Abstract
Background & Aims
Non-alcoholic fatty liver disease (NAFLD) is characterised by accelerated lipid deposition, aberrant inflammation, and excessive extracellular matrix production in the liver. Short of effective intervention, NAFLD can progress to cirrhosis and hepatocellular carcinoma. In the present study we investigated the involvement of the C–C motif ligand 11 (CCL11) in NAFLD pathogenesis.
Methods
NAFLD was induced by feeding mice with a high-fat high-carbohydrate diet. CCL11 targeting was achieved by genetic deletion or pharmaceutical inhibition. The transcriptome was analysed using RNA-seq.
Results
We report that CCL11 expression was activated at the transcription level by free fatty acids (palmitate) in hepatocytes. CCL11 knockdown attenuated whereas CCL11 treatment directly promoted production of pro-inflammatory/pro-lipogenic mediators in hepatocytes. Compared with wild-type littermates, CCL11 knockout mice displayed an ameliorated phenotype of NAFLD when fed a high-fat high-carbohydrate diet as evidenced by decelerated body weight gain, improved insulin sensitivity, dampened lipid accumulation, reduced immune cell infiltration, and weakened liver fibrosis. RNA-seq revealed that interferon regulatory factor 1 as a mediator of CCL11 induced changes in hepatocytes. Importantly, CCL11 neutralisation or antagonism mitigated NAFLD pathogenesis in mice. Finally, a positive correlation between CCL11 expression and NAFLD parameters was identified in human patients.
Conclusions
Our data suggest that CCL11 is a novel regulator of NAFLD and can be effectively targeted for NAFLD intervention.
Impact and implications
Non-alcoholic fatty liver disease (NAFLD) precedes cirrhosis and hepatocellular carcinoma. In this paper we describe the regulatory role of CCL11, a C–C motif ligand chemokine, in NAFLD pathogenesis. Our data provide novel insights and translational potential for NAFLD intervention.
Keywords: Non-alcoholic fatty liver disease, Hepatocytes, Transcriptional regulation, Chemokine, Inflammation
Graphical abstract
Highlights
-
•
CCL11 levels are elevated in hepatocytes by pro-NAFLD stimuli.
-
•
CCL11 deletion attenuates NAFLD in mice.
-
•
CCL11 neutralisation attenuates NAFLD in mice.
-
•
Antagonism of CCL11 receptor CCR3 attenuates NAFLD in mice.
-
•
Correlation between CCL11 and NAFLD markers is identified in human patients.
Introduction
Owing to combinatorial changes in dietary choices and lifestyles, non-alcoholic fatty liver disease (NAFLD) has recently become the most important cause for cirrhosis and hepatocellular carcinoma worldwide.[1], [2], [3] It is estimated that NAFLD affects more than one-third of the global population with ∼1% year-to-year increase in new diagnoses in the past decade.4 A host of risk factors, including obesity, diabetes, hyperlipidaemia, and sleep apnoea, have been identified for NAFLD.[5], [6], [7] NAFLD encompasses a continuum of pathologies, which typically manifests as aberrant accumulation of lipid droplets, extensive inflammatory foci, and widespread interstitial fibrosis in the liver.8 There are currently no effective interventional regimens specifically designed for NAFLD although several are being tested in clinical trials.[9], [10], [11], [12] Short of effective intervention, 5–12% of all patients with NAFLD may eventually proceed to develop some form of end-stage liver diseases (e.g. cirrhosis), in which case liver transplant is indicated.13 The economic burden for patients with NAFLD is significant and reaches 103 billion dollars in the USA alone.14 Studies aimed at unravelling the mechanisms of NAFLD pathogenesis further the understanding of this highly prevalent disease and allow identification of key molecules involved in NAFLD development for drug targeting.
It is believed that several inter-dependent pathophysiological events collectively drive NAFLD pathogenesis. Excessive influx of nutrients plus accelerated de novo lipid synthesis in hepatocytes leads to fat accumulation in the liver. Indeed, pro-lipogenic transcription factors such as sterol response element binding protein (SREBP) are activated in the liver during NAFLD pathogenesis in rodents and humans contributing to elevated production of triglycerides and cholesterol.15 Consistently, SREBP overexpression promotes whereas SREBP deletion ameliorates NAFLD development in different animal models.[16], [17], [18], [19] Dysregulated hepatic metabolism, either independently or in conjunction with other factors (e.g. unfolded protein stress response or reactive oxygen species), cultivates a pro-inflammatory environment by boosting the production of inflammatory mediators in hepatocytes and by attracting immune infiltrates from the circulation.20 The ensuing ‘meta-inflammation’ triggers a variety of death programmes in hepatocytes but further skews hepatic metabolism thereby creating a vicious feedback loop that perpetuates the loss of key liver functions.21 Accompanying these pathophysiological events there is extensive fibrosis in the liver with the emergence of a specialised cell type (myofibroblast) that mediates the synthesis and deposition of extracellular matrix proteins. Liver fibrosis during NAFLD pathogenesis, often serving as a benchmark against which the efficacies of anti-NAFLD therapeutic regimens are measured, exacerbates the disruption of liver architecture and pivots NAFLD towards more malignant stages.22
The C–C motif ligand (CCL) chemokines represent a large family of immunomodulatory proteins that play important roles in both the maintenance and disruption of hepatic homeostasis.23 Contribution to NAFLD pathogenesis has been documented for several different CCL proteins. For instance, CCL2 (also known as MCP1) levels are upregulated in animal models of NAFLD and in NAFLD patients.24,25 Concordantly, a milder NAFLD phenotype, including diminished inflammation and fibrogenesis, was observed in CCL2 knockout (KO) mice compared with wild-type (WT) mice when fed a methionine-and-choline deficient (MCD) diet.26 Mice with deficiency in CCL3 also display an amelioration of NAFLD when challenged with a high-fat high-cholesterol (HFHC) diet probably attributable to an altered macrophage phenotype.27
C–C motif ligand 11 (CCL11), also known as eotaxin, is a polypeptide of 73 amino acids initially isolated and characterised by the Williams laboratory as a chemoattractant for eosinophils.28 Elevated CCL11 levels have been observed in both chronic29,30 and acute[31], [32], [33], [34] liver diseases in humans and animal models. Of note, CCL11 levels were reported to correlate with obesity and metabolic syndrome in humans in a study led by Ballantyne35 and a study led by Powell36 but a study led by Kolb37 failed to identify such correlation. More recently, Wong et al. have reported a positive correlation between plasma CCL11 levels and severity of steatosis in a cohort of NAFLD patients.38 In the present study we investigated the involvement of CCL11 in NAFLD pathogenesis, the underlying mechanism, and translational potential. Our data show that CCL11 is essential for NAFLD pathogenesis and can be effectively targeted for NAFLD intervention.
Materials and methods
Animals
All the animal experiments were reviewed and approved by the Nanjing University Ethics Committee on Humane Treatment of Experimental Animals. The mice were maintained in a specific-pathogen free environment with 12-h light/dark cycles and access to food and water ad libitum. The Ccl11-/- mice have been described previously.39 NAFLD was induced in mice by one of the following feeding schemes: (1) C57/B6 mice on a HFHC diet (D12492, Research Diets, New Brunswick, NJ, USA) for 12 consecutive wk; (2) Apoe-/- mice on a Western diet (D12079B, Research Diets) for 12 wk; (3) db/db mice on an MCD diet (A02082002BR, Research Diets) for 4 wk. In certain experiments, the mice were injected three times a wk with either a CCL11 neutralising antibody (R&D, MAB420, 20 μg/kg), or a chemokine receptor 3 (CCR3) antagonist (Selleck, S0129, 50 μg/kg). For calorimetry analysis, the mice were housed in individual calorimeter chambers (Oxymax). After 24 h of acclamation, calorimetric data were collected over a period of 48 h as previously described.40,41
Cell culture, plasmids, and transient transfection
Human hepatoma cells (HepG2) were maintained in DMEM supplemented with 10% FBS (Hyclone, Marlborough, MA, USA). Primary murine hepatocytes were isolated and cultured as previously described.42 Primary human hepatocytes were purchased from Sigma. CCL11 promoter–luciferase constructs43 have been previously described. An IRF1 promoter–luciferase construct was generated by amplifying genomic DNA spanning the proximal promoter and the first exon of IRF1 gene (-1000/+10) and ligating into a pGL3-basic vector (Promega). Truncation mutants were made using a QuikChange kit (Thermo Fisher Scientific, Waltham, MA, USA) and verified by direct sequencing. Small-interfering RNAs were purchased from Dharmacon. Transient transfections were performed with Lipofectamine 2000 (Invitrogen, Waltham, MA, USA). Luciferase activities were assayed 24–48 h after transfection using a luciferase reporter assay system (Promega) as previously described.44
Enzyme-linked immunosorbent assay
Secreted protein levels were examined by ELISA as previously described using commercially available kits (for murine CCL11, R&D, catalogue# MME00; for human CCL11, R&D, catalogue# DY320; for murine IL-1β, R&D, catalogue# MLB00C; for murine IL-6, R&D, catalogue# M6000B; for murine MCP-1, R&D, catalogue# MJE00B) according to vendor’s recommendations. Data were normalised by cell number.
