Abstract
Purpose of review
Hematopoietic stem cells (HSC) are functionally heterogeneous in a clone-specific manner. The complexity of that heterogeneous mix of cells is progressively lost with age as a myeloid-dominant hematopoietic system is established. Yet, the function of this diversity, as well as the consequences of its loss, remains unknown. This review will bring together recent advances in HSC diversity and novel insights into myeloid heterogeneity and specification in order to bring focus on how this may affect the ageing individual.
Recent findings
The ageing haematopoietic system is dominated by a low number of active HSC clones that produce an excess of myeloid cells. In addition, individual myeloid progenitors and their mature progeny are proving to be more functionally restricted than previously recognized. The presence or absence of a particular type of myeloid cell can greatly affect the outcome of various pathological processes.
Summary
Myeloid cells are important drivers of many ageing-associated diseases. The loss of HSC heterogeneity, with a possible concomitant restriction of myeloid cell diversity, could significantly impact health during ageing.
Keywords: ageing, haematopoietic stem cell heterogeneity, myeloid cell diversity
All haematopoietic stem cells (HSCs) were thought to have been created equal. That has now fundamentally changed. As our ability to analyse and track individual HSCs has evolved, so has the concept of what cells comprise the group bundled under the term HSC. It is now widely acknowledged that individual HSCs differ in their lineage potency as well as their production rate [1]. An aspect of HSC biology particularly impacted by stem cell heterogeneity is ageing. The ageing haematopoietic system is primarily characterized by a skewing towards myelopoiesis and a loss of lymphocyte differentiation [2]. This imbalance is thought to contribute to increased cardiovascular disease and cancer in ageing wherein myeloid-driven inflammation and reduced immune surveillance, respectively, play central roles [3,4].
The implications of declining HSC diversity do however go beyond imbalances between blood cell lineages. It is now apparent that clonal diversity among HSCs changes with time and dominant clones become progressively evident with frequencies estimated to range from 10 to 95% by age 70 [5,6]. Loss of HSC heterogeneity is therefore not just qualitative but quantitative. This is also seen in more mature cell populations such as lymphoid cells wherein clonality is readily measured. Lymphoid populations as a whole may not change, but diversity [7]. The myeloid compartment has generally been viewed as more simple, limited to a few cell types with innate immune function and high adaptability. This is however changing. Single-cell technologies have revealed an unexpected degree of heterogeneity among myeloid progenitors as well as mature myeloid cells. In addition, functional testing has demonstrated that different myeloid subsets significantly impact various disease states [8,9]. Although myeloid heterogeneity is unlikely to be as great as lymphoid, ongoing definition of diversity among myeloid populations raises that prospect that they too will undergo changes in relative complexity with ageing contributing to specific vulnerabilities. Advances in HSC and myeloid cell biology with a particular focus on changes with age will be the focus of this essay.
HAEMATOPOIETIC STEM CELL HETEROGENEITY AND AGEING
Single-cell transplantation studies as well in-vivo lineage tracing experiments have shown that individual HSCs in the young adult display a wide range of characteristics. HSC clones display a remarkable consistency when it comes to mature lineage output across multiple bone marrow transplant recipients, as well as, throughout serial transplantations [10-12]. Indicating that lineage commitment is mainly a cell-intrinsic trait. These studies have defined at least four subtypes of HSCs. Balanced HSCs that are capable of production of both myeloid and lymphoid cell types, megakaryocytic or megakaryocytic/erythroid stem cells and myeloid-biased stem cells [10,11,13,14■,15,16]. Lymphoid-dominant stem cell clones have been observed in primary bone marrow transplantation recipients, but these cells mostly fail to propagate in secondary recipients suggesting that they are lineage-restricted progenitors rather than true HSCs [10,11,14■,16]. Proliferation and self-renewal are additional stem cell features that appear to be hardwired in individual clones [11,12]. In fact, HSCs exhibit stereotypical behaviours even in the face of systemic stressors such as genotoxic injury or an inflammatory challenge where the output and lineage of descendent cells were clone specific and stable across multiple individuals [12].
