Abstract
Blocking androgen receptor signaling is the mainstay of therapy for advanced prostate cancer (PCa). However, acquired resistance to single agents targeting this pathway results in the development of lethal castration-resistant PCa. Combination therapy approaches represent a promising strategy for the treatment of advanced disease. Here, we explore a therapeutic strategy for PCa based on the ability of shRNAs/siRNAs to function essentially as miRNAs and, via seed sequence complementarity, induce RNA interference of numerous targets simultaneously. We developed a library that contained shRNAs with all possible seed sequence combinations to identify those ones that most potently reduce cell growth and viability when expressed in PCa cells. Validation of some of these RNAi sequences indicated that the toxic effect is associated with seed sequence complementarity to the 3′ UTR of AR coregulatory and essential genes. In fact, expression of siRNAs containing the identified toxic seed sequences led to global inhibition of AR-mediated gene expression and reduced expression of cell-cycle genes. When tested in mice, the toxic shRNAs also inhibited castration-resistant PCa and exhibited therapeutic efficacy in pre-established tumors. Our findings highlight RNAi of androgen signaling networks as a promising therapeutic strategy for PCa.
Keywords: MT: Oligonucleotides: Therapies and Applications, prostate cancer, RNA interference, small RNA, seed sequence, androgen signaling, gene networks, androgen receptor coregulators, prostate cancer essential genes
Graphical abstract

Ruiz Echevarría and colleagues describe a random seed-based RNAi selection screen identifying shRNAs/siRNAs that inhibit PCa cell growth/viability. The toxic shRNAs/siRNAs function as specialized miRNAs to simultaneously target multiple androgen receptor (AR) coregulators, essential for AR signaling and PCa cell survival. This suggests RNAi of hormone signaling networks as therapeutic strategy for PCa.
Introduction
Prostate cancer (PCa) is the second leading cause of cancer-related death in men, and a major public health concern.1 Mortality is largely due to development of resistance to current therapies for advanced disease. Androgen deprivation therapy (ADT), which prevents androgen-mediated stimulation, exploits the dependency of PCa cells on androgen signaling for growth and survival, and it is the standard of care for metastatic disease.2 However, while ADT is initially effective, most patients relapse with castration-resistant PCa (CRPC), a lethal form of the disease in which the PCa cells acquire the ability to grow in a low-androgen environment while remaining dependent on androgen receptor (AR) activity.3,4 This reliance has led to the development of second-generation antiandrogens, or androgen biosynthesis blockers, which show effective but short-lived responses, leading to secondary resistance, and even to complete AR independence.5,6
Critical to AR-mediated signaling is the recruitment of AR coregulators, which modulate and specify its transcriptional response, pointing to an important role for AR coregulators in PCa. Accordingly, mechanisms of resistance to hormonal therapies often involve not only genetic and epigenetic alterations in the AR itself (i.e., amplification, mutations, and splice variants), but also changes in the expression of AR coregulators.3,4 In fact, increased expression of AR coregulators generally correlates with aggressive disease and poor clinical outcome,7 suggesting that targeting AR coregulators, or their interaction with the AR, are plausible therapeutic alternatives to block androgen signaling. However, our understanding of the composition and the function of the AR coregulator complexes is incomplete, and targeting individual AR coregulators has been largely unsuccessful. This is likely due to functional redundancies and compensatory mechanisms, arguing for finding therapeutic approaches that target multiple AR coregulators.
RNA interference (RNAi) via small non-coding RNAs, exemplified by miRNAs, may provide such an approach. miRNAs bind and recruit the RNA-induced silencing complex to their mRNA targets (preferentially in the 3′ UTR) through seed sequence (nucleotides 2–7 or 8 of the guide strand of the miRNA) complementarity, resulting in translation inhibition and/or degradation of the mRNA.8,9,10,11,12,13,14 miRNAs are predominantly implicated in regulating critical biological pathways and it has been proposed that the targets of a single miRNA are generally functionally associated (networks).15,16 Recently published studies from our lab identified shRNAs/siRNAs that reduce the expression of AR coregulatory and essential gene networks in PCa cells through an miRNA-like seed-mediated mechanism, resulting in androgen signaling inhibition and cell death.17 Together with other studies demonstrating the ability of RNAi to target essential gene networks to induce cell death in other cancer cell types,18,19,20 our work provides a rationale for pursuing RNAi of AR signaling networks as a potential therapeutic strategy in PCa.
In this study, we develop a novel unbiased, seed-based, shRNA-negative selection screen to identify shRNAs that most effectively inhibit PCa cell growth (toxic). Our results indicate that the toxic shRNAs inhibit global androgen signaling and suggest a clear link between the effect on PCa cell growth and viability and RNAi targeting of AR coregulatory and essential gene networks. Importantly, our results validate the therapeutic potential of the toxic shRNAs in vivo and point to a therapeutic advantage with respect to individual coregulator targeting in PCa cells. Overall, our results suggest that RNAi-mediated targeting of androgen signaling regulatory and essential gene networks might present a promising therapeutic strategy for the treatment of PCa.
Results
A novel unbiased RNAi screen for the identification of negatively selected shRNAs in PCa cells
To identify novel RNAi sequences that potently inhibit PCa cell growth, we designed a novel unbiased negative selection seed-based shRNA screen in PCa cell lines. We used a lentiviral-based library expressing 15,572 unique shRNAs from a doxycycline (Dox)-inducible promoter that differ in the 7-nucleotide seed sequence (nucleotides 2–8 of the guide strand). All possible seven nucleotide permutations within the guide strand 7mer seed sequence were included, while the sequence context surrounding the seed was identical for all shRNAs (Figure 1A) and essentially mimics the sequence of a known non-toxic siRNA backbone19,21 (see materials and methods). We selected an androgen-dependent PCa cell line (LNCaP) and three CRPC cell lines (LNCaP abl, LNCaP-95, and 22Rv1) for the screen. 22Rv1 and LNCaP-95 cells express the constitutively active AR splice isoform, AR-V7. 22Rv1 cells were grown in the presence and absence of 10 nM dihydrotestosterone (DHT). Therefore, the screen was designed to identify RNAi sequences that are inhibitory even in low-androgen settings and advanced disease models. Cells were transduced with the lentiviral library, and grown in the presence of Dox for 10 doublings to allow for negative selection prior to next-generation sequencing (NGS) for quantification and identification of depleted sequences. Z scores were calculated as previously described,22 and 88 depleted shRNAs were selected (Figures 1B, S1, and S2). The 7mer seeds from the 88 depleted shRNAs had a significant tendency toward being G/T rich (Figures 1C, 1D, and S3), differing from the approximated 25% frequency for each nucleotide at each position of the seed when all the 15,572 shRNAs used in the screen where analyzed (Figure S3). We identified 11 6mer seed sequences (nucleotides 2–7 of the guide strand) that were present within multiple depleted shRNAs (Figures 1A and 1D); a finding that is of significance, as seed sequences can also function as 6mers.10,11,12,13,14 These seeds include that of the miR-34 tumor suppressor miRNA19,23,24 (GGCAGT), which has been found to target the AR in PCa cell lines,25 confirming the utility of the screen to identify shRNAs that target genes that are essential for PCa growth.
Figure 1.
Identification of negatively selected shRNA sequences
(A) Schematic of the shRNA construct used for the screen. (B) Heatmaps showing Z scores of the shRNAs (n = 15,572) after 10 doublings (left). Average Z scores of selected depleted shRNAs (n = 88) are shown (right). Red or blue indicates that the corresponding shRNA was positively or negatively selected respectively. (C) Motif stack plot showing the relative frequency of each nucleotide at each position for the 7mer seed sequences of the 88 depleted shRNAs. (D) Stacked bar graph showing the frequency of each nucleotide in the 7mer seed for the 88 depleted and for all other shRNAs (top). p < 0.05 was considered a significant association as calculated by Pearson’s chi-squared test. List of 6mer seeds that appear multiple times within the 7mer seeds of the selected 88 depleted shRNAs (bottom).
Depleted shRNAs reduce viability and growth of PCa cells and xenografts
To validate and more thoroughly determine the effect of the depleted shRNAs on PCa cell growth and viability, we used siRNAs corresponding to the guide strand sequences of 8 selected depleted shRNAs from the initial 88 (siRNA mimics of the toxic shRNAs) and determined their effects on PCa cell growth and viability. Two non-target siRNAs (siNT-1 and siNT-2) and “no siRNA” were used as negative controls. LNCaP and 22Rv1 cells transfected with any of the toxic siRNA showed significantly reduced growth (Figures 2A and 2B) and viability (Figures 2E and 2F) compared with cells not transfected, or transfected with the control siRNAs. The toxic siRNAs induced PARP cleavage and H2AX phosphorylation, markers of cell death and DNA damage, respectively (Figures 2I and 2J).
Figure 2.
Validation of toxic RNAi sequences
Cell growth (A–D) and viability (E–H) graphs, and western blots (I–L) from lysates, of LNCaP (A, E, and I), 22Rv1 (B, F, and J), LNCaP abl (C, G, and K), and LNCaP-95 (D, H, and L) cells transfected with the designated siRNAs. Relative confluency was used as a surrogate for cell growth and was determined via IncuCyte. Relative viability was assessed by trypan blue 6 days after siRNA transfection (8 days after transfection for LNCaP-95 cells). Western blots show AR (top band, AR full length; bottom band, AR-V7 in 22Rv1 and LNCaP-95), androgen-responsive proteins (PSA, FKBP5, CDK1), and cell death and DNA damage markers (cleaved PARP and γH2AX); lysates were collected 72 h after siRNA transfection and Calnexin was used as a loading control (I–L). n = 3 or 4 for each experiment, ∗q < 0.05 compared with pooled siNT-1, siNT-2, and no-siRNA (A, B, E, and F) or siNT-1 (C, D, G, and H) transfection was considered significant as determined by t test.
