Abstract
Proteostasis reinforcement is a promising approach in the design of therapeutic interventions against proteinopathies, including Alzheimer's disease. Understanding how and which parts of the proteostasis network should be enhanced is crucial in developing efficient therapeutic strategies. The ability of specific tissues to induce proteostatic responses in distal ones (cell non-autonomous regulation of proteostasis) is attracting interest. Although the proteasome is a major protein degradation node, nothing is known on its cell non-autonomous regulation. We show that proteasome activation in the nervous system can enhance the proteasome activity in the muscle of Caenorhabditis elegans. Mechanistically, this communication depends on Small Clear Vesicles, with glutamate as one of the neurotransmitters required for the distal regulation. More importantly, we demonstrate that this cell non-autonomous proteasome activation is translated into efficient prevention of amyloid-beta (Αβ)-mediated proteotoxic effects in the muscle of C. elegans but notably not to resistance against oxidative stress. Our in vivo data establish a mechanistic link between neuronal proteasome reinforcement and decreased Aβ proteotoxicity in the muscle. The identified distal communication may have serious implications in the design of therapeutic strategies based on tissue-specific proteasome manipulation.
Keywords: C. elegans, Cell non-autonomous regulation, Proteasome, Proteinopathies, Proteostasis
Graphical abstract
Highlights
-
•
Proteasome function is subjected to cell non-autonomous regulation.
-
•
Neuron-specific proteasome activation enhances the muscular proteasome activity.
-
•
This distal communication depends on Small Clear Vesicles and glutamate.
-
•
This distal regulation results in protection against Aβ-induced proteotoxicity.
-
•
This distal regulation is not effective for oxidative stress-related toxicity.
Abbreviations
- AD
Alzheimer’s disease
- ALP
autophagy lysosomal pathway
- APP
Amyloid precursor protein
- Aβ
amyloid-beta
- Aβ1-42
amyloid-beta amino acids 1 to 42
- C. elegans
Caenorhabditis elegans
- DCVs
dense core vesicles
- dFP
dual fluorescent protein
- DMSO
dimethyl sulfoxide
- GABA
γ-aminobutyric acid
- HSR
heat shock response
- mFP
monomeric fluorescent protein
- OE
overexpression
- RNAi
RNA interference
- SCVs
small clear vesicles
- UPRER
unfolded protein response related to the endoplasmic reticulum
- UPRmit
unfolded protein response related to mitochondria
- UPS
ubiquitin-proteasome system
- wt
wild type
1. Introduction
Organisms are exposed to various stresses throughout their whole lifespan. To preserve their protein homeostasis (proteostasis) and therefore ensure the integrity of the proteome, cells have developed a complex proteostasis network regulating protein synthesis, folding and protein degradation through either the ubiquitin-proteasome system (UPS) or the autophagy lysosomal pathway (ALP). There are also surveillance stress response pathways that detect and respond to protein damage; the heat shock response (HSR) and the unfolded protein response in mitochondria (UPRmit) and endoplasmic reticulum (UPRER). The proteostasis mechanisms have been shown to become deregulated during ageing and age-related diseases, including proteinopathies [1]. Given this failure of proteostasis along with the fact that ageing is the major risk factor for neurodegenerative diseases, it is reasonable to assume that the induction of proteostatic mechanisms and the elucidation of the involved pathways are central to design preventive/therapeutic strategies against both ageing and neurodegenerative diseases [2].
The proteasome is one of the major molecular regulators of cellular proteostasis and one of the main secondary antioxidant mechanisms subjected to tight redox control [3]. It is responsible for the rapid degradation of misfolded and damaged proteins but also the proteolysis of normal cellular proteins [4]. The 26S/30S proteasome consists of two different sub-complexes, the 20S core and the 19S regulatory particle. The 20S complex can be capped by one (26S) or two (30S) 19S regulatory particle(s). The 20S particle is composed of 28 subunits (14 α-type and 14 β-type) and exerts its proteolytic activities through the β1, β2 and β5 subunits that host the caspase-like, trypsin-like, and chymotrypsin-like activities, respectively [5]. There is increasing interest in the upregulation of proteasome function through various means, including the direct stereochemical alterations of the 20S core that might lead to enhanced activity levels due to the opening of the α-gated channel of the core, the activity-modifying phosphorylation of the proteasome itself or the enhanced expression of proteasome subunits; the translational ramifications for proteinopathies are obvious [6,7]. We and others have previously shown that overexpression of 20S [β5 subunit and its orthologs/homologs in the nematode C. elegans (pbs-5), Drosophila (Prosβ5) and mice (PSMB5)], or 19S [PSMD11 subunit and its homolog in the nematode (rpn-6)] subunits is sufficient to enhance proteasome activity in human cells, C. elegans, Drosophila and mice and to positively modulate ageing progression and protect against proteotoxicity in Αβ-, amyloid precursor protein (APP)- or polyglutamine-expressing animal models [[8], [9], [10], [11], [12]].
The cell autonomous regulation of various key proteostasis players is increasingly revealed but there are still many molecular processes that need to be elucidated. Notably, the cell non-autonomous regulation of a few, but importantly not all, key players has also emerged the past few years, further pinpointing the need to understand how tissues can exchange signals, thus affecting phenomenically distal tissues and impacting on organismal physiology. In this regard, C. elegans has played a central role in the investigation of the proteostatic cell non-autonomous responses in vivo. More specifically, regulation of the cellular heat shock response of somatic cells in C. elegans is regulated in a cell non-autonomous manner by thermosensory neurons [13]. Mitochondrial deficiency in neurons activates the UPRmit in the intestine of C. elegans [14,15]. Likewise, germline proteostasis affects mitochondrial function and protein aggregation across multiple tissues via the UPRmit and long-range Wnt signaling pathway [16], while binding of an aggregation-prone protein expressed in the neurons of C. elegans to the mitochondria elicits a global induction of UPRmit, affecting whole-animal physiology [17]. This inter-tissue communication also extends to the UPRER; overexpression of a UPRER-related protein in the nervous system of C. elegans leads to the activation of UPRER in non-neuronal tissues of the animal [18,19]. Neuronal and intestinal overexpression of the autophagy-related gene atg-18 in C. elegans leads to lifespan extension in a cell non-autonomous manner [20], while a microRNA produced and secreted by the intestine can regulate the autophagy levels in the body wall muscle of the animal [21]. Cell non-autonomous proteostasis regulation has been also reported in Drosophila. Neuron-specific upregulation of AMPK (AMP-activated protein kinase) can elevate autophagy in the intestine of the flies [22]. Similarly, neuronal overexpression of Gclc (glutamate-cysteine ligase catalytic subunit) increases longevity in Drosophila and causes longevity-associated gene transcription changes in the thorax [23]. Despite the highly important role of the proteasome, it is noteworthy that no reports exist so far on its potential cell non-autonomous regulation and the potential downstream effects on animal physiology.
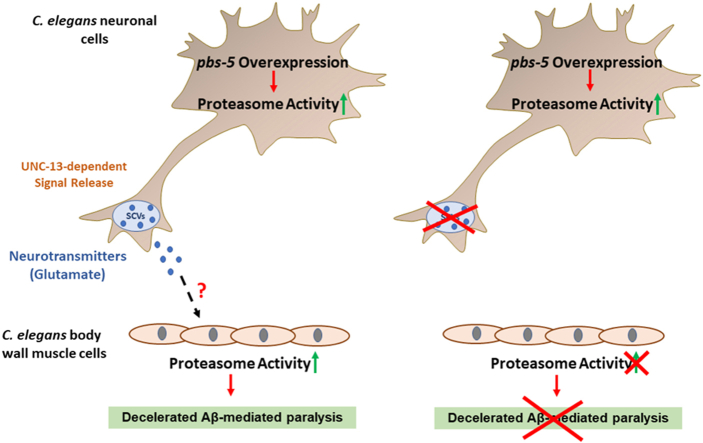
With the knowledge that proteasome activity can be pharmacologically modulated and, therefore, may be exploited for novel therapeutic approaches, we sought to unravel the potential cell-non autonomous regulation of the proteasome and the downstream effects on proteotoxicity in a distal tissue. To this end, we generated transgenic nematodes overexpressing the pbs-5 proteasome subunit in the nervous system and examined its effects in the muscle tissue of the animal. Our results reveal that pbs-5 overexpression in the nervous system is sufficient to cell non-autonomously increase the proteasome activity and function in the muscle. We also provide evidence that this interconnection occurs via a specific type of synaptic vesicles, the small clear vesicles (SCVs). Furthermore, we identify glutamate as at least one of the responsible neurotransmitters and we reveal that the dense core vesicles (DCVs), a different type of synaptic vesicles, are also involved in the proteasome-regulating signal transmission. Together, our in vivo findings may significantly contribute to shaping our current perception of how the proteasome should be manipulated in therapeutic strategies targeting proteinopathies.
2. Materials and Methods
2.1. C. elegans strains and culture
All strains were grown and maintained at 20°C, unless otherwise specified, using standard techniques for maintenance [24] and RNAi experiments [25]. The following C. elegans strains were obtained from the Caenorhabditis Genetics Center (CGC): wild type (wt) N2 (Bristol), CL4176 dvIs27[myo-3p:Αβ1-42:let-851 3’UTR + rol-6(su1006)]), DLM9 uwaEx5[myo-3p::CERULEAN-VENUSlgg-1 + unc-119(+)], GMC101 dvIs100[unc-54p::Αβ1-42::unc-54 3'UTR + mtl-2p::GFP], CB246 unc-64(e246), CB928 unc-31(e928), KR1787 unc-13(e51). The modified ZsProSensor-1 reporter strains YD114 xzIs2[unc-54p::UIM2ZsProSensor-1] and YD70 xzIs1[vha-6p::UIM2::ZsProSensor-1] were a kind gift from Dr. Carina I. Holmberg (University of Helsinki). The strain expressing Aβ in glutamatergic neurons UA198 baIn34[eat-4p::ssAβ42 + myo-2p::mCherry]; adIs1240[eat-4p::GFP] [26] was a kind gift from Prof. Guy A. Caldwell (The University of Alabama). The following strains carrying extrachromosomal arrays were also used: Ex[myo-2p::GFP], Ex[let-858p::pbs-5 + myo-2p::GFP], Ex[rgef-1p:pbs-5 + myo-2p::GFP], Ex[rol-6(su1006)], Ex[let-858p::pbs-5 + rol-6(su1006)] and Ex[rgef-1p:pbs-5 + rol-6(su1006)]. A full list of the strains used in this work is documented in Table S1.
2.2. Cloning and C. elegans transgenesis
For the construct encoding the rgef-1p::pbs-5 transgene, the Gateway® Cloning (Invitrogen life technologies, CA, USA) was used. The rgef-1p was amplified with a PCR on gDNA of wt nematodes with the attB primers Fw: GGGGACAAGTTTGTACAAAAAAGCAGGCTTActtt actgctcatcgtcgtcgt and Rev: GGGGACAACTTTTGTATACAAAGTTGTcgtttccgatacccccttatatca gc and recombined with BP recombinase in pDONR™ 221 P1-P5r. The sequence of pbs-5 was amplified also from wt gDNA with the attB primers Fw: GGGGACAACTTTGTATACAAAAGTTGTAATG TGGGGCGAGACATTCG and Rev: GGGGACCACTTTGTACAAGAAAGCTGGGTTACGT CATCAACACCCAGCC and recombined with BP recombinase in pDONR221™ P5-P2. The two entry clones carrying rgef-1p and pbs-5 were recombined with pGC93 with the LR recombinase to produce an expression clone. This vector was microinjected in the gonads of wt nematodes at 50 ng/μL together with either 5 ng/μL of the co-injection marker myo-2p::GFP or 50 ng/μL of the pRF4 plasmid rol-6(su1006). The nematode strain overexpressing pbs-5 under let-858p was generated using a construct made previously [10] and microinjected at 20 ng/μL. Control strains were microinjected with either co-injection marker only, at the same concentrations. All transgenes were carried in extrachromosomal arrays.
2.3. C. elegans treatment with neurotransmitters
All neurotransmitters (acetylcholine; Alfa Aesar, MA, USA, #L02168, GABA; Acros Organics, Belgium, #103280250, glutamate; Alfa Aesar, MA, USA, #A12919, octopamine; Thermo Scientific, MA, USA, #J61281.03, serotonin; Sigma-Aldrich, MO, USA, #H7752, dopamine; Alfa Aesar, MA, USA, #A11136.06, tyramine; Fluorochem, UK, #078887), were dissolved in M9 buffer (42 mM Na2HPO4, 22 mM KH2PO4, 85 mM NaCl, 1 mM MgSO4). The stock solutions were diluted with M9 and 200 μL were spread onto plates seeded with UV-inactivated OP50. Nematodes of L3 stage were transferred on these NGM plates containing acetylcholine, GABA, glutamate, octopamine, serotonin, dopamine or tyramine at final concentrations 50 μM - 100 mM, 150 μM - 30 mM, 15 μM - 15 mM, 40 μΜ - 20 mM, 1.5 μΜ - 73 μΜ, 5.2 μΜ - 158 μΜ and 14 μΜ - 143 μΜ respectively. The nematodes were treated for 20 h and imaged at the stage of young adult for unc-13(e51) and of L4 for unc-64(e246) mutants.
2.4. C. elegans treatment with epoxomicin
Epoxomicin (UBPBio, USA, #F1440-5) was dissolved in DMSO (180 mM stock solution). C. elegans embryos were obtained following treatment of gravid adults with a hypochlorite solution and seeding onto NGM plates with UV-killed bacteria. Once the nematodes reached the L3 stage they were transferred into a microcentrifuge tube with 200 μL M9 in which the pellet of 4 mL heat-inactivated OP50 culture had been dissolved, containing 1 mM epoxomicin [27] or DMSO (vehicle). In the case of epoxomicin-treated Muscle ZsProSensor-1 reporter animals, the tubes were stirred for 12 h at 20°C before the animals were imaged. In the case of paralysis experiments with CL4176 animals, the epoxomicin treatment was started at the time of the temperature upshift from 16°C to 25°C and after 12 h the animals were transferred on regular NGM plates seeded with UV-killed OP50. The paralysis assessment was then performed as described below.
