Abstract
Fluorescein isothiocyanate (FITC) is widely used to fluorescently label reactive lysine residues on proteins, including antibodies. The rate and extent of labeling varies with reaction conditions, concentration of label, and the concentration and nature of the protein. Fluorescently labeled proteins are very useful, and one use for FITC labeled mAbs is development of assays to measure anti-mAb antibodies produced in vivo during treatment with antibody therapeutics. Our laboratory has developed a humanized anti-cocaine mAb (h2E2) intended for the treatment of cocaine use disorders. Thus, a well characterized FITC labeled h2E2 mAb is needed to quantitate possible anti-mAb antibodies. The time course of labeling and the relative incorporation of FITC into the heavy and light chains, as well as into the Fab and Fc portions of the mAb, was assessed. A novel use of differential scanning fluorimetry in the absence of any extrinsic fluorophore was developed and demonstrated to be capable of measuring antigen (cocaine) binding. In addition, the effect of increasing degrees of labeling by FITC on the thermodynamic parameters driving the binding of cocaine to the mAb was assessed via isothermal titration calorimetry (ITC). This binding technique, unlike others developed recently to measure cocaine binding, is not dependent on, or subject to interference by, the absorbance or fluorescence of the incorporated FITC label. The methods and results reported herein guide the optimization of FITC labeling needed for anti-mAb assays and other assays important for the development of therapeutic mAbs, which are some of the most specific and clinically useful drugs available.
Keywords: Monoclonal antibody, Fluorescein isothiocyanate, Cocaine, Differential scanning fluorimetry, Isothermal titration calorimetry, Thermodynamics of antibody-antigen binding
Graphical abstract
Highlights
-
•
Anti-cocaine h2E2 mAb was labeled with 2–5 mol FITC/mol mAb.
-
•
FITC labeling of mAb heavy and light chains, and Fab and Fc, were compared.
-
•
DSF was performed without an extrinsic dye to demonstrate cocaine binding.
-
•
ITC analysis revealed small changes in cocaine binding thermodynamics by FITC.
-
•
FITC labeling caused a small decrease in the number of mAb cocaine binding sites.
1. Introduction
We have developed a high affinity humanized anti-cocaine IgG1 mAb, named h2E2, for the treatment of cocaine use disorders. The h2E2 mAb has a fully human sequence in its Fc region, as well as a fully human sequence in the portion of the Fab region that does not bind the antigen, cocaine (i.e., the heavy chain CH1 region and the light chain CL region are both humanized). As a part of the mAb characterization process, several antigen (cocaine and cocaine metabolite) binding assays were established, including ligand quenching of intrinsic tyrosine and tryptophan mAb fluorescence [1], the use of extrinsic fluorescent dyes and differential scanning fluorimetry (DSF) [2], and both absorbance [3] and fluorescence [4] methods using the DASPMI dye to quantitate cocaine and metabolite binding affinities. We also demonstrated that oxidation of tryptophan residues located in the h2E2 mAb drug binding site abolished high affinity binding [5], and that tyrosine labeling near the cocaine binding site modifies mAb ligand binding selectivity [6].
There is a need for the development of assays to detect and quantitate the possible formation of anti-anti-cocaine mAb antibodies, which assays are known as anti-drug antibody assays (ADA assays). These anti-antibodies can be formed in vivo in response to administration of therapeutic mAbs, and can be detrimental to the success of mAb therapy by neutralizing or reducing the effective concentration of the therapeutic antibody, thereby decreasing its therapeutic efficacy. One of the most commonly used ADA assays employs FITC labeled therapeutic mAbs as one of its critical components (ADA assays reviewed in Ref. [7]). Lysine labeling by FITC modification is not able to be directed to specific lysine residues, but the desired labeling outcome is to maximize the resultant labeling and fluorescence, while minimizing the effects on antigen binding and mAb structure and function. Thus, in this study, we varied the stoichiometry of FITC labeling of the h2E2 mAb by varying the labeling reaction time, and analyzed the labeled mAb for differential labeling of heavy and light chains, as well as for differential labeling of Fab and Fc fragments. We also assessed the utility of FITC labeled mAb for use in differential scanning fluorimetry (DSF) assays of protein stability and cocaine binding, in the absence of any extrinsic fluorescent dye such as Sypro orange, which is normally required for DSF analyses. In addition, we analyzed the FITC labeled mAb samples for any possible changes in cocaine binding affinity and thermodynamics by analysis with isothermal scanning titration calorimetry (ITC). Taken together, these experiments indicated that labeling stoichiometries between approximately 2 and 5 FITC molecules incorporated per mAb did not substantially affect mAb antigen (cocaine) binding, indicating the probable utility of these labeled samples in ADA assays, as well as in other assays which require fluorescently labeled antibodies.