Yet, interclonal variation was considerable with some clones being very sensitive to particular stresses leading to their depletion [12]. Given that age imposes many and diverse genotoxic and inflammatory stresses, clonal complexity will invariably change. In addition, the ageing bone marrow microenvironment displays changes in availability of secreted factors [17,18] and cellular composition [19,20] likely adding more selection pressure on HSC clones. Finally, cell intrinsic changes such as progressive deficits in DNA repair and cell polarity [21-23] appear to accumulate with age adding selection pressure. The result is broadly understood on a population basis as a shift towards myeloid-producing cells [13,24-26,27■■] and with advances in tracking such as fluorescent tracing of erythroid-megakaryocytic lineages, an increase in platelet-biased HSC [27■■,28]. Although the immunophenotypic HSC pool size is thought to increase, the regenerative ability on a per cell basis is reduced [24,26,29,30] and the self-renewing ability of HSCs is impaired [24,27■■]. These features are not dissimilar to the lymphoid system wherein age does not compromise overall T cell number but does decrease clonal diversity [7]. As more clone-specific functional tracing of HSC becomes available, it is likely this will demonstrate more precisely which functions are lost or enhanced as clonal compression evolves.
Results from human studies suggest that clonal shifts do occur with age with fewer HSCs active in older individuals [31-33]. Some clones clearly become dominant and mosaicism emerges [6]. Those who acquire a mutation in a known leukaemia-associated gene, clonal haematopoiesis of indeterminate potential (CHIP), have a significantly increased risk of developing a haematological malignancy [34-36]. CHIP also increases the incidence of cardiovascular disease. In about 10% of CHIP patients, this is caused by mutations in the epigenetic regulator TET2, which augments the inflammatory phenotype of monocytes and macrophages [37,38]. In the remaining fraction, the driver of cardiovascular disease is unknown. Many of these individuals will have mutations in other leukaemia-associated genes such as DNMT3a and ASXL1 and it is possible that they confer a similar pro-inflammatory profile to myeloid cells [34-36]. Still, approximately 40% of all patients with a dominant clone have no detectable candidate driver mutations [34]. The shift in mosaicism itself may impose risks as from ecology to adaptive immunity, a high degree of diversity ensures system resilience. Evidence for diverse subpopulations among innate immune cells of the myeloid lineage further raises the prospect of shifts in clonal complexity being accompanied by shifts in resilience with age.
MYELOID CELL SUBSETS
The myeloid system has long been known to have some degree of variability, as granulocytes, macrophages and monocytes have all been divided into subsets. Some, like the granulocytic series, have highly distinctive cell types from distinctive differentiation programmes, neutrophils, eosinophils and basophils, and at least neutrophils acquire further functional diversity based on their relative age [9]. An ontogenic distinction also appears to occur among macrophages. Tissue-resident macrophages originally derived yolk-sac localized embryonic precursors that seed the tissue before or just after birth [39-41]. In some tissues such as brain and liver, these remain for long periods and are capable of self-replenishment. Others such as in the intestine, turnover more rapidly and are replenished by bone marrow derived monocytes. The tissue resident macrophages have common molecular features yet do acquire tissue-specific transcriptional programmes that becomes established in the tissue. When these macrophages are replaced by monocyte-derived cells from the adult bone marrow [39,42], they acquire a molecular programme resembling but not always matching that of the endogenous cells [43]. Therefore, diversity in the myeloid system can be ontologically imposed, shaped by the environment and acquired.
Macrophage diversity beyond that of the professional tissue-resident cells is also evident. The M1 and M2 macrophage subsets have been defined in the mouse based on the factors they secrete and the functions they execute, though this crude bundling of cell states is thought by many to underrepresent the complexity of the cell type. The M1 and M1 cell state has been largely regarded as a common cell responding to different microenvironmental cues rather than inherent properties [44]. Also, circulating monocytes have been categorized into two groups, classical, pro-inflammatory cells and non-inflammatory, patrolling monocytes [45,46]. The mature cell diversity within monocyte/macrophages is therefore largely regarded as an acquired state. A common population of cells responding to cues and transitioning into a modified state in response. The caveat here is that the studies have been conducted with cell populations, similar to the historic studies characterizing HSC. As technologies for resolving HSC activity to the single cell level have emerged, concepts of the HSC pool have changed with them. This certainly raises the possibility that what are currently viewed as a highly plastic set of monocyte/macrophages acquiring different states is actually a mix of cells with preferential outgrowth of the subsets primed to respond to a particular stimulus. What is currently viewed as cell induction cannot be definitively proven until clonal analyses enable exclusion of clonal selection.