2.5. C. elegans treatment with chloroquine
Chloroquine (TCI Chemicals, Tokyo Japan, #C2301) was dissolved in M9 buffer (20 mM final working solution). Synchronized day 1 adult nematodes were transferred into a microcentrifuge tube with 200 μL M9 or 200 μL M9+chloroquine. In either case the pellet of 4 mL heat-inactivated OP50 culture had been dissolved in the tube. The tubes were stirred for 16 h at 20°C before the animals were rinsed and collected for protein extraction and western blotting.
2.6. C. elegans treatment with oxidative stressors
Paraquat (Sigma-Aldrich, MO, USA, #856177) and NaN3 (Merck, Darmstadt, Germany, #1.06688) were dissolved in ddH2O and M9 buffer, respectively. Synchronized L4 nematodes were transferred to plates containing 2 mM paraquat and transferred to freshly prepared plates every other day. Survival was scored until all animals were dead (∼8 days after the beginning of the treatment). Synchronized young adult nematodes were transferred to plates supplemented with 180 μM freshly prepared NaN3 for 24 h and survival was scored after 24 h recovery.
2.7. RNA extraction
∼200 young adult nematodes/sample were resuspended in 1 mL Trizol™ (Invitrogen, MA, USA, #15596-026) and underwent 3–4 cycles of freezing (liquid N2) and thawing (37°C) until intact bodies were no longer visible. Following addition of 200 μL chloroform (Sigma-Aldrich, MO, USA, #372978) and vigorous mixing, the samples were centrifuged at 13000 rpm for 15 min at 4°C. The upper aqueous phase was pipetted and mixed with 500 μL of chilled isopropanol and centrifuged again at 13000 rpm for 10 min at 4°C. The RNA pellet was washed with 300 μL of 75% ethanol and centrifuged again at 13000 rpm for 10 min at 4°C. Once air dried, the pellet was resuspended in 25 μL RNase-free water and quantified using a spectrophotometer (Quawell, CA, USA, Q5000).
2.8. cDNA synthesis and qRT-PCR
1 μg of RNA was used for reverse transcription using the iScript cDNA synthesis kit (Bio-Rad, CA, USA, #1708891) according to manufacturer instructions. After cDNA synthesis, the reactions were diluted with 80 μL of nuclease-free water and 1 μL of this was used for the qRT-PCR reaction with the iTaq Universal SYBR Green Supermix (Bio-Rad, CA, USA, #17251224) on a Bio-Rad CFX Connect Real-Time System (Bio-Rad, CA, USA). Each set of primers was verified through a two-step protocol that included (i) testing of nine combinations of primer concentrations against 10 ng of wt cDNA, choosing the combination which gave the lowest CT value and only the expected product when the qRT-PCR reaction was run on a 2% agarose gel, and (ii) testing of the linearity of CT values given by the primer combination identified in step (i) with a serial dilution of cDNA ranging from 50 to 1.56 ng. The following primers were used in this study at 400 nM: pbs-5, Fw: GAGATCGTATGGTTGCCACG, pbs-5, Rev: GGAGCTCGTACAAAGTGCAG, cdc-42, Fw: CGTTGACGCAGAAGGGACTG, cdc-42, Rev: CTTCTTCTCCTGTTGTGGTGGGT, Aβ, Fw: GCAGAATTCCGACATGACTCAGG, Αβ, Rev: GCCCACCATGAGTCCAATGATT, dod-24, Fw: CAAACACTTCACCGAGCCAG, dod-24, Rev: AGTCATCGTTCTGGAGCAATTC nit-1, Fw: GCATAGGTGGCCAGATTCAAAA, nit-1, Rev: CCAGCTGCTTCTTCCACATTT, skn-1, Fw: GAGACGAGACGATAA skn-1, Rev: CAGATGAATATGGACGACACTC, gst-4, Fw: AAGTTGTTGAACCAGCCC, gst-4, Rev: AATCACAATATCAGCCCAAGTC.
2.9. Protein extraction, dot and western blotting
Nematodes were lysed via sonication in RIPA buffer supplemented with proteinase inhibitors. Protein concentration was determined with DC assay (Bio-Rad, CA, USA, #500-0113, -0114, -0115) and 15–20 μg of protein were separated with SDS-PAGE or 5–7 μg of protein were directly loaded on a nitrocellulose membrane mounted on a dot blotter. In the case of western blotting, the separated proteins were transferred onto a nitrocellulose membrane. All membranes were blocked with 5% BSA or 5% non-fat milk for 1 h at RT and incubated overnight with a primary antibody. The secondary antibodies were incubated at RT for 1 h and the membranes were developed using an ECL kit (Bio-Rad, CA, USA, #170–5051) and a Bio-Rad XRS+ Chemidoc (Bio-Rad, CA, USA). Coomassie Brilliant Blue staining was used for equal loading control. The primary antibodies used in this study were: anti-6E10 (BioLegend, CA, USA, #803001), anti-amyloid oligomer (EMD Millipore Corp., MA, USA, #AB9234) and anti-GFP (Cell Signaling, MA, USA, #2956). The secondary antibodies used in this study were: anti-rabbit (Santa Cruz, TX, USA, sc-2357) and anti-mouse (Santa Cruz, sc-516102).
2.10. Proteasome activities
To measure the chymotrypsin-like (CT-L) activity in cell-free assays, 0.5 μg of human purified proteasome (Proteasome Protein Center Kibbutz Ramat-Yohanan, Israel) was mixed with glutamate (Alfa Aesar, MA, USA, #A12919) in concentrations ranging from 15 μM to 15 mM and with Succinyl-Leucine-Leucine-Valine-Tyrosine-7-amino-4-methylcoumarin (Suc-LLVY-AMC, UBPBio, TX, USA, #G1100) at a 100 μM final concentration in a final volume of 100 μL in the wells of a black 96 well plate. 5 technical replicates per sample were used for each assay. SDS at final concentration 0.01% was used as a positive control and the proteasome inhibitor MG132 (UBPBio, TX, USA, #F1101) at a final concentration 20 μM was used as a negative control. The fluorescence was measured at 380/460 nm excitation/emission every 5 min for 1 h in a TECAN (Tecan Ltd., Switzerland) plate reader at 37°C.
2.11. Paralysis assays
For transgenic animals crossed with the CL4176 strain: 150–300 animals of L3 stage were shifted from 16°C to 25°C and 16 h later they were scored over the span of 10-14 h for paralysis until the entire population was paralyzed. For transgenic animals crossed with the GMC101 strain: 80–120 animals of L4 stage were transferred on NGM plates with 40 μΜ FUdR (Acros Organics, Belgium, #227601000) seeded with UV-inactivated OP50. Once they reached the stage of young adult, the animals were shifted from 20°C to 25°C. Scoring of paralyzed nematodes was initiated 12 h after temperature upshift and lasted until the entire population was paralyzed. For both Αβ models, the animals were scored as paralyzed if they failed to undergo a full body wave propagation upon prodding and each paralysis assay was repeated at least three times by two blinded experimenters.
2.12. Microscopy and image analysis
The animals were mounted on 2% agarose pads on glass slides and immobilized using 50 mM levamisole (Sigma-Aldrich, MO, USA, #L9756) dissolved in M9 buffer. The imaging was performed with a Leica TCS SPE confocal laser scanning microscope. For the animals expressing ZsProSensor-1, images from the anterior part of the animal were acquired using a 20x/0.70 objective and Z-stacks were obtained with stable laser intensity and gain throughout all conditions of every experiment. Regions of interest were determined on maximum intensity projection images of the Z-stacks and their intensity was determined with ImageJ. For transgenic animals crossed with UA198 animals, a 40x/1.15 oil immersion objective was used and the number of neurons in the tail was determined with Z-stacks and maximum intensity projection images.
2.13. Statistical analysis
All statistical analyses were performed with GraphPad Prism Software (San Diego, USA). Depending on the experiment, a variety of statistical tests was used and each is mentioned in the respective figure legend together with the sample size for each experiment. For analyses comparing the mean of two groups, we used the parametric two-tailed Student’s t-test, provided that each group comprised of at least 10 values. For fewer than 10 values we used the non-parametric test Mann-Whitney. When fewer than 10 replicates were included in an experimental condition, individual points representing each replicate are shown in the bar charts. For the comparison of more than 2 groups with fewer than 10 values per group, we used a non-parametric ANOVA-single factor with a post-hoc correction for multiple comparisons (Kruskal-Wallis test with Dunn’s correction). For the comparison of more than 2 groups with more than 10 values per group, we used a parametric ANOVA-single factor with a post-hoc correction for multiple comparisons (Tukey’s multiple comparisons test). For paralysis curves we used the log-rank Mantel-Cox test. All bar charts indicate the mean of the independent experiments ± SEM.
3. Results
3.1. Neuron-specific proteasome overexpression increases proteasome activity in the muscle
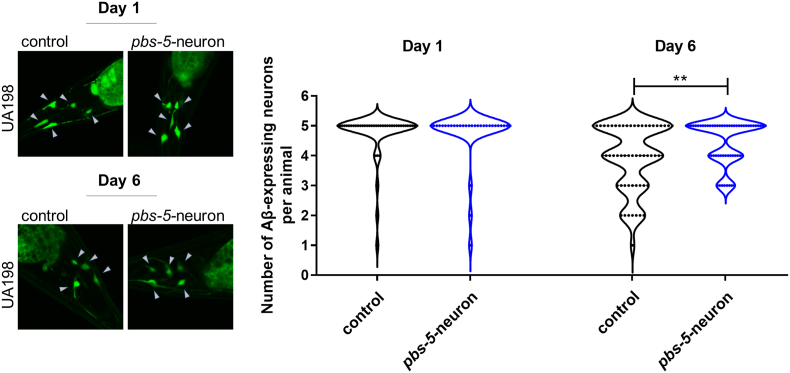
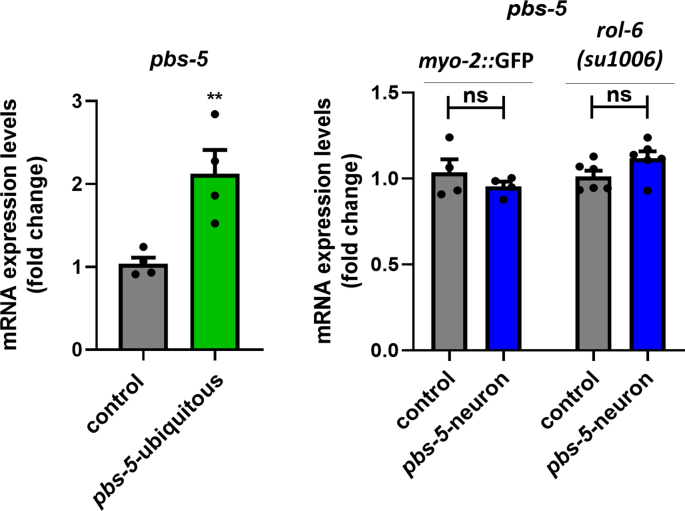
We have previously shown that ubiquitous overexpression of pbs-5 proteasome subunit is sufficient to increase the overall proteasome activity and to promote enhanced resistance against Aβ proteotoxicity in C. elegans [10]. Given that there are no reports on the potential cell non-autonomous regulation of the proteasome and since the nervous system is one of the main tissues communicating with other tissues, we sought to investigate whether neuron-specific pbs-5 overexpression affects (a) the proteasome activity in distal tissues and, (b) their resistance/response to proteotoxicity in a cell non-autonomous manner. To generate transgenic animals with proteasome overexpression, we took advantage of our previously published pbs-5 transgene [10]. To drive proteasome overexpression in the nervous system, we introduced in wt N2 nematodes a transgene (as an extrachromosomal array) composed of the promoter of the neuron-specific gene rgef-1 [28] fused to pbs-5 (rgef-1p::pbs-5) (referred to pbs-5-neuron hereafter; for simplicity we use simplified names for the various strains throughout the text, for complete genotypes please refer to the Materials and Methods section as well as to Table S1). We also created transgenic nematodes overexpressing pbs-5 under the ubiquitous promoter of the gene let-858 (let-858p::pbs-5 strain; referred to pbs-5-ubiquitous hereafter). The relative control strain was only overexpressing the co-injection marker (myo-2p::GFP; referred to control hereafter). In accordance with our previous results [10], we detected increased transcript levels of pbs-5 subunit in pbs-5-ubiquitous animals (Fig. S1). With regard to the pbs-5-neuron animals, although we generated two independent lines using different transformation markers (myo-2p::GFP and rol-6(su1006)), at most we only observed a slight increase in the levels of the pbs-5 transcript (Fig. S1), probably due to the tissue-specific expression only in the neurons. Given that overexpression of β5 subunit in all tissues in C. elegans (pbs-5 [10]) and in neurons in Drosophila (Prosβ5 [8,29]) and mouse (PSMB5 [8]) have been shown to enhance the resistance against proteotoxicity and to prevent cognitive deficits, we validated the neuronal proteasome overexpression with a functional assay. To this end, we crossed the pbs-5-neuron strain with the UA198 strain that expresses the human Aβ1-42 peptide in its GFP-tagged glutamatergic neurons, showing progressive, age-dependent neurodegeneration [26,30,31]. We scored significantly more surviving neurons in UA198 pbs-5-neuron animals compared to control nematodes on day 6 of adulthood (Fig. 1), thus suggesting a positive effect of neuronal pbs-5 overexpression. Furthermore, these results also reveal an anti-proteotoxic effect of proteasome overexpression in a cell autonomous manner (modulation in neurons, positive effects in neurons).
Fig. 1.
Delayed Aβ-mediated glutamatergic neurodegeneration in pbs-5-neuron animals. Representative fluorescence micrographs of UA198 control and pbs-5-neuron animals expressing the human Aβ1-42 peptide in their GFP-tagged glutamatergic neurons, on days 1 and 6 of adulthood. The arrows point to intact neurons (left panel). Violin graph showing the number of tail glutamatergic neurons expressing the human Aβ1-42 peptide in each examined animal from UA198 control and pbs-5-neuron populations, on days 1 and 6 of adulthood. Each dot represents an examined animal (right panel). Graph shows the distribution of pooled values from 3 independent experiments, >30 animals/condition, **p value < 0.01 (two-tailed Student’s t-test).
