Abstract
Barley yellow dwarf luteovirus (BYDV) generates three 3′-coterminal subgenomic RNAs (sgRNAs) in infected cells. The promoter of sgRNA1 is a putative hot spot for RNA recombination in luteovirus evolution. The sgRNA1 transcription start site was mapped previously to either nucleotide 2670 or nucleotide 2769 of BYDV genomic RNA (gRNA) in two independent studies. Our data support the former initiation site. The boundaries of the sgRNA1 promoter map between nucleotides 2595 and 2692 on genomic RNA. Computer prediction, phylogenetic comparison, and structural probing revealed two stem-loops (SL1 and SL2) in the sgRNA1 promoter region on the negative strand. Promoter function was analyzed by inoculating protoplasts with a full-length infectious clone of the BYDV genome containing mutations in the sgRNA promoter. Because the promoter is located in an essential coding region of the replicase gene, we duplicated it in a nonessential part of the genome from which a new sgRNA was expressed. Mutational analysis revealed that secondary structure, but not the nucleotide sequence, was important at the base of SL1. Regions with both RNA primary and secondary structural features that contributed to transcription initiation were found at the top of SL1. Primary sequence, but not the secondary structure, was required in SL2, which includes the initiation site. Disruption of base pairing near the sgRNA1 start site increased the level of transcription three- to fourfold. We propose that both primary and secondary structures of the sgRNA1 promoter of BYDV play unique roles in sgRNA1 promoter recognition and transcription initiation.
Many positive-strand RNA viruses express genes via RNA-templated transcription of subgenomic mRNAs. Several mechanisms of subgenomic RNA (sgRNA) synthesis have been proposed for various viruses, including internal initiation of the replicase at subgenomic promoters, premature termination of negative-sense RNA synthesis with subsequent independent replication, 5′ leader-primed synthesis, and RNA recombination (reviewed in references 19 and 27). So far, only internal initiation on the negative-strand template has been demonstrated as a mechanism of sgRNA synthesis in plant RNA viruses (16, 29, 47), although premature termination during negative-strand synthesis has been suggested as an alternative mechanism (27, 43). Despite the lack of direct evidence for internal initiation of transcription in most viruses, we will adhere to convention and refer to cis elements responsible for synthesis of sgRNAs as sgRNA promoters.
Boundaries of sgRNA promoters have been determined for several RNA viruses in vivo (3, 4, 15, 17, 23, 45, 47, 48). Their sizes vary from 24 nucleotides (nt) in Sindbis virus (23) and 27 nt in cucumber necrosis virus (17) to nearly 100 nt in turnip crinkle virus (TCV) (47) and over 100 nt in beet necrotic yellow vein virus (3). With the exception of beet necrotic yellow vein virus (3), the larger portions of subgenomic promoters are located upstream of the transcription initiation site (in the positive sense).
In vitro analysis of the sgRNA4 promoter of brome mosaic virus (BMV) and its comparison with other alpha-like virus subgenomic promoters revealed four major structural elements: a core promoter, an AU tract downstream of the initiation site, an oligo(A) tract, and an enhancer element located upstream of the core (24). However, wild-type levels of RNA4 synthesis in vivo require an additional upstream element that contains repeats of sequences from the core (15). The core promoter of BMV RNA4 has been characterized extensively in vitro, revealing sequence requirements for transcription initiation and suggesting that the primary and not the secondary structure of RNA is critical for specific and accurate initiation, much like in DNA-dependent RNA polymerase promoters (42). However, RNA secondary and tertiary structures play important roles in various processes of the virus life cycle including RNA replication, recombination, translation, and others. These processes involve RNA-protein interactions (1, 6, 10, 35, 38). The combination of primary and secondary RNA structure requirements is common in control of virus replication (21, 36, 38, 40). Therefore, we set out to determine the role of RNA structure in sgRNA promoter function.
The object of this study is the PAV strain of barley yellow dwarf virus (BYDV), which belongs to the genus Luteovirus (formerly called subgroup I) of the family Luteoviridae (formerly the luteovirus group) (9, 26). BYDV has a positive-sense, 5.7-kb genomic RNA (gRNA). In the infected cell, three 3′-coterminal sgRNAs are produced (12, 18). They are not encapsidated. All three sgRNAs play different roles in virus replication. sgRNA1 is the mRNA for the coat protein, a readthrough extension of the coat protein involved in aphid transmission, and a 17-kDa protein required for plant systemic infection (8). sgRNA2 codes for a small 4.3- to 7.0-kDa peptide that is dispensable for virus replication in protoplasts (33). sgRNA2 may also regulate translation from gRNA and sgRNA1. The role of sgRNA3 which lacks open reading frames (ORFs) is unclear (27, 28, 31).
The family Luteoviridae is split into two major genera, Luteovirus and Polerovirus, based on differences in the 5′ halves of their genomes (9, 26). The genes in these regions, including the RNA-dependent RNA polymerase (RdRp), are unrelated between the genera (28). The border between divergent and homologous regions is located between ORF2 and ORF3 (RdRp and coat protein genes) (28). This region also includes the 5′ end of sgRNA1. Based on this observation, we proposed that recombination has occurred during luteovirus evolution by replicase strand switching at the subgenomic promoters (28, 30).
As a first step in testing the recombination model, we have begun mapping the sgRNA promoters of BYDV. In this study, we mapped the primary and secondary structures required for sgRNA1 synthesis in detail. We show that both primary and secondary RNA structure play unique roles in promoter recognition by viral replicase in vivo.
MATERIALS AND METHODS
Plasmids.
pPAV6 is a full-length cDNA clone of BYDV-PAV described in reference 11. pGK-1 was constructed by cloning the AvaI (2456)-SspI (2737) fragment of pPAV6 into pGEM-3Z digested with AvaI and SspI. Mutants pKel-6, pKel-f, and p2670M were constructed by two-step PCR (20). To make pKel-6 and pKel-f, in the first round of PCR we used an upstream mutagenic primer, 5′-GCCCAACTCCAGTC[G/C/A]GT[T/C]AAAGTGACGACTCCACAT-3′ (altered bases in boldface), spanning bases 2655 to 2690 and a downstream primer (5′-CTGAATTCGTTCACCACC-3′) complementary to bases 2867 to 2850. For 2670M, we used an upstream mutagenic primer (5′-CCAGAGTCTGAAGGTGACGACT-3′) complementary to bases 2663 to 2684 and the above downstream primer. The product of the first round was gel purified and used in the second round of PCR as the downstream primer with the upstream primer RB1100 (5′-TGGCTCTTGCACTTGAAC-3′) spanning bases 1927 to 1945. The resulting PCR product was digested with Bst1107 I and Tth111 I and cloned into pPAV6 cut with Bst1107 I and Tth111 I. Mutants with the duplicated sgRNA1 and sgRNA3 promoters were constructed by PCR amplification of the promoter region with the primers listed in Table 1, containing flanking KpnI restriction sites, and by cloning KpnI-digested PCR products into the unique KpnI site of pPAV6 (4154). For the sgRNA1 promoter secondary structure probing, pT7SGP1 was constructed by amplifying a region of pPAV6 between nt 2595 and 2716 with primers 2595 (Table 1) and T7SGP1 (CCGGAATTCTAATACGACTCACTATAGGGATGGAAAGCAGTATTGATT, EcoRI site underlined, T7 promoter italicized), which contained flanking KpnI and EcoRI restriction sites, respectively, and cloning the PCR product into the plasmid pUC118/1180 described in reference 11.
TABLE 1.