MYELOID PROGENITOR DIVERSITY
Classical haematology teaches us that a multipotent HSC sits atop a pyramid of hierarchically ordered progenitors that will eventually give rise to mature cells. Although functional studies of single HSCs have already overturned one part of this paradigm, single-cell sequencing technologies of HSCs and myeloid progenitors might upend the other half. In humans, epigenetic and transcriptional programmes confirm the existence of lineage primed HSC; previously, well defined myeloid progenitor subsets have however been put into question [12,47■,48]. In the established haematopoietic tree, common myeloid progenitors (CMPs) can generate all types of myeloid cells from platelets and erythrocytes to granulocytes and monocytes, whereas the granulocyte-monocyte progenitor (GMP) has been restricted to only two lineages [49,50]. On a molecular level, on the contrary, these populations displayed a significant degree of diversity, with unipotent cell states being more common than multipotent [47■,48]. Transcriptomic analysis of mouse myeloid progenitors demonstrated a similarly diverse array of transcriptional states. Cells phenotypically defined as CMPs were found to mostly consist of unipotent progenitors, including platelet and dendritic cell dominant progenitor subtypes [51]. Contrarily, interrogation of the GMP compartment suggests the existence of transient bipotent as well as unipotent progenitors capable of either just granulocyte or monocyte differentiation [52]. Some of the information from these data sets appear to be in contradiction [47■,48,51,52]. What can be concluded, however, is that populations of progenitors previously considered as uniform are actually heterogeneous, potentially specialized enough to give rise to distinct myeloid subtypes.
Functional testing of myeloid progenitors suggests that functional specialization beyond the broad categories of cell type exist. Also, lineage sources are less unilinear than historic models reflected, as, for example, monocytes have been shown to come from two types of progenitor sources, GMPs and monocyte-DC progenitors (MDPs) [49,50,53]. At the transcriptional level, the two types of monocytes also differ, GMP-derived cells share gene expression signatures with granulocytes, whereas MDP progeny has more in common with macrophages and dendritic cells. Accordingly, different stimuli elicit divergent responses. GMPs only respond to LPS stimulation, whereas MDPs specifically respond to CpG administration implying that different types of infections activate distinct subsets of monocytic cells [54■■]. Further evidence for progenitor diversity comes from a model of lung fibrosis. The progression of the disease is linked to a particular type of monocyte (Ceacam1+Msr1+Lc6C-F4/80-Mac1+) that shares both transcriptional and morphological features with granulocytes. This fibrosis-inducing monocyte was produced by a dedicated GMP and disease could be completely abrogated when this differentiation axis was inhibited [55■■]. Therefore, myeloid progenitor heterogeneity is not just a curiosity found in sequencing data; it is a very real property of the haematopoietic system that can have a significant impact on disease outcomes.
MATURE MYELOID CELL DIVERSITY
Single-cell RNA sequencing of human peripheral blood is indicating a substantially greater number of cell states among monocytic and dendritic cell populations than previously recognized [56]. Similarly, murine proinflammatory monocytes are now thought to include two novel subtypes that differ in their ability to differentiate into dendritic cells and macrophages, respectively [57]. Further, the types of macrophages present in atherosclerotic lesions seem to vary with stage of disease and go beyond the M1/M2 paradigm. Active disease progression is associated with multiple proinflammatory macrophage subtypes, whereas cells with an anti-inflammatory profile arise in regression [58]. Also, a specific subset of monocytes has been associated with limiting injury-induced fibrosis. If Lyve1hiMHCIIlo macrophages are absent in the lung, experimental fibrosis is exacerbated [59■■].