RNA isolation and real-time PCR
RNA was extracted with the RNeasy RNA isolation kit (Qiagen) as previously described.45,46 Reverse transcriptase reactions were performed using a SuperScript First-strand Synthesis System (Invitrogen). Real-time PCR reactions were performed on an ABI Prism 7500 system. The primers are listed in Table S1. Ct values of target genes were normalised to the Ct values of housekeeping control gene (18s, 5′-CGCGGTTCTATTTTGTTGGT-3′ and 5′-TCGTCTTCGAAACTCCGACT-3′ for both human and mouse genes) using the ΔΔCt method and expressed as relative mRNA expression levels compared with the control group which is arbitrarily set as 1.
Protein extraction and Western blot
Whole cell lysates were obtained by re-suspending cell pellets in RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Triton X-100) with freshly added protease and phosphatase inhibitors (Roche) as previously described.47 Antibodies used for Western blotting are listed in Table S2. For densitometrical quantification, densities of target proteins were normalised to those of β-actin. Data are expressed as relative protein levels compared with the control group which is arbitrarily set as 1.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described before.48,49 Chromatin was cross-linked with 1% formaldehyde for 8 min at room temperature, and then sequentially washed with ice-cold PBS, Solution I (10 mM HEPES, pH 7.5, 10 mM EDTA, 0.5 mM EGTA, 0.75% Triton X-100), and Solution II (10 mM HEPES, pH 7.5, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA). Cells were incubated in lysis buffer (150 mM NaCl, 25 mM Tris pH 7.5, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholate) supplemented with protease inhibitor tablet. DNA was fragmented into 500-bp pieces using a Branson 250 sonicator. Aliquots of lysates containing 100 μg of protein were used for each immunoprecipitation reaction with indicated antibodies followed by adsorption to protein A/G PLUS-agarose beads (Santa Cruz Biotechnology). Precipitated DNA-protein complexes were washed sequentially with RIPA buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, 1 mM EDTA), high-salt buffer (50 mM Tris, pH 8.0, 500 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, 1 mM EDTA), LiCl buffer (50 mM Tris, pH 8.0, 250 mM LiCl, 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, 1 mM EDTA), and TE buffer (10 mM Tris, 1 mM EDTA pH 8.0), respectively. DNA-protein cross-linking was reversed by heating the samples to 65 °C overnight. Proteins were digested with proteinase K (Sigma), and DNA was phenol/chloroform-extracted and precipitated by 100% ethanol. Precipitated genomic DNA was amplified by real-time PCR using the primers listed in Table S2. A total of 10% of the starting material is also included as the input.
Human NASH biopsy specimens
Liver biopsies were collected from patients with non-alcoholic steatohepatitis (NASH) who were referred to the First People's Hospital of Changzhou. Control liver samples were collected from donors without NASH but who were deemed unsuitable for transplantation. Written informed consent was obtained from individuals or families of liver donors. All procedures that involved human samples were approved by the Ethics Committee of the First People's Hospital of Changzhou and adhered to the principles outlined in the Declaration of Helsinki. Patient information is summarised in Table S3.
Glucose tolerance assays
Glucose tolerance assays were performed as previously described.[50], [51], [52] For the glucose tolerance test (GTT), mice fasted overnight were injected intraperitoneally with 2 g/kg glucose and blood samples were taken at the indicated intervals. For the insulin tolerance test (ITT), mice were fasted for 6 h and injected intraperitoneally with 0.75 IU/kg soluble insulin and blood samples were taken at the indicated intervals. For the pyruvate tolerance test (PTT), mice were fasted for 6 h and injected intraperitoneally with 2.5 g/kg pyruvate dissolved in PBS and blood samples were taken at the indicated intervals. Blood glucose was measured using an Accu-Chek compact glucometer (Roche).
Histology
Histological analyses were performed essentially as described before.53 Pictures were taken using an Olympus IX-70 microscope (Center Valley, PA, USA). Quantifications were performed with Image J. For each mouse, at least three slides were stained and at least five different fields were analysed for each slide.
RNA sequencing and data analysis
RNA-seq was performed as previously described.54 Total RNA was extracted using the TRIzol reagent according to the manufacturer’s protocol. RNA purity and quantification were evaluated using the NanoDrop 2000 spectrophotometer (Thermo Scientific). RNA integrity was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Then the libraries were constructed using TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer's instructions and sequenced on an Illumina HiSeq X Ten platform and 150-bp paired-end reads were generated. Raw data (raw reads) of fastq format were firstly processed using Trimmomatic and the low-quality reads were removed to obtain the clean reads. The clean reads were mapped to the mouse genome (Mus_musculus.GRCm38.99) using HISAT2. FPKM of each gene was calculated using Cufflinks, and the read counts of each gene were obtained by HTSeqcount. Differential expression analysis was performed using the DESeq (2012) R package (R Foundation for Statistical Computing, Vienna, Austria). A p value <0.05 and fold change >2 or fold change <0.5 was set as the threshold for significant differential expression. Hierarchical cluster analysis of differentially expressed genes (DEGs) was performed to demonstrate the expression pattern of genes in different groups and samples. Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of DEGs were performed, respectively, using R based on the hypergeometric distribution.
Statistical analysis
For comparison between two groups, a two-tailed t test was performed. For comparison among three or more groups, one-way ANOVA or two-way ANOVA with post-hoc Tukey analyses were performed using an SPSS package (IBM; Armonk, NY, USA). The assumptions of normality were checked using Shapiro-Wilks test and equal variance was checked using Levene’s test; both were satisfied. Values of p <0.05 were considered statistically significant (∗). All in vitro experiments were repeated at least three times and three replicates were estimated to provide 80% power.
Results
CCL11 expression was upregulated by NF-κB/RelA in hepatocytes by pro-NAFLD stimuli in vivo and in vitro
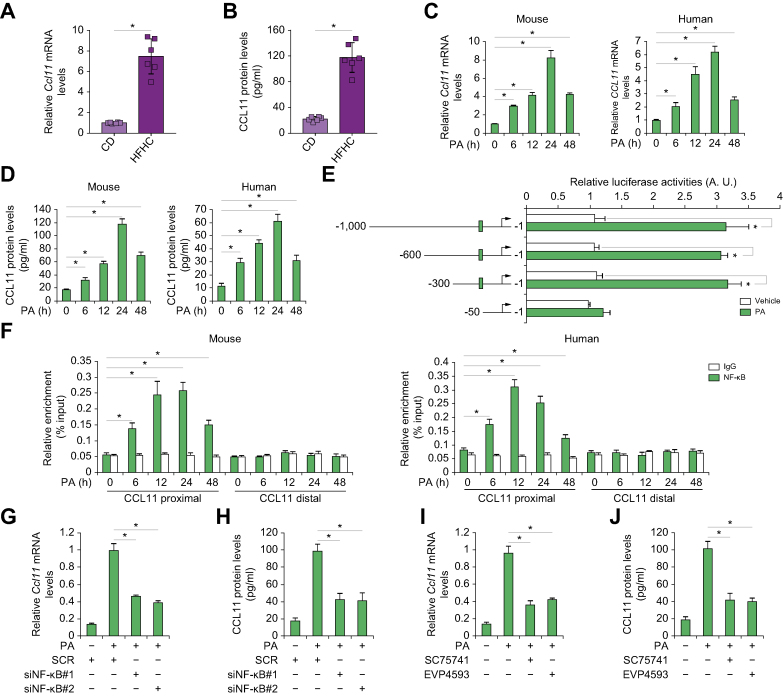
We first sought to determine whether and, if so, how CCL11 expression might be altered by pro-NAFLD stimuli. In a classic animal model of NAFLD in which 8-wk-old male C57/B6 mice were fed a HFHC or a control diet (CD) for 12 wk, it was discovered that hepatocytes isolated from the HFHC-fed mice expressed much higher levels of CCL11, as measured by qPCR and ELISA, than the CD-fed mice (Fig. 1A and B). Of note, CCL11 expression was also upregulated in hepatic stellate cells and Kupffer cells isolated from the HFHC-fed mice than from the CD-fed mice although the level of augmentation was not as high as it was in hepatocytes (Fig. S1). Similarly, in two alterative NAFLD models, one in which 8-wk-old male Apoe-/- mice were fed a Western diet for 12 wk (Fig. S2) and the other in which 8-wk-old male db/db mice were fed an MCD diet for 4 wk (Fig. S3), it was confirmed that significantly more CCL11 was produced in the hepatocytes from the NAFLD mice than from the control mice. Interestingly, CCL11 expression was positively correlated with liver injury (measured by plasma alanine aminotransferase [ALT] levels), steatosis (hepatic triglyceride levels as a proxy), and inflammation (determined by hepatic tumour necrosis factor-alpha [TNF-α] expression) in all three NAFLD models (Fig. S4).
