Summary
Mammalian peptide transporters, PepT1 and PepT2, mediate uptake of small peptides and are essential for their absorption. PepT also mediates absorption of many drugs and prodrugs to enhance their bioavailability. PepT has twelve transmembrane (TM) helices that fold into an N-terminal domain (NTD, TM1–6) and a C-terminal domain (CTD, TM7–12), and has a large extracellular domain (ECD) between TM9–10. It is well-recognized that peptide transport requires movements of the NTD and CTD, but the role of the ECD in PepT1 remains unclear. Here we report the structure of horse PepT1 encircled in lipid nanodiscs and captured in the inward-open apo conformation. The structure shows that the ECD bridges the NTD and CTD by interacting with TM1. Deletion of ECD or mutations to the ECD-TM1 interface impairs the transport activity. These results demonstrate an important role of ECD in PepT1 and enhance our understanding of the transport mechanism in PepT1.
eTOC blurb
Shen et al. report the structure of a mammalian PepT1 in an inward-facing conformation. The structure reveals interaction between the extracellular domain (ECD) and transmembrane helix 1 (TM1), suggesting obligatory engagement of the ECD in peptide transport.
Graphical Abstract
Introduction
Mammalian peptide transporters, PepT1 and PepT2, are members of the solute carrier (SLC) transporter family 15 (Daniel and Kottra, 2004). PepT1 is mainly found on intestinal brush border membranes and mediates uptake of small peptides (Matthews, 1983; Qandeel et al., 2009). PepT2 is found in epithelial cells in kidney and retrieves peptides from the glomerulus filtrate (Daniel and Kottra, 2004; Kamal et al., 2009; Shen et al., 1999). Due to their broad substrate spectrum, PepT1 and PepT2 also transport a large variety of small-molecule drugs such as antibiotics, antiviral drugs, and prodrugs with amino acids as part of the scaffold (Zhang et al., 2013). The concentrative uptake of peptides or small-molecule drugs by PepT is achieved by co-transport of protons and is thermodynamically driven by both the pH gradient and the membrane potential (Daniel and Kottra, 2004).
Structures of human PepT1 and PepT2 (Killer et al., 2021), rat PepT2 (Parker et al., 2021), and several bacterial homologs have been reported previously (Boggavarapu et al., 2015; Doki et al., 2013; Guettou et al., 2014; Guettou et al., 2013; Lyons et al., 2014; Martinez Molledo et al., 2018a; Martinez Molledo et al., 2018b; Minhas and Newstead, 2019; Newstead et al., 2011; Parker et al., 2017; Quistgaard et al., 2017; Ural-Blimke et al., 2019). The structures show that PepTs have twelve transmembrane (TM) helices (TM1–12) which fold into two well-defined domains. TM1–6 form the N-terminal domain (NTD), and TM7–12 form the C-terminal domain (CTD). A peptide binding site is located approximately in the middle of the membrane and lined by residues from TM1, 2, 4, and 5 of the NTD and TM7, 8, and 10 of the CTD. PepT structures have been captured in various conformations that can be categorized based on solvent accessibility of the peptide binding site: the outward-open conformation in which the substrate binding site is solvent accessible to the extracellular side; the inward-open conformation in which the substrate binding site is solvent accessible to the intracellular side; and the inward- or outward-occluded conformations when the substrate binding site is not accessible to solvent (Killer et al., 2021; Newstead, 2017; Parker et al., 2021). These conformations likely represent snapshots of the transporter in different stages of substrate transport and led to the recognition that structural changes during substrate translocation mainly come from rigid-body motions of the NTD and CTD. This mechanism is often referred to as the rocker-switch model of alternating access, and is shared by many transporters of the major facilitator superfamily (MFS) fold (Drew et al., 2021).
Mammalian PepT1 and PepT2 have an extracellular domain (ECD) composed of ~200 amino acid residues and located between TM9 and TM10. The molecular weight of ECD is comparable to that of the NTD or CTD. The ECD has a well-defined structure composed mainly of β-strands folded into two immunoglobin-like subdomains (Beale et al., 2015; Killer et al., 2021; Parker et al., 2021). The structure of human PepT2 captured in the inward-occluded conformation (PDB ID: 7PMY) shows that the ECD is in close proximity to the NTD, but no specific interactions were identified (Killer et al., 2021). A previous mutational study showed that peptide transport in PepT2 is not perturbed by deletion or swapping of its ECD with that of PepT1 (Beale et al., 2015), however, the role of ECD in PepT1 has not been resolved, because PepT1, with its ECD deleted or swapped with that of PepT2, does not have sufficient level of expression in the Xenopus oocytes system (Beale et al., 2015).
In this study, we determined the structure of PepT1 from horse (Equus caballus) in the inward-open conformation. The structure shows that its ECD interacts with the NTD, which prompted us to examine the interaction in the function of PepT1. We measured the transport activities of PepT1 in human embryonic kidney (HEK) cells and found that deletion of the ECD or mutations to the interface of ECD and NTD significantly reduce the rate of substrate transport. These results led us to conclude that interactions between ECD and NTD are functionally relevant and may stabilize the inward-open conformation or facilitate transition of PepT1 from the inward-occluded to inward-open conformations.