There is also clear diversity among neutrophils. This cell type was historically viewed as a short-lived foot soldier akin to a chess pawn. A distinct subset of neutrophils is however specifically recruited to areas of hypoxia. Once at the site, these neutrophils increase angiogenesis, thus facilitating engraftment of transplanted Islets of Langerhans [60,61]. Also, lung cancer induces changes in the bone marrow microenvironment that in turn drive the development of a specialized SiglecFhi neutrophil population essential for tumour progression [62■]. The composition of the mature myeloid cell pool can consequently have a significant impact on various aspects of health and disease.
CONCLUSION
We are born with an HSC population with diverse properties, a diversity that appears to be gradually lost as we go through life. If model organisms and gene therapy studies are correct, reduced numbers of mostly myeloid biased stem cells remain active over time. Myeloid cell populations themselves are heterogenous and decidedly specialized. Some of these populations are directly generated from distinctive progenitor populations as is the case with yolk-sac derived tissue resident macrophages. However, these lineage-specified cells are also shaped by the microenvironment in which they reside leading to increased diversity within a subpopulation of cells [63]. Therefore, heterogeneity is both prescripted by cell origin and influenced by the tissue environment.
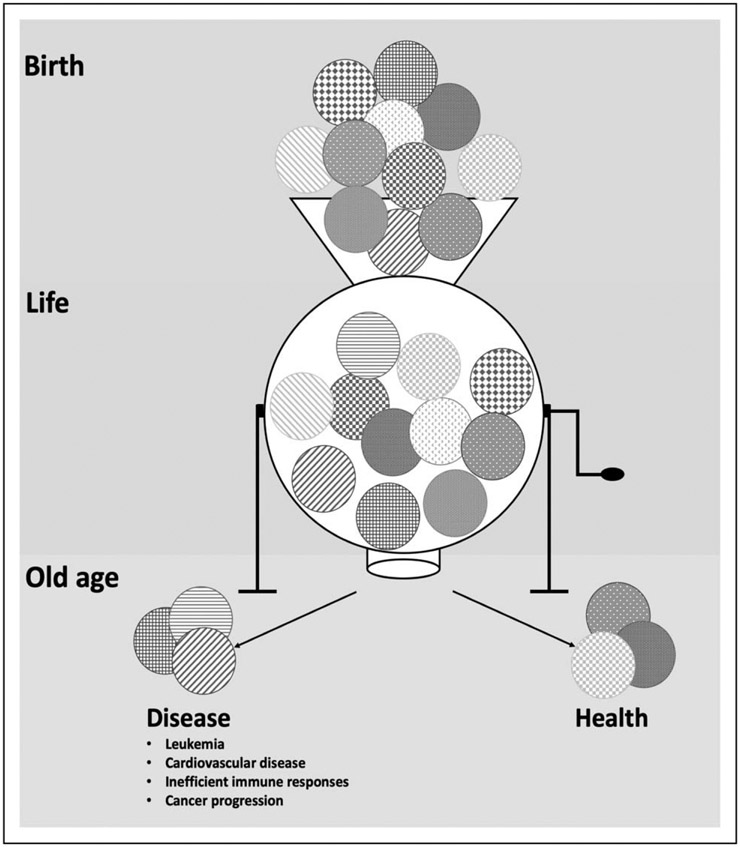
The consequences of myeloid cell diversity can be profound. This is evident in experimental models wherein specific cell subsets determine fibrosis and scarring and differential handling of inflammatory stimuli. These findings suggest that the ability to respond to injuries, pathogens and mutagenic events will be influenced by the abundance of specific subsets of myeloid cells. Therefore, the degree of heterogeneity may determine the relative vulnerability or resilience against particular challenges (Fig. 1). What we do not know is just how much of myeloid diversity is established and bounded by stable epigenetic determinants set at the progenitor stage, compared with that which is induced by the particular challenges at hand. The case of tissue resident macrophages suggests that both types of fate determination are present. However, the fibrosis models suggest that for at least some key subsets and cell functions, plasticity is far more limited. The cells appear determined by their development and a compensatory shift in fate by other myeloid cells does not occur when the subset is lost. If this is correct, then a broad plasticity among myeloid cells is no longer a defensible intellectual construct. Rather, specific subsets of cells have specific functions established in their development. When a particular subset is seen in greater or lesser abundance in a disease setting, it is likely that the subset is selectively recruited or expanded, not that generic myeloid cells are induced to have the phenotype observed. Selection may account for the particular tumour-associated macrophages, for example. If with age and with particular challenges we are selecting for and against particular subsets of myeloid cells, then the diversity we have may determine our relative susceptibility to challenges going forward (Fig. 1). Further, if particular subsets do indeed represent a stable cell state, the possibility of adoptive transfer of such cells or select killing of them for therapy can be envisioned. It may be that we can define what arrows remain in the quiver as we age and selectively replenish or remove them based on what lies ahead.
