Abstract
The intracellular enveloped form of vaccinia virus (IEV) induces the formation of actin tails that are strikingly similar to those seen in Listeria and Shigella infections. In contrast to the case for Listeria and Shigella, the vaccinia virus protein(s) responsible for directly initiating actin tail formation remains obscure. However, previous studies with recombinant vaccinia virus strains have suggested that the IEV-specific proteins A33R, A34R, A36R, B5R, and F13L play an undefined role in actin tail formation. In this study we have sought to understand how these proteins, all of which are predicted to have small cytoplasmic domains, are involved in IEV assembly and actin tail formation. Our data reveal that while deletion of A34R, B5R, or F13L resulted in a severe reduction in IEV particle assembly, IEVs formed by the ΔB5R and ΔF13L deletion strains, but not ΔA34R, were still able to induce actin tails. The ΔA36R deletion strain produced normal amounts of IEV particles, although these were unable to induce actin tails. Using several different approaches, we demonstrated that A36R is a type Ib membrane protein with a large, 195-amino-acid cytoplasmic domain exposed on the surface of IEV particles. Finally, coimmunoprecipitation experiments demonstrated that A36R interacts with A33R and A34R but not with B5R and that B5R forms a complex with A34R but not with A33R or A36R. Using extracts from ΔA34R- and ΔA36R-infected cells, we found that the interaction of A36R with A33R and that of A34R with B5R are independent of A34R and A36R, respectively. We conclude from our observations that multiple interactions between IEV membrane proteins exist which have important implications for IEV assembly and actin tail formation. Furthermore, these data suggest that while A34R is involved in IEV assembly and organization, A36R is critical for actin tail formation.
Viruses succeed as intracellular pathogens because they are able to invade cells and appropriate the cellular machinery required for their life cycle. In many cases the actin cytoskeleton of the host cell is used or disrupted by the virus to facilitate the infection process (reviewed in reference 7). Although many different viruses are capable of interacting with and modifying the host actin cytoskeleton, vaccinia virus, a large DNA virus that is a close relative of variola virus, the causative agent of smallpox, induces the most dramatic effects on the actin cytoskeleton (1, 5, 6, 15, 17, 18, 22, 33, 41). As early as 1976, vaccinia virus particles were observed on the tips of large microvilli extending from the cell surface (41). Subsequently these virus-tipped microvilli were shown to contain actin, as well as the actin-cross-linking proteins α-actinin, filamin, and fimbrin, but not myosin or tropomyosin (15, 18). More recently, the effects of vaccinia virus on the actin cytoskeleton were reexamined (1, 5, 6, 33). Vaccinia virus infection results in the dramatic reorganization of actin from stress fibers into virus-tipped actin tails (5). By using mutant viruses and drugs that inhibit viral morphogenesis, it was shown that the intracellular enveloped form of vaccinia virus (IEV) is responsible for nucleating actin tails (5). IEV particles arise from a small proportion of the intracellular mature form of vaccinia virus (IMV) which becomes enveloped by a membrane cisterna derived from the trans-Golgi network (38). With actin polymerization as the driving force, IEV particles are propelled in vivo on the tips of actin tails at a speed of ∼3.0 μm/min (5). Upon contact with the cell surface, virus particles extend outward on actin projections at a similar rate, to contact and infect neighboring cells. IEV is thought to leave the host cell by fusion of its outermost membrane with the plasma membrane, giving rise to the extracellular enveloped form of vaccinia virus (EEV) (2, 26, 31). During this fusion event, a small number of EEV particles are not released into the medium but remain associated with the outer surface of the plasma membrane (3). These virus particles are termed cell-associated enveloped viruses (CEV) (3). While actin tail assembly is not essential for virus spread between cells, it does enhance the efficiency of the process, as recombinant viral strains that do not induce actin tails have a small-plaque phenotype compared to wild-type strains (37).
Vaccinia virus-induced actin tails are strikingly similar to those seen in infections with the bacterial pathogens Listeria, Shigella, and Rickettsia, suggesting that intracellular pathogens have developed a common mechanism to exploit the actin cytoskeleton to facilitate their spread (5). However, in contrast to the case for Listeria and Shigella, the viral protein(s) responsible for initiating the cascade of events that leads to actin tail formation by IEV particles is still unknown. To date only six virus-encoded IEV-specific proteins, termed A33R (35), A34R (8), A36R (29), A56R (32, 39), B5R (9, 20), and F13L (19), have been identified. Studies using vaccinia virus mutants in which these IEV-specific genes were deleted or repressed have examined their roles in IEV assembly and actin tail formation. These studies have shown that A56R, the viral hemagglutinin, is not required for IEV assembly or actin tail formation (37), while F13L (2) and B5R (10, 47) are involved in morphogenesis of IEV particles. Deletion of A33R reduces the number of IEV particles that are completely wrapped within trans-Golgi network-derived membrane cisternae, and those particles that assemble are unable to form actin tails (36). Studies with viruses lacking A34R (37, 48) or A36R (37, 49) showed that these proteins are essential for actin tail formation, but the mechanism remains obscure. A34R (8) and A36R (29) have been described as type II integral membrane proteins with predicted cytoplasmic domains of 12 and 2 amino acids, respectively, exposed on the surface of IEV particles. Type II integral membrane proteins are defined as having a single hydrophobic domain near the cytoplasmic N terminus that anchors the protein in the membrane, while the bulk of the protein, including the C terminus, is exposed to the luminal side of the membrane (40, 45). While it is clear that A34R and A36R are important in vaccinia virus-induced actin tail formation, it is difficult to envisage how such short cytoplasmic domains exposed on the surface of IEV can play a direct role in recruiting host cytoskeletal factors required for actin tail formation.
In this study we have further examined the roles of A36R and A34R in IEV assembly and actin tail formation. We show that the A36R protein has a type Ib membrane topology, according to the classification described previously (40), with a 195-amino-acid cytosolic domain, rather than a type II topology as reported previously (29). Our observations suggest that A34R is involved in IEV assembly, whereas A36R is critical for actin tail formation but not required for IEV assembly.
MATERIALS AND METHODS
Generation of antibodies against A33R, A34R, and A36R.
The DNAs encoding residues Met2 to Gly34 and Asp146 to Val181 of A33R, Tyr101 to Ala140 of A34R, and Thr142 to Glu214 of A36R were amplified by PCR from the genome of vaccinia virus strain Western Reserve (WR) by using the Expand system (Boehringer, Mannheim, Germany). The resulting PCR products were cloned into the BamHI and EcoRI sites of the glutathione S-transferase (GST) fusion vector pGEX-2T (Pharmacia, Freiburg, Germany). The fidelity of the resulting expression constructs was verified by DNA sequencing prior to expression in XL-1 Blue by induction with isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 0.6 mM. Three hours after induction, bacteria were harvested and the soluble fraction was prepared as described previously (46). GST fusion proteins were purified by affinity chromatography on glutathione-Sepharose resin according to the instructions of the manufacturer (Pharmacia). Proteins were eluted from the resin with 50 mM glutathione in phosphate-buffered saline (PBS) at 4°C, and the protein concentration was determined by using the Bio-Rad (Munich, Germany) protein assay. Rabbits and rats were initially injected with 200 and 100 μg of GST fusion protein, respectively, mixed with RIBI adjuvant (RIBI ImmunoChem Research, Hamilton, Mont.). Animals were boosted subsequently with 50 μg (rabbits) or 25 μg (rats) of GST fusion protein. Antibodies against residues Met2 to Gly34 of A33R were affinity purified on the peptide CTPENDEEQTSVFSATVYGDKIQGKNKRKRVIG, corresponding to the cytoplasmic domain of the protein, that had been coupled to a SulfoLink column (Pierce Chemical Co., Rockford, Ill.) via the N-terminal Cys residue. All other antibodies were affinity purified on their respective fusion proteins bound to N-hydroxysuccinimide-activated HiTrap columns (Pharmacia) after the serum had been depleted of contaminating GST antibodies by using GST bound to glutathione-Sepharose resin.
Infection and immunofluorescence analysis.
