Abstract
The nucleocapsid core protein of hepatitis C virus (HCV) has been shown to trans-act on several viral or cellular promoters. To get insight into the trans-action mechanism of HCV core protein, a yeast two-hybrid cloning system was used for identification of core protein-interacting cellular protein. One such cDNA clone encoding the DEAD box family of putative RNA helicase was obtained. This cellular putative RNA helicase, designated CAP-Rf, exhibits more than 95% amino acid sequence identity to other known RNA helicases including human DBX and DBY, mouse mDEAD3, and PL10, a family of proteins generally involved in translation, splicing, development, or cell growth. In vitro binding or in vivo coimmunoprecipitation studies demonstrated the direct interaction of the full-length/matured form and C-terminally truncated variants of HCV core protein with this targeted protein. Additionally, the protein’s interaction domains were delineated at the N-terminal 40-amino-acid segment of the HCV core protein and the C-terminal tail of CAP-Rf, which encompassed its RNA-binding and ATP hydrolysis domains. Immunoblotting or indirect immunofluorescence analysis revealed that the endogenous CAP-Rf was mainly localized in the nucleus and to a lesser extent in the cytoplasm, and when fused with FLAG tag, it colocalized with the HCV core protein either in the cytoplasm or in the nucleus. Similar to other RNA helicases, this cellular RNA helicase has nucleoside triphosphatase-deoxynucleoside triphosphatase activity, but this activity is inhibited by various forms of homopolynucleotides and enhanced by the HCV core protein. Moreover, transient expression of HCV core protein in human hepatoma HuH-7 cells significantly potentiated the trans-activation effect of FLAG-tagged CAP-Rf or untagged CAP-Rf on the luciferase reporter plasmid activity. All together, our results indicate that CAP-Rf is involved in regulation of gene expression and that HCV core protein promotes the trans-activation ability of CAP-Rf, likely via the complex formation and the modulation of the ATPase-dATPase activity of CAP-Rf. These findings provide evidence that HCV may have evolved a distinct mechanism in alteration of host cellular gene expression regulation via the interaction of its nucleocapsid core protein and cellular putative RNA helicase known to participate in all aspects of cellular processes involving RNA metabolism. This feature of core protein may impart pleiotropic effects on host cells, which may partially account for its role in HCV pathogenesis.
Hepatitis C virus (HCV) is a major causative agent of non-A, non-B hepatitis (11, 46), and its infections often progress to chronic hepatitis, cirrhosis, and hepatocellular carcinoma (6, 79). The HCV viral genome is a positive-strand RNA about 9.5 kb long that encodes a single large polyprotein of about 3,010 to 3,033 amino acids (10, 12, 42, 66, 90). This polyprotein undergoes proteolytic processing to yield 10 mature viral proteins (27, 28, 58). Among these, the core protein of HCV located at the N terminus of the polyprotein is a component of viral capsid, which is highly basic and is capable of binding RNA (82).
Apart from its role in nucleocapsid assembly, emerging evidence suggests that the core protein of HCV has additional biological properties. For example, it is phosphorylated (86) and has both cytoplasmic and nuclear localizations (61, 82, 87). Additionally, the core protein has effects on cell growth and proliferation (9, 71–73, 78, 102). Recently, studies from several laboratories including ours have identified several cellular factors that can associate with the HCV core protein. Specifically, it interacts with lymphotoxin-β receptor (9, 65), tumor necrosis factor alpha type 1 receptor (102), apolipoprotein AII (2), and heterogeneous nuclear ribonucleoprotein K (35) and influences their biological functions. Furthermore, the core protein of HCV has been shown to trans-act on several viral or cellular promoters (43, 70, 74, 87). More specifically, it activates the promoter of c-myc, simian virus 40 early promoter, and Rous sarcoma virus long terminal repeat but suppresses the promoters for hepatitis B virus, c-fos, p53, the beta interferon gene, β-actin, and the human immunodeficiency virus type 1 long terminal repeat (43, 70, 74, 87). At present, the exact mechanism for this plethoric effect of core protein on several promoters is still elusive. To understand more about the trans-action mechanism of the core protein, we have adopted the yeast two-hybrid system to identify cellular proteins that interact with the core protein. One such candidate as reported in this study belongs to a DEAD box family protein-RNA helicase (for reviews, see references 21, 83, and 95).
The DEAD family proteins have been found in a wide range of organisms, ranging from Escherichia coli to mammals (for reviews, see references 21, 83, and 95). They are divided into several subgroups based on the DEAD box motif (e.g., DEAD, DEAH, and DEXH subfamilies), and many of them appear to have a modulatory effect on RNA secondary structure and thus participate in disparate cellular functions including RNA splicing, translation, ribosome assembly, spermatogenesis, embryogenesis, cell growth, and division (21, 83, 95). This family is characterized by a core region of 300 to 360 amino acids that contains eight domains of strong peptide sequence conservation (21). Among them, the first domain, AXXGXGKT, has been described as the A motif of ATPase (92) which is involved in ATP binding (77). The fifth domain (DEAD) is the B motif of ATPase (92) that is also involved in ATP binding and/or ATP hydrolysis (26, 59). Its adjacent SAT motif (sixth motif) is required for RNA unwinding and may also couple ATP hydrolysis to RNA unwinding (68). The eighth domain (HRIGRXXR) is part of a basic region and is involved in RNA binding and ATP hydrolysis (67). Notably, sequence comparison of these DEAD family proteins reveals that the conserved core region in most cases is flanked by N-terminal and C-terminal extensions that have little in common but may determine the specific function of the individual protein. Thus, an expanding number of proteins in the DEAD family may reflect different functions, RNA substrates, or tissue specificities of RNA helicases. Intriguingly, while nucleoside triphosphatase (NTPase) activities have been demonstrated in nearly all members of purified RNA helicases, for most members of the family except eIF-4A/4B, p68, An3, Vasa, or viral helicases (32, 34, 41, 44, 49, 56, 84, 89, 94), an RNA helicase activity has not yet been demonstrated.
In this study, we show that the HCV core protein interacts with a putative cellular RNA helicase closely related to a subgroup of DEAD box proteins including human DBX and DBY found in the X and Y chromosomes, respectively (48), murine mDEAD3 (24), and PL10 expressed in the male germ line (54). The biological functions of these RNA helicases except for PL10 have not yet been established. We show here that this particular RNA helicase targeting by the HCV core protein has a role in the process of gene expression and that the interaction with the core protein can influence its NTPase-dNTPase activity or modulate its trans-activation ability.
MATERIALS AND METHODS
Yeast two-hybrid cloning system.
All yeast strains and plasmids for two-hybrid experiments (Matchmaker two-hybrid system) were obtained from Clontech (Palo Alto, Calif.). Saccharomyces cerevisiae HF7c was used for library screening and for assay of protein and protein interactions as described previously (9). Plasmids pGBT/HCVc195, pGBT/HCVc122, and pGBT/HCVc101, which encode various lengths of HCV core protein, were used as baits in two-hybrid screens of human liver cDNA libraries (Clontech) according to the Matchmaker two-hybrid system protocol (Clontech) (9). Positive yeast clones were selected by histidine prototrophy and β-galactosidase expression. A total of 2 × 106 transformed colonies were plated. The library clones that were LacZ+ His+ were isolated. The library clones that activated the lacZ reporter gene only in the presence of the pGBT9 derivative of HCV core gene were chosen for sequencing. The cDNA insert of approximately 2.1 kb from one hybrid clone, pGAD/CAP-Rd, was used as a probe to screen human HepG2 cDNA libraries in λgt11 (Stratagene) by standard methods (80). The cDNA insert (2.5-kb SalI-BglII fragment) of the positive clone was subcloned into SalI/BamHI-digested pGEM-3Zf(+) (Promega), the resulting construct was designated pGEM-3Zf(+)/CAP-Rf, and its nucleotide sequence was determined by standard methods (80).
Plasmid construction.
Plasmids pSRα/HCVc122 and pSRα/HCVc101, derivatives of pSRα vector (91), were constructed by insertion of the 0.4-kb PstI-ClaI (filled-in) or 0.3-kb PstI-SacII (filled-in) fragment of HCV core gene derived from pGM1/HCVC construct (86) into the PstI/KpnI (T4 DNA polymerase-digested)-treated pSRα. Plasmids pGAD/CAP-RdΔNcoI and pGAD/CAP-RdΔPstI were constructed by deletion of the 0.97-kb NcoI-BglII (filled in with Klenow enzyme) or 1.7-kb PstI-BglII (T4 DNA polymerase-digested) fragment of the CAP-Rd gene from pGAD/CAP-Rd. The resulting plasmids can direct the expression of fusion protein containing amino acid residues 106 to 472 or 106 to 230 of CAP-Rf protein fused in frame with the GAL4 DNA-binding domain. Plasmid pFLAG/Rf, the mammalian expression construct for FLAG-tagged CAP-Rf, was constructed by in-frame insertion of the full-length 2.4-kb KpnI-BamHI fragment of the CAP-Rf gene (nucleotides 130 to 2516) into the KpnI/BamHI-digested pFLAG-CMV-2 (Kodak). The resulting pFLAG/Rf can direct the expression of FLAG · CAP-Rf with an additional 25 amino acid residues (FLAG peptide plus 17 amino acid residues derived from the polylinker region of the vector) at the N terminus of CAP-Rf. Plasmid pECE/Rf, the mammalian expression construct for CAP-Rf driven by the simian virus 40 early promoter, contains a full-length 2.4-kb EcoRI fragment of the CAP-Rf gene that was excised from pGEM-3Zf(+)/CAP-Rf and inserted into the EcoRI site of pECE vector (16). Plasmids pET/His · CAP-Rf and pET/His · CAP-Rd for bacterial expression of His-tagged CAP-Rf or His-tagged CAP-Rd were constructed by in-frame insertion of the CAP-Rf gene fragment harboring either its full-length coding region (660-amino-acid fragment without the initiation codon encoded by 2.35-kb BamHI fragment of CAP-Rf gene) or the fragment containing amino acids 106 to 661 (2.1-kb XhoI-BglII fragment excised from pGAD/CAP-Rd) into the BamHI-digested or XhoI/BamHI-digested pET-15b plasmid (Novagen), respectively. The resulting His · CAP-Rf or His · CAP-Rd recombinant protein has an additional 21 amino acid residues (6 His residues plus 15 amino acid residues from the polylinker region) at the N terminus of CAP-Rf or CAP-Rd. Plasmid pCMV-LUC, a derivative of pGL2-Basic (Promega) consisting of a luc gene driven by the cytomegalovirus immediate-early promoter (nucleotides +3 to −760), was kindly provided by Y.-S. Chang (8).
Cell culture and subcellular fractionation.
