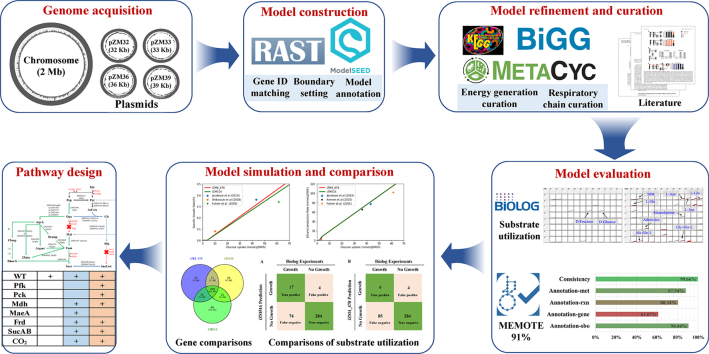
Abstract
High-quality genome-scale metabolic models (GEMs) could play critical roles on rational design of microbial cell factories in the classical Design-Build-Test-Learn cycle of synthetic biology studies. Despite of the constant establishment and update of GEMs for model microorganisms such as Escherichia coli and Saccharomyces cerevisiae, high-quality GEMs for non-model industrial microorganisms are still scarce. Zymomonas mobilis subsp. mobilis ZM4 is a non-model ethanologenic microorganism with many excellent industrial characteristics that has been developing as microbial cell factories for biochemical production. Although five GEMs of Z. mobilis have been constructed, these models are either generating ATP incorrectly, or lacking information of plasmid genes, or not providing standard format file. In this study, a high-quality GEM iZM516 of Z. mobilis ZM4 was constructed. The information from the improved genome annotation, literature, datasets of Biolog Phenotype Microarray studies, and recently updated Gene-Protein-Reaction information was combined for the curation of iZM516. Finally, 516 genes, 1389 reactions, 1437 metabolites, and 3 cell compartments are included in iZM516, which also had the highest MEMOTE score of 91% among all published GEMs of Z. mobilis. Cell growth was then predicted by iZM516, which had 79.4% agreement with the experimental results of the substrate utilization. In addition, the potential endogenous succinate synthesis pathway of Z. mobilis ZM4 was proposed through simulation and analysis using iZM516. Furthermore, metabolic engineering strategies to produce succinate and 1,4-butanediol (1,4-BDO) were designed and then simulated under anaerobic condition using iZM516. The results indicated that 1.68 mol/mol succinate and 1.07 mol/mol 1,4-BDO can be achieved through combinational metabolic engineering strategies, which was comparable to that of the model species E. coli. Our study thus not only established a high-quality GEM iZM516 to help understand and design microbial cell factories for economic biochemical production using Z. mobilis as the chassis, but also provided guidance on building accurate GEMs for other non-model industrial microorganisms.
Keywords: Genome-scale metabolic models (GEMSs); Non-model industrial microorganism; Zymomonas mobilis; Biolog phenotype microarray; Succinate; 1,4-Butanediol
Graphical abstract
Highlights
-
•
A high-quality genome-scale metabolic model iZM516 built for non-model industrial microorganism Zymomonas mobilis.
-
•
Cell growth predicted by iZM516 using multiple substrates had 79.4% agreement with experimental results.
-
•
Potential endogenous succinate synthesis pathway of Z. mobilis ZM4 proposed.
-
•
Metabolic pathways to produce succinate and 1,4-butanediol designed and simulated using iZM516.
-
•
1.68 mol/mol succinate and 1.07 mol/mol 1,4-BDO can be achieved in Z. mobilis.
1. Introduction
Robust and efficient microbial cell factories are crucial for biomanufacturing and the transformation of petroleum-based economy to sustainable bioeconomy [1]. The rational design of cell factories is becoming reality with the development of systems and synthetic biology. Before the practices of metabolic engineering, it is important to investigate the metabolic network within the chassis cells to identify metabolic pathways and genetic targets associated with them for genetic manipulation. Genome-scale metabolic model (GEM) is an effective tool to simulate metabolic fluxes in silico with algorithms such as flux balance analysis (FBA) [2]. Meanwhile, GEM can be used to perform metabolic analyses under different conditions, explore new scientific findings, and guide the design and modification of metabolic pathways and synthetic microorganisms for economic biochemical production [3].
High-quality GEMs have been constantly established and updated for model microorganisms such as E. coli [2] and S. cerevisiae [4]. For example, the model iJE660 was the first GEM of E. coli after its genome was sequenced [5]. After that, a series of GEMs were reconstructed with different degrees of quality improvement [2]. iML1515 was the latest stoichiometric model of E. coli, of which ∼50% of function-known genes (1515 genes) were included in the model [6]. Meanwhile, iML1515 integrated with 1515 protein structural information, enabling analysis of structural proteome comparison and evaluation of mutational impact across strains [6]. Recently, multiple constraints of thermodynamic and enzymatic were integrated into iML1515, making the pathway analysis and phenotype prediction more realistic [7]. The same development route happened with the yeast model [4], into which the enzyme constraints were introduced recently [8].
However, similar work is still scarce for non-model microorganisms with excellent characteristics from industrial practices. Zymomonas mobilis is a gram-negative ethanologenic bacterium with unique physiological properties and extraordinary ethanol production characteristics such as high sugar uptake rate, high ethanol yield and ethanol tolerance [9], which is the only known microorganism that can utilize the Entner-Doudoroff (ED) pathway under anaerobic conditions [10]. Therefore, Z. mobilis has been engineered for economic production of a variety of biofuels and biochemicals such as lignocellulosic ethanol [11,12], poly-3-hydroxybutyrate (PHB) [13], d-lactic acid [14,15], 2,3-butanediol [16], sorbitol [17], acetaldehyde [18], and isobutanol [19].
Although five GEMs of Z. mobilis have been published, no standard format files were provided before 2020 with only the reaction sets involved in the model attached, which cannot be directly reused for simulation [20,21]. Recently, two models iHN446 and iZM4_478 were published with standard SBML files, but still suffered from issues. For example, amino acids are needed to support cell growth for the iHN446 model, although previous results exhibited that Z. mobilis is not an amino acid deficient strain and amino acids are not necessary for its growth in minimal medium [22], which suggests that wrong pathway(s) may be included in model iHN446 [23]. Meanwhile, instead of 1 mol ATP per 1 mol glucose under anaerobic condition, iHN446 produced incorrect amount of 3 mol ATP per 1 mol glucose. Furthermore, most published GEMs of Z. mobilis ZM4 lack plasmid gene information. The genome of Z. mobilis ZM4 was sequenced and released in 2005, which contained a circular chromosome with 1998 open reading frames (ORF) [24]. In 2009, the genome annotation of ZM4 was improved by combining systems biology approaches of genome resequencing and proteomics study [25]. The accurate genome sequence of Z. mobilis containing gene information of a revised chromosome and four native plasmids was further updated in 2018 [26], which could help revisit and improve the GEM of Z. mobilis. However, only iHN446 included four plasmid genes, and other models did not contain any plasmid information.
In contrast, the iZM516 constructed in this study for Z. mobilis ZM4 combined the information of the improved genomic annotation including plasmid, datasets of Biolog Phenotype Microarrays experiments, literature, and Gene-Protein-Reaction information of the recent models. Meanwhile, we further solved the mass balance and ATP infinite generation problems to ensure the quality of model, and used the standard genome-scale metabolic model test suite MEMOTE to improve the model quality. The construction of high-quality iZM516 model can then be applied to help design the effective metabolic pathways for the production of succinate and 1,4-butanediol in Z. mobilis.