HeLa cells were grown and infected with vaccinia virus strain WR or vaccinia virus mutants as described previously (5). The mutant viruses used in this study that lack the A34R, A36R, B5R, and F13L genes are referred to as ΔA34R, ΔA36R, ΔB5R, and ΔF13L (vRB12) for simplicity (2, 10, 25, 29). Infected cells were fixed for 1 min in methanol at −20°C or for 10 min in 3% paraformaldehyde in cytoskeletal buffer (CB) (10 mM MES [morpholineethanesulfonic acid], 150 mM NaCl, 5 mM EGTA, 5 mM MgCl2, 5 mM glucose, pH 6.1) and subsequently processed as described previously (13). Infected cells were labeled with combinations of A33R, A34R, and A36R antibodies as well as with the mouse monoclonal antibody C3 against the 14-kDa peripheral membrane protein (A27L) of IMV (34) and with the rat monoclonal antibody 19C2 against B5R (16, 38). Actin was visualized with either the anti-β-actin antibody AC-74 (Sigma, Deisenhofen, Germany) or Bodipy-phallacidin obtained from Molecular Probes (Eugene, Oreg.). Following immunolabeling, cells were mounted in MOWIOL supplemented with DABCO (13). All images were recorded on a DMRXA microscope (Leica, Bensheim, Germany) by using a high-performance charge-coupled digital camera (Cohu, San Diego, Calif.) and NIH Image (version 1.62). Acquired images were processed and annotated by using the Adobe software package (Adobe Systems Incorporated, San Jose, Calif.).
Microinjection of infected HeLa cells.
HeLa cells were infected with vaccinia virus strain WR for 12 to 14 h and then microinjected by using the Zeiss (Oberkochen, Germany) automated injection system. Antibodies used for microinjection were dialyzed into microinjection buffer (100 mM KCl, 5 mM sodium phosphate, pH 7.5) by using Amicon (Witten, Germany) microconcentrators. After injection, cells were left for 1 h to recover at 37°C before being fixed with 3% paraformaldehyde in CB for 10 min, permeabilized with 0.1% Triton X-100 in CB for 2 min, and processed for immunofluorescence as described above (13).
Immunolabeling of semithin cryosections.
HeLa cells were infected with vaccinia virus strain WR, ΔA34R, ΔA36R, or ΔB5R at a multiplicity of infection (MOI) of 1 for 8 to 12 h. Subsequently, cells were fixed for 3 h in 2% paraformaldehyde and 0.2% glutaraldehyde in 0.2 M sodium phosphate buffer (pH 7.4) and embedded in 10% gelatin in PBS. Small pieces of pellet were infiltrated with 2.1 M sucrose and frozen in liquid nitrogen. Semithin cryosections were cut with a Reichert FCS ultramicrotome (Leica, Vienna, Austria) at −95°C and picked up in 2.3 M sucrose. Sections were kept on PBS prior to immunolabeling. Nonspecific antibody binding sites were blocked with 1% fish skin gelatin and 0.8% bovine serum albumin in PBS. Sections were incubated with affinity-purified rabbit polyclonal antibodies against A33R, A34R, or A36R. Sections were then washed several times in PBS, followed by incubation with protein A coupled to 10-nm gold particles and several washes in PBS. Finally, sections were washed in distilled water and subsequently positively stained and embedded with 2% methyl cellulose containing 0.3% uranyl acetate (42). After labeling and embedding, the grids were air dried. Sections were viewed in a Zeiss EM10 electron microscope at an accelerating voltage of 80 kV.
Preembedding labeling.
HeLa cells were infected with the vaccinia virus WR strain at an MOI of 1 for 8 h, washed three times with PBS, osmotically swollen in distilled water for 2 min, and fixed with 4% paraformaldehyde in CB for 10 min. The cells were then washed with PBS containing 200 mM glycine, and nonspecific binding sites were blocked with 5% fetal calf serum in PBS containing 200 mM glycine. Cells were incubated with affinity-purified antibodies against A33R, A34R, or A36R overnight at 4°C. The cells were then washed several times with PBS containing 200 mM glycine, incubated with protein A coupled to 5-nm gold particles for 2 h at 4°C, washed several times in PBS containing 200 mM glycine, and fixed with 1% glutaraldehyde. After fixation, cells were treated with 1% osmium tetroxide in 1.5% potassium ferrocyanide followed by saturated uranyl acetate in 70% ethanol and embedded in Epon. Sections cut from embedded cells were contrasted with 3% uranyl acetate in water followed by Reynold’s solution (80 mM lead nitrate, 120 mM sodium citrate, 0.64% NaOH). Sections were viewed as described above.
Immunoprecipitation and immunoblot analyses.
HeLa cells were infected with the vaccinia virus WR strain at an MOI of 0.5 for 24 h and then washed with ice-cold PBS, scraped in the same buffer, and spun down for 10 min at 2,500 × g and 4°C. Cells were resuspended in extraction buffer (25 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1 mM EGTA, 100 mM NaCl, 1% Triton X-100, 0.5% Nonidet P-40, and protease inhibitor cocktail [0.2 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, 10 μg of chymostatin per ml, 10 μg of pepstatin A per ml, and 10 μg of antipain per ml]), extracted for 1.5 h at 4°C, and centrifuged for 15 min each at 16,000 × g and subsequently at 150,000 × g at 4°C. Supernatants were diluted to a protein concentration of 5 mg/ml and incubated with antibodies against A33R, A34R, or A36R or with the rat monoclonal antibody 17C4 against B5R (16, 38) overnight at 4°C. Protein A-Sepharose beads were added to the cell extracts and incubated for 1 h. After centrifugation at 500 × g and 4°C, the supernatants were collected and the Sepharose beads were washed extensively with cell extraction buffer. Proteins present in both supernatants and beads were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 15% gels. After semidry blotting, IEV-specific proteins were detected with antibodies against A33R, A34R, or A36R or antibody 17C4 against B5R. The IMV-associated proteins A27L (p14) and D8L (p32) were visualized with monoclonal antibody C3 (34) and a rabbit polyclonal antiserum against D8L (27), respectively. Western blots were developed by using the ECL system according to the instructions of the manufacturer (Amersham International, Braunschweig, Germany).
EEV purification, protease digestion, and immunolabeling.
Confluent RK13 cells were infected with vaccinia virus International Health Department J strain at an MOI of 5 for 24 h. At 2 h postinfection, cells were washed three times with PBS and incubated with serum-free minimal essential medium. At 24 h postinfection, the culture supernatants were collected and cellular debris was spun down for 10 min at 1,000 × g and 4°C. Supernatants were then centrifuged for 30 min at 100,000 × g (4°C), and the pellets were resuspended in 10 mM Tris-HCl, pH 7.5. Proteinase K (300 μg/ml) or trypsin (5 μg/ml) was added, and samples were incubated for 30 min at 4 or 37°C, respectively. Virus particles were spun for 5 min at 16,000 × g and 4°C, and the pellets were resuspended in sample buffer and subjected to immunoblotting as described above. The same EEV preparations were also immunolabeled and viewed by electron microscopy. Resuspended EEV preparations were incubated with 300-mesh copper grids for 15 min and immunolabeled as described for cryosections. After immunolabeling, virus particles were negatively stained with a mixture of 2% uranyl acetate and 0.7% methylcellulose for 10 min. Sections were viewed as described above.
RESULTS
Characterization of A33R, A34R, and A36R antibodies.