Human hepatocellular carcinoma cell line HuH-7 or HepG2 and human cervical carcinoma cell line HeLa were cultured as described elsewhere (9). For preparation of total cell lysates, cells were lysed in the buffer containing 10 mM Tris-HCl (pH 7.1), 1 mM EDTA, and 1% Triton X-100 and protease inhibitor cocktail (Complete; Boehringer), and the extracts were recovered by centrifugation (12,000 rpm; Kubota 1720 RA-50J rotor). For preparation of the cytoplasmic or nuclear fractions, cell pellets were suspended in a hypotonic buffer (20 mM HEPES [pH 7.4], 1 mM MgCl2, 10 mM KCl, 0.5% Nonidet P-40, 0.5 mM dithiothreitol [DTT], and protease inhibitor cocktail [Complete; Boehringer]) at 4°C for 30 min. After centrifugation at 6,500 rpm (Kubota 1720 RA-50J rotor) for 4 min (4°C), the supernatants recovered were treated as the cytoplasmic fractions, while the resulting pellets containing the nuclei were resuspended in a high-salt buffer (20 mM HEPES [pH 7.4], 20% glycerol, 0.4 M NaCl, 1 mM MgCl2, 10 mM KCl, 0.5 mM DTT, and protease inhibitor cocktail [Complete; Boehringer]) for 15 min, then vortexed 10 s, and kept on ice for another 15 min. The resulting samples were centrifuged (12,000 rpm; Kubota 1720 RA-50J rotor) for 15 min (4°C), and the supernatants recovered were treated as the nuclear extracts.
RNA preparation and Northern blotting.
Total cellular RNA was isolated by using TRI reagent RNA-DNA-protein isolation reagent (Molecular Research Center) according to the instructions of the supplier. Total RNA was dissolved in H2O, and poly(A)+ RNA was purified with an mRNA separation kit (Clontech). Poly(A)+ RNA samples (5 μg/lane) were electrophoresed on a 6% formaldehyde–1% agarose RNA gel and then transferred into a nylon filter. The filter was prehybridized and hybridized according to the standard method (80). DNA probes were prepared by the Rediprime DNA labeling method (Amersham).
Purification of His · CAP-Rd or His · CAP-Rf fusion protein and preparation of antiserum against CAP-Rf.
E. coli host BL21(DE3) harboring the CAP-Rf expression vector pET/His · CAP-Rf or pET/His · CAP-Rd was cultured in Luria-Bertani medium with 50 μg of ampicillin per ml and induced by 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h. Cells were harvested and lysed with the binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl, 6 M urea, pH 7.9), and the lysates were loaded onto Ni2+ prebound His-Bind resin (Novagen). The column was washed with 25 ml of binding buffer and 15 ml of 20 mM imidazole buffer (combination of 11 ml of binding buffer with 4.1 ml of wash buffer [60 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl, 6 M urea, pH 7.9]). The bound proteins were eluted with the elution buffer (0.5 M imidazole, 0.5 M NaCl, 20 mM Tris-HCl, 6 M urea, pH 7.9). The eluants were dialyzed first in 10 mM Tris-HCl buffer (pH 7.9) containing 6 M urea, and then the dialysis buffer was successively changed to contain 4 M, 2 M, and then no urea every 12 h. After dialysis, the proteins were concentrated by lyophilization. The His · CAP-Rd hybrid proteins after purification through the His-Bind resin column (Fig. 2C) were used for immunization of rabbits. The immunospecificities of antisera against His · CAP-Rd for detection of CAP-Rf were examined by immunoblotting, as shown in Fig. 2B, 2C, 3B, and 3E.
FIG. 2.
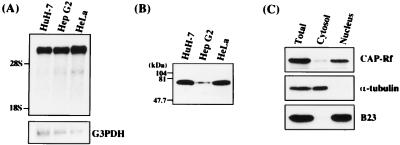
Northern blot and immunoblot analysis of CAP-Rf expression in various cell lines. (A) The poly(A)+ mRNAs (5 μg) extracted from cell lines HuH-7 (lane 1), HepG2 (lane 2), and HeLa (lane 3) were used for Northern blot analysis with the 32P-labeled CAP-Rd DNA fragment (2.1-kb EcoRI fragment of pGAD/CAP-Rd) as a probe. The same blot was rehybridized with glyceraldehyde-3-phosphate dehydrogenase DNA probe and served as the control. (B) Immunoblot analysis of CAP-Rf expression. Total cell pellets from various cell lines were solubilized by the sample buffer (47) and analyzed by immunoblotting with rabbit anti-His · CAP-Rd antiserum for detection. The protein amount loaded in the gel is 20 μg in each lane. (C) Analysis of the subcellular localization of CAP-Rf by immunoblotting. The total cell extracts, cytoplasmic fractions, or nuclear extracts (50 μg each) prepared from various cell lines (see Materials and Methods) were analyzed by immunoblotting with mouse monoclonal anti-chicken α-tubulin (Amersham; 1:6,700 dilution), goat anti-human B23 (Santa Cruz; 1:2,000 dilution), or rabbit anti-His · CAP-Rd (1:2,000 dilution) antibodies for detection.
FIG. 3.
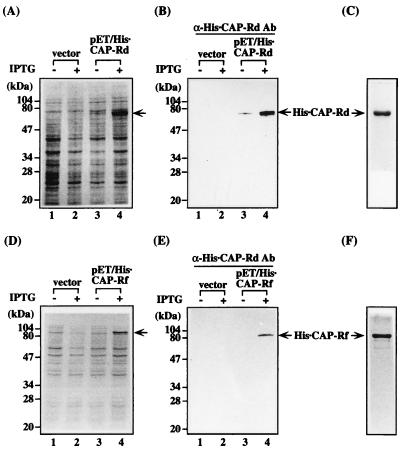
Expression and purification of His · CAP-Rd and His · CAP-Rf proteins. (A, B, D, and E) Analysis of the His · CAP-Rd or His · CAP-Rf expression in E. coli. E. coli BL21(DE3) harboring the vector pET15b or CAP-Rf expression vector pET/His · CAP-Rd or pET/His · CAP-Rf was induced with (lanes 2 and 4) or without (lanes 1 and 3) 1 mM IPTG for 3 h. Cells were harvested, solubilized in the sample buffer (47), subsequently analyzed by SDS-PAGE, and stained with Coomassie brilliant blue (A and D) or processed for immunoblotting with anti-His · CAP-Rd antisera (lanes 1 to 4) with the ECL detection system. The CAP-Rd (C) and CAP-Rf (F) proteins affinity purified by Ni2+ prebound His-Bind resin (see Materials and Methods) were analyzed by SDS-PAGE and detected by Coomassie brilliant blue staining. The position for His · CAP-Rd or His · CAP-Rf is indicated.
In vitro binding analysis of GST-HCV core fusion protein and endogenous CAP-Rf.
HCV core proteins expressed as glutathione S-transferase (GST) fusion proteins from the expression vector pGST/HCVc122 or pGST/HCVcΔ(41-107) were purified as described elsewhere (9, 87). For the in vitro binding assay, 20 μl of glutathione-Sepharose 4B beads (Pharmacia) containing various GST fusion proteins (10 μg) were incubated with HepG2 cell extracts (500 μg) (9) at 4°C overnight under gentle rotation. The beads were washed four times with 1 ml of NETNT (150 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl [pH 7.5], 0.5% Nonidet P-40, 0.5% Tween 20). Proteins bound on the beads were eluted by the sample buffer (47), fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% polyacrylamide gel), and processed for Western blot analysis. Detection of CAP-Rf was performed with rabbit anti-His · CAP-Rd antiserum as the primary antibody and horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG) (DAKO) as the secondary antibody by the enhanced chemiluminescence detection method (ECL; Amersham).
In vitro binding assay of HCV core protein and His · CAP-Rd.
In vitro translation products (C195, C122, and C101 variants) of HCV core protein prepared from pGM1/HCVc DNA templates (86) were precipitated by His-Bind resin that was prebound with His · CAP-Rd fusion protein (4 μg) in the binding buffer without urea at 4°C for 3 h. The resins were washed four times with NETNT buffer, and the bound proteins were eluted with the sample buffer (47), fractionated by SDS-PAGE (13.5% polyacrylamide gel), and detected by autoradiography.
NTPase-dNTPase activity analysis of His · CAP-Rf.
For detection of nucleotide hydrolyzing activity, the purified His · CAP-Rf proteins were incubated in NTPase buffer (50 mM MOPS [morpholinepropanesulfonic acid]-KCl [pH 6.5], 2 mM EDTA, 10 mM NaCl) containing 3 μM α-32P-labeled NTP-dNTP substrate for 1 h at 37°C. The reaction was stopped by adding EDTA to a final concentration of 20 mM, and a 1/10 volume of the reaction mixture was applied onto a polyethyleneimine thin-layer chromatography (TLC) plate (J. T. Baker Co.). The buffer systems used for TLC were 0.5 M potassium phosphate buffer (pH 3.5) for ATP, dATP, and dGTP; 0.1 M potassium phosphate buffer (pH 3.5) for CTP, UTP, dCTP, and dTTP; and 0.75 M potassium phosphate buffer (pH 3.5) for CTP. The polyethyleneimine plate was air dried and scanned with a PhosphorImager. The percentage of nucleoside diphosphate-deoxynucleoside diphosphate conversion was quantitated in each sample.
In vivo coimmunoprecipitation.
HuH-7 cells (density of 2 × 106 cells/10-cm-diameter plate) were seeded and grown in 5% CO2 at 37°C for 24 h before transfection by the calcium phosphate precipitation method (87). Cells were cotransfected with the CAP-Rf expression construct pFLAG/Rf together with the HCV core construct (pSRα/HCVc195, pSRα/HCVc122, or pSRα/HCVc101) or its control vector pSRα (10 μg each). After 48 h, cells were washed twice with ice-cold phosphate-buffered saline, trypsinized, and collected by centrifugation. After three cycles of freezing on dry ice and thawing at 37°C, cells were lysed in 400 μl of lysis buffer (25 mM Tris-HCl [pH 7.8], 70 mM potassium phosphate buffer [pH 7.8], 2.1 mM MgCl2, 0.7 mM DTT, 0.1% Nonidet P-40, and protease inhibitor cocktail [Complete; Boehringer]). Cell extracts (200 μl) recovered from the centrifugation were incubated overnight at 4°C with the protein A-Sepharose-bound anti-FLAG antibody (Kodak) (20-μl packed volume). The immunoprecipitate was recovered by centrifugation and washed four times with NETN buffer. Bound proteins were separated by SDS-PAGE and processed for immunoblot analysis with rabbit anti-HCV core antiserum (87) with the ECL detection system.
Assay of reporter plasmid activity.