We analyzed the effect of these siRNAs on the expression of the AR as well as on the expression of a representative AR target. Although several toxic siRNAs reduced the expression of the full-length AR, AR-V7, and androgen-responsive proteins (Figures 2I and 2J), transfection of the same cell lines with an siRNA pool that targets the AR, and strongly reduced its expression (Figures 2I and 2J), was significantly less effective at reducing PCa cell growth and viability compared with the toxic siRNAs (Figures 2A, 2B, 2E, and 2F). This indicates that the effects of our selected siRNAs are not primarily mediated by reduced expression of the AR. In fact, some of the selected siRNAs (e.g., siUACUGGC) did not affect AR expression, but still significantly reduced expression of PSA (an androgen-responsive protein), as well as growth and viability of LNCaP cells (Figures 2A, 2E, and 2I).
Three siRNAs that consistently demonstrated potent effects on growth and viability of LNCaP and 22Rv1 cells (siGUGUGUG, siGUGUGUA, siUGUUUGC) were further validated in additional PCa cell lines. Transfection with any of these three siRNAs decreased growth (Figures 2C and 2D) and viability (Figures 2G and 2H) of LNCaP abl and LNCaP-95 cells to varying degrees, and induced PARP cleavage and H2AX phosphorylation (Figures 2K and 2L). They also reduced expression of AR and AR targets. siGUGUGUG and siGUGUGUA were shown to reduce the expression of AR-V7 in LNCaP-95 cells (Figure 2L), similar to the results seen in the AR-V7+ 22Rv1 cells. The same three siRNAs also reduced the growth of 22Rv1 cells cultured in the presence or absence of DHT to a similar degree (Figure S4). These data suggest that the identified siRNAs are toxic across different PCa cell lines and growth conditions.
We then examined the effect of one of the toxic shRNAs on the growth of subcutaneous tumors in mice. To this end, we cloned the shRNA containing the UGUUUGC seed sequence or a shNT control in the Dox-inducible lentiviral vector used for library construction and expressed them in 22Rv1 cells. In cell culture, Dox treatment significantly reduced growth and viability of 22Rv1 cells expressing the shUGUUUGC shRNA compared with shNT-expressing cells (Figures S5A and S5B). For in vivo experiments, these two cell lines were grown in the absence of Dox—to prevent cell death—mixed with 50% basement membrane extract and inoculated subcutaneously into flanks of NRG mice that were pre-fed (2 days) and kept in a Dox-containing diet for the duration of the experiment. Consistent with the in vitro observations, expression of the shUGUUUGC shRNA, significantly reduced tumor growth in this murine model (Figure S5C). These results validate our novel seed-based shRNA screen as a method to identify toxic RNAi molecules that inhibit growth and viability of PCa cells. Importantly, while many of these shRNAs/siRNAs reduce AR expression, they do not exclusively rely on, and result in more potent viability/growth effects than, targeting the AR alone.
Depleted shRNA seed sequences are predicted to target AR coregulatory and PCa essential genes
The selected toxic shRNAs/siRNAs only differ among them, and from non-toxic ones, in their 7mer seed sequences. Therefore, we anticipated that the toxic effect of the selected shRNAs/siRNAs was mediated by the seed sequence, as we and others have previously shown for other small toxic RNAs.17,18,19 To confirm this prediction, we designed siRNAs derived from the toxic siGUGUGUG, siGUGUGUA, or siUGUUUGC, in which the seed sequence was shifted out of the seed region, or the order of nucleotides within the seed region was altered (Figure S6C). Transfection of LNCaP cells with these modified siRNAs did not affect their growth or viability compared with the non-target siRNA control (siNT-1; Figures S6A and S6B), suggesting that altering the seed reverses the toxic phenotype, and therefore that the observed toxic phenotypes are seed mediated. These results supported the use of the miRNA database, miRDB (www.mirdb.org) (which uses the seed sequence), to identify the predicted targets of the selected 88 depleted shRNAs.
Based on our previous work17 and the essential role of the AR in PCa cell growth, we hypothesized that AR coregulatory and PCa essential genes would be enriched among the predicted targets of the depleted seeds. In fact, the predicted targets of 32 out of the 88 toxic shRNAs were significantly enriched for AR coregulatory26 and LNCaP essential27 genes (Figure 3A, Table S1). More broadly, analysis with the predicted targets of all the 88 toxic shRNAs indicated a more significant enrichment of AR coregulatory26 and LNCaP essential genes27 than of the non-PCa essential18,28,29 genes (Figures S7A and S7B) and the significance of their enrichment (log10q values) was strongly correlated (Figure S7C). The enrichment of AR coregulatory and LNCaP essential genes (log10q) within the predicted targets of the 88 toxic shRNAs was also correlated with their respective target scores (Figures S8A and S8B). The target score represents the abundance of sequences complementary to the seed sequences of the toxic shRNAs in the 3′ UTRs of AR coregulatory or LNCaP essential genes relative to their abundance in the 3′ UTRs of the whole transcriptome. More strikingly, a positive correlation was found between the AR coregulatory and the LNCaP essential gene target scores when considering the sequences complementary to the seeds of all the shRNAs in the screen (n = 15,572; Figure S8C). This suggests that the similar abundance of specific 6 and 7 nucleotide sequences in the 3′ UTR may account for the paired enrichment of AR coregulatory and PCa essential genes observed within the predicted targets of the depleted shRNAs. Importantly, for the depleted shRNAs, predicted targeting of the AR coregulatory genes is significantly associated with predicted targeting of the AR (Figure 3B), suggesting that RNAi of hormone receptors and their coregulators may happen concurrently.
Figure 3.
Depleted shRNAs are predicted to target AR coregulatory and PCa essential genes
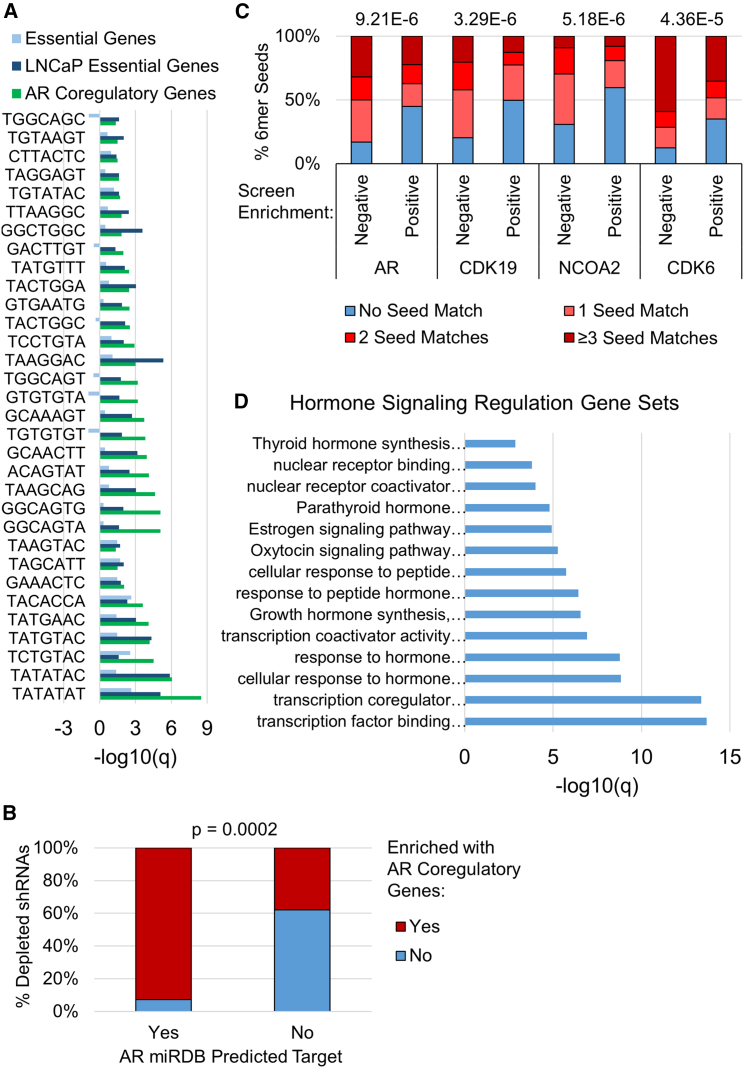
(A) Bar graph showing the –log10(q) of enrichment of AR coregulatory, LNCaP essential (PCa essential genes), and essential (obtained from non-PCa cells) genes among the predicted targets for depleted shRNAs. Only the ones with significant AR coregulatory and LNCaP essential gene enrichment are shown (n = 32). shRNAs are named by their 7mer seed sequence. Negative values indicate that gene sets are under-represented among predicted targets. –log10(q) > 1.3 (q < 0.05) was considered significant, as determined by hypergeometric distribution. (B) Stacked bar graph showing that a high percentage of depleted shRNAs predicted to target the AR are also predicted to target AR coregulatory genes (red). p < 0.05 was considered a significant association as calculated by Fisher’s exact test. (C) Stacked bar graph showing the percentage of negatively selected (depleted, n= 88) and positively selected (enriched, n= 474) shRNAs with 0, 1, 2, or 3 or more 6mer seed matches (complementarity) in the 3′ UTR of each of the indicated target genes (AR, CDK19, NCOA2, CDK6). p values are above the plot for each association, and p < 0.05 was considered a significant association as calculated by Pearson’s chi-square test. (D) Bar graph showing the significance (–log10q) of GO and reactome gene sets involved in hormone signaling regulation enriched among genes predicted to be targeted by >10% of depleted seeds. GO and reactome gene sets were analyzed using www.metascape.org. -log10(q) > 1.3 (q < 0.05) was considered significant, as determined by hypergeometric distribution.
Genes predicted to be targeted by at least 10% (≥9) of the depleted shRNAs were also enriched in AR coregulatory and LNCaP essential genes (Figure S9), and included several genes known to be important for PCa cell growth and survival, for example, the AR, the AR coregulators NCOA2 and CDK6, and the LNCaP essential gene CDK19. Seed sequence complementarity in the 3′ UTR of these four genes was more strongly associated with depleted shRNAs than with positively selected shRNAs (Figure 3C). Moreover, genes targeted by 9 or more depleted shRNAs were also enriched in several hormone signaling GO and reactome gene sets (Figure 3D), suggesting that hormone receptor signaling inhibition could be a common mechanism through which RNAi seed sequences reduce growth and/or viability of hormone-driven cancer cells. Other gene sets with significantly enriched genes targeted by nine or more depleted shRNAs are listed in Table S2.