Fig. 1.
CCL11 expression was upregulated by NF-κB/RelA in hepatocytes by pro-NAFLD stimuli in vivo and in vitro.
(A, B) C57/BL6 mice were fed a high-fat high carbohydrate diet (HFHC) or a control diet (CD) for 12 wk. Hepatocytes were isolated and CCL11 expression was examined by qPCR and ELISA. N = 6 mice for each group. Data are expressed as mean ± SD ∗p <0.05, two-tailed Student t test. (C, D) Primary murine and human hepatocytes were treated with or without palmitate (PA, 0.2 mM) and harvested at indicated time points. CCL11 expression was examined by qPCR and ELISA. N = 3 biological replicates. Data are expressed as mean ± SD ∗p <0.05, one-way ANOVA with the post-hoc Scheffe test. (E) CCL11 promoter–luciferase constructs were transfected into HepG2 cells followed by treatment with PA (0.2 mM) for 24 h. Luciferase constructs were normalised by protein concentration and green fluorescence protein fluorescence. N = 3 biological replicates. Data are expressed as mean ± SD ∗p <0.05, two-tailed Student t test. (F) Primary murine and human hepatocytes were treated with or without PA (0.2 mM) and harvested at indicated time points. ChIP assays were performed with anti–NF–κB or IgG. N = 3 biological replicates. Data are expressed as mean ± SD ∗p <0.05, one-way ANOVA with the post-hoc Scheffe test. (G, H) Primary murine hepatocytes were transfected with siRNA targeting NF-κB or scrambled siRNA (SCR) followed by treatment with PA (0.2 mM) for 24 h. CCL11 expression was examined by qPCR and ELISA. N = 3 biological replicates. Data are expressed as mean ± SD ∗p <0.05, one-way ANOVA with the post-hoc Scheffe test. (I, J) Primary murine hepatocytes were treated with PA (0.2 mM) in the presence or absence of SC75741 (1 μM) or EVP4593 (0.5 μM) for 24 h. CCL11 expression was examined by qPCR and ELISA. N = 3 biological replicates. Data are expressed as mean ± SD ∗p <0.05, one-way ANOVA with the post-hoc Scheffe test. CCL11, C–C motif ligand 11; ChIP, chromatin immunoprecipitation; NAFLD, non-alcoholic fatty acid disease; NF-κB, nuclear factor kappa B.
Next, primary murine and human hepatocytes were exposed to palmitate (PA), a well-documented risk factor for NAFLD.20 PA treatment led to robust induction of CCL11 synthesis at both mRNA (Fig. 1C) and protein (Fig. 1D) levels. The following experiments were performed to determine the transcriptional mechanism whereby pro-NAFLD stimuli regulate CCL11 expression. Reporter assay (Fig. 1E) showed that PA treatment augmented the activity of CCL11 promoter to the extent comparable with the changes in CCL11 mRNA/protein levels. However, when serial deletions introduced to the CCL11 promoter extended beyond -300 relative to the transcription start site, PA treatment failed to elicit an augmentation of the CCL11 promoter activity (Fig. 1E). Because a conserved NF-κB site could be located between -300 and -50 of the CCL11 promoter, we hypothesised that NF-κB might mediate CCL11 transactivation by PA treatment. A ChIP assay showed that occupancy of the CCL11 promoter by NF-κB was enhanced when primary hepatocytes were exposed to PA (Fig. 1F). On the contrary, NF-κB depletion by siRNAs (Fig. 1G and H) or inhibition by small-molecule compounds (Fig. 1I and J) abrogated CCL11 induction by PA treatment in hepatocytes. Because signalling triggered by pathogen-associated molecular patterns (PAMPs) plays a key role in NAFLD pathogenesis, the relevance of lipopolysaccharide (LPS) in CCL11 regulation in hepatocytes was evaluated. Similar to PA treatment, LPS treatment led to robust recruitment of NF-κB to the CCL11 promoter (Fig. S5A). On the contrary, NF-κB knockdown abrogated CCL11 induction by LPS in primary murine hepatocytes (Fig. S5B and C). Together, these data suggest that CCL11, possibly activated at the transcriptional level by NF-κB in hepatocytes, might be a pathogenic factor for NAFLD.
CCL11 promotes a pro-inflammatory/pro-lipogenic phenotype of hepatocytes in vitro
The next set of experiments was performed to determine the effect of CCL11 on hepatocytes. To this end, primary hepatocytes were isolated from CCL11 KO mice and control (WT) mice. When the cells were exposed to palmitate, significant upregulation of pro-inflammatory and pro-lipogenic mediators was observed in WT cells but less markedly in KO cells as measured by qPCR (Fig. 2A), ELISA (Fig. 2B), and Western blotting (Fig. 2C). Oil Red O staining detected fewer lipid droplets in the KO cells than in the WT cells exposed to PA (Fig. 2D). On the contrary, recombinant CCL11 (rCCL11) directly stimulated expression of pro-inflammatory and pro-lipogenic genes (Fig. 2E–G) and promoted lipid accumulation (Fig. 2H).
Fig. 2.
CCL11 promotes a pro-inflammatory/pro-lipogenic phenotype of hepatocytes in vitro.
(A–D) Primary murine hepatocytes were treated with recombinant CCL11 (50 ng/ml) for 24 h. Gene expression levels were examined by qPCR, ELISA, and Western blotting. Lipid accumulation was measured by Oil Red O staining. n = 3 biological replicates. Data are expressed as mean ± SD ∗p <0.05, one-way ANOVA with the post-hoc Scheffe test. (E–H) Primary hepatocytes were isolated from wild-type (WT) and CCL11 knockout (KO) mice and treated with or without PA (0.2 mM) for 24 h. Gene expression levels were examined by qPCR, ELISA, and Western blotting. Lipid accumulation was measured by Oil Red O staining. n = 3 biological replicates. Data are expressed as mean ± SD ∗p <0.05, one-way ANOVA with the post-hoc Scheffe test. (I–L) Primary murine hepatocytes were treated with PA (0.2 mM) in the presence or absence of SB297006 (5 μM) for 24 h. Gene expression levels were examined by qPCR, ELISA, and Western blotting. Lipid accumulation was measured by Oil Red O staining. n = 3 biological replicates. Data are expressed as mean ± SD ∗p <0.05, one-way ANOVA with the post-hoc Scheffe test. CCL11, C–C motif ligand 11; PA, palmitate.
CCL11 typically exerts its biological functions via binding to the trans-membrane receptor CCR3. Blockade of CCR3 engagement by a small-molecule antagonist (SB297006) attenuated the upregulation of pro-inflammatory and pro-lipogenic genes (Fig. 2I–K) and intracellular lipid accumulation (Fig. 2L). Likewise, silencing of CCR3 expression by siRNAs led to the same conclusion that intercellular CCL11-CCR3 signalling might be essential for a pro-inflammatory/pro-lipogenic phenotype in hepatocytes (Fig. S6).
CCL11 deficiency mitigates NAFLD pathogenesis in mice
To determine the effect of CCL11 on NAFLD pathogenesis in vivo, CCL11 KO mice and WT mice were fed a HFHC diet for 12 wk. HFHC feeding increased the body weight significantly in both WT and KO mice whereas the WT mice gained BW faster than the KO mice (Fig. 3A). Notably, no difference in body weight was observed between the WT mice and the KO mice fed the control diet (CD). Calorimetric analysis indicated that whereas food consumption (Fig. S7A) and locomotor activity (Fig. S7B) were comparable between the WT and KO mice, energy expenditure (Fig. S7C) was significantly higher in the KO mice than in the WT mice. Next, GTT (Fig. 3B), ITT (Fig. 3C), and PTT (Fig. 3D) were performed to determine insulin resistance (IR), a key pathophysiological characteristic of NAFLD in humans. Results of all three tests indicated that CCL11 deficiency significantly improved insulin sensitivity in mice. Plasma ALT (Fig. 3E) and aspartate aminotransferase (AST) (Fig. 3F) values indicated that steatotic injuries were ameliorated in the KO mice compared with the WT mice. Levels of triglyceride and cholesterol in the plasma (Fig. 3G and H) and in the liver (Fig. 3I and J) were significantly lower in the KO mice than in the WT mice. Histological examination of the liver pathology revealed that there was a reduction in lipid accumulation, immune infiltrates, and interstitial fibrosis in the KO mice compared with the WT mice (Fig. 3K). Pro-inflammatory mediators, pro-lipogenic molecules, and pro-fibrogenic molecules were collectively downregulated in the KO livers compared with the WT livers (Fig. 3L). Finally, hydroxylproline levels were lower in the KO livers than in the WT livers (Fig. 3M) confirming attenuation of liver fibrosis owing to CCL11 deletion. Combined, these data demonstrate that CCL11 might play an essential role for NAFLD pathogenesis.