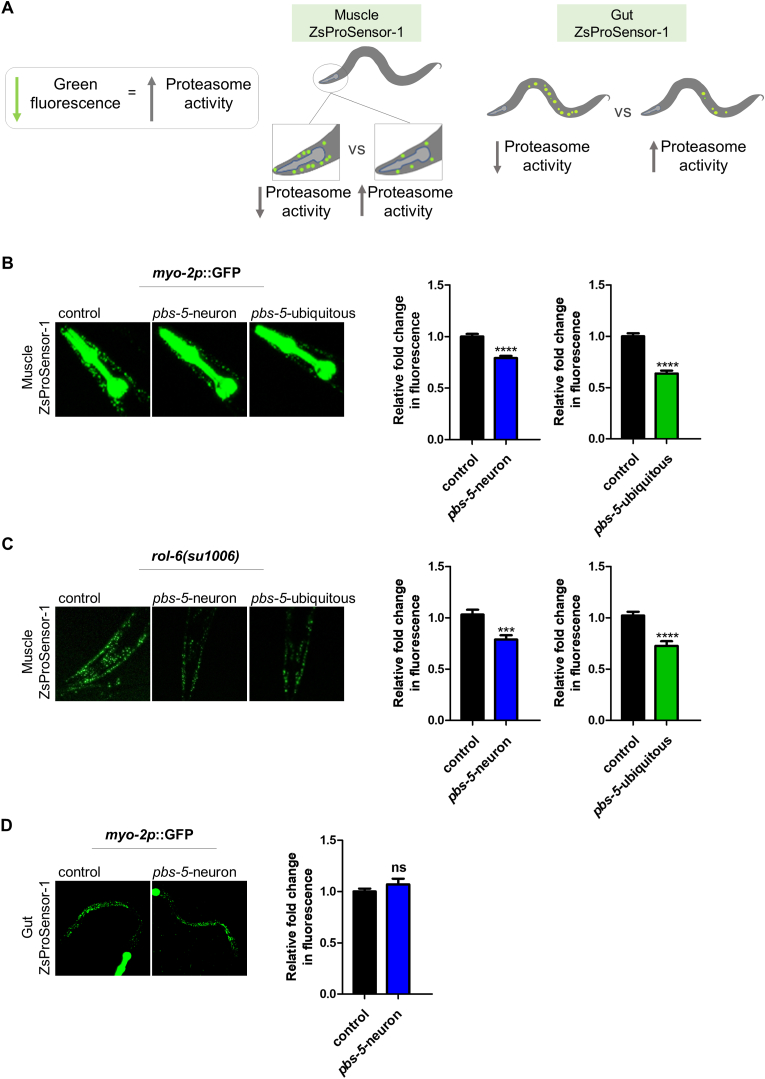
Using the pbs-5-neuron strain, we initially addressed the question whether neuron-specific pbs-5 overexpression affects the proteasome activity in a distal tissue. We took advantage of a well-characterized nematode strain expressing a modified ZsProSensor-1 fluorescent reporter under a muscle-specific promoter (referred to Muscle ZsProSensor-1 hereafter). More specifically, the modified ZsProSensor-1 reporter is fused to ubiquitin-interacting motifs derived from the C. elegans proteasome subunit RPN-10. Therefore, decreased fluorescence indicates increased proteasome-mediated degradation and vice versa (Fig. 2A) [32]. We crossed our control, pbs-5-neuron and pbs-5-ubiquitous animals with the Muscle ZsProSensor-1 strain. The fluorescence levels of ZsProSensor-1 reporter were significantly reduced in the muscle of both pbs-5-neuron and pbs-5-ubiquitous animals (Fig. 2B), thus revealing proteasome activation. To confirm these results and exclude potential effects due to the co-injection marker, we crossed the Muscle ZsProSensor-1 strain with pbs-5-neuron and pbs-5-ubiquitous animals carrying the rol-6(su1006) dominant transformation marker as a co-injection marker. We detected a consistent phenotype with the previous transgenic animals (Fig. 2C). We further sought to investigate whether neuron specific proteasome activation was also affecting other tissues. To this end, we crossed our pbs-5-neuron animals with the nematode strain expressing the modified ZsProSensor-1 fluorescent reporter under a gut-specific promoter (referred to Gut ZsProSensor-1); we did not detect enhanced proteasome activity in the gut, thus suggesting a tissue-specific cell non-autonomous effect (Fig. 2D). In total, our results reveal a cell non-autonomous regulation of the proteasome, since pbs-5 overexpression in the nervous system led to enhanced proteasome activity levels in the muscle.
Fig. 2.
Enhanced proteasome activity in the muscle of pbs-5-neuron animals. (A) Schematic describing the interpretation of the changes in the ZsProSensor-1 signal while monitoring proteasome activity in the muscle (Muscle ZsProSensor-1) or the gut (Gut ZsProSensor-1) of transgenic nematodes. Decreased fluorescence indicates increased proteasome activity and vice versa. (B, C, D) Representative fluorescence micrographs of animals expressing the ZsProSensor-1 reporter (left panel) and quantification of the ZsProSensor-1 signal (right panel) in (B, C) the muscle and (D) the gut, crossed with control, pbs-5-neuron or pbs-5-ubiquitous animals (only control and pbs-5-neuron animals in D) carrying (B, D) the myo-2p::GFP co-injection marker and, (C) the rol-6(su1006) co-injection marker. The mean value of fluorescence in control animals was set to 1. All values are reported as the mean of at least 3 independent experiments ± SEM, >20 animals/condition, ***p value < 0.001, ****p value < 0.0001, ns, not significant (two-tailed Student’s t-test).
3.2. Neuron-specific proteasome overexpression confers anti-proteotoxic effects in a cell non-autonomous manner
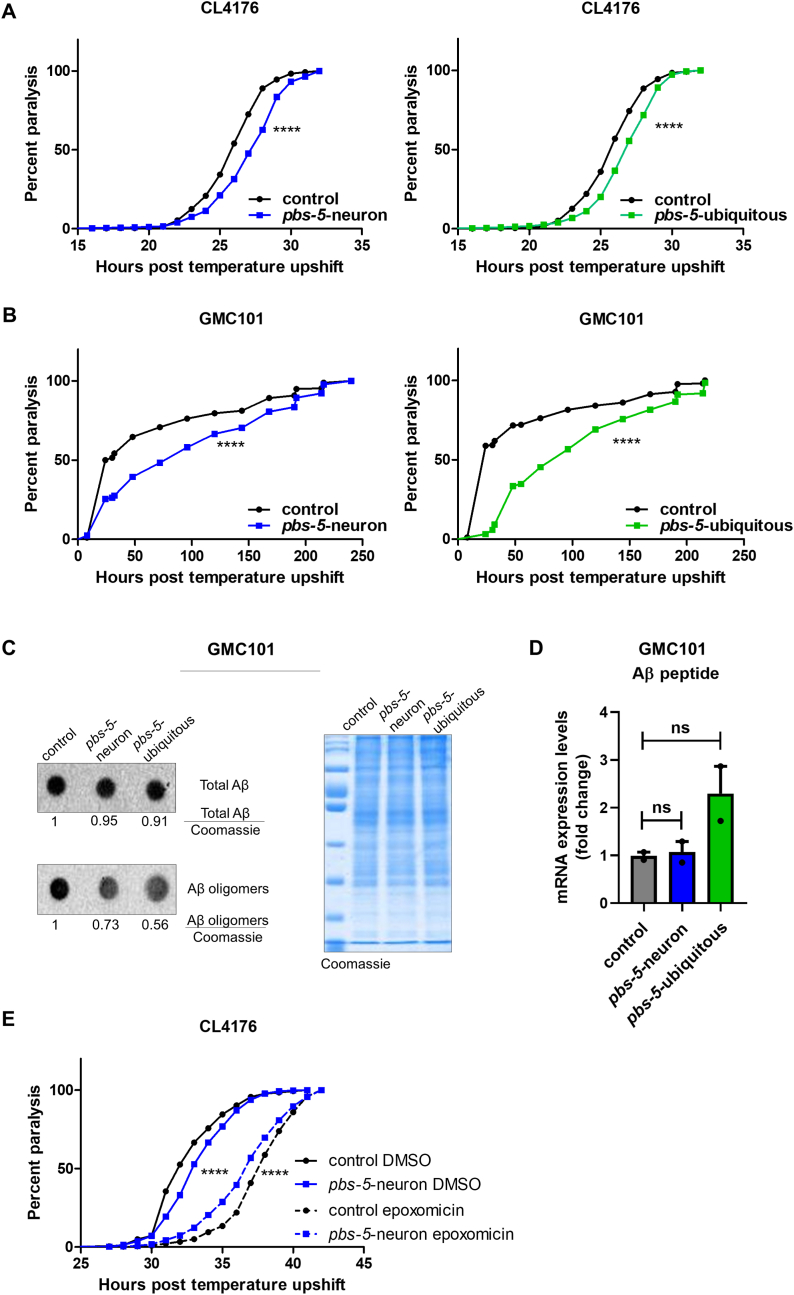
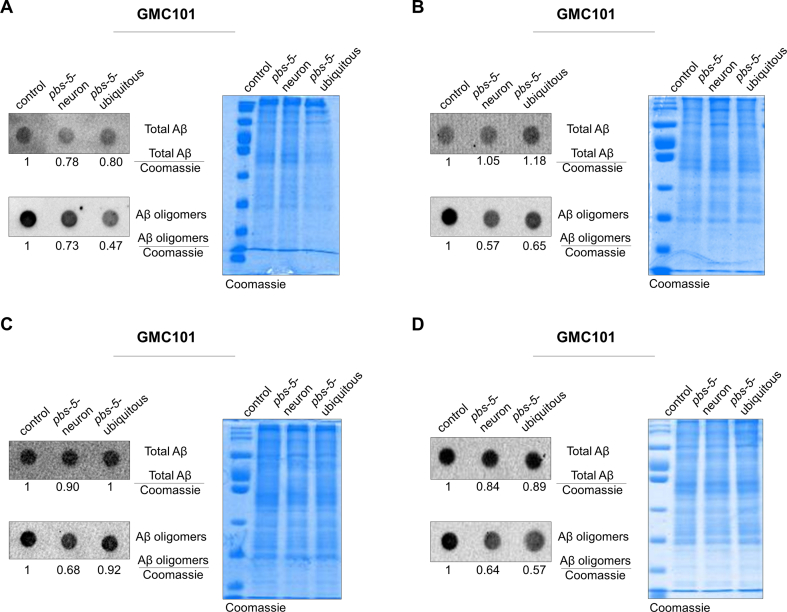
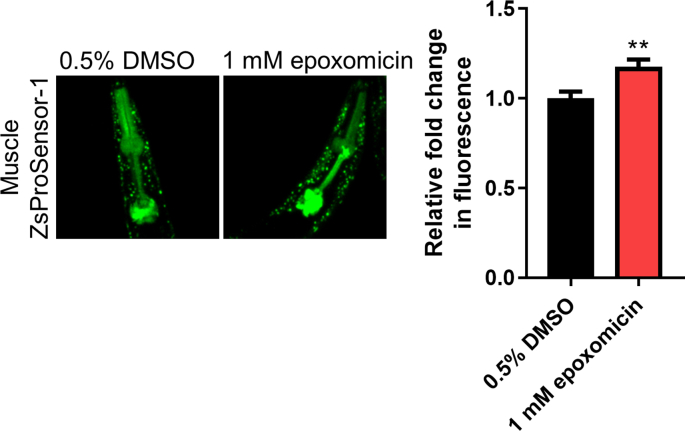
We then sought to investigate whether the cell non-autonomous proteasome activation in the muscle may counteract a proteotoxic threat as it was previously shown for the ubiquitous pbs-5 overexpression [10]. To this end, we crossed our pbs-5-neuron and pbs-5-ubiquitous animals with the CL4176 strain, an Aβ C. elegans model. This strain expresses the human Aβ1-42 peptide under a muscle-specific promoter and upon temperature upshift, they accumulate Aβ peptide in their body wall muscle cells and become paralyzed in a few hours [33]. Ubiquitous as well as neuron-specific proteasome overexpression significantly delayed the proteotoxic effects of Αβ peptide leading to delayed paralysis (Fig. 3A, Table S2). To confirm this protective effect, we evaluated the effects of ubiquitous and pan-neuronal proteasome overexpression in an additional Αβ nematode model, the GMC101 strain. These animals constitutively express the human Aβ1-42 peptide under a muscle-specific promoter and manifest age-progressive paralysis [34]. Consistent with our previous data, paralysis was also significantly delayed in GMC101 pbs-5-neuron and pbs-5-ubiquitous animals (Fig. 3B, Table S2). Moreover, ubiquitous and neuron-specific proteasome activation significantly reduced the levels of Αβ oligomers (Fig. 3C, Fig. S2). A tendency for reduced total Aβ levels was also recorded. Lack of significant changes in the transcript levels of Aβ peptide in our transgenic animals verified that the observed differences in the protein levels of Aβ forms are unlikely to arise from changes in the transcription of the transgene (Fig. 3D). To verify that the activation of the proteasome is responsible for the observed positive outcomes, we have then sought to inhibit this activation. We attempted to pharmacologically alter the proteasome activity by using epoxomicin, a highly specific proteasome inhibitor [35]. Following a series of dose-response experiments, we found that 1 mM epoxomicin (constant treatment for up to 12 h) could decrease proteasome activity in the Muscle ZsProSensor-1 strain (Fig. S3) without compromising survival. We then treated both CL4176 pbs-5-neuron and control animals with DMSO (vehicle) and 1 mM epoxomicin and paralysis assays were performed. Despite a small developmental delay exhibited by the animals treated with epoxomicin, we found that epoxomicin not only abrogated the delayed paralysis of pbs-5-neuron animals but it accelerated it compared to control animals treated with epoxomicin (Fig. 3E). This strongly suggests that proteasome activation underlies the observed beneficial effects on Aβ-mediated paralysis.
Fig. 3.