Downregulation of AR coregulatory and PCa essential genes is associated with sequences in their 3′ UTR complementary to the GUGUGUA and UGUUUGC seeds
To confirm downregulation of AR coregulators and PCa essential genes by the toxic siRNAs, we conducted RNA-seq with RNA from 22Rv1, LNCaP, and LNCaP abl cells, transfected with siNT-1, and the toxic siGUGUGUA or siUGUUUGC siRNAs, and analyzed gene expression changes. RNA was also obtained from LNCaP abl and 22Rv1 cells after transfection with AR-targeted siRNA (siAR), but not for LNCaP, since AR-regulated genes in LNCaP cells are well defined in the literature and reported by us.17 RNA was extracted prior to loss in viability triggered by the toxic siRNAs, which was at 40 h post-transfection in LNCaP cells, 48 h for LNCaP abl, and 64 h for 22Rv1 cells (data not shown). Genes that exhibited ±0.5 log2 fold change and adjusted p value <0.05 in samples from cells transfected with toxic siRNAs relative to siNT control were considered to be significantly differentially expressed genes (DEGs).
Consistent with the data from the predicted target analyses using the miRDB software (Figure 3A), the RNA-seq data showed that genes downregulated by siGUGUGUA and siUGUUUGC in LNCaP and other PCa cell lines were significantly enriched in AR coregulatory and LNCaP essential genes (Figure S10). Enrichment of essential genes was further validated using CRISPR screening data across multiple PCa cell lines profiled by DepMap30 (Figure S11). To verify that downregulation occurs via a seed-mediated mechanism, we performed a series of tests: (1) we used the cWords software31 to identify 6–7 nucleotide 3′ UTR sequences that were most associated with mRNA downregulation in the PCa cells transfected with siGUGUGA, siUGUUUGC, and then determined whether the enriched sequences were complementary to the seed sequence of those siRNAs. The analyses demonstrated that the 6–7 nucleotide 3′ UTR sequences most significantly associated with downregulated genes were in fact complementary to the siGUGUGUA, siUGUUUGC seeds, (2) The Kolmogorov-Smirnov (KS) test was used to confirm that genes with 3′ UTR 7mer complementary sequences to the GUGUGUA or UGUUUGC seeds were significantly downregulated by the respective siRNAs, and (3) GSEA showed that miRDB-predicted targets (which are based on the 7mer seed sequence) for both siRNAs were significantly downregulated (Figures 4A–4D, S10A–S10D, S12A–S12D, S13A–S13D, S14A–S14D). Similarly, seed sequence complementarity in the 3′ UTR was significantly associated with AR coregulatory and LNCaP essential gene downregulation (Figures 4E, S12E, S13E, and S14E). These data show that siGUGUGUA and siUGUUUGC downregulate multiple AR coregulatory and PCa essential genes through a seed-mediated mechanism.
Figure 4.
Global, AR coregulatory, and PCa essential gene downregulation in LNCaP cells is associated with sequences complementary to the GUGUGUA and UGUUUGC seeds in the 3′ UTR
(A and B) cWords plots displaying the top 300 words (6 and 7 nucleotide 3′ UTR sequences) associated with gene downregulation in LNCaP cells transfected with siGUGUGUA (A) or siUGUUUGC (B). The top seed-related sequences are indicated; “seed cluster” includes other seed-related sequences. Genes were ranked ordered by signal-to-noise values from GSEA. (C and D) eCDF plots from LNCaP cells transfected with siGUGUGUA (C, left) or siUGUUUGC (D, left) showing the cumulative fraction of genes across the signal-to-noise rank ordered gene expression list, for genes with and without 7mer seed match sequences in their 3′ UTR; p < 0.05 as determined by KS test, D and p values are labeled on plots. GSEA enrichment plots for miRDB-predicted target gene sets in LNCaP cells transfected with siGUGUGUA (C, right) or siUGUUUGC (D, right). (E) Bar graphs showing the percentage of AR coregulatory (left) and LNCaP essential (right) genes with 7mer and/or multiple 6mer seed matches to siGUGUGUA or siUGUUUGC in the 3′ UTR, stratified by whether the genes are downregulated (yes) or not (no) by the corresponding siRNAs in LNCaP cells. ∗p < 0.05, ∗∗p < 0.0001, as determined by Fisher’s exact test.
siGUGUGUA and siUGUUUGC siRNAs reduce the expression of androgen-responsive and cell-cycle genes in PCa cells
Because AR coregulatory and essential gene networks were direct targets of siGUGUGUA and siUGUUUGC, we hypothesized that siGUGUGUA, siUGUUUGC, and siAR would elicit similar changes in gene expression. Indeed, the RNA-seq data indicated that a large number of upregulated and downregulated DEGs were common in response to siGUGUGUA, siUGUUUGC, and siAR transfection in all cell lines (Figures 5A, 5B, S15A, and S15B). However, downregulated DEGs generally demonstrated a stronger overlap than upregulated genes (Figure S15C), suggesting that siGUGUGUA and siUGUUUGC may block the activation of similar genes and pathways, including AR-mediated gene activation. We next used GSEA to identify pathways affected by the toxic siRNAs. GSEA using the HALLMARK gene sets within the molecular signature database (http://www.gsea-msigdb.org) indicate that siGUGUGUA and siUGUUUGC significantly downregulate androgen signaling (HALLMARK_ANDROGEN_RESPONSE) and several cell-cycle-related gene sets (HALLMARK_E2F_TARGETS, HALLMARK_G2M_CHECKPOINT and HALLMARK_MITOTIC_SPINDLE) (Figures 5C, 5D, S15D, and S15E). Furthermore, GSEA with other AR signaling-related gene sets, such as AR score,32,33 AR-V734 regulated genes, genes upregulated and downregulated by siAR in 22Rv1 and LNCaP abl cells in our current RNA-seq, and genes upregulated and downregulated by DHT in LNCaP control cells from a previous publication by our lab17 showed significantly altered expression in siGUGUGUA- and siUGUUUGC-transfected cells in all cell lines (Figures 5E, 5F, S15F, S15G, S16A, S16B, S17A–S17B, and S18A–S18D).
Figure 5.
siGUGUGUA and siUGUUUGC reduce the expression of androgen-responsive and cell-cycle genes in PCa cells
(A and B) Venn diagrams showing the overlap of significantly upregulated and downregulated genes as determined by RNA-seq ([log2(FC)] > 0.5, padj < 0.05) in LNCaP cells (A) and 22Rv1 cells grown in the absence of DHT (B) transfected with siGUGUGUA-, siUGUUUGC-, or AR-targeted siRNA (siAR) compared with cells transfected with siNT-1. (C and D) Plots showing NES and –log10(q) for significantly upregulated and downregulated Hallmark gene sets in response to transfection with siGUGUGUA and siUGUUUGC in LNCaP (C) and 22Rv1 (D) cells. (E and F) GSEA enrichment plots showing the enrichment of the “LNCaP DHT up” gene set in LNCaP cells (E), and the enrichment of the “22Rv1 −DHT siAR down” gene set in 22Rv1 −DHT cells (F) transfected with siGUGUGUA and siUGUUUGC. Positive and negative NES indicate upregulated and downregulated gene sets respectively. FDR < 0.1 was considered significant.
Using qRT-PCR, we validated the significant downregulation of several AR responsive and cell-cycle genes, as well as AR and AR coregulatory genes, in LNCaP, LNCaP abl, and 22Rv1 cells transfected with the siGUGUGUA and siUGUUUGC siRNAs. (Figures S19A, S19B, S20A, and S20B). Importantly, the AR and AR coregulatory genes contained 3′ UTR sequences complementary to the seeds, while the AR regulated and cell-cycle genes did not (Figures S19C and S20C). Together these data suggest that siGUGUGUA and siUGUUUGC directly target androgen signaling regulatory networks to indirectly inhibit androgen signaling and modulate cell cycle in ADPC and CRPC cells. We used DepMap data to confirm the association between essential gene downregulation and androgen signaling. We selected the top 1,000 most essential genes from PCa and from hormone-independent cancer cell lines (from the DepMap database) and determined that (1) essential gene downregulation by siGUGUGUA and siUGUUUGC is in fact associated with DHT regulation (Figure S21), (2) 3′ UTR seed complementarity is associated with downregulation of essential genes that are not DHT regulated, but not with downregulation of DHT-regulated genes (Figures S22A and S22B), and (3) DHT regulation is strongly associated with downregulation of essential genes that do not have 3′ UTR seed complementarity to siGUGUGUA and siUGUUUGC (Figure S22C). These data strongly indicate that hormone regulation is involved in toxic siRNA-mediated downregulation of essential genes. Based on the enrichment in seed matched AR coregulatory targets of these toxic siRNAs, we propose that the toxic siRNAs function by direct targeting of multiple AR coregulators leading to global inhibition of AR signaling, including downregulation of essential DHT-regulated genes and cell death. Direct seed-mediated targeting of essential genes likely plays a role in growth and viability reductions by the toxic siRNAs; however, a much larger percentage of downregulated essential genes are regulated by androgens.