Fig. 3.
CCL11 deletion attenuates NAFLD in mice.
Wild-type (WT) and CCL11 knockout (KO) mice were fed a HFHC or a control diet (CD) for 12 wk. (A) Body weight. (B) Glucose tolerance test (GGT). (C) Insulin tolerance test (ITT). (D) Pyruvate tolerance test (PTT). (E) Plasma ALT levels. (F) Plasma AST levels. (G) Plasma triglyceride levels. (H) Plasma total cholesterol levels. (I) Hepatic triglyceride levels. (J) Hepatic total cholesterol levels. (K) Paraffin sections were stained with H&E, Oil Red O, Picrosirius Red, and anti-CD68. The NAS score was calculated based on pathological evaluations of stained slides. (L) Gene expression levels were examined by qPCR. (M) Hepatic hydroxylproline levels. N = 8–10 mice for each group. Data are expressed as mean ± SD ∗p <0.05, two-tailed Student t test. ALT, alanine aminotransferase; AST, aspartate aminotransferase; CCL11, C–C motif ligand 11; HFHC, high-fat high-carbohydrate; NAFLD, non-alcoholic fatty acid disease; NAS, NAFLD activity score.
RNA-seq identifies IRF1 as a downstream target of CCL11 signalling
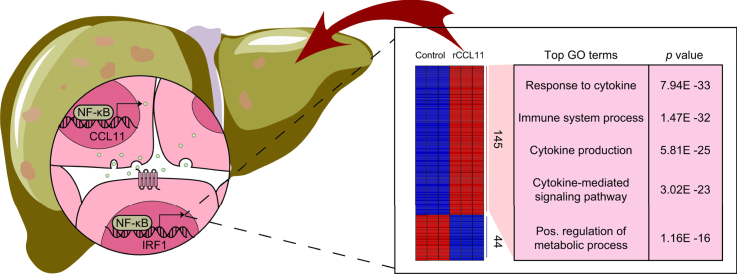
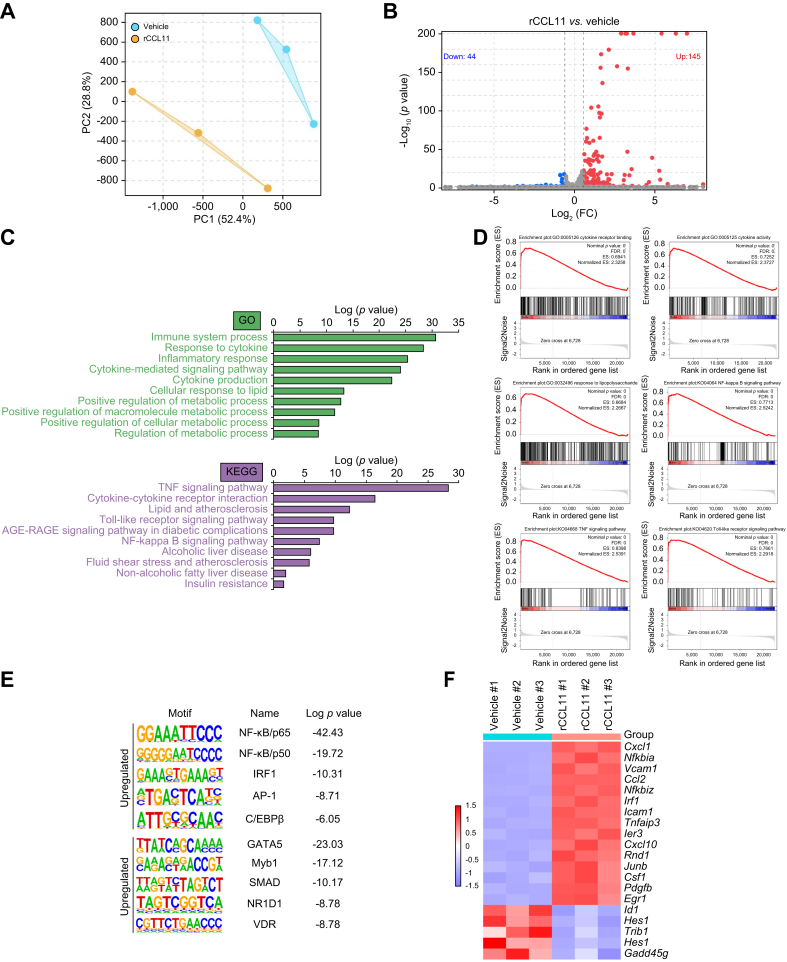
To understand the mechanism whereby CCL11 might contribute to the alteration of hepatocyte phenotype, RNA-seq was performed to identify potential downstream target(s) of CCL11. Stimulation with recombinant CCL11 markedly altered the transcriptome of primary murine hepatocytes (Fig. 4A). More genes were upregulated (145) than downregulated (44) in response to CCL11 treatment using 1.5-fold change and p <0.05 as threshold (Fig. 4B). GO and KEGG analysis suggested that DEGs were mostly involved in inflammation/immune response and metabolism (Fig. 4C). Gene Set Enrichment Analysis confirmed that CCL11 stimulation was positively correlated with alterations of genes related to the immune response (Fig. 4D). Hypergeometric optimisation of motif enrichment analysis pointed to several transcription factors, including NF-κB and AP-1, the activities of which were influenced by CCL11 (Fig. 4E). Interferon regulatory factor 1 (IRF1) was found to be the top-ranked transcription factor among the most significantly upregulated genes by CCL11 stimulation (Fig. 4F). We therefore focused on the IRF1 for the remainder of the study.
Fig. 4.
RNA-seq identifies IRF1 as a downstream target of CCL11 signalling.
Murine primary hepatocytes were treated with recombinant CCL11 for 24 h. RNA-seq was performed as described in the Materials and methods. (A) PCA plot. (B) Volcano plot. (C) GO and KEGG analysis. (D) Gene Set Enrichment |Analysis. (E) Hypergeometric optimisation of motif enrichment analysis. (F) Heat map of differentially expressed genes. CCL11, C–C motif ligand 11; GO, gene ontology; IRF1, interferon regulatory factor 1; KEGG, Kyoto Encyclopedia of Genes and Genomes; PCA, principal component analysis; TNF, tumour necrosis factor.
CCL11 regulates IRF1 transcription through NF-κB
Quantitative PCR (Fig. 5A) and Western blotting (Fig. 5B) confirmed that exposure of hepatocytes to recombinant CCL11 stimulated IRF1 expression. Elevation of IRF1 expression was also observed in the livers of the HFHC-fed mice compared with the CD-fed mice (Fig. S8). On the contrary, CCL11 deficiency dampened the upregulation of IRF1 expression by PA treatment in primary hepatocytes (Fig. 5C and D) and by HFHC feeding in murine livers (Fig. 5E and F). Additional experiments were designed to determine the mechanism whereby CCL11 might regulate IRF1 transcription. A reporter assay indicated that CCL11 could stimulate the IRF1 promote activity, which relied on a proximal κB motif (Fig. 5G). A ChIP assay confirmed that NF-κB was recruited to the proximal, but not the distal, IRF1 promoter in response to CCL11 stimulation (Fig. 5H). Either NF-κB knockdown by siRNAs (Fig. 5I and J) or inhibition by small-molecule compounds (Fig. 5K and L) blocked induction of IRF1 expression by CCL11.
Fig. 5.
CCL11 regulates IRF1 transcription through NF-κB.
