Abstract
The correct coupling of amino acids with transfer RNAs (tRNAs) is vital for translating genetic information into functional proteins. Errors during this process lead to mistranslation, where a codon is translated using the wrong amino acid. While unregulated and prolonged mistranslation is often toxic, growing evidence suggests that organisms, from bacteria to humans, can induce and use mistranslation as a mechanism to overcome unfavorable environmental conditions. Most known cases of mistranslation are caused by translation factors with poor substrate specificity or when substrate discrimination is sensitive to molecular changes such as mutations or posttranslational modifications. Here we report two novel families of tRNAs, encoded by bacteria from the Streptomyces and Kitasatospora genera, that adopted dual identities by integrating the anticodons AUU (for Asn) or AGU (for Thr) into the structure of a distinct proline tRNA. These tRNAs are typically encoded next to a full-length or truncated version of a distinct isoform of bacterial-type prolyl-tRNA synthetase. Using two protein reporters, we showed that these tRNAs translate asparagine and threonine codons with proline. Moreover, when expressed in Escherichia coli, the tRNAs cause varying growth defects due to global Asn-to-Pro and Thr-to-Pro mutations. Yet, proteome-wide substitutions of Asn with Pro induced by tRNA expression increased cell tolerance to the antibiotic carbenicillin, indicating that Pro mistranslation can be beneficial under certain conditions. Collectively, our results significantly expand the catalog of organisms known to possess dedicated mistranslation machinery and support the concept that mistranslation is a mechanism for cellular resiliency against environmental stress.
Keywords: aminoacyl-tRNA synthetase, genetic code, mistranslation, protein synthesis, streptomyces, translation, tRNA
Mistranslation of the genetic code, wherein a codon is decoded with the wrong amino acid, is inherent to organisms from all domains of life. In basic cellular conditions, mistranslation is estimated to occur once every 103-104 translated codons, although environmental and metabolic factors can increase this rate (1, 2, 3, 4, 5, 6). Elevated and uncontrolled mistranslation can cause irreparable damage (6, 7, 8, 9, 10, 11, 12). However, in recent years, our understanding of mistranslation has been transformed by the discoveries of cellular conditions in which mistranslation is used as a mechanism to overcome unfavorable circumstances. For instance, human cancer cells can withstand immune response challenges by translating Trp codons as Tyr (13). Similarly, global mistranslation with Met can protect mammalian and bacterial cells against oxidative stress (14, 15), while the opportunistic human pathogen Candida albicans mistranslates Leu codons with Ser as a mechanism to escape the host’s immune response (16, 17, 18, 19). Several other examples of mistranslation as a mechanism of cellular resiliency and survival have been described (20, 21, 22, 23, 24, 25, 26, 27, 28, 29).
Aminoacyl-tRNA synthetases (aaRSs) and ribosomes are the primary sources of mistranslation. aaRSs catalyze the ligation of amino acids to tRNAs to form the aminoacyl-tRNAs, the substrates for ribosomal protein synthesis. Ribosomes and aaRSs are inherently error-prone, and their specificities are also susceptible to molecular changes (e.g., mutations or modification). Consequently, most reported cases of mistranslation are caused by the ligation of amino acids to the wrong tRNAs or incorrect pairing of aminoacyl-tRNAs with mRNA codons on the ribosomes (27, 30). However, in a few cases, dedicated mistranslation factors exist. For example, C. albicans encodes a unique tRNA with a dual identity. As a result, this tRNA is acylated with either Leu or Ser by the corresponding aaRSs, leading to the translation of CUG codons as either Ser or Leu (16, 17, 18).