Primers used to construct mutants of the duplicated sgRNA promotersa
| Construct | 5′ primer (sequence) | 3′ primer (sequence) |
|---|---|---|
| pPAVSG1A | ||
| p2SL | 2595 (ATAGGTACCGGAGGTGACCCTAAGAT) | SL (ATAGGTACCAGATGTGGAGTCGTCAC) |
| pI | 2611 (ATAGGTACCTACAGCAAATCGTCGAG) | SL |
| pJ | 2595 | 2679 (ATAGGTACCTCACCTTCACACTCTGG) |
| pK | 2611 | 2679 |
| pSL11A | 2595 | SL11A (ATAGGTACCAGATGTGGAGTCGTCACCTTCACACTCTGCTCATGGGCACTTACCGTA) |
| pSL11B | SL11B (ATAGGTACCGCTCATGACCCTAAGATACA) | SL |
| pSL11C | SL11B | SL11A |
| pSL12A | SL12A (ATAGGTACCGGAGTTGACCCTAAGATACAGCAAATCGTCGAGAGGTACTAGCTCGGTCTTACGGTAAGT) | SL |
| pSL12B | SL12B (ATAGGTACCGGAGTTGACCCTAAGATACAGCAAATCGAGCAGAGGTACTACG) | SL |
| pSL12C | SL12C (ATAGGTACCGGAGTTGACCCTAAGATACAGCAAATCGAGCAGAGGTACTAGCTCGGTCTTACGGTAAGT) | SL |
| pSL1D | SL1D (ATAGGTACCGGAGTTG_CGTCGAGAGGTACTACGACG_CAACTCCAGAGTG) | SL |
| pSL1U | SL1U (ATAGGTACCGGAGTTGACCCTAAGATACAGCAAATCGTCGAGAGGTACT_CGACGGTCTTAC) | SL |
| pSL13 | SL13 (ATAGGTACCGGAGTTGACCCTAAGATACAGCAAATCGTCGAGTCCATCTACGACGGTCTTAC) | SL |
| p2670G | 2595 | 2670G (ATAGGTACCAGATGTGGAGTCGTCACCTTCAGACTCTGGAGTT) |
| p2688C | 2595 | 2688C (ATAGGTACCAGATCTGGAGTCGTCACCTTCACACTCTGGAGTT) |
| pComp1 | 2595 | comp1 (ATAGGTACCAGATCTGGAGTCGTCACCTTCAGACTCTGGAGTT) |
| pTrpl1 | 2595 | trpl1 (ATAGGTACCAGATGTGGAGTCGTCACCAGGACACTCTGGAGTT) |
| pTrpl2 | 2595 | trpl2 (ATAGGTACCAGATGTCTTGTCGTCACCTTCACACTCTGGAGTT) |
| pTrplc | 2595 | trplc (ATAGGTACCAGATGTCTTGTCGTCACCAGGACACTCTGGAGTT) |
| pSL21 | 2595 | SL21 (ATAGGTACCAGATGTGGAGAGCAGTGCTTCACACTCTGGAGTT) |
| pSL2SG3 | 2595 | SL2SG3 (ATAGGTACCAGATGTGGAGTCGTCACGTCGTCACTCTGGAGTTGG) |
| pSGP300 | 5150 (ATAGGTACCACATAAATAACCCGCTA) | 5450 (ATAGGTACCGTGGCTCCAAGAGACCC) |
KpnI recognition sequences are underlined, mutant nucleotides are in boldface, and deletions are shown as underlined spaces.
Infection of protoplasts and Northern blot analysis.
Infectious RNA transcripts were obtained by transcription in vitro of plasmids linearized with SmaI, by using T7 RNA polymerase (RiboMax kit; Promega, Madison, Wis.). Oat protoplasts were prepared and inoculated with 10 μg of RNA as described in reference 12. Total RNA was extracted from inoculated protoplasts by using the Qiagen (Los Angeles, Calif.) RNeasy plant RNA isolation kit. RNA (5 to 10 μg) was analyzed by Northern blot hybridization essentially as described in reference 41. A probe complementary to the 3′-terminal 1.5 kb of the PAV genome was used to detect viral gRNA and sgRNA accumulation. This probe was obtained by in vitro transcription of the plasmid pSP10 (12) linearized with HindIII with T7 RNA polymerase. Membranes hybridized with this probe were exposed to PhosphorImager screens for 1 to 2 days. The subgenomic promoter activity was quantified as the ratio of the sgRNA1A signal intensity to that of the gRNA. Because of the significant accumulation of the lower-molecular-mass RNA due to RNA degradation, the value of the sgRNA1A signal was determined by subtracting the background value (region under the sgRNA1A band) from the sgRNA1A signal. All mutants were evaluated in two to five separate experiments.
RNA structure analysis.
pT7SGP1 was linearized with KpnI prior to transcription with T7 RNA. Transcripts were 5′ end labeled with [γ-32P]ATP as described in references 14 and 46. RNA was purified by denaturing 5% polyacrylamide–8 M urea gel electrophoresis. Structural probing with imidazole was performed in 0.04 mM NaCl–1 mM EDTA–10 mM MgCl2 with 0, 0.8, and 1.6 M imidazole for 17 and 22 h at 25°C as described in references 14 and 46. Partial digestion with T1 RNase was done as described in reference 32. Reaction products were separated by using denaturing 6% polyacrylamide–8 M urea gel electrophoresis. The gels were dried and exposed to PhosphorImager screens for 1 to 3 days and visualized with a STORM 840 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). The U2 and T1 RNA sequencing ladders were generated as described in references 14 and 46.
RESULTS
Reexamining the 5′ end of sgRNA1.
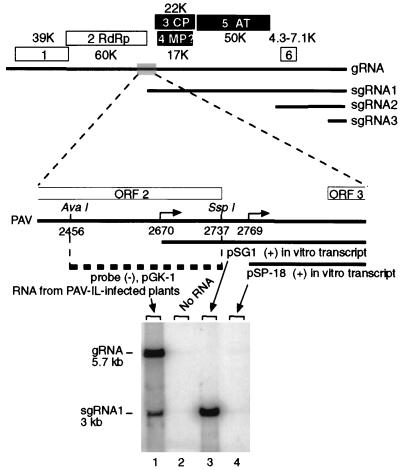
The 5′ end of sgRNA1 of BYDV-PAV was mapped to position 2769 of gRNA by Dinesh-Kumar et al. (12) and to position 2670 by Kelly et al. (18). The 99-nt discrepancy has been attributed to the two different isolates of BYDV-PAV used in these studies: Australia and Illinois isolates were used by Kelly et al. and by Dinesh-Kumar et al., respectively. However, the high homology of the two isolates as well as a conserved hexanucleotide, GUGAAG, present at the 5′ ends of the gRNA and sgRNA1 and sgRNA2, revealed in the study by Kelly et al. (18), prompted us to revisit this issue. We constructed a probe, pGK-1, that is complementary to the region of BYDV gRNA (bases 2456 to 2737) that spans the sgRNA1 start site as mapped by Kelly et al. (2670) but should not detect sgRNA1 as mapped by Dinesh-Kumar et al. (2769 [Fig. 1]). This probe hybridized with sgRNA1 of BYDV-PAV-Illinois and an in vitro transcript that contains the 5′ end at 2670. It did not detect an in vitro transcript with the 5′ end at 2769. Therefore, the 5′ extremity of sgRNA1 of this isolate is in fact well upstream of 2769 and consistent with the 5′ end at 2670 in BYDV-PAV-Australia (Fig. 1). Promoter mapping described herein supports an initiation site at or near 2670.
FIG. 1.