FIGURE 1.
Schematic overview of how loss of heterogeneity may affect the haematopoietic system. The diversity of cell populations within the haematopoietic stem and progenitor pools changes with age. Progressively clonal diversity will change and mosaicism from the clones with shift. Depending on the composition of the clones that comprise our haematopoietic system as we age, our resilience or vulnerability to particular challenges will shift.
KEY POINTS.
Haematopoietic cell plasticity has been progressively called into question with new clonal analytic techniques indicating clone-specific behaviours.
Ageing results in clonal aberrations altering the complexity of cells comprising the blood.
The myeloid system is increasingly recognized as heterogeneous and functional restrictions exist among some myeloid cell types.
The relative abundance of specific subsets of myeloid cells may determine resilience and vulnerability to immune challenges as we age.
Financial support and sponsorship
This study was supported by grants from NIH DK107784 and HL131477 (DTS), the Swedish Research Council (KG) and the Gerald and Darlene Jordan Professor of Medicine Chair (DTS).
Footnotes
Conflicts of interest
DTS is a director and stockholder of Magenta Therapeutics, Agios Pharmaceuticals, Editas Medicines, Clear Creek Bio, Red Oak Medicines and LifeVaultBio. He is a stockholder of Fate Therapeutics and a SAB member/consultant for FOG Pharma, Magenta Therapeutics, Bone Therapeutics and a consultant for VCanBio.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
■ of special interest
■■ of outstanding interest
- 1.Haas S, Trumpp A, Milsom MD. Causes and consequences of hematopoietic stem cell heterogeneity. Cell Stem Cell 2018; 22:627–638. [DOI] [PubMed] [Google Scholar]
- 2.Akunuru S, Geiger H. Aging, clonality, and rejuvenation of hematopoietic stem cells. Trends Mol Med 2016; 22:701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frodermann V, Nahrendorf M. Macrophages and cardiovascular health. Physiol Rev 2018; 98:2523–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akbar AN, Henson SM, Lanna A. Senescence of T Lymphocytes: implications for enhancing human immunity. Trends Immunol 2016; 37:866–876. [DOI] [PubMed] [Google Scholar]
- 5.Steensma DP. Clinical consequences of clonal hematopoiesis of indeterminate potential. Blood Adv 2018; 2:3404–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun 2016; 7:12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yanes RE, Gustafson CE, Weyand CM, Goronzy JJ. Lymphocyte generation and population homeostasis throughout life. Semin Hematol 2017; 54:33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guilliams M, Mildner A, Yona S. Developmental and functional heterogeneity of monocytes. Immunity 2018; 49:595–613. [DOI] [PubMed] [Google Scholar]
- 9.Ng LG, Ostuni R, Hidalgo A. Heterogeneity of neutrophils. Nat Rev Immunol 2019; 19:255–265. [DOI] [PubMed] [Google Scholar]
- 10.Dykstra B, Kent D, Bowie M, et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell 2007; 1:218–229. [DOI] [PubMed] [Google Scholar]
- 11.Muller-Sieburg CE, Cho RH, Thoman M, et al. Deterministic regulation of hematopoietic stem cell self-renewal and differentiation. Blood 2002; 100:1302–1309. [PubMed] [Google Scholar]
- 12.Yu VWC, Yusuf RZ, Oki T, et al. Epigenetic memory underlies cell-autonomous heterogeneous behavior of hematopoietic stem cells. Cell 2016; 167:1310–1322; e1317. [DOI] [PubMed] [Google Scholar]
- 13.Beerman I, Bhattacharya D, Zandi S, et al. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc Natl Acad Sci U S A 2010; 107:5465–5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.■. Carrelha J, Meng Y, Kettyle LM, et al. Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature 2018; 554:106–111. A study demonstarting the existence of platlet-restricted HSCs contributing to posttransplantation as well as steady-state haematopoieisis.