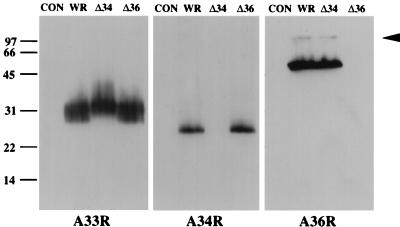
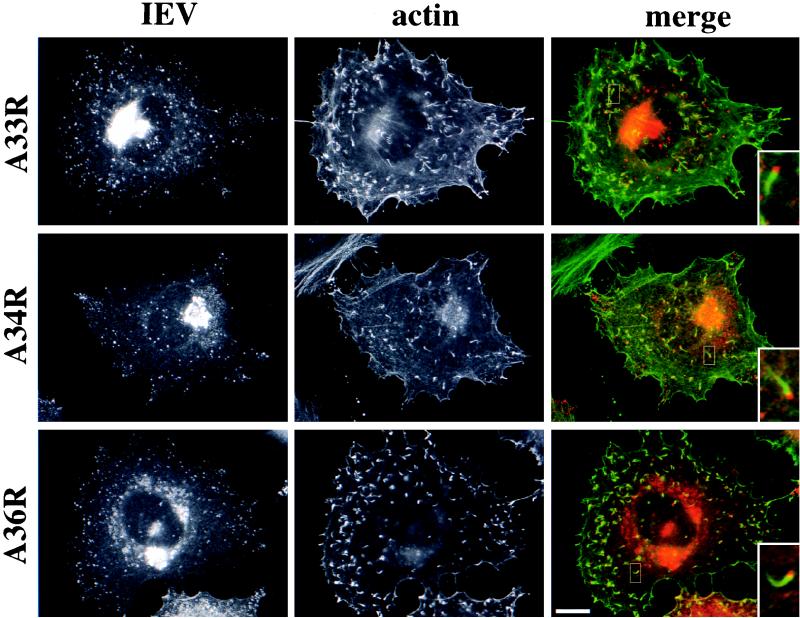
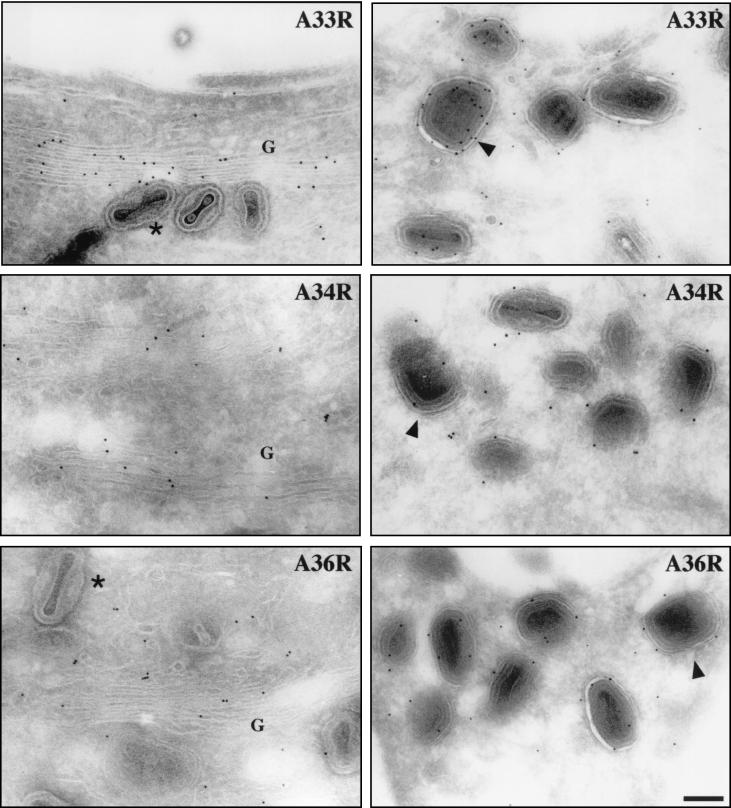
To further investigate the roles of A33R, A34R, and A36R in IEV assembly and actin tail formation, polyclonal antibodies were raised against these three proteins. Immunoblot analysis revealed that specific antibodies had been generated against the predicted luminal domains of A33R, A34R, and A36R as well as the cytoplasmic domain of A33R (Fig. 1). For unknown reasons, the mobility of A33R was always found to be slower in the absence of A34R. Identical results were obtained with antibodies against the cytoplasmic or the luminal domain of A33R. By indirect immunofluorescence all antibodies showed strong labeling of IEV particles that were readily identified by their association with actin tails. We also observed a juxtanuclear staining typical of the Golgi apparatus, which is the cellular site where IMV particles become enveloped to form IEV (Fig. 2). Immunoelectron microscopy of semithin cryosections confirmed that in addition to being localized to IEV particles, A33R, A34R, and A36R are present in the Golgi apparatus (Fig. 3) and are also found in endosomes and the plasma membrane (data not shown). In contrast, IMV particles and viral factories were not labeled with these antibodies.
FIG. 1.
Immunoblot analysis of extracts prepared from uninfected HeLa cells (CON) or HeLa cells infected with vaccinia virus strain WR or deletion mutant ΔA34R (Δ34) or ΔA36R (Δ36). Western blots were probed with antisera against A33R, A34R, and A36R. The arrowhead indicates the position of a possible A36R homodimer. Molecular mass markers are indicated in kilodaltons.
FIG. 2.
Localization of A33R, A34R, and A36R in vaccinia virus-infected HeLa cells. WR-infected cells were labeled with antibodies against A33R, A34R, or A36R as indicated on the left and with an antiactin antibody. Inserts in the merged images clearly show that all three IEV proteins (red) are localized to viral particles at the tips of actin tails (green). Bar, 10 μm.
FIG. 3.
Cryosections of vaccinia virus-infected HeLa cells were immunogold labeled with antibodies against A33R, A34R, and A36R as indicated in each panel. All three IEV proteins are present in the Golgi apparatus (G in left panels) as well as in IEV particles (right panels). Arrowheads point to IEV particles where the surrounding membranes are clearly visible. Unlabeled IMV particles are indicated by asterisks. Bar, 150 nm.
A36R is a type Ib membrane protein.
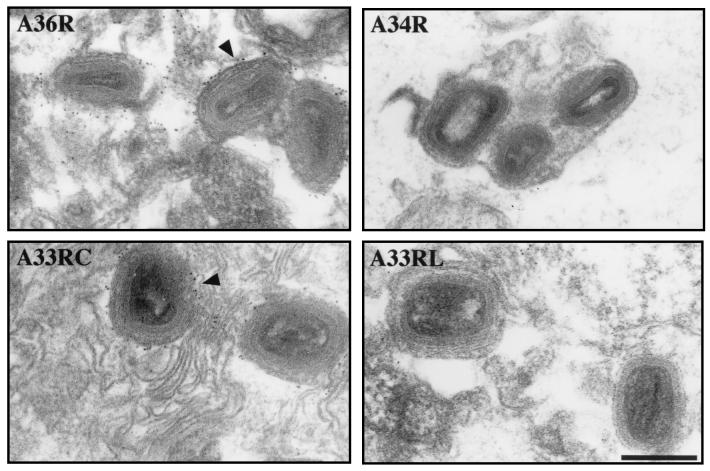
During our characterization of antibodies against A33R, A34R, and A36R, we observed that IEV particles were immunolabeled by antibodies against the cytoplasmic domain of A33R and the predicted luminal domain of A36R when a preembedding technique was employed for immunoelectron microscopy (Fig. 4). In contrast, the antibodies against the luminal domains of A33R and A34R failed to give any labeling, indicating that IEV membranes were intact and that their respective antigens were not accessible (Fig. 4). The simplest explanation for the unexpected labeling by antibodies against the predicted luminal domain of A36R is that the protein is a type Ib (40) and not a type II integral membrane protein as suggested previously (29). This conclusion was confirmed by microinjection of antibodies against A33R, A34R, and A36R into vaccinia virus-infected cells followed by staining for actin and processing for immunofluorescence. Antibodies against the cytoplasmic domain of A33R (Fig. 5) and the predicted luminal domain of A36R (Fig. 5) label IEV particles associated with actin tails, thus revealing that their respective epitopes are exposed to the cytoplasm. In contrast, antibodies against the A33R (data not shown) and A34R (Fig. 5) luminal domains are found diffusely throughout the cytoplasm and show no specific localization, indicating that their epitopes are not accessible.
FIG. 4.