Toxic siRNAs reduce PCa cell growth and viability more effectively than the knockdown of AR or individual AR coregulatory genes and inhibit pre-established PCa xenografts
Our data show that siGUGUGUA and siUGUUUGC target networks of AR coregulatory and essential genes. We therefore hypothesized that siGUGUGUA and siUGUUUGC would inhibit the growth and viability of PCa cells more significantly than siRNAs that target individual AR coregulators. To test this hypothesis, LNCaP and 22Rv1 cells were individually transfected with siRNAs targeting EP300 (p300),35 HOXB13,34 and KMT2A (MLL) mRNAs,36—which have been previously demonstrated to function as AR coregulators, the toxic siGUGUGUA, siUGUUUGC, or the siNT-1. siRNA targeting the AR was used as a control since we previously showed that is less efficient than the siGUGUGUA or siUGUUUGC siRNAs at inhibiting growth and viability of PCa cells (Figures 2A, 2B, 2E, and 2F). Both siGUGUGUA and siUGUUUGC reduced the growth and viability of LNCaP and 22Rv1 cells more efficiently than the individual targeting of AR or AR coregulators mRNAs (Figures 6A–6D). Based on qRT-PCR analysis of a few targets selected for validation, siGUGUGUA and siUGUUUGC were in general more consistent at reducing the expression of androgen-responsive genes in LNCaP cells than the targeting of individual coregulators (Figure 6E). Interestingly, siGUGUGUA and siUGUUUGC also significantly decreased the expression of EP300, HOXB13, and AR at different levels (Figure 6F).
Figure 6.
siGUGUGUA and siUGUUUGC reduce PCa cell growth and viability more effectively than the knockdown individual AR coregulatory genes
(A–D) Cell growth (A and B) and viability (C and D) of LNCaP (A and C) and 22Rv1 (B and D) cells transfected with the indicated siRNA. Relative confluency was used as a surrogate for cell growth and was determined via IncuCyte. Relative viability was assessed by trypan blue 6 days after siRNA transfection. (E and F) Relative mRNA expression of androgen-responsive genes, AR coregulators, and AR in response to transfection of the indicated siRNA in LNCaP cells. Relative viability and gene expression are expressed as fold and log2(fold) of siNT-1 respectively. n= 3, ∗q < 0.05 was considered significant as determined by t test.
Importantly, while transfection with siGUGUGUA and siUGUUUGC negatively affected PCa cell growth and viability, they had negligible effects on the growth and viability of benign prostate cell lines, NHPre1 and BHPre1 (Figure S23). This suggests that the seed-mediated toxicity is specific to cancer cells, as described previously,17 which is a desirable therapeutic characteristic. To determine the therapeutic potential of this mechanism, we examined the effect of toxic shRNAs on the growth of pre-established tumors. To this end, we established xenografts with 22Rv1 cells expressing the Dox-inducible shUGUUUGC or shNT as control. After xenografts reached a volume of approximately 120 mm3, mice were switched to a Dox-containing diet. Dox induction of shUGUUUGC significantly reduced xenograft growth rate compared with xenografts expressing shNT (Figure S24), suggesting that shUGUUUGC can inhibit pre-established growth of advanced PCa in vivo. Overall our data suggest that, through a multi-target mechanism, siRNAs such as siGUGUGUA and siUGUUUGC may offer a therapeutic advantage over the targeting AR or individual AR coregulators in the treatment of PCa.
Discussion
Development of resistance to AR signaling targeted therapies is the main cause of PCa-related mortality and the reason PCa remains the second leading cause of cancer-related death in men.1,2 Identification of novel therapeutic modalities that prevent and/or cure resistant disease are therefore essential for the effective management of PCa. The broad range of adaptations that account for development of resistance to AR signaling targeted therapies, have prompted recent clinical trials that asses the efficacy of double or triple combination therapies for castration-sensitive and CRPC.37,38,39,40,41,42 The results of most of these trials are currently limited, but some demonstrated a significant overall survival benefit.37,42 While these developments point to the promise of combination therapy for the treatment of this disease, they also underline the difficulties of finding the appropriate combinations for maximum efficacy and limited interactions.
AR coregulators modulate AR transcriptional output, and therefore simultaneously targeting multiple coregulators, alone or in combination with AR-directed therapies, could represent a very effective synergistic therapeutic approach for PCa. Unfortunately, the functional redundancy among coregulators, our incomplete knowledge of the AR coregulatory arsenal and function, and the lack of specific inhibitors complicate this approach. A potential path to overcome these limitations is the use of a seed-mediated RNAi approach. Similar to miRNAs, a single exogenous small RNA can target multitude of mRNAs via their seed sequence,43,44,45 ultimately preventing the expression of the corresponding proteins. While miRNAs often have a modest effect on individual targets, the effect is amplified due to the fact that the targets of a single miRNA frequently encode proteins that participate in common pathways,15,16 resulting in an additive or synergistic effect. This aspect is especially relevant for PCa where growth and survival of cancer cells is largely dominated by the AR signaling pathway.
We began assessing the potential for seed-mediated RNAi as a therapeutic approach for PCa by running a negative selection screen using a novel shRNA library expressing all the potential 7mer seed sequences—within an otherwise identical stem-loop backbone—to identify those shRNAs with deleterious effect to PCa cells. Using this unbiased methodology, we were able to identify 88 shRNAs with distinct 7mer seed sequences whose expression in PCa cells led to growth inhibition and/or loss of viability (depleted from the population). Predicted target analyses demonstrated an enrichment for AR coregulatory and hormone signaling-related gene sets among the genes predicted to be targeted by numerous depleted shRNAs, supporting the hypothesis that androgen signaling regulatory networks can be targeted by RNAi to inhibit the growth of PCa cells. Furthermore, siRNA with two selected seed sequences, GUGUGUA and UGUUUGC, were shown to (1) reduce PCa cell growth and viability in vitro, (2) lower the expression of the AR (including AR-V7), as well as AR coregulatory and essential gene networks associated with 3′ UTR sequences complementary to the seed, and (3) inhibit global androgen signaling and cell-cycle gene expression. Validation of some of the targets of the toxic siRNAs indicated that, while the AR and AR coregulatory genes contained 3′ UTR sequences complementary to the seeds of the toxic siRNAs, the AR-regulated genes did not. Based on these and other data indicating a toxic siRNA seed-mediated mechanism associated with the loss of cell growth/viability, we conclude that it is the direct targeting of a network of AR coregulators that leads to inhibition of global AR signaling and subsequent downregulation of AR-regulated essential genes and inhibition of cell growth/viability. The effects in PCa cells were evident in both ADPC and CRPC cells. Importantly, we demonstrated that both siGUGUGUA and siUGUUUGC reduced PCa cell growth and viability to a greater extent than individually targeting AR or single AR coregulators using siRNA. Finally, in vivo, expression of a shRNA with the UGUUUGC seed sequence reduced xenograft tumor growth. These data support the validity and potential for using RNAi to target networks of AR coregulatory and essential genes for the treatment of PCa.
Several studies have investigated AR targeted microRNAs (miRNAs) in PCa,25,46,47 but little work has been done exploring the potential of miRNAs to modulate androgen signaling regulatory networks. One study revealed that several tumor-suppressive miRNAs that are downregulated in CRPC directly target the AR in addition to some of the NCOA family coregulators.48 Given the function of miRNAs to target numerous genes, we hypothesize that some endogenous miRNAs may function to regulate hormone signaling through lowering the expression of receptors and coregulators. In fact, several of the siRNAs with seed sequences identified in our screen and that we validated to reduce PCa cell growth and viability (Figure 2) share seed sequences with a known miRNA, miR-34 (GGCAGUG), and with other miRNA candidates: miR-6867-5p, GUGUGU; miR-7150, UGGCAG; miR-3924, UAUGUA; miR-3652/4430/4505/5787, GGCUGG. In addition to targeting the AR, siRNAs containing all of the above seeds showed an enrichment of AR coregulatory genes among their predicted targets. In fact, while miR-34 has been shown to have a role in PCa through regulating AR expression,24,25 our data suggest that it regulates androgen signaling likely through targeting several AR coregulators as well. Together our results support the concept that miRNAs control hormone signaling networks.
The success of our approach relies on the ability of small RNAs to directly affect the expression of many target mRNAs by acting essentially as miRNAs. While the main caveat of this kind of approach is the potential for unwanted toxicities, i.e., immune stimulation,49,50,51,52 our results indicated that the selected siRNAs have minimal effects on the growth/viability of benign prostate cells, suggesting minimal toxicity and consistent with previous results.17,53 We did not observe toxicity in studies in which the seed sequence was moved out of place or rearranged (Figure S6) while still conserving putative immunostimulatory motifs.49,50,51 While this suggests that the toxicity of the selected siRNAs is not due to immune response, strategies to mitigate this effect have been described and could be implemented.54,55,56,57 Moreover, most toxic shRNAs identified in our screen do not seem to primarily target general essential genes as we did not observe enrichment of the non-PCa essential gene set within their predicted targets. In fact, when considering the group of eight siRNAs selected to validate our screen, there was no correlation between their effect on the observed PCa cell viability and on the viability of other cancer cells as determined in silico using the 6merdb (www.6merdb.org).19 This points to PCa-specific toxicity for a subset of the selected siRNAs.
While our approach was aimed to identify therapeutics for PCa, its applications go far beyond. It is possible that many of the sequences identified in the screen could also have therapeutic potential in other hormone-driven cancers, such as breast and ovarian cancers. In fact, several hormone signaling regulatory gene sets involving different receptors (i.e., thyroid, parathyroid, oxytocin, and estrogen receptors) were enriched among genes predicted to be targeted by several depleted shRNAs (Figure 3D), likely due to similarity in RNAi relevant sequences in the 3′ UTR of hormone signaling regulatory genes. Similarly, this novel screen could be conducted in cell lines of other cancer types to identify potentially therapeutic seed sequences. Finally, since our screen revealed enrichment of androgen signaling regulatory networks, it is possible that predicted target analyses from the depleted shRNAs in combination with other biochemical and bioinformatic approaches could be helpful to identify novel AR coregulators.
In summary, in this study, using a novel seed-based shRNA screen in PCa cancer cell lines, we identified small RNAs that inhibit androgen signaling through seed-mediated targeting of androgen regulatory and essential gene networks, resulting in a reduction in cell growth and viability. These results warrant further investigation into the use of RNAi to target hormone signaling networks as a potential therapeutic strategy for PCa.