2. Materials and methods
2.1. Materials
The generation, production, and purification of the h2E2 anti-cocaine monoclonal antibody by the manufacturer, Catalent, was previously described [8]. Fluorescein isothiocyanate (FITC) was purchased from Invitrogen (isomer 1, cat #F1907). Sephadex G-50-150 was purchased from Sigma. Fab and Fc fragments of this mAb were generated by proteolysis using Endo-Lys-C immobilized on agarose beads (SignalChem, cat #L585-31AN-1). A 10 mM stock solution of cocaine in distilled water was made from solid cocaine HCl as described [9]. PBS buffer was made by diluting PBS buffer concentrate (10X) purchased from Cambrex (BioWhittaker, without calcium or magnesium, catalog number 17-517Q). The Applied Biosystems 48 well RT PCR plates (cat. 4375816) and the MicroAmp plate sealing optical adhesive transparent film (cat. 4375928) used in the StepOne RT PCR instrument used to perform the DSF analyses were purchased from ThermoFisher Scientific. Electrophoresis chemicals and SDS-PAGE reagents were purchased from BioRad. The SDS-PAGE gel protein standards were from SMOBIO (cat. PM1600).
2.2. Methods
2.2.1. Labeling and quantitation of FITC modified h2E2 mAb
Anti-cocaine h2E2 mAb at 5 mg/ml was labeled with 2.0 mM FITC at 22 °C in 100 mM NaBO3 pH = 8.0 buffer for 15, 30, 60, 90, and 120 min. Labeling was stopped at each time point by applying 0.8 ml of the reaction mixture to an approximately 5 ml Sephadex G-50 sizing column equilibrated in PBS buffer. Fractions of 0.4 ml were collected and analyzed for protein and FITC content and degree of labeling by scanning 1.5 μl of each fraction from 900 nm to 200 nm using an Implen N60 NanoPhotometer. The extinction coefficients used for the h2E2 mAb (at 280 nm) and FITC (at 494 nm) were 219,500 and 70,000 M−1cm−1, respectively, and a correction factor for FITC absorbance at 280 nm of 0.30 was used to calculate the concentration of mAb and incorporated FITC label, and thus the stoichiometry of labeling. G-50 column fractions with the same calculated stoichiometry of FITC labeling from each labeled time point sample were pooled and re-quantitated for concentration of mAb and degree of labeling with FITC, using the same nanophotometer.
2.2.2. SDS-PAGE gel analysis: Coomassie staining, and fluorescent band quantitation of FITC labeled h2E2 mAb
The mAb samples were analyzed on either 7% or 10% acrylamide SDS-PAGE gels according to Laemmli [10], after boiling in reducing or non-reducing sample buffer for 3 min. The gels were photographed for fluorescence using a 365 nm light source and an Azure 280 gel imager. Fluorescence exposures of images were optimized to ensure that no gel bands used for quantification of fluorescence with the ImageJ software displayed any saturation. Subsequently, the gels were stained for 1 h using Quick Blue protein Coomassie stain from IBI Scientific (cat # IB01034), and destained with water prior to photography using white light transillumination.
2.2.3. Differential scanning fluorimetry analysis of h2E2 mAb labeled for various times with FITC
Differential scanning fluorimetry (DSF) analysis of 20 μl of 2.5 μM mAb labeled from 15 to 120 min with FITC was performed in PBS buffer (with no Sypro orange or other extrinsic fluorescent dye added). For FITC labeled mAb, the StepOne PCR FAM dye calibration was used as the detector, setting the passive reference to none (excitation at 470 nm, emission at 520 nm). The concentration of cocaine was varied from 0 to 1000 μM, and samples were thermally scanned from 35 °C to 95 °C using a temperature ramp rate of 0.45 °C per minute.