Results
Proton-coupled peptide transport by horse PepT1
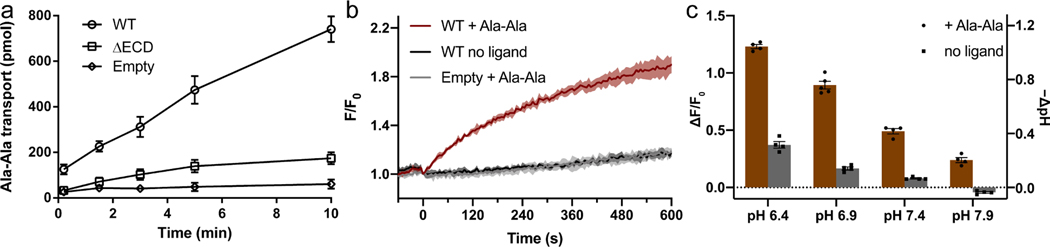
To examine the transport activity of horse PepT1, we expressed it in HEK293 cells and measured the uptake of peptide and proton. As shown in Figure 1a, cells expressing horse PepT1 accumulate a labeled di-peptide (3H-Ala-Ala) while cells transfected with a control vector do not accumulate a significant amount of the peptide. We also measured co-transport of protons with peptide substrates by first loading the cells with a pH-sensitive dye and monitoring intracellular pH changes. Proton uptake occurs in the presence of a di-peptide substrate (Ala-Ala, 1 mM), and lower the external pH, higher the amount of proton uptake (Figure 1b & 1c and Figure S1). As a further test that the observed proton uptake is induced by peptide transport, we measured the pH changes in the presence of two additional di-peptides, Glu-Glu, and Gly-Sar, and found that although all three di-peptides are transported, Ala-Ala induces the largest fluorescence increase (Figure S1d). The expression level of PepT1 in different batches of transfection was estimated by western blot (Method details and Figure S2) and the amount of di-peptide or proton uptake was normalized accordingly. These results indicate that horse PepT1 is a proton coupled peptide co-transporter similar to human PepT1 (Steel et al., 1997).
Figure 1. Functional characterization of horse PepT1 with cell-based assays.
a, Peptide uptake activity of horse PepT1 WT and ΔECD in HEK293 cells. Error bars represent standard error of the mean (SEM) from three repeats. b, pH changes induced by proton-coupled peptide uptake. Ligand Ala-Ala (red) or blank buffer (dark gray) was added to HEK293 cells expressing horse PepT1 WT. Cells transfected with empty vector were added with the same ligand (light gray). The fluorescence changes (F/F0) of a pH-sensitive dye were used to indicate cytosolic pH changes. The time courses are shown as solid line (mean) with shaded region (standard deviation, SD) from four repeats. c, Intracellular pH changes after 10 min of transport in the outside buffer of different pHs. Two-way ANOVA: among different pHs, p < 0.0001; with or without ligand, p < 0.0001; interaction, p < 0.0001. For all bar graphs in this paper, a scatter plot is overlaid on each bar and the height is the mean of at least three repeats with the error bar representing the SEM.
Cryo-EM structure of horse PepT1 in nanodisc
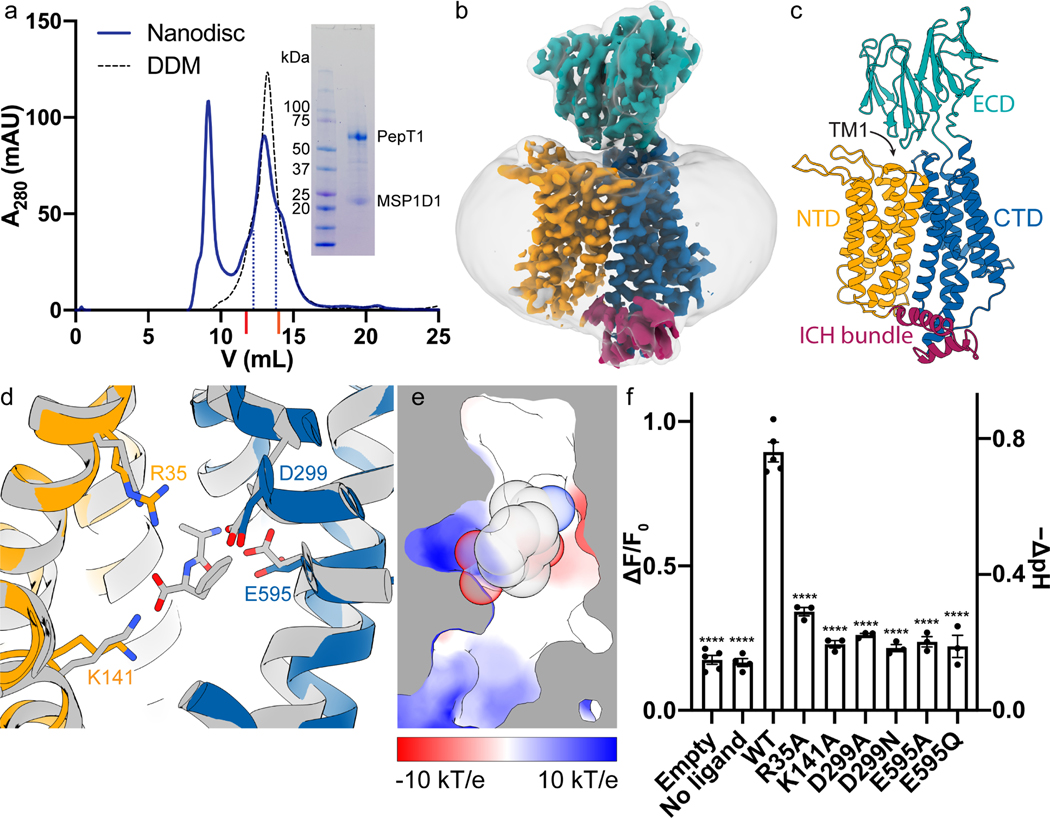
We expressed and purified the horse PepT1 from Sf9 (Spodoptera frugiperda) cells and reconstituted the protein into lipid nanodiscs for structure determination by cryo-electron microscopy (cryo-EM) (Figure 2 and Method details). The elution volume of horse PepT1 from a size-exclusion chromatography column is consistent with it being a monomer (Figure 2a). Human and rat PepTs also exist as a monomer after purification (Killer et al., 2021; Parker et al., 2021). The amino acid sequence of horse PepT1 is 84% identical and 96% similar to that of human PepT1. Although PepT1 has a molecular weight of ~70 kDa, the ECD served as a fiducial marker for particle alignment during 3D reconstruction, and we were able to obtain a density map with an overall resolution of ~3.7 Å (Figure 2b and Figure S3). The map of the TM region is of higher resolution and sufficient quality for de novo model building, while that of the ECD, which is mainly composed of β-strands and not constrained by nanodisc, has a more modest resolution. However, the two subdomains of the ECD and individual β-strands are readily recognizable. The overall structure of horse PepT1 is shown in Figure 2c, and fit of individual TMs and ECD to the density map is shown in Figure S4.