- 15.Sanjuan-Pla A, Macaulay IC, Jensen CT, et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature 2013; 502:232–236. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto R, Morita Y, Ooehara J, et al. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell 2013; 154:1112–1126. [DOI] [PubMed] [Google Scholar]
- 17.Ergen AV, Boles NC, Goodell MA. Rantes/Ccl5 influences hematopoietic stem cell subtypes and causes myeloid skewing. Blood 2012; 119:2500–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guidi N, Sacma M, Standker L, et al. Osteopontin attenuates aging-associated phenotypes of hematopoietic stem cells. EMBO J 2017; 36:840–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kusumbe AP, Ramasamy SK, Itkin T, et al. Age-dependent modulation of vascular niches for haematopoietic stem cells. Nature 2016; 532:380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh L, Brennan TA, Russell E, et al. Aging alters bone-fat reciprocity by shifting in vivo mesenchymal precursor cell fate towards an adipogenic lineage. Bone 2016; 85:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flach J, Bakker ST, Mohrin M, et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 2014; 512:198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Florian MC, Dorr K, Niebel A, et al. Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. Cell Stem Cell 2012; 10:520–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossi DJ, Seita J, Czechowicz A, et al. Hematopoietic stem cell quiescence attenuates DNA damage response and permits DNA damage accumulation during aging. Cell Cycle 2007; 6:2371–2376. [DOI] [PubMed] [Google Scholar]
- 24.Dykstra B, Olthof S, Schreuder J, et al. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J Exp Med 2011; 208:2691–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossi DJ, Bryder D, Zahn JM, et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci U S A 2005; 102:9194–9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sudo K, Ema H, Morita Y, Nakauchi H. Age-associated characteristics of murine hematopoietic stem cells. J Exp Med 2000; 192:1273–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.■■. Yamamoto R, Wilkinson AC, Ooehara J, et al. Large-scale clonal analysis resolves aging of the mouse hematopoietic stem cell compartment. Cell Stem Cell 2018; 22:600–607; e604. This study reveals that myeloid-biased stem cells, in general, and platelet-restricted stem cells, in particular, increase in numbers with age.
- 28.Grover A, Sanjuan-Pla A, Thongjuea S, et al. Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat Commun 2016; 7:11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janzen V, Forkert R, Fleming HE, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature 2006; 443:421–426. [DOI] [PubMed] [Google Scholar]
- 30.Morrison SJ, Wandycz AM, Akashi K, et al. The aging of hematopoietic stem cells. Nat Med 1996; 2:1011–1016. [DOI] [PubMed] [Google Scholar]
- 31.Busque L, Mio R, Mattioli J, et al. Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood 1996; 88:59–65. [PubMed] [Google Scholar]
- 32.Gale RE, Fielding AK, Harrison CN, Linch DC. Acquired skewing of X-chromosome inactivation patterns in myeloid cells of the elderly suggests stochastic clonal loss with age. Br J Haematol 1997; 98:512–519. [DOI] [PubMed] [Google Scholar]
- 33.Holstege H, Pfeiffer W, Sie D, et al. Somatic mutations found in the healthy blood compartment of a 115-yr-old woman demonstrate oligoclonal hematopoiesis. Genome Res 2014; 24:733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Genovese G, Kahler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014; 371:2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014; 371:2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014; 20:1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuster JJ, MacLauchlan S, Zuriaga MA, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 2017; 355:842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaiswal S, Natarajan P, Silver AJ, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med 2017; 377:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hashimoto D, Chow A, Noizat C, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 2013; 38:792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulz C, Gomez Perdiguero E, Chorro L, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 2012; 336:86–90. [DOI] [PubMed] [Google Scholar]
- 41.Yona S, Kim KW, Wolf Y, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 2013; 38:79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van de Laar L, Saelens W, De Prijck S, et al. Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity 2016; 44:755–768. [DOI] [PubMed] [Google Scholar]
- 43.Bain CC, Hawley CA, Garner H, et al. Long-lived self-renewing bone marrow-derived macrophages displace embryo-derived cells to inhabit adult serous cavities. Nat Commun 2016; 7:ncomms11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gordon S. Alternative activation of macrophages. Nat Rev Immunol 2003; 3:23–35. [DOI] [PubMed] [Google Scholar]
- 45.Auffray C, Fogg D, Garfa M, et al. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science 2007; 317:666–670. [DOI] [PubMed] [Google Scholar]
- 46.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 2003; 19:71–82. [DOI] [PubMed] [Google Scholar]
- 47.■. Buenrostro JD, Corces MR, Lareau CA, et al. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell 2018; 173:1535–1548; e1516. A comprehensive analysis of epigenitic as well as transcriptional states of haematopoietic stem and progenitors.