Materials and methods
Cell culture
LNCaP (CRL-1740) and 22Rv1 (CRL-2505) cell lines were obtained from American Type Culture Collection (ATCC) (Manassas, VA). LNCaP abl cells were obtained from Dr. Zoran Culig (University of Innsbruck). LNCaP-95 cells were obtained from Dr. Jun Luo (from the Genetic Resources Core Facility at Johns Hopkins University). BHPre1 and NHPre1 cells were obtained from Dr. S. Hayward (NorthShore Research Institute). LentiX-293T packaging cells were obtained from Clontech/Takara Bio (Mountain View, CA). None of these cell lines are on the list of contaminated and misidentified cell lines reported by ICLAC (https://iclac.org/databases/cross-contaminations/). Cells tested negative for mycoplasm using the Mycosensor PCR assay kit (Agilent, Santa Clara, CA) or the LookOut Mycoplasma PCR detection kit (Sigma-Aldrich, St. Louis, MO). LNCaP abl were subjected to short tandem repeat DNA profiling by IDEXX BioAnalytics (Columbia, MO).
LNCaP abl and LNCaP-95 cell lines were maintained in RPMI 1640 phenol red-free medium (Gibco, Gaithersburg, MD) supplemented with 10% charcoal-stripped serum (CSS) (Corning, Corning, NY), 100 units/mL penicillin, 100 μg/mL streptomycin, amphotericin B, and 2 mM L-glutamine. LNCaP and 22Rv1 cell lines were maintained in RPMI GlutaMAX growth medium (Gibco) supplemented with 10% fetal bovine serum (Corning), 100 units/mL penicillin, 100 μg/mL streptomycin, amphotericin B, and 2 mM L-glutamine. BHPre1 and NHPre1 cell lines were maintained in HPrE-conditional medium, as described previously.58 For growth in presence or absence of DHT (1 or 10 nM DHT; 0.0001% or 0.00001% EtOH vehicle control) 22Rv1 cells were maintained in RPMI GlutaMAX growth medium supplemented with 10% CSS.
shRNA library
The custom lentiviral shRNA library (Cellecta, Mountain View, CA) consisted of 15,572 unique shRNAs. It contains shRNAs with all possible combinations of 7mer seed sequences (except for 812 7mer sequences that could not be cloned due to restriction enzyme and Pol III transcription termination sequence incompatibility). The rest of the sequence (non-seed sequence) is common for all shRNAs (see Figure 1A for shRNA sequence). The backbone of the shRNAs was designed considering studies that indicate that the precision of Dicer cleavage is a result of an optimal distance to an upstream noncomplementary structure (loop).59 Our shRNAs consist of a 21 bp stem and 9 nt loop previously shown to yield consistent processing via Dicer, 2 nt away from the loop.59 Furthermore, the specific sequence surrounding the seed in our mature guide strand essentially mimic an siRNA backbone previously demonstrated to be non-toxic,19,21 except for being 1 nt shorter to accommodate the extra nucleotide in our 7mer seed vs. the 6mer seed used in those studies. shRNAs were cloned in the pRSIT16-U6Tet-sh-CMV-TetRep-2A-TagRFP-2A-Puro vector (SVSHU6T16-L, Cellecta), which allows Dox induction. Each individual shRNA was linked to a unique sequence barcode for shRNA read identification with NGS.
shRNA negative selection screen
For the shRNA screen, all cell lines were maintained in complete RPMI growth medium, except for 22Rv1 cells, which were grown in the presence and absence of 10 nM DHT for 6 days before transductions and throughout the screen. Cells were transduced with the shRNA library at an MOI of 0.35 and 500-fold representation (two repeats per cell line). After puromycin selection, half of the surviving cells (>1,000-fold representation) were collected as “day 0” control samples and stored at −80°C for genomic isolation at a later time. Flow cytometry was used to confirm that greater than 90% of cells were RFP positive. The rest of the cells were kept in low-dose puromycin and 500 ng/mL Dox was added to induce shRNA expression. These cells were continuously passaged at 50%–60% confluency to maintain a minimum of 1,000-fold representation for 10 doublings and then collected as the endpoint sample (for 22Rv1 –DHT and 22Rv1 +DHT cells we also obtained a sample after two doublings). Genomic DNA was extracted from at least 2 × 107 cells (>1,000-fold representation) from the day 0 and the endpoint samples using DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany) followed by DNA precipitation to concentrate the DNA. DNA samples were sequenced by NGS, and sequence reads were normalized as described previously.22 Z scores were then calculated for shRNA normalized read counts in 2 and 10 doubling samples relative to baseline (day 0). Significantly depleted shRNAs were then identified as shown in Figure S1. Average Z score > 0.75 and q < 0.3 across all cell lines was used as a cutoff for selecting a pool of positively selected shRNAs.
Selection of the eight shRNAs—out of the 88 depleted from the screen—for validation using siRNA mimics was based on one or more of the following criteria: (1) shRNAs with predicted targets significantly enriched in AR coregulators, (2) shRNAs with 6mer seeds that conform to the sequence motif, and/or that were present within multiple depleted shRNAs, and (3) shRNAs with the lowest Z scores across the five cell lines used for the screen. Those with the most consistent effects on growth and viability of PCa cell lines (siGUGUGUG, siGUGUGUA, siUGUUUGC) were selected for further validation.
Predicted target and 3′ UTR sequence analyses
The miRNA database (www.mirdb.org) custom prediction tool was used for predicted target analyses. Enrichment of AR coregulatory,26 LNCaP essential,27 and essential18,28,29 gene sets among predicted targets was determined by hypergeometric distribution, and q values were calculated to correct for multiple comparisons. Enrichment of GO and reactome gene sets was analyzed using metascape (www.metascape.org). Fasta files containing Ensembl 103 MANE select transcript 3′ UTR sequences were obtained from ensembl Biomart, and seankross/warppipe (https://rdrr.io/github/seankross/warppipe/) and stringr R packages were used for sequence analyses. ARCTS, LEGTS, and EGTS indicated the enrichment of 6mer and 7mer seed complementary sequences (seed match) in the 3′ UTR of AR coregulatory,26 LNCaP essential,27 and essential18,28,29 genes, respectively, relative to their occurrence across the whole transcriptome (seed match/kbexpected) (Z scores calculated from normalized residuals: (seed match/kbobserved − seed match/kbexpected)/sqrt(seed match/kbexpected)).
For studies using predicted targets of 10% (≥9) of the depleted shRNAs (Figures 3D and S9; Table S2), the cutoff was used to ensure the reliability of the results. Based on a 10% cutoff and assuming a 50% prediction accuracy, there is a >99.8% probability that at least one of the predicted gene-shRNA pairs is correct. Note that 50% prediction accuracy reflects a non-informative prediction model, or random guess, and we expect >50% prediction accuracy, which will further increase the probability that at least one predicted gene-shRNA pair is correct.
siRNA transfections and lentiviral shRNA vector transductions
Unmodified custom siRNAs and ON-TARGET-Plus pool siRNAs were obtained from Dharmacon (Horizon Discovery, Waterbeach, UK). Non-target siRNA seeds were selected due to the low occurrence of complementary sequences in 3′ UTRs across the transcriptome (Ensembl 103 MANE select transcript 3′ UTR sequences) and their lack of toxicity in the screen.
Unmodified siRNA guide strand sequences
siNT-1: UCGUACGAACAUGUAACCG
siNT-2: UUUCGCGAACAUGUAACCG
siGGCAGUG: UGGCAGUGACAUGUAACCG
siUGGCAGA: UUGGCAGAACAUGUAACCG
siGUGUGUG: UGUGUGUGACAUGUAACCG
siGUGUGUA: UGUGUGUAACAUGUAACCG
siUAUGUAC: UUAUGUACACAUGUAACCG
siUGUUUGC: UUGUUUGCACAUGUAACCG
siUACUGGC: UUACUGGCACAUGUAACCG
siGGCUGGC: UGGCUGGCACAUGUAACCG
siCGUGUUU: UCGUGUUUACAUGUAACCG
siNT-1-UGUGUGUG: UCGUACGAUGUGUGUGACAΔ
siCGCAUGU-GUGUG: UCGCAUGUGUGUGACAUGU.
ON-TARGET-plus pool siRNAs: AR (L-003400-00-0005), EP300 (p300) (L-003486-00-0005), HOXB13 (L-012226-00-0005), KMT2A (MLL) (L-009914-00-0005), GAPDH (L-004253-00-0005), NT-pool (D-001810-10-05) from Horizon Discovery.
Dharmafect siRNA transfection protocol was used for transfections (https://horizondiscovery.com/-/media/Files/Horizon/resources/Protocols/basic-dharmafect-protocol.pdf). siRNA (30 nM) was used for each transfection. dharmafect reagent no. 3 (0.2%) was used for transfections in LNCaP, 22Rv1, LNCaP abl, and LNCaP-95 cell lines. dharmafect reagent no. 1 (0.2%) was used for transfections in BHPre1 and NHPre1 cell lines. For experiments involving DHT treatment, 22Rv1 cells were grown in the presence and absence of 1 nM DHT for 6 days before siRNA transfections.
Individual shRNAs were cloned into the pRSIT16-U6Tet-sh-CMV-TetRep-2A-TagRFP-2A-Puro vector (SVSHU6T16-L, Cellecta) using the following oligonucleotides:
shNT For: ACCGGCGGTTACATGTTCGTACGACTCCTGACCCAAGTCGTACGAACATGTAACCGTTTTTGAA
shNT Rev:
CGAATTCAAAAAACGGTTACATGTTCGTACGACTTGGGTCAGGAGTCGTACGAACATGTAACCGC
shGUGUGUA For:
ACCGGCGGTTACATGTTACACACACTCCTGACCCAAGTGTGTGTAACATGTAACCGTTTTTTGAA
shGUGUGUA Rev:
CGAATTCAAAAAACGGTTACATGTTACACACACTTGGGTCAGGAGTGTGTGTAACATGTAACCGC
shUGUUUGC For:
ACCGGCGGTTACATGTGCAAACAACTCCTGACCCAAGTTGTTTGCACATGTAACCGTTTTTTGAA
shUGUUUGC Rev:
CGAATTCAAAAAACGGTTACATGTGCAAACAACTTGGGTCAGGAGTTGTTTGCACATGTAACCGC.