2. Materials and methods
2.1. Construction of the draft model using ModelSEED
The latest genomic information of Z. mobilis subsp. mobilis ZM4 (chromosome: NZ_CP023715.1/CP023715.1, plasmid pZM32: NZ_CP023716.1/CP023716.1, plasmid pZM33: NZ_CP023717.1/CP023717.1, plasmid pZM36: NZ_CP023718.1/CP023718.1, and plasmid pZM39: NZ_CP023719.1/CP023719.1) was used for model construction, and the online webserver Rapid Annotation using Subsystem Technology (RAST) was used to annotate genome with option “Build metabolic model”, which will automatically build draft model with ModelSEED [27]. The temporary gene IDs annotated by RAST were transformed into specific IDs and names of Z. mobilis ZM4 by BLASTp with the latest Z. mobilis genome annotation from NCBI as reference [28]. In this process, the threshold of homologues was set as e-value ≤10−5, and the identity ≥40%. In addition, the information of genes, reactions, and metabolites of the draft model was improved by integrating the information from MetaCyc [29], Biochemical, Genetic and Genomic (BiGG) knowledge base [30], Kyoto Encyclopedia of Genes and Genomes (KEGG), BRaunschweig ENzyme DAtabase (BRENDA) [31], and other databases [32].
2.2. Biomass synthesis curation and automatic gap filling
Biomass composition usually includes DNA, RNA, proteins, lipids, peptidoglycan, carbohydrates, and small molecules. In this study, biomass composition was set referred to the published literature [20,21], and was made up of 15 sub-reactions. Macromolecules are important in the biomass synthesis. When there is a synthetic or a putative synthetic pathway for a macromolecule, relevant reactions are involved in the model, such as the biosynthesis of fatty acids and hopanes in Z. mobilis [33]. Hopane is an important membrane component of Z. mobilis contributing to ethanol tolerance, and the possible metabolic pathway of hopane was reported in literature [33]. Combining the hopane biosynthesis pathway PWYN-7072 from the MetaCyc database and the literature, the hopane biosynthesis module was cured in iZM516 constructed in this study. If the detailed knowledge of some macromolecule synthesis was not very clear, a total reaction equation was used for these macromolecules, and the sub-reactions were ignored.
Metabolic gaps usually occur in the draft model, resulting in the inability of model to synthesize biomass. To achieve automatic gap filling, a weight-added pFBA algorithm was used [34]. Briefly, biomass compositions that cannot be synthesized in the model were identified firstly. Then, the gaps in each composition synthesis pathway were confirmed using FBA calculation. Finally, the weight-added pFBA gap-filling algorithm was employed to fill the gaps. The reactions in the draft model were all set weight as 1000, and the upper limit of the biomass equation was set as 0.1 to minimize the filling reaction numbers introduced from ModelSEED database [34]. In this way, the reactions in the draft model will be preferentially used in calculation. When there is a gap in the calculational pathway, the missing reactions are automatically retrieved from the reactions of ModelSEED database. To ensure the correction of added reactions and determine the Gene-Protein-Reaction (GPR) relations, manual checks were performed if necessary.
2.3. ATP synthesis and respiratory chain correction
Except for a few well-known ATP generation reactions, general principles were used for ATP generation correction including [35]: 1) The reaction of ATP to AMP conversion is irreversible. 2) If the reaction generating ATP from ADP does not possess carbon-containing metabolites, the direction is set to consume ATP except for the known ATP synthesis reactions. 3) Reaction with oxygen generation is generally irreversible. 4) Transamination is usually the decomposition reaction of amino acids. 5) If the reaction does not involve in above principles, manual check is performed combined with MetaCyc, KEGG, and eQuilibrator [32] databases.
2.4. Biolog Phenotype Microarray experiments and data analysis
The Biolog Phenotype Microarrays (PMs) were employed to test the substrates utilization of different carbon, nitrogen, sulfur, and phosphorus sources with the Omnilog PM automatic system using the wild-type strain Z. mobilis ZM4. It was reported that the phenotype of Z. mobilis was similar to that of yeast in the previous study [36]. Therefore, the experimental process was referred to the PMs Procedure for S. cerevisiae and other Yeast. The image data of PMs were processed into values using OmniLog PM program suite of data processing and analysis based on the operational manual. Then, the FBA calculation was performed with the biomass equation as the objective function, either the metabolites in PM1 and PM2 were set as the only carbon source, or the metabolites in PM3, PM4 A1-E12, or PM4 F1–H12 were set as the only nitrogen, sulfur, or phosphorus source, respectively.
2.5. Model simulation analysis, evaluation, and visualization
The GEM iML1515 of E. coli was used to simulate chemical production in E. coli [6]. COBRApy is a Python-based toolkit package, which can be used to modify model, generate executable file, and simulate analyses such as FBA, Parsimonious flux balance analysis (pFBA), flux variability analysis (FVA), and gene necessity analysis [37]. The model was evaluated by MEMOTE to check the mass balance, charge balance, and model annotation [38]. All simulations were performed under anaerobic condition in minimal media with oxygen uptake rate set as 0, glucose as sole C sources with 40 mmol/gDW/h uptake rate, ammonia as sole N sources with 1000 mmol/gDW/h uptake rate unless otherwise indicated. When the cell growth was taken into consideration, the biomass growth rate was set as 0.1 h−1.
3. Results
3.1. Construction of the GEM iZM516 and comparison of published models
In this study, a new genome-scale metabolic model for Z. mobilis ZM4 was constructed using modelSEED, and then model curation and verification were performed based on the experimental data and latest information. Finally, the new model was named as iZM516 (Supplementary file 1: SBML version file of iZM516), which contains 1389 reactions, 1437 metabolites, and 516 genes with 6 genes located on native plasmids corresponding to 80 reactions (Table S1). iZM516 also contains 3 cell compartments of intracellular, periplasmic, and extracellular.
Z. mobilis uses ED pathway to ferment glucose under anaerobic condition, which results in only 1 mol of ATP yielded per mol glucose. Using the ATP maintenance (ATPM) reactions (atp_c + h2o_c → adp_c + h_c + pi_c) as objective function, three models (iZM516, iZM4_478 and iHN446) were compared with glucose uptake rate set as 10 mmol gDW−1 h−1. 10 mmol gDW−1 h−1 ATP was generated using either iZM516 or iZM4_478 while 30 mmol gDW−1 h−1 ATP was generated using iHN446, which means iHN446 used incorrect pathway under anaerobic condition (Table 1). The pentose phosphate pathway (PPP) was incomplete in Z. mobilis, which lacks of 6-phosphogluconate (6 PG) dehydrogenase catalyzing 6 PG to form ribulose-5-phosphate (Ru5P) (6 pg_c + nad_c → ru5p_c + co2_c + nadh_c) [39,40]. However, iHN446 contains a reaction R10221 (6 pg_c + nad_c → ru5p_c + co2_c + nadh_c) with GPR annotation as ZMO0942, which is inaccurate and results in the PPP available in the model. Meanwhile, iHN446 uses ATP synthesis reaction (R00086: adp_c + 4.0 h_e + pi_c ≤> atp_c + h2o_c + 3.0 h_c) to generate ATP under anaerobic condition, which contributes to the incorrection of simulation results of iHN446.
Table 1.
Comparisons of ATP generation using different models with ATP maintain reaction as the objective function under aerobic or anaerobic conditions. The glucose uptake rate was set as 10 mmol gDW−1h−1.
| iZM516 | iZM4_478 | iHN446 | |
|---|---|---|---|
| Anaerobic | 10 | 10 | 30 |
| Aerobic | 30 | 20 | 30 |
For aerobic condition, Z. mobilis uses a structural respiratory chain that mainly includes type II NADH dehydrogenase (Ndh, ZMO1113), coenzyme Q10, terminal oxidase cytochrome bd (CydAB, ZMO1571-ZMO1572), and other unidentified components [41]. Meanwhile, Z. mobilis is one of the few bacteria known to use NADH and NADPH as electron donors for the respiratory type II NADH dehydrogenase [42]. However, iZM4_478 does not contain the reaction using NADPH as the electron donor. Glucose and lactate can also contribute electrons to the respiratory chain by donating electrons to the membrane-bound glucose dehydrogenase (Gdh, ZMO0558) and lactate dehydrogenase (Ldh, ZMO0256), respectively, which are transferred to coenzyme Q10 and subsequently to terminal oxidases that reduce oxygen to water [43]. In addition, because the respiratory chain of Z. mobilis may be completed in the periplasm [43], periplasm was introduced into the model as the third compartment with suffix “_p”. Except for iZM4_478, iHN446 and all other published models do not include periplasm compartment. Instead, the above genes and reactions involved in the respiratory chain are included in the current model iZM516.