Neuron-specific proteasome overexpression confers protection against Αβ proteotoxicity in the muscle of pbs-5-neuron animals (cell non-autonomous effect). (A, B) Paralysis curves of control, pbs-5-neuron (left panels) and pbs-5-ubiquitous (right panels) animals expressing the human Aβ1-42 peptide in their body wall muscle cells, (A) CL4176 strain (temperature-dependent expression of the human Aβ1-42 peptide in the muscle with progressive paralysis within hours), 8 independent experiments with >800 animals, ****p value < 0.0001 (log-rank Mantel-Cox test) and, (B) GMC101 strain (constitutive expression of the human Aβ1-42 peptide in the muscle with age-progressive paralysis), 3 independent experiments with >140 animals, ****p value < 0.0001 (log-rank Mantel-Cox test). Curves are the pooled result of the indicated independent experiments. (C) Dot blot analysis (representative blots from 4 independent experiments) of total Aβ and Aβ oligomers in GMC101 control, pbs-5-neuron and pbs-5-ubiquitous animals expressing the human Aβ1-42 peptide in their body wall muscle cells, collected when 50% of the control population was paralyzed. A gel stained with Coomassie brilliant blue was used as a loading control in each experiment. The mean value of signal of the protein of interest to the Coomassie signal in control animals was set to 1. Quantification that appears underneath the blots represents the mean of 4 independent experiments. (D) mRNA expression levels of Aβ peptide in GMC101 control, pbs-5-neuron and pbs-5-ubiquitous animals expressing the human Aβ1-42 peptide in their body wall muscle cells on day 1 of adulthood. The mean value of Aβ peptide mRNA expression in control animals was set to 1. All values are reported as the mean of 2 independent experiments ± SEM, ns, not significant (Kruskal-Wallis test). (E) Paralysis curves of CL4176 control and pbs-5-neuron animals expressing the human Aβ1-42 peptide in their body wall muscle cells treated with DMSO or 1 mM epoxomicin, 4 independent experiments with >450 animals, ****p value < 0.0001 (log-rank Mantel-Cox test). Curves are the pooled result of 4 independent experiments. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
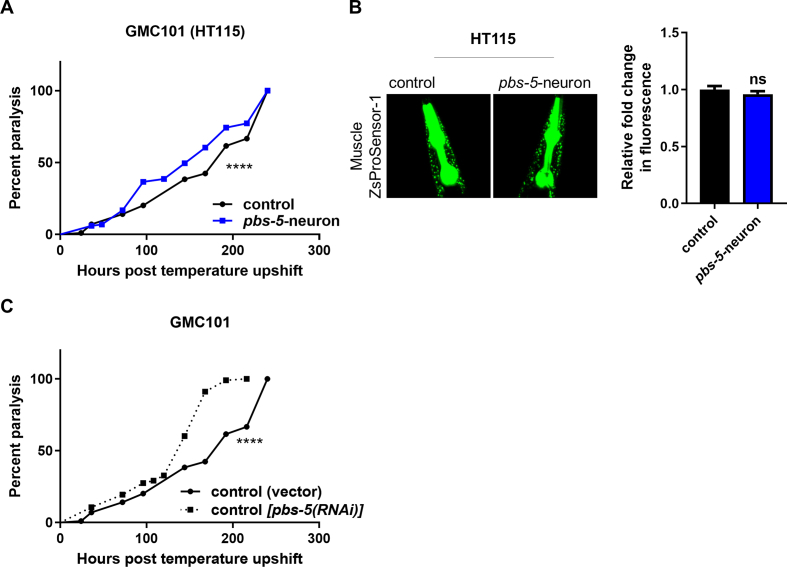
To further prove the link between proteasome activation and protection against proteotoxicity, we attempted to perform RNAi of pbs-5 subunit in GMC101 pbs-5-neuron animals. Nonetheless, this proved technically impossible as the GMC101 pbs-5-neuron animals lost their delayed paralysis rates over their control counterparts, once grown on HT115 bacteria used for RNAi experiments (Fig. S4A). To cross-validate these results, we evaluated the proteasome activity in Muscle ZsProSensor-1 pbs-5-neuron and control animals grown on HT115 bacteria. Indeed, the signal was not reduced anymore (Fig. S4B), as opposed to the observed reduction when the animals are grown on OP50 bacteria (Fig. 2B), thus suggesting that HT115 bacteria interfere with either the function of the proteasome and/or the communication between the neurons and the muscle. Nevertheless, we performed paralysis assays in GMC101 control animals in the presence of pbs-5 RNAi; the paralysis was significantly accelerated (Fig. S4C), thus verifying the importance of the proteasome function on the paralysis phenotype.
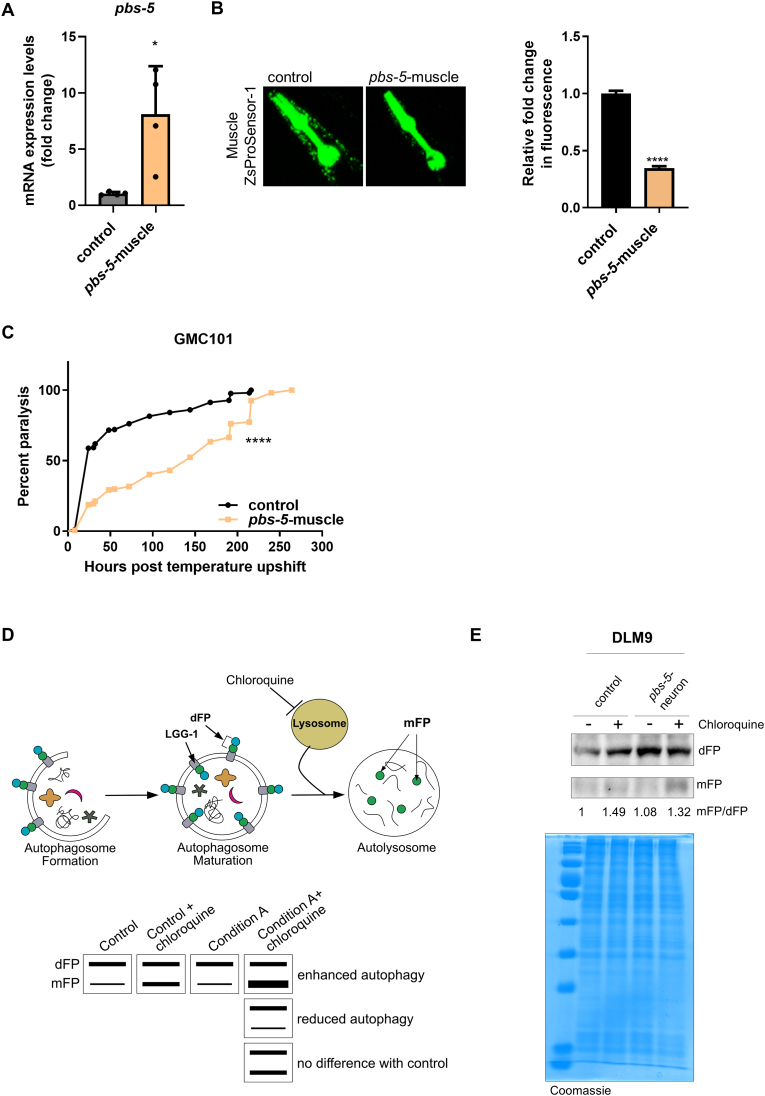
As a proof of concept and to verify the link between proteasome activation in the muscle and the protection against Aβ proteotoxicity in the muscle, we created transgenic nematodes overexpressing pbs-5 subunit under the promoter of the muscle-specific gene myo-3 (myo-3p::pbs-5 strain; referred to pbs-5-muscle hereafter). The relative control strain carried only the co-injection marker (myo-2p::GFP; referred to control hereafter). Quantitative PCR analysis confirmed pbs-5 overexpression (Fig. 4A). To test whether muscle-specific pbs-5 overexpression results in enhanced proteasome activity in the same tissue (i.e. cell autonomously), we initially crossed the pbs-5-muscle strain with the strain expressing the modified ZsProSensor-1 fluorescent reporter in the muscle. As expected, we found reduced levels of fluorescence indicating proteasome activation in Muscle ZsProSensor-1 pbs-5-muscle animals (Fig. 4B). We then crossed the pbs-5-muscle strain with the GMC101 animals. Paralysis was significantly delayed in GMC101 pbs-5-muscle animals, thus verifying that proteasome activation is sufficient for protection against Aβ proteotoxicity (Fig. 4C). Moreover, this result reveals a cell autonomous anti-proteotoxic effect (modulation in the muscle, positive effects in the muscle).
Fig. 4.
Proteasome activation in the muscle confers protection against Αβ proteotoxicity in the muscle (cell autonomous effect). (A) mRNA expression levels of pbs-5 proteasome subunit in control and pbs-5-muscle animals. The mean value of pbs-5 mRNA expression in control animals was set to 1. All values are reported as the mean of 4 independent experiments ± SEM, *p value < 0.05 (Mann-Whitney test). (B) Representative fluorescence micrographs of control and pbs-5-muscle animals carrying the Muscle ZsProSensor-1 reporter (left panel) and quantification of the Muscle ZsProSensor-1 signal (right panel). The mean value of fluorescence in control animals was set to 1. All values are reported as the mean of at least 3 independent experiments ± SEM, >25 animals/condition, ****p value < 0.0001 (two-tailed Student’s t-test). (C) Paralysis curves of GMC101 control and pbs-5-muscle animals expressing the human Aβ1-42 peptide in their body wall muscle cells, 3 independent experiments with >140 animals, ****p value < 0.0001 (log-rank Mantel-Cox test). Curves are the pooled result of 3 independent experiments. (D) Schematic describing the potential outcomes and their interpretation when performing a flux assay to a strain expressing a dual fluorescent protein (dFP) fused to LGG-1. LGG-1dFP is localized on the membrane of the autophagosome. During autophagosome maturation some mFP is released but most of the mFP is released by the lysosomal proteases once the autophagosome fuses with the lysosome. However, the mFP released in the autolysosome is rapidly degraded, hence very little is detected in steady-state levels. The steady-state amount of mFP can reflect both the efficiency of the lysosomal proteolysis as well as the rate of autophagosome formation. Chloroquine inhibits the acidification of the lysosome, thus inhibiting lysosomal proteases and blocking lysosomal proteolysis. Hence addition of chloroquine can induce the accumulation of mFP. Depending on the accumulated mFP levels in the sample compared to control, the results from the sample treated with chloroquine indicate the origin of mFP, thus the status of autophagic flux. (E) Immunoblot analysis (representative blots from 5 independent experiments) of dFP and mFP in lysates from control and pbs-5-neuron animals expressing LGG-1 tagged with dFP, in the absence (-) or presence (+) of chloroquine for evaluation of the autophagic flux. A gel stained with Coomassie brilliant blue was used as a loading control. The mean value of signal of mFP/dFP in control (-) animals was set to 1. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Mounting evidence suggests that the ALP is implicated in the clearance of misfolded or aggregated Αβ forms in various models, including C. elegans [36,37]. Moreover, the cross-talk between the UPS and ALP is well-accepted although not fully elucidated [38]. We therefore examined the potential involvement of autophagy in our setting. We took advantage of DLM9 strain that expresses under the muscle-specific promoter myo-3p a dual fluorescent protein (dFP) fused to LGG-1, a key protein in autophagosome formation. Upon docking and fusion to the lysosome, a monomeric fluorescent protein (mFP) is released, easily distinguishable from dFP via immunoblotting [39]. The ratio mFP/dFP gives an indication of the rate of autophagosome fusion with lysosomes, while a flux assay in the presence of chloroquine that inhibits the acidification of the lysosome thus, blocking the turnover of autolysosomes, reveals if the increased mFP/dFP ratio originates from increased autophagosome formation or block of autolysosome turnover (Fig. 4D). We crossed this strain with pbs-5-neuron and control animals. Autophagic flux was found unchanged in the muscle of DLM9 pbs-5-neuron animals (Fig. 4E), thus suggesting that the anti-proteotoxic effect of neuron-specific proteasome overexpression was independent of ALP activation.
In conclusion, neuron-specific proteasome activation can exert protection against Aβ proteotoxicity in the muscle in a cell non-autonomous manner.
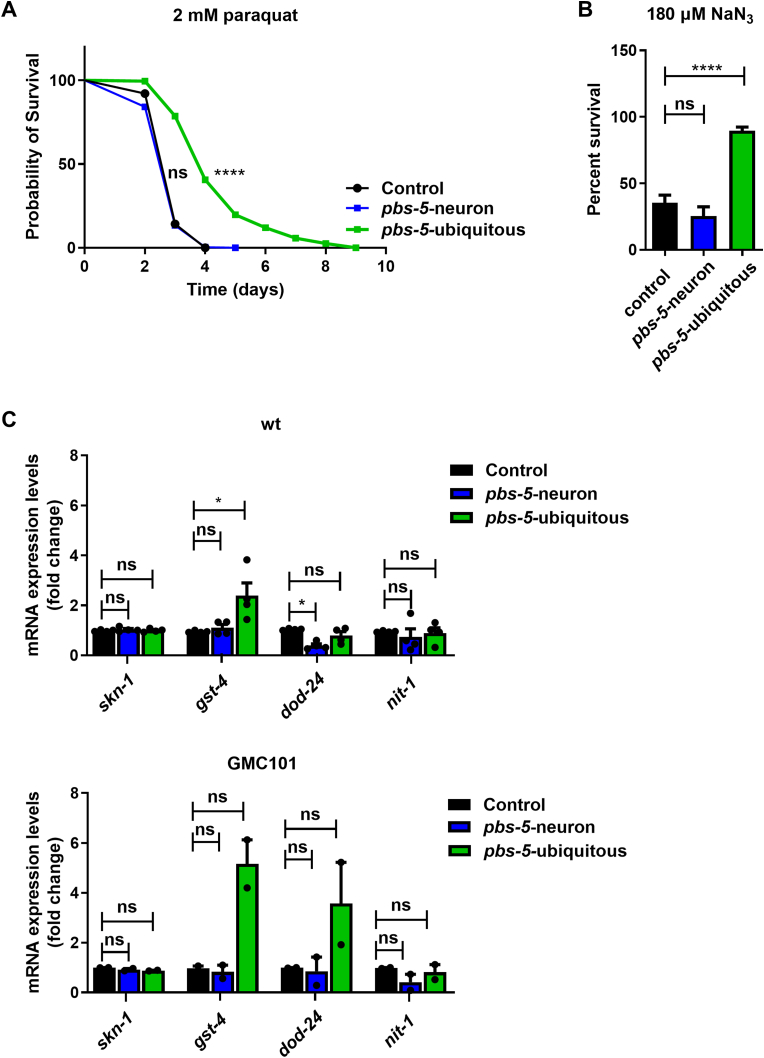
3.3. The cell non-autonomous protective effects following neuronal proteasome activation are not related to protection against oxidative stress
Given the established link between oxidative stress and AD [40,41] along with the interplay between the proteasome and oxidative stress response [[42], [43], [44]], we sought to investigate whether the observed beneficial effects against Aβ proteotoxicity were linked to enhanced oxidative stress response. We thus challenged our pbs-5-neuron and pbs-5-ubiquitous animals with two different oxidative stressors, namely paraquat and NaN3. Although proteasome activation in all tissues induced protection against oxidative stress (Fig. 5A-B; pbs-5-ubiquitous) in accordance with our previous results [10], notably, no resistance against oxidative stress was recorded in the pbs-5-neuron animals (Fig. 5A-B; pbs-5-neuron). Our results suggest that the neuronal proteasome activation confers cell non-autonomous protective effects in the muscle specifically against proteotoxic stress and not against oxidative stress. Given that the transcription factor SKN-1 is pivotal in oxidative stress resistance while it triggers strong responses to proteasome perturbations [[45], [46], [47]], we also investigated whether SKN-1 activation could account for the protective effects against Aβ-proteotoxicity. We therefore revealed the mRNA expression levels of skn-1 as well as of three of its target genes, namely gst-4, dod-24 and nit-1 [47], in control, pbs-5-neuron and pbs-5-ubiquitous animals in wt and GMC101 backgrounds. Although in the pbs-5-ubiquitous animals, gst-4 was significantly upregulated (statistically significant in wt background, exhibiting a tendency in GMC101 background without reaching statistical significance), none of the skn-1 target genes or skn-1 itself were upregulated in pbs-5-neuron animals in either background (Fig. 5C). In total our results suggest that the distal protective effects we score upon neuronal proteasome activation are specific for the Αβ proteotoxic stress and not for the related oxidative stress.