Cells were transduced with lentiviruses containing the above plasmids at an MOI of 20, and were selected for 7 days in 1 μg/mL puromycin.
Cell growth and viability analyses
For cell growth assays, cells were seeded in 96-well plates at a concentration of 3,000 cells per well (LNCaP cells were seeded 4,000 cell per well) 24 h after siRNA transfections or 48 h after Dox treatment for cells expressing shRNAs. Cell growth was assessed using the IncuCyte Live Cell Imaging System. Percent confluence was measured every 6 h, and values were normalized to percent confluence at the first reading (relative confluence). Trypan blue was utilized to calculate cell viability using the Nexcelom Cellometer Auto T4 after the completion of growth assays (132 or 186 h after siRNA transfections, and 156 h after Dox treatment for cells expressing shRNAs). Relative percent viability was calculated by dividing percent viability of cells transfected/transduced with experimental siRNA/shRNA by that of cells transfected/transduced with non-target control.
Statistics: two-tailed t tests were used to calculate significant differences in growth and viability; n = 3 or 4. p < 0.05 was considered to be statistically significant (FDR (q) was calculated when indicated to correct for multiple comparisons, q < 0.05 was considered significant).
Mouse xenografts
Animal procedures were approved by the Oklahoma Medical Research Foundation Institutional Animal Care and Use Committee. Immunodeficient NRG (no. 007799) or NOD/Scid (no. 001303) mice (The Jackson Laboratory, Bar Harbor, ME) were used for this study. 22Rv1 cells expressing Dox-inducible shNT or shUGUUUGC shRNAs were mixed with 50% basement membrane extract (R&D Systems, Minneapolis, MN) and injected subcutaneously into the flanks (3 × 106 cell per injection). Mice were fed Dox-containing chow (200 mg/kg, Bioserv, Flemington, NJ) 2 days before the injection and for the remaining of the experiment and tumor volume was measured three times per week using calipers. Tumor volume was calculated using the formula ½(length × width2). Significant differences in tumor growth between shUGUUUGC- and shNT-expressing cells were determined using a segmented linear mixed model. Specifically, we first identified the transition point and then we fitted a linear mixed model for the data before and after the transition point. R packages segmented, Ime4, ImerTest, nlme, and emmeans were used. p < 0.05 was considered significant. For therapeutic experiments, after injections, mice were fed with normal chow until the tumors reached a volume of 100–120 mm3 and then transferred to Dox-containing diet until the end of the experiment.
Western blot analysis and antibodies
Whole-cell lysates were obtained using Cell Signaling lysis buffer (Cell Signaling Technology, Danvers, MA, 9803). Stain free mini-PROTEAN TGX gels (Bio-Rad, Hercules, CA) were used for SDS-PAGE protein separation, and proteins were transferred to 0.2 μm nitrocellulose membranes using the turbo transfer system (Bio-Rad). Membranes were incubated in 5% nonfat dry milk (NFDM) in Tris-buffered saline with 0.1% Tween 20 (TBST) for 1 h for blocking, and then incubated overnight in primary antibodies diluted in 5% NFDM TBST. The following antibodies and dilutions were used: anti-PSA (Abcam, Cambridge, UK, 76113) 1:1,000, anti-FKBP5 (Abcam, 2901) 1:1,000, anti-CDK1 (Abcam, A17) 1:1,000, anti-AR ( Active Motif, Carlsbad, CA, 39781) 1:1,000, anti-Cleaved PARP (Asp214) (Cell Signaling Technology, 5625S) 1:1,000, anti-phospho-H2AX Ser139 (Cell Signaling Technology, 9718S) 1:1,000, anti-Calnexin (Abcam, 22595) 1:4,000, anti-GAPDH (Cell Signaling Technology, 2118).
Membranes were incubated for 1 h in secondary antibody diluted in 5% NFDM TBST at a concentration of 80 ng/mL: goat anti-rabbit or goat anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibody (Thermo Fisher Scientific, Waltham, MA, 31460 and 31460, respectively). Clarity Western ECL (Bio-Rad) or SuperSignal West Femto (Thermo Fisher Scientific) and the ChemiDoc Touch Imaging System (Bio-Rad) were used for chemiluminescent detection.
qRT-PCR
RNeasy mini kit (QIAGEN) with on-column DNase incubation was used for RNA extractions. iScript reverse transcription supermix (Bio-Rad) was used for reverse transcription reactions (500 ng RNA per reaction). SSO Advanced or iTaq Universal SYBR Green supermixes (Bio-Rad) were used for qRT-PCR reactions in 96-well plates (25 ng cDNA per well), and the Biorad CFX96 touch RT-PCR detection system was used. The ΔΔCT method was used for quantification, and the geometric mean of the CT values of three housekeeping genes (RPL8, RPL38, PSMA1) was used for normalization. The following primers were used:
KLK3: For (5′-CTTACCACCTGCACCCGGAG-3′)
Rev (5′-TGCAGCACCAATCCACGTCA-3′)
KLK2: For (5′-AGAGGAGTTCTTGCGCCCC-3′)
Rev (5′-CCCAGCACACAACATGAACTCT-3′)
TMPRSS2: For (5′-CCTCTGACTTTCAACGACCTAGTG-3′)
Rev (5′-TCTTCCCTTTCTCCTCGGTGG-3′)
NKX3-1: For (5′-CAGAGACCGAGCCAGAAAGG-3′)
Rev (5′-ACTCGATCACCTGAGTGTGGG-3′)
AR: For (5′-CCAGGGACCATGTTTTGCC-3′)
Rev (5′-CGAAGACGACAAGATGGACAA-3′)
AR Full: For (5′-AGACAACCCAGAAGCTGACAGTG-3′)
Rev (5′-GTGTAAGTTGCGGAAGCCAGG-3′)
AR-V7: For (5′-AATTGTCCATCTTGTCGTCTTCGG-3′)
Rev (5′-GAGTCAGCAATCAAGAGAGTAGCC-3′)
CDC25A: For (5′-CATGAGAACTACAAACCTTGACAACC-3′)
Rev (5′-CCCAGACATGCTCTTCCTCCTC-3′)
EXO1: For (5′-CTGCAGAGTTCAAATGCATCA-3′)
Rev (5′-CGTAGCTTGGAGGTCTGGTC-3′)
CENPN: For (5′-TACACCGCTTCTGGGTCAGG-3′)
Rev (5′-CTGTAGAGGTGTCGTAGAGTTGTGAG-3′)
CDC45: For (5′-CATGACAGCCTGTGCAACAC-3′)
Rev (5′-GGGAAGACCCATGTCTGCAA-3′)
SENP1: For (5′-TTAGTACAGCAGAAGAGACAGTTCAAG-3′)
Rev (5′-ACTGGAACTAAGACATCGAGACAGG-3′)
CDK6: For (5′-GATGGCTCTAACCTCAGTGGTCG-3′)
Rev (5′-AGTTGATCAACATCTGAACTTCCACG-3′)
CCND1: For (5′-ATGCCAACCTCCTCAACGACC-3′)
Rev (5′-CTGTTCCTCGCAGACCTCCAG-3′)
RNF6: For (5′-GAGAGATGGAACGAATTACAGAGACTC-3′)
Rev (5′-CCAAACTAAACCGAAACTCTCCATTG-3′)
PPP1CC: For (5′-GGAGACGATCTGCCTCTTACTGG-3′)
Rev (5′-TGAAGATCTGGTGATAAACCTCCATG-3′)
EP300: For (5′-CCAGATGGGAGGACAAACAGGA-3′)
Rev (5′-CTGGCTGTTGACCCATGTTGG-3′)
HOXB13: For (5′-GGAAGGCAGCATTTGCAGACTC-3′)
Rev (5′-CGCCTCTTGTCCTTGGTGATG-3′)
KMT2A: For (5′-ATGGTGATGACAGTGCTAATGATGC-3′)
Rev (5′-GTTGCTGGTGCAGGATGTGAGAC-3′)
RPL8: For (5′-CACCGTTATCTCCCACAACCCT-3′)
Rev (5′-AGCCACCACACCAACCACAG-3′)
RPL38: For (5′-ACTTCCTGCTCACAGCCCGA-3′)
Rev (5′-TCAGTTCCTTCACTGCCAAACCG-3′)
PSMA1: For (5′-CTGCCTGTGTCTCGTCTTGTATC-3′)
Rev (5′-GGCCCATATCATCATAACCAGCA-3′)
Statistics: two-tailed t tests were used to calculate significant differential gene expression; n = 3. p < 0.05 was considered to be statistically significant (FDR (q) was calculated when indicated to correct for multiple comparisons, q < 0.05 was considered significant).
RNA-seq
LNCaP, LNCaP abl, and 22Rv1 −/+ DHT cells were transfected with siGUGUGUA, siUGUUUGC, siNT-1, or siAR pool. Three repeats were conducted per siRNA in each cell line. RNA was extracted as for qRT-PCR at 40, 48, and 64 h after siRNA transfections in LNCaP, LNCaP abl, and 22Rv1 −/+DHT cells, respectively. RNA integrity was ensured using Agilent 2100, and mRNAs were isolated using poly(T)-coated beads and subsequently fragmented. Deoxythymidine triphosphate to deoxyuridine triphosphate containing cDNA libraries were generated, ligated to NEBNext Adaptor, PCR amplified, and purified using AMPure XP beads. Sequencing was carried out with the Illumina Next-Generation sequencer at a depth of 20 million reads per sample. FastQC (Novogene) was used for quality control and the elimination of low-quality reads. STAR was used to map reads to the human genome/transcriptome and the DESeq2 R package was used to calculate differential gene expression. The Benjamini and Hochberg method was used for p value adjustment, and average absolute log2 fold change >0.5 relative to siNT transfected cells and adjusted p < 0.05 were used as cut offs for differential gene expression. All the data have been deposited in NCBI GEO under accession no. GSE227987.