Since ATP generation under aerobic condition was associated with proton motive force (PMF), the reaction involved with h_p production and consumption in the model may affect ATP generation. However, there is no enough data to support the exact reactions that are associated with h_p in Z. mobilis. The comparison results of three models shown that iZM516 and iHN446 generate 30 mmol gDW−1 h−1 ATP, and iZM4_478 generates 20 mmol gDW−1 h−1 ATP (Table 1). This difference resulted from different h_p containing reactions involved in different models. In the simulation, iHN446 does not contain periplasm compartment, and must supply amino acids in the medium for cell growth, which is not the essential nutritional requirement for Z. mobilis [22]. Hence, iHN446 was not included for further simulation comparison.
Although the latest published model iZM4_478 has improved GPR association through pooled transposon mutant fitness experiments [44], iZM516 further corrected GPR association based on experiment results such as 13C-labelled metabolomics data [39] and the latest literature, such as ZMO1754 encoding an NADP + -dependent acetaldehyde dehydrogenase that was determined recently [45]. All manual curation reaction list can be found in supplementary materials (Supplementary file 2). Compared with five published GEMs of Z. mobilis, iZM516 has the largest datasets of genes, reactions, and metabolites (Table 2) containing 25.8% (516/2001) of totally annotated genes of ZM4. Based on the enrichment results, these 516 genes were mainly distributed in metabolism, transporter, and tRNA synthesis, while amino acid metabolism contains the largest gene number of 139 (Fig. S1). The number of genes in iZM516 was also 8% larger than that in iZM4_478 without genes from native plasmids. Based on the gene number comparisons, 380 genes were all included in iZM516, iZM4_478, and iHN446. However, there are 60 unique genes in iZM516, 27 unique genes in iZM4_478, and 28 unique genes in iHN446, respectively (Fig. S2). The reactions associated with these unique genes in iZM4_478 or iHN446 are either already presented in iZM516 but associated with other genes, or uncertain whether they exist but not affect biomass synthesis. Therefore, these genes were not introduced into iZM516.
Table 2.
Comparisons of genome-scale metabolic models of Z. mobilis.
| Model | iZM516 | iZM4_478 | iHN446 | iEM439 | iZM363 | ZmoMBEL601 |
|---|---|---|---|---|---|---|
| Strain | ZM4 | ZM4 | ZM4 | ZM1 | ZM4 | ZM4 |
| SBML filea | + | + | + | – | – | – |
| Compartment | 3 | 3 | 2 | 2 | 2 | 2 |
| Genes | 516 | 478 | 446 | 439 | 363 | 348 |
| Plasmid genes | 6 | 0 | 4 | 0 | 0 | 0 |
| Reactions | 1389 | 857 | 859 | 692 | 747 | 601 |
|
Metabolites |
1437 |
799 |
894 |
658 |
704 |
579 |
|
Year |
2022 |
2020 |
2020 |
2016 |
2011 |
2010 |
| Ref. | This study | [44] | [23] | [46] | [21] | [20] |
+ SBML file available, - SBML file not available.
3.2. Biolog Phenotype Microarrays experiments and data correction
Biolog Phenotype Microarrays (PMs) experiments were employed to evaluate and confirm the metabolic capabilities of Z. mobilis. The phenotype plates of carbon sources (PM1, PM2A), nitrogen sources (PM3B), phosphorus sources (PM4 A1-E12), and sulfur sources (PM4 F1–H12) were measured in the PM system to determine the utilization of different C, N, P, and S related substrates as the sole source. The 2 replicate of Biolog's PMs experiments data have high consistency with R2 value of 0.96 (Fig. S3). For quantitative analysis, the max-height data of each well based on the average value of 2 replicates were used to compare with those of the control wells. If the value of test well was 1.5 times higher than that of the control well, the substrate was considered as responsive to Z. mobilis. The results showed that Z. mobilis responded to 91 substrates including 13 Carbon, 19 Nitrogen, 43 Phosphorus, and 16 Sulfur substrates (Table S2), which was consistent with the previous report [36]. All responsive substrates were simulated using the draft model as the sole carbon, nitrogen, or sulfur sources and some of which were included in iZM516 by adding the corresponding transport reactions.
Wild-type Z. mobilis ZM4 can only utilize glucose, fructose, and sucrose as sole carbon sources, and cannot use pentoses such as xylose and arabinose. In both PM1 and PM2A plates, Z. mobilis not only responded to glucose and fructose, but also responded to xylose, arabinose, and other carbon sources weakly as shown in the kinetic curves (Fig. 1), which is consistent with a previous study [36]. In addition, sucrose (PM1_D11) in PM1 plate had no response, which is also consistent with a previous study that no response was observed even at high initial cell concentrations, and the previous study indicated that sucrose may subject to strong catabolite repression in the PM system [36]. The weak response to pentose carbon sources and no response to sucrose may be related to the mechanism of the PM system or unknown redox reaction of Z. mobilis.
Fig. 1.
Biolog Phenotype Microarrays profiling of Z. mobilis under different carbon (A, B), nitrogen (C), phosphorus and sulfur (D) sources. Each plate contains A-H rows and 01–12 columns, and the detail substrate information of each cell can be found in Table S2. L-Asn: l-Asparagine; L-Asp: l-Aspartic Acid; L-Cys: l-Cysteine; L-Glu: l-Glutamic Acid; L-Met: l-Methionine; Ala-Gln-L: L-Alanyl-glutamine; Gly-Gln-L: L-glycyl-glutamine. The blue font in the graph indicates the substrate in the cell.
It was reported that Z. mobilis can utilize NH4+, glutamic acid, glutamine, aspartate, and asparagine as nitrogen sources [36]. iZM516 can utilize more than 10 different nitrogen sources including NH4+, asparagine, cysteine, glutamic acid, serine, ethanolamine, adenosine, and some peptides (Table S2). The result of PM4A phenotype plates (A1-E12) indicated that Z. mobilis can utilize 39 phosphorus sources except for pyrophosphate and tripolyphosphate (Fig. 1, Table S2), which is consistent with the previous study too [36]. iZM516 simulation showed that 12 substrates can be used as the sole phosphorus source when used the sink reaction (Table S2). The result of PM4A phenotype plate (F1–H12) showed that 16 substrates can be the sole sulfur source (Fig. 1, Table S2). Shake flask experiments confirmed that sulfate, thiosulfate, cysteine, and methionine can be utilized as the sole sulfur source for Z. mobilis [47]. Correspondingly, the transport and exchange reactions were added to the model.
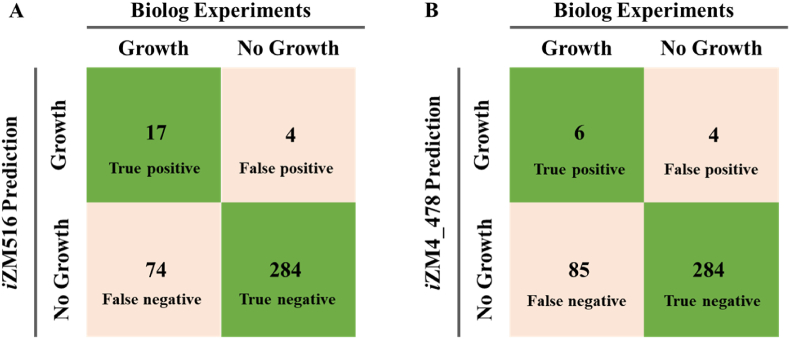
Furthermore, total 379 substrates were used to compare the different substrate utilization capabilities of iZM516 and iZM4_478. The results suggested that iZM516 has an agreement of 79.4% (17 true positives and 284 true negatives) with experimental data, which is higher than that of 76.5% obtained by iZM4_478, especially in true positive results (Fig. 2, Table S2). As for the remaining 74 false negative results with unknown encoding genes and pathways, no reactions were added into iZM516 to predict growth. Optimization will be updated accordingly in the future studies.
Fig. 2.