2.2.4. Isothermal titration calorimetry analysis of control and FITC labeled h2E2 mAb
Isothermal titration calorimetry (ITC) experiments were performed at 20 °C in 0.22 μm filtered PBS buffer using a MicroCal VP-ITC instrument. Twenty 14 μl injections of 0.1 mM cocaine (in filtered PBS buffer) were performed for each experiment, adding to the 1.4227 ml cell sample containing approximately 0.005 mM mAb (approximately 0.01 mM mAb cocaine binding sites). The ITC sample cell was stirred at 329 rpm, and there was a delay of 240 s between each injection of cocaine. The data were blank corrected using an average of the last 5 injections of the mAb sample ITC data (after the mAb was saturated with cocaine) to subtract from all the integrated mAb injection ITC peaks, prior to fitting of the data. The data were then analyzed using the one binding site model incorporated into the Origin 7.0 software supplied with the instrument. Kd (dissociation constant) values were calculated by taking the reciprocal of the ITC determined K (association constant) values, and ΔG values reported in the Table were calculated using the Origin fitted values of ΔH and program calculated values of ΔS, and the Gibbs free energy equation, ΔG = ΔH – TΔS. Approximately 100 μL of each diluted protein sample loaded into the ITC cell was used to determine the protein concentrations of all ITC samples, using the N60 nanophotometer and correcting for FITC absorbance at 280 nm as described above.
3. Results
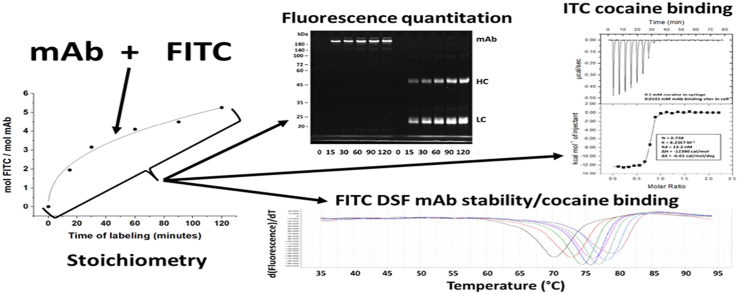
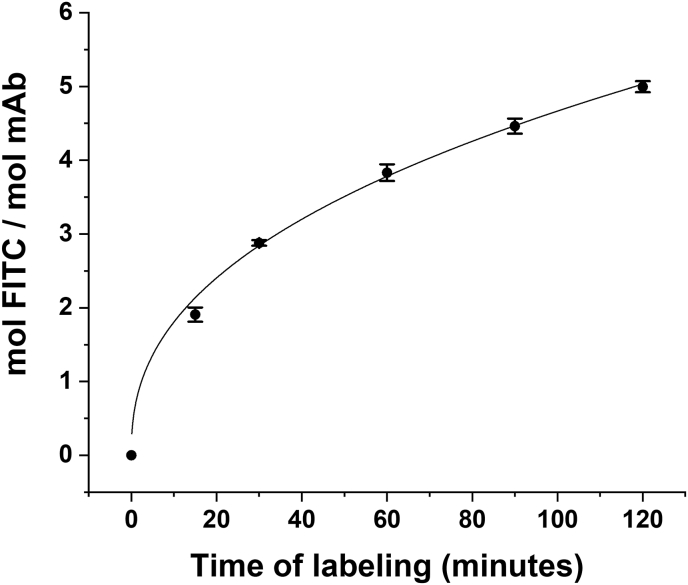
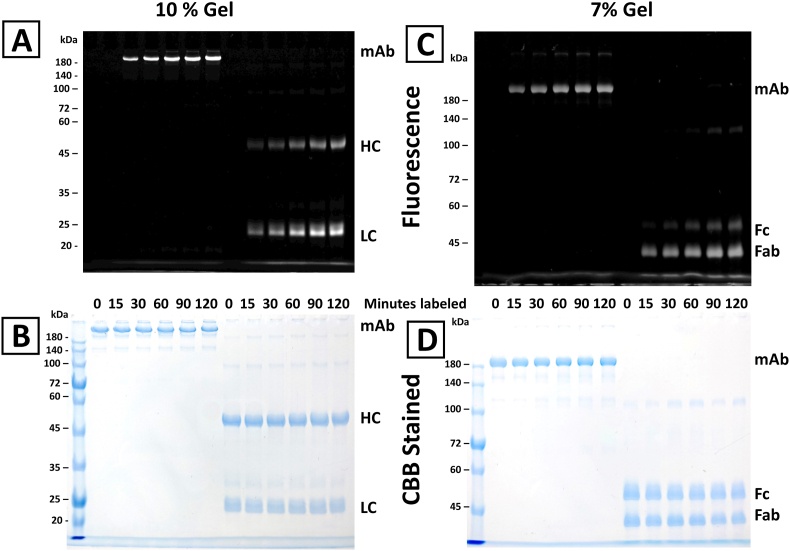
The time course of h2E2 mAb labeling with FITC in pH = 8.0 buffer at 22 °C is shown in Fig. 1. The stoichiometry of labeling varied from approximately 2.0 to 5.0 mol FITC/mol mAb. These samples were then analyzed on a 10% SDS-PAGE gel, both with and without reduction with dithiothreitol, as shown in Fig. 2, Panel A (fluorescence) and Panel B (Coomassie blue staining). Aliquots of these same samples were also digested with Endo-Lys C to generate Fab and Fc fragments, and both the intact mAb and the digested mAb samples were analyzed on a 7% SDS-PAGE gel (without reduction), as shown in Fig. 2, Panel C (fluorescence) and Panel D (Coomassie blue staining). The fluorescent bands shown in Fig. 2A and C were then quantitated using ImageJ software and the relative fluorescence labeling ratios were calculated and plotted in Fig. 3A (for the LC/HC labeling ratio) and Fig. 3B (for the Fab/Fc labeling ratio), as a function of time of labeling with FITC.