Fig. 5.
Neuron-specific proteasome overexpression does not confer protection against oxidative stress as opposed to ubiquitous proteasome overexpression. (A) Lifespan curve of control, pbs-5-neuron and pbs-5-ubiquitous animals treated with 2 mM paraquat, 3 independent experiments with >190 animals per condition, ****p value < 0.0001, ns, not significant (log-rank Mantel-Cox test). Curves are the pooled result of 3 independent experiments. (B) Survival percentage of control, pbs-5-neuron and pbs-5-ubiquitous animals 24 h after exposure to 180 μM NaN3, 3 independent experiments with >350 animals per condition, ****p value < 0.0001, ns, not significant (one-way ANOVA, Tukey’s multiple comparisons test). (C) mRNA expression levels of skn-1 and skn-1 target genes (gst-4, dod-24, nit-1) in wt (upper panel) and GMC101 (lower panel) control, pbs-5-neuron and pbs-5-ubiquitous animals in the young adult stage. The mean value of the mRNA expression of each gene in control animals was set to 1. All values are reported as the mean of 4 (for the wt animals) and 2 (for the GMC101 animals) independent experiments ± SEM, *p value < 0.05, ns, not significant (Mann-Whitney test). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
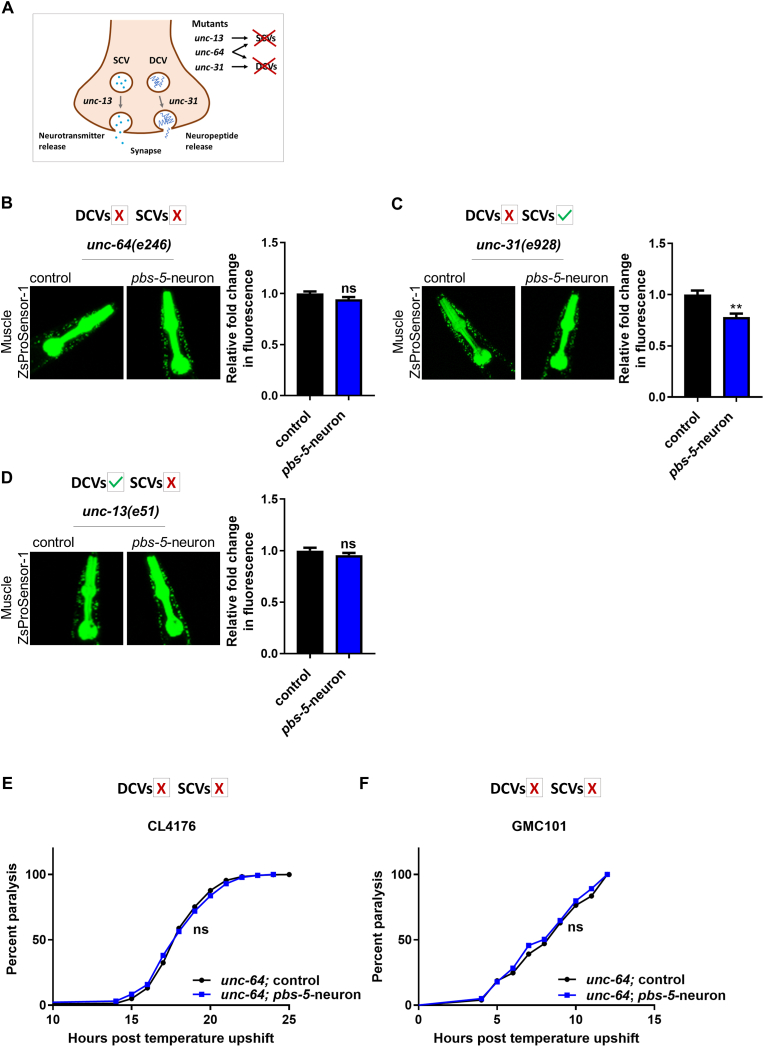
3.4. Τhe cell non-autonomous activation of the proteasome in the muscle is mediated by SCVs
Our results suggested that one or more neuronal-derived signals prompt proteasome reinforcement in a non-neuronal tissue such as the muscle. The communication between the nervous and the muscle tissues is accomplished via synaptic transmission [48]. In C. elegans, neurotransmitters are secreted from neurons in two types of vesicles classified as SCVs and DCVs, based on their size and appearance in electron micrographs [49]; Fig. 6A). We hypothesized that neurons that overexpress the pbs-5 subunit may affect proteasome activation in the muscle through a neuroendocrine signaling pathway mediated by SCVs and/or DCVs. To address this hypothesis, we crossed our pbs-5-neuron animals with unc-64 mutants that are deficient in the priming or exocytosis of both DCVs and SCVs [50]. To obtain a fast readout, the derived transgenic animals (unc-64(e246); pbs-5-neuron) and their controls were further crossed with the Muscle ZsProSensor-1 strain. We found that the loss-of-function unc-64(e246) allele completely abolished the proteasome-activating effect of neuronal pbs-5 overexpression on muscular proteasome activity (Fig. 6B), indicating that the activating signal is most likely contained in SCVs and/or DCVs.
Fig. 6.
The cell non-autonomous effects of neuron-specific proteasome overexpression is dependent on SCVs. (A) Schematic describing the involvement of UNC-13 and UNC-31 in the release of neurosecretory vesicles. UNC-13 participates in the priming of SCVs containing small-molecule neurotransmitters. UNC-31 participates in the exocytosis of DCVs primarily containing neuropeptides. (B, C, D) Representative fluorescence micrographs of animals expressing the Muscle ZsProSensor-1 reporter crossed with control or pbs-5-neuron animals carrying the myo-2p::GFP co-injection marker in (B) unc-64, (C) unc-31 and, (D) unc-13 mutant backgrounds (left panels) and quantification of the Muscle ZsProSensor-1 signal for each set of experiments (right panels). The mean value of fluorescence in control animals was set to 1. All values are reported as the mean of at least 3 independent experiments ± SEM, >20 animals/condition, **p value < 0.01, ns, not significant (two-tailed Student’s t-test). (E, F) Paralysis curves of control and pbs-5-neuron animals expressing the human Aβ1-42 peptide in their body wall muscle cells in an unc-64 mutant background, (E) CL4176 strain (temperature-dependent expression of the human Aβ1-42 peptide in the muscle with progressive paralysis within hours), 9 independent experiments with >1000 animals per condition, ns, not significant (log-rank Mantel-Cox test) and, (F) GMC101 strain (constitutive expression of the human Aβ1-42 peptide in the muscle with age-progressive paralysis), 3 independent experiments with >250 animals per condition, ns, not significant (log-rank Mantel-Cox test). Curves are the pooled result of the indicated independent experiments. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We then sought to tease apart the roles of the two vesicle types and to reveal which one is responsible for the cell non-autonomous communication of neuronal proteasome activation. The unc-31 gene is known to participate in the exocytosis of DCVs while the unc-13 in the priming of SCVs [51] (Fig. 6A). Although the unc-31(e928) loss-of-function allele did not inhibit the cell non-autonomous effect of proteasome activation in the muscle (Fig. 6C), the unc-13(e51) loss-of-function allele completely abolished the proteasome activation occurring in the muscle of ZsProSensor-1 pbs-5-neuron animals (Fig. 6D). Our results show that SCVs are responsible for the cell non-autonomous proteasome activation in the muscle following neuronal proteasome overexpression.
Next, we asked whether disruption of the SCVs-mediated communication between neurons and muscle is also translated into loss of protection against Aβ proteotoxicity. Since unc-13(e51) mutants are almost completely immobile, performing paralysis assays in unc-13(e51); pbs-5-neuron animals was impossible. Nevertheless, unc-64(e246) mutants are not immobile. They have defects in both DCVs and SCVs but our results had already excluded the involvement of DCVs in the cell non-autonomous proteasome activation in the muscle (Fig. 6C). The unc-64(e246); pbs-5-neuron animals did not exhibit decreased paralysis rates in neither CL4176 (Fig. 6E) nor GMC101 (Fig. 6F) backgrounds (Table S2). Together, our findings demonstrate that pbs-5 overexpression in neurons stimulates proteasome activation in the muscle through SCVs, resulting in protection against Aβ proteotoxicity.
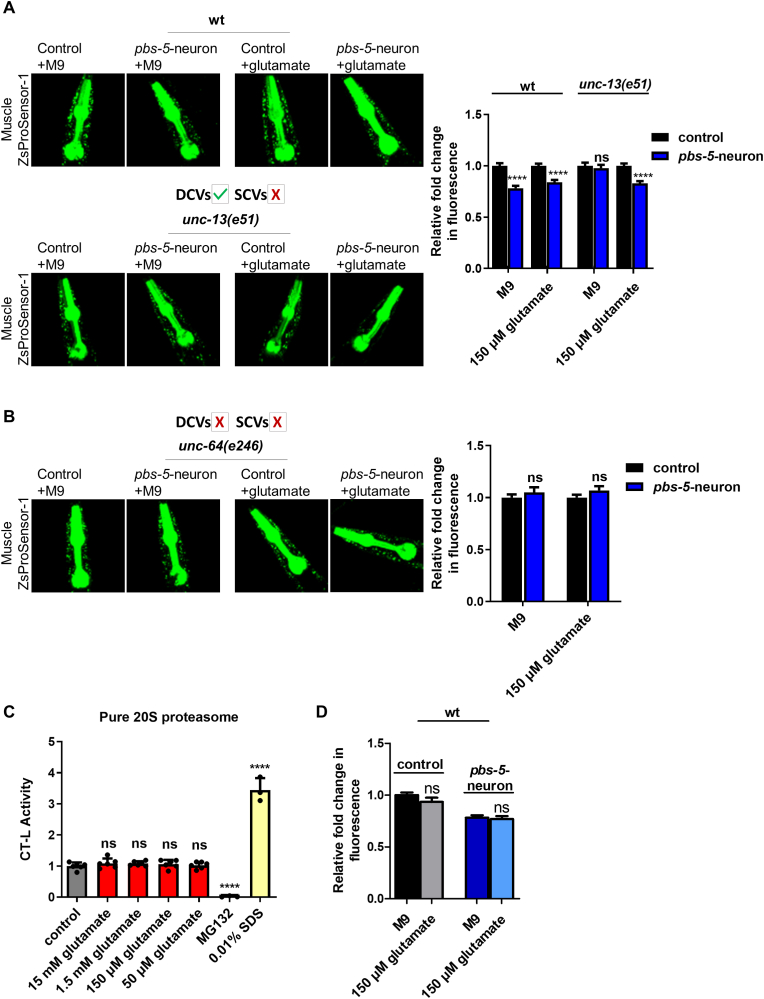
3.5. Glutamate can restore proteasome activity levels in the muscle of pbs-5-neuron animals lacking SCVs
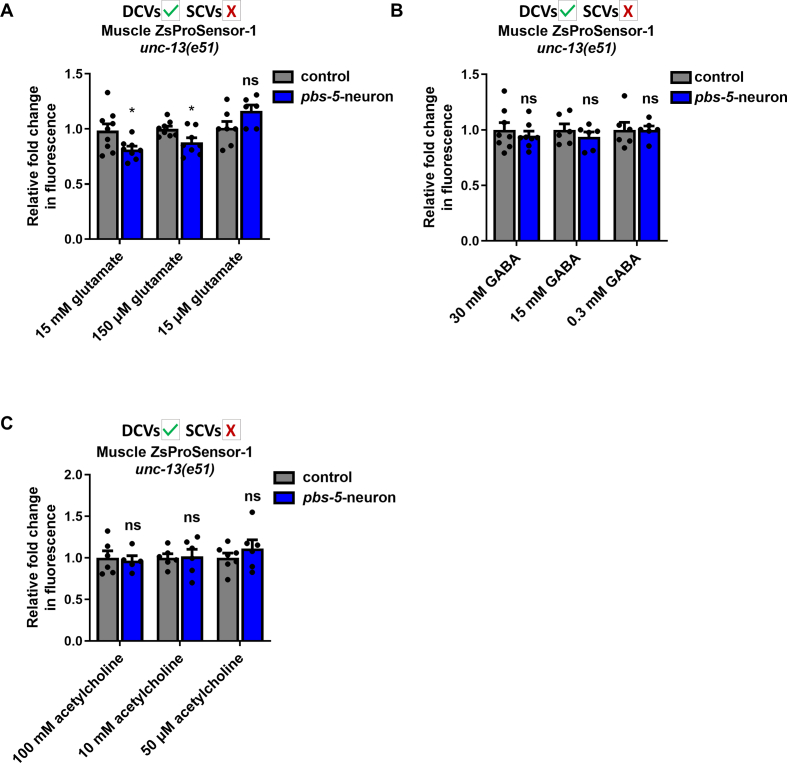
We then sought to identify the potential neurotransmitter(s) contained in SCVs that could transmit the signal from the neurons leading to proteasome activation in the muscle. SCVs contain classical neurotransmitters such as glutamate, γ-aminobutyric acid (GABA) and acetylcholine [49]. Since the unc-13(e51) mutants are unable to produce SCVs, we anticipated that external supplementation of the responsible molecule(s) would restore the effects of neuronal pbs-5 overexpression in the muscle of unc-13(e51); pbs-5-neuron animals. We therefore tested various concentrations of the three main neurotransmitters, namely glutamate, GABA and acetylcholine in Muscle ZsProSensor-1 unc-13; pbs-5-neuron animals and their controls (Fig. S5). Proteasome activity was enhanced only upon glutamate supplementation (Fig. 7A, Fig. S5A).