Mapping the 5′ end of sgRNA1 of BYDV. The upper part of the figure shows the genome organization of BYDV. Boxes represent ORFs (1 to 6) with the sizes of protein products indicated in kilodaltons. Thick horizontal lines represent gRNAs and sgRNAs. The lower part of the figure shows the putative sgRNA1 promoter region with the reported initiation sites indicated by right-angled arrows. sgRNA transcripts (which extend to the 3′ end of the genome) that represent sgRNA1s with 5′ ends determined by Kelly et al. (18) (pSG1) and by Dinesh-Kumar et al. (12) (pSP-18) are indicated by thick lines below the map. The probe for the Northern blot hybridization is complementary to the region of gRNA between nt 2456 and 2737 (pGK-1, dashed line). The Northern blot contains total RNA from infected plants (lane 1) or the indicated in vitro transcripts.
Mutations near the sgRNA1 start site affect transcription.
The sgRNA1 initiation site (2670) is located within ORF2, which encodes viral RdRp (Fig. 1), which is required for replication (33). To examine the possibility of using deletion mutagenesis to map the subgenomic promoter, we introduced a stop codon at position 2650 of the infectious clone, pPAV6, which truncated ORF2 by 30 3′-terminal codons. No replication of this mutant transcript in oat protoplasts was detected by Northern blot analysis (data not shown). Thus, deletion mapping of the subgenomic promoter in this region was not possible.
To determine the importance of individual nucleotides around the start site for sgRNA synthesis, we introduced point mutations in this region of pPAV6. Two mutants with five base changes around the start site replicated but synthesized no sgRNA1 (Fig. 2). One had a Val→Asp amino acid change; the other had no amino acid changes (Fig. 2). To test the importance of the G at the initiation site, it alone was mutated to a C in mutant 2670M. This resulted in an unavoidable amino acid substitution (Val→Leu). This mutant did not replicate at all (Fig. 2). This is surprising considering that the more radical change of Val→Asp at this site did not knock out replication. Thus, either the RdRp tolerated Asp but not Leu at amino acid position 825, or the particular base that coincides with the 5′ end of sgRNA1 is essential for gRNA replication. Mutants Kel-6 and Kel-f demonstrated the sensitivity of sgRNA1 transcription to changes in the conserved hexanucleotide GUGAAG and immediately upstream of the initiation site. Mutant 2670M further emphasized the difficulty of the promoter characterization due to potential undesired alterations in RdRp function.
FIG. 2.
Effect of point mutations around the sgRNA1 start site. Northern blot analysis shows gRNA and sgRNA1 accumulation in protoplasts inoculated with mutant full-length genomic transcripts. Total RNA from oat protoplasts (24 hpi) was hybridized with a 32P-labeled transcript complementary to bases 2737 to 2985 of BYDV gRNA. Each mutant was analyzed in duplicate. Altered nucleotides and amino acids are in boldface. Arrows above each sequence show the sgRNA1 transcription start site identified by Kelly et al. (18) (position 2670).
Mapping the boundaries of the sgRNA1 promoter.
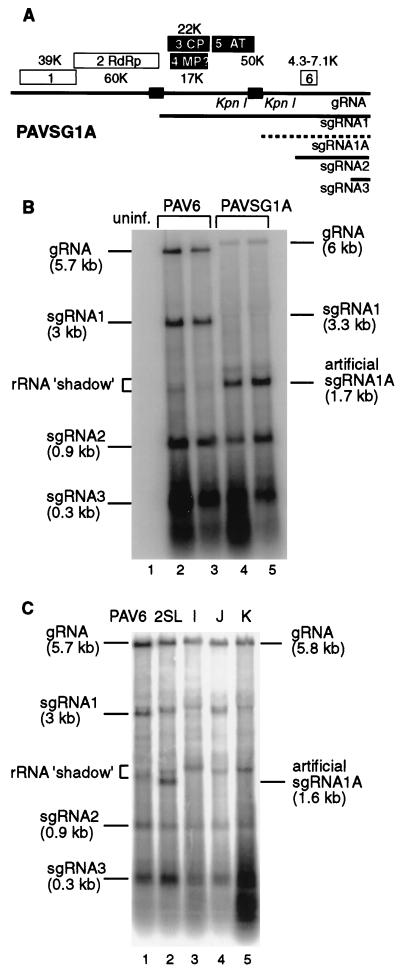
To allow more mutagenesis of the sgRNA1 promoter, we moved a copy of it to a nonessential portion of the genome. This duplication of subgenomic promoters and this synthesis of sgRNAs from ectopic locations have been demonstrated elsewhere for several RNA viruses (3, 4, 15, 22, 45, 47). We duplicated a 314-nt region flanking the putative sgRNA1 initiation site (nt 2503 to 2816) and introduced it into a unique KpnI site in ORF5 (position 4154 [Fig. 3A]). This ORF is not required for virus replication in protoplasts (33). As expected, the promoter duplication resulted in the expression of an additional 1.7-kb sgRNA (sgRNA1A [Fig. 3B]). This caused a dramatic drop in accumulation of sgRNA1 from its natural setting and also reduced gRNA levels. This phenomenon of reduced synthesis of sgRNAs from upstream promoters when additional promoters are inserted downstream has been observed elsewhere with other RNA viruses (15, 47). Reduced gRNA accumulation is likely due to lack of coat protein synthesis (33) caused by reduced levels of its mRNA (sgRNA1). This ectopic expression of sgRNA1 set the stage for the deletion mapping and detailed characterization of the sgRNA1 promoter without interfering with the RdRp coding region.
FIG. 3.
Ectopic expression of the sgRNA1 promoter. (A) Map of the construct PAVSG1A that contains a duplicated subgenomic promoter region (gray box) inserted in the unique KpnI site of PAV6. The KpnI site was duplicated in the cloning process. The dashed line represents the expected artificial sgRNA1A produced from the duplicated promoter. (B) Northern blot shows viral RNAs from protoplasts (24 hpi) inoculated with PAV6 (lanes 2 and 3) and PAVSG1A which has the 314-nt region expected to contain the sgRNA1 promoter duplicated in the KpnI site (sgRNA1A, lanes 4 and 5). uninf., uninoculated protoplasts (lane 1). RNA degradation products formed a band just below the position of the 18S rRNA (rRNA “shadow”) caused by the very abundant rRNA. The probe is transcript from pSP10, complementary to the 3′-terminal 1.5 kb of the viral gRNA. (C) Northern blot analysis of RNAs from protoplasts (24 hpi) infected with full-length transcripts containing the following portions of the sgRNA1 promoter region duplicated in the KpnI site: 2SL, nt 2595 to 2692; I, nt 2611 to 2692; J, nt 2595 to 2679; K, nt 2611 to 2679.
We tested a set of mutants containing various portions of the 314-nt promoter region for sgRNA1A synthesis in oat protoplasts. The smallest construct capable of directing sgRNA1A transcription, 2SL, consisted of the 98-nt RNA sequence from bases 2595 to 2692. Three smaller constructs, I (nt 2611 to 2692), J (nt 2595 to 2679), and K (nt 2611 to 2679), were incapable of producing the artificial sgRNA1A (Fig. 3C). The 2SL construct did not cause such a large reduction in sgRNA1 and gRNA accumulation as did construct PAVSG1A, perhaps because it was not as strong a promoter as the 314-nt insert. Furthermore, the 314-nt duplication in PAVSG1A may have given a gRNA too large to be encapsidated, while the smaller overall size of 2SL gRNA (98-nt duplication) may have permitted encapsidation.
Secondary structure prediction for the sgRNA1 promoter.