HuH-7 cells at a density of 8 × 104 cells/well were seeded on 24-well plates and grown in 5% CO2 at 37°C for 24 h before transfection with the SuperFect transfection reagent (Qiagen, Hilden, Germany). Cells were transfected with appropriate amounts of luciferase reporter plasmid pCMV-LUC (8), CAP-Rf expression construct pFLAG/Rf (or pECE/Rf), or its control vector pFLAG-CMV-2 (or pECE), if applicable, together with the HCV core construct (pSRα/HCVc122 or pSRα/HCVc101) or its control vector pSRα. After 48 h, cells were collected by centrifugation and lysed by freezing-thawing in 150 μl of lysis buffer. Cell extracts (80 μl) recovered from the centrifugation (Microfuge, 15 min at 4°C) were then mixed with 250 μl of luciferase assay buffer (43.2 mM glycylglycine [pH 7.8], 22 mM MgSO4, 2.4 mM EDTA, 7.4 mM ATP, 1 mM DTT, and 0.4 mg of bovine serum albumin), and the resulting mixtures were assayed for luciferase activity with 100 μl of 0.5 mM luciferin (Sigma) as substrate, activity being measured with an AutoLumat LB953 luminometer (Berthold, Bad Wildbad, Germany).
Confocal immunofluorescence microscopy.
The localizations of HCV core proteins and FLAG-tagged CAP-Rf (FLAG · CAP-Rf) in transfected cells were examined by confocal laser scanning microscopy (Leica TCS-NT). For immunofluorescence staining, cells were fixed with acetone-methanol (1:1) (−20°C) and probed with rabbit anti-HCV core protein antiserum (86) or mouse monoclonal anti-FLAG M2 antibody (Kodak), followed by fluorescein isothiocyanate-conjugated goat anti-rabbit IgG or rhodamine-conjugated goat anti-mouse IgG (Transduction Laboratories).
RESULTS
Identification of cellular putative RNA helicase CAP-Rf interacting with HCV core protein by use of the yeast two-hybrid system.
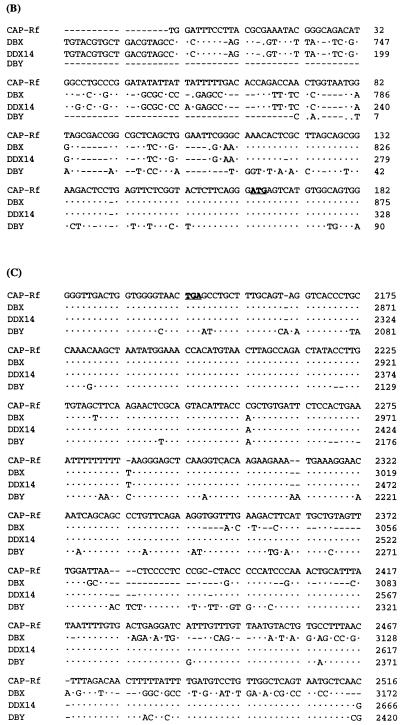
When using the yeast two-hybrid system to identify cellular factors that directly interact with HCV core protein, the GAL4 DNA-binding domain fused with the full-length (pGBT/HCVc195) or truncated (pGBT/HCVc122 and pGBT/HCVc101) versions of core protein was used as bait for yeast two-hybrid screening in a human liver cDNA library (Matchmaker; Clontech) (see Materials and Methods). Ten positive clones (as pGAD fusion constructs) were obtained by using truncated versions, but not the full-length version, of core protein as baits (Table 1). The cDNA sequences of positive clones were determined and aligned with sequences in the EMBL data bank. Three of 10 cDNA clones, designated pGAD/CAP-Rd, were found to encode the C-terminal portion of cellular putative RNA helicase encompassing amino acid residues 106 to 661 (CAP-Rf numbering; see below), since its deduced amino acid sequence has more than 95% homology to that of human putative RNA helicase (DBX or DBY) or mouse putative RNA helicase (mDEAD3 [also named mouse DDX3 by SWISSPROT data bank] or PL10) as reported previously (24, 48, 54) (Fig. 1A). This cDNA fragment (2.1 kb) was used to rescreen for its full-length cDNA clone in a human HepG2 cDNA library (see Materials and Methods). One such cDNA clone, containing a 2.5-kb insert, was obtained, and its encoded full-length putative RNA helicase was thus designated CAP-Rf (core-associated protein-RNA helicase full-length). The entire nucleotide sequence of the CAP-Rf gene including its 5′ and 3′ noncoding regions was determined. Alignment of its deduced amino acid sequence (661 amino acid residues) with those of several RNA helicases (DBX, DBY, PL10, and mDEAD3) revealed the presence of eight highly conserved functional motifs characteristic of the DEAD box family (residues 224 to 534, CAP-Rf numbering [Fig. 1A]). Moreover, these amino acid sequence comparisons also suggested that the CAP-Rf protein is most similar to DBX and mDEAD3 (98% identity) and less closely related to PL10 (95% identity). Judging from this high level of homology, it appears that CAP-Rf is a homologue of DBX, DBY, or mDEAD3. Interestingly, the 5′ and 3′ noncoding regions of CAP-Rf are similar but not identical to the corresponding regions of human DBX, DBY, or another CAP-Rf-related gene, DDX14 (also named human DDX3 by the SWISSPROT data bank) (14) (Fig. 1B), suggesting that more than one transcript for CAP-Rf or a CAP-Rf-related gene was expressed. Northern blot analysis of CAP-Rf expression supports this notion, because apart from one major CAP-Rf transcript with a size larger than that of 28S rRNA, a minor species of smaller transcript of about 3 to 4 kb was also present in HepG2, HeLa, or HuH-7 cells investigated (Fig. 2A). These RNAs may result from different promoters, alternative splicing, or different polyadenylation states.
TABLE 1.
Yeast two-hybrid system for analysis of the interaction between HCV core protein and CAP-Rd and its deletion mutants
| pGBT fusion | pGAD fusion | His− Leu− Trp− plating assaya | X-Gal filter assay resulta |
|---|---|---|---|
| pGBT/HCVc195 | − | White | |
| pGBT/HCVc122 | − | White | |
| pGBT/HCVc101 | − | White | |
| pGBT/HCVc195 | pGAD/CAP-Rd | − | White |
| pGBT/HCVc195 | pGAD/CAP-RdΔNcoI | − | White |
| pGBT/HCVc195 | pGAD/CAP-RdΔPstI | − | White |
| pGBT/HCVc122 | pGAD/CAP-Rd | + | Blue |
| pGBT/HCVc122 | pGAD/CAP-RdΔNcoI | − | White |
| pGBT/HCVc122 | pGAD/CAP-RdΔPstI | − | White |
| pGBT/HCVc101 | pGAD/CAP-Rd | + | Blue |
| pGBT/HCVc101 | pGAD/CAP-RdΔNcoI | − | White |
| pGBT/HCVc101 | pGAD/CAP-RdΔPstI | − | White |
| pGAD/CAP-Rd | − | White | |
| pGAD/CAP-RdΔNcoI | − | White | |
| pGAD/CAP-RdΔPstI | − | White |
See Materials and Methods. +, transformant growth on His− Leu− Trp− triple-dropout plate; −, no growth. X-Gal, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside.
FIG. 1.
Alignment of amino acid sequence or 5′ and 3′ noncoding region sequences of CAP-Rf with those of the DEAD family of RNA helicases. (A) Alignment of amino acid sequence of CAP-Rf with those of other known RNA helicases. The amino acid sequence of CAP-Rf is aligned with those of human DBX and DBY (48) (GenBank accession no. AF000982 and AF000985, respectively), mouse PL10 (54) (GenBank accession no. J04847), and mDEAD3 (24) (GenBank accession no. L25126). A consensus sequence is shown at the bottom of the alignment. Positions of identical amino acids in the six proteins are boxed. The roman numerals indicate the positions of conserved motifs found in RNA helicases. (B and C) Alignment of 5′ (B) and 3′ (C) noncoding regions of CAP-Rf cDNA with those of other known RNA helicases. The 5′ or 3′ noncoding sequence of CAP-Rf is compared with the corresponding region of human RNA helicases from DBX and DBY (48) (GenBank accession no. AF000982 and AF000985, respectively) or DDX14 (14) (accession no. U50553). The initiation or stop codon for these four RNA helicases is boldface and underlined. Only the different positions are indicated. The dotted lines represent identical nucleotides, while the dashed lines indicate deletions.
To facilitate the purification of CAP-Rd and CAP-Rf proteins for biochemical characterization, their N termini were fused in frame with His tag (designated His · CAP-Rd or His · CAP-Rf, respectively), and the purified His · CAP-Rd protein was used as antigen for immunization of rabbits (see Materials and Methods) (Fig. 3C). Immunoblotting analysis suggested that the antibody generated could detect the bacterially expressed His · CAP-Rd or His · CAP-Rf (Fig. 3A, B, D, and E). A strong 73-kDa immunospecific protein band, as expected for the size of a full-length CAP-Rf, was readily detected in the total cell extracts of HuH-7 and HeLa cells but at a low level in HepG2 cells (Fig. 2B), suggesting differential expression of CAP-Rf in various cell lines. Moreover, it was noted that the protein and mRNA levels of CAP-Rf in HepG2 cells were not correlated with those of HuH-7 and HeLa cells (Fig. 2A and B). This discrepancy may be caused by the differences in translation efficiency or protein stability in particular cell lines. Subcellular fractionation in combination with immunoblotting analysis with the cytoplasmic α-tubulin and nuclear B23 as controls revealed that, similar to B23, the full length of CAP-Rf was predominantly found in the nuclear fraction of HeLa cells (Fig. 2C).
In vitro binding analysis confirms that the truncated versions of HCV core protein can associate with CAP-Rf.
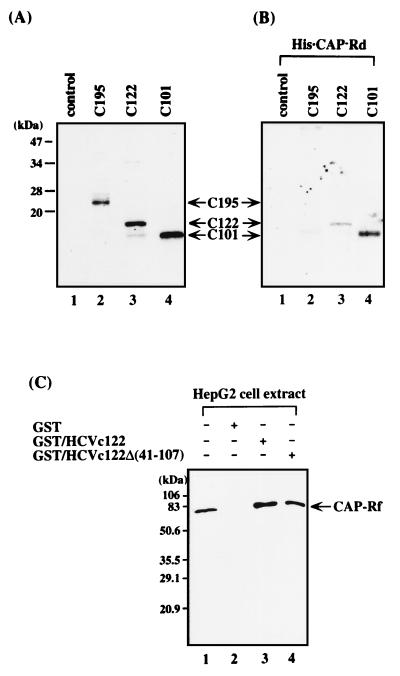
To confirm the yeast two-hybrid results, the in vitro binding properties of the HCV core protein and CAP-Rf were examined. When the in vitro-translated HCV core proteins were tested for binding to a His-Bind resin prebound with His · CAP-Rd, results indicated that, consistent with the yeast two-hybrid data, His · CAP-Rd could associate with the truncated versions (C122 and C101) of in vitro-translated HCV core protein but not with the full-length one (C195) (Fig. 4A and B). Additionally, when the GST-HCV core fusion proteins were used for pull-down analysis of the interaction between HCV core protein and CAP-Rf, results showed that the endogenous CAP-Rf from HepG2 cells could associate with the HCV core protein harboring the N-terminal 122 amino acid residues or the same fragment but with an internal deletion of amino acid residues 41 to 107 (Fig. 4C). This result suggested that the N-terminal 40-amino-acid fragment of HCV core protein is sufficient for interaction with CAP-Rf.