Comparisons of in silico simulation and Biolog Phenotype Microarrays results of iZM516 (A) and iZM4_478 (B) for the utilization of various substrates. True positive and true negative means that the in silico simulation results were same as the in vivo experiments, while false positive means the substrate can be used in simulation but cannot be used in vivo. False negative is in contrast with false positive.
3.3. Model evaluation by MEMOTE and cell growth validation
MEMOTE is a set of standardized metabolic model tests, which evaluates the metabolic model from multiple perspectives, and the resultant score could help identify the possible problems to improve the quality of the model [38]. Among the published models with simulation ready format files, only iZM4_478 can be tested by MEMOTE, and iHN446 cannot be tested by MEMOTE with unknown reason. According to MEMOTE scoring (Fig. 3A and B), the standardized score of iZM516 (91%, Supplementary file 3) was higher than that of iZM4_478 (80%, Supplementary file 4). The accuracy of the mass and charge balance is better than 99% for both models, while only a few reactions in macromolecular synthesis were imbalanced. This demonstrated the comprehensiveness of iZM516 for subsequent simulations.
Fig. 3.
Comparisons of MEMOTE test scores of iZM516 (A) and iZM4_478 (B) as well as the predictions for specific cell growth rates (C), and ethanol production fluxes (D) under anaerobic condition using iZM516 (green line) or iZM4_478 (red line) models. iZM4_478 is a model published recently. iZM516 is a new model constructed in this study. The reference data in C and D were experimental data from published literature [39,48,49].
As mentioned above, wild-type Z. mobilis can only utilize glucose, fructose, and sucrose. The growth rate of Z. mobilis in minimal medium (MM) is 0.13–0.45 h−1, and the maximum uptake rate of glucose can be 60 mmol·gDW−1·h−1 [48]. When FBA was performed to evaluate the accuracy of iZM516 and the glucose uptake rate was set from 0 to 70 mmol·gDW−1·h−1, the simulation results showed that the simulated specific growth and ethanol productivities of iZM516 and iZM4_478 were both consistent with experimental data (Fig. 3C and D) from literature reports [39,48,49]. The relevant analysis codes are provided in the Supplementary file 5.
3.4. Exploration of succinate biosynthesis pathway in Z. mobilis
Succinate is an important platform chemical for the synthesis of numerous chemical products, such as 1,4-butanediol, tetrahydrofuran, and bio-polyesters. It was experimentally confirmed that Z. mobilis can produce succinate under anaerobic condition [50,51], but the exact succinate synthetic pathway in wild-type Z. mobilis was not illustrated completely yet although the incomplete TCA cycle in Z. mobilis lacking two key genes encoding 2-oxoglutarate dehydrogenase complex (SucAB) and malate dehydrogenase (Mdh) could be used for succinate biosynthesis [40]. Previous research proposed that succinate was synthesized via the fumarate reduction reaction with the succinate precursor converted from pyruvate through malic enzyme (MaeA) (Fig. 4A) [20,50,52]. However, the [6–13C] and [1–13C] labelled fluxes analysis results exhibited that PEP was the predominant anaplerotic source of carbon for TCA [39], which suggested that succinate was not synthesized from pyruvate naturally in Z. mobilis (Fig. 4A). The 13C labelled fluxes analysis also indicated that succinate was not produced from α-ketoglutarate via oxidative TCA cycle with the truncated TCA cycle, which suggested that there must exist substitutional pathway for succinate production in Z. mobilis.
Fig. 4.
The potential pathway of succinate generation in wild-type Z. mobilis (A), and the transcriptome expression box plot of genes in succinate synthesis pathway (B). The red cross lines represent the missing genes in the genome of Z. mobilis. The black and blue dot lines indicate there may have multiple reaction steps. The green lines represent the succinate synthetic pathway suggested by simulation of iZM516. 25aics: 5′-phosphoribosyl-4-(N-succinocarboxamide)-5-aminoimidazole, AcCoA: acetyl-CoA, Akg: 2-oxoglutarate (2-ketoglutaric acid), Argsuc: L-arginino-succinate, Asp-L: l-aspartate, Cbasp: N-carbamoyl-l-aspartate, Cit: citrate, Dcamp: adenylosuccinate, Dhor-S: (S)-dihydroorotate, Fum: fumarate, Mal: l-Malate, Oaa: oxaloacetate, Pep: phosphoenolpyruvate, Pyr: pyruvate, Succ: Succinate, SucCoA: succinyl-CoA.
Using the genome-scale model iZM516, in silico analysis was performed to simulate anaerobic succinate production in Z. mobilis. Taking both the biomass synthesis and succinate production into consideration, the glucose uptake rate was set as 40 mmol/gDW/h, the biomass growth rate was set as 0.1 h−1, then the maximization of succinate production was set as the objective function. The pFBA result suggested that succinate production in Z. mobilis may go through a complicated pathway (Fig. 4A). The flux starts from phosphoenolpyruvate (Pep) to oxaloacetate (Oaa) by a phosphoenolpyruvate carboxylase (ZMO1_ZMO1496, Ppc), which is then converted to aspartate (Asp) through an aminotransferase. Subsequently, Asp is catalyzed to fumarate (Fum) by different routes, which is finally reduced to succinate (Succ) through dihydroorotate dehydrogenase (ZMO1_ZMO0120) (Fig. 4A). Due to the complicated network for succinate generation, the simulation yield of succinate was pretty low at 0.03 mol Succ/mol glucose (0.017 g/g glucose) in wild-type Z. mobilis with ethanol or lactate as the primary products. This result is partially consistent with the 13C fluxes analysis result that aspartate was produced from PEP and then to succinate [39]. However, simulation using iZM4_478, succinate was produced from Akg through the reaction of OXGDC (akg_c + h_c ≤> co2_c + sucsal_c) with GPR correlation gene of ZMO0687, which does not catalyze this reaction with the annotation of acetolactate synthase large subunit. Thus, the succinate synthesis pathway in iZM4_478 may not be correct.
To confirm that genes involved in the complicated succinate biosynthesis pathway in iZM516 were truly expressed in vivo, the transcriptional expression levels of these genes under different conditions were analyzed using the one-stop database ZymOmics (http://zymomics.cn/), which suggested that these genes were indeed expressed with low to middle level under different conditions (Fig. 4B). Meanwhile, compared with natural and engineered succinate producers, the pathway used for succinate synthesis in wild-type Z. mobilis is complicated and inefficient, and metabolic engineering strategies are needed to introduce heterologous pathways into Z. mobilis for efficient succinate biosynthesis.