(A, B) Primary murine hepatocytes were treated with or without recombinant CCL11 for 24 h. IRF1 expression levels were examined by qPCR and Western blotting. N = 3 biological replicates. Data are expressed as mean ± SD ∗p <0.05, two-tailed Student t test. (C, D) Primary hepatocytes were isolated from wild-type (WT) and CCL11 knockout (KO) mice and treated with or without PA (0.2 mM) for 24 h. IRF1 expression levels were examined by qPCR and Western blotting. N = 3 biological replicates. Data are expressed as mean ± SD ∗p <0.05, one-way ANOVA with the post-hoc Scheffe test. (E, F) WT and CCL11 KO mice were fed a HFHC or a control diet (CD) for 12 wk. IRF1 expression levels were examined by qPCR and Western blotting. N = 4–6 mice for each group. Data are expressed as mean ± SD ∗p <0.05, two-tailed Student t test. (G) IRF1 promoter–luciferase constructs were transfected into HepG2 cells followed by treatment with PA (0.2 mM) for 12 h. Luciferase constructs were normalised by protein concentration and green fluorescent protein fluorescence. N = 3 biological replicates. Data are expressed as mean ± SD ∗p <0.05, two-tailed Student t test. (H) Primary murine hepatocytes were treated with or without CCL11 and harvested at indicated time points. ChIP assays were performed with anti–NF–κB or IgG. N = 3 biological replicates. Data are expressed as mean ± SD ∗p <0.05, one-way ANOVA with the post-hoc Scheffe test. (I, J) Primary murine hepatocytes were transfected with siRNA targeting NF-κB or scrambled siRNA (SCR) followed by treatment with CCL11 for 24 h. IRF1 expression was examined by qPCR and Western blotting. N = 3 biological replicates. Data are expressed as mean ± SD ∗p <0.05, one-way ANOVA with the post-hoc Scheffe test. (K, L) Primary murine hepatocytes were treated with CCL11 in the presence or absence of SC75741 (1 μM) or EVP4593 (0.5 μM) for 24 h. IRF1 expression was examined by qPCR and Western blotting. N = 3 biological replicates. Data are expressed as mean ± SD ∗p <0.05, one-way ANOVA with the post-hoc Scheffe test. CCL11, C–C motif ligand 11; ChIP, chromatin immunoprecipitation; HFHC, high-fat high-carbohydrate; IRF1, interferon regulatory factor 1; NF-κB, nuclear factor kappa B; PA, palmitate.
CCR3 antagonism attenuates NAFLD in mice
We then asked whether targeting the CCL11-CCR3 axis could mitigate steatotic injury in mice. To address this issue, two different interventional regimens were devised. In the first scenario, male C57/B6 mice were fed the HFHC diet for 4 wk before i.p. injection of SB297006 was administered thrice-weekly for the duration of HFHC feeding (Fig. 6A). CCR3 blockade by SB297006 significantly decelerated BW increase in the HFHC-fed mice (Fig. 6B). Calorimetric analysis showed that CCR3 antagonism enhanced energy expenditure without altering food intake or locomotor activity in mice (Fig. S9). Tolerance tests showed that CCR3 blockade ameliorated insulin resistance (Fig. 6C–E). Steatotic injuries were less prominent in the SB-injected mice than the mock-injected mice as shown by plasma ALT levels (Fig. 6F), plasma AST levels (Fig. 6G), plasma/hepatic triglyceride levels (Fig. 6H and J), and plasma/hepatic cholesterol levels (Fig. 6I and K). H&E staining and Oil Red O staining confirmed that a less severe form of steatosis was developed as result of CCR3 antagonism (Fig. 6L). This was probably attributable to the downregulation of pro-lipogenic genes, measured by qPCR, in the liver (Fig. 6M). In addition, CD68 histochemical staining (Fig. 6L) and qPCR measurements of pro-inflammatory mediators (Fig. 6M) pointed to the dampening of hepatic inflammation in the SB-injected mice compared with the vehicle-injected mice. Further, Sirius Red staining (Fig. 6L), qPCR determination of pro-fibrogenic genes (Fig. 6M), and hydroxylproline levels (Fig. 6N) suggested that there was weakened liver fibrosis following CCR3 blockade.
Fig. 6.
CCR3 antagonism attenuates NAFLD in mice.
C57/BL6 mice were fed a HFHC diet for 12 wk to develop NAFLD. Starting at Week 4, the mice were injected i.p. three times a wk with a CCR3 antagonist (SB297006). (A) Scheme of protocol. (B) Body weight. (C) Glucose tolerance test (GGT). (D) Insulin tolerance test (ITT). (E) Pyruvate tolerance test (PTT). (F) Plasma ALT levels. (G) Plasma AST levels. (H) Plasma triglyceride levels. (I) Plasma total cholesterol levels. (J) Hepatic triglyceride levels. (K) Hepatic total cholesterol levels. (L) Paraffin sections were stained with H&E, Oil Red O, Picrosirius Red, and anti-CD68. The NAS was calculated based on pathological evaluations of stained slides. (M) Gene expression levels were examined by qPCR. (N) Hepatic hydroxylproline levels. N = 10 mice for each group. Data are expressed as mean ± SD ∗p <0.05, two-tailed Student t test. ALT, alanine aminotransferase; AST, aspartate aminotransferase; CCR3, chemokine receptor 3; HFHC, high-fat high-carbohydrate; NAFLD, non-alcoholic fatty acid disease; NAS, NAFLD activity score.
In an alternative experimental scheme, SB297006 administration was started at 9 wk, instead of 4 wk, after HFHC feeding (Fig. S10A). It was observed that the body weight of the mice was not appreciably altered (Fig. S10B). However, short-term SB297006 administration did improve insulin sensitivity (Fig. S10C) and ameliorate NAFLD (Fig. S10D–J).
CCL11 neutralisation attenuates NAFLD in mice
The possibility of targeting CCL11 directly in NAFLD intervention was tested using a CCL11 neutralising antibody.55,56 Efficiency of CCL11 neutralisation was verified by ELISA (Fig. S11). Similar to the CCR3 blockade regimen, thrice-weekly injection of anti-CCL11 started at the fifth week of HFHC feeding and continued until the mice were sacrificed (Fig. 7A). CCL11 neutralisation significantly boosted insulin sensitivity in the mice as shown by GTT (Fig. 7B), ITT (Fig. 7C), and PTT (Fig. 7D) data. Biochemical assays demonstrated that blocking CCL11 could alleviate steatotic injuries in the HFHC-fed mice (Fig. 7E–J). Histological stainings (Fig. 7K), qPCR measurements of gene expression (Fig. 7L), and hydroxylproline quantification (Fig. 7M) provided supporting evidence that CCL11 neutralisation was associated with dampened hepatic inflammation, lipogenesis, and fibrogenesis.
Fig. 7.
CCL11 neutralisation attenuates NAFLD in mice.
C57/BL6 mice were fed an HFHC for 12 wk to develop NAFLD. Starting at Week 4, the mice were injected i.p. three times a wk with a CCL11-neutralising antibody. (A) Scheme of protocol. (B) Glucose tolerance test (GTT). (C) Insulin tolerance test (ITT). (D) Pyruvate tolerance test (PTT). (E) Plasma ALT levels. (F) Plasma AST levels. (G) Plasma triglyceride levels. (H) Plasma total cholesterol levels. (I) Hepatic triglyceride levels. (J) Hepatic total cholesterol levels. (K) Paraffin sections were stained with H&E, Oil Red O, Picrosirius Red, and anti-CD68. NAS score was calculated based on pathological evaluations of stained slides. (L) Gene expression levels were examined by qPCR. (M) Hepatic hydroxylproline levels. N = 10 mice for each group. Data are expressed as mean ± SD ∗p <0.05, two-tailed Student t test. ALT, alanine aminotransferase; AST, aspartate aminotransferase; CCL11, C–C motif ligand 11; NAFLD, non-alcoholic fatty acid disease; NAS, NAFLD activity score.
CCL11 levels correlate with NAFLD parameters in humans
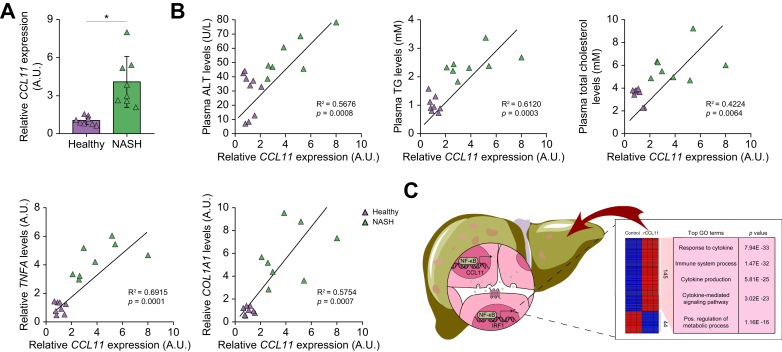
Finally, we asked whether the findings using cell culture and model animals could be extrapolated to humans. Liver specimens collected from NASH patients and healthy donors were examined for CCL11 expression by qPCR. As shown in Fig. 8A, CCL11 levels were markedly elevated in individuals diagnosed with NAFLD compared with healthy individuals. More importantly, a positive correlation was identified between CCL11 levels and liver injury (plasma ALT levels), between CCL11 levels and dyslipidaemia (plasma triglyceride and cholesterol levels), between CCL11 levels and hepatic inflammation (determined by TNF-α expression), and between CCL11 levels and liver fibrosis (determined by collagen type I expression) (Fig. 8B), suggesting that elevation of CCL11 expression might play a role in NAFLD pathogenesis in humans.
Fig. 8.
CCL11 levels correlate with NAFLD parameters in humans.