FIG. 4.
Analysis of the interaction between CAP-Rf and HCV core variants. (A) Analysis of in vitro translation products of HCV core protein. The 35S-labeled HCV core proteins of various lengths (C195, C122, and C101) prepared by in vitro transcription and translation (lanes 2 to 4) (86) (also see Materials and Methods) (5 to 10 μl) were individually precipitated by HCV-positive patients’ sera and analyzed by SDS-PAGE (13.5% polyacrylamide gel) and autoradiography. (B) Analysis of the interaction between His · CAP-Rf and in vitro-translated HCV core protein. The in vitro-translated HCV core proteins (10 to 20 μl) were loaded onto His-Bind resin prebound with His · CAP-Rd (4 μg in 20-μl resins) (see Materials and Methods). The bound proteins were boiled in the sample buffer, analyzed by SDS-PAGE (13.5% polyacrylamide gel), and detected by autoradiography. Lane 1, in vitro-translated products without HCV core mRNA; lane 2, C195; lane 3, C122; lane 4, C101. (C) In vitro binding analysis of endogenous CAP-Rf and the various truncated forms of HCV core protein. HepG2 cell lysates were incubated with glutathione-Sepharose 4B beads which were prebound with GST, GST/HCVc122, or GST/HCVc122Δ(41-107) (see Materials and Methods). The bound proteins retained on the resins were immunoblotted with antiserum against His · CAP-Rf (see Materials and Methods).
The C-terminal integrity of CAP-Rf is essential for interaction with the HCV core protein.
Based on the yeast two-hybrid data, the integrity of the N-terminal portion of CAP-Rf was found to be dispensable for binding with the HCV core protein (Table 1). To further delineate the functional domain of CAP-Rf responsible for interaction with the HCV core protein, the GAL4 trans-activation domain fused in frame with amino acid residues 106 to 472 (pGAD/CAP-RdΔNcoI) and 106 to 230 (pGAD/CAP-RdΔPstI) of CAP-Rf was examined for its interaction with the GAL4 DNA-binding domain and HCV core fusion proteins (pGBT/HCVc195, pGBT/HCVc122, and pGBT/HCVc101) in the yeast two-hybrid assay. As shown in Table 1, in contrast to that of wild-type CAP-Rd (pGAD/CAP-Rd), these two C-terminally truncated forms of CAP-Rd lost the interaction with the truncated forms of HCV core protein. Therefore, the integrity of the C-terminal portion of CAP-Rf, but not that of the N terminus, is essential for interaction with the HCV core protein. Notably, this C-terminal region (residues 473 to 611) of CAP-Rf identified as essential for interaction with the HCV core protein comprises domains VII and VIII of the helicase core region and a C-terminal end (last 80 residues), which is rich in glycine, serine, arginine, or aromatic residues (phenylalanine and tyrosine) (Fig. 1A).
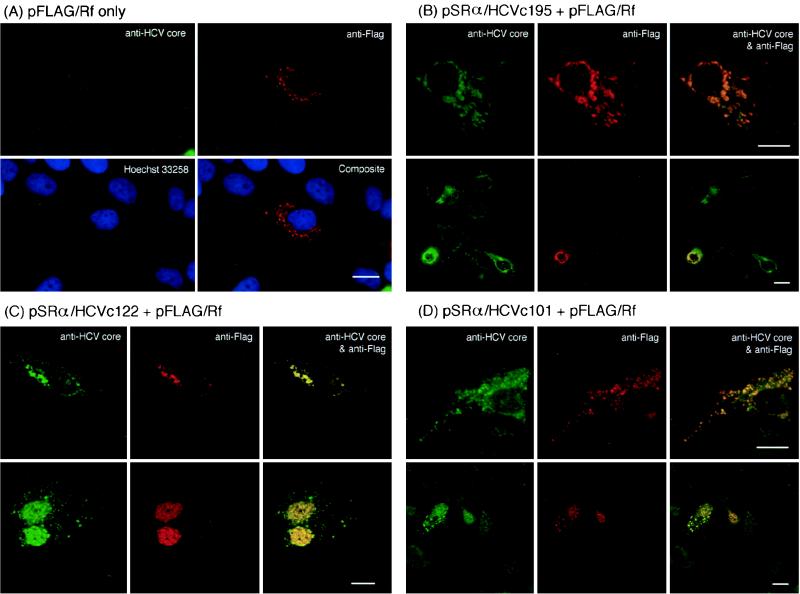
HCV core protein colocalizes with FLAG-tagged CAP-Rf in the nucleus and cytoplasm.
We next examined whether the transiently expressed HCV core protein and FLAG-tagged CAP-Rf (FLAG · CAP-Rf) colocalize in HuH-7 cells (see Materials and Methods). A typical example of immunofluorescence staining observed under a confocal microscope is shown in Fig. 5. The cells were typically stained to reveal the HCV core protein (anti-HCV core), FLAG-tagged transfected proteins (anti-FLAG), and DNA (Hoechst 33258). Note that in the cells transfected with FLAG · CAP-Rf only, the FLAG · CAP-Rf protein was mainly found in the cytoplasm (Fig. 5A), which is inconsistent with the nuclear localization of endogenous CAP-Rf as described above (Fig. 2C). The discrepancy between these analyses may be due to the overexpressed FLAG · CAP-Rf protein resulting in the bright cytoplasmic staining and thus obscuring the nuclear protein staining. Alternatively, this may result from the N-terminal fusion with FLAG tag on CAP-Rf. Nevertheless, in cells cotransfected with either the full-length construct (pSRα/HCVc195 [Fig. 5B]) or the truncated HCV core expression construct (pSRα/HCVc122 or pSRα/HCVc101 [Fig. 5C and D, respectively]) and FLAG · CAP-Rf, both proteins could be identified either in the cytoplasm (top panel in Fig. 5B, C, and D) or in the nucleus (bottom panel in Fig. 5B, C, and D). Interestingly, although the localization of proteins varied, the various forms of HCV core protein always colocalized with FLAG · CAP-Rf. Since FLAG · CAP-Rf alone was predominantly detected in the cytoplasm (Fig. 5A), the appearance of nuclear FLAG · CAP-Rf in these cotransfected cells was presumably due to the cotransport of FLAG · CAP-Rf with the HCV core protein into the nucleus. Notably, although both the yeast two-hybrid experiment (Table 1) and the in vitro binding analysis (Fig. 4) did not show significant interaction between the full-length HCV core protein and CAP-Rf, the localization study appeared to have an inconsistent result. The failure to detect the interaction between the full-length core protein and CAP-Rf is probably due to the strong hydrophobicity of the C-terminal fragment of the core protein, which may hinder its function in the yeast two-hybrid screen or the in vitro binding analysis as noted previously (9, 35, 65). Alternatively but not exclusively, since the full-length 22-kDa (191 amino acid residues, designated p22 species) species of HCV core protein is readily matured into the 20-kDa species (173 to 174 amino acid residues, designated p20) (Fig. 6A) (also see references 60 and 98) or degraded into smaller forms (18 kDa or smaller) intracellularly (61, 86, 88) and since these matured and degraded forms, but not the full-length form, of core protein are mainly located in the nucleus (60, 61, 88, 98), the molecular species colocalizing with FLAG · CAP-Rf in the nuclear compartment of the full-length core construct-transfected cells (bottom panel in Fig. 5B) is likely the matured or degraded form of core protein.
FIG. 5.
Both FLAG · CAP-Rf and HCV core protein colocalize inside a cell. HuH-7 cells were transfected with FLAG-tagged CAP-Rf construct pFLAG/Rf (A) or together with various forms of HCV core construct (pSRα/HCVc195, pSRα/HCVc122, and pSRα/HCVc101) (B to D, respectively). The distributions of CAP-Rf and HCV core protein were assessed by indirect immunofluorescence staining (see Materials and Methods). For double immunofluorescence staining, cells were stained with rabbit anti-HCV core protein antiserum (1:1,000 dilution) and mouse anti-FLAG-tagged M2 monoclonal antibody (1:250 dilution), followed by fluorescein isothiocyanate-conjugated goat anti-rabbit IgG or rhodamine-conjugated goat anti-mouse IgG. Cell nuclei were also visualized by Hoechst 33258 staining (blue). These immunofluorescence patterns were recorded by confocal laser scanning microscopy. As noted, the top and bottom sections of panels B to D show the colocalization of core protein with FLAG · CAP-Rf in the cytoplasmic or nuclear compartment, respectively.
FIG. 6.
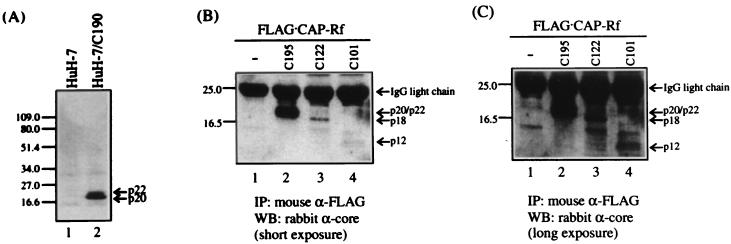
In vivo coimmunoprecipitation of FLAG · CAP-Rf and HCV core protein. (A) Immunoblot analysis of the HCV core protein expression in HCV core-producing HuH-7 cells (HuH-7/C190). The total cell extracts (20 μg) prepared from HuH-7 and HuH-7/C190 cells (9) were examined for the expression of HCV core protein by immunoblotting with rabbit antisera against HCV core protein. (B and C) HuH-7 cells were transfected with FLAG · CAP-Rf construct pFLAG/Rf (lanes 1 to 4) together with various forms of HCV core construct (pSRα/HCVc195 [C195], pSRα/HCVc122 [C122], and pSRα/HCVc101 [C101]) (lanes 2 to 4, respectively) or its vector pSRα (lanes 1) (see Materials and Methods). Two days after transfection, cells were lysed and subjected to immunoprecipitation (IP) with anti-FLAG monoclonal antibody (M2) (see Materials and Methods). Immunoprecipitates were analyzed by SDS-PAGE followed by Western blotting (WB) assay with rabbit antibody against HCV core protein. Panels B and C show identical sets of experiments but with different exposure times. Numbers at left of each panel indicate molecular mass in kilodaltons.
HCV core protein interacts with CAP-Rf in vivo.