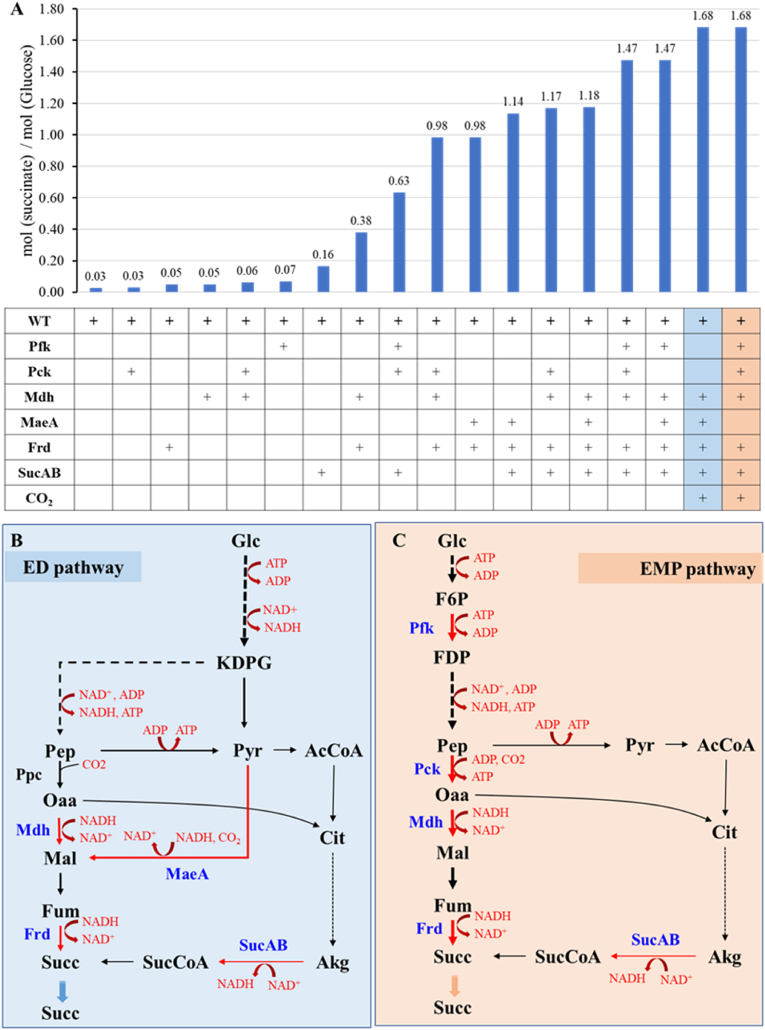
3.5. Simulation of succinate production at high yield in Z. mobilis using iZM516
To the best of our knowledge, the maximum yield of succinate synthesis pathway was from the reductive branch of the TCA cycle (redTCA) with 1.71 mol/mol theoretical yield, which was from Pep to Oaa, then followed by the malate reduced pathway [53]. Based on above analyses, the related genes (mdh and frd) and reactions (rxn00248 and rxn00284) within redTCA were introduced into iZM516 to explore the optimal succinate production pathway. Meanwhile, other genes from literature for succinate production were also introduced into iZM516, such as pck, sucAB in oxTCA and pfk in Embden-Meyerhof-Parnas (EMP) pathway to enhance the precursor supply. The simulation results suggested that the introduction of only one key gene into Z. mobilis cannot improve succinate production significantly (Fig. 5A), therefore the combination simulation strategies were conducted (Fig. 5A).
Fig. 5.
Comparisons of different strategies for succinate production in Z. mobilis using the in-silico simulation (A) as well as the heterologous genes needed for succinate production through ED pathway (B) or EMP pathway (C). AcCoA: acetyl-CoA, Akg: 2-oxoglutarate (2-ketoglutaric acid), Cit: citrate, F6P: fructose-6-phosphate, Frd: fumarate reductase, Fum: fumarate, Glc: glucose, KDPG: 2-Keto-3-deoxy-6-phosphogluconate, MaeA: malate dehydrogenase, Mal: l-Malate, Mdh: malate dehydrogenase, Oaa: oxaloacetate, Pep: phosphoenolpyruvate, Pyr: pyruvate, Pck: PEP carboxykinase, Pfk: 6-phosphofructokinase, SucAB: α-ketoglutarate dehydrogenase complex, Succ: Succinate, SucCoA: succinyl-CoA. The blue fonts in B and C represent the enzyme catalyzing the reactions.
In the succinate synthesis pathway, two important precursors are Pep and pyruvate. Therefore, enhancing the fluxes from Pep or pyruvate to redTCA is an effective strategy to improve the succinate production. For example, the fluxes can be enhanced from Pep into redTCA by introducing Pck, Mdh combining with the downstream reaction of “Fum + NADH - > Succ + NAD+” catalyzed by fumarate reductase (Frd). In this way, the yield of succinate was able to achieve 0.98 mol/mol (Fig. 5A), which can also be achieved with the same yield by introducing MaeA and Frd. When the redTCA and oxTCA pathways were combined to improve precursors in different strategies, the yield of succinate can be achieved to 1.14–1.18 mol/mol. However, the native ED pathway in Z. mobilis usually produces one mol Pep and two mol pyruvate from one mol glucose. Therefore, if the native ED pathway is used for succinate production, the key enzyme MaeA will not only compete NADH with ethanol generation pathway, but also fixes one mol CO2 in each reaction. The simulation result exhibited that the succinate yield could reach 84% of theoretical yield to 1.68 mol/mol coupling with CO2 fixation (Fig. 5A and B).
In other way, the succinate yield can be enhanced to 1.47 mol/mol with the combination of redTCA and oxTCA fluxes by supplying the precursor Pep using EMP pathway through the introduction of pfk gene. If CO2 was supplied infinitely to meet the maximum CO2 fixation by Pck, the yield can be further improved 15% up to 1.68 mol/mol (Fig. 5A and C), which was consisted with 83.3% from the redTCA and 16.7% from the oxTCA. Above simulation thus suggested that the strategy to combine redTCA and oxTCA with CO2 fixation is effective for succinate production in Z. mobilis, either using ED pathway or EMP pathway. Since succinate production under anaerobic condition has economic advantages, the in-silico simulation using industrial microorganisms such as Z. mobilis can provide guidance on designing metabolic pathways and microbial cell factories for succinate production under anaerobic conditions.
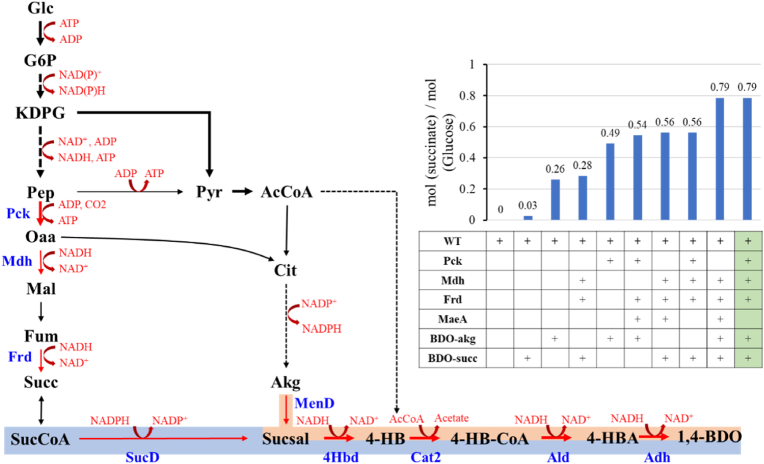
3.6. Exploration of 1,4-butanediol biosynthesis strategy in Z. mobilis using iZM516
1,4-Butanediol (1,4-BDO) is another platform chemical used for numerous applications. There has no natural microorganisms for 1,4-BDO production, and it is majorly produced by recombinant microorganisms by introducing artificial biosynthesis pathways [54]. The pathway proposed by Kim et al. through biopathway prediction algorithms had high 1,4-BDO yield compared with other published pathways [54,55]. Since 1,4-BDO is the downstream product of succinate or Akg from TCA cycle, which was defined as BDO-succ (the precursor is from succinate) and BDO-akg (the precursor is from 2-oxoglutarate) pathway in this study, the production of 1,4-BDO was further simulated based on the succinate results using iZM516 to help design and construct the metabolic pathways for 1,4-BDO production in Z. mobilis.
The simulation results suggested that the yield of 1,4-BDO was low if introducing either BDO-succ or BDO-akg 1,4-BDO production pathway only. The complicated biosynthesis pathway of succinate in wild-type Z. mobilis resulted in low yield of 1,4-BDO using BDO-succ pathway (Fig. 4, Fig. 6). To optimize the production of 1,4-BDO, different combination strategies were tested using iZM516 for efficient precursor supply or redox balance (Fig. 6). Through introducing Pck into BDO-akg pathway to enhance Oaa and energy supply, the 1,4-BDO yield was improved 88% compared with introducing BDO-akg pathway only. When combining Mdh and Frd as well as Pck or MaeA into the two pathways respectively, the yield was increased as expected because of the precursor enrichment, but the yield was limited to 0.54–0.56 mol (1,4-BDO)/mol (glucose). The modeling result demonstrated that the BDO-succ pathway consumes NADPH, while BDO-akg pathway produces NADPH (Fig. 6). Therefore, two pathways were combined with each route contributing 50% fluxes for redox balance during the simulation, the optimal yield can then be achieved to 0.79 mol (1,4-BDO)/mol (glucose) that is as high as the yield reported before [56]. It is worth noting that the optimal pathway to synthesize 1,4-BDO was not based on the highest succinate production pathway due to the imbalance of cofactor, energy, and biomass, which indicated that in silico simulation is beneficial before carrying out experiments.