Fig. 7.
Glutamate supplementation restores the cell non-autonomous activation of the proteasome in unc-13 mutants. (A, B) Representative fluorescence micrographs of animals expressing the Muscle ZsProSensor-1 reporter crossed with control or pbs-5-neuron animals carrying the myo-2p::GFP co-injection marker, in (A) wt and unc-13 mutant backgrounds treated with M9 (vehicle) or 150 μM glutamate (left panel) and quantification of the Muscle ZsProSensor-1 signal (right panel). The mean value of fluorescence in control animals was set to 1. All values are reported as the mean of 3 independent experiments ± SEM, >20 animals/condition, ****p value < 0.0001, ns, not significant (two-tailed Student’s t-test) and, (B) unc-64 mutant background treated with M9 (vehicle) or 150 μM glutamate (left panel) and quantification of the Muscle ZsProSensor-1 signal (right panel). The mean value of fluorescence in control animals was set to 1. All values are reported as the mean of 3 independent experiments ± SEM, >20 animals/condition, ns, not significant (two-tailed Student’s t-test). (C) CT-L proteasome activity of pure 20S proteasome in the presence of various concentrations of glutamate in cell-free assays. MG132 (proteasome inhibitor) was used as a negative and SDS (proteasome activator) as a positive control. The mean value of CT-L activity in control sample (no glutamate) was set to 1. All values are reported as the mean of 2 independent experiments ± SEM, ****p value < 0.0001, ns, not significant (one-way ANOVA with Dunnett's Multiple Comparison). (D) Quantification of data from (A) comparing the relative fold change in fluorescence between wt control animals carrying the myo-2p::GFP co-injection marker (left columns) and pbs-5-neuron animals (right columns) supplemented with M9 (vehicle) or 150 μM glutamate. The mean value of fluorescence in control animals supplemented with M9 was set to 1. All values are reported as the mean of 3 independent experiments ± SEM, >20 animals/condition, ns, not significant (two-tailed Student’s t-test). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
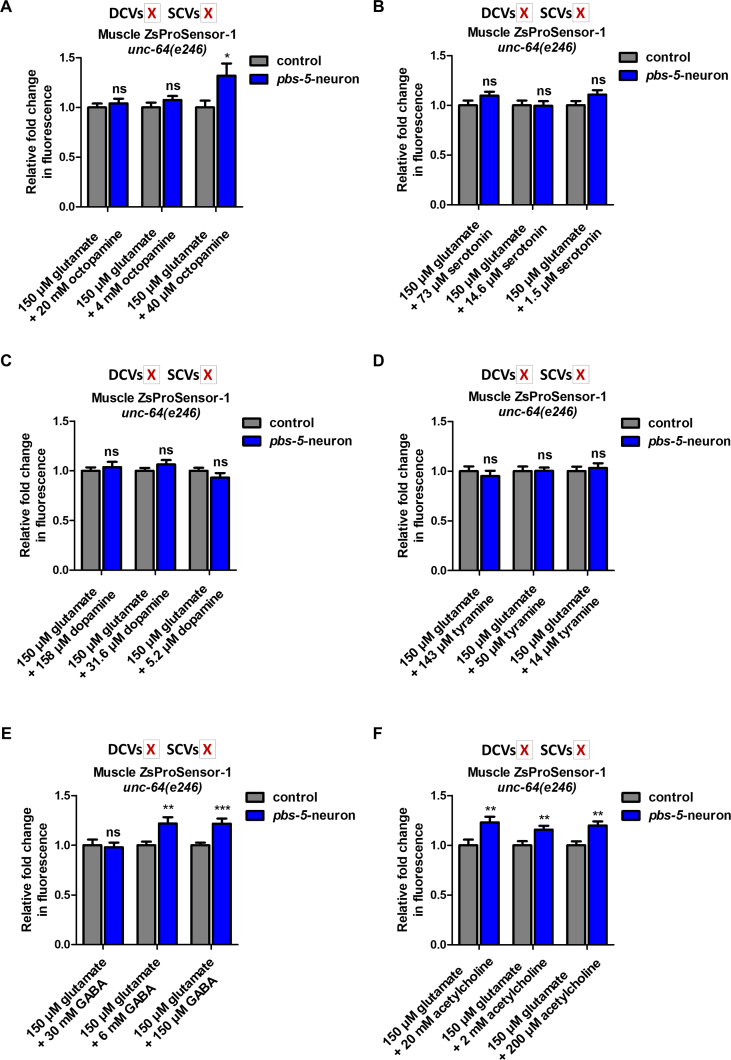
Since we could not perform paralysis assays in unc-13(e51) mutants due to their immobile phenotype, we tested whether glutamate supplementation could restore the proteasome activity in the muscle of unc-64(e246) mutants. Surprisingly, we found that external glutamate supplementation did not restore the proteasome activity in the muscle of pbs-5-neuron animals in an unc-64 mutant background (Fig. 7B). Given that the unc-64 mutants have defects in both SCVs and DCVs, the inability of glutamate alone to restore the proteasome activity in the muscle in these mutants suggests that glutamate can compensate for the lack of SCVs only in the presence of functional DCVs, thus attributing a role to the DCVs and their cargo in this inter-tissue communication. We therefore investigated whether combined supplementation of glutamate (mainly found in SCVs) and other neurotransmitters that are most probably found in DCVs, could restore the proteasome activity in the muscle of unc-64 mutants and thus the neuron-muscle communication. To this end, we treated Muscle ZsProSensor-1 unc-64; pbs-5-neuron animals, with glutamate and various concentrations of the 4 biogenic amines that are packaged in DCVs, namely octopamine, serotonin, dopamine and tyramine [49]. We did not detect any restoration of the proteasome activity with the combined supplementation of glutamate and biogenic amines in unc-64 mutants (Figs. S6A–D). Consequently, the observed results might be due to the action of neuropeptides found in DCVs and therefore the potential cargo of DCVs that plays a role in the neuron-muscle communication needs further investigation. Although GABA and acetylcholine did not restore muscular proteasome activity in unc-13 mutants (SCVs-defective animals; Figs. S5B–C), unc-64 mutants have defects in both SCVs and DCVs and therefore, we reasoned that in these animals, neurotransmitters commonly found in SCVs could take over and exhibit additional regulatory roles. We therefore treated Muscle ZsProSensor-1 unc-64; pbs-5-neuron animals with a combination of glutamate and GABA or acetylcholine. Interestingly, we found that the double supplementation with glutamate and either GABA or acetylcholine results in reduction of proteasome activity in Muscle ZsProSensor-1 unc-64; pbs-5-neuron animals (Figs. S6E–F). This suggests that GABA and acetylcholine may have proteasome inhibitory roles that are counteracted by DCVs-dependent signals, as we never observed proteasome inhibition by GABA or acetylcholine in animals that are defective for SCVs but have intact DCVs (Figs. S5B–C).
To dissect the mechanism behind glutamate-mediated proteasome activation, we assessed whether glutamate may have direct stereochemical effects on the proteasome complex resulting in its activation. Incubation of purified proteasome with different concentrations of glutamate did not reveal any proteasome activating properties (Fig. 7C), thus suggesting that the glutamate effect on proteasome activity is most likely indirect. We have also checked whether glutamate addition in animals can phenocopy the results of neuronal pbs-5 overexpression on proteasome activity. Glutamate supplementation in Muscle ZsProSensor-1 control animals did not affect their proteasome activity in the muscle (Fig. 2, Fig. 7 first columns). Likewise, glutamate supplementation of Muscle ZsProSensor-1 pbs-5-neuron animals did not further alter the already enhanced proteasome activity due to the pbs-5 overexpression (Fig. 2, Fig. 7 last columns). These results suggest that proteasome activation in the muscle of pbs-5-neuron animals is not due to a generic or unspecific proteasome-activating effect of glutamate but rather to a specific regulated process.
In conclusion, we show that proteasome activity is subjected to cell non-autonomous regulation since pbs-5 overexpression in the neurons promotes proteasome activation in the muscle of C. elegans. Glutamate contained in SCVs is necessary for this activation but the presence of intact DCVs is also required. This cell non-autonomous proteasome activation is translated into efficient prevention of Αβ-mediated proteotoxic effects in the muscle of C. elegans.
4. Discussion
Proteinopathies including AD have been linked to inefficient protein degradation machineries, such as the UPS or the ALP [52,53]. This inefficiency is further accentuated during ageing which in turn is a top risk factor for aggregation-related diseases [2,54]. Consequently, reinforcement of the proteolytic part of proteostasis during ageing and upon disease progression has been suggested as a pivotal potential preventive and/or therapeutic approach. Here, we report the cell non-autonomous communication of neuronal proteasome activation. We show that overexpression of the pbs-5 proteasome subunit in the nervous system can promote proteasome activation in the muscle tissue of C. elegans. The result is enhanced resistance to Aβ proteotoxicity in the distal tissue (muscle) but not enhanced resistance to oxidative stress. This communication depends on intact SCVs and glutamate, thus establishing a mechanistic link between neuronal proteasome reinforcement and decreased Aβ proteotoxicity in the muscle. Furthermore, we show that in the absence of SCVs, glutamate can compensate only in the presence of functional DCVs, thus suggesting a role of the DCVs’ cargo in this distal tissue communication as well. Cell autonomous anti-proteotoxic effects are also revealed upon neuron- and muscle-specific proteasome activation.
Our in vivo approach conclusively demonstrates the cell non-autonomous control of proteasome function as previously described for other players of proteostasis such as the HSR [13], UPRER [18,19,55], UPRmit [17] and autophagy [20]. Among the proteolytic pathways, the lysosome-mediated degradation [18] has been already shown to be affected in other tissues once proteostatic mechanisms such as the UPRER are modulated in neurons [18,55] or in the intestine [18]. Neuron-specific Prosβ5 overexpression in Drosophila has been shown to slow down age-related deficits in learning, memory, and circadian rhythmicity [29]. A follow-up of this study showed that neuronal-specific overexpression of β5 subunit in Drosophila (Prosβ5) and in mice (PSMB5) was sufficient to ameliorate disease-related phenotypes in the respective AD models [8]. Notably, despite the attempts for tissue-specific proteasome activation, no data exists on its cell non-autonomous regulation and the outcomes in distal tissues.
Tissue-specific proteotoxic stress has been shown to trigger various proteostatic mechanisms cell non-autonomously in distal tissues as a compensatory response to stress. For example, neuronal overexpression of an aggregation-prone protein (polyQ40) and its binding on neuronal mitochondria has been shown to elicit a global induction of a mitochondrial-specific unfolded protein response (UPRmit), affecting whole-animal physiology [17]. With regard to the proteasome, silencing of various proteasome subunits (i.e. proteasome stress) in the skeletal muscle of Drosophila has been shown to mount an adaptive response in the brain that finally hinders retinal and brain ageing [56]. Nevertheless, our study is the first to show that a positive modulation of the proteasome (activation) is communicated to a distal tissue resulting in tissue protection (increased resistance to Aβ proteotoxicity).
Our data clearly show that neuronal overexpression of the pbs-5 subunit leads to proteasome activation in a distal tissue such as the muscle and its resistance to Aβ proteotoxicity. Although we excluded the implication of the intestine since we did not detect enhanced proteasome activity in this tissue upon neuronal proteasome overexpression, we cannot rule out that other tissues like hypodermis or the somatic gonad could be also affected by the proteasome activation in the nervous system. Ιt would be interesting to assess if non-neuronal tissues that are affected by the nervous system also respond to the neuronal proteasome activation and further communicate with other tissues thereby creating an inter-tissue network of higher complexity.
One could ask how proteasome activation confers protection against Aβ proteotoxicity. Our results revealed significantly lower oligomeric Aβ levels (and a tendency for reduced total Aβ levels) in pbs-5-neuron animals. This suggests that proteasome activation may promote the degradation of Aβ species before they become highly aggregated and thus inaccessible and inhibitory to the proteasome [57,58]. In our previous work, proteasome activation through genetic means (systemic pbs-5 overexpression) [10] or through natural compounds with proteasome-activating properties [59,60], had elicited similar results. Likewise, in a very recent work, genetic and pharmacological proteasome augmentation also resulted in enhanced clearance of APP in flies, cultured cells and mice [8]. Even more importantly, a negative correlation between proteasome activity and protein levels of APP and soluble Aβ42 levels in human hippocampal tissues of AD patients was revealed, thus strengthening the biological relevance of the findings in the organismal models [8]. Finally, combined supplementation of 18α-glycyrrhetinic acid (that activates the proteasome through enhanced transcription of several proteasome subunits [59,61]) and omega-3 fatty acids (that have been shown to directly positively modulate proteasome activity [62]) also resulted in a significant reduced percentage of Aβ42 coverage in the parietal cortex as well as in the hippocampus of 5xFAD (transgenic mice carrying 5 familial AD-associated mutations) mice [62]. It is well established in yeast and mammalian cells that the proteasome crosstalks with autophagy [63] while the same has been demonstrated in C. elegans [64,65]. Although we did not detect ALP activation in our setting, thus attributing a major role to the proteasome system for the observed results, one cannot rule out the potential implication of the ALP possibly in a later stage, when heavily aggregated proteins are formed. This was also suggested recently by an elegant study on small molecule VCP/p97 activators where both proteasome and autophagy routes shared a role in the clearance of different forms of misfolded species [66]. In total, our findings suggest that proteasome-related protection against Αβ proteotoxicity emerges from enhanced clearance of various Aβ forms.