To further determine whether the HCV core protein is capable of binding to CAP-Rf in cells, coimmunoprecipitation was performed on HuH-7 cells transiently transfected with the expression constructs for various forms of HCV core protein (pSRα/HCVc195, pSRα/HCVc122, or pSRα/HCVc101) and FLAG · CAP-Rf (pFLAG/Rf) (see Materials and Methods). When cell lysates from transfected cells were immunoprecipitated by anti-FLAG monoclonal antibody (M2) and immunoprecipitates were analyzed by Western blotting assay with antibody against HCV core protein, results indicated that the full-length/matured form (p22/20) and the N-terminal 122- or 101-amino-acid fragment of core protein (p18 and p12 species, respectively) were differentially coprecipitated by the anti-FLAG antibody (Fig. 6B and C), suggesting that FLAG · CAP-Rf, presumably CAP-Rf, could interact with both full-length/matured and truncated forms of HCV core protein in cells, a result consistent with colocalization study (Fig. 5). Surprisingly, this in vivo experiment suggested that the two truncated forms of core protein exhibited only weak interaction with CAP-Rf compared to that of the full-length/matured form of core protein (Fig. 6B and C), which is different from the results obtained by the yeast two-hybrid experiment (Table 1) and in vitro binding analysis (Fig. 4). Apart from the possible explanation given above, the exact reason for this inconsistency is not clear. Nevertheless, based on several independent approaches, our results indicated that HCV core protein and CAP-Rf can interact with each other both in vivo and in vitro.
The purified His · CAP-Rf has NTPase-dNTPase activity.
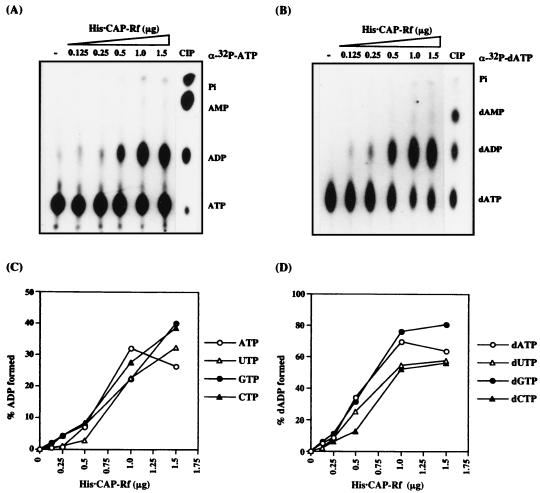
Since, similar to other RNA helicases CAP-Rf has the functional motif of ATPase (Fig. 1A and Introduction), it is pertinent to know whether the purified His · CAP-Rf has this enzymatic activity. We performed in vitro NTPase-dNTPase assays on four different α-32P-NTPs or -dNTPs, and the nucleoside diphosphate or deoxynucleoside diphosphate release was measured with increasing amounts of purified CAP-Rf (see Materials and Methods). As shown in Fig. 7, the purified CAP-Rf displayed comparably strong NTPase activities on four different NTP substrates, but with a higher enzymatic activity on the substrate dNTPs, especially dATP and dGTP. Most intriguingly, this NTPase-dNTPase activity of His · CAP-Rf was RNA independent. Moreover, it was inhibited to various levels (87 to <0.1% of the control value measured without polynucleotide) practically by all kinds of synthetic polynucleotides examined including poly(A), poly(U), poly(G), poly(C), and poly(dI-dC) (Table 2). Of these, poly(G) or poly(dI-dC) had a more profound effect on the extent of inhibition (Table 2). The ability of polynucleotides to inhibit the NTPase-dNTPase activity of CAP-Rf is similar to that of the DEAD box protein An3 (32) but is rather distinct from most of the known RNA helicases, while their ATPase activity is found to be polynucleotide dependent or stimulated (21, 53, 83).
FIG. 7.
NTP and dNTP hydrolysis activities of His · CAP-Rf. For detection of the nucleotide hydrolysis activity of the purified His · CAP-Rf, four α-32P-labeled ribonucleotides (3 μM) (A) and four α-32P-labeled deoxyribonucleotides (3 μM) (B) were used as substrates. All the enzymatic activities were assayed at indicated concentrations of purified His · CAP-Rf as described in Materials and Methods. The top of each panel indicates the representative TLC plate used for analyzing the ADP or dATP conversion from [α-32P]ATP or [α-32P]dATP, respectively. CIP, the reaction mixtures of [α-32P]ATP or [α-32P]dATP treated with calf intestinal phosphatase as a control. Graphs of the data from panels A and B are shown in panels C and D, respectively.
TABLE 2.
Effects of different polynucleotides on the NTPase-dNTPase activity of His · CAP-Rf
| Substrate | Relative NTPase-dNTPase activitya
|
|||||
|---|---|---|---|---|---|---|
| None | poly(A) | poly(U) | poly(G) | poly(C) | poly(dI-dC) | |
| ATP | 1.000 | 0.401 | 0.795 | 0.154 | 0.593 | 0.195 |
| UTP | 1.000 | <0.001 | 0.255 | 0.004 | 0.384 | <0.001 |
| GTP | 1.000 | 0.489 | 0.865 | 0.395 | 0.601 | 0.454 |
| CTP | 1.000 | 0.220 | 0.318 | 0.129 | 0.248 | 0.042 |
| dATP | 1.000 | 0.265 | 0.363 | 0.059 | 0.216 | 0.076 |
| dTTP | 1.000 | 0.281 | 0.344 | 0.077 | 0.238 | 0.088 |
| dGTP | 1.000 | 0.417 | 0.365 | 0.076 | 0.327 | 0.116 |
| dCTP | 1.000 | 0.118 | 0.280 | 0.085 | 0.064 | 0.083 |
For the nucleotide hydrolysis assay, four different α-32P-labeled ribonucleotides or α-32P-labeled deoxyribonucleotides were used as substrates for purified His · CAP-Rf (0.5 μg) enzymatic assay (see Materials and Methods). All of the enzymatic activities for specific substrates were determined in the absence or presence of the indicated polynucleotides (25 μM), and the data are represented as relative activities compared to those in the absence of polynucleotides.
HCV core protein enhances the ATPase-dATPase activity of His · CAP-Rf.
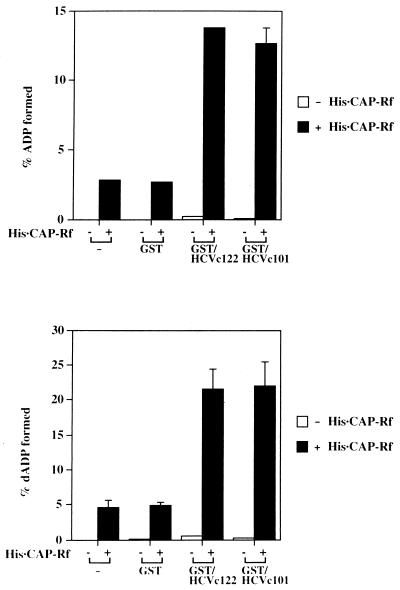
To determine whether the complex association between the HCV core protein and CAP-Rf can modulate the enzymatic activity of CAP-Rf, the purified GST or GST-HCV core fusion protein (GST/HCVc122 and GST/HCVc101) (0.25 μg) was incubated with His · CAP-Rf (0.5 μg) and the ATPase-dATPase activity in this protein mixture was examined. Remarkably, both GST-HCV core proteins, but not GST, could stimulate both ATPase and dATPase activities of His · CAP-Rf four- to fivefold (Fig. 8). As the GST/HCVc122 or GST/HCVc101 protein did not possess ATPase-dATPase activity by itself (Fig. 8), the increased ATPase-dATPase activity in the protein mixtures of GST-HCV core fusion proteins and His · CAP-Rf should be a consequence of HCV core protein binding to CAP-Rf, presumably through influencing the interaction domains resident within the ATP hydrolysis and RNA-binding regions of CAP-Rf (domains VII and VIII of helicase core region [Fig. 1A]) as defined in the yeast two-hybrid assay (Table 1).
FIG. 8.
HCV core protein enhances the NTPase-dNTPase activities of His · CAP-Rf. The purified His · CAP-Rf (0.5 μg) was preincubated in NTPase buffer (50 mM MOPS-KCl [pH 6.5], 2 mM EDTA, 10 mM NaCl) alone or with 0.25 μg of GST, GST/HCVc122, or GST/HCVc101, respectively, for 30 min at 37°C and subsequently assayed for nucleotide hydrolyzing activity with [α-32P]ATP (A) or [α-32P]dATP (B) (3 μM) as the substrate.
CAP-Rf plays a role in gene expression regulation, and HCV core protein can modulate its trans-activation ability.
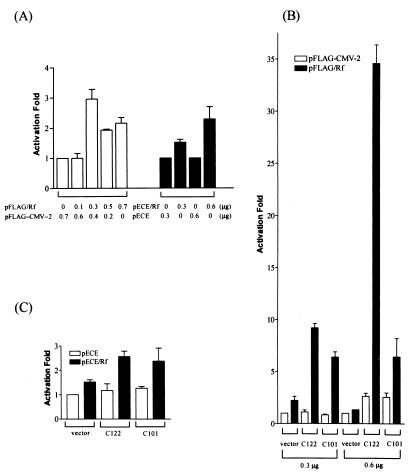
Since the biochemical property of CAP-Rf is rather distinct from the typically known RNA helicase and the biological function of its homologue DBX or mDEAD3 has not yet been characterized, the exact biological function of CAP-Rf is not clear. However, since most RNA helicases are involved in all aspects of cellular RNA metabolism including splicing, translation, RNA stability and degradation, and ribosomal assembly (see Introduction and Discussion), CAP-Rf may be involved in regulation of gene expression as well. To test this possibility, the trans-action ability of CAP-Rf on luciferase reporter plasmid (pCMV-Luc) was examined (see Materials and Methods). As shown in Fig. 9A, introducing various amounts (0.1 to 0.7 μg) of CAP-Rf (pECE/Rf) or FLAG · CAP-Rf (pFLAG/Rf) expression construct into HuH-7 cells could lead to about 1.5- to 3.0-fold enhancement of reporter plasmid activity, indicating clearly that CAP-Rf is involved in gene expression regulation. Additionally, in transient transfection of HuH-7 cells the HCV truncated core construct (pSRα/HCVc122 or pSRα/HCVc101) (0.3 or 0.6 μg) or FLAG · CAP-Rf expression construct pFLAG/Rf (0.3 μg) activated the luciferase reporter plasmid activity about 0.9- to 2.6-fold, whereas cotransfecting similar amounts of core and FLAG · CAP-Rf expression constructs together could provoke a significant enhancement (5.5- to 34-fold) of reporter plasmid activity (Fig. 9B). Of these, pSRα/HCVc101 (C101)-cotransfected cells had a moderate effect, whereas pSRα/HCVc122 (C122)-cotransfected cells had a strong effect (Fig. 9B). Moreover, at least in the case of pSRα/HCVc122 (C122)-transfected cells, the level of enhancement (9.6- to 34-fold) increased in a core construct dose-responsive manner (Fig. 9B). In examining the effect of the full-length core construct (pSRα/HCVc195) on the trans-activation ability of FLAG · CAP-Rf, essentially the same conclusion was obtained (data not shown). Therefore, the core protein of HCV can potentiate the trans-activation ability of FLAG · CAP-Rf. A similar conclusion is applicable to pECE/Rf and HCV core construct-cotransfected cells, albeit the modulatory effect elicited by the HCV core construct (0.3 μg) on the pECE/Rf-transfected cells was not so profound (2.4- to 2.6-fold) (Fig. 9C) as those of pFLAG/Rf-transfected cells (Fig. 9B). This variation on the extent of modulatory effect by the HCV core protein may reflect the difference in expression level or nature (recombinant or native) of CAP-Rf protein expressed in these two different cotransfection experiments. Despite this and whatever the mechanism is, CAP-Rf is clearly involved in the process of gene expression, and the core protein of HCV can modulate its trans-activation ability.