Figure 2. Cryo-EM structure and ligand binding sites of horse PepT1.
a, SEC profile and SDS-PAGE gel image (inset) show the reconstitution of horse PepT1 into MSP1D1 nanodisc (solid blue line). Fractions within the blue dotted lines were collected for Cryo-EM grids. The SEC profile of horse PepT1 in DDM (black dash line) is shown for the comparison of its elution volume to that of a dimeric (~75 kDa per protomer, UniProt ID: Q9FY46) and monomeric transporter (~63 kDa, UniProt ID: A0A1U7U6F1) marked with red and orange ticks, respectively, on X-axis. b, Cryo-EM map of horse PepT1 in nanodisc (contoured at 6σ) colored as described in c. A gaussian smoothened map (translucent gray) is overlaid to display the contour of nanodisc density around the transporter. c, Structure of horse PepT1 in cartoon representation shows the inward-open conformation with NTD in orange, intracellular domain helix (ICH) bundle in red, and CTD in blue. d, Structural alignment of horse PepT1 with human PepT2 bound with an Ala-Phe ligand (PDB ID: 7PMY) colored in gray. Conserved charged residues in the ligand binding pocket are shown as stick. e, Cut-away view of the electrostatic surface in the binding pocket of horse PepT1 show the opposite charges on the NTD (positive) and CTD (negative). The ligand Ala-Phe shown in sphere are superimposed. f, Functional characterization of mutations in the ligand binding pocket. The Dunnett’s test was used as a post hoc test following one-way analysis of variance (ANOVA) with the WT group as control. Statistical significances are indicated: ****, p < 0.0001.
Comparison of horse PepT1 to existing PepT structures
Horse PepT1 is captured in the inward-open conformation, with the extracellular sides of its NTD and CTD making contacts and leaving the substrate binding site solvent accessible from the intracellular side. When the TM domain of horse PepT1 is aligned to that of human PepT2 in the inward-occluded conformation (PDB ID: 7PMY) (Killer et al., 2021), the overall root-mean-square deviation (RMSD) of Cα atoms is 2.5 Å. Larger deviations are seen at the intracellular ends of TM1, TM5 and TM10, which opens the substrate binding site to the intracellular side (Figure S5a) and suggest that flexibility of these helices could fine tune accessibility to the substrate binding site while the rigid-body motions of the NTD and CTD convert the transporter from the outward- to inward-facing state.
The NTD or CTD of horse PepT1 aligns well to the NTD or CTD of human PepT1 or PepT2, or rat PepT2 (Killer et al., 2021; Parker et al., 2021) with RMSD of Cα atoms at 1.0 to 1.4 Å. TM1–2 and TM5 in the NTD (Figure S5b), and the TM7 and TM11 in the CTD (Figure S5c) show larger deviations in the alignments, suggesting that these structural elements are more dynamic. The ECD in horse PepT1 aligns well to that of human PepT1 or PepT2, or rat PepT2 with an RMSD of 1.1 to 2.5 Å, however, the relative position of ECD to CTD varies in these structures as evident when the CTDs are aligned (Figure S5d). While the orientation of ECD of horse PepT1 differs from those of human PepT1 or PepT2 by less than 30° rotation, the difference is more prominent when aligned to that of rat PepT2 (~79° rotation). While the modest difference of ECD to CTD between horse and human PepT1s could be due to differences in amino acid sequence or sample environment (lipid nanodisc vs detergent), the larger difference of ECD orientation in rat PepT2 is likely caused by the presence of a nanobody used to facilitate structure determination (Parker et al., 2021). Regardless, these results suggest that the linkage between ECD and CTD is not rigid and allows flexibility between the two domains.
The substrate binding pocket of horse PepT1 is similar to that of human PepT2 (Killer et al., 2021), and composed of conserved positive residues, Arg35 and Lys141, from the NTD, and negative residues, Asp299 and Glu595, from the CTD (Figure 2d). A peptide substrate would be clamped in a cavity of opposite electric charges with its N-terminus facing negative charged surface in the CTD and C-terminus facing the positively charged surface in the NTD (Figure 2e).
We mutated residues that line the substrate binding site in horse PepT1 and measured their transport activities (Figure 2f and Figure S2). Significant reduction in transport activity was observed for all the mutations, and this result is consistent with similar experiments in previous reports (Terada et al., 2004; Xu et al., 2009). These results highlight the importance of these conserved charged residues in the recognition of the N- and C-terminus of the peptide substrates.
Extracellular gates in PepT1
The inward-open conformation of horse PepT1 shows interactions between the extracellular sides of NTD and CTD, which serve as a gate to limit access of the substrate binding site to the external side. NTD and CTD make contact in three places. Asn51 from TM2 interacts with the Arg304 from TM7 (Figure S6a). His58 from TM2 interacts with Asn630 from TM11, and the latter also interacts with Asp299 in the substrate binding pocket (Figure S6b). Arg186 at the end of TM5 interacts with Gln322 and Asp324 in the loop connecting TM7 and TM8 (Figure S6c). Similar interactions are also observed in the structure of human PepT2 captured in the inward-occluded conformation (Killer et al., 2021). Some of these interactions are known to be important for peptide transport, for example, His58 was reported to mediate proton transport in human PepT1 (Parker et al., 2017).