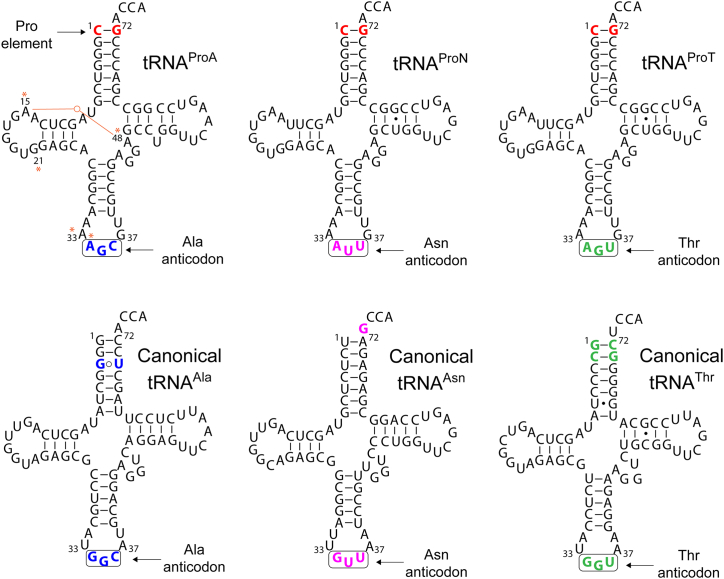
We recently discovered a unique family of proline tRNAs in a group of Streptomyces bacteria that includes agriculturally important plant pathogens (31). This peculiar tRNAPro has adopted a dual identity by replacing its proline anticodon GGG with an AGC alanine anticodon (Fig. 1). To indicate its tRNAPro structure and alanine anticodon this tRNA family was named tRNAProA. tRNAProA is encoded with a distinct isoform of bacterial-type prolyl-tRNA synthetase (ProRS; called ProRSx), which appears to have coevolved with tRNAProA. When expressed recombinantly in Escherichia coli, tRNAProA mistranslates Ala codons with Pro, suggesting that organisms encoding tRNAProA can deliberately mistranslate their genetic code (31). tRNAProA is one of the first examples of a bacterial tRNA that evolved to mistranslate a sense codon.
Figure 1.
The predicted cloverleaf structures of the tRNAProXfamily. The three members of this family share the proline identity element, C1:G72 (indicated in red) but contain different anticodons. tRNAProA contains the AGC alanine anticodon (shown in blue), while the newly discovered tRNAProT and tRNAProN have AGU threonine and AUU asparagine anticodons, respectively. Members of the tRNAProX family have a combination of elements rarely found in canonical tRNAs (see Fig. S1), including G21, A33, A34, and the A15:A48 tertiary pair, which are indicated with orange asterisks in tRNAProA. The predicted secondary structures of canonical tRNAAla, tRNAAsn, and tRNAThr are shown with their predicted identity elements indicated in colored and bolded fonts (32). The tRNA sequences are from Streptomyces turgidiscabies (tRNAAla and tRNAProA), Streptomyces sp. OK228 (tRNAProN and tRNAAsn), and Kitasatospora setae KM-6054 (tRNAProT and tRNAThr).
In the present work, we report two new families of noncanonical tRNAs encoded by gram-positive bacteria from the Streptomyces and Kitasatospora genera. Both tRNA families have a tRNAProA-like structure but have adopted either an AUU anticodon (Asn) or an AGU anticodon (Thr) (Fig. 1). Like tRNAProA, the tRNAs are encoded next to a putative ProRSx gene, although a truncated version of ProRSx is found in some cases. We show that these novel tRNAs mistranslate their corresponding codons with Pro, leading to proteome-wide Asn-to-Pro and Thr-to-Pro changes. Pro mistranslation caused varying degrees of growth defects in E. coli based on the nature of the mistranslated codon. Despite the harmful impact on cell fitness, we observed that the expression of the tRNA mistranslators increased the resistance of E. coli to the antibiotic carbenicillin.