FIG. 9.
HCV core protein affects the luciferase reporter activity trans activated by CAP-Rf. (A) HuH-7 cells were transfected with a reporter plasmid, pCMV-Luc (0.15 μg), and various amounts of pFLAG/Rf or pECE/Rf or their control vector. After day 2 posttransfection, cells were assayed for luciferase activity as described in Materials and Methods. The luciferase activity is represented as fold induction relative to that for cells transfected with reporter plasmid and control vector. (B) All experimental conditions were similar to those described for panel A except that HuH-7 cells were transfected with pCMV-Luc (0.15 μg) and 0.3 μg of pFLAG/Rf (or pFLAG-CMV-2) together with 0.3 or 0.6 μg of HCV core construct (pSRα/HCVc122 or pSRα/HCVc101) or their control vector pSRα. (C) All experimental conditions were similar to those described for panel A except that HuH-7 cells were transfected with pCMV-Luc (0.15 μg) and pECE/Rf (or pECE) together with the HCV core construct (pSRα/HCVc122 or pSRα/HCVc101) or their control vector pSRα (0.3 μg each). Values shown in all panels are averages (means ± standard deviations) of one representative experiment in which each transfection was performed in triplicate.
DISCUSSION
DEAD box family proteins, with ATPase and RNA helicase activity, participate in a wide range of cellular processes where manipulation of RNA structure is involved (for reviews, see references 21, 59, 83, and 95). In all living organisms, a diverse array of DEAD box proteins has evolved, implying that control of RNA secondary structure is a major preoccupation for the cell. This family of proteins characterized so far participates in translation, spliceosome assembly, splicing reaction, rRNA maturation, ribosome assembly, mRNA transport, mRNA stability, germ line development, and embryogenesis (21, 37, 57, 59, 64, 69, 83, 95). This wide spectrum of functions can be explained by the fact that these proteins differ in size and that their additional N- and C-terminal sequences confer the specificity of each polypeptide, including subcellular localization, RNA binding specificity, and regions required for the interaction with accessory proteins (for reviews, see references 21 and 83).
In this study, we have identified one member of the DEAD box family of proteins as the HCV core protein-interacting target by the yeast two-hybrid system, the in vitro and in vivo binding assay, and the immunocolocalization analysis (Table 1; Fig. 4 to 6). This DEAD box protein, CAP-Rf, is a homologue of human DEAD box proteins DBX and DBY (48) or mouse DEAD3 (24). Compared to other members of the DEAD box protein family, CAP-Rf is most similar to mouse PL10 (95% identity and 98% similarity) (54) or Xenopus An3 (81% identity and 90% similarity) (30) and, to a lesser extent, to yeast Ded1 and Dbp1 (49 to 50% identity and 74 to 75% similarity) (38, 39) or Drosophila Vasa (39% identity and 63% similarity) (33, 52). Strikingly, among these, DBX, DBY, PL10, An3, and Vasa are all germ cell-specific or developmentally regulated genes (30, 31, 33, 48, 52, 54), suggesting that they may impart a developmental or tissue-specific level of regulation to RNA translation, RNA splicing, or yet uncharacterized RNA helicase function. Additionally, the similarity of CAP-Rf to these aforementioned DEAD box proteins offers a framework for considering the function of CAP-Rf. Along this line, testis-specific PL10 protein from mice is implicated in spermatogenesis (54) and is a functional homologue of Ded1 (13). An3 from Xenopus has a dynamic pattern of subcellular localization during oogenesis and is in close association with the amplified copies of ribosomal DNA that make up the multiple extrachromosomal nucleoli in oocytes (30, 31). Based on its unusual RNA unwindase property, this protein has been proposed to play a role in rRNA or pre-mRNA processing (32). Vasa is a component of Drosophila polar granules which governs the posterior group maternal effect gene and thus is required for oocyte maturation (33, 52, 56). Yeast Ded1 and Dbp1 or very likely mouse PL10 is directly involved in the process of translation (13, 15), albeit genetic studies have shown that Ded1 can act as a suppressor for the splicing defect in PRP8 mutation (39). In the present work, we demonstrated that overexpression of CAP-Rf led to activation of the luciferase reporter plasmid activity (Fig. 9A), supporting the notion that CAP-Rf is involved in gene expression regulation. Moreover, since CAP-Rf is present predominantly as a nuclear protein (Fig. 2C), it probably has a role in nuclear events such as transcriptional control and RNA processing. Alternatively, but not exclusively, since within the DEAD box superfamily CAP-Rf (or DBX/DBY) is most similar to PL10, the role of CAP-Rf in the mRNA translational process should not yet be discounted.
All DEAD box proteins possess ATPase activity, and most of their ATPase activities have been shown to be RNA dependent (21, 53, 83). In general, a large variety of RNAs that include homopolynucleotides like poly(U), poly(A), poly(C), and poly(G) stimulate ATPase activity and different RNA species are not equivalent activators for specific DEAD box proteins (53, 85). In some cases, such as E. coli DbpA and yeast Slt22, their ATPase activities are stimulated only by a very specific RNA ligand (23S rRNA and U2/U6 small nuclear RNA, respectively) (22, 97). In contrast to most DEAD box proteins, CAP-Rf hydrolyzed NTP-dNTP even without RNA (Fig. 7 and Table 2). RNA-independent ATPase activity is not unique to CAP-Rf. The ATPase activity of recombinant Drosophila Vasa protein or plant PRH75 protein is RNA independent (56, 62). Also, the ATPase activity of An3 is inhibited at least 10-fold by the duplex RNA known to be a substrate for its helicase activity (32). While studying CAP-Rf, we noted almost a complete reduction in NTP-dNTP hydrolysis in certain synthetic RNA ligands (Table 2). However, until the natural RNA substrate for CAP-Rf is defined, it remains possible that stimulation of ATPase activity in this particular DEAD box protein, as with the DbpA or Slt22 protein, may require a specific RNA.
Apart from viral RNA helicases (41, 49, 84), only a few members (eIF-4A/4B, An3, p68, and Vasa) of the DEAD box proteins have now been shown to have RNA helicase activity (31, 34, 56, 67, 68, 76). Although CAP-Rf has all functional motifs of RNA helicase (Fig. 1A), we were unable to detect any DNA- or RNA-unwinding activity associated with this recombinant CAP-Rf when we used the artificial RNA or DNA substrate (53) known to be a standard substrate for other RNA helicases (data not shown). This lack of RNA-DNA-unwinding activity in recombinant CAP-Rf could be caused simply by an artificial fusion of the His tag at the N terminus of CAP-Rf. Alternatively, CAP-Rf may require an additional protein to exhibit the RNA-unwinding activity as shown for eIF-4A (67, 68, 76) or E. coli RhlB (69), where it exhibits ATPase or/and RNA-unwinding activity only in the presence of the second protein, eIF-4B, or with another accessory protein in the RNA degradosome. Moreover, it is also likely that CAP-Rf exhibits its RNA-unwinding activity only for a specific RNA ligand as found in the case of An3 (32). Therefore, to resolve this issue one probably needs to identify the CAP-Rf-associated cellular factor or its natural RNA ligand, if possible.
In this study, we demonstrate that CAP-Rf with a C-terminal deletion, but not with the N-terminal deletion, failed to interact with the HCV core protein (Table 1). The C-terminal regions of CAP-Rf deleted (residues 473 to 661) comprise the helicase domains VII and VIII of central core region and the C terminus, with the characteristic 80 amino acids at the very end, referred to as the RS/GYR domain, which is enriched in serine, arginine, glycine, and phenylalanine (or tyrosine) residues (Fig. 1A). The helicase domain VIII (HRIGRXXR) is important for RNA-binding and ATPase activities and thus is essential for RNA unwinding in some viral RNA helicases (17, 75) and in the DEAD box protein eIF-4A (67, 68) and may be in CAP-Rf as well. The C-terminal RS/GYR domain of CAP-Rf is reminiscent of the basic RS domains found in Drosophila splicing factors Sx1 (3, 36) and Tra (55) and mammalian splicing factors ASF/SF2 (23, 45), SC35 (19, 20), and SR protein (99, 100). This domain also has some similarity to the glycine-rich regions present in nucleolus proteins such as nucleolin (50), fibrillarin (51), and SSB1 protein (40). In light of the facts that the glycine-rich domain has been postulated to form unusual secondary structures with RNA binding and RNA duplex destabilizing properties (7, 25) and that most RS-rich proteins are involved in constitutive pre-mRNA splicing reactions and also can regulate alternative splicing (1, 4, 18, 29, 63, 93, 96, 99–101), CAP-Rf may have a role in RNA processing, particularly in the assembly or disassembly of the RNA duplexes in splicing complexes or ribosomal particles. In keeping with this, since the HCV core protein also targets the C-terminal region of CAP-Rf, the core protein may either alter the conformation of CAP-Rf through the complex formation or compete for the RNA ligand bound in CAP-Rf, which in turn affects these above-mentioned biological activities of CAP-Rf. Consistent with this notion is our result which indicates that GST-core protein, but not GST protein, can enhance the ATPase-dATPase activity of CAP-Rf (Fig. 8). Given the importance of ATPase activity in the functions of most RNA helicases, if not all, the core protein conceivably may affect the cellular events in which CAP-Rf participates. Our data in this work corroborates this hypothesis, since the core protein can modulate the trans-activation effect of CAP-Rf in a transient-transfection experiment (Fig. 9). However, our understanding of the exact role of core protein in CAP-Rf activity probably requires further study to distinguish whether the CAP-Rf protein is associated with translation machinery or with transcription-posttranscriptional complexes.