Enrichment analyses
GSEA software version 4.2.3 was used for GSEAs60,61 of Hallmark gene sets (50 gene sets) from the Molecular Signature Database (MSigDB) (http://www.gsea-msigdb.org/gsea/index.jsp), as well as AR-regulated and predicted target gene sets. AR-regulated gene sets were as follows: siAR up and down gene sets were generated from the top 150 significantly upregulated and downregulated genes, respectively, for each cell line; siAR-V7 up and down gene sets were obtained from Chen et al.34 (GSE99378) and were selected by absolute log2 fold change >1 and p-adjust <0.05; LNCaP DHT up and down gene sets were obtained from Corbin et al.17 (GSE165249) and were selected by the top 150 significantly upregulated and downregulated genes; AR score was also utilized.32,33 For the analyses, genes were rank ordered from upregulated to downregulated for each comparison relative to siNT transfected cells using the signal-to-noise ratio. One thousand gene set permutations were used for each analysis and FDR < 0.1 was considered significant enrichment.
Essential gene lists were obtained from Fei et al. (999 genes27) and Blomen et al. and Wang et al. (1,210 genes18,28,29), and the AR coregulatory gene list was obtained from DePriest et al. (274 genes26). p values for enrichment among downregulated genes were calculated by hypergeometric distribution and q values were calculated to correct for multiple comparisons:
p is the p value, N is the number of total genes, M is the number of genes in gene set, n is the number of DEGs, and i is the number of overlapped genes of M and n.
DepMap validation analysis: mean PCa CRISPR gene effect scores were generated from data downloaded from depmap.org. Genes were rank ordered from low mean PCa CRISPR gene effect score (essential) to high (non-essential). It was then determined whether siGUGUGUA and siUGUUUGC downregulated genes identified via RNA-seq had significantly different cumulative distributions within the rank ordered list compared with all other genes. Significance was determined by KS test.
cWords and 3′ UTR sequence analyses
Genes were rank ordered from downregulated to upregulated by signal-to-noise metric for each comparison based on the RNA-seq data, and cWords31 webserver (http://servers.binf.ku.dk/cwords/) was used to determine 6 or 7 nucleotide 3′ UTR sequences most associated with downregulation. Ensembl 103 MANE select transcript 3′ UTR sequences were used for analysis. The downregulation of genes containing 3′ UTR sequences complementary to the siRNA seeds was confirmed by Kolmogorov–Smirnov (KS) test, p < 0.05 was considered significant.
AR coregulatory and LNCaP essential gene sets were stratified by significant downregulation compared with siNT transfected cells (yes or no) and by the presence of 7mer and/or multiple 6mer seed match sequences in the 3′ UTR.
Statistics: Significant associations between downregulation and 3′ UTR seed match sequences were determined by Fisher’s exact test, p < 0.05 was considered significant.
Immunohistochemistry
Four μm sections were prepared from paraffin embedded xenografts for immunohistochemistry (IHC) with a Ki67 antibody. IHC was performed according to the manufacturer’s protocol using the Leica Bond-RX Polymer Refine Detection system (DS 9800). In brief, the slides with the sections were deparaffinized and rehydrated in an automated Multistainer (Leica ST5020) and transferred to the Leica Bond-RX for antigen retrieval at 100°C (20 min). Endogenous peroxidase was blocked using peroxidase-blocking reagent, followed by incubation with the Ki67 antibody (Abcam, ab16667) for 30 min, and then post-primary IgG-linker and/or Poly-HRP IgG reagents. For image analysis, the tumors were divided into 4 quadrants and pictures were taken of each quadrant with a Nikon Eclipse Ni-E microscope. Positively stained cells were quantified in selected areas (200–300 cells) using ImageJ.
Acknowledgments
We are grateful to Dr. Zoran Culig (University of Innsbruck), Dr. Jun Luo (Johns Hopkins University), and Dr. S. Hayward (NorthShore Research Institute) for sharing cell lines. We appreciate Cody B. Bullock’s help with testing growth and viability of benign prostate cell lines and Swati Choudhary for help with a western blot added during the review process. This work was supported by the Oklahoma IDeA Network of Biomedical Research Excellence (OK-INBRE) (P20GM103447) and the National Institutes of Health (NIH) (5U54GM104938 to J.D.W.). Research reported in this publication was supported in part by the National Cancer Institute Cancer Center Support Grant (P30CA225520, COBRE P20GM103639), and the Oklahoma Tobacco Settlement Endowment Trust contract awarded to the University of Oklahoma Stephenson Cancer Center, and used the Tissue Pathology and the Molecular Biology and Cytometry Research Shared Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Oklahoma Tobacco Settlement Endowment Trust.
Author contributions
Conceptualization, J.M.C. and M.J.R.E.; methodology, J.M.C. and M.J.R.E.; validation, J.M.C. and M.J.R.E.; formal analysis, J.M.C., L.W., C.G., J.D.W., C.X., and M.J.R.E.; investigation, J.M.C. and M.J.R.E.; resources, C.G., J.D.W., C.X., and M.B.; writing – original draft, J.M.C. and M.J.R.E.; writing – review & editing, J.M.C., C.G., J.D.W., C.X., M.B., A.S.A., and M.J.R.E; supervision, M.B., J.D.W., and M.J.R.E.; funding acquisition, J.D.W. and M.J.R.E.
Declaration of interests
The authors declared no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2023.06.021.
Contributor Information
Joshua M. Corbin, Email: jmcorbin@email.unc.edu.
Maria J. Ruiz Echevarría, Email: maria-ruizechevarria@ouhsc.edu.
Supplemental information
Data availability
RNA sequencing data is publicly available (deposited in NCBI GEO).
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA. Cancer J. Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Davey R.A., Grossmann M. Androgen Receptor Structure, Function and Biology: From Bench to Bedside. Clin. Biochem. Rev. 2016;37:3–15. [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan X., Cai C., Chen S., Chen S., Yu Z., Balk S.P. Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene. 2014;33:2815–2825. doi: 10.1038/onc.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y., Jiang X., Liang X., Jiang G. Molecular and cellular mechanisms of castration resistant prostate cancer. Oncol. Lett. 2018;15:6063–6076. doi: 10.3892/ol.2018.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharp A., Welti J., Blagg J., de Bono J.S. Targeting androgen receptor aberrations in castration-resistant prostate cancer. Clin. Cancer Res. 2016;22:4280–4282. doi: 10.1158/1078-0432.CCR-16-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bluemn E.G., Coleman I.M., Lucas J.M., Coleman R.T., Hernandez-Lopez S., Tharakan R., Bianchi-Frias D., Dumpit R.F., Kaipainen A., Corella A.N., et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell. 2017;32:474–489.e6. doi: 10.1016/j.ccell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coutinho I., Day T.K., Tilley W.D., Selth L.A. Androgen receptor signaling in castration-resistant prostate cancer: a lesson in persistence. Endocr. Relat. Cancer. 2016;23:T179–T197. doi: 10.1530/ERC-16-0422. [DOI] [PubMed] [Google Scholar]
- 8.Jackson A.L., Burchard J., Schelter J., Chau B.N., Cleary M., Lim L., Linsley P.S. Widespread siRNA "off-target" transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zamore P.D., Tuschl T., Sharp P.A., Bartel D.P. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 10.Peters L., Meister G. Argonaute proteins: mediators of RNA silencing. Mol. Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Kehl T., Backes C., Kern F., Fehlmann T., Ludwig N., Meese E., Lenhof H.P., Keller A. About miRNAs, miRNA seeds, target genes and target pathways. Oncotarget. 2017;8:107167–107175. doi: 10.18632/oncotarget.22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 14.Fang Z., Rajewsky N. The impact of miRNA target sites in coding sequences and in 3'UTRs. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satoh J., Tabunoki H. Comprehensive analysis of human microRNA target networks. BioData Min. 2011;4:17. doi: 10.1186/1756-0381-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ebert M.S., Sharp P.A. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corbin J.M., Georgescu C., Wren J.D., Xu C., Asch A.S., Ruiz-Echevarria M.J. Seed-mediated RNA interference of androgen signaling and survival networks induces cell death in prostate cancer cells. Mol. Ther. Nucleic Acids. 2021;24:337–351. doi: 10.1016/j.omtn.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Putzbach W., Gao Q.Q., Patel M., van Dongen S., Haluck-Kangas A., Sarshad A.A., Bartom E.T., Kim K.Y.A., Scholtens D.M., Hafner M., et al. Many si/shRNAs can kill cancer cells by targeting multiple survival genes through an off-target mechanism. Elife. 2017;6:e29702. doi: 10.7554/eLife.29702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Q.Q., Putzbach W.E., Murmann A.E., Chen S., Sarshad A.A., Peter J.M., Bartom E.T., Hafner M., Peter M.E. 6mer seed toxicity in tumor suppressive microRNAs. Nat. Commun. 2018;9:4504. doi: 10.1038/s41467-018-06526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murmann A.E., McMahon K.M., Haluck-Kangas A., Ravindran N., Patel M., Law C.Y., Brockway S., Wei J.J., Thaxton C.S., Peter M.E. Induction of DISE in ovarian cancer cells in vivo. Oncotarget. 2017;8:84643–84658. doi: 10.18632/oncotarget.21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murmann A.E., Gao Q.Q., Putzbach W.E., Patel M., Bartom E.T., Law C.Y., Bridgeman B., Chen S., McMahon K.M., Thaxton C.S., Peter M.E. Small interfering RNAs based on huntingtin trinucleotide repeats are highly toxic to cancer cells. EMBO Rep. 2018;19:e45336. doi: 10.15252/embr.201745336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q.V., Dixon G., Verma N., Rosen B.P., Gordillo M., Luo R., Xu C., Wang Q., Soh C.L., Yang D., et al. Genome-scale screens identify JNK-JUN signaling as a barrier for pluripotency exit and endoderm differentiation. Nat. Genet. 2019;51:999–1010. doi: 10.1038/s41588-019-0408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L., Liao Y., Tang L. MicroRNA-34 family: a potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 2019;38:53. doi: 10.1186/s13046-019-1059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagman Z., Larne O., Edsjö A., Bjartell A., Ehrnström R.A., Ulmert D., Lilja H., Ceder Y. miR-34c is downregulated in prostate cancer and exerts tumor suppressive functions. Int. J. Cancer. 2010;127:2768–2776. doi: 10.1002/ijc.25269. [DOI] [PubMed] [Google Scholar]
