBO J. 1989;8:4251–4258. doi: 10.1002/j.1460-2075.1989.tb08610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmid I, Uittenbogaart C H, Giorgi J V. A gentle fixation and permeabilization method for combined cell surface and intracellular staining with improved precision in DNA quantification. Cytometry. 1991;12:279–285. doi: 10.1002/cyto.990120312. [DOI] [PubMed] [Google Scholar]
- 61.Schwartz R, Helmich B, Spector D H. CREB and CREB-binding proteins play an important role in the IE2 86-kilodalton protein-mediated transactivation of the human cytomegalovirus 2.2-kilobase RNA promoter. J Virol. 1996;70:6955–6966. doi: 10.1128/jvi.70.10.6955-6966.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scully A L, Sommer M H, Schwartz R, Spector D H. The human cytomegalovirus IE2 86-kDa protein interacts with an early gene promoter via site-specific DNA binding and protein-protein interactions. J Virol. 1995;69:6533–6540. doi: 10.1128/jvi.69.10.6533-6540.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sekiguchi T, Miyata T, Nishimoto T. Molecular cloning of the cDNA of human X chromosomal gene (CCG1) which complements the temperature-sensitive G1 mutants, tsBN462 and ts13 of the BHK cell line. EMBO J. 1988;7:1683–1687. doi: 10.1002/j.1460-2075.1988.tb02996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sekiguchi T, Nakashima T, Hayashida T, Kuraoka A, Hashimoto S, Tsuchida N, Shibata Y, Hunter T, Nishimoto T. Apoptosis is induced in BHK cells by the tsBN462/13 mutation in the CCG1/TAFII250 subunit of the TFIID basal transcription factor. Exp Cell Res. 1995;218:490–498. doi: 10.1006/excr.1995.1183. [DOI] [PubMed] [Google Scholar]
- 65.Sekiguchi T, Noguchi E, Hayashida T, Nakashima T, Toyoshima H, Nishimoto T, Hunter T. D-type cyclin expression is decreased and p21 and p27 CDK inhibitor expression is increased when tsBN462 CCG1/TAFII250 mutant cells arrest in G1 at the restrictive temperature. Genes Cells. 1996;1:687–705. doi: 10.1046/j.1365-2443.1996.00259.x. [DOI] [PubMed] [Google Scholar]
- 66.Sekiguchi T, Nohiro Y, Nakamura Y, Hisamoto N, Nishimoto T. The human CCG1 gene, essential for progression of the G1 phase, encodes a 210-kilodalton nuclear DNA-binding protein. Mol Cell Biol. 1991;11:3317–3325. doi: 10.1128/mcb.11.6.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen Y, Zhu H, Shenk T. Human cytomegalovirus IE1 and IE2 proteins are mutagenic and mediate “hit-and-run” oncogenic transformation in cooperation with the adenovirus E1A proteins. Proc Natl Acad Sci USA. 1997;94:3341–3345. doi: 10.1073/pnas.94.7.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sommer M H, Scully A L, Spector D H. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J Virol. 1994;68:6223–6231. doi: 10.1128/jvi.68.10.6223-6231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Speir E, Modali R, Huang E-S, Leon M B, Shawl F, Finkel T, Epstein S E. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science. 1994;265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 70.Staprans S I, Rabert D K, Spector D H. Identification of sequence requirements and trans-acting functions necessary for regulated expression of a human cytomegalovirus early gene. J Virol. 1988;62:3463–3473. doi: 10.1128/jvi.62.9.3463-3473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stasiak P C, Mocarski E S. Transactivation of the cytomegalovirus ICP36 gene promoter requires the α gene product TRS1 in addition to IE1 and IE2. J Virol. 1992;66:1050–1058. doi: 10.1128/jvi.66.2.1050-1058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stenberg R M, Depto A S, Fortney J, Nelson J A. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate-early gene region. J Virol. 1989;63:2699–2708. doi: 10.1128/jvi.63.6.2699-2708.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stenberg R M, Fortney J, Barlow S W, Magrane B P, Nelson J A, Ghazal P. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J Virol. 1990;64:1556–1565. doi: 10.1128/jvi.64.4.1556-1565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stenberg R M, Witte P R, Stinski M F. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J Virol. 1985;56:665–675. doi: 10.1128/jvi.56.3.665-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suzuki-Yagawa Y, Guermah M, Roeder R G. The ts13 mutation in the TAFII250 subunit (CCG1) of TFIID directly affects transcription of D-type cyclin genes in cells arrested in G1 at the nonpermissive temperature. Mol Cell Biol. 1997;17:3284–3294. doi: 10.1128/mcb.17.6.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Talavera A, Basilico C. Temperature sensitive mutants of BHK cells affected in cell cycle progression. J Cell Physiol. 1977;92:425–436. doi: 10.1002/jcp.1040920310. [DOI] [PubMed] [Google Scholar]
- 77.Tevethia M J, Spector D J, Leisure K M, Stinski M F. Participation of two human cytomegalovirus immediate early gene regions in transcriptional activation of adenovirus promoters. Virology. 1987;161:276–285. doi: 10.1016/0042-6822(87)90119-x. [DOI] [PubMed] [Google Scholar]
- 78.Wade M, Kowalik T F, Mudryj M, Huang E-S, Azizkhan J C. E2F mediates dihydrofolate reductase promoter activation and multiprotein complex formation in human cytomegalovirus infection. Mol Cell Biol. 1992;12:4364–4374. doi: 10.1128/mcb.12.10.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walker S, Hagemeier C, Sissons J G P, Sinclair J H. A 10-base-pair element of the human immunodeficiency virus type 1 long terminal repeat (LTR) is an absolute requirement for transactivation by the human cytomegalovirus 72-kilodalton IE1 protein but can be compensated for by other LTR regions in transactivation by the 80-kilodalton IE2 protein. J Virol. 1992;66:1543–1550. doi: 10.1128/jvi.66.3.1543-1550.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang E H, Tjian R. Promoter-selective transcriptional defect in cell cycle mutant ts13 rescued by hTAFII250. Science. 1994;263:811–814. doi: 10.1126/science.8303298. [DOI] [PubMed] [Google Scholar]
- 81.Wang E H, Zou S, Tjian R. TAFII250-dependent transcription of cyclin A is directed by ATF activator proteins. Genes Dev. 1997;11:2658–2669. doi: 10.1101/gad.11.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yeung K C, Stoltzfus C M, Stinski M F. Mutations of the human cytomegalovirus immediate-early 2 protein define regions and amino acid motifs important in transactivation of transcription from the HIV-1 LTR promoter. Virology. 1993;195:786–792. doi: 10.1006/viro.1993.1431. [DOI] [PubMed] [Google Scholar]
- 83.Yoo Y D, Chiou C-J, Choi K S, Yi Y, Michelson S, Kim S, Hayward G S, Kim S-J. The IE2 regulatory protein of human cytomegalovirus induces expression of the human transforming growth factor B1 gene through an Egr-1 binding site. J Virol. 1996;70:7062–7070. doi: 10.1128/jvi.70.10.7062-7070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yurochko A D, Kowalik T F, Huong S-M, Huang E-S. Human cytomegalovirus upregulates NF-κB activity by transactivating the NF-κB p105/p50 and p65 promoters. J Virol. 1995;69:5391–5400. doi: 10.1128/jvi.69.9.5391-5400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]