Another important issue that deserves attention is the biological significance of interaction between CAP-Rf and the HCV core protein in HCV-infected cells. One possibility is that the HCV core protein interacts with CAP-Rf in ribonucleoprotein particles (spliceosome or ribosome) to recruit the particles for the benefit of the virus, such as translation of HCV polyprotein or replication of the HCV RNA genome. Consistent with this view is the finding that, at least in the cases of herpes simplex virus or adenovirus, redistribution of small nuclear ribonucleoprotein particle or SR proteins occurs in virus-infected cells, which may constitute one of the ways in which viruses alter the host to favor viral replication (5, 81). Alternatively, by virtue of high-level conservation of the DEAD box superfamily proteins, it is also very likely that the core protein may interact with other members of the DEAD box superfamily and thereby impose a pleiotropic effect on the host cellular gene regulation events, including translation, splicing, mRNA stability, and nucleocytoplasmic transport, which have been shown to be DEAD box protein dependent. Likewise, since HCV also possesses ATP-dependent RNA helicase activity in its nonstructural protein NS3, which belongs to the DEAH subfamily of DEAD box proteins (44, 89), it is thus attractive to propose that the core protein may also target the NS3 protein, modulate its RNA-unwinding activity, and consequently affect the HCV viral genome replication or translation by its own structural protein. We are currently examining this possibility. All together, our study thus provides a clue to explain the diverse effects of core protein in cellular or viral promoter activity and moreover the role of core protein in HCV pathogenesis.
ACKNOWLEDGMENTS
We are grateful to L.-H. Hwang for providing the purified HCV NS3 helicase for the control experiment.
This work was supported by grants NSC84-2331-B010-016MH, NSC86-2315-B010-001-MH, NSC87-2315-B010-001MH, and NSC88-2315-B010-001MH from the National Science Council and in part by grant DOH88-HR-502 from the National Health Research Institute of the Republic of China to Y.-H.W.L.
REFERENCES
- 1.Adams M D, Rudner D Z, Rio D C. Biochemistry and regulation of pre-mRNA splicing. Curr Opin Cell Biol. 1996;8:331–339. doi: 10.1016/s0955-0674(96)80006-8. [DOI] [PubMed] [Google Scholar]
- 2.Barba G, Harper F, Harada T, Kohara M, Goulinet S, Matsuura Y, Eder G, Schaff Z, Chapman M J, Miyamura T, Brechot C. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc Natl Acad Sci USA. 1997;94:1200–1205. doi: 10.1073/pnas.94.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell L R, Horabin J I, Schedl P, Cline T W. Positive autoregulation of sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell. 1991;65:229–239. doi: 10.1016/0092-8674(91)90157-t. [DOI] [PubMed] [Google Scholar]
- 4.Blencowe B J, Issner R, Nickerson J A, Sharp P. A coactivator of pre-mRNA splicing. Genes Dev. 1998;12:996–1009. doi: 10.1101/gad.12.7.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bridge E, Xia D-X, Carmo-Fonseca M, Cardinali B, Lamond A I, Pettersson U. Dynamic organization of splicing factors in adenovirus-infected cells. J Virol. 1995;69:281–290. doi: 10.1128/jvi.69.1.281-290.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruix J, Barrera J M, Calvet X, Ercilla G, Costa J, Sanchez-Tapias J M, Ventura M, Vall M, Bruguera M, Bru C, Castillo R, Rodes J. Prevalence of antibodies to hepatitis C virus in Spanish patients with hepatocellular carcinoma and hepatic cirrhosis. Lancet. 1989;ii:1004–1006. doi: 10.1016/s0140-6736(89)91015-5. [DOI] [PubMed] [Google Scholar]
- 7.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 8.Chang P-J, Chang Y-S, Liu S-T. Role of Rta in the translation of bicistronic BZLF1 of Epstein-Barr virus. J Virol. 1998;72:5128–5136. doi: 10.1128/jvi.72.6.5128-5136.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C-M, You L-R, Hwang L-H, Lee Y-H W. Direct interaction of hepatitis C virus core protein with the cellular lymphotoxin-β receptor modulates the signal pathway of the lymphotoxin-β receptor. J Virol. 1997;71:9417–9426. doi: 10.1128/jvi.71.12.9417-9426.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen P-J, Lin M-H, Tai K-F, Liu P-C, Lin C-J, Chen D-S. The Taiwanese hepatitis C virus genome: sequence determination and mapping the 5′-termini of viral genome and antigenomic RNA. Virology. 1992;188:102–113. doi: 10.1016/0042-6822(92)90739-c. [DOI] [PubMed] [Google Scholar]
- 11.Choo Q-L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 12.Choo Q-L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby A, Barr P J, Weiner A J, Bradley D W, Kuo G, Houghton M. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chuang R-Y, Weaver P L, Liu Z, Chang T-H. Requirement of the DEAD-box protein Ded1p for messenger RNA translation. Science. 1997;275:1468–1471. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- 14.Chung J, Lee S, Song K. Identification of a human homolog of a putative RNA helicase gene (mDEAD3) expressed in mouse erythroid cells. Korean J Biochem. 1995;27:193–197. [Google Scholar]
- 15.Cruz J, Iost I, Kressler D, Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis L, Clauser E, Morgan D O, Edery M, Roth R A, Rutter W J. Replacement of insulin receptor tyrosine residues 1162 and 1163 compromises insulin-stimulated kinase activity and uptake of 2-deoxyglucose. Cell. 1986;45:721–732. doi: 10.1016/0092-8674(86)90786-5. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez A, Lain S, Garcia J A. RNA helicase activity of the plum pox potyvirus CI protein expressed in Escherichia coli. Mapping of an RNA binding domain. Nucleic Acids Res. 1995;23:1327–1332. doi: 10.1093/nar/23.8.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu X-D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 19.Fu X-D, Maniatis T. Isolation of a complementary DNA that encodes the mammalian splicing factor SC35. Science. 1992;256:535–538. doi: 10.1126/science.1373910. [DOI] [PubMed] [Google Scholar]
- 20.Fu X-D, Mayeda A, Maniatis T, Krainer A R. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci USA. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuller-Pace F V. RNA helicases: modulators of RNA structure. Trends Cell Biol. 1994;4:271–274. doi: 10.1016/0962-8924(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 22.Fuller-Pace F V, Nicol S M, Reid A D, Lane D P. DbpA: a DEAD box protein specifically activated by 23S rRNA. EMBO J. 1993;12:3619–3626. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ge H, Zuo P, Manley J L. Primary structure of the human splicing ASF reveals similarities with Drosophila regulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- 24.Gee S L, Conboy J G. Mouse erythroid cells express multiple putative RNA helicase genes exhibiting high sequence conservation from yeast to mammals. Gene. 1994;140:171–177. doi: 10.1016/0378-1119(94)90541-x. [DOI] [PubMed] [Google Scholar]
- 25.Ghisolfi L, Joseph G, Amalric F, Erard M. The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J Biol Chem. 1992;267:2955–2959. [PubMed] [Google Scholar]
- 26.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Two related superfamilies of putative helicase involved in replication, recombination, repair, and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4729. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grakoui A, Wychowski C, Lin C, Feinstone S M, Rice C M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graveley B R, Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol Cell. 1998;1:765–771. doi: 10.1016/s1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- 30.Gururajan R, Perry-O’Keefe H, Melton D A, Weeks D L. The Xenopus localized messenger RNA An3 may encode an ATP-dependent RNA helicase. Nature. 1991;349:717–719. doi: 10.1038/349717a0. [DOI] [PubMed] [Google Scholar]
- 31.Gururajan R, Mathews L, Longo F J, Weeks D L. An3 mRNA encodes an RNA helicase that colocalizes with nucleoli in Xenopus oocytes in a stage-specific manner. Proc Natl Acad Sci USA. 1994;91:2056–2060. doi: 10.1073/pnas.91.6.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gururajan R, Weeks D L. An3 protein encoded by a localized maternal mRNA in Xenopus laevis is an ATPase with substrate-specific RNA helicase activity. Biochim Biophys Acta. 1997;1350:169–182. doi: 10.1016/s0167-4781(96)00155-8. [DOI] [PubMed] [Google Scholar]
- 33.Hay B, Jay L Y, Jan Y N. Localization of Vasa, a component of Drosophila polar granules, in maternal-effect mutants that alter embryonic anteroposterior polarity. Cell. 1988;55:577–587. doi: 10.1242/dev.109.2.425. [DOI] [PubMed] [Google Scholar]
- 34.Hirling H, Scheffner M, Restle T, Stahl H. RNA helicase activity associated with human p68 protein. Nature. 1989;339:562–564. doi: 10.1038/339562a0. [DOI] [PubMed] [Google Scholar]
- 35.Hsieh T-Y, Matsumoto M, Chou H-C, Schneider R, Hwang S B, Lee A S, Lai M M C. Hepatitis C virus core protein interacts with heterologous nuclear ribonucleoprotein K. J Biol Chem. 1998;273:17651–17659. doi: 10.1074/jbc.273.28.17651. [DOI] [PubMed] [Google Scholar]
- 36.Inoue K, Hoshijima K, Sakamoto H, Shimura Y. Binding of the Drosophila sex-lethal gene product to the alternative splice site of transformer primary transcript. Nature. 1990;344:461–463. doi: 10.1038/344461a0. [DOI] [PubMed] [Google Scholar]
- 37.Iost I, Dreyfus M. mRNA can be stabilized by DEAD-box proteins. Nature. 1994;372:193–196. doi: 10.1038/372193a0. [DOI] [PubMed] [Google Scholar]
- 38.Jamieson D J, Beggs J D. A suppressor of yeast spp81/ded1 mutations encodes a very similar putative ATP-dependent RNA helicase. Mol Microbiol. 1991;5:805–812. doi: 10.1111/j.1365-2958.1991.tb00753.x. [DOI] [PubMed] [Google Scholar]
- 39.Jamieson D J, Rahe B, Pringle J, Beggs J D. A suppressor of a yeast splicing mutation (prp8-1) encodes a putative ATP-dependent RNA helicase. Nature. 1991;349:715–717. doi: 10.1038/349715a0. [DOI] [PubMed] [Google Scholar]
- 40.Jong A Y-S, Clark M W, Gilbert M, Oehm A, Campbell J L. Saccharomyces cerevisiae SSB1 protein and its relationship to nucleolar RNA-binding proteins. Mol Cell Biol. 1987;7:2947–2955. doi: 10.1128/mcb.7.8.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kadare G, Haenni A-L. Virus-encoded RNA helicase. J Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D W, Suzuki R, Harada T, Saito I, Miyamura T. Trans-suppression of gene expression by hepatitis C viral core protein. Jpn J Med Sci Biol. 1994;47:211–220. doi: 10.7883/yoken1952.47.211. [DOI] [PubMed] [Google Scholar]