Fig. 1.
The time course of the reaction of FITC with the h2E2 mAb at 22°C in borate buffer, pH=8.0. Aliquots were taken and the excess FITC reagent was removed, and the buffer exchanged to PBS by size exclusion chromatography at the times indicated, and the degree of labeling (mol FITC/mol mAb) was determined spectroscopically as described in Methods. Three independent experiments were performed, and the means and standard deviations for each time point are shown in the figure. The curve shown is a power function fit to the data means at each time point.
Fig. 2.
SDS-PAGE gel analysis of the time course of FITC labeling of the h2E2 mAb shown inFig. 1. Panels A and B are the fluorescence and the Coomassie staining, respectively, of the intact mAb (2 μg per lane, not reduced) and the heavy chain (HC) and light chain (LC), after reduction of 4 μg per lane of the intact mAb. Panels C and D are the fluorescence and the Coomassie staining, respectively, of the intact mAb (2 μg per lane, not reduced) and the Fab and Fc fragments after proteolysis of mAb with Endo Lys-C (4 μg per lane, not reduced).
Fig. 3.
Dependence on the time of FITC reaction of the relative FITC labeling of the light and heavy mAb chains (Panel A) and the Fab and Fc portions of the mAb (Panel B). Non-overexposed images of the fluorescent bands in the gels shown in Fig. 2, Panels A and C, were quantitated using ImageJ software and expressed as ratios of labeling as a function of reaction time with FITC. Three independent FITC labeling experiments were performed, and the means and standard deviations for the fluorescent band intensity ratios for each time point for these three experiments are shown in the figure.
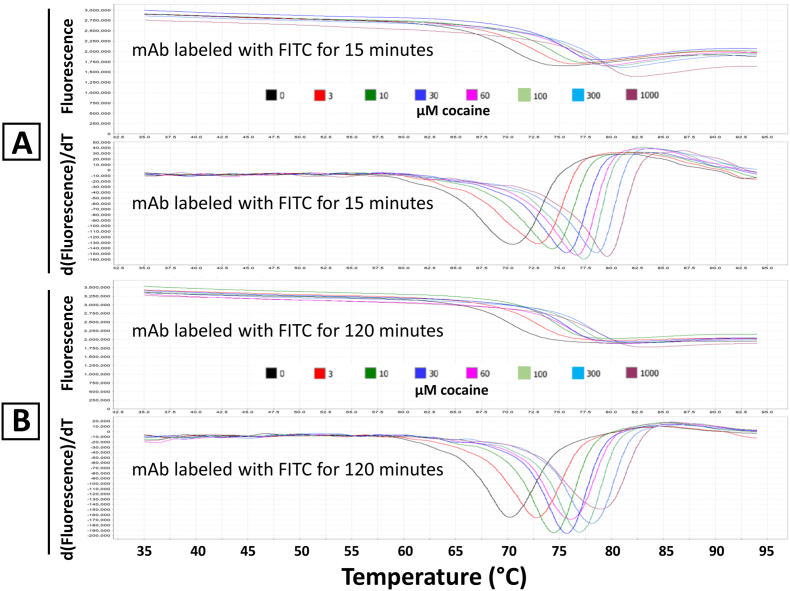
The fluorescence of FITC precluded standard DSF analysis of the labeled mAb samples using Sypro orange or DASPMI dyes we had previously used [2] for the purpose of measuring the affinity of the mAb for cocaine and cocaine metabolites (due to spectral overlap and fluorescence interference between the FITC and Sypro orange or DASPMI dyes). However, we were able to demonstrate cocaine binding, and the concentration dependence of cocaine binding, to all the FITC labeled mAb samples using the covalently attached FITC fluorescence, in the absence of any extrinsic fluorescent dye. Results of the titration of the 15 and 120 min labeled FITC mAb samples are shown in Fig. 4. The two samples gave similar, but not identical, results and both demonstrated the increasing thermal stability reported by the FITC label with increasing concentrations of cocaine, similar to that observed using unlabeled mAb and the DASPMI dye [2]. However, the FITC fluorescence decreases when the mAb is thermally denatured, unlike the Sypro orange or DASPMI dye fluorescence, which increases upon protein thermal denaturation, due to exposure of non-covalent dye-binding hydrophobic sites on the mAb upon unfolding of the antibody as a function of increasing temperature [2].