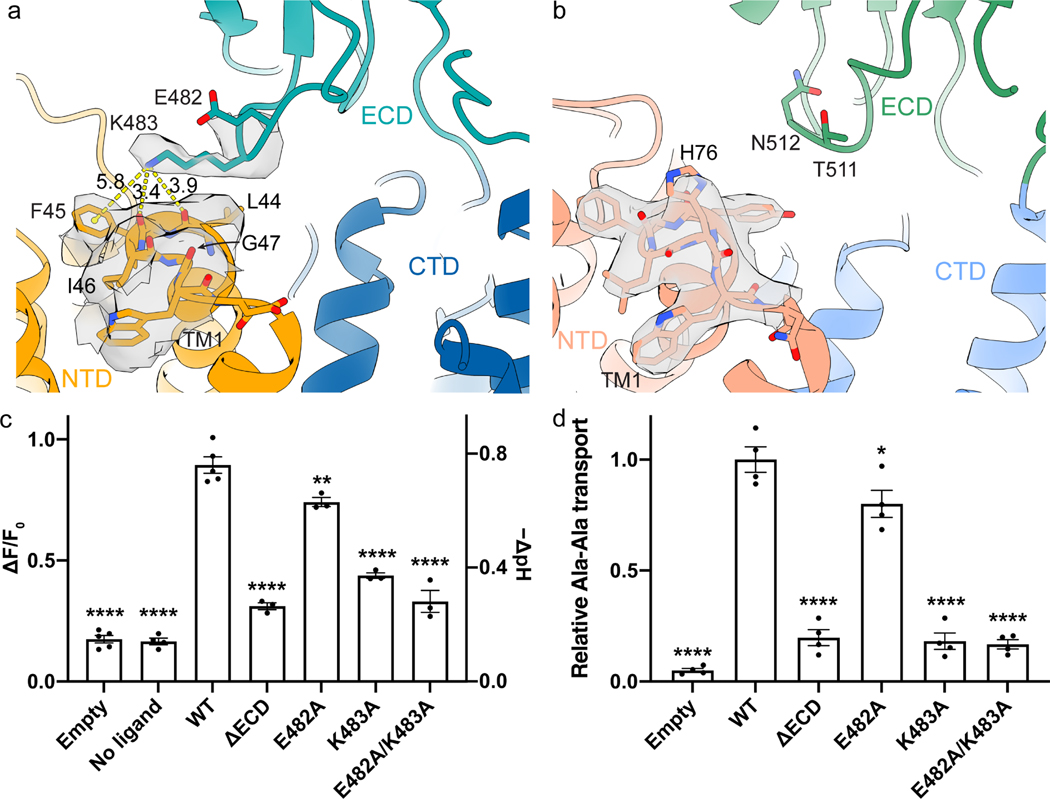
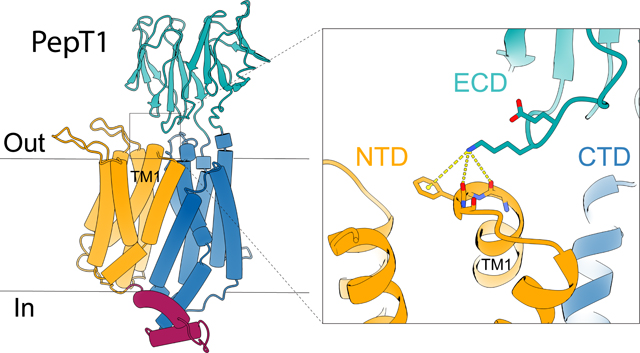
The structure of horse PepT1 reveals interactions between ECD and NTD that were not observed in the structure of human PepT2 in an inward-occluded conformation. Lys483 on the ECD interacts with the backbone carbonyl oxygen atoms of Leu44 and Phe45 at the C-terminal end of TM1 (Figure 3a). The side chain of Lys483 is well-resolved in the density map. The positively charged Lys483 could be further stabilized by a cation-π interaction with Phe45 and by the helical dipole moment of TM1. These residues are not conserved in PepT2. In human PepT2, the equivalent residue to Lys483 in PepT1 is an Asn512 or Thr511 in NTD (Figure S7), while the extracellular end of TM1 is capped by a histidine (His76) on the TM1-TM2 loop (Figure 3b) (Killer et al., 2021).
Figure 3. Interactions between the ECD and NTD in horse PepT1.
a, The structure of horse PepT1 shows interactions between ECD and NTD mediated by K483 and TM1. E482 and K483 from NTD and residues on the extracellular side of TM1 and TM2 are shown as stick with surrounding electron density shown as translucent gray surface at 4σ. Distance measurements are indicated as yellow dash lines with labeled distances in Å. b, The structure of human PepT2 (PDB ID: 7PMY) shows that H76 cap the C-terminus of TM1. The electron density around H76 is shown at 6.5σ. Transport activities of horse PepT1 with mutations that affect the ECD-NTD interactions measured in the cell-based proton transport assay (c) and peptide uptake assay (d). The Dunnett’s test was used as a post hoc test following one-way ANOVA with the WT group as control. Statistical significances are indicated: *, p < 0.05; **, p < 0.01; ****, p < 0.0001.
We then examined functional impact of the interactions between ECD and NTD. We first constructed a mutant horse PepT1 with its ECD deleted (ΔECD). ΔECD can be expressed in HEK cells and mediates proton and peptide transport, however, the rate of transport is significantly lower than these of the wild type (WT) (Figure 1a and Figure 3c–d). We then mutated Lys483 to Ala and found that Lys483Ala also has a significantly reduced transport activity (Figure 3c–d). As a control, we made an alanine mutation to Glu482 which is close to Lys483 but does not interact with TM1. Glu482Ala has a slightly reduced rate of transport compared to that of the WT, but is substantially faster than that of Lys483Ala or the ΔECD. These results are consistent with the hypothesis that interactions between ECD and NTD facilitate peptide transport. Further studies are required to understand how the interactions change the energy landscape of substrate transport in PepT1.
Discussion
The structure of horse PepT1 embedded in lipid nanodiscs is captured in the inward-open apo conformation. The structure reinforces our knowledge of substrate recognition in PepT and enhances our understanding of structural changes involved in substrate translocation. Although lipids are not resolved in the structure, the lipidic environment likely contributes to stabilization of the current conformation to reveal new interactions between ECD and NTD. Mutational studies show that the ECD-NTD interaction has a significant functional impact. Lys483, a key residue on ECD involved in the interaction, is highly conserved in mammalian PepT1 but not in mammalian PepT2 (Figure S7). This may explain why the interaction was not observed in the structure of human PepT2 captured in the inward-occluded state (Killer et al., 2021). The interactions between Lys483 and extracellular end of TM1 are state-dependent as they are not observed in the outward-facing human PepT1 structure (PDB ID: 7PMX). Therefore, these interactions may act as a unique extracellular gate that stabilize the inward-facing conformation of PepT1.