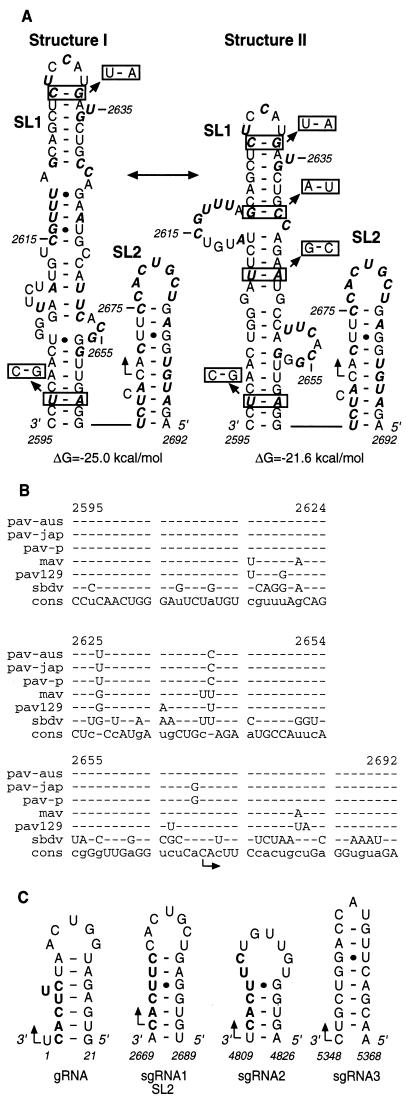
To explore the possible role of RNA secondary structure in sgRNA1 synthesis, we analyzed the 98-nt promoter sequence for potential RNA folding patterns by using the MFOLD program (49). Because sgRNA synthesis has been shown to occur by internal initiation of transcription on the negative-strand template in other viruses, we used the complement of the mapped subgenomic promoter region for the secondary structure predictions. Most of the suboptimal folding patterns contained two stem-loop structures (SL1 and SL2). There were two major variations of the SL1 folding: structure I and structure II (Fig. 4A). To establish which one is more likely to exist, we compared the sgRNA1 promoter regions of other BYDV isolates and the related soybean dwarf virus (SbDV). The BYDV sequences were too highly conserved to shed light on the secondary structure, while SbDV diverged significantly (Fig. 4B). The predicted structure of the SbDV subgenomic promoter region resembled only that of BYDV-PAV structure II. Sequence covariations found in SbDV revealed four sites at which base changes retained base pairing (boxed base pairs [Fig. 4A]) in structure II. Thus, structure II is more likely to exist in BYDV, even though it is calculated to be slightly less stable than structure I (Fig. 4A).
FIG. 4.
RNA sequence and secondary structure analysis of the sgRNA1 promoter of BYDV. (A) Two secondary structures (I and II), with the calculated free energies predicted with MFOLD (49), contain two stem-loops, SL1 and SL2. Bases in boldface italics differ among Luteovirus members (mostly SbDV [B]). Boxed base pairs indicate covariations that preserve the predicted secondary structure. The right-angled arrow indicates the initiation site (nt 2670). (B) Alignment of RNA sequences in the subgenomic promoter regions of five BYDV strains and SbDV. The BYDV strains are PAV-Australia (also known as PAV-Vic) (pav-aus), PAV-Japan (pav-jap), PAV-Purdue (pav-p), PAV-129 (pav129), and MAV (mav). The bottom row shows consensus sequence (cons). Dashes indicate bases that do not differ from consensus. (C) Computer-predicted stem-loop structures in the genomic and subgenomic sgRNA1 promoter regions of BYDV. The sequence is negative sense; numbering is positive sense. The conserved hexanucleotides at the initiation sites are in boldface.
Comparison of the sgRNA1 promoter with those of sgRNA2 and sgRNA3 and the 3′ end of the negative strand of gRNA did not show any significant sequence homology except for the earlier identified hexanucleotide 3′-CACUUC-5′ in sgRNA1 and sgRNA2 and gRNA (5′-GUGAAG-3′ in the positive sense). RNA secondary structure predictions of the negative-strand RNA in those regions revealed a hairpin with the initiation site in its stem similar to SL2 (Fig. 4C). No structure like SL1 was predicted in the other sgRNA promoter regions. This suggested that SL2-like structure may be a common structural element of RdRp recognition regions.
Nuclease probing of the sgRNA1 promoter secondary structure.
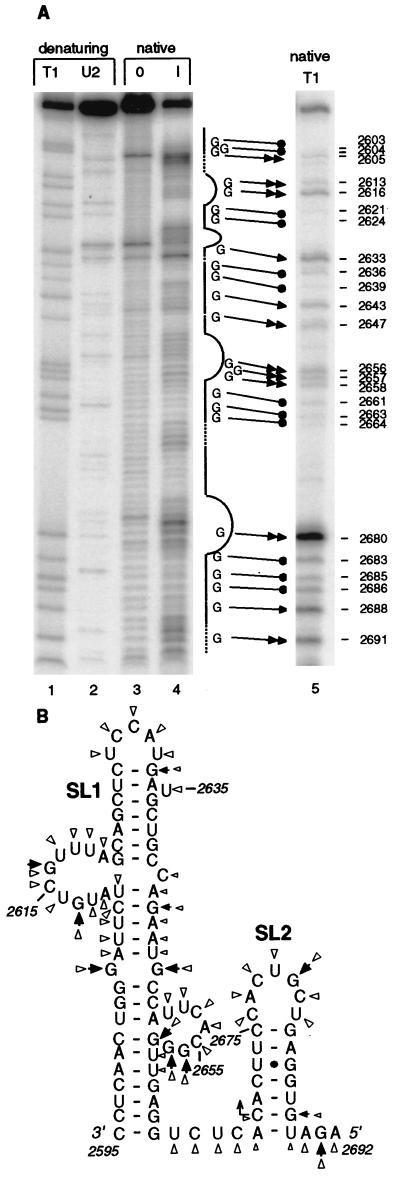
To test the existence of the two computer-predicted stem-loops in the sgRNA1 promoter, we constructed a plasmid, pT7SGP1, which contained a region of the negative-strand gRNA spanning the minimal sgRNA1 promoter (nt 2595 to 2716) cloned behind the bacteriophage T7 promoter. For RNA secondary structure analysis, we used 5′-end-labeled in vitro transcripts of the sgRNA1 promoter obtained from KpnI-linearized pT7SGP1. To detect double- and single-stranded RNA regions within the subgenomic promoter, we performed partial digests of the 5′-end-labeled transcripts with RNase T1 (cuts single-stranded G’s) and imidazole, a chemical RNase that cleaves RNA at all single-stranded bases. Imidazole has been used elsewhere to resolve secondary structure of various RNAs (14, 46).
Both the T1 and the imidazole analyses identified most of the single-stranded RNA regions corresponding to those predicted by computer (Fig. 5A). The lower part of SL2 was sensitive to nuclease, suggesting the existence of a single-stranded junction between the two major stem-loop domains. The upper and the lower regions of SL1 were well protected from both T1 and imidazole digestion, implying stable RNA helices. Highly protected C2648/C2649/A2650 and U2602/G2603/G2604 appeared paired to each other, whereas both G2605 and G2647 were nuclease sensitive and consistent with the bulge in structure II (Fig. 4A and 5A). Therefore, despite the lower predicted stability, structure II appears more likely to form in solution than does structure I.
FIG. 5.