(A) CCL11 expression levels in the livers of NASH patients and healthy individuals were examined by qPCR. N = 8 cases for each group. Data are expressed as mean ± SD ∗p <0.05, two-tailed Student t test. (B) Correlations between CCL11 and plasma ALT/TG/cholesterol levels were evaluated by linear regression. (C) A schematic model depicting the major finding of the present study. ALT, alanine aminotransferase; CCL11, C–C motif ligand 11; GO, gene ontology; IRF1, interferon regulatory factor 1; NAFLD, non-alcoholic fatty acid disease; NASH, non-alcoholic steatohepatitis; NF-κB, nuclear factor kappa B; TG, triglyceride.
Discussion
Chronic inflammation is considered the linchpin of NAFLD pathogenesis linking metabolic disorders to the disruption of hepatic homeostasis.57 Here we present evidence to show that CCL11, once activated by pro-NAFLD stimuli, elicits a strong pro-inflammatory transcription programme in hepatocytes (Fig. 8C). In accordance, genetic deletion or pharmaceutical inhibition of CCL11 confers protection against NAFLD in mice. Both CCL11 and its apparent downstream effector IRF1 are activated by NF-κB, the master regulator of inflammatory response. Despite overwhelming evidence that NF-κB integrates multiple pathophyiological processes to promote NAFLD, direct targeting NF-κB for NAFLD intervention has been elusive likely because of the fact that long-term indiscriminate NF-κB inhibition could increase the susceptibility to infections and weaken cancer immune surveillance.58 Our data thus provide proof-of-concept that CCL11 may be considered as a druggable alternative to NF-κB for NAFLD intervention.
Liver fibrosis is a hallmark event in NAFLD progression and a benchmark against which interventional regimens are gauged.59 Our data show that CCL11 deletion, neutralisation, and antagonism are all met with attenuation of liver fibrosis in mice. These observations could easily be explained by the fact that CCL11 deficiency leads to reduced macrophage infiltration and restores metabolic homeostasis thereby dampening the transition of hepatic stellate cells to myofibroblasts. However, a myofibroblast-autonomous role of CCL11 that directly contributes to liver fibrosis independent of intrahepatic meta-inflammatory milieu cannot be excluded at this point. Previously, Berkman and colleagues have demonstrated that CCL11 treatment promotes fibroblast–myofibroblast transition in the lungs.60 It would be of great interest to determine whether myofibroblast-specific deletion of CCL11 in mice could impact liver fibrosis in the NAFLD models.
Our data identify IRF1 as a downstream mediator of CCL11 and suggest that IRF1 depletion may be beneficial to restore hepatic homeostasis at least in vitro. IRF1 knockout mice are viable and indistinguishable from the WT littermates under physiological conditions.61 However, the Irf1-/- mice are protected from liver injuries in several different animal models, including ischaemia–reperfusion injury62,63 and fulminant hepatitis,64,65 largely attributable to ameliorated hepatic inflammation. Targeting transcription factors for clinical intervention has proven to be a notoriously difficult but not entirely futile endeavour; successful examples include regulators of carcinogenesis p5366 and Myc.67 Future studies using new and preferably hepatocytes-specific IRF1 transgenic models would hopefully solidify its role in NAFLD pathogenesis and provide more rationale for IRF1-based therapeutic development.
Although the present study provides compelling evidence to show a causal relationship between CCL11 and NAFLD, several limitations need to be addressed. First, focus was placed on how CCL11 influenced phenotype and transcriptome of hepatocytes but not non-parenchymal cells (e.g. Kupffer cells and liver sinusoidal endothelial cells) or extrahepatic cells (e.g. adipocytes). A cell type specific role of CCL11 in NAFLD pathogenesis awaits further clarification. In addition, because CCL11 deficiency or inhibition improved insulin sensitivity, the regulatory role of CCL11 and CCR3 in extrahepatic insulin target tissues, that is, adipose tissue and skeletal muscle, deserves to be investigated. This issue is especially crucial given that our conclusion that CCL11 contributes to insulin resistance was based entirely on systemic gene deletion or pharmaceutical inhibition models. Elevated CCL11 and CCR3 levels have been detected in adipose tissues from patients with metabolic syndrome and/or obesity compared to lean healthy individuals.68 CCL11 has also been shown to regulate skeletal muscle phenotypes in both physiological and disease settings.69,70 Targeted deletion of CCL11 and/or CCR3 in adipocytes/skeletal muscle cells would shed further light on how CCL11 signalling contributes to insulin resistance. Second, it was observed that CCL11 deletion or long-term (9-wk) CCR3 antagonism simultaneously suppressed body weight gain and attenuated NAFLD in mice whereas CCL11 neutralisation or short-term (3-wk) CCR3 antagonism ameliorated NAFLD without altering body weight. These observations argue that regulation of liver pathology and body weight by CCL11 might be dose-dependent (complete loss of CCL11 in the CCL11 KO mice vs. partial CCL11 loss attributable to CCL11 neutralisation), temporally controlled (long-term CCR3 intervention vs. short-term CCR3 intervention), and separable. Third, we have previously shown that CCL11 deficiency impairs liver regeneration in mice.43 Because patients with NAFLD display heightened risk of liver failure owing to failure of cell cycle entry by hepatocytes under metabolic stress,71 the potential benefit of targeting CCL11 needs to be weighed against the caveat of delayed liver regeneration. Finally, off-target effects of CCL11 antagonism should be considered. CCR3, in addition to functioning as a receptor for CCL11, also relays the signals initiated by CCL5, CCL7, CCL13, and CCL26.72 Some of these chemokines, for instance CCL5, have been implicated in NAFLD pathogenesis and can be potentially targeted.45,73,74 It may be desirable to screen for small-molecule compounds that more specifically blocks the effect of CCL11.
In summary, our data provide genetic and pharmaceutical evidence that CCL11 is a novel regulator and an effective target of non-alcoholic fatty liver disease. Further studies are clearly warranted so that these findings can be seamlessly translated to real-world therapies against NAFLD.
Financial support
This work was supported by grants from the National Natural Science Foundation of China (82170592, 82200684, 82170609, 81970545, 82072652, and 81871947) and the Natural Science Foundation of Jiangsu Province (BK20221032).
Authors’ contributions
Conceived the project: ZLL, ZWF, XPZ, J Li. Designed experiments: all authors. Performed experiments, collected data, and analysed data: XS, XC, HL, XLM, YG, ZWF, ZLL. Drafted the manuscript: YX. Edited and finalised the manuscript: all authors. Secured funding: ZF, XPZ, JL, ZLL.
Data availability statement
The data that support the findings of this study are available upon reasonable request.
Conflicts of interest
The authors have no conflicts of interest to declare.
Please refer to the accompanying ICMJE disclosure forms for further details.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2023.100805.
Contributor Information
Jie Li, Email: lijier@sina.com.
Xiaoping Zou, Email: zouxp@nju.edu.cn.
Zilong Li, Email: lzl1114@cpu.edu.cn.
Supplementary data
The following are the supplementary data to this article.