The ECD-NTD interaction in PepT1 likely contribute to different transport kinetics of PepT1 and PepT2. It is well documented that the PepT1 has higher transport capacity, i.e., higher rate of transport, than PepT2 (Ramamoorthy et al., 1995). The stoichiometry of the coupled protons is also different in PepT1 and PepT2 as PepT1 requires less protons to complete a transport cycle (Chen et al., 1999; Steel et al., 1997). Since the substrate binding pocket of PepT1 and PepT2 are highly conserved (Killer et al., 2021), differences in the transport kinetics may arise from the extra ECD-NTD interactions unique to PepT1. Consistent with this notion, removal of ECD in the human PepT2 does not affect substrate transport significantly (Beale et al., 2015), while our studies showed that deletion of ECD in PepT1 reduces the rate of transport. Further analysis will help pinpoint the contribution of ECD to the function of PepT1.
STAR Methods
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ming Zhou (mzhou@bcm.edu).
Materials availability
Protein expression plasmids for horse PepT1 are available upon request.
Data and code availability
The atomic coordinate file of horse PepT1 in nanodisc has been deposited in the PDB (http://www.rcsb.org) under the accession codes 7S8U. The corresponding cryo-electron microscopy map has been deposited in the Electron Microscopy Data Bank (https://www.ebi.ac.uk/pdbe/emdb/) under the accession codes EMD-24922. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Sf9 and HEK293F cells were purchased form Thermo Fisher and were maintained following manufacturer’s instructions as described in Method details.
Methods details
Cloning, expression, and purification of horse PepT1
The PepT1 gene from horse (Equus caballus, UniProt ID: F6SG69) was codon-optimized and cloned into a pFastBac dual vector for production of baculovirus by the Bac-to-Bac method (Invitrogen). Sf9 cells (Invitrogen/Thermo Fisher) at a density of ~3×106 cells/ml were infected with baculovirus and grown at 27 °C for 60–70 hour before harvesting. Cell pellets were frozen in liquid nitrogen and stored in −80 °C fridge before purification.
Cell pellets were thawed and homogenized in lysis buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol) and 2 mM β-mercaptoethanol, and then solubilized with 1% (w/v) Lauryl Maltose Neopentyl Glycol (LMNG, Anatrace) at 4 °C for 2 h. After centrifugation (55,000 g, 45 min, 4 °C), supernatants were applied to pre-equilibrated cobalt-based affinity resins (Talon, Clontech) and allowed to bind at 4 °C for 1 h under gentle rotation. The His6-tag was cleaved by TEV protease at room temperature for 1 hour. The flow-through containing the target protein was concentrated (Amicon 50 kDa cutoff, Millipore) and loaded onto a size-exclusion column (SRT-3C SEC-300, Sepax Technologies, Inc.) equilibrated with the FPLC buffer (20 mM HEPES, pH7.5, 150 mM NaCl) plus 1 mM (w/v) n-dodecyl-β-D-maltoside (DDM, Anatrace).
Nanodisc reconstitution
MSP1D1 was expressed and purified following as described before (Martens et al., 2016). A mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac)-choline (POPC, Avanti Polar Lipids), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac)-ethanolamine (POPE, Avanti Polar Lipids) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac)-glycerol (POPG, Avanti Polar Lipids) at a molar ratio of 3:1:1, were dried and resuspended in the FPLC buffer plus 14 mM DDM (Autzen et al., 2018) by sonication. Horse PepT1, MSP1D1 and the lipid mixture were mixed at a molar ratio of 1:(2.5):(62.5) and incubated on ice for 1 hour (Pan et al., 2020). Detergents were removed by incubation with Biobeads SM2 (Bio-Rad) overnight at 4 °C. The sample was loaded onto a size-exclusion column equilibrated with the FPLC buffer. The purified nanodisc sample was assessed by SDS-PAGE and concentrated for grid preparation.
Cryo-EM sample preparation and data collection
Quantifoil R1.2/1.3 Cu grids were applied with 3.5 μL of purified horse PepT1 in nanodisc at concentration of 5 or 10 mg/mL after glow-discharged with air for 15 seconds. The grids were plunged and frozen into liquid ethane cooled by liquid nitrogen after 6 second blotting at condition of 8 °C, 100% humidity using Vitrobot Mark IV (Thermo Fisher). A total of 2,531 multi-frame stacks were automatically collected with SerialEM (Mastronarde, 2005) on Titan Krios at 300 kV equipped with a K2 Summit direct electron detector (Gatan), a Quantum energy filter (Gatan) and Cs corrector (Thermo Fisher), at a nominal magnification of 105,000× with defocus range set to −2.1 μm to −1.9 μm. Each stack was exposed in super-resolution mode for 5.6 s with an exposure time of 0.175 s per frame, resulting in 32 frames per stack. The total dose was approximately 50 e−/Å2 for each stack.
Cryo-EM data processing
The 2,531 super-resolution movies were motion-corrected in Relion (Kimanius et al., 2016; Scheres, 2012, 2015; Zheng et al., 2017) with 2-fold binning to generate dose-weighted micrographs (pixel size 1.114 Å). The defocus values were estimated with Gctf (Zhang, 2016). Those micrographs were then imported to cryoSPARC (Punjani et al., 2017). The blob picker was performed to 20 randomly selected micrographs and followed by 2D classification to generate templates for auto-picking. 3,437,234 particles were picked by template picker and extracted to perform another round of 2D classification for particle screening. 1,504,753 particles were selected from 2D classification and used by ab initio reconstruction (set to 5 classes). Three 3D references (one good class and two bad classes) generated by ab initio reconstruction were used to perform heterogenous refinement with the original extracted 3,437,234 particles. 833,983 particles in good class were transferred to Relion (Kimanius et al., 2016; Scheres, 2012, 2015) (by csparc2star.py in pyem package (Asarnow et al., 2019)) to perform 3D classification with 2 references (one good and one bad). Relion 3D auto-refine and post-processing then provided a 5 Å map with 438,001 selected particles. Those particles were imported into cryoSPARC again to perform non-uniform (NU) refinement (Punjani et al., 2020). Refined particles were polished in Relion with Bayesian polishing and then were imported into cryoSPARC for two rounds of heterogenous refinement with 2 classes. Finally, 315,767 particles were selected for the final NU-refinement. Resolutions were estimated using the gold-standard Fourier shell correlation (GSFSC) with a 0.143 cut-off (Rosenthal and Henderson, 2003). Local resolution was estimated using cryoSPARC (Punjani et al., 2017).