Fig. 6.
The strategies for 1,4-BOD production in Z. mobilis based on in silico GEM simulation. 1,4-BDO: 1,4-butanediol, 4-HB: 4-hydroxybutyrare, 4-HBA: 4-hydroxybutyraldehyde, 4-HB-CoA: 4-hydroxybutyryl-CoA, AcCoA: acetyl-CoA, Akg: 2-oxoglutarate (2-ketoglutaric acid), Cit: citrate, Frd: fumarate reductase, Fum: fumarate, Glc: glucose, KDPG: 2-Keto-3-deoxy-6-phosphogluconate, Mal: l-Malate, Mdh: malate dehydrogenase, Oaa: oxaloacetate, Pep: phosphoenolpyruvate, Pck: phosphoenolpyruvate carboxykinase, Pyr: pyruvate, Succ: succinate, SucCoA: succinyl-CoA, SucD: succinate semialdehyde dehydrogenase, Sucsal: succinate semialdehyde. The blue fonts in B and C represent the enzyme catalyzed the reaction.
3.7. Comparisons of succinate and 1,4-BDO production simulated with the models of Z. mobilis and E. coli
Since the model species E. coli is the most studied prokaryotic chassis cell for biochemical production and has been successfully engineered to produce succinate and 1,4-BDO, the complete GEM iML1515 of E. coli [6] was used to compare the yields of succinate and 1,4-BDO with Z. mobilis. While E. coli naturally harbors the succinate synthesis pathway, no heterologous gene was introduced into the model iML1515. The simulation results suggested that the maximum yield of succinate can be achieved to 1.68 mol/mol with cell growth rate set as 0.1 h−1 under anaerobic condition, which is as high as the optimal yield in Z. mobilis (Table 3), and was higher than that of 1.5 mol/mol for the previous recombinant strain [57]. When the 1,4-BDO synthesis pathway was introduced into iML1515, 0.99 mol/mol 1,4-BDO will be produced in E. coli, which was higher than that of 0.79 mol/mol in Z. mobilis (Table 3).
Table 3.
Comparisons of succinate and 1,4-butanediol (1,4-BDO) production using genome-scale metabolic models of Z. mobilis and E. coli.
| Model | Chassis cell | Succinate (mol/mol) | 1,4-BDO (Acetate not recycle) (mol/mol) | 1,4-BDO (Acetate recycle) (mol/mol) |
|---|---|---|---|---|
| iZM516 | Z. mobilis | 1.68 | 0.79 | 1.07 |
| iML1515 | E. coli | 1.68 | 0.79 | 0.99 |
By comparing metabolic pathways, we found that the primary byproduct acetate was utilized in E. coli, which shed light on enhancing 1,4-BDO yield in Z. mobilis. Therefore, the pathway that recycling acetate to generate acetyl-CoA by ATP: acetate phosphotransferase (AckA) and acetyl-CoA: phosphate acetyltransferase (Pta) was introduced into iZM516. As a result, the yield of 1,4-BDO can be increased to 1.07 mol/mol in Z. mobilis (Table 3), which was higher than that of E. coli due to the lower ATP maintenance requirement of Z. mobilis. It should be noted that the reaction of converting acetate to acetyl-phosphate by AckA was an ATP consuming reaction. Thus, it is necessary to complete EMP pathway in Z. mobilis to supply more ATP, which was confirmed by simulation using iZM516.
Although the E. coli was successfully engineered to produce succinate [57] and 1,4-BDO [55], there still has the space to enhance the yield based on the simulation. Compared with E. coli, Z. mobilis has the same yield of succinate and higher optimal yield of 1,4-BDO when acetate recycle was applied under anaerobic condition. Since Z. mobilis possesses industrial characteristics such as anaerobic fermentation at a broad range of pH and temperature conditions, few byproducts, free of phage infection, and robustness against lignocellulosic inhibitors [40], it will be an excellent chassis cell to be developed as microbial cell factories for the industrial-scale production of lignocellulosic biofuels and biochemicals.
Meanwhile, our results also exhibited that the in silico modeling and analysis is an effective approach to evaluate the design of metabolic pathways and microbial cell factories before experimentation, which is not only important for model microorganisms such as E. coli with tremendous amount of experimental data and comprehensive databases, but also vital for those non-model microorganisms with specific industrial characteristics such as Z. mobilis in this study [58]. Therefore, high-quality GEMs and comprehensive strain specific databases are necessary to reduce the time and cost associated with the classical trial-and-error research approach, and lay the foundation to facilitate the construction of digital cells in the future.
4. Conclusion
In this work, a high-quality GEM iZM516 of Z. mobilis ZM4 was constructed based on the improved genomic information, experimental datasets of Biolog Phenotype Microarrays, databases, and literature reports. The model iZM516 contains 1389 reactions, 1437 metabolites, 516 genes, and 3 cell compartments, and has the highest MEMOTE evaluation score 91% among all published models of Z. mobilis. Based on iZM516, the native succinate synthesis pathway in Z. mobilis was examined and proposed, and the potential of Z. mobilis to produce succinate and 1,4-BDO with high yield under anaerobic condition was then evaluated using iZM516. The modeling results suggested that 1.68 mol/mol succinate and 1.07 mol/mol 1,4-BDO can be achieved through metabolic engineering in Z. mobilis. Therefore, this high-quality genome-scale metabolic model iZM516 of Z. mobilis developed in this study not only offers a new tool to expend the product spectrum using Z. mobilis as the chassis cell, but also provides a guidance on developing high-quality GEMs to facilitate the rational design of metabolic pathways and microbial cell factories using non-model industrial microorganisms. This study provided a theoretical analysis for different products, and the experimental validation is needed in the future work.
Funding
This work was supported by the National Key Technology Research and Development Program of China (2018YFA0900300 and 2022YFA0911800), the National Natural Science Foundation of China (21978071 and U1932141), the Key Science and Technology Innovation Project of Hubei Province (2021BAD001), 2022 Joint Projects between Chinese and CEEC's Universities (202004), the Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang Province (2018R01014), and the Innovation Base for Introducing Talents of Discipline of Hubei Province (2019BJH021). Funding was also supported by State Key Laboratory of Biocatalysis and Enzyme Engineering.
CRediT authorship contribution statement
Yalun Wu: Data curation, Visualization, Writing – original draft. Qianqian Yuan: Data curation, Visualization, Writing – original draft. Yongfu Yang: Experimentation, Data curation, Visualization, Writing – original draft. Defei Liu: Methodology, and, Experimentation. Shihui Yang: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. Hongwu Ma: Conceptualization, Funding acquisition, Supervision, Writing – review & editing, All authors have read and approved the final manuscript.
Declaration of competing interest
All authors declare that they have no conflicts of interest.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2023.07.001.
Contributor Information
Yalun Wu, Email: aaronwu@stu.hubu.edu.cn.
Qianqian Yuan, Email: yuan_qq@tib.cas.cn.
Yongfu Yang, Email: yongfu.yang@stu.hubu.edu.cn.
Defei Liu, Email: liudf@tib.cas.cn.
Shihui Yang, Email: Shihui.Yang@hubu.edu.cn.