- 48.Velten L, Haas SF, Raffel S, et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat Cell Biol 2017; 19:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 2000; 404:193–197. [DOI] [PubMed] [Google Scholar]
- 50.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 1997; 91:661–672. [DOI] [PubMed] [Google Scholar]
- 51.Paul F, Arkin Y, Giladi A, et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell 2015; 163:1663–1677. [DOI] [PubMed] [Google Scholar]
- 52.Olsson A, Venkatasubramanian M, Chaudhri VK, et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 2016; 537:698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fogg DK, Sibon C, Miled C, et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science 2006; 311:83–87. [DOI] [PubMed] [Google Scholar]
- 54.■■. Yanez A, Coetzee SG, Olsson A, et al. Granulocyte-monocyte progenitors and monocyte-dendritic cell progenitors independently produce functionally distinct monocytes. Immunity 2017; 47:890–902; e894. The authors show that monocytes are derived from two distinct progenitor sources. These monocytes also differ at the transcriptional level and in their response to infectious stimuli.
- 55.■■. Satoh T, Nakagawa K, Sugihara F, et al. Identification of an atypical monocyte and committed progenitor involved in fibrosis. Nature 2017; 541:96–101. A distinct population of GMPs produce a monocyte subtype that drive lung fibrosis.
- 56.Villani AC, Satija R, Reynolds G, et al. Single-cell RNA-seq reveals newtypes of human blood dendritic cells, monocytes, and progenitors. Science 2017; 356:pii: eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Menezes S, Melandri D, Anselmi G, et al. The heterogeneity of Ly6C(hi) monocytes controls their differentiation into iNOS(+) macrophages or monocyte-derived dendritic cells. Immunity 2016; 45:1205–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin JD, Nishi H, Poles J, et al. Single-cell analysis of fate-mapped macrophages reveals heterogeneity, including stem-like properties, during atherosclerosis progression and regression. JCI Insight 2019; 4:pii: 124574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.■■. Chakarov S, Lim HY, Tan L, et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 2019; 363:pii: eaau0964. Two monocyte-derived tissue-resident macrophage subsets were identified using scRNA seq. The absence of one of these subsets was shown to aggravate tissue fibrosis.
- 60.Christoffersson G, Vagesjo E, Vandooren J, et al. VEGF-A recruits a proangiogenic MMP-9-delivering neutrophil subset that induces angiogenesis in transplanted hypoxic tissue. Blood 2012; 120:4653–4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Massena S, Christoffersson G, Vagesjo E, et al. Identification and characterization of VEGF-A-responsive neutrophils expressing CD49d, VEGFR1, and CXCR4 in mice and humans. Blood 2015; 126:2016–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.■. Engblom C, Pfirschke C, Zilionis R, et al. Osteoblasts remotely supply lung tumors with cancer-promoting SiglecF(high) neutrophils. Science 2017; 358:pii: eaal5081. Lung cancer tumours intsruct the bone marrow to produce a specific, tumour-promoting neutrophil.
- 63.Mass E, Ballesteros I, Farlik M, et al. Specification of tissue-resident macrophages during organogenesis. Science 2016; 353:pii: aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]