Model building and refinement
The initial model of horse PepT1 was generated by AlphaFold2 (Jumper et al., 2021). The TMD and ECD were individually docked into the cryo-EM density map in Chimera (Pettersen et al., 2004). The map was sharpened in a convolutional neural network-based algorithm, DeepEMhancer (Sanchez-Garcia et al., 2021), with the highRes model. Processed map has significantly better resolvability of side chains and was used for model refinement. The docked model was manually adjusted in COOT (Emsley et al., 2010), and subjected to real-space refinement with secondary structure and geometry restraints in Phenix (Adams et al., 2010). The EMRinger Score (Barad et al., 2015) was calculated. All structure figures were prepared in ChimeraX (Pettersen et al., 2021).
Expression of horse PepT1 in HEK cells
The cDNA of horse PepT1 was cloned into a pEG BacMam vector with a C-terminal GFP tag. Mutations to horse PepT1 were generated using the QuikChange method (Stratagene) and the entire cDNA was sequenced to verify the mutation. The primers information is provided in Table S2.
The HEK 293S cells in FreeStyle 293 media (Invitrogen/Thermo Fisher) supplemented with 2% fetal bovine serum (FBS; Sigma) were maintained at 37 °C with 8% CO2 in suspension culture at 100 rpm. Cells were plated one day before transfection to reach >90% confluency. Transfection of horse PepT1 plasmid or empty plasmid was performed with 293fectin transfection reagent (Invitrogen/Thermo Fisher) as per manufacturer’s instruction. Transfected cells were incubated at 37 °C with 8% CO2 for 2 days. Cells on the plate were washed in PBS before scraping. Cell membranes were solubilized in the lysis buffer plus 1% LMNG and Protease Inhibitor Cocktail (Roche) for 1 h at 4°C. Insoluble fractions were pelleted by centrifugation and supernatants were run in SDS-PAGE. Bands of target proteins were visualized by western blotting with mouse anti-GFP (Invitrogen/Thermo Fisher) antibodies as primary antibodies and IRDye-800CW anti-mouse IgG (Licor) as secondary antibody. Images were taken on an Odyssey infrared scanner (Licor).
Peptide uptake assay
The HEK 293S cells were plated on 6-well plates and transfected with the pEG BacMam plasmids with horse PepT1 as described above. Transfected cells were harvested after 2 days and transferred to 1.5 mL Eppendorf tubes. Cells were resuspended in transport buffer (20 mM HEPES, pH 6.9, 140 mM NaCl, 2.5 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2) shortly before assays. The ligand Ala-Ala was added to 1 mM with trace amount of 3H-Ala-Ala to start the peptide transport. The transport was stopped by quickly diluting the cells into cold transport buffer and filtering through 0.45 μm nitrocellulose filters (Millipore). Cells were lysed in 0.1 M NaOH and 1% SDS for 10 min. The radioactivity retained on the filters was determined by liquid scintillation counting. Counts per minute were converted to pmol by comparing to a standard curve plotted with known amounts of 3H-Ala-Ala. One-way ANOVA followed by the Dunnett’s post hoc test was performed. All statistical analyses were performed in GraphPad Prism 8.2.1.
Proton transport assay
HEK293S cells from suspension culture were adjusted to a density of 0.75×106 mL−1 with culture media. Transfection mixtures of the pEG BacMam plasmids with horse PepT1 were slowly added to cells under agitation. Transfected cells were aliquoted to poly-lysine-coated 96-well plates and incubated for 2 days. Cells were loaded with pHrodo Red dye (Invitrogen/Thermo Fisher) as per manufacturer’s instruction. Fluorescence was recorded in FlexStation 3 Multi-Mode Microplate Reader (Molecular Devices) at 37 °C. Shortly before the start of recording, cells on assay plates were exchanged to the pre-warmed transport buffer. The transport of peptide was initiated by adding 10 μL of ligands in the transport buffer to 90 μL of buffer in assay plates. Fluorescence readings at equilibrium (550 – 600 s) were averaged to represent intrasellar pH changes. Two-way ANOVA was used to compare the effect of outside pH. Two-tailed Student’s t-test was performed for comparison of mutants to the WT.
Quantification and statistical analysis
The statistics for cryo-EM map and model validation were calculated in cryoSPARC and Phenix and reported in Table S1. Statistical analyses for transport assays are described in Method details or figure legends.