References
- 1.Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease – meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.Bianco C., Casirati E., Malvestiti F., Valenti L. Genetic predisposition similarities between NASH and ASH: identification of new therapeutic targets. JHEP Rep. 2021;3 doi: 10.1016/j.jhepr.2021.100284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Younossi Z.M., Henry L. Epidemiology of non-alcoholic fatty liver disease and hepatocellular carcinoma. JHEP Rep. 2021;3 doi: 10.1016/j.jhepr.2021.100305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le M.H., Yeo Y.H., Li X., Li J., Zou B., Wu Y., et al. 2019 Global NAFLD prevalence: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2022;20:2809–2817. doi: 10.1016/j.cgh.2021.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Younossi Z., Anstee Q.M., Marietti M., Hardy T., Henry L., Eslam M., et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15:11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 6.Ayonrinde O.T. Historical narrative from fatty liver in the nineteenth century to contemporary NAFLD – reconciling the present with the past. JHEP Rep. 2021;3 doi: 10.1016/j.jhepr.2021.100261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meijnikman A.S., Herrema H., Scheithauer T.P.M., Kroon J., Nieuwdorp M., Groen A.K. Evaluating causality of cellular senescence in non-alcoholic fatty liver disease. JHEP Rep. 2021;3 doi: 10.1016/j.jhepr.2021.100301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedossa P. Pathology of non-alcoholic fatty liver disease. Liver Int. 2017;37(Suppl 1):85–89. doi: 10.1111/liv.13301. [DOI] [PubMed] [Google Scholar]
- 9.Dufour J.F., Anstee Q.M., Bugianesi E., Harrison S., Loomba R., Paradis V., et al. Current therapies and new developments in NASH. Gut. 2022;71:2123–2134. doi: 10.1136/gutjnl-2021-326874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazilescu L.I., Selzner M., Selzner N. Defatting strategies in the current era of liver steatosis. JHEP Rep. 2021;3 doi: 10.1016/j.jhepr.2021.100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen R., Du J., Zhu H., Ling Q. The role of cGAS-STING signalling in liver diseases. JHEP Rep. 2021;3 doi: 10.1016/j.jhepr.2021.100324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parlati L., Regnier M., Guillou H., Postic C. New targets for NAFLD. JHEP Rep. 2021;3 doi: 10.1016/j.jhepr.2021.100346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ratziu V. The painful reality of end-stage liver disease in NASH. Lancet Gastroenterol Hepatol. 2018;3:8–10. doi: 10.1016/S2468-1253(17)30365-5. [DOI] [PubMed] [Google Scholar]
- 14.Younossi Z.M., Blissett D., Blissett R., Henry L., Stepanova M., Younossi Y., et al. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology. 2016;64:1577–1586. doi: 10.1002/hep.28785. [DOI] [PubMed] [Google Scholar]
- 15.Shimano H., Sato R. SREBP-regulated lipid metabolism: convergent physiology – divergent pathophysiology. Nat Rev Endocrinol. 2017;13:710–730. doi: 10.1038/nrendo.2017.91. [DOI] [PubMed] [Google Scholar]
- 16.Liang G., Yang J., Horton J.D., Hammer R.E., Goldstein J.L., Brown M.S. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J Biol Chem. 2002;277:9520–9528. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
- 17.Shimano H., Shimomura I., Hammer R.E., Herz J., Goldstein J.L., Brown M.S., et al. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest. 1997;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimomura I., Shimano H., Horton J.D., Goldstein J.L., Brown M.S. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Invest. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimano H., Horton J.D., Shimomura I., Hammer R.E., Brown M.S., Goldstein J.L. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parthasarathy G., Revelo X., Malhi H. Pathogenesis of nonalcoholic steatohepatitis: an overview. Hepatol Commun. 2020;4:478–492. doi: 10.1002/hep4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brenner C., Galluzzi L., Kepp O., Kroemer G. Decoding cell death signals in liver inflammation. J Hepatol. 2013;59:583–594. doi: 10.1016/j.jhep.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 22.Wallace S.J., Tacke F., Schwabe R.F., Henderson N.C. Understanding the cellular interactome of non-alcoholic fatty liver disease. JHEP Rep. 2022;4 doi: 10.1016/j.jhepr.2022.100524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagata N., Chen G., Xu L., Ando H. An update on the chemokine system in the development of NAFLD. Medicina (Kaunas) 2022;58:761. doi: 10.3390/medicina58060761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertola A., Bonnafous S., Anty R., Patouraux S., Saint-Paul M.C., Iannelli A., et al. Hepatic expression patterns of inflammatory and immune response genes associated with obesity and NASH in morbidly obese patients. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westerbacka J., Kolak M., Kiviluoto T., Arkkila P., Siren J., Hamsten A., et al. Genes involved in fatty acid partitioning and binding, lipolysis, monocyte/macrophage recruitment, and inflammation are overexpressed in the human fatty liver of insulin-resistant subjects. Diabetes. 2007;56:2759–2765. doi: 10.2337/db07-0156. [DOI] [PubMed] [Google Scholar]
- 26.Galastri S., Zamara E., Milani S., Novo E., Provenzano A., Delogu W., et al. Lack of CC chemokine ligand 2 differentially affects inflammation and fibrosis according to the genetic background in a murine model of steatohepatitis. Clin Sci (Lond) 2012;123:459–471. doi: 10.1042/CS20110515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu L., Chen Y., Nagashimada M., Ni Y., Zhuge F., Chen G., et al. CC chemokine ligand 3 deficiency ameliorates diet-induced steatohepatitis by regulating liver macrophage recruitment and M1/M2 status in mice. Metabolism. 2021;125 doi: 10.1016/j.metabol.2021.154914. [DOI] [PubMed] [Google Scholar]
- 28.Jose P.J., Griffiths-Johnson D.A., Collins P.D., Walsh D.T., Moqbel R., Totty N.F., et al. Eotaxin: a potent eosinophil chemoattractant cytokine detected in a Guinea pig model of allergic airways inflammation. J Exp Med. 1994;179:881–887. doi: 10.1084/jem.179.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tacke F., Trautwein C., Yagmur E., Hellerbrand C., Wiest R., Brenner D.A., et al. Up-regulated eotaxin plasma levels in chronic liver disease patients indicate hepatic inflammation, advanced fibrosis and adverse clinical course. J Gastroenterol Hepatol. 2007;22:1256–1264. doi: 10.1111/j.1440-1746.2006.04621.x. [DOI] [PubMed] [Google Scholar]
- 30.Burke M.L., McManus D.P., Ramm G.A., Duke M., Li Y., Jones M.K., et al. Temporal expression of chemokines dictates the hepatic inflammatory infiltrate in a murine model of schistosomiasis. PLoS Negl Trop Dis. 2010;4:e598. doi: 10.1371/journal.pntd.0000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pham B.N., Bemuau J., Durand F., Sauvanet A., Degott C., Prin L., et al. Eotaxin expression and eosinophil infiltrate in the liver of patients with drug-induced liver disease. J Hepatol. 2001;34:537–547. doi: 10.1016/s0168-8278(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 32.Jaruga B., Hong F., Sun R., Radaeva S., Gao B. Crucial role of IL-4/STAT6 in T cell-mediated hepatitis: up-regulating eotaxins and IL-5 and recruiting leukocytes. J Immunol. 2003;171:3233–3244. doi: 10.4049/jimmunol.171.6.3233. [DOI] [PubMed] [Google Scholar]
- 33.Duran A., Rodriguez A., Martin P., Serrano M., Flores J.M., Leitges M., et al. Crosstalk between PKCzeta and the IL4/Stat6 pathway during T-cell-mediated hepatitis. EMBO J. 2004;23:4595–4605. doi: 10.1038/sj.emboj.7600468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proctor W.R., Chakraborty M., Chea L.S., Morrison J.C., Berkson J.D., Semple K., et al. Eosinophils mediate the pathogenesis of halothane-induced liver injury in mice. Hepatology. 2013;57:2026–2036. doi: 10.1002/hep.26196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasudevan A.R., Wu H., Xydakis A.M., Jones P.H., Smith E.O., Sweeney J.F., et al. Eotaxin and obesity. J Clin Endocrinol Metab. 2006;91:256–261. doi: 10.1210/jc.2005-1280. [DOI] [PubMed] [Google Scholar]
- 36.Loughrey B.V., McGinty A., Young I.S., McCance D.R., Powell L.A. Increased circulating CC chemokine levels in the metabolic syndrome are reduced by low-dose atorvastatin treatment: evidence from a randomized controlled trial. Clin Endocrinol (Oxf) 2013;79:800–806. doi: 10.1111/cen.12113. [DOI] [PubMed] [Google Scholar]
- 37.Herder C., Haastert B., Muller-Scholze S., Koenig W., Thorand B., Holle R., et al. Association of systemic chemokine concentrations with impaired glucose tolerance and type 2 diabetes: results from the Cooperative Health Research in the Region of Augsburg Survey S4 (KORA S4) Diabetes. 2005;54(Suppl 2):S11–S17. doi: 10.2337/diabetes.54.suppl_2.s11. [DOI] [PubMed] [Google Scholar]
- 38.Wong S.W., Ting Y.W., Yong Y.K., Tan H.Y., Barathan M., Riazalhosseini B., et al. Chronic inflammation involves CCL11 and IL-13 to facilitate the development of liver cirrhosis and fibrosis in chronic hepatitis B virus infection. Scand J Clin Lab Invest. 2021;81:147–159. doi: 10.1080/00365513.2021.1876245. [DOI] [PubMed] [Google Scholar]