Fig. 4.
Differential scanning fluorimetry (DSF) analysis of FITC labeled mAb. The raw fluorescence data (top plots) and the first derivative of the raw fluorescence data (bottom plots) are shown for the mAb labeled with FITC for 15 min (Panel A) and for 120 min (Panel B). For both panels, the curves are color coded for the concentration of cocaine present, with concentrations varying from 0 to 1000 μM cocaine. No extrinsic dye was present, as is usually required for DSF analyses. Note a decrease, rather than an increase, in fluorescence with temperature, resulting in inverted first derivative peaks (relative to the positive (upward) first derivative peaks typically observed using extrinsic DSF dyes), due to the decrease in mAb covalently-bound FITC fluorescence with temperature observed in all samples. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
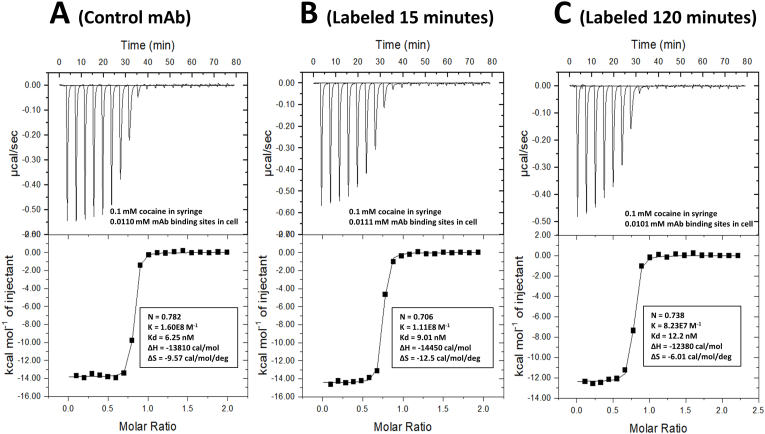
The effects of FITC labeling on the thermodynamics of cocaine binding and the number of mAb cocaine binding sites was assessed using isothermal titration calorimetry (ITC), a technique which is not subject to interference caused by the FITC label. Data (upper plots) and fitting of the data (lower plots) to a one binding site model are shown for unlabeled control mAb, 15 min FITC labeled mAb, and 120 min FITC labeled mAb in Fig. 5 (Panels A, B, and C, respectively). ITC data for all control and FITC labeled mAb samples are summarized in Table 1.
Fig. 5.
Isothermal titration calorimetry (ITC) analysis of FITC labeled mAb. Shown are data and curve fits for the control, unlabeled mAb, as well as the mAb labeled for 15 and 120 min with FITC (resulting in 1.8 and 4.9 mol FITC/mol mAb, respectively). The baseline-leveled raw ITC data is shown in each panel on the top, while the one-binding site fitting to the resultant titration curve is shown on the bottom of each of the three panels. The concentrations of cocaine in the syringe and of the h2E2 mAb in the sample cell are given in the top section of each panel, and the fitted parameters describing the binding for each sample are given in the bottom sections of each panel.
Table 1.