Hongwu Ma, Email: ma_hw@tib.cas.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Yilmaz S., Nyerges A., van der Oost J., Church G.M., Claassens N.J. Towards next-generation cell factories by rational genome-scale engineering. Nat Catal. 2022;5(9):751–765. [Google Scholar]
- 2.Fang X., Lloyd C.J., Palsson B.O. Reconstructing organisms in silico: genome-scale models and their emerging applications. Nat Rev Microbiol. 2020;18(12):731–743. doi: 10.1038/s41579-020-00440-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu C.D., Kim G.B., Kim W.J., Kim H.U., Lee S.Y. Current status and applications of genome-scale metabolic models. Genome Biol. 2019;20:121. doi: 10.1186/s13059-019-1730-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y., Li F., Nielsen J. Genome-scale modeling of yeast metabolism: retrospectives and perspectives. FEMS Yeast Res. 2022;22(1):1–9. doi: 10.1093/femsyr/foac003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards J.S., Palsson B.O. The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities. Proc Natl Acad Sci USA. 2000;97(10):5528–5533. doi: 10.1073/pnas.97.10.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monk J.M., Lloyd C.J., Brunk E., Mih N., Sastry A., King Z., Takeuchi R., Nomura W., Zhang Z., Mori H., et al. iML1515, a knowledgebase that computes Escherichia coli traits. Nat Biotechnol. 2017;35(10):904–908. doi: 10.1038/nbt.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X., Mao Z., Zhao X., Wang R., Zhang P., Cai J., Xue C., Ma H. Integrating thermodynamic and enzymatic constraints into genome-scale metabolic models. Metab Eng. 2021:133–144. doi: 10.1016/j.ymben.2021.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Li F., Yuan L., Lu H., Li G., Chen Y., Engqvist M.K.M., Kerkhoven E.J., Nielsen J. Deep learning-based kcat prediction enables improved enzyme-constrained model reconstruction. Nat Catal. 2022;5:662–672. [Google Scholar]
- 9.Xia J., Yang Y.F., Liu C.G., Yang S.Y., Bai F.W. Engineering Zymomonas mobilis for robust cellulosic ethanol production. Trends Biotechnol. 2019;37(9):960–972. doi: 10.1016/j.tibtech.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Wang X., He Q., Yang Y., Wang J., Haning K., Hu Y., Wu B., He M., Zhang Y., Bao J., et al. Advances and prospects in metabolic engineering of Zymomonas mobilis. Metab Eng. 2018;50:57–73. doi: 10.1016/j.ymben.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Yan Z., Zhang J., Bao J. Increasing cellulosic ethanol production by enhancing phenolic tolerance of Zymomonas mobilis in adaptive evolution. Bioresour Technol. 2021;329 doi: 10.1016/j.biortech.2021.124926. [DOI] [PubMed] [Google Scholar]
- 12.Geng B., Liu S., Chen Y., Wu Y., Wang Y., Zhou X., Li H., Li M., Yang S. A plasmid-free Zymomonas mobilis mutant strain reducing reactive oxygen species for efficient bioethanol production using industrial effluent of xylose mother liquor. Front Bioeng Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.1110513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y., Wang Y., Wang R., Yan X., Wang J., Wang X., Chen S., Bai F., He Q., Yang S. Metabolic engineering of Zymomonas mobilis for continuous co-production of bioethanol and poly-3-hydroxybutyrate (PHB) Green Chem. 2022;24(6):2588–2601. [Google Scholar]
- 14.Lawford H.G., Rousseau J.D. Steady-state measurements of lactic acid production in a wild-type and a putative D-lactic acid dehydrogenase-negative mutant of Zymomonas mobilis: influence of glycolytic flux. Appl Biochem Biotechnol. 2002;98–100:215–228. doi: 10.1385/abab:98-100:1-9:215. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y., Ghosh I.N., Martien J., Zhang Y., Amador-Noguez D., Landick R. Regulated redirection of central carbon flux enhances anaerobic production of bioproducts in Zymomonas mobilis. Metab Eng. 2020;61:261–274. doi: 10.1016/j.ymben.2020.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Yang S., Mohagheghi A., Franden M.A., Chou Y.C., Chen X., Dowe N., Himmel M.E., Zhang M. Metabolic engineering of Zymomonas mobilis for 2,3-butanediol production from lignocellulosic biomass sugars. Biotechnol Biofuels. 2016;9(1):189. doi: 10.1186/s13068-016-0606-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Folle A.B., Baschera V.M., Vivan L.T., Carra S., Polidoro T.A., Malvessi E., da Silveira M.M. Assessment of different systems for the production of aldonic acids and sorbitol by calcium alginate-immobilized Zymomonas mobilis cells. Bioproc Biosyst Eng. 2018;41(2):185–194. doi: 10.1007/s00449-017-1856-1. [DOI] [PubMed] [Google Scholar]
- 18.Kalnenieks U., Balodite E., Strahler S., Strazdina I., Rex J., Pentjuss A., Fuchino K., Bruheim P., Rutkis R., Pappas K.M., et al. Improvement of acetaldehyde production in Zymomonas mobilis by engineering of its aerobic metabolism. Front Microbiol. 2019;10:2533. doi: 10.3389/fmicb.2019.02533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu M., Shen W., Yan X., He Q., Cai D., Chen S., Wei H., Knoshaug E.P., Zhang M., Himmel M.E., et al. Metabolic engineering of Zymomonas mobilis for anaerobic isobutanol production. Biotechnol Biofuels. 2020;13:15. doi: 10.1186/s13068-020-1654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee K.Y., Park J.M., Kim T.Y., Yun H., Lee S.Y. The genome-scale metabolic network analysis of Zymomonas mobilis ZM4 explains physiological features and suggests ethanol and succinic acid production strategies. Microb Cell Factories. 2010;9:94. doi: 10.1186/1475-2859-9-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Widiastuti H., Kim J.Y., Selvarasu S., Karimi I.A., Kim H., Seo J.S., Lee D.Y. Genome-scale modeling and in silico analysis of ethanologenic bacteria Zymomonas mobilis. Biotechnol Bioeng. 2011;108(3):655–665. doi: 10.1002/bit.22965. [DOI] [PubMed] [Google Scholar]
- 22.Goodman A.E., Rogers P.L., Skotnicki M.L. Minimal medium for isolation of auxotrophic Zymomonas mutants. Appl Environ Microbiol. 1982;44(2):496–498. doi: 10.1128/aem.44.2.496-498.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nouri H., Fouladiha H., Moghimi H., Marashi S.A. A reconciliation of genome-scale metabolic network model of Zymomonas mobilis ZM4. Sci Rep. 2020;10(1):7782. doi: 10.1038/s41598-020-64721-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seo J.S., Chong H., Park H.S., Yoon K.O., Jung C., Kim J.J., Hong J.H., Kim H., Kim J.H., Kil J.I., et al. The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nat Biotechnol. 2005;23(1):63–68. doi: 10.1038/nbt1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang S., Pappas K.M., Hauser L.J., Land M.L., Chen G.L., Hurst G.B., Pan C., Kouvelis V.N., Typas M.A., Pelletier D.A., et al. Improved genome annotation for Zymomonas mobilis. Nat Biotechnol. 2009;27(10):893–894. doi: 10.1038/nbt1009-893. [DOI] [PubMed] [Google Scholar]