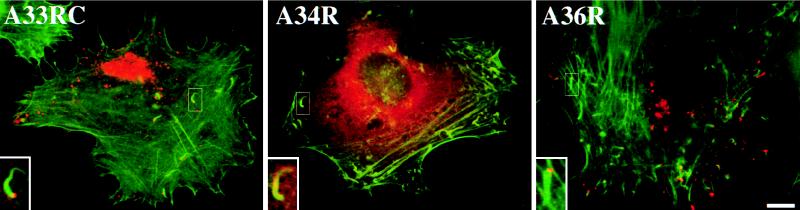
Preembedding labeling of vaccinia virus-infected cells reveals that the predicted luminal domain of A36R is exposed on the surface of IEV particles. Arrowheads indicate IEV particles labeled by antibodies against the predicted luminal domain of A36R (A36R) and the cytoplasmic domain of A33R (A33RC). Antibodies against the luminal domains of A34R (A34R) and A33R (A33RL) gave no labeling. Bar, 200 nm.
FIG. 5.
Microinjection of antibodies into vaccinia virus-infected cells shows domain accessibility of IEV proteins. Cells were injected with antibodies against the cytoplasmic domain of A33R (A33RC), the luminal domain of A34R (A34R), or the predicted luminal domain of A36R (A36R) as indicated in each panel. In the merged images, injected antibodies are visualized in red and the actin cytoskeleton is visualized in green. The inserts clearly demonstrate that antibodies against the cytoplasmic domain of A33R and the predicted luminal domain of A36R label IEV particles associated with actin tails. Antibodies against the luminal domain of A34R or A33R (data not shown) show only a diffuse cytoplasmic staining. Bar, 10 μm.
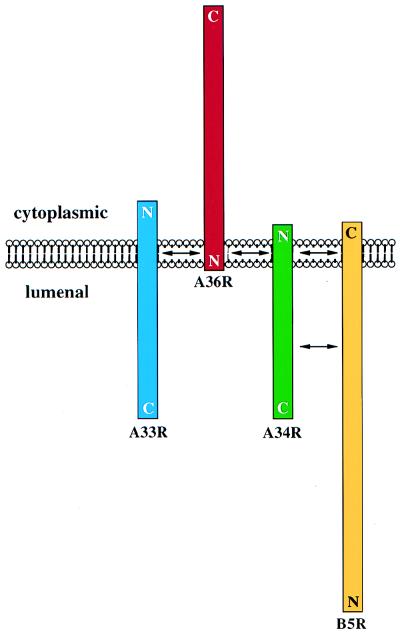
The orientation of A36R in freshly prepared, unfrozen EEV preparations was also examined to see if it was consistent with a type Ib membrane topology. Electron microscopic analysis of immunolabeled preparations of EEV revealed that the luminal domains of A33R and A34R were exposed, while the cytoplasmic domain of A33R and the predicted luminal domain of A36R were not accessible to labeling (Fig. 6). Proteolytic digestion of the same preparations followed by Western blot analysis revealed that the luminal domains of A33R and A34R were degraded, while a substantial proportion of A36R remained intact (Fig. 7). The degradation of a significant proportion of A36R is presumably attributable to the rupture of the outer envelopes of some EEV particles during purification. The rupture of the EEV envelope, which is enhanced by freeze-thawing cycles, may explain why Parkinson and Smith (29) observed sensitivity of A36R to proteases, leading them to conclude that A36R had a type II topology. Collectively, the data presented here establish that A36R is a type Ib integral membrane protein with ∼195 amino acid residues exposed on the cytoplasmic surface of IEV particles (Fig. 8).
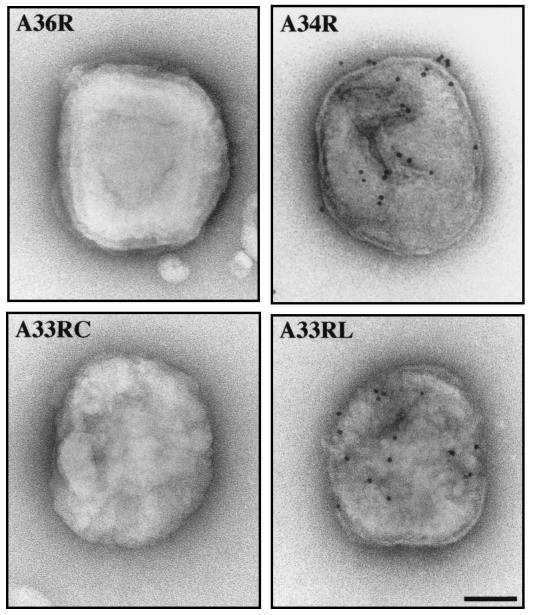
FIG. 6.
Immunolabeling of purified EEV confirms that A36R is a type Ib integral membrane protein. While the luminal domains of A33R (A33RL) and A34R (A34R) are accessible to antibody labeling, the predicted luminal domain of A36R (A36R) and the cytoplasmic domain of A33R (A33RC) are not. Bar, 80 nm.
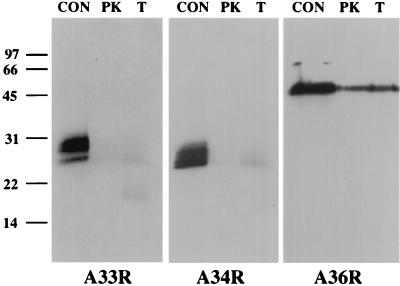
FIG. 7.
Immunoblot analysis of protease-treated EEV. Purified EEV particles were digested with proteinase K (PK) or trypsin (T), and Western blot analysis was performed with the antibody indicated at the bottom of each panel. While A33R and A34R were sensitive to proteolytic digestion, A36R was largely protease resistant. Undigested EEV samples were used as controls (CON). Molecular mass markers are indicated in kilodaltons.
FIG. 8.
Schematic representation of the topology and interactions between A33R, A34R, A36R, and B5R in the outer membrane of IEV particles based on our observations. The cytoplasmic and luminal faces of the outer IEV membrane as well as the positions of the N and C termini of A33R, A34R, A36R, and B5R are indicated. Proteins are shown to scale, and arrows denote possible sites of interaction.
B5R and F13L are not required for actin tail formation.