Summary of isothermal titration calorimetry (ITC) cocaine binding data for control and FITC labeled h2E2 mAb.
| Sample | Average mol FITC/mol mAb | N (cocaine sites/Fab) | K (M−1) | ΔH (cal/mol) | ΔS (cal/mol/deg) | ΔG (cal/mol) | Kd (nM) |
|---|---|---|---|---|---|---|---|
| control h2E2 mAb | 0 | 0.782 | 1.60E+08 | −13800 | −9.57 | −11000 | 6.25 |
| 15 min FITC-h2E2 mAb | 1.91 | 0.706 | 1.11E+08 | −14400 | −12.5 | −10800 | 9.01 |
| 30 min FITC-h2E2 mAb | 2.88 | 0.789 | 1.15E+08 | −12900 | −7.26 | −10800 | 8.70 |
| 60 min FITC-h2E2 mAb | 3.83 | 0.707 | 7.58E+08 | −13200 | −8.98 | −10600 | 13.2 |
| 90 min FITC-h2E2 mAb | 4.46 | 0.748 | 8.45E+07 | −12700 | −6.91 | −10600 | 11.8 |
| 120 min FITC-h2E2 mAb | 5.00 | 0.738 | 8.23E+07 | −12400 | −6.01 | −10600 | 12.2 |
4. Discussion
Fig. 1 shows the time dependence of FITC mAb labeling in pH 8.0 borate buffer for three independent experiments. The 5 mg/ml mAb used for labeling is equivalent to 34.6 μM mAb, and since there are 86 lysine residues per mAb, 2.98 mM of lysine residues. FITC was used at 2 mM, which is about a 58 fold molar excess over the mAb, translating to about 67% of the equimolar value with respect to the total number of lysine residues present in the mAb (many of which will not be accessible to, or reactive with, FITC). These conditions resulted in a stoichiometry of labeling of about 2.0–5.0 lysine residues per mAb from 15 to 120 min of reaction time, with the data being adequately fit using a power function, as shown in Fig. 1.
It is of interest, as well as relevant to this study, to determine the relative labeling of the mAb heavy and light chains, as well as the labeling of Fab and Fc fragments, with respect to the time/degree of labeling. In Fig. 2, Fig. 3 it is apparent that the light chain is selectively labeled relative to the heavy chain, and to a much greater extent, the Fab fragment is selectively labeled relative to the Fc fragment. It is also apparent in Fig. 3 that this selectivity of labeling decreases with increasing time/degree of FITC labeling for both of these comparisons. The light chains contain only 24 out of 86 of the total lysine residues (28%) present in the mAb, indicating that the light chain lysine residues are modified more efficiently by FITC under these conditions. This is also consistent with the ratio of FITC labeling observed for the Fab vs Fc fragments, where the Fab fragments contain 56% (48 of 86 total) of the lysine residues, but the Fab/Fc labeling ratio starts at approximately 8.4, decreasing to about 4.3 with increasing times/degrees of FITC labeling.
Since the selectivity of labeling of the Fab fragments (that bind cocaine) is high, especially after only 15 min of labeling, we reasoned that FITC labeling might serve as a good reporter of cocaine binding, since most of the FITC fluorescence originates from the part of the mAb that binds the cocaine antigen, i.e., the Fab fragment. Thus, we employed a novel modification of differential scanning fluorimetry (DSF), leaving out the usually required extrinsic fluorescent dye, and relying on the covalently attached fluorescein fluorescence, and using the appropriate excitation and emission wavelengths for detection of FITC in the RT PCR instrument used for the DSF experiments. We previously used DSF and the extrinsic DASPMI dye to quantitatively measure cocaine and cocaine metabolite affinities for the h2E2 anti-cocaine mAb [2]. As can be seen in Fig. 4, the FITC fluorescence decreased upon mAb unfolding, unlike the DSF results seen with Sypro or DASPMI dyes, where the extrinsic dye fluorescence increases due to the change in dye microenvironment caused by binding to hydrophobic patches on the mAb exposed upon unfolding of the protein domain. We attempted to quantitate this FITC DSF cocaine titration data from all the FITC labeled samples in this study using Boltzmann fitting of the raw fluorescence curves shown in the upper traces of Fig. 4A and B, but the determined apparent affinities (Kds calculated using the methods previously described [2]) did not result in reasonable values for cocaine affinity (about 300–500 nM apparent Kds at the melting temperature of the FITC labeled mAb in the absence of cocaine (i.e., about 70 °C), compared to about 20 nM calculated for the unlabeled mAb using the DASPMI dye and run on the same plate in the same experiment (data not shown)). This may be due to the change in FITC fluorescence also reporting on the thermal stability of other parts of the mAb besides the part of the antibody that binds cocaine. Regardless, although the data shown in Fig. 4 clearly show that the unfolding of the FITC labeled mAb can be analyzed by DSF, and that the stabilization of the unfolding of the FITC labeled protein is a function of the concentration of added cocaine (and thus the binding of cocaine to the mAb), this technique is not quantitative for determining affinity (Kd), unlike the previously published cocaine binding data using DSF and the extrinsic DASPMI data [2].