Nuclease probing of the sgRNA1 promoter secondary structure. (A) Imidazole and T1 RNase partial digests of 5′-end-labeled transcript of pT7SGP1 containing the sgRNA1 promoter region (negative sense). Gel-purified, end-labeled RNA was incubated in 0 M (0) and 0.8 M (I) imidazole under nondenaturing (native) conditions for 17 h (lanes 3 and 4). The nondenaturing T1 digest was performed with 0.01 U of the enzyme for 5 min at 37°C (lane 5) (see Materials and Methods). Denaturing digests with the T1 (cuts after G) and U2 (cuts A > U) RNases generated markers in lanes 1 and 2. The products were separated on a 6% polyacrylamide gel containing 8 M urea. The straight line beside lane 4 indicates predicted base-paired regions; dashed lines represent predicted single-stranded junctions and ambiguous regions; curved lines show predicted loops and bulges. Double and single arrows represent G’s that were cleaved strongly and weakly, respectively, by T1 nuclease. Filled circles indicate uncut or very weakly cut G’s. (B) Solution structure of the sgRNA1 promoter. Arrowheads represent the T1 analysis data; triangles represent the imidazole digestion data. Larger and smaller symbols indicate strong and weak cuts, respectively.
The middle portion of SL1 exhibited ambiguous base pairing. Both the base-paired and the single-stranded conformations may coexist in dynamic equilibrium (breathing), reflecting the weak base pairing of the AU-rich AUUCU:AGAAU helix. Based on their nuclease sensitivities, both terminal loops of SL1 and SL2 seemed well defined and consistent with the computer prediction (Fig. 5A). The RNase probing analysis superimposed on the phylogenetically conserved, computer-generated secondary structure allowed us to propose the solution structure of the sgRNA1 promoter (Fig. 5B).
Primary and secondary RNA structures of SL1 are required for transcription of sgRNA1.
To test the involvement of primary and secondary RNA structure elements of SL1 in sgRNA synthesis, we introduced a series of mutations into the duplicated sgRNA1 promoter. Oat protoplasts were inoculated with the mutants, and total RNA was isolated 24 h postinoculation (hpi) and analyzed by Northern blot hybridization. We used the ratio of steady-state levels of sgRNA1A to those of gRNA as a measure of the promoter activity.
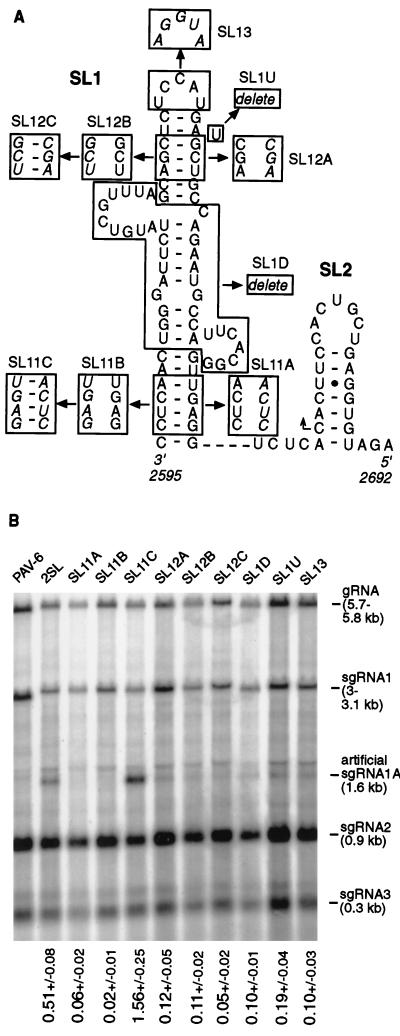
To examine the role of the helix at the base of SL1, we introduced nucleotide alterations (blocks of 4 nt) in both strands of the helix that disrupted and restored the base pairing (mutants SL11A, SL11B, and SL11C [Fig. 6A]). Transcription of sgRNA1A was eliminated when the base pairing was disrupted in mutants SL11A (sgRNA1A/gRNA ratio = 0.06 ± 0.02) and SL11B (0.02 ± 0.01) and was restored to a level higher than that of wild type (construct 2SL, 0.51 ± 0.08 [Fig. 6B]) in the compensatory mutant SL11C (1.56 ± 0.25 [Fig. 6B]). These results indicated that the secondary structure, and not the nucleotide sequence at the bottom of SL1, is required for transcription.
FIG. 6.
Effect of site-specific mutations in the SL1 region on sgRNA1A accumulation. (A) Mutations in the SL1 region of the duplicated sgRNA1 promoter. Altered structures are boxed; mutant sequences are italicized. The names of mutant constructs are above the diagrammed mutations. (B) sgRNA synthesis by the mutants as determined by Northern blot analysis. Total RNA from oat protoplasts (24 hpi) was blotted and probed with labeled T7 transcript from pSP10. The names of the mutants are shown above individual lanes. The promoter activity values calculated as the ratio of sgRNA1A to gRNA level are shown under each lane (± standard deviation). Data represent averages from three separate experiments for each mutant.
To test the role of the upper part of SL1, we introduced similar mutations into the upper helix (SL12A, SL12B, and SL12C [Fig. 6A]). Both mutants SL12A and SL12B, which disrupted the base pairing, exhibited low levels of sgRNA1A accumulation (0.12 ± 0.05 and 0.11 ± 0.02, respectively [Fig. 6B]). The compensatory mutant SL12C failed to restore the promoter activity (0.05 ± 0.02 [Fig. 6B]), indicating that specific nucleotide sequences on both sides of the helix (and possibly RNA secondary structure as well) are important in this region.
To examine the role of the single-stranded and ambiguous regions of SL1 in sgRNA1A synthesis, mutant SL1D was constructed with two sequences tracts deleted: U2602-A2620 and C2641-G2657 (Fig. 6A). Surprisingly, this mutant produced low but significant levels of sgRNA1A (0.10 ± 0.01 [Fig. 6B]), indicating that the middle portion of SL1 could be deleted while SL1 still retained a low level of transcription. Deletion of the bulged U2635 (SL1U [Fig. 6A]) reduced transcription (0.19 ± 0.04 [Fig. 6B]), indicating its importance, but not its absolute requirement, for the promoter activity. Changing the sequence of the terminal loop of SL1 to its complement (Fig. 6A) yielded low levels of sgRNA1A (mutant SL13, 0.10 ± 0.03 [Fig. 6B]). Thus, the specific sequence of the terminal loop is also important for transcription.
Nucleotide sequence but not the secondary structure of SL2 is required for transcription of sgRNA1.
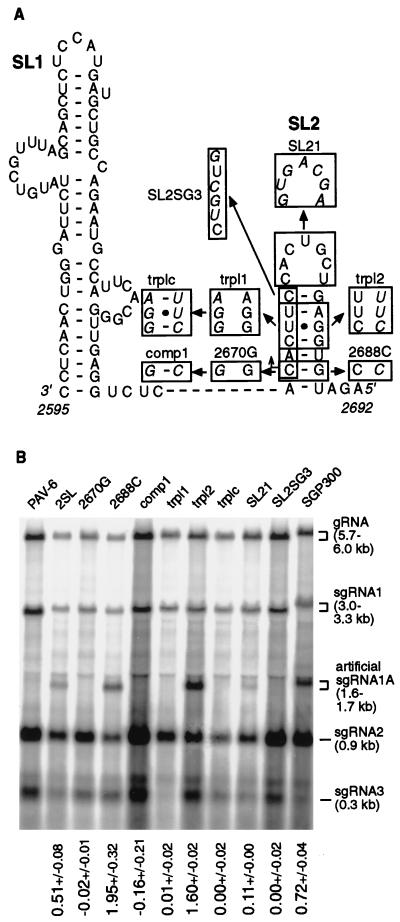
We next characterized the sequence and structural elements of SL2 that are involved in sgRNA synthesis. As reported for initial mutants, sequence alterations within five bases of the transcription start site in its natural location in ORF2 knocked out sgRNA1 synthesis (Fig. 2). Mutations in Kel-6, Kel-f, and 2670M were predicted to disrupt the secondary structure of SL2. To separate the influence of the nucleotide sequence alteration from that of the RNA secondary structure disruption, we designed a series of sgRNA1 promoter mutants that disrupted and restored the secondary structure of SL2 in the duplicated promoter. Three nucleotides near the start site were altered to disrupt base pairing in either strand of SL2 in mutants trpl1 and trpl2 (Fig. 7A). The mutant trplc contained both of the above sets of substitutions to restore the structure of SL2 (Fig. 7A). No sgRNA1A synthesis was detected in trpl1 (0.01 ± 0.02) and trplc (0.0 ± 0.02), both of which are mutated in the conserved hexanucleotide at the start of sgRNA1 (Fig. 7B). However, trpl2 exhibited a level of transcription three times higher than that of wild type (1.60 ± 0.02). Thus, the primary structure adjacent to the start is required for sgRNA synthesis (Fig. 7B), and SL2 secondary structure may actually inhibit transcription.