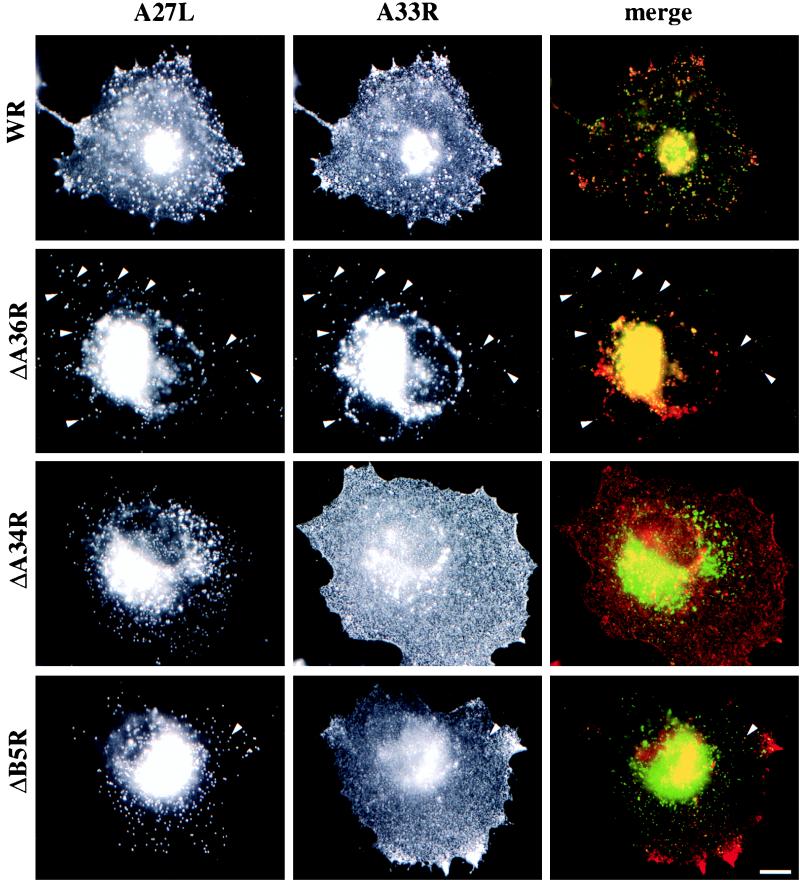
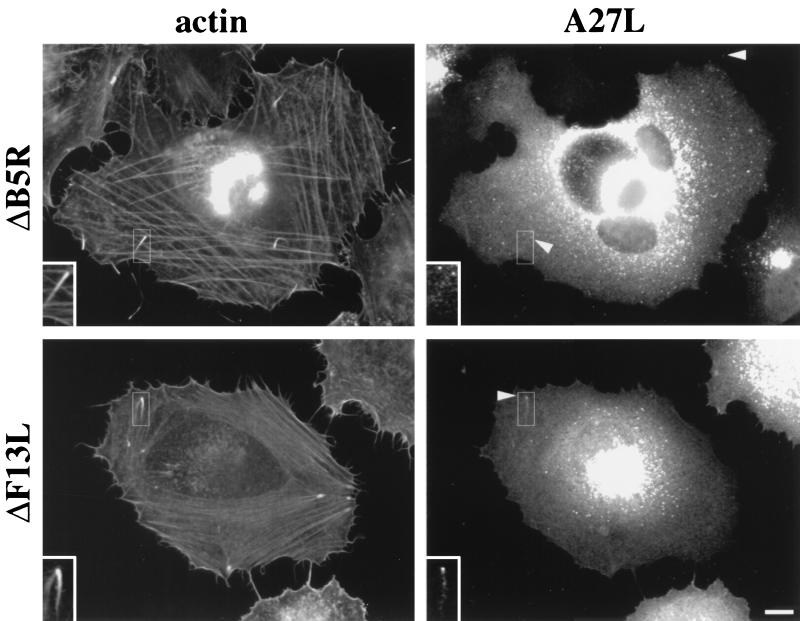
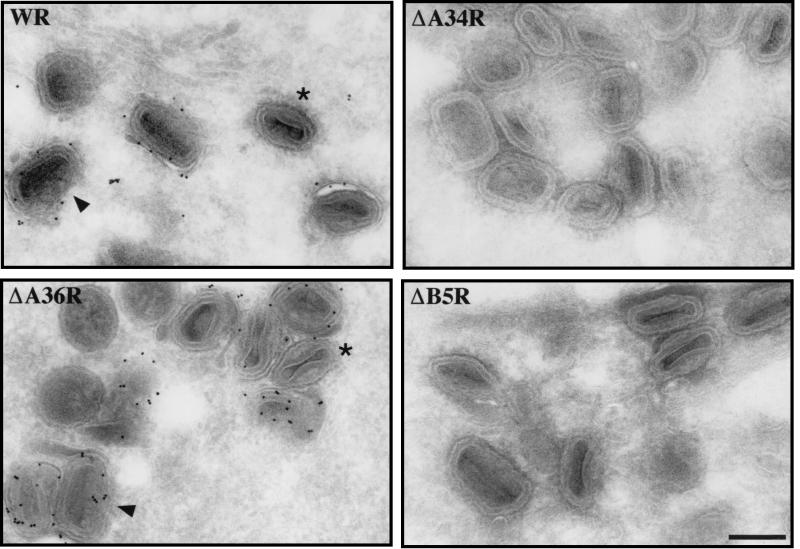
Indirect immunofluorescence revealed that in ΔA36R-infected cells the A33R protein is found associated with IEV particles and in the juxtanuclear region in a fashion identical to that for WR (Fig. 9). However, in the absence of A34R, A33R is present throughout the cell and shows no specific localization (Fig. 9). In contrast, we occasionally observe A33R associated with viral particles, as judged by colocalization with the viral marker A27L (p14), when cells are infected with the virus strain ΔB5R (Fig. 9) or ΔF13L (vRB12) (data not shown). We assume that these double-labeled particles represent IEV. Consistent with this suggestion, we found that the few IEV particles formed by ΔB5R and ΔF13L (vRB12) are able to induce actin tails (Fig. 10). As we have previously observed that the ability to form actin tails is highly cell type dependent (37), we examined a number of different cell types for the ability of ΔB5R to induce actin tails at different infection times. We found that the severe reduction in both IEV assembly and actin tail formation by ΔB5R compared to WR was common to all cell types tested (data not shown). These observations indicate that B5R and F13L are not required for actin tail formation and that their principal role is in IEV assembly, as reported previously (2, 10, 47). Immunofluorescence labeling patterns similar to those with anti-A33R antibody were obtained when cells infected with ΔA34R, ΔA36R, ΔB5R, or ΔF13L were stained with antibodies against A34R, A36R, and B5R where appropriate (data not shown). Immunoelectron microscopy labeling with antibodies against A33R confirmed that IEV assembly by ΔA34R or ΔB5R viruses is an extremely rare event. In the case of ΔA34R, we could find no evidence for A33R labeling of virus particles in over 300 cells that were examined (Fig. 11). In contrast to the case for ΔA34R and ΔB5R, viral particles produced by the ΔA36R deletion strain are readily labeled by antibodies against A33R, indicating that A36R is not required for IEV assembly (Fig. 11).
FIG. 9.
Deletion of A34R or B5R has severe effects on IEV assembly. Cells were infected with the virus strain indicated on the left and labeled with antibodies against the viral marker A27L and the IEV protein A33R as indicated at the top. In all cases, arrowheads indicate IEV particles that label for both A27L and A33R. While comparable numbers of IEV particles are seen in WR- and ΔA36R-infected cells, IEV particle assembly is strongly reduced in ΔA34R- and ΔB5R-infected cells. Bar, 10 μm.
FIG. 10.
B5R and F13L are not required for actin tail formation. Cells were infected with ΔB5R or ΔF13L and labeled with the antibody against A27L as a marker of viral particles and with phalloidin to visualize the actin cytoskeleton as indicated at the top. The inserts show viral particles associated with the tips of actin tails. Bar, 10 μm.
FIG. 11.
Immunogold labeling of cryosections from HeLa cells infected with vaccinia virus strain WR, ΔA36R, ΔA34R, or ΔB5R as indicated in each panel. Sections were labeled with an antibody against A33R as a marker of IEV particles. In WR- and ΔA36R-infected cells, comparable numbers of labeled IEV particles were observed, whereas no viral particles positive for A33R could be found in ΔA34R- and ΔB5R-infected cells. Arrowheads point to IEV particles where surrounding membranes are clearly visible, and examples of unlabeled IMV are indicated by asterisks. Bar, 200 nm.
Interactions between IEV membrane proteins.