Supplementary Material
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Cell Strains | ||
|
| ||
| FreeStyle™ 293-F Cells | Gibco | Cat#R79007 |
| Sf9 Cells | Gibco | Cat# 11496015 |
|
| ||
| Chemicals, Peptides, and Recombinant Protein | ||
|
| ||
| FreeStyle™ 293 Expression Medium | Gibco | Cat# 12338018 |
| Hyclone SFX-Insect | GE Healthcare | Cat#SH30278.02 |
| Grace’s Insect Medium Supplemented | Gibco | Cat#11605–094 |
| Grace’s Insect Medium Unsupplemented |
Gibco | Cat#11595030 |
| Fetal Bovine Serum | Gibco | Cat#A4736301 |
| n-Dodecyl-β-D-Maltopyranoside | Anatrace | Cat#D310 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac)-choline (POPC) | Avanti Polar Lipids | Cat#850457C |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac)-ethanolamine (POPE) | Avanti Polar Lipids | Cat#850457C |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac)-glycerol (POPG) | Avanti Polar Lipids | Cat#840457C |
| Ala-Ala | Sigma | Cat# A9502 |
| 3H-Ala-Ala | American Radiolabeled Chemicals |
Cat#ART0893 |
|
| ||
| Critical Commercial Assays | ||
|
| ||
| TALON Metal Affinity Resin | TaKaRa | Cat#635504 |
| SRT-10C SEC-300 | Sepax Technologies | Cat#239300–10030 |
| Biobeads SM2 | Bio-Rad | Cat#1528920 |
| KOD Hot Start DNA polymerase | Novagen | Cat#71086–3 |
| pHrodo™ Red AM | Invitrogen | Cat# P35372 |
|
| ||
| Deposited Data | ||
|
| ||
| Cryo-EM map of horse PepT1 | This study | EMD-24922 |
| Coordinates of horse PepT1 | This study | PDB: 7S8U |
| Coordinates of inward-occluded human PepT2 |
Killer et al., 2021 | PDB: 7PMY |
| Coordinates of outward-open human PepT1 |
Killer et al., 2021 | PDB: 7PMX |
| Coordinates of outward-open rat PepT2 | Parker et al., 2021 | PDB: 7NQK |
|
| ||
| Software and Algorithms | ||
|
| ||
| Serial EM | Mastronarde, 2005 | http://bio3d.colorado.edu/SerialEM |
| MotionCorr2 | Zheng et al., 2017 | https://msg.ucsf.edu/em/software/motioncor2.html |
| Gctf | Zhang, 2016 | https://www.mrc-lmb.cam.ac.uk/kzhang/Gctf/ |
| Relion 3.0 | Kimanius et al., 2016 | https://www3.mrclmb.cam.ac.uk/relion |
| CryoSPARC | Punjani et al., 2017 | https://cryosparc.com/ |
| Chimera | Pettersen et al., 2004 | http://www.cgl.ucsf.edu/chimera/ |
| ChimeraX | Pettersen et al., 2021 | https://www.cgl.ucsf.edu/chimerax/ |
| COOT | Emsley et al., 2010 | https://www2.mrclmb.cam.ac.uk/personal/pemsley/coot/ |
| PHENIX | Adams et al., 2010 | http://www.phenix0online.org/ |
| EMringer | Barad et al., 2015 | http://fraserlab.com/2015/02/18/EMringer/ |
| DeepEMhancer | Sanchez-Garcia et al., 2021 | https://github.com/rsanchezgarc/deepEMhancer |
|
| ||
| Other | ||
|
| ||
| Grids: R1.2/1.3 Cu 300 mesh | Quantifoil | Cat#Q325CR1.3 |
Highlights.
Cryo-EM structure of a mammalian PepT1 encircled in nanodisc
PepT1 is captured in an inward-open apo conformation
The extracellular domain (ECD) interacts with the transmembrane helix 1 (TM1)
Mutations to the ECD-TM1 interface reduce PepT1 activity
Acknowledgments
This work was supported by grants from NIH (DK122784, GM145416, HL086392, and GM098878 to M.Z.), and Cancer Prevention and Research Institute of Texas (R1223 to M.Z.). We acknowledge the use of Princeton’s Imaging and Analysis Center, which is partially supported by the Princeton Center for Complex Materials, and the National Science Foundation (NSF)-MRSEC program (DMR-1420541).
Footnotes
Declaration of interests
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asarnow D, Palovcak E, and Cheng Y. (2019). UCSF pyem v0.5. Zenodo. [Google Scholar]
- Autzen HE, Myasnikov AG, Campbell MG, Asarnow D, Julius D, and Cheng Y. (2018). Structure of the human TRPM4 ion channel in a lipid nanodisc. Science (New York, NY) 359, 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barad BA, Echols N, Wang RY-R, Cheng Y, DiMaio F, Adams PD, and Fraser JS (2015). EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nature methods 12, 943–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale JH, Parker JL, Samsudin F, Barrett AL, Senan A, Bird LE, Scott D, Owens RJ, Sansom MSP, Tucker SJ, et al. (2015). Crystal Structures of the Extracellular Domain from PepT1 and PepT2 Provide Novel Insights into Mammalian Peptide Transport. Structure 23, 1889–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggavarapu R, Jeckelmann JM, Harder D, Ucurum Z, and Fotiadis D. (2015). Role of electrostatic interactions for ligand recognition and specificity of peptide transporters. BMC Biol 13, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XZ, Zhu T, Smith DE, and Hediger MA (1999). Stoichiometry and kinetics of the high-affinity H+-coupled peptide transporter PepT2. J Biol Chem 274, 2773–2779. [DOI] [PubMed] [Google Scholar]
- Daniel H, and Kottra G. (2004). The proton oligopeptide cotransporter family SLC15 in physiology and pharmacology. Pflugers Arch 447, 610–618. [DOI] [PubMed] [Google Scholar]
- Doki S, Kato HE, Solcan N, Iwaki M, Koyama M, Hattori M, Iwase N, Tsukazaki T, Sugita Y, Kandori H, et al. (2013). Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT. Proc Natl Acad Sci U S A 110, 11343–11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew D, North RA, Nagarathinam K, and Tanabe M. (2021). Structures and General Transport Mechanisms by the Major Facilitator Superfamily (MFS). Chem Rev 121, 5289–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K. (2010). Features and development of Coot. Acta crystallographica Section D, Biological crystallography 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guettou F, Quistgaard EM, Raba M, Moberg P, Low C, and Nordlund P. (2014). Selectivity mechanism of a bacterial homolog of the human drug-peptide transporters PepT1 and PepT2. Nat Struct Mol Biol 21, 728–731. [DOI] [PubMed] [Google Scholar]