In cases where tissue-specific manipulation of a proteostatic mechanism has been achieved, studies mainly focused on the overall organismal fitness and longevity without focusing on the effects on specific distal tissues. Only a few studies have dealt with the effects of proteostasis manipulation in specific tissues and the downstream distal effects in models for protein aggregation [18,67]. Most importantly, none of them has investigated the role of the proteasome. We clearly show the beneficial effects of neuronal proteasome overexpression against Αβ proteotoxicity in the neuronal tissue but also in the periphery. Mechanistically, the “nervous system-to-periphery” cell non-autonomous communication usually occurs via extracellular signaling molecule release [68,69]. The distal activation of stress responses upon interventions in the nervous system has been shown to be mediated by neurons [13,19,55] and recently by glial cells [67]. Depending on the proteostatic mechanism that is modulated, either SCVs [55,70] or DCVs [17,67] have been shown to be responsible for the distal effects. We clearly show that SCVs are required for the cell non-autonomous communication of the neuronal proteasome activation which was rather surprising. Given that DCVs contain neuropeptides that are potential proteasome substrates and since in our setting the proteasome activity is enhanced, we had speculated that neuropeptides (and thus DCVs) could be the ones to be responsible for the observed effects. Moreover, a 20S proteasome form that is tightly associated with the neuronal plasma membrane and exposed to the extracellular space has been suggested to exist. This proteasome type has been shown to mediate the 20S-dependent degradation of intracellular proteins into bioactive extracellular peptides that modulate neuronal function [71,72]. As our results indicate the implication of DCVs as well, it would be interesting to investigate further their role and to examine whether the observed enhanced proteasome activity in our pbs-5-neuron animals affects this particular type of proteasomes and therefore the extracellular peptides produced by those proteasomes.
We also identified glutamate as a central player, without however being able to exclude the involvement of other neurotransmitters. How can glutamate restore this “proteasome activating” signal in pbs-5-neuron animals that lack SCVs? A direct or an indirect impingement of glutamate on proteasome activity could be speculated. We ruled out a direct effect on the proteasome complex since glutamate was not able to activate the 20S core proteasome in a cell-free assay. Moreover, direct administration of glutamate in wt background animals was not sufficient to promote proteasome activation whereas glutamate supplementation in animals with neuronal proteasome activation did not further enhance the proteasome activity. It seems that neuron-specific proteasome overexpression should precede before glutamate can function as a messenger provided however that DCVs are also functional. Given the multiple roles of glutamate, primarily as a neurotransmitter ([73] but also as a precursor to the antioxidant glutathione [74] or as a modulator of the tricarboxylic acid cycle once converted into α-ketoglutarate [74], glutamate could contribute to redox or energy status alterations that may significantly impact proteasome activity and function. More extensive studies on the metabolic effects of pbs-5 neuronal overexpression are needed.
There are limitations in our study mainly linked to constraints imposed by our model. For example, although we partially respond to the question, we were not able to fully elucidate the mechanistic details through which glutamate triggers the increase of the proteasome activity in the muscle. This is mainly due to the fact that in C. elegans it is virtually impossible to isolate and analyse the different involved tissues in order to understand tissue-specific dysregulation in an inter-tissue communication context. Finally, we cannot investigate the role of the nervous system in the regulation of the proteasome activity of the muscle under more physiological conditions, i.e. in the absence of pbs-5 OE. This would suggest the comparison of wt nematodes to mutants for the genes we have identified as responsible for the distal communication, i.e. unc-64 and unc-13. Since these mutants have severe feeding defects (reduced pharyngeal pumping), this comparison is not adequate as these defects may significantly affect the proteasome activity. Nevertheless, our results emphasize on functional insights and are important for potential future applications.
In this study, we show that cell non-autonomous proteasome activation can contribute to anti-proteotoxic effects in nematode models for Αβ toxicity. This could be a common regulatory mechanism in other metazoans, as well as in other protein aggregation-related diseases. This point is of particular significance as it demonstrates how cell non-autonomous proteostasis regulation can have a functional outcome in distal tissues. It is highly important since various neurodegenerative diseases are rarely restrained to a single tissue and are rather accompanied by peripheral maladies exhibited as metabolic alterations and deficiencies in non-neuronal tissues [75,76]. Deciphering this cell non-autonomous communication further in invertebrates along with complementary studies in vertebrates, may have serious implications in the design of tissue-specific proteasome manipulation strategies with therapeutic interest. Our results may contribute significantly to the optimization of proteostatic manipulation to fight proteinopathies through tissue-targeting. It is however also important to reveal all the potential cell non-autonomous affected aspects of proteostatic mechanisms, not only to optimize a tissue-specific manipulation per se but also to guarantee that one takes into account all the anticipated side effects, whether positive or negative. Moreover, the identification of the proteasome cell non-autonomous regulation may affect the translation of results of groups working in various fields where the proteasome is affected or manipulated, including the cancer and malaria fields where proteasome inhibition is aimed and tissue-specific manipulation is targeted as anti-cancer and anti-malaria therapeutic avenues, respectively. Finally, our results are also important for physiological processes in which the proteasome and its manipulation are important such as ageing. In total, our study provides the context for elucidating the precise mechanism through which cell non-autonomous proteasome regulation occurs and indicates potential tissue-specific avenues against, but not exclusively, proteinopathies.
5. Conclusions
Our study demonstrates that proteasome activation in the nervous system can enhance the proteasome activity in the muscle of C. elegans, thus revealing for the first time a cell non-autonomous regulation of the proteasome function. We show that this distal communication is mediated through SCVs and glutamate. More importantly, we demonstrate that this cell non-autonomous proteasome activation results in effective prevention of the human Αβ-mediated proteotoxicity in the distal tissue (muscle). Notably, this distal effect does not apply to all kinds of stress since neuronal proteasome activation did not confer resistance to oxidative stress. These distal effects should be considered when designing tissue-specific interventions targeting the proteasome as they might dampen the effects that are anticipated through the tissue-specific manipulation.
Author contributions
NC conceived the idea. AG and NC conceptualized and designed the work. AG and DB designed the constructs for pbs-5 overexpression. EP and AG performed experiments and analyzed the data. EP, AG and NC interpreted the data. AG and NC prepared the first manuscript and all authors reviewed and edited the text. AG and NC supervised the work.
Declaration of competing interest
None.
Acknowledgements
Nematode strains used in this study were provided by the Caenorhabditis Genetics Center (CGC) which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We are grateful to Prof. Guy A. Caldwell for the UA198 strain and to Dr. Carina I. Holmberg for the ZsProSensor-1 strains. EP was supported by the Hellenic Foundation for Research and Innovation (HFRI) under the HFRI PhD Fellowship grant (Fellowship Number: 1540) (salaries only). With the exception of the HFRI PhD Fellowship grant to EP, this research did not receive any other specific grant from funding agencies in the public, commercial, or, not-for-profit sectors.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102817
Contributor Information
Eleni Panagiotidou, Email: epanagiotidou@eie.gr.
Anna Gioran, Email: agioran@eie.gr.
Daniele Bano, Email: Daniele.Bano@dzne.de.
Niki Chondrogianni, Email: nikichon@eie.gr.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
figs5.
figs6.
Data availability
Data will be made available on request.
References
- 1.Balch W.E., Morimoto R.I., Dillin A., Kelly J.W. Adapting proteostasis for disease intervention. Science (New York, N.Y.) 2008;319(5865):916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 2.Taylor R.C., Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harbor Perspect. Biol. 2011;3(5) doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lefaki M., Papaevgeniou N., Chondrogianni N. Redox regulation of proteasome function. Redox Biol. 2017;13:452–458. doi: 10.1016/j.redox.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins G.A., Goldberg A.L. The logic of the 26S proteasome. Cell. 2017;169(5):792–806. doi: 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chondrogianni N., Petropoulos I., Grimm S., Georgila K., Catalgol B., Friguet B., Grune T., Gonos E.S. Protein damage, repair and proteolysis. Mol. Aspect. Med. 2014;35:1–71. doi: 10.1016/j.mam.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Boland B., Yu W.H., Corti O., Mollereau B., Henriques A., Bezard E., Pastores G.M., Rubinsztein D.C., Nixon R.A., Duchen M.R., Mallucci G.R., Kroemer G., Levine B., Eskelinen E.-L., Mochel F., Spedding M., Louis C., Martin O.R., Millan M.J. Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 2018;17(9):660–688. doi: 10.1038/nrd.2018.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Njomen E., Tepe J.J. Proteasome activation as a new therapeutic approach to target proteotoxic disorders. J. Med. Chem. 2019;62(14):6469–6481. doi: 10.1021/acs.jmedchem.9b00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chocron S.E., Munkácsy E., Harper S.K., Karpowicz P., Jiang N., Van Skike C., DeRosa N., Banh A.Q., Palavicini J.P., Wityk P., Kalinowski L., Galvan V., Osmulski P.A., Jankowska E., Gaczynska M., Pickering A.M. Genetic and pharmacologic proteasome augmentation ameliorates Alzheimer’s-like pathology in mouse and fly APP overexpression models. Sci. Adv. 2022;8(23):eabk2252. doi: 10.1126/sciadv.abk2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chondrogianni N., Tzavelas C., Pemberton A.J., Nezis I.P., Rivett A.J., Gonos E.S. Overexpression of proteasome beta5 assembled subunit increases the amount of proteasome and confers ameliorated response to oxidative stress and higher survival rates. J. Biol. Chem. 2005;280(12):11840–11850. doi: 10.1074/jbc.M413007200. [DOI] [PubMed] [Google Scholar]
- 10.Chondrogianni N., Georgila K., Kourtis N., Tavernarakis N., Gonos E.S. 20S proteasome activation promotes life span extension and resistance to proteotoxicity in Caenorhabditis elegans. FASEB (Fed. Am. Soc. Exp. Biol.) J. : Off. Publ.Fed. Am. Soc. Exp. Biol. 2015;29(2):611–622. doi: 10.1096/fj.14-252189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen N.N., Rana A., Goldman C., Moore R., Tai J., Hong Y., Shen J., Walker D.W., Hur J.H. Proteasome β5 subunit overexpression improves proteostasis during aging and extends lifespan in Drosophila melanogaster. Sci. Rep. 2019 doi: 10.1038/s41598-019-39508-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vilchez D., Morantte I., Liu Z., Douglas P.M., Merkwirth C., Rodrigues A.P.C., Manning G., Dillin A. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489(7415):263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 13.Prahlad V., Cornelius T., Morimoto R.I. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science (New York, N.Y.) 2008;320(5877):811–814. doi: 10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durieux J., Wolff S., Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144(1):79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q., Wu X., Chen P., Liu L., Xin N., Tian Y., Dillin A. The mitochondrial unfolded protein response is mediated cell-non-autonomously by retromer-dependent Wnt signaling. Cell. 2018;174(4):870–883.e17. doi: 10.1016/j.cell.2018.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calculli G., Lee H.J., Shen K., Pham U., Herholz M., Trifunovic A., Dillin A., Vilchez D. Systemic regulation of mitochondria by germline proteostasis prevents protein aggregation in the soma of C. elegans. Sci. Adv. 2021;7(26) doi: 10.1126/sciadv.abg3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berendzen K.M., Durieux J., Shao L.-W., Tian Y., Kim H.-E., Wolff S., Liu Y., Dillin A. Neuroendocrine coordination of mitochondrial stress signaling and proteostasis. Cell. 2016;166(6):1553–1563.e10. doi: 10.1016/j.cell.2016.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imanikia S., Özbey N.P., Krueger C., Casanueva M.O., Taylor R.C. Neuronal XBP-1 activates intestinal lysosomes to improve proteostasis in C. elegans. Curr. Biol. : CB. 2019;29(14):2322–2338.e7. doi: 10.1016/j.cub.2019.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor R.C., Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013;153(7):1435–1447. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minnerly J., Zhang J., Parker T., Kaul T., Jia K. The cell non-autonomous function of ATG-18 is essential for neuroendocrine regulation of Caenorhabditis elegans lifespan. PLoS Genet. 2017;13(5) doi: 10.1371/journal.pgen.1006764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Yifei, Wang X., Song M., He Z., Cui G., Peng G., Dieterich C., Antebi A., Jing N., Shen Y. A secreted microRNA disrupts autophagy in distinct tissues of Caenorhabditis elegans upon ageing. Nat. Commun. 2019;10(1):4827. doi: 10.1038/s41467-019-12821-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulgherait M., Rana A., Rera M., Graniel J., Walker D.W. AMPK modulates tissue and organismal aging in a non-cell-autonomous manner. Cell Rep. 2014;8(6):1767–1780. doi: 10.1016/j.celrep.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moskalev A., Guvatova Z., Shaposhnikov M., Lashmanova E., Proshkina E., Koval L., Zhavoronkov A., Krasnov G., Kudryavtseva A. The neuronal overexpression of Gclc in Drosophila melanogaster induces life extension with longevity-associated transcriptomic changes in the thorax. Front. Genet. 2019;10:149. doi: 10.3389/fgene.2019.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. https://pubmed.ncbi.nlm.nih.gov/4366476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamath R.S., Martinez-Campos M., Zipperlen P., Fraser A.G., Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2000;2(1) doi: 10.1186/gb-2000-2-1-research0002. research0002.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez B.A., Kim H., Ray A., Caldwell G.A., Caldwell K.A. A bacterial metabolite induces glutathione-tractable proteostatic damage, proteasomal disturbances, and PINK1-dependent autophagy in C. elegans. Cell Death Dis. 2015;6(10) doi: 10.1038/cddis.2015.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nayak S., Fiaschi M., King D., Tabakin E.R., Wood L., Hunt D.A. Development of small molecular proteasome inhibitors using a Caenorhabditis elegans screen. Int. J. Med. Chem. 2014 doi: 10.1155/2014/237286. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altun-Gultekin Z., Andachi Y., Tsalik E.L., Pilgrim D., Kohara Y., Hobert O. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development. 2001;128(11):1951–1969. doi: 10.1242/dev.128.11.1951. [DOI] [PubMed] [Google Scholar]