- 44.Kim D W, Gwack Y, Han J H, Choe J. C-terminal domain of the hepatitis virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 45.Krainer A R, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 46.Kuo G, Choo Q-L, Alter H J, Gitnick G L, Redeker A G, Purcell R H, Miyamura T, Dienstag L, Alter M J, Stevens C E, Tegtmeier G E, Bonino F, Colombo M, Lee W-S, Kuo C, Berger K, Shuster J R, Overby L R, Bradely D W, Houghton M. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- 47.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 48.Lahn B T, Page D C. Functional coherence of the human Y chromosome. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- 49.Lain S, Riechmann J L, Garcia J A. RNA helicase: a novel activity associated with a protein encoded by a positive strand RNA virus. Nucleic Acids Res. 1991;18:7003–7006. doi: 10.1093/nar/18.23.7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lapeyre B, Mariottini P, Mathieu C, Ferrer P, Amaldi F, Amalric F, Caizergues-Ferrer M. Molecular cloning of Xenopus fibrillarin, a conserved U3 small nuclear ribonucleoprotein recognized by antisera from humans with autoimmune disease. Mol Cell Biol. 1990;10:430–434. doi: 10.1128/mcb.10.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lapeyre B, Bourbon H, Amalric F. Nucleolin, a major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by the nucleic sequence. Proc Natl Acad Sci USA. 1987;84:1472–1476. doi: 10.1073/pnas.84.6.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lasko P F, Ashburner M. Posterior localization of Vasa protein correlates with, but is not sufficient for, pole cell development. Nature. 1988;335:611–617. doi: 10.1101/gad.4.6.905. [DOI] [PubMed] [Google Scholar]
- 53.Lee C-G, Hurwitz J. A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3′ to 5′ direction. J Biol Chem. 1992;267:4398–4407. [PubMed] [Google Scholar]
- 54.Leroy P, Alzari P, Sassoon D, Wolgemuth D, Fellous M. The protein encoded by a murine male germ cell-specific transcript is a putative ATP-dependent RNA helicase. Cell. 1989;57:549–559. doi: 10.1016/0092-8674(89)90125-6. [DOI] [PubMed] [Google Scholar]
- 55.Li H, Bingham P M. Arginine/serine-rich domains of the su(wa) and tra RNA processing regulator target proteins to subnuclear compartment implicated in splicing. Cell. 1991;67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- 56.Liang L, Diehl-Jones W, Lasko P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development. 1994;120:1201–1211. doi: 10.1242/dev.120.5.1201. [DOI] [PubMed] [Google Scholar]
- 57.Liang S, Hitomi M, Hu Y-H, Liu Y, Tartakoff A M. A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Mol Cell Biol. 1996;16:5139–5146. doi: 10.1128/mcb.16.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin C, Lindenbach B D, Pragai B M, McCourt D W, Rice C M. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J Virol. 1994;68:5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Linder P, Lasko P F, Leroy P, Nielsen P J, Nishi K, Schnier J, Slonimski P P. Birth of the D-E-A-D box. Nature. 1989;337:121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- 60.Liu Q, Tackney C, Bhat R A, Prince A M, Zhang P. Regulated processing of hepatitis C virus core protein is linked to subcellular localization. J Virol. 1997;71:657–662. doi: 10.1128/jvi.71.1.657-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lo S-Y, Masiarz F, Hwang S B, Lai M M C, Ou J-H. Differential subcellular localization of hepatitis C virus core gene products. Virology. 1995;213:455–461. doi: 10.1006/viro.1995.0018. [DOI] [PubMed] [Google Scholar]
- 62.Lorkovic Z J, Herrmann R G, Oelmuller R. PRH75, a new nucleus-localized member of the DEAD-box protein family from higher plants. Mol Cell Biol. 1997;17:2257–2265. doi: 10.1128/mcb.17.4.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 64.Margossian S P, Li H, Zassenhaus H P, Butow R A. The DEXH box protein Suv3p is a component of a yeast mitochondrial 3′-to-5′ exoribonuclease that suppresses group I intron toxicity. Cell. 1996;84:199–209. doi: 10.1016/s0092-8674(00)80975-7. [DOI] [PubMed] [Google Scholar]
- 65.Matsumoto M, Hsieh T-Y, Zhu N, Vanarsdale T, Hwang S B, Jeng K-S, Gorbalenya A E, Lo S-Y, Ou J-H, Ware C F, Lai M M. Hepatitis C virus core protein interacts with the cytoplasmic tail of lymphotoxin-β receptor. J Virol. 1997;71:1301–1309. doi: 10.1128/jvi.71.2.1301-1309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Okamoto H, Kurai K, Okada S-I, Yamamoto K, Lizuka H, Tanaka T, Fukuda S, Tsuda F, Mishiro S. Full-length sequence of a hepatitis C virus genome having poor homology to reported isolates: comparative study of four distinct genotypes. Virology. 1992;188:331–341. doi: 10.1016/0042-6822(92)90762-e. [DOI] [PubMed] [Google Scholar]
- 67.Pause A, Methot N, Sonenberg N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol Cell Biol. 1993;13:6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Py B, Higgins C F, Krisch H M, Carpousis A J. A DEAD-box RNA helicase in the Escherichia coli degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 70.Ray R B, Lagging L M, Meyer K, Steele R, Ray R. Transcriptional regulation of cellular and viral promoters by the hepatitis C virus core protein. Virus Res. 1995;37:209–220. doi: 10.1016/0168-1702(95)00034-n. [DOI] [PubMed] [Google Scholar]
- 71.Ray R B, Lagging L M, Meyer K, Ray R. Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J Virol. 1996;70:4438–4443. doi: 10.1128/jvi.70.7.4438-4443.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ray R B, Meyer K, Ray R. Suppression of apoptotic cell death by hepatitis C virus core protein. Virology. 1996;226:176–182. doi: 10.1006/viro.1996.0644. [DOI] [PubMed] [Google Scholar]
- 73.Ray R B, Meyer K, Shrivastava A, Aggarwaland B B, Ray R. Inhibition of tumor necrosis factor (TNF-alpha)-mediated apoptosis by hepatitis C virus core protein. J Biol Chem. 1998;273:2256–2259. doi: 10.1074/jbc.273.4.2256. [DOI] [PubMed] [Google Scholar]
- 74.Ray R B, Steele R, Meyer K, Ray R. Transcriptional repression of p53 promoter by hepatitis C virus core protein. J Biol Chem. 1997;272:10983–10986. doi: 10.1074/jbc.272.17.10983. [DOI] [PubMed] [Google Scholar]
- 75.Rodriguez P L, Carrasco L. Poliovirus protein 2C contains two regions involved in RNA binding activity. J Biol Chem. 1995;270:10105–10112. doi: 10.1074/jbc.270.17.10105. [DOI] [PubMed] [Google Scholar]
- 76.Rozen F, Edery I, Meerovitch K, Dever T E, Merrick W C, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rozen F, Pelletier J, Trachsel H, Sonenberg N. A lysine substitution in the ATP-binding site of eukaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1989;9:4061–4063. doi: 10.1128/mcb.9.9.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ruggieri A, Harada T, Matsuura Y, Miyamura T. Fas-mediated apoptosis by hepatitis C virus core protein. Virology. 1997;229:68–76. doi: 10.1006/viro.1996.8420. [DOI] [PubMed] [Google Scholar]
- 79.Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama S, Kikuchi S, Watanabe Y, Koi S, Onji M, Choo Q-L, Houghton M, Kuo G. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 81.Sandri-Goldin R M, Hibbard M K, Hardwicke M A. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J Virol. 1995;69:6063–6076. doi: 10.1128/jvi.69.10.6063-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Santolini E, Migliaccio G, Monica N L. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol. 1994;68:3631–3641. doi: 10.1128/jvi.68.6.3631-3641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schmid S R, Linder P. D-E-A-D protein family of putative RNA helicase. Mol Microbiol. 1992;6:283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 84.Schuman S. Vaccinia virus RNA helicase. J Biol Chem. 1993;268:11798–11802. [PubMed] [Google Scholar]
- 85.Schwer B, Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;349:494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- 86.Shih C-M, Chen C-M, Chen S-Y, Lee Y-H W. Modulation of the trans-suppression activity of hepatitis C virus core protein by phosphorylation. J Virol. 1995;69:1160–1171. doi: 10.1128/jvi.69.2.1160-1171.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shih C-M, Lo S J, Miyamura T, Chen S-Y, Lee Y-H W. Suppression of hepatitis B virus expression and replication by hepatitis C virus core protein in HuH-7 cells. J Virol. 1993;67:5823–5832. doi: 10.1128/jvi.67.10.5823-5832.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Suzuki R, Matsuura Y, Suzuki T, Ando A, Chiba J, Harada S, Saito I, Miyamura T. Nuclear localization of the truncated hepatitis C virus core protein with its hydrophobic C terminus deleted. J Gen Virol. 1995;76:53–61. doi: 10.1099/0022-1317-76-1-53. [DOI] [PubMed] [Google Scholar]
- 89.Tai C-L, Chi W-K, Chen D-S, Hwang L-H. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3) J Virol. 1996;70:8477–8484. doi: 10.1128/jvi.70.12.8477-8484.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Takebe Y, Seike M, Fujisawa J-I, Hoy P, Yokota K, Arai K-I, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and R-U5 segment of human T-cell leukemia virus type 1 long terminal reapeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequence in the α- and β-subunits of ATP synthase, myosin, kinases, and other ATP requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang J, Manley J L. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- 94.Warrener P, Collett M S. Pestivirus NS3 (p80) protein possesses RNA helicase activity. J Virol. 1995;69:1720–1726. doi: 10.1128/jvi.69.3.1720-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wassarman D A, Steitz J A. Alive with DEAD proteins. Nature. 1991;349:463–464. doi: 10.1038/349463a0. [DOI] [PubMed] [Google Scholar]
- 96.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 97.Xu D, Nouraini S, Field D, Tang S-J, Friesen J D. An RNA-dependent ATPase activity associated with U2/U6 snRNAs in pre-mRNA splicing. Nature. 1996;381:709–713. doi: 10.1038/381709a0. [DOI] [PubMed] [Google Scholar]
- 98.Yasui K, Wakita T, Tsukiyama-Kohara K, Funahashi S-I, Ichikawa M, Kajita T, Moradpour D, Wands J R, Kohara M. The native form and maturation process of hepatitis C virus core protein. J Virol. 1998;72:6048–6055. doi: 10.1128/jvi.72.7.6048-6055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zahler A M, Lane W S, Stolk J A, Roth M B. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 100.Zahler A M, Neugebauer K M, Lane W S, Roth M B. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 101.Zahler A M, Roth M B. Distinct functions of SR proteins in recruitment of U1 small ribonucleoprotein to alternative 5′ splice sites. Proc Natl Acad Sci USA. 1995;92:2642–2646. doi: 10.1073/pnas.92.7.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhu N, Khoshnan A, Schneider R, Matsumoto M, Dennert G, Ware C, Lai M M C. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J Virol. 1998;72:3691–3697. doi: 10.1128/jvi.72.5.3691-3697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]