FIG. 7.
Effect of site-specific mutations in the SL2 region on the subgenomic promoter activity. (A) Mutations in the SL2 region of the duplicated subgenomic promoter. (B) Activity of the subgenomic promoter mutants. All designations and methods are as described for Fig. 6.
In order to examine the role of the initiating C (G2670 in the positive strand), we changed it to a G (mutant 2670G [Fig. 7A]). We also weakened the bottom of SL2 by changing the complementary G to a C (2688C [Fig. 7A]). In the double mutant, comp1, the base pairing was restored while the alteration of the primary structure was maintained (Fig. 7A). The results were similar to those in the previous experiment: no transcription in 2670G (−0.02 ± 0.01) and comp1 (−0.16 ± 0.21), and transcription almost four times higher than the wild-type level of transcription in 2688C (1.95 ± 0.32) (Fig. 7B). Finally, changing the sequence of the SL2 terminal loop to that of its complement in the mutant SL21 (Fig. 7A) reduced the sgRNA1A level to 0.11 ± 0.01 (Fig. 7B), indicating the importance, but not the absolute requirement, of the nucleotide sequence of the SL2 terminal loop. These results stressed the importance of the primary RNA sequence and the negative effect of the secondary structure at the initiation site.
In pursuit of the nucleotides in SL2 that could be altered without affecting transcription, we replaced the 5′-terminal conserved hexanucleotide of sgRNA1 (GUGAAG in the positive strand) with the 5′ terminus of sgRNA3 (GACGAC in the positive strand). Interestingly, this mutant, SL2SG3 (Fig. 7A), did not produce detectable levels of sgRNA1A (0.0 ± 0.02 [Fig. 7B]). We also constructed a mutant, SGP300, with the duplicated putative sgRNA3 promoter region (301 nt, 5150 to 5450) spanning the sgRNA3 start site (nt 5348) inserted in the KpnI site in ORF5 in order to test if this promoter could function outside its wild-type location. The mutant produced sgRNA1A (0.72 ± 0.04 [Fig. 7B]), which indicated that the sgRNA3-specific 5′-terminal hexanucleotide could function in the context of its own promoter. This shows that the replicase recognizes very different promoters.
DISCUSSION
Reevaluation of the 5′ end of sgRNA1.
Here we show that the 5′ end of sgRNA1 is most likely at position 2670 as reported by Kelly et al. (18) and not at our originally reported site of 2769 (12). The error may have occurred due to a variety of technical difficulties, involving mismatches between the probe (from BYDV-PAV Australia) and the viral RNA (BYDV-PAV Illinois), and the unexpectedly far-upstream location of the 5′ end. The difficulty of mapping 5′ ends of Luteoviridae sgRNAs is further indicated by discrepancies reported for potato leafroll virus. Initially, the 5′ end of sgRNA1 of potato leafroll virus was reported to be only 40 nt upstream of the coat protein start codon (44). However, subsequent analysis mapped it to 212 nt upstream at a region which, like BYDV (nt 2670 to 2675), shows homology to the 5′ end of the gRNA (25). Phylogenetic comparisons support this latter start site for other members of the Luteoviridae (28).
Structure and function of sgRNA1.
Mutations in the subgenomic promoter unveiled roles for three types of structures: (i) one in which the secondary and not the primary structure is important (base of SL1), (ii) an ambiguous region where the primary structure and possibly secondary structure both may influence the transcription efficiency (upper stem and loop of SL1), and (iii) a region where the primary and not the secondary structure is required (stem of SL2). The nuclease probing and phylogenetic analysis support the existence of the base of SL1. Stem-loops have been predicted in sgRNA promoters of other RNA viruses (24, 47, 48), with the initiation site usually located within a single-stranded region. To our knowledge, our data represent the first actual demonstration of a requirement for a specific helical domain in a viral sgRNA promoter. The requirement for primary and not secondary structure at the sgRNA1 initiation site result is consistent with other studies showing the role of single-stranded regions for specific protein recognition (2, 35, 36).
The ambiguous results of the structure probing of the distal portion of SL1 lead us to propose that this portion of the subgenomic promoter forms metastable structures. Alternative conformations have been demonstrated for other viral RNAs (32, 39); therefore, a portion of molecules may also fold as predicted in structure I, or the entire AUUCU:AGAAU stem of structure II may be unpaired, giving a very large bulge between stems at the top and the base of SL1. The 36-base deletion mutant (SL1D) lacking this ambiguously paired region still retained 20% of wild-type promoter activity. This indicates that the deleted region probably does not contact the viral replicase directly but may provide favorable spatial localization of the essential elements.
Recognition of the sgRNA1 promoter by replicase.
We propose that SL1 acts as a replicase recognition site, placing the replicase in proximity to the start site at the base complementary to 2670. The double-stranded proximal end (bottom) of SL1 may serve only to provide the structural foundation that ensures that the sequence at the distal end (top) of SL1 (Fig. 6) is presented in the proper orientation for specific binding by the replicase. The ambiguously structured, nonessential, central portion of SL1 may play only an auxiliary role in spacing between the distal region and the initiation site.
This model of a separate RNA binding site and adjacent initiation site resembles those proposed for a recombination site in TCV satellite RNAs, subgenomic promoter recognition by BMV replicase (42), and recognition of bacteriophage Qβ RNA by its replicase (5). The TCV satellite RNA sequence includes a bulged stem-loop as the putative replicase binding site adjacent to the actual site at which RNA synthesis takes place (35). The well-characterized BMV promoter differs by its lack of requirement for the secondary structure in replicase recognition (42). However, this difference may be explained in part by the use of a cell-free transcription system which may have less stringent requirements for cis elements in vitro than those observed in vivo (15). The divergent sequences of the three BYDV promoters may allow differential expression of each. Each sgRNA promoter may have a separate recognition site on the replicase holoenzyme, each of which may be on a separate protein factor as shown for positive- and negative-strand recognition by Qβ replicase (5), or the sequences may have different affinities for the same site on the replicase.
Possible alternative mechanisms of sgRNA1 synthesis.
Our data leave open the possibility that sgRNAs could be synthesized by premature termination during negative-strand synthesis on genomic template followed by independent replication of the sgRNA (27). Evidence suggesting such a mechanism has been provided for coronaviruses (7) and dianthoviruses (43). Upstream of the sgRNA1 5′ end, stem-loops complementary to SL1 and SL2 are predicted to exist in the positive strand by MFOLD (49). The mutations that support a role for the helix at the base of SL1 in the negative strand could equally well support the role of the complementary helix in the positive strand. Such a structure in the positive strand could inhibit replicase migration along the template, favoring termination. The resulting 3′ end of the truncated negative strand would resemble that of full-length negative strand owing to the CACUUC homology, allowing it to be recognized and replicated by the replicase. Thus, that essential sequence would still be serving the promoter function.