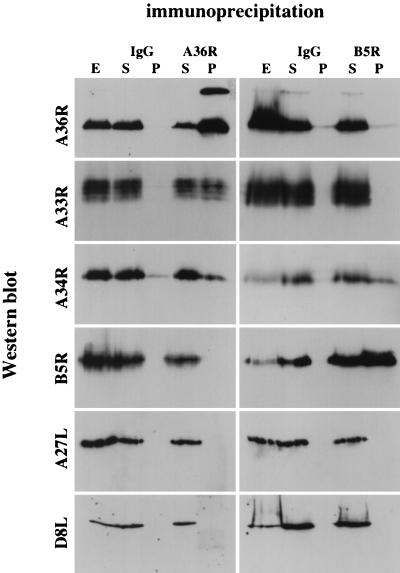
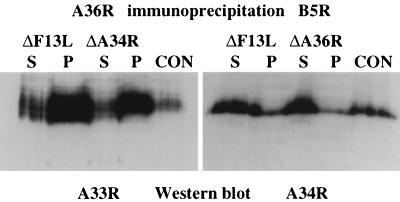
The redistribution of A33R in the absence of A34R and B5R suggested that there might be protein-protein interactions between these IEV proteins which may play an important role in both IEV assembly and actin tail formation. Furthermore, previous studies have shown that B5R and F13L form a complex linked by disulfide bonds (30), while F13L has been reported to interact noncovalently with A56R (28). We therefore examined the possibility of interactions between A33R, A34R, A36R, and B5R by performing coimmunoprecipitation experiments using extracts prepared from cells at both 8 and 24 h postinfection. Unfortunately, the antibodies against A33R and A34R failed to work for immunoprecipitation. In contrast, antibodies against A36R coimmunoprecipitated A33R and A34R but not B5R (Fig. 12). Interestingly, in A36R immunoprecipitations we observed an additional higher-molecular-weight signal that may correspond to an A36R homodimer (Fig. 12). This signal was most prominent in immunoprecipitations but was also observed on Western blots of extracts prepared from infected cells, albeit to a lesser degree (Fig. 1). Additional immunoprecipitation experiments demonstrated that B5R is complexed with A34R but not A33R or A36R (Fig. 12). However, it should be noted that we cannot exclude the possibility of interactions between B5R and A33R or A36R, as the monoclonal antibody against B5R may interfere with weak interactions between these proteins or the experimental conditions may not have been optimal to detect interactions. Alternatively, the epitope for the monoclonal antibody against B5R may not be accessible in B5R-A33R or B5R-A36R complexes if they exist. While the interaction of A34R with A36R or B5R appears to be weaker than the interaction between A33R and A36R, comparison with control immunoglobulin G immunoprecipitations performed in parallel with the same cell extracts demonstrates that the interaction of A34R with A36R or B5R is significant. Nevertheless, it is impossible to tell from these experiments whether this observation reflects a real difference in binding affinities between the proteins. The interaction of A36R with A33R as well as the interaction of B5R with A34R is not dependent on IEV formation, as identical results were obtained with extracts prepared from cells infected with ΔF13L (vRB12) (Fig. 13). We assume that these extracts are essentially free of IEV, as assembly of IEV particles by the ΔF13L deletion strain is a rare event that occurs only late during infection. Immunoprecipitations from extracts of cells infected with the ΔA34R and ΔA36R deletion strains also demonstrated that the interaction of A36R with A33R as well as the interaction of B5R with A34R is independent of A34R and A36R, respectively (Fig. 13).
FIG. 12.
Western blot analysis of immunoprecipitates reveals that IEV proteins interact with each other but not with the IMV-associated proteins A27L and D8L. Immunoprecipitations were performed on cell extracts prepared at 24 h postinfection with either control immunoglobulin G (IgG) or antibodies against A36R and B5R as indicated at the top. S and P, supernatant and pellet from the immunoprecipitation, respectively; E, untreated control extract. Western blots were probed with antibodies against A36R, A33R, A34R, B5R, A27L, and D8L as indicated on the left. Identical results were obtained with cell extracts prepared at 8 h postinfection (data not shown).
FIG. 13.
Western blot analysis of immunoprecipitates with extracts prepared from ΔF13L-, ΔA34R-, or ΔA36R-infected cells. Immunoprecipitations were performed with antibodies against A36R (left panel) or B5R (right panel). S and P, supernatant and pellet, respectively; E, untreated extract prepared from WR-infected cells. Western blots were probed with antibodies against A33R (left panel) or A34R (right panel), showing that the interaction of A36R with A33R as well as the interaction of B5R with A34R is not dependent on IEV formation as they are obtained in extracts from ΔF13L-infected cells. Both interactions are also preserved in the absence of A34R and A36R, respectively, as demonstrated with extracts from ΔA34R- and ΔA36R-infected cells.
DISCUSSION
In the bacterial pathogens Listeria and Shigella, the proteins ActA and IcsA, respectively, have been shown to be both necessary and sufficient for actin tail formation (14). To date, the viral protein(s) responsible for initiating actin tail formation by the IEV form of vaccinia virus is unknown. However, vaccinia virus strains deficient in the IEV-specific proteins A33R, A34R, A36R, B5R, and F13L were reported not to produce actin tails (5, 36, 37, 48, 49). It is unlikely that all of these proteins are directly involved in actin tail formation, especially given that many of them have only small domains exposed on the surface of the IEV. Actin tail formation is likely to be a complex process that depends on many factors, including correct IEV assembly. Distinguishing between the absence of actin tails due to direct effects versus secondary effects, such as IEV assembly, represents the major problem in identifying the viral actin tail nucleator. By examining both IEV assembly and actin tail formation together with the interactions of EEV proteins with each other, we have obtained data that contribute to our understanding of the roles of the A34R, A36R, B5R, and F13L proteins in IEV assembly and actin tail formation (summarized in Table 1).
TABLE 1.
Summary of the roles of known IEV-specific proteins in IEV assembly and actin tail formation
| Protein | Topology | Deletion of corresponding gene allows:
|
|
|---|---|---|---|
| IEV assembly | Actin tail formation | ||
| A56R | Type Ia | Yes | Yes |
| A33R | Type II | Yes, but rare | No |
| A34R | Type II | Yes, but rare | No |
| A36R | Type Ib | Yes | No |
| B5R | Type Ia | Yes, but rare | Yes |
| F13L | Peripheral | Yes, but rare | Yes |
B5R and F13L are not required for actin tail formation.
Previous reports suggested that deletion of all or part of B5R (12, 24, 37) or F13L (5, 37) prevented actin tail formation. Our observations reveal that while deletion of B5R or F13L severely reduces the number of IEV particles, these mutants are capable of forming actin tails (Fig. 10). The fact that these rare actin tails were missed previously for B5R and F13L may be due to short infection times or the cell types used. For instance, BS-C-1 cells assemble fewer actin tails than HeLa cells when infected with an equivalent amount of virus (for example, compare Fig. 1 and 3 in reference 37). Severe changes in the actin cytoskeleton occur during vaccinia virus infection, leading to structures that could easily be misinterpreted as evidence that vaccinia virus-induced actin tails are formed (see Fig. 10D in reference 12 and Fig. 10H in reference 24). Furthermore, endogenous vesicles are capable of inducing actin tail-like structures in the absence of vaccinia virus infection (11). We believe that actin tails induced by vaccinia virus can be identified definitively only when they show virus particles at their tips (Fig. 10). Thus, of the six known IEV proteins, A56R, B5R, and F13L are not essential for actin tail formation (Table 1).
A34R is involved in IEV assembly.
Previous work showed that either repression (8) or deletion (48) of the A34R gene resulted in fewer IEV particles. Data presented here confirmed a very severe reduction in IEV particles, suggesting an important role for A34R in IEV assembly. However, in contrast to the case for ΔB5R and ΔF13L, the few IEV particles that still assemble in the absence of A34R do not form actin tails. Given this phenotype, is A34R involved directly in actin tail formation, or is the lack of actin tails merely a secondary consequence of incorrectly assembled IEV particles? Our coimmunoprecipitation experiments demonstrate that A34R interacts independently with both A36R and B5R. The fact that ΔB5R can assemble actin tails indicates that interactions between A34R and B5R are not essential for actin tail formation. Deletion of either A34R or A36R, however, results in the absence of actin tails, indicating either that both of these proteins are required directly or that interactions between them are important for actin tail formation. Apart from A56R, A36R is the only known IEV protein that can be deleted without a considerable reduction in IEV assembly, and yet the ΔA36R virus does not induce actin tails (29, 37, 49). This suggests immediately that in contrast to A34R, A36R has a primary role in actin tail formation and not in IEV assembly. Taken together, the most straightforward interpretation of the available data is that A34R is important in IEV assembly, organization, and release of EEV particles and their infectivity (4, 8, 25, 37, 48) rather than in actin tail formation.