Thus, we sought a method that would allow quantitative evaluation of the effects of FITC labeling on cocaine binding affinity of the labeled mAb, as well as any possible changes in the thermodynamic parameters supporting that binding. In addition, we also assessed the possibility of the loss of functional cocaine binding sites on the mAb as a result of FITC labeling. This potential loss of binding sites might be due to labeling of lysine residues in the cocaine binding site or lysine residues close to the binding site which could result in a change in ligand affinity of the binding sites. Isothermal titration calorimetry (ITC) fulfills these requirements very well. Therefore, we analyzed the control, unlabeled mAb, as well as the mAb labeled for 15, 30, 60, 90 and 120 min using ITC. Representative data is shown in Fig. 5, and data resulting from fitting of the data from all analyzed samples is included in Table 1. As can be seen from Table 1, the enthalpic driving force (ΔH values) supporting the high affinity binding of cocaine to the mAb decreased slightly with increasing labeling, leading to a trend towards slightly higher Kd values (slightly lower affinities) with increasing levels of FITC labeling. Thus, the highest levels of labeling resulted in an approximate doubling of the Kd (Kd at 20 °C increasing from about 6 to 12 nM). The ITC parameters determined for the control, unlabeled mAb agree well with those recently published for cocaine binding to this unlabeled mAb measured by ITC (i.e., Kd = 8.1 nM, N = 0.79, [11]). However, there is a small (5–10%) decrease in the number of cocaine binding sites compared to the unlabeled control in some of the FITC labeled samples. (It should be noted that the ITC reported number of sites, i.e., the “N” value, is expressed in number of cocaine molecules bound per mAb Fab present, not per molecule of mAb.) The lack of a substantial decrease in cocaine binding sites, even at the highest level of FITC labeling, is consistent with no lysine residues in the crystal structure of the Fab portion of this h2E2 mAb that are in the cocaine binding site, i.e., there are no lysine residues in direct contact with the co-crystalized cocaine metabolite, benzoylecgonine [12]. However, there is one solvent-exposed lysine residue near the cocaine binding site (lysine 57 in the heavy chain [12]), whose modification with FITC may be the explanation for the small decrease in affinity for cocaine, which is evident in the ITC data.
In summary, we assessed the effects of FITC labeling of the anti-cocaine h2E2 mAb, and the distribution of this labeling on various parts of the mAb, as a function of the degree of labeling. We discovered, and report for the first time, the use of FITC labeled antibody to assess thermal stability and antigen binding of an antibody via DSF (in the absence of any extrinsic dyes normally required for protein DSF analyses). We clearly demonstrated the utility of ITC analysis for the quantitative assessment of the effects of FITC labeling on antigen (cocaine) binding. FITC labeling caused slight changes in the enthalpy of cocaine binding, resulting in only a small decrease in cocaine binding affinity (a minor increase in Kd), along with a slight decrease in the number of cocaine binding sites. FITC labeling was shown to be very selective for the Fab portion of the antibody at lower levels of labeling, and labeling of up to 5 mol FITC/mol mAb gave rise to only minor changes in antigen binding. This approach and these results may be useful for the labeling of other mAbs with FITC and other molecules of interest, including biotin, which is also commonly used in anti-drug assays (ADAs). ADA assays are needed to monitor the production of antibodies produced in vivo against the therapeutic mAb, which is a potential immunological concern, as well as a possible threat to the clinical efficacy of all therapeutic mAbs. Thus, these findings are relevant to the characterization and usage of mAbs in modern medical practice, the importance of which is evidenced by the numbers of recently FDA approved therapeutic mAbs, as well as the commercial sales and profitability of such mAbs.