- 29.Munkácsy E., Chocron E.S., Quintanilla L., Gendron C.M., Pletcher S.D., Pickering A.M. Neuronal-specific proteasome augmentation via Prosβ5 overexpression extends lifespan and reduces age-related cognitive decline. Aging Cell. 2019 doi: 10.1111/acel.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffin E.F., Yan X., Caldwell K.A., Caldwell G.A. Distinct functional roles of Vps41-mediated neuroprotection in Alzheimer’s and Parkinson’s disease models of neurodegeneration. Hum. Mol. Genet. 2018;27(24):4176–4193. doi: 10.1093/hmg/ddy308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffin E.F., Scopel S.E., Stephen C.A., Holzhauer A.C., Vaji M.A., Tuckey R.A., Berkowitz L.A., Caldwell K.A., Caldwell G.A. ApoE-associated modulation of neuroprotection from Aβ-mediated neurodegeneration in transgenic Caenorhabditis elegans. Disease Models & Mechanisms. 2019;12(2) doi: 10.1242/dmm.037218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matilainen O., Jha S., Holmberg C.I. Fluorescent tools for in vivo studies on the ubiquitin-proteasome system. Methods Mol. Biol. 2016;1449:215–222. doi: 10.1007/978-1-4939-3756-1_12. [DOI] [PubMed] [Google Scholar]
- 33.Link C.D., Taft A., Kapulkin V., Duke K., Kim S., Fei Q., Wood D.E., Sahagan B.G. Gene expression analysis in a transgenic Caenorhabditis elegans Alzheimer’s disease model. Neurobiol. Aging. 2003;24(3):397–413. doi: 10.1016/s0197-4580(02)00224-5. [DOI] [PubMed] [Google Scholar]
- 34.McColl G., Roberts B.R., Pukala T.L., Kenche V.B., Roberts C.M., Link C.D., Ryan T.M., Masters C.L., Barnham K.J., Bush A.I., Cherny R.A. Utility of an improved model of amyloid-beta (Aβ₁₋₄₂) toxicity in Caenorhabditis elegans for drug screening for Alzheimer’s disease. Mol. Neurodegener. 2012;7:57. doi: 10.1186/1750-1326-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim K.B., Myung J., Sin N., Crews C.M. Proteasome inhibition by the natural products epoxomicin and dihydroeponemycin: insights into specificity and potency. Bioorg. Med. Chem. Lett. 1999;9(23):3335–3340. doi: 10.1016/s0960-894x(99)00612-5. [DOI] [PubMed] [Google Scholar]
- 36.Lin H., Gao Y., Zhang C., Ma B., Wu M., Cui X., Wang H. Autophagy regulation influences β-amyloid toxicity in transgenic Caenorhabditis elegans. Front. Aging Neurosci. 2022;14 doi: 10.3389/fnagi.2022.885145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahman M.A., Rahman M.D.H., Mamun-Or-Rashid A.N.M., Hwang H., Chung S., Kim B., Rhim H. Autophagy modulation in aggresome formation: emerging implications and treatments of Alzheimer’s disease. Biomedicines. 2022;10(5) doi: 10.3390/biomedicines10051027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korolchuk V.I., Menzies F.M., Rubinsztein D.C. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS (Fed. Eur. Biochem. Soc.) Lett. 2010;584(7):1393–1398. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- 39.Chapin H.C., Okada M., Merz A.J., Miller D.L. Tissue-specific autophagy responses to aging and stress in C. elegans. Aging. 2015;7(6):419–434. doi: 10.18632/aging.100765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butterfield D.A., Swomley A.M., Sultana R. Amyloid β-peptide (1-42)-induced oxidative stress in Alzheimer disease: importance in disease pathogenesis and progression. Antioxidants Redox Signal. 2013;19(8):823–835. doi: 10.1089/ars.2012.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forman H.J., Zhang H. Targeting oxidative stress in disease: promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021;20(9):689–709. doi: 10.1038/s41573-021-00233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korovila I., Hugo M., Castro J.P., Weber D., Höhn A., Grune T., Jung T. Proteostasis, oxidative stress and aging. Redox Biol. 2017;13:550–567. doi: 10.1016/j.redox.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pickering A.M., Koop A.L., Teoh C.Y., Ermak G., Grune T., Davies K.J.A. The immunoproteasome, the 20S proteasome and the PA28αβ proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem. J. 2010;432(3):585–594. doi: 10.1042/BJ20100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pickering A.M., Staab T.A., Tower J., Sieburth D., Davies K.J.A. A conserved role for the 20S proteasome and Nrf2 transcription factor in oxidative stress adaptation in mammals, Caenorhabditis elegans and Drosophila melanogaster. J. Exp. Biol. 2013;216(Pt 4):543–553. doi: 10.1242/jeb.074757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kahn N.W., Rea S.L., Moyle S., Kell A., Johnson T.E. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem. J. 2008;409(1):205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- 46.Li X., Matilainen O., Jin C., Glover-Cutter K.M., Holmberg C.I., Blackwell T.K. Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 2011;7(6) doi: 10.1371/journal.pgen.1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oliveira R.P., Porter Abate J., Dilks K., Landis J., Ashraf J., Murphy C.T., Blackwell T.K. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell. 2009;8(5):524–541. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richmond J. In: WormBook. Jorgensen E.M., Kaplan J.M., editors. 2007. Synaptic function. (The C. elegans Research Community, WormBook). [DOI] [Google Scholar]
- 49.Calahorro F., Izquierdo P.G. The presynaptic machinery at the synapse of C. elegans. Invertebr. Neurosci. 2018;18(2):4. doi: 10.1007/s10158-018-0207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tien C.-W., Yu B., Huang M., Stepien K.P., Sugita K., Xie X., Han L., Monnier P.P., Zhen M., Rizo J., Gao S., Sugita S. Open syntaxin overcomes exocytosis defects of diverse mutants in C. elegans. Nat. Commun. 2020;11(1):5516. doi: 10.1038/s41467-020-19178-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Speese S., Petrie M., Schuske K., Ailion M., Ann K., Iwasaki K., Jorgensen E.M., Martin T.F.J. UNC-31 (CAPS) is required for dense-core vesicle but not synaptic vesicle exocytosis in <em>Caenorhabditis elegans</em>. J. Neurosci. 2007;27(23):6150. doi: 10.1523/JNEUROSCI.1466-07.2007. LP – 6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hong L., Huang H.-C., Jiang Z.-F. Relationship between amyloid-beta and the ubiquitin-proteasome system in Alzheimer’s disease. Neurol. Res. 2014;36(3):276–282. doi: 10.1179/1743132813Y.0000000288. [DOI] [PubMed] [Google Scholar]
- 53.Zhang W., Xu C., Sun J., Shen H.-M., Wang J., Yang C. Impairment of the autophagy-lysosomal pathway in Alzheimer’s diseases: pathogenic mechanisms and therapeutic potential. Acta Pharm. Sin. B. 2022;12(3):1019–1040. doi: 10.1016/j.apsb.2022.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ilyinsky N.S., Nesterov S.V., Shestoperova E.I., Fonin A.V., Uversky V.N., Gordeliy V.I. On the role of normal aging processes in the onset and pathogenesis of diseases associated with the Abnormal accumulation of protein Aggregates. Biochemistry. Biokhimiia. 2021;86(3):275–289. doi: 10.1134/S0006297921030056. [DOI] [PubMed] [Google Scholar]
- 55.Daniele J.R., Higuchi-Sanabria R., Durieux J., Monshietehadi S., Ramachandran V., Tronnes S.U., Kelet N., Sanchez M., Metcalf M.G., Garcia G., Frankino P.A., Benitez C., Zeng M., Esping D.J., Joe L., Dillin A. UPR(ER) promotes lipophagy independent of chaperones to extend life span. Sci. Adv. 2020;6(1) doi: 10.1126/sciadv.aaz1441. eaaz1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rai M., Coleman Z., Curley M., Nityanandam A., Platt A., Robles-Murguia M., Jiao J., Finkelstein D., Wang Y.-D., Xu B., Fan Y., Demontis F. Proteasome stress in skeletal muscle mounts a long-range protective response that delays retinal and brain aging. Cell Metabol. 2021;33(6):1137–1154.e9. doi: 10.1016/j.cmet.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bence N.F., Sampat R.M., Kopito R.R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science (New York, N.Y.) 2001;292(5521):1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 58.Thibaudeau T.A., Anderson R.T., Smith D.M. A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat. Commun. 2018;9(1):1097. doi: 10.1038/s41467-018-03509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Papaevgeniou N., Sakellari M., Jha S., Tavernarakis N., Holmberg C.I., Gonos E.S., Chondrogianni N. 18α-Glycyrrhetinic acid proteasome activator decelerates aging and Alzheimer’s disease progression in Caenorhabditis elegans and neuronal cultures. Antioxidants Redox Signal. 2016;25(16):855–869. doi: 10.1089/ars.2015.6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vasilopoulou M.A., Gioran A., Theodoropoulou M., Koutsaviti A., Roussis V., Ioannou E., Chondrogianni N. Healthspan improvement and anti-aggregation effects induced by a marine-derived structural proteasome activator. Redox Biol. 2022;56 doi: 10.1016/j.redox.2022.102462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kapeta S., Chondrogianni N., Gonos E.S. Nuclear erythroid factor 2-mediated proteasome activation delays senescence in human fibroblasts. J. Biol. Chem. 2010;285(11):8171–8184. doi: 10.1074/jbc.M109.031575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mladenovic Djordjevic A.N., Kapetanou M., Loncarevic-Vasiljkovic N., Todorovic S., Athanasopoulou S., Jovic M., Prvulovic M., Taoufik E., Matsas R., Kanazir S., Gonos E.S. Pharmacological intervention in a transgenic mouse model improves Alzheimer’s-associated pathological phenotype: involvement of proteasome activation. Free Radical Biol. Med. 2021;162:88–103. doi: 10.1016/j.freeradbiomed.2020.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun-Wang J.L., Ivanova S., Zorzano A. The dialogue between the ubiquitin-proteasome system and autophagy: implications in ageing. Ageing Res. Rev. 2020;64 doi: 10.1016/j.arr.2020.101203. [DOI] [PubMed] [Google Scholar]
- 64.Jha S., Holmberg C.I. Tissue-specific impact of autophagy genes on the ubiquitin-proteasome system in C. elegans. Cells. 2020;9(8) doi: 10.3390/cells9081858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keith S.A., Maddux S.K., Zhong Y., Chinchankar M.N., Ferguson A.A., Ghazi A., Fisher A.L. Graded proteasome dysfunction in Caenorhabditis elegans activates an adaptive response involving the conserved SKN-1 and ELT-2 transcription factors and the autophagy-lysosome pathway. PLoS Genet. 2016;12(2) doi: 10.1371/journal.pgen.1005823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wrobel L., Hill S.M., Djajadikerta A., Fernandez-Estevez M., Karabiyik C., Ashkenazi A., Barratt V.J., Stamatakou E., Gunnarsson A., Rasmusson T., Miele E.W., Beaton N., Bruderer R., Feng Y., Reiter L., Castaldi M.P., Jarvis R., Tan K., Bürli R.W., Rubinsztein D.C. Compounds activating VCP D1 ATPase enhance both autophagic and proteasomal neurotoxic protein clearance. Nat. Commun. 2022;13(1):4146. doi: 10.1038/s41467-022-31905-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frakes A.E., Metcalf M.G., Tronnes S.U., Bar-Ziv R., Durieux J., Gildea H.K., Kandahari N., Monshietehadi S., Dillin A. Four glial cells regulate ER stress resistance and longevity via neuropeptide signaling in C. elegans. Science (New York, N.Y.) 2020;367(6476):436–440. doi: 10.1126/science.aaz6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bishop N.A., Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447(7144):545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 69.Wolkow C.A., Kimura K.D., Lee M.S., Ruvkun G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science (New York, N.Y.) 2000;290(5489):147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- 70.Higuchi-Sanabria R., Durieux J., Kelet N., Homentcovschi S., de Los Rios Rogers M., Monshietehadi S., Garcia G., Dallarda S., Daniele J.R., Ramachandran V., Sahay A., Tronnes S.U., Joe L., Dillin A. Divergent nodes of non-autonomous UPR(ER) signaling through serotonergic and dopaminergic neurons. Cell Rep. 2020;33(10) doi: 10.1016/j.celrep.2020.108489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He H.-Y., Ahsan A., Bera R., McLain N., Faulkner R., Ramachandran K.V., Margolis S.S., Cline H.T. Neuronal membrane proteasomes regulate neuronal circuit activity in vivo and are required for learning-induced behavioral plasticity. Proc. Natl. Acad. Sci. U. S. A. 2023;120(3) doi: 10.1073/pnas.2216537120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramachandran K.V., Margolis S.S. A mammalian nervous-system-specific plasma membrane proteasome complex that modulates neuronal function. Nat. Struct. Mol. Biol. 2017;24(4):419–430. doi: 10.1038/nsmb.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou Y., Danbolt N.C. Glutamate as a neurotransmitter in the healthy brain. J. Neural. Transm. 2014;121(8):799–817. doi: 10.1007/s00702-014-1180-8. Vienna, Austria : 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Magi S., Piccirillo S., Amoroso S., Lariccia V. Excitatory amino acid transporters (EAATs): glutamate transport and beyond. Int. J. Mol. Sci. 2019;20(22) doi: 10.3390/ijms20225674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ilieva H., Polymenidou M., Cleveland D.W. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J. Cell Biol. 2009;187(6):761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liddelow S.A. Modern approaches to investigating non-neuronal aspects of Alzheimer’s disease. FASEB (Fed. Am. Soc. Exp. Biol.) J. : Off. Publ.Fed. Am. Soc. Exp. Biol. 2019;33(2):1528–1535. doi: 10.1096/fj.201802592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.