The premature termination model has been invoked for red clover necrotic mosaic dianthovirus (RCNMV), owing to a remarkable base pairing between the positive strands of the two gRNAs of the virus that is essential for formation of sgRNA from gRNA1 (43). The polymerase of RCNMV is closely related to that of BYDV (29, 31), and so a similar replication mechanism might apply to BYDV. Because BYDV has only one gRNA, the termination structure would form either as the complement of SL1, as discussed above, or intermolecularly, in which two gRNA molecules dimerize by base pairing at the complementary sequences that would otherwise form the stem-loop. Alternatively, it is possible that other inter- or intramolecular interactions could generate a transcription termination structure.
Some observations argue against a premature termination model. The disruption of the secondary structure at the initiation site (SL2) that increases sgRNA1A synthesis (Fig. 7) should have had the opposite effect if sgRNA1A was synthesized by premature termination during the negative-strand synthesis because stable stems should increase termination (37). Furthermore, the role of SL1 is more than just providing a stem-loop structure, which would (in the positive strand) block replicase migration, because mutations at the distal end of SL1 reduced sgRNA1A accumulation independently of their effect on secondary structure.
Role in recombination.
The presence of an sgRNA promoter at the 3′ end of ORF2 is consistent with our proposed model in which Luteoviridae genomes recombine at a subgenomic promoter in the vicinity of the intergenic region between ORF2 and ORF3 (28). This model requires that sgRNAs are generated by internal initiation of the replicase on the negative strand. A premature termination mechanism, followed by independent replication of the sgRNA, is difficult to reconcile with the sgRNA promoter being a recombination hot spot, if a stem-loop in the positive strand facilitates termination. However, if base pairing between gRNAs occurs as with RCNMV, one could imagine replicase occasionally switching gRNA strands, rather than terminating at this base-paired region. This type of recombination, mediated by base pairing between template strands, has been demonstrated for BMV (34).
Evolution of sgRNA promoters.
The small sizes of viral genomes often require the overlapping of protein coding regions with cis-acting RNA elements. It is an intriguing question, how genetic information coding for a protein and an RNA cis element with which it interacts coevolved on the same region of viral genome. ORF2 is very sensitive to deletions and point mutations (Fig. 2), while the sgRNA promoters tolerate changes and consist of quite diverse sequences. Thus, we propose that sgRNA promoters evolved independently at the appropriate genomic locations while allowing overlapping ORFs to maintain their function. The size of the BYDV sgRNA1 promoter is comparable to that in other RNA viruses, but no apparent sequence homology can be found with subgenomic promoters of members of other virus groups. This diversity among sgRNA promoters of related virus taxa and ability to tolerate movement to different regions of the genome (3, 4, 15, 22, 45, 47) further support the hypothesis of multiple, independent origins of sgRNA promoters. The only conserved secondary structures among the BYDV promoters are the SL2-like hairpins flanking all initiation sites (Fig. 4C), yet the SL2 stem structure inhibits sgRNA1 transcription (Fig. 7). Perhaps the SL2-like stems serve as negative regulatory elements to prevent too much transcription of the sgRNAs at inappropriate stages in RNA replication. Obviously, much additional research is necessary to unveil this complex interplay of replicase-RNA interactions.
ACKNOWLEDGMENTS
We thank Brice Felden for advice on RNA structure probing methods.
We thank the USDA Biotechnology Risk Assessment Research Grants Program (no. 94-39210-0531) and the USDA National Research Initiative (grant no. 98-35303-6447) for funding. This work was part of the Iowa State University Agricultural and Home Economics Experiment Station Project 3545 and supported by Hatch Act and State of Iowa funds.
Footnotes
Paper no. J-18114 of the Iowa State University Agricultural and Home Economics Experiment Station Project 3545.
REFERENCES
- 1.Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 2.Ansel-McKinney P, Gehrke L. RNA determinants of a specific RNA-coat protein peptide interaction in alfalfa mosaic virus: conservation of homologous features in ilarvirus RNAs. J Mol Biol. 1998;278:767–785. doi: 10.1006/jmbi.1998.1656. [DOI] [PubMed] [Google Scholar]
- 3.Balmori E, Gilmer D, Richards K, Guilley H, Jonard G. Mapping the promoter for subgenomic RNA synthesis on beet necrotic yellow vein virus RNA 3. Biochimie. 1993;75:517–521. doi: 10.1016/0300-9084(93)90056-x. [DOI] [PubMed] [Google Scholar]
- 4.Boccard F, Baulcombe D. Mutational analysis of cis-acting sequences and gene function in RNA3 of cucumber mosaic virus. Virology. 1993;193:563–578. doi: 10.1006/viro.1993.1165. [DOI] [PubMed] [Google Scholar]
- 5.Brown D, Gold L. RNA replication by Qβ replicase: a working model. Proc Natl Acad Sci USA. 1996;93:11558–11562. doi: 10.1073/pnas.93.21.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carpenter C D, Oh J-W, Zhang C, Simon A E. Involvement of a stem-loop structure in the location of junction sites in viral RNA recombination. J Mol Biol. 1995;245:608–622. doi: 10.1006/jmbi.1994.0050. [DOI] [PubMed] [Google Scholar]
- 7.Chang R-Y, Krishnan R, Brian D A. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J Virol. 1996;70:2720–2729. doi: 10.1128/jvi.70.5.2720-2729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chay C A, Gunasinge U B, Dinesh-Kumar S P, Miller W A, Gray S M. Aphid transmission and systemic plant infection determinants of barley yellow dwarf luteovirus-PAV are contained in the coat protein readthrough domain and 17-kDa protein, respectively. Virology. 1996;219:57–65. doi: 10.1006/viro.1996.0222. [DOI] [PubMed] [Google Scholar]
- 9.D’Arcy, C., L. Domier, and M. A. Mayo. Luteoviridae. In Seventh report of the International Committee on the Taxonomy of Viruses, in press. Elsevier, New York, N.Y.