While actin tail formation may enhance cell-to-cell spread, it is not essential for long-range dissemination, as recombinant vaccinia virus strains that do not produce actin tails are able to release EEV with a three- to fivefold reduction (ΔA36R) (29) or even at greatly enhanced levels (ΔA34R) (25) compared to wild-type virus. Consistent with this observation, we find that actin tails move in a random fashion within the cell and not in a directed manner toward the cell periphery (5). Furthermore, it is clear that other transport mechanisms must exist, as virus particles are able to reach the cell periphery in the absence of actin tails (Fig. 9). In the case of ΔA36R, one can envisage that any IEV that reaches the cell surface would be able to fuse with the plasma membrane in the normal way. In contrast, in the case of ΔA34R, the majority of virus particles that come into contact with the plasma membrane would be IMV, which cannot fuse in the same fashion as IEV. Earlier observations have shown that IMV particles are able to bud through the plasma membrane to release extracellular enveloped virus particles (43). It is possible that EEV particles formed by ΔA34R, in contrast to those formed by WR or ΔA36R, are the consequence of an IMV budding event. Such a direct IMV budding route by ΔA34R might provide a simple explanation for the large numbers of EEV particles with an altered infectivity (25), as EEV formed by IMV budding rather than an IEV fusion event may have a different membrane composition or structural organization. However, this hypothesis does not explain why deletion of B5R, which also severely reduces IEV assembly, results in a 10-fold reduction in EEV formation (10, 47), nor does it explain why short consensus repeat deletion mutants of B5R, which do not form actin tails, produce 50-fold more EEV with normal infectivity (24). While it is evident that actin tail formation is not required for EEV formation or infectivity, a correlation exists between the ability to form actin tails and plaque size, with the exception of the B5R mutant lacking all short consensus repeat domains (12). This observation suggests a role for actin-based motility of vaccinia virus in efficient cell-to-cell spread. It is clear that further analysis of available recombinant virus strains is required to understand the role of IEV assembly and actin tail formation in direct cell-to-cell spread and production and infectivity of EEV and CEV, all of which affect plaque size.
Is A36R the actin tail nucleator of vaccinia virus?
It was difficult to reconcile the possibility that A36R was involved directly in vaccinia virus-induced actin tail formation with its previously reported topology, which predicted a cytoplasmic domain of only two methionine residues (29). In contrast to the earlier report of Parkinson and Smith (29), we have found, by using a number of different approaches, that A36R is a type Ib integral membrane protein with a large, ∼195-amino-acid domain exposed on the surface of IEV (Fig. 8). Type Ib membrane proteins have a single hydrophobic domain near the luminal N terminus that anchors the protein in the membrane, but in contrast to type II membrane proteins, the bulk of the protein, including its C terminus, is exposed to the cytoplasm (40). The previous identification of A36R as a type II membrane protein was based only on its sensitivity to proteolytic digestion in EEV preparations (29). However, it has recently become clear that the outer membrane of EEV is highly susceptible to rupture (44). Indeed, in our experiments we also see that A36R is not fully protected from proteolytic digestion (Fig. 7), consistent with the presence of disrupted EEV particles in our fresh preparations. We believe that the difference between the observations of Parkinson and Smith and our data is probably due to the degree of structural integrity of EEV in the preparation, which is reduced by cycles of freeze-thawing.
Is A36R the viral actin tail nucleator, given that it is the only IEV protein essential for actin tail formation which has a large domain exposed on the surface of IEV particles? The fact that deletion of A36R affects only actin tail formation and not IEV assembly tends to support this hypothesis. In the rare case in which IEV particle assembly occurs in the absence of A34R, we find that A36R is present together with B5R on the particles (data not shown). So why do these IEV particles fail to induce actin tails? The most likely explanation is that in the absence of A34R, which would normally interact with both A36R and B5R, the structural organization of the few IEV particles that still assemble is altered and this alteration prevents actin tail formation. If A36R is the actin tail nucleator of vaccinia virus, then the regions of possible interaction between A34R and A36R required for actin tail formation are restricted to the small cytoplasmic domain of A34R or the transmembrane regions of the two proteins (Fig. 8). The fact that addition of a peptide corresponding to the cytoplasmic domain of A34R did not affect the ability of A36R to coimmunoprecipitate A34R suggests that the interaction between the two proteins occurs in the transmembrane domains. Such interactions are also known to occur for other membrane proteins, such as glycophorin A (23). Interactions in the transmembrane domain of B5R probably also account for the ability of a 42-amino-acid sequence containing the cytoplasmic and transmembrane domains of B5R to target the ectodomain of the human immunodeficiency virus type 1 Env glycoprotein to EEV particles (21).
In conclusion, we have shown that there are multiple interactions between IEV membrane proteins that are clearly important for both IEV assembly and actin tail formation. This intimate relationship makes it very difficult to investigate IEV assembly separately from actin tail formation. However, to understand the mechanism of vaccinia virus-induced actin tail assembly, we require a definitive demonstration of whether A36R is indeed the viral actin tail nucleator, as we suspect.
ACKNOWLEDGMENTS
We thank Gerhardt Hiller (Boehringer, Mannheim, Germany) for the rat monoclonal antibodies 17C4 and 19C2 against B5R, Mariano Esteban (Madrid, Spain) for the mouse monoclonal antibody C3 against A27L, Edward Niles (Buffalo, N.Y.) for the rabbit antiserum against D8L, and Rafael Blasco (Madrid, Spain) for the viral strain vRB12. We are also grateful to Pauli Peräsalmi (EMBL animal house) for injection of animals for antibody production and to Anja Habermann (EMBL Cell Biology Programme) for excellent photographic assistance. We also thank Jacomine Krijnse-Locker (EMBL) and Christopher Sanderson (Oxford) for many helpful suggestions and critical reading of the manuscript.
S.R. and F.F. are recipients of an EMBL predoctoral fellowship.
ADDENDUM IN PROOF
Regarding the role of the B5R transmembrane domain, a recent report by Lorenzo et al. (M. M. Lorenzo, E. Herrera, R. Blasco, and S. N. Isaacs, Virology 252:450–457, 1999) demonstrated that it is required for both targeting of B5R to EEV particles and EEV formation.
REFERENCES
- 1.Blasco R, Cole N B, Moss B. Sequence analysis, expression, and deletion of a vaccinia virus gene encoding a homolog of profilin, a eukaryotic actin-binding protein. J Virol. 1991;65:4598–4608. doi: 10.1128/jvi.65.9.4598-4608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasco R, Moss B. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J Virol. 1991;65:5910–5920. doi: 10.1128/jvi.65.11.5910-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasco R, Moss B. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J Virol. 1992;66:4170–4179. doi: 10.1128/jvi.66.7.4170-4179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasco R, Sisler J R, Moss B. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J Virol. 1993;67:3319–3325. doi: 10.1128/jvi.67.6.3319-3325.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 6.Cudmore S, Reckmann I, Griffiths G, Way M. Vaccinia virus: a model system for actin-membrane interactions. J Cell Sci. 1996;109:1739–1747. doi: 10.1242/jcs.109.7.1739. [DOI] [PubMed] [Google Scholar]
- 7.Cudmore S, Reckmann I, Way M. Viral manipulations of the actin cytoskeleton. Trends Microbiol. 1997;5:142–148. doi: 10.1016/S0966-842X(97)01011-1. [DOI] [PubMed] [Google Scholar]
- 8.Duncan S A, Smith G L. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J Virol. 1992;66:1610–1621. doi: 10.1128/jvi.66.3.1610-1621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelstad M, Howard S T, Smith G L. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology. 1992;188:801–810. doi: 10.1016/0042-6822(92)90535-w. [DOI] [PubMed] [Google Scholar]