- 39.Huaux F., Gharaee-Kermani M., Liu T., Morel V., McGarry B., Ullenbruch M., et al. Role of Eotaxin-1 (CCL11) and CC chemokine receptor 3 (CCR3) in bleomycin-induced lung injury and fibrosis. Am J Pathol. 2005;167:1485–1496. doi: 10.1016/S0002-9440(10)61235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koorneef L.L., van den Heuvel J.K., Kroon J., Boon M.R., t Hoen P.A.C., Hettne K.M., et al. Selective glucocorticoid receptor modulation prevents and reverses nonalcoholic fatty liver disease in male mice. Endocrinology. 2018;159:3925–3936. doi: 10.1210/en.2018-00671. [DOI] [PubMed] [Google Scholar]
- 41.de Souza D.N., Teixeira C.J., Veronesi V.B., Murata G.M., Santos-Silva J.C., Hecht F.B., et al. Dexamethasone programs lower fatty acid absorption and reduced PPAR-gamma and fat/CD36 expression in the jejunum of the adult rat offspring. Life Sci. 2021;265 doi: 10.1016/j.lfs.2020.118765. [DOI] [PubMed] [Google Scholar]
- 42.Kong M., Zhu Y., Shao J., Fan Z., Xu Y. The chromatin remodeling protein BRG1 regulates SREBP maturation by activating SCAP transcription in hepatocytes. Front Cel Dev Biol. 2021;9 doi: 10.3389/fcell.2021.622866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan Z., Kong M., Dong W., Dong C., Miao X., Guo Y., et al. Trans-activation of eotaxin-1 by Brg1 contributes to liver regeneration. Cell Death Dis. 2022;13:495. doi: 10.1038/s41419-022-04944-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lv F., Shao T., Xue Y., Miao X., Guo Y., Wang Y., et al. Dual regulation of tank binding kinase 1 (TBK1) by BRG1 in hepatocytes contributes to ROS production. Front Cel Dev Biol. 2021;9 doi: 10.3389/fcell.2021.745985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kong M., Dong W., Zhu Y., Fan Z., Miao X., Guo Y., et al. Redox-sensitive activation of CCL7 by BRG1 in hepatocytes during liver injury. Redox Biol. 2021;46 doi: 10.1016/j.redox.2021.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kong M., Dong W., Xu H., Fan Z., Miao X., Guo Y., et al. Choline kinase alpha is a novel transcriptional target of the Brg1 in hepatocyte: implication in liver regeneration. Front Cel Dev Biol. 2021;9 doi: 10.3389/fcell.2021.705302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan Z., Kong M., Miao X., Guo Y., Ren H., Wang J., et al. An E2F5-TFDP1-BRG1 complex mediates transcriptional activation of MYCN in hepatocytes. Front Cel Dev Biol. 2021;9 doi: 10.3389/fcell.2021.742319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong W., Zhu Y., Zhang Y., Fan Z., Zhang Z., Fan X., et al. BRG1 links TLR4 trans-activation to LPS-induced SREBP1a expression and liver injury. Front Cel Dev Biol. 2021;9 doi: 10.3389/fcell.2021.617073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shao T., Xue Y., Fang M. Epigenetic repression of chloride channel accessory 2 transcription in cardiac fibroblast: implication in cardiac fibrosis. Front Cel Dev Biol. 2021;9 doi: 10.3389/fcell.2021.771466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gillard J., Clerbaux L.A., Nachit M., Sempoux C., Staels B., Bindels L.B., et al. Bile acids contribute to the development of non-alcoholic steatohepatitis in mice. JHEP Rep. 2022;4 doi: 10.1016/j.jhepr.2021.100387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanyal A.J., Ling L., Beuers U., DePaoli A.M., Lieu H.D., Harrison S.A., et al. Potent suppression of hydrophobic bile acids by aldafermin, an FGF19 analogue, across metabolic and cholestatic liver diseases. JHEP Rep. 2021;3 doi: 10.1016/j.jhepr.2021.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Becattini B., Breasson L., Sardi C., Zani F., Solinas G. PI3Kgamma promotes obesity-associated hepatocellular carcinoma by regulating metabolism and inflammation. JHEP Rep. 2021;3 doi: 10.1016/j.jhepr.2021.100359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong W., Kong M., Liu H., Xue Y., Li Z., Wang Y., et al. Myocardin-related transcription factor A drives ROS-fueled expansion of hepatic stellate cells by regulating p38-MAPK signalling. Clin Transl Med. 2022;12:e688. doi: 10.1002/ctm2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu X., Dong W., Kong M., Ren H., Wang J., Shang L., et al. Down-regulation of CXXC5 de-represses MYCL1 to promote hepatic stellate cell activation. Front Cel Dev Biol. 2021;9 doi: 10.3389/fcell.2021.680344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang Z., Zhong L., Lee J.T.H., Zhang J., Wu D., Geng L., et al. The FGF21-CCL11 axis mediates beiging of white adipose tissues by coupling sympathetic nervous system to type 2 immunity. Cell Metab. 2017;26:493–508.e494. doi: 10.1016/j.cmet.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 56.Chandra G., Rangasamy S.B., Roy A., Kordower J.H., Pahan K. Neutralization of RANTES and eotaxin prevents the loss of dopaminergic neurons in a mouse model of Parkinson disease. J Biol Chem. 2016;291:15267–15281. doi: 10.1074/jbc.M116.714824. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Sutti S., Albano E. Adaptive immunity: an emerging player in the progression of NAFLD. Nat Rev Gastroenterol Hepatol. 2020;17:81–92. doi: 10.1038/s41575-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elsharkawy A.M., Mann D.A. Nuclear factor-kappaB and the hepatic inflammation-fibrosis-cancer axis. Hepatology. 2007;46:590–597. doi: 10.1002/hep.21802. [DOI] [PubMed] [Google Scholar]
- 59.Sanyal A.J., Van Natta M.L., Clark J., Neuschwander-Tetri B.A., Diehl A., Dasarathy S., et al. Prospective study of outcomes in adults with nonalcoholic fatty liver disease. N Engl J Med. 2021;385:1559–1569. doi: 10.1056/NEJMoa2029349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Puxeddu I., Bader R., Piliponsky A.M., Reich R., Levi-Schaffer F., Berkman N. The CC chemokine eotaxin/CCL11 has a selective profibrogenic effect on human lung fibroblasts. J Allergy Clin Immunol. 2006;117:103–110. doi: 10.1016/j.jaci.2005.08.057. [DOI] [PubMed] [Google Scholar]
- 61.Matsuyama T., Kimura T., Kitagawa M., Pfeffer K., Kawakami T., Watanabe N., et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- 62.Ueki S., Dhupar R., Cardinal J., Tsung A., Yoshida J., Ozaki K.S., et al. Critical role of interferon regulatory factor-1 in murine liver transplant ischemia reperfusion injury. Hepatology. 2010;51:1692–1701. doi: 10.1002/hep.23501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yokota S., Yoshida O., Dou L., Spadaro A.V., Isse K., Ross M.A., et al. IRF-1 promotes liver transplant ischemia/reperfusion injury via hepatocyte IL-15/IL-15Ralpha production. J Immunol. 2015;194:6045–6056. doi: 10.4049/jimmunol.1402505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rani R., Kumar S., Sharma A., Mohanty S.K., Donnelly B., Tiao G.M., et al. Mechanisms of concanavalin A-induced cytokine synthesis by hepatic stellate cells: distinct roles of interferon regulatory factor-1 in liver injury. J Biol Chem. 2018;293:18466–18476. doi: 10.1074/jbc.RA118.005583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao Z., Dhupar R., Cai C., Li P., Billiar T.R., Geller D.A. A critical role for IFN regulatory factor 1 in NKT cell-mediated liver injury induced by alpha-galactosylceramide. J Immunol. 2010;185:2536–2543. doi: 10.4049/jimmunol.1000092. [DOI] [PubMed] [Google Scholar]
- 66.Lane D.P., Cheok C.F., Lain S. p53-based cancer therapy. Cold Spring Harb Perspect Biol. 2010;2:a001222. doi: 10.1101/cshperspect.a001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Llombart V., Mansour M.R. Therapeutic targeting of “undruggable” MYC. EBioMedicine. 2022;75 doi: 10.1016/j.ebiom.2021.103756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huber J., Kiefer F.W., Zeyda M., Ludvik B., Silberhumer G.R., Prager G., et al. CC chemokine and CC chemokine receptor profiles in visceral and subcutaneous adipose tissue are altered in human obesity. J Clin Endocrinol Metab. 2008;93:3215–3221. doi: 10.1210/jc.2007-2630. [DOI] [PubMed] [Google Scholar]
- 69.Kim D.A., Park S.J., Lee J.Y., Kim J.H., Lee S., Lee E., et al. Effect of CCL11 on in vitro myogenesis and its clinical relevance for sarcopenia in older adults. Endocrinol Metab (Seoul) 2021;36:455–465. doi: 10.3803/EnM.2020.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beyer I., Njemini R., Bautmans I., Demanet C., Bergmann P., Mets T. Inflammation-related muscle weakness and fatigue in geriatric patients. Exp Gerontol. 2012;47:52–59. doi: 10.1016/j.exger.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 71.Caldez M.J., Bjorklund M., Kaldis P. Cell cycle regulation in NAFLD: when imbalanced metabolism limits cell division. Hepatol Int. 2020;14:463–474. doi: 10.1007/s12072-020-10066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murphy P.M., Baggiolini M., Charo I.F., Hebert C.A., Horuk R., Matsushima K., et al. International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol Rev. 2000;52:145–176. [PubMed] [Google Scholar]
- 73.Kim B.M., Abdelfattah A.M., Vasan R., Fuchs B.C., Choi M.Y. Hepatic stellate cells secrete Ccl5 to induce hepatocyte steatosis. Sci Rep. 2018;8:7499. doi: 10.1038/s41598-018-25699-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li B.H., He F.P., Yang X., Chen Y.W., Fan J.G. Steatosis induced CCL5 contributes to early-stage liver fibrosis in nonalcoholic fatty liver disease progress. Transl Res. 2017;180:103–117.e4. doi: 10.1016/j.trsl.2016.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available upon reasonable request.