- 25.Östling P., Leivonen S.K., Aakula A., Kohonen P., Mäkelä R., Hagman Z., Edsjö A., Kangaspeska S., Edgren H., Nicorici D., et al. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011;71:1956–1967. doi: 10.1158/0008-5472.CAN-10-2421. [DOI] [PubMed] [Google Scholar]
- 26.DePriest A.D., Fiandalo M.V., Schlanger S., Heemers F., Mohler J.L., Liu S., Heemers H.V. Regulators of Androgen Action Resource: a one-stop shop for the comprehensive study of androgen receptor action. Database. 2016;2016 doi: 10.1093/database/bav125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fei T., Chen Y., Xiao T., Li W., Cato L., Zhang P., Cotter M.B., Bowden M., Lis R.T., Zhao S.G., et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc. Natl. Acad. Sci. USA. 2017;114:E5207–E5215. doi: 10.1073/pnas.1617467114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blomen V.A., Májek P., Jae L.T., Bigenzahn J.W., Nieuwenhuis J., Staring J., Sacco R., van Diemen F.R., Olk N., Stukalov A., et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- 29.Wang T., Birsoy K., Hughes N.W., Krupczak K.M., Post Y., Wei J.J., Lander E.S., Sabatini D.M. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsherniak A., Vazquez F., Montgomery P.G., Weir B.A., Kryukov G., Cowley G.S., Gill S., Harrington W.F., Pantel S., Krill-Burger J.M., et al. Defining a Cancer Dependency Map. Cell. 2017;170:564–576. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasmussen S.H., Jacobsen A., Krogh A. cWords - systematic microRNA regulatory motif discovery from mRNA expression data. Silence. 2013;4:2. doi: 10.1186/1758-907X-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hieronymus H., Lamb J., Ross K.N., Peng X.P., Clement C., Rodina A., Nieto M., Du J., Stegmaier K., Raj S.M., et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Hu C., Fang D., Xu H., Wang Q., Xia H. The androgen receptor expression and association with patient's survival in different cancers. Genomics. 2020;112:1926–1940. doi: 10.1016/j.ygeno.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Chen Z., Wu D.Y., Thomas-Ahner J.M., Lu C.X., Zhao P., Zhang Q.F., Geraghty C., Yan P.S., Hankey W., Sunkel B., et al. Diverse AR-V7 cistromes in castration- resistant prostate cancer are governed by HoxB13 (vol 115, pg 6810, 2018) Proc. Natl. Acad. Sci. USA. 2018;115:E9025. doi: 10.1073/pnas.1814741115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin L., Garcia J., Chan E., de la Cruz C., Segal E., Merchant M., Kharbanda S., Raisner R., Haverty P.M., Modrusan Z., et al. Therapeutic Targeting of the CBP/p300 Bromodomain Blocks the Growth of Castration-Resistant Prostate Cancer. Cancer Res. 2017;77:5564–5575. doi: 10.1158/0008-5472.CAN-17-0314. [DOI] [PubMed] [Google Scholar]
- 36.Malik R., Khan A.P., Asangani I.A., Cieślik M., Prensner J.R., Wang X., Iyer M.K., Jiang X., Borkin D., Escara-Wilke J., et al. Targeting the MLL complex in castration-resistant prostate cancer. Nat. Med. 2015;21:344–352. doi: 10.1038/nm.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powers E., Karachaliou G.S., Kao C., Harrison M.R., Hoimes C.J., George D.J., Armstrong A.J., Zhang T. Novel therapies are changing treatment paradigms in metastatic prostate cancer. J. Hematol. Oncol. 2020;13:144. doi: 10.1186/s13045-020-00978-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clarke N., Wiechno P., Alekseev B., Sala N., Jones R., Kocak I., Chiuri V.E., Jassem J., Fléchon A., Redfern C., et al. Olaparib combined with abiraterone in patients with metastatic castration-resistant prostate cancer: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 2018;19:975–986. doi: 10.1016/S1470-2045(18)30365-6. [DOI] [PubMed] [Google Scholar]
- 39.Sharma P., Pachynski R.K., Narayan V., Fléchon A., Gravis G., Galsky M.D., Mahammedi H., Patnaik A., Subudhi S.K., Ciprotti M., et al. Nivolumab Plus Ipilimumab for Metastatic Castration-Resistant Prostate Cancer: Preliminary Analysis of Patients in the CheckMate 650 Trial. Cancer Cell. 2020;38:489–499.e3. doi: 10.1016/j.ccell.2020.08.007. [DOI] [PubMed] [Google Scholar]
- 40.Shenderov E., Boudadi K., Fu W., Wang H., Sullivan R., Jordan A., Dowling D., Harb R., Schonhoft J., Jendrisak A., et al. Nivolumab plus ipilimumab, with or without enzalutamide, in AR-V7-expressing metastatic castration-resistant prostate cancer: A phase-2 nonrandomized clinical trial. Prostate. 2021;81:326–338. doi: 10.1002/pros.24110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Attard G., Murphy L., Clarke N.W., Cross W., Jones R.J., Parker C.C., Gillessen S., Cook A., Brawley C., Amos C.L., et al. Abiraterone acetate and prednisolone with or without enzalutamide for high-risk non-metastatic prostate cancer: a meta-analysis of primary results from two randomised controlled phase 3 trials of the STAMPEDE platform protocol. Lancet. 2022;399:447–460. doi: 10.1016/S0140-6736(21)02437-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saad F., Bögemann M., Suzuki K., Shore N. Treatment of nonmetastatic castration-resistant prostate cancer: focus on second-generation androgen receptor inhibitors. Prostate Cancer Prostatic Dis. 2021;24:323–334. doi: 10.1038/s41391-020-00310-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackson A.L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J.M., Lim L., Karpilow J., Nichols K., et al. Position-specific chemical modification of siRNAs reduces "off-target" transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson A.L., Linsley P.S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 45.Kamola P.J., Nakano Y., Takahashi T., Wilson P.A., Ui-Tei K. The siRNA non-seed region and its target sequences are auxiliary determinants of off-target effects. PLoS Comput. Biol. 2015;11:e1004656. doi: 10.1371/journal.pcbi.1004656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fletcher C.E., Sulpice E., Combe S., Shibakawa A., Leach D.A., Hamilton M.P., Chrysostomou S.L., Sharp A., Welti J., Yuan W., et al. Androgen receptor-modulatory microRNAs provide insight into therapy resistance and therapeutic targets in advanced prostate cancer. Oncogene. 2019;38:5700–5724. doi: 10.1038/s41388-019-0823-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bielska A., Skwarska A., Kretowski A., Niemira M. The role of androgen receptor and microRNA interactions in androgen-dependent diseases. Int. J. Mol. Sci. 2022;23:1553. doi: 10.3390/ijms23031553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coarfa C., Fiskus W., Eedunuri V.K., Rajapakshe K., Foley C., Chew S.A., Shah S.S., Geng C., Shou J., Mohamed J.S., et al. Comprehensive proteomic profiling identifies the androgen receptor axis and other signaling pathways as targets of microRNAs suppressed in metastatic prostate cancer. Oncogene. 2016;35:2345–2356. doi: 10.1038/onc.2015.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meng Z., Lu M. RNA interference-induced innate immunity, off-target effect, or immune adjuvant? Front. Immunol. 2017;8:331. doi: 10.3389/fimmu.2017.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hornung V., Guenthner-Biller M., Bourquin C., Ablasser A., Schlee M., Uematsu S., Noronha A., Manoharan M., Akira S., de Fougerolles A., et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 51.Forsbach A., Nemorin J.G., Montino C., Müller C., Samulowitz U., Vicari A.P., Jurk M., Mutwiri G.K., Krieg A.M., Lipford G.B., Vollmer J. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J. Immunol. 2008;180:3729–3738. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- 52.Bartoszewski R., Sikorski A.F. Editorial focus: understanding off-target effects as the key to successful RNAi therapy. Cell. Mol. Biol. Lett. 2019;24:69. doi: 10.1186/s11658-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hadji A., Ceppi P., Murmann A.E., Brockway S., Pattanayak A., Bhinder B., Hau A., De Chant S., Parimi V., Kolesza P., et al. Death induced by CD95 or CD95 ligand elimination. Cell Rep. 2014;7:208–222. doi: 10.1016/j.celrep.2014.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sioud M. Single-stranded small interfering RNA are more immunostimulatory than their double-stranded counterparts: a central role for 2'-hydroxyl uridines in immune responses. Eur. J. Immunol. 2006;36:1222–1230. doi: 10.1002/eji.200535708. [DOI] [PubMed] [Google Scholar]
- 55.Sajid M.I., Moazzam M., Kato S., Yeseom Cho K., Tiwari R.K. Overcoming barriers for siRNA therapeutics: from bench to bedside. Pharmaceuticals. 2020;13 doi: 10.3390/ph13100294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee Y., Urban J.H., Xu L., Sullenger B.A., Lee J. 2'Fluoro modification differentially modulates the ability of RNAs to activate pattern recognition receptors. Nucleic Acid Therapeut. 2016;26:173–182. doi: 10.1089/nat.2015.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deleavey G.F., Watts J.K., Damha M.J. Chemical modification of siRNA. Curr. Protoc. Nucleic Acid Chem. 2009;Chapter 16 doi: 10.1002/0471142700.nc1603s39. Unit 16.3. [DOI] [PubMed] [Google Scholar]
- 58.Jiang M., Strand D.W., Fernandez S., He Y., Yi Y., Birbach A., Qiu Q., Schmid J., Tang D.G., Hayward S.W. Functional remodeling of benign human prostatic tissues in vivo by spontaneously immortalized progenitor and intermediate cells. Stem Cell. 2010;28:344–356. doi: 10.1002/stem.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu S., Jin L., Zhang Y., Huang Y., Zhang F., Valdmanis P.N., Kay M.A. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 61.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data is publicly available (deposited in NCBI GEO).