- 10.Engelstad M, Smith G L. The vaccinia virus 42kD envelope protein is required for the envelopement and egress of extracellular virus and for virus virulence. Virology. 1993;194:627–637. doi: 10.1006/viro.1993.1302. [DOI] [PubMed] [Google Scholar]
- 11.Frischknecht F, Cudmore S, Moreau V, Reckmann I, Röttger S, Way M. Tyrosine phosphorylation is required for vaccinia but not Listeria or Shigella actin based motility. Curr Biol. 1999;9:89–92. doi: 10.1016/s0960-9822(99)80020-7. [DOI] [PubMed] [Google Scholar]
- 12.Herrera E, del Mar Lorenzo M, Blasco R, Isaacs S N. Functional analysis of vaccinia virus B5R protein: essential role in virus envelopment is independent of a large portion of the extracellular domain. J Virol. 1998;72:294–302. doi: 10.1128/jvi.72.1.294-302.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herzog M, Draeger A, Ehler E, Small J V. Immunofluorescence microscopy of the cytoskeleton: double and triple immunofluorescence. 1st ed. San Diego, Calif: Academic Press; 1994. [Google Scholar]
- 14.Higley S, Way M. Actin and cell pathogenesis. Curr Opin Cell Biol. 1997;9:62–69. doi: 10.1016/s0955-0674(97)80153-6. [DOI] [PubMed] [Google Scholar]
- 15.Hiller G, Jungwirth C, Weber K. Fluorescence microscopical analysis of the life cycle of vaccinia virus in chick embryo fibroblasts. Virus-cytoskeleton interactions. Exp Cell Res. 1981;132:81–87. doi: 10.1016/0014-4827(81)90085-9. [DOI] [PubMed] [Google Scholar]
- 16.Hiller G, Weber K. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J Virol. 1985;55:651–659. doi: 10.1128/jvi.55.3.651-659.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiller G, Weber K. A phosphorylated vaccinia virion polypeptide of molecular weight 11,000 is exposed on the surface of mature particles and interacts with actin-containing cytoskeletal elements. J Virol. 1982;44:647–657. doi: 10.1128/jvi.44.2.647-657.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiller G, Weber K, Schneider L, Parajsz C, Jungwirth C. Interaction of assembled progeny pox viruses with the cellular cytoskeleton. Virology. 1979;98:142–153. doi: 10.1016/0042-6822(79)90533-6. [DOI] [PubMed] [Google Scholar]
- 19.Hirt P, Hiller G, Wittek R. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J Virol. 1986;58:757–764. doi: 10.1128/jvi.58.3.757-764.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isaacs S N, Wolffe E J, Payne L G, Moss B. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J Virol. 1992;66:7217–7224. doi: 10.1128/jvi.66.12.7217-7224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katz E, Wolffe E J, Moss B. The cytoplasmic and transmembrane domains of vaccinia virus B5R protein target a chimeric human immunodeficiency virus type 1 glycoprotein to the outer envelope of nascent vaccinia virions. J Virol. 1997;71:3178–3187. doi: 10.1128/jvi.71.4.3178-3187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krempien U, Schneider L, Hiller G, Weber K, Katz E, Jungwirth C. Conditions for pox virus-specific microvilli formation studied during synchronized virus assembly. Virology. 1981;113:556–564. doi: 10.1016/0042-6822(81)90183-5. [DOI] [PubMed] [Google Scholar]
- 23.MacKenzie K R, Prestegard J H, Engelman D M. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 24.Mathew E, Sanderson C M, Hollinshead M, Smith G L. The extracellular domain of vaccinia virus protein B5R affects plaque phenotype, extracellular enveloped virus release, and intracellular actin tail formation. J Virol. 1998;72:2429–2438. doi: 10.1128/jvi.72.3.2429-2438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McIntosh A A, Smith G L. Vaccinia virus glycoprotein A34R is required for infectivity of extracellular enveloped virus. J Virol. 1996;70:272–281. doi: 10.1128/jvi.70.1.272-281.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan C. Vaccinia virus reexamined: development and release. Virology. 1976;73:43–58. doi: 10.1016/0042-6822(76)90059-3. [DOI] [PubMed] [Google Scholar]
- 27.Niles E G, Seto J. Vaccinia virus gene D8 encodes a virion transmembrane protein. J Virol. 1988;62:3772–3778. doi: 10.1128/jvi.62.10.3772-3778.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oie M, Shida H, Ichihashi Y. The function of the vaccinia hemagglutinin in the proteolytic activation of infectivity. Virology. 1990;176:494–504. doi: 10.1016/0042-6822(90)90019-n. [DOI] [PubMed] [Google Scholar]
- 29.Parkinson J E, Smith G L. Vaccinia virus gene A36R encodes a M(r) 43–50 K protein on the surface of extracellular enveloped virus. Virology. 1994;204:376–390. doi: 10.1006/viro.1994.1542. [DOI] [PubMed] [Google Scholar]
- 30.Payne L G. Characterization of vaccinia virus glycoproteins by monoclonal antibody precipitation. Virology. 1992;187:251–260. doi: 10.1016/0042-6822(92)90313-e. [DOI] [PubMed] [Google Scholar]
- 31.Payne L G. Significance of extracellular enveloped virus in the in vitro and in vivo dissemination of vaccinia. J Gen Virol. 1980;50:89–100. doi: 10.1099/0022-1317-50-1-89. [DOI] [PubMed] [Google Scholar]
- 32.Payne L G, Norrby E. Presence of hemagglutinin in the envelope of extracellular vaccinia virus particles. J Gen Virol. 1976;32:63–72. doi: 10.1099/0022-1317-32-1-63. [DOI] [PubMed] [Google Scholar]
- 33.Reckmann I, Higley S, Way M. The vaccinia virus F17R protein interacts with actin. FEBS Lett. 1997;409:141–146. doi: 10.1016/s0014-5793(97)00450-x. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez J F, Janeczko R, Esteban M. Isolation and characterization of neutralizing monoclonal antibodies to vaccinia virus. J Virol. 1985;56:482–488. doi: 10.1128/jvi.56.2.482-488.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roper R L, Payne L G, Moss B. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J Virol. 1996;70:3753–3762. doi: 10.1128/jvi.70.6.3753-3762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roper R L, Wolffe E J, Weisberg A, Moss B. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J Virol. 1998;72:4192–4204. doi: 10.1128/jvi.72.5.4192-4204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanderson C M, Frischknecht F, Way M, Hollinshead M, Smith G L. Roles of vaccinia virus EEV-specific proteins in intracellular actin tail formation and low pH-induced cell-cell fusion. J Gen Virol. 1998;79:1415–1425. doi: 10.1099/0022-1317-79-6-1415. [DOI] [PubMed] [Google Scholar]
- 38.Schmelz M, Sodeik B, Ericsson M, Wolffe E J, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans-Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shida H. Nucleotide sequence of the vaccinia virus hemagglutinin gene. Virology. 1986;150:451–462. doi: 10.1016/0042-6822(86)90309-0. [DOI] [PubMed] [Google Scholar]
- 40.Singer S J. The structure and insertion of integral proteins in membranes. Annu Rev Cell Biol. 1990;6:247–296. doi: 10.1146/annurev.cb.06.110190.001335. [DOI] [PubMed] [Google Scholar]
- 41.Stokes G V. High-voltage electron microscope study of the release of vaccinia virus from whole cells. J Virol. 1976;18:636–643. doi: 10.1128/jvi.18.2.636-643.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tokuyasu K T. A study of positive staining of ultrathin frozen sections. J Ultrastruct Res. 1978;63:287–307. doi: 10.1016/s0022-5320(78)80053-7. [DOI] [PubMed] [Google Scholar]
- 43.Tsutsui K. Release of vaccinia virus from FL cells infected with IHD-W strain. J Electron Microsc. 1983;32:125–140. [PubMed] [Google Scholar]
- 44.Vanderplasschen A, Smith G L. A novel virus binding assay using confocal microscopy: demonstration that the intracellular and extracellular vaccinia virions bind to different cellular receptors. J Virol. 1997;71:4032–4041. doi: 10.1128/jvi.71.5.4032-4041.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Von Heijne G, Gavel Y. Topogenic signals in integral membrane proteins. Eur J Biochem. 1988;174:671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- 46.Way M, Pope B, Weeds A G. Evidence for functional homology in the F-actin binding domains of gelsolin and alpha-actinin: implications for the requirements of severing and capping. J Cell Biol. 1992;119:835–842. doi: 10.1083/jcb.119.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolffe E J, Isaacs S N, Moss B. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J Virol. 1993;67:4732–4741. doi: 10.1128/jvi.67.8.4732-4741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolffe E J, Katz E, Weisberg A, Moss B. The A34R glycoprotein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J Virol. 1997;71:3904–3915. doi: 10.1128/jvi.71.5.3904-3915.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolffe E J, Weisberg A S, Moss B. Role for the vaccinia virus A36R outer envelope protein in the formation of virus-tipped actin-containing microvilli and cell-to-cell virus spread. Virology. 1998;25:20–26. doi: 10.1006/viro.1998.9103. [DOI] [PubMed] [Google Scholar]