Conflict–of–interest and financial disclosure statement
Dr. Norman is named as a co-inventor on a portfolio of patents for the matter and use of the h2E2 humanized anti-cocaine monoclonal antibody.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Andrew B. Norman reports financial support was provided by National Institute on Drug Abuse. Andrew B. Norman has patent #10501556 issued to University of Cincinnati, E. R. Squibb & Sons, L.L.C. Andrew B. Norman has patent #9957332 issued to University of Cincinnati. Andrew B. Norman has patent #9758593 issued to University of Cincinnati, E. R. Squibb & Sons, L.L.C. None.
Acknowledgments
This work was supported in part by the National Institutes of Health National Institute on Drug Abuse Grant U01DA050330. We are grateful to Catalent PharmaSolutions, Inc. (Madison, WI) for providing the recombinant humanized h2E2 anti-cocaine mAb protein expressed using their GPex® technology. We acknowledge Dr. Guochang Fan in this Department for the use of the StepOne RT PCR instrument for DSF measurements. We thank Dr. Rhett Kovall and Dr. Zhenyu Yuan in the Department of Molecular and Cellular Biosciences at the University of Cincinnati College of Medicine for the use of the MicroCal VP-ITC instrument.
References
- 1.Kirley T.L., Norman A.B. Characterization of a recombinant humanized anti-cocaine monoclonal antibody and its Fab fragment. Hum. Vaccines Immunother. 2015;11:458–467. doi: 10.4161/21645515.2014.990856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirley T.L., Norman A.B., Wetzel H.N. A novel differential scanning fluorimetry analysis of a humanized anti-cocaine mAb and its ligand binding characteristics. J. Immunol. Methods. 2020;476 doi: 10.1016/j.jim.2019.112676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirley T.L., Norman A.B. Ligand binding to a humanized anti-cocaine mAb measured by dye absorption spectroscopy. Biochem. Biophys. Res. Commun. 2021;535:93–98. doi: 10.1016/j.bbrc.2020.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirley T.L., Norman A.B. Cocaine binding to the Fab fragment of a humanized anti-cocaine mAb quantitated by dye absorption and fluorescence spectroscopy. J. Immunol. Methods. 2021;496 doi: 10.1016/j.jim.2021.113103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirley T.L., Norman A.B., Greis K.D. Oxidation of specific tryptophan residues inhibits high-affinity binding of cocaine and its metabolites to a humanized anticocaine mAb. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.101689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirley T.L., Greis K.D., Norman A.B. Tyrosine nitration of a humanized anti-cocaine mAb differentially affects ligand binding of cocaine and its metabolites. Biochem. Biophys. Rep. 2022;30 doi: 10.1016/j.bbrep.2022.101278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collet-Brose J., Couble P.J., Deehan M.R., Nelson R.J., Ferlin W.G., Lory S. Evaluation of multiple immunoassay technology platforms to select the anti-drug antibody assay exhibiting the most appropriate drug and target tolerance. J. Immunol. Res. 2016;2016 doi: 10.1155/2016/5069678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norman A.B., Gooden F.C., Tabet M.R., Ball W.J. A recombinant humanized anti-cocaine monoclonal antibody inhibits the distribution of cocaine to the brain in rats. Drug Metab. Dispos.: the biological fate of chemicals. 2014;42:1125–1131. doi: 10.1124/dmd.114.057034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wetzel H.N., Webster R.P., Saeed F.O., Kirley T.L., Ball W.J., Norman A.B. Characterization of a recombinant humanized anti-cocaine monoclonal antibody produced from multiple clones for the selection of a master cell bank candidate. Biochem. Biophys. Res. Commun. 2017;487:690–694. doi: 10.1016/j.bbrc.2017.04.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Kirley T.L., Norman A.B. Isothermal titration calorimetry determination of thermodynamics of binding of cocaine and its metabolites to humanized h2E2 anti-cocaine mAb. Biochem. Biophys. Rep. 2022;32 doi: 10.1016/j.bbrep.2022.101354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan K., Zhou M., Ahrendt A.J., Duke N.E.C., Tabaja N., Ball W.J., Kirley T.L., Norman A.B., Joachimiak A., Schiffer M., Wilton R., Pokkuluri P.R. Structural analysis of free and liganded forms of the Fab fragment of a high-affinity anti-cocaine antibody, h2E2, Acta crystallographica. Section F, Structural biology communications. 2019;75:697–706. doi: 10.1107/S2053230X19013608. [DOI] [PMC free article] [PubMed] [Google Scholar]