- Guettou F, Quistgaard EM, Tresaugues L, Moberg P, Jegerschold C, Zhu L, Jong AJ, Nordlund P, and Low C. (2013). Structural insights into substrate recognition in proton-dependent oligopeptide transporters. EMBO Rep 14, 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal MA, Jiang H, Hu Y, Keep RF, and Smith DE (2009). Influence of genetic knockout of Pept2 on the in vivo disposition of endogenous and exogenous carnosine in wild-type and Pept2 null mice. Am J Physiol Regul Integr Comp Physiol 296, R986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killer M, Wald J, Pieprzyk J, Marlovits TC, and Low C. (2021). Structural snapshots of human PepT1 and PepT2 reveal mechanistic insights into substrate and drug transport across epithelial membranes. Sci Adv 7, eabk3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimanius D, Forsberg BO, Scheres SH, and Lindahl E. (2016). Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons JA, Parker JL, Solcan N, Brinth A, Li D, Shah ST, Caffrey M, and Newstead S. (2014). Structural basis for polyspecificity in the POT family of proton-coupled oligopeptide transporters. EMBO Rep 15, 886–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens C, Stein RA, Masureel M, Roth A, Mishra S, Dawaliby R, Konijnenberg A, Sobott F, Govaerts C, and McHaourab HS (2016). Lipids modulate the conformational dynamics of a secondary multidrug transporter. Nature structural & molecular biology 23, 744–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Molledo M, Quistgaard EM, Flayhan A, Pieprzyk J, and Low C. (2018a). Multispecific Substrate Recognition in a Proton-Dependent Oligopeptide Transporter. Structure 26, 467–476 e464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Molledo M, Quistgaard EM, and Low C. (2018b). Tripeptide binding in a proton-dependent oligopeptide transporter. FEBS Lett 592, 3239–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Matthews DM (1983). Intestinal absorption of peptides. Biochem Soc Trans 11, 808–810. [DOI] [PubMed] [Google Scholar]
- Minhas GS, and Newstead S. (2019). Structural basis for prodrug recognition by the SLC15 family of proton-coupled peptide transporters. Proc Natl Acad Sci U S A 116, 804–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newstead S. (2017). Recent advances in understanding proton coupled peptide transport via the POT family. Curr Opin Struct Biol 45, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newstead S, Drew D, Cameron AD, Postis VL, Xia X, Fowler PW, Ingram JC, Carpenter EP, Sansom MS, McPherson MJ, et al. (2011). Crystal structure of a prokaryotic homologue of the mammalian oligopeptide-proton symporters, PepT1 and PepT2. EMBO J 30, 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Ren Z, Gao S, Shen J, Wang L, Xu Z, Yu Y, Bachina P, Zhang H, Fan X, et al. (2020). Structural basis of ion transport and inhibition in ferroportin. Nat Commun 11, 5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JL, Deme JC, Wu Z, Kuteyi G, Huo J, Owens RJ, Biggin PC, Lea SM, and Newstead S. (2021). Cryo-EM structure of PepT2 reveals structural basis for proton-coupled peptide and prodrug transport in mammals. Sci Adv 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JL, Li C, Brinth A, Wang Z, Vogeley L, Solcan N, Ledderboge-Vucinic G, Swanson JMJ, Caffrey M, Voth GA, et al. (2017). Proton movement and coupling in the POT family of peptide transporters. Proc Natl Acad Sci U S A 114, 13182–13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Punjani A, Zhang H, and Fleet DJ (2020). Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat Methods 17, 1214–1221. [DOI] [PubMed] [Google Scholar]
- Qandeel HG, Duenes JA, Zheng Y, and Sarr MG (2009). Diurnal expression and function of peptide transporter 1 (PEPT1). J Surg Res 156, 123–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quistgaard EM, Martinez Molledo M, and Low C. (2017). Structure determination of a major facilitator peptide transporter: Inward facing PepTSt from Streptococcus thermophilus crystallized in space group P3121. PLoS One 12, e0173126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy S, Liu W, Ma YY, Yang-Feng TL, Ganapathy V, and Leibach FH (1995). Proton/peptide cotransporter (PEPT 2) from human kidney: functional characterization and chromosomal localization. Biochim Biophys Acta 1240, 1–4. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, and Henderson R. (2003). Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333, 721–745. [DOI] [PubMed] [Google Scholar]
- Sanchez-Garcia R, Gomez-Blanco J, Cuervo A, Carazo JM, Sorzano COS, and Vargas J. (2021). DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun Biol 4, 874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2015). Semi-automated selection of cryo-EM particles in RELION-1.3. J Struct Biol 189, 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Smith DE, Yang T, Huang YG, Schnermann JB, and Brosius FC 3rd (1999). Localization of PEPT1 and PEPT2 proton-coupled oligopeptide transporter mRNA and protein in rat kidney. Am J Physiol 276, F658–665. [DOI] [PubMed] [Google Scholar]
- Steel A, Nussberger S, Romero MF, Boron WF, Boyd CA, and Hediger MA (1997). Stoichiometry and pH dependence of the rabbit proton-dependent oligopeptide transporter PepT1. J Physiol 498 (Pt 3), 563–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada T, Irie M, Okuda M, and Inui K. (2004). Genetic variant Arg57His in human H+/peptide cotransporter 2 causes a complete loss of transport function. Biochem Biophys Res Commun 316, 416–420. [DOI] [PubMed] [Google Scholar]
- Ural-Blimke Y, Flayhan A, Strauss J, Rantos V, Bartels K, Nielsen R, Pardon E, Steyaert J, Kosinski J, Quistgaard EM, et al. (2019). Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1. J Am Chem Soc 141, 2404–2412. [DOI] [PubMed] [Google Scholar]
- Xu L, Haworth IS, Kulkarni AA, Bolger MB, and Davies DL (2009). Mutagenesis and cysteine scanning of transmembrane domain 10 of the human dipeptide transporter. Pharm Res 26, 2358–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. (2016). Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sun J, Sun Y, Wang Y, and He Z. (2013). Prodrug design targeting intestinal PepT1 for improved oral absorption: design and performance. Curr Drug Metab 14, 675–687. [DOI] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinate file of horse PepT1 in nanodisc has been deposited in the PDB (http://www.rcsb.org) under the accession codes 7S8U. The corresponding cryo-electron microscopy map has been deposited in the Electron Microscopy Data Bank (https://www.ebi.ac.uk/pdbe/emdb/) under the accession codes EMD-24922. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.