- 26.Yang S., Vera J.M., Grass J., Savvakis G., Moskvin O.V., Yang Y., McIlwain S.J., Lyu Y., Zinonos I., Hebert A.S., et al. Complete genome sequence and the expression pattern of plasmids of the model ethanologen Zymomonas mobilis ZM4 and its xylose-utilizing derivatives 8b and 2032. Biotechnol Biofuels. 2018;11:125. doi: 10.1186/s13068-018-1116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brettin T., Davis J.J., Disz T., Edwards R.A., Gerdes S., Olsen G.J., Olson R., Overbeek R., Parrello B., Pusch G.D., et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sayers E.W., Beck J., Brister J.R., Bolton E.E., Canese K., Comeau D.C., Funk K., Ketter A., Kim S., Kimchi A., et al. Database resources of the National Center for biotechnology information. Nucleic Acids Res. 2020;48(D1):D9–D16. doi: 10.1093/nar/gkz899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caspi R., Billington R., Keseler I.M., Kothari A., Krummenacker M., Midford P.E., Ong W.K., Paley S., Subhraveti P., Karp P.D. The MetaCyc database of metabolic pathways and enzymes - a 2019 update. Nucleic Acids Res. 2020;48(D1):D445–D453. doi: 10.1093/nar/gkz862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norsigian C.J., Pusarla N., McConn J.L., Yurkovich J.T., Drager A., Palsson B.O., King Z. BiGG Models 2020: multi-strain genome-scale models and expansion across the phylogenetic tree. Nucleic Acids Res. 2020;48(D1):D402–D406. doi: 10.1093/nar/gkz1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang A., Jeske L., Ulbrich S., Hofmann J., Koblitz J., Schomburg I., Neumann-Schaal M., Jahn D., Schomburg D. BRENDA, the ELIXIR core data resource in 2021: new developments and updates. Nucleic Acids Res. 2021;49(D1):D498–D508. doi: 10.1093/nar/gkaa1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beber M.E., Gollub M.G., Mozaffari D., Shebek K.M., Flamholz A.I., Milo R., Noor E. eQuilibrator 3.0: a database solution for thermodynamic constant estimation. Nucleic Acids Res. 2022;50(D1):D603–D609. doi: 10.1093/nar/gkab1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brenac L., Baidoo E.E.K., Keasling J.D., Budin I. Distinct functional roles for hopanoid composition in the chemical tolerance of Zymomonas mobilis. Mol Microbiol. 2019;112(5):1564–1575. doi: 10.1111/mmi.14380. [DOI] [PubMed] [Google Scholar]
- 34.Luo J., Yuan Q., Mao Y., Wei F., Zhao J., Yu W., Kong S., Guo Y., Cai J., Liao X., et al. Reconstruction of a genome-scale metabolic network for Shewanella oneidensis MR-1 and analysis of its metabolic potential for bioelectrochemical systems. Front Bioeng Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.913077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma H., Zeng A.P. Reconstruction of metabolic networks from genome data and analysis of their global structure for various organisms. Bioinformatics. 2003;19(2):270–277. doi: 10.1093/bioinformatics/19.2.270. [DOI] [PubMed] [Google Scholar]
- 36.Bochner B., Gomez V., Ziman M., Yang S., Brown S.D. Phenotype microarray profiling of Zymomonas mobilis ZM4. Appl Biochem Biotechnol. 2010;161(1–8):116–123. doi: 10.1007/s12010-009-8842-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ebrahim A., Lerman J.A., Palsson B.O., Hyduke D.R. COBRApy: COnstraints-based reconstruction and analysis for Python. BMC Syst Biol. 2013;7:74. doi: 10.1186/1752-0509-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lieven C., Beber M.E., Olivier B.G., Bergmann F.T., Ataman M., Babaei P., Bartell J.A., Blank L.M., Chauhan S., Correia K., et al. MEMOTE for standardized genome-scale metabolic model testing. Nat Biotechnol. 2020;38(3):272–276. doi: 10.1038/s41587-020-0446-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobson T.B., Adamczyk P.A., Stevenson D.M., Regner M., Ralph J., Reed J.L., Amador-Noguez D. 2H and 13C metabolic flux analysis elucidates in vivo thermodynamics of the ED pathway in Zymomonas mobilis. Metab Eng. 2019;54:301–316. doi: 10.1016/j.ymben.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 40.Wang X., He Q.N., Yang Y.F., Wang J.W., Haning K., Hu Y., Wu B., He M.X., Zhang Y.P., Bao J., et al. Advances and prospects in metabolic engineering of Zymomonas mobilis. Metab Eng. 2018;50:57–73. doi: 10.1016/j.ymben.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 41.Kalnenieks U., Balodite E., Rutkis R. Metabolic engineering of bacterial respiration: high vs. low P/O and the case of Zymomonas mobilis. Front Bioeng Biotechnol. 2019;7:327. doi: 10.3389/fbioe.2019.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalnenieks U., Galinina N., Strazdina I., Kravale Z., Pickford J.L., Rutkis R., Poole R.K. NADH dehydrogenase deficiency results in low respiration rate and improved aerobic growth of Zymomonas mobilis. Microbiology (Read) 2008;154(Pt 3):989–994. doi: 10.1099/mic.0.2007/012682-0. [DOI] [PubMed] [Google Scholar]
- 43.Martien J.I., Hebert A.S., Stevenson D.M., Regner M.R., Khana D.B., Coon J.J., Amador-Noguez D. Systems-level analysis of oxygen exposure in Zymomonas mobilis: implications for isoprenoid production. mSystems. 2019;4(1):1–23. doi: 10.1128/mSystems.00284-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ong W.K., Courtney D.K., Pan S., Andrade R.B., Kiley P.J., Pfleger B.F., Reed J.L. Model-driven analysis of mutant fitness experiments improves genome-scale metabolic models of Zymomonas mobilis ZM4. PLoS Comput Biol. 2020;16(8) doi: 10.1371/journal.pcbi.1008137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Felczak M.M., TerAvest M.A. Zymomonas mobilis ZM4 utilizes an NADP(+)-dependent acetaldehyde dehydrogenase to produce acetate. J Bacteriol. 2022 doi: 10.1128/jb.00563-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Motamedian E., Saeidi M., Shojaosadati S.A. Reconstruction of a charge balanced genome-scale metabolic model to study the energy-uncoupled growth of Zymomonas mobilis ZM1. Mol Biosyst. 2016;12(4):1241–1249. doi: 10.1039/c5mb00588d. [DOI] [PubMed] [Google Scholar]
- 47.Yan X., Wang X., Yang Y., Wang Z., Zhang H., Li Y., He Q., Li M., Yang S. Cysteine supplementation enhanced inhibitor tolerance of Zymomonas mobilis for economic lignocellulosic bioethanol production. Bioresour Technol. 2022 doi: 10.1016/j.biortech.2022.126878. [DOI] [PubMed] [Google Scholar]
- 48.Fuhrer T., Fischer E., Sauer U. Experimental identification and quantification of glucose metabolism in seven bacterial species. J Bacteriol. 2005;187(5):1581–1590. doi: 10.1128/JB.187.5.1581-1590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kremer T.A., LaSarre B., Posto A.L., McKinlay J.B. N2 gas is an effective fertilizer for bioethanol production by Zymomonas mobilis. Proc Natl Acad Sci U S A. 2015;112(7):2222–2226. doi: 10.1073/pnas.1420663112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shui Z., Wang J., Qin H., Wu B., Tan F., Mingxiong H. Construction and preliminary fermentation of succinate-producing recombinant ethanologenic Zymomonas mobilis. Chin J Appl Environ Biol. 2015;21:657–664. [Google Scholar]
- 51.Swings J., De Ley J. The biology of Zymomonas. Bacteriol Rev. 1977;41(1):1–46. doi: 10.1128/br.41.1.1-46.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Widiastuti H., Lee N.-R., Karimi I., Lee D.-Y. Genome-scale in silico analysis for enhanced production of succinic acid in Zymomonas mobilis. Processes. 2018;6(4):30. [Google Scholar]
- 53.Thoma F., Schulze C., Gutierrez-Coto C., Hadrich M., Huber J., Gunkel C., Thoma R., Blombach B. Metabolic engineering of Vibrio natriegens for anaerobic succinate production. Microb Biotechnol. 2022;15(6):1671–1684. doi: 10.1111/1751-7915.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng J., Li J., Zheng L. Achievements and perspectives in 1,4-butanediol production from engineered microorganisms. J Agric Food Chem. 2021;69(36):10480–10485. doi: 10.1021/acs.jafc.1c03769. [DOI] [PubMed] [Google Scholar]
- 55.Yim H., Haselbeck R., Niu W., Pujol-Baxley C., Burgard A., Boldt J., Khandurina J., Trawick J.D., Osterhout R.E., Stephen R., et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol. 2011;7(7):445–452. doi: 10.1038/nchembio.580. [DOI] [PubMed] [Google Scholar]
- 56.Burgard A., Burk M.J., Osterhout R., Van Dien S., Yim H. Development of a commercial scale process for production of 1,4-butanediol from sugar. Curr Opin Biotechnol. 2016;42:118–125. doi: 10.1016/j.copbio.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 57.Zhu X., Tan Z., Xu H., Chen J., Tang J., Zhang X. Metabolic evolution of two reducing equivalent-conserving pathways for high-yield succinate production in Escherichia coli. Metab Eng. 2014;24:87–96. doi: 10.1016/j.ymben.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 58.Helmy M., Smith D., Selvarajoo K. Systems biology approaches integrated with artificial intelligence for optimized metabolic engineering. Metab Eng Commun. 2020;11 doi: 10.1016/j.mec.2020.e00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.