- 10.Deiman B A L M, Kortlever R M, Pleij C W A. The role of the pseudoknot at the 3′ end of turnip yellow mosaic virus RNA in minus-strand synthesis by the viral RNA-dependent RNA polymerase. J Virol. 1997;71:5990–5996. doi: 10.1128/jvi.71.8.5990-5996.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di R, Dinesh-Kumar S P, Miller W A. Translational frameshifting by barley yellow dwarf virus RNA (PAV serotype) in Escherichia coli and in eukaryotic cell-free extracts. Mol Plant-Microbe Interact. 1993;6:444–452. doi: 10.1094/mpmi-6-444. [DOI] [PubMed] [Google Scholar]
- 12.Dinesh-Kumar S P, Brault V, Miller W A. Precise mapping and in vitro translation of a trifunctional subgenomic RNA of barley yellow dwarf virus. Virology. 1992;187:711–722. doi: 10.1016/0042-6822(92)90474-4. [DOI] [PubMed] [Google Scholar]
- 13.Dinesh-Kumar S P, Miller W A. Control of start codon choice on a plant viral RNA encoding overlapping genes. Plant Cell. 1993;5:679–692. doi: 10.1105/tpc.5.6.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felden B, Himeno H, Muto A, McCutcheon J, Atkins J, Gesteland R F. Probing the structure of the Escherichia coli 10Sa RNA (tmRNA) RNA. 1997;3:89–103. [PMC free article] [PubMed] [Google Scholar]
- 15.French R, Ahlquist P. Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA. J Virol. 1988;62:2411–2420. doi: 10.1128/jvi.62.7.2411-2420.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gargouri R, Joshi R L, Bol J F, Astier-Manifacier S, Haenni A-L. Mechanism of synthesis of turnip yellow mosaic virus coat protein subgenomic RNA in vivo. Virology. 1989;171:386–393. doi: 10.1016/0042-6822(89)90606-5. [DOI] [PubMed] [Google Scholar]
- 17.Johnston J C, Rochon D M. Deletion analysis of the promoter for the cucumber necrosis virus 0.9-kb subgenomic RNA. Virology. 1995;214:100–109. doi: 10.1006/viro.1995.9950. [DOI] [PubMed] [Google Scholar]
- 18.Kelly L, Gerlach W L, Waterhouse P M. Characterisation of the subgenomic RNAs of an Australian isolate of barley yellow dwarf luteovirus. Virology. 1994;202:565–573. doi: 10.1006/viro.1994.1378. [DOI] [PubMed] [Google Scholar]
- 19.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landt O, Grunert H-P, Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990;96:125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- 21.Lauber E, Guilley H, Richards K, Jonard G, Gilmer D. Conformation of the 3′-end of beet necrotic yellow vein benevirus RNA 3 analysed by chemical and enzymatic probing and mutagenesis. Nucleic Acids Res. 1997;25:4723–4729. doi: 10.1093/nar/25.23.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehto K, Grantham G L, Dawson W O. Insertion of sequences containing the coat protein subgenomic RNA promoter and leader in front of the tobacco mosaic virus 30K ORF delays its expression and causes defective cell-to-cell movement. Virology. 1990;174:145–157. doi: 10.1016/0042-6822(90)90063-w. [DOI] [PubMed] [Google Scholar]
- 23.Levis R, Schlesinger S, Huang H V. Promoter for Sindbis virus RNA-dependent subgenomic RNA transcription. J Virol. 1990;64:1726–1733. doi: 10.1128/jvi.64.4.1726-1733.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsh L E, Dreher T W, Hall T C. Mutational analysis of the core and modulator sequences of the BMV RNA3 subgenomic promoter. Nucleic Acids Res. 1988;16:981–995. doi: 10.1093/nar/16.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J S, Mayo M A. The location of the 5′ end of the potato leafroll luteovirus subgenomic coat protein mRNA. J Gen Virol. 1991;72:2633–2638. doi: 10.1099/0022-1317-72-11-2633. [DOI] [PubMed] [Google Scholar]
- 26.Miller, W. A. Luteoviridae. In R. G. Webster and A. Granoff (ed.), Encyclopedia of virology, 2nd ed., in press. Academic Press, London, United Kingdom.
- 27.Miller W A, Brown C M, Wang S. New punctuation for the genetic code: luteovirus gene expression. Semin Virol. 1997;8:3–13. [Google Scholar]
- 28.Miller W A, Dinesh-Kumar S P, Paul C P. Luteovirus gene expression. Crit Rev Plant Sci. 1995;14:179–211. [Google Scholar]
- 29.Miller W A, Dreher T W, Hall T C. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−) sense genomic RNA. Nature. 1985;313:68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- 30.Miller W A, Koev G, Mohan B R. Are there risks associated with transgenic resistance to luteoviruses? Plant Dis. 1997;81:700–710. doi: 10.1094/PDIS.1997.81.7.700. [DOI] [PubMed] [Google Scholar]
- 31.Miller W A, Rasochova L. Barley yellow dwarf viruses. Annu Rev Phytopathol. 1997;35:167–190. doi: 10.1146/annurev.phyto.35.1.167. [DOI] [PubMed] [Google Scholar]
- 32.Miller W A, Silver S L. Alternative tertiary structure attenuates self-cleavage of the ribozyme in the satellite RNA of barley yellow dwarf virus. Nucleic Acids Res. 1991;19:5313–5320. doi: 10.1093/nar/19.19.5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohan B R, Dinesh-Kumar S P, Miller W A. Genes and cis-acting sequences involved in replication of barley yellow dwarf virus-PAV RNA. Virology. 1995;212:186–195. doi: 10.1006/viro.1995.1467. [DOI] [PubMed] [Google Scholar]
- 34.Nagy P D, Bujarski J J. Targeting the site of RNA-RNA recombination in brome mosaic virus with antisense sequences. Proc Natl Acad Sci USA. 1993;90:6390–6394. doi: 10.1073/pnas.90.14.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagy P D, Zhang C, Simon A E. Dissecting RNA recombination in vitro: role of RNA sequences and the viral replicase. EMBO J. 1998;17:2392–2403. doi: 10.1093/emboj/17.8.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parsley T B, Towner J S, Blyn L B, Ehrenfeld E, Semler B L. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 37.Platt T. Termination of transcription and its regulation in the tryptophan operon of E. coli. Cell. 1981;24:10–23. doi: 10.1016/0092-8674(81)90496-7. [DOI] [PubMed] [Google Scholar]
- 38.Pogue G P, Hall T C. The requirement for a 5′ stem-loop structure in brome mosaic virus replication supports a new model for viral positive-strand RNA initiation. J Virol. 1992;66:674–684. doi: 10.1128/jvi.66.2.674-684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Proutski V, Gould E A, Holmes E C. Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 1997;25:1194–1202. doi: 10.1093/nar/25.6.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohll J B, Moon D H, Evans D J, Almond J W. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J Virol. 1995;69:7835–7844. doi: 10.1128/jvi.69.12.7835-7844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seeley K A, Byrne D H, Colbert J T. Red light-independent instability of oat phytochrome mRNA in vivo. Plant Cell. 1992;4:29–38. doi: 10.1105/tpc.4.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siegel R W, Adkins S, Kao C C. Sequence-specific recognition of a subgenomic RNA promoter by a viral RNA polymerase. Proc Natl Acad Sci USA. 1997;94:11238–11243. doi: 10.1073/pnas.94.21.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sit T L, Vaewhongs A A, Lommel S A. RNA-mediated transactivation of transcription from a viral RNA. Science. 1998;281:829–832. doi: 10.1126/science.281.5378.829. [DOI] [PubMed] [Google Scholar]
- 44.Tacke E, Prufer D, Salamini F, Rohde W. Characterization of a potato leafroll luteovirus subgenomic RNA: differential expression by internal translation initiation and UAG suppression. J Gen Virol. 1990;71:2265–2272. doi: 10.1099/0022-1317-71-10-2265. [DOI] [PubMed] [Google Scholar]
- 45.van der Vossen E A, Notenboom T, Bol J F. Characterization of sequences controlling the synthesis of alfalfa mosaic virus subgenomic RNA in vivo. Virology. 1995;212:663–672. doi: 10.1006/viro.1995.1524. [DOI] [PubMed] [Google Scholar]
- 46.Vlassov V V, Zuber G, Felden B, Behr J P, Giege R. Cleavage of tRNA with imidazole and spermine imidazole constructs: a new approach for probing RNA structure. Nucleic Acids Res. 1995;23:3161–3167. doi: 10.1093/nar/23.16.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Simon A E. Analysis of the two subgenomic RNA promoters for turnip crinkle virus in vivo and in vitro. Virology. 1997;232:174–186. doi: 10.1006/viro.1997.8550. [DOI] [PubMed] [Google Scholar]
- 48.Zavriev S K, Hickey C M, Lommel S A. Mapping of the red clover necrotic mosaic virus subgenomic RNA. Virology. 1996;216:407–410. doi: 10.1006/viro.1996.0076. [DOI] [PubMed] [Google Scholar]
- 49.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]