Results
Identification of tRNAs with dual identity
We previously determined that tRNAProA genes are annotated as alanine tRNAs (tRNAAla) in genomic databases due to automated annotation algorithms’ reliance on the anticodon sequence to establish the identity of tRNA genes. Consequently, the AGC anticodon of tRNAProA causes its annotation as tRNAAla, which hindered a complete assessment of the phylogenetic distribution of tRNAProA genes. To circumvent this, we performed searches based on sequence homology using the Streptomyces turgidiscabies tRNAProA as a query. The search identified 18 new genomes containing a region with a predicted tRNA gene and high homology to tRNAProA. While all the identified tRNA genes contained the signature C1:G72 base pair of tRNAProA and tRNAPro, only six tRNAs had the AGC anticodon of tRNAProA. Surprisingly, the remaining tRNA genes had either an Asn AUU (1 genome) or a Thr AGU (11 genomes) anticodon (Figs. 1 and S1). These genes are annotated as tRNAAsn and tRNAThr according to their anticodon sequence. However, like tRNAProA, the tRNAs lack the known key elements for aminoacylation by their cognate aaRS (32), suggesting that they are not paired with the amino acids corresponding to their anticodon. Given the Asn or Thr anticodon combination with the Pro identity element, we named the tRNAs tRNAProN and tRNAProT (Fig. 1). Using the newly discovered tRNAProN and tRNAProT sequences as queries, we performed new searches that unearthed 21 additional organisms with tRNAProA (7), tRNAProN (1), and tRNAProT (13). Notably, tRNAProN and tRNAProT contain the same unusual and unique structural features of tRNAProA (33), including adenosine at positions 33 (A33) and 34 (A34), an A15:A48 tertiary pair, and a G21 (although several tRNAProT species contain A21) (Figs. 1 and S1). Because of the conservation of these elements, tRNAProA, tRNAProN, and tRNAProT appear to have originated from a common ancestral tRNA and belong to a single class of noncanonical tRNAs, which we named the tRNAProX family.
A unique ProRS isoform encoded with tRNAProX
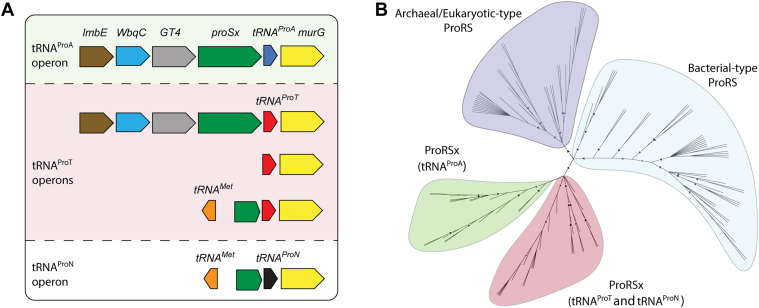
The discovery of tRNAProN and tRNAProT prompted us to examine their genomic context. We previously showed that tRNAProA is encoded next to a proSx gene encoding ProRSx, a distinct form of bacterial ProRS (31). Consistent with this observation, we found that every newly identified tRNAProA gene is flanked by a proSx gene except for Streptomyces vinaceus, which lacks this gene (Figs. 2A and S2). Furthermore, almost half of the bacteria that encode tRNAProT also possess a proSx gene (Figs. 2A and S2). However, in the other half of organisms with tRNAProT and the two encoding tRNAProN, a truncated proSx gene encoding roughly 200 residues is found (Figs. 2A and S2). Sequence alignments showed that the remaining section of the gene matches the C-terminal region of the ProRSx protein, which comprises the tRNA anticodon-binding domain (Figs. 2A and S3). A phylogenetic analysis of the ProRS enzymes revealed two distinct clades for ProRSx, indicating that ProRSx emerged from the bacterial-type ProRS and continued to evolve divergently (Fig. 2B). The divergence of the ProRSx enzyme family appears to be related to the identity of their accompanying tRNAProX gene as ProRSx located with the tRNAProN and tRNAProT cluster separately from ProRSx sequences encoded with tRNAProA (Fig. 2B). This suggests that ProRSx has coevolved with the corresponding tRNAs to adapt to changes in the anticodon. Thus, we surmise that the last base of the anticodon may have been the evolutionary driving force since tRNAProN and tRNAProT share U36 and tRNAProA contains C36. A multiple sequence alignment guided by the bacterial ProRS-tRNAPro complex showed that the residues predicted to interact with N36 vary between the two ProRSx subfamilies (Fig. S3) (31).
Figure 2.
The tRNAProXoperon.A, the predicted operon organization of the tRNAProA, tRNAProT, and tRNAProN families. All families are encoded upstream of a putative murG gene. tRNAProA and some tRNAProT genes are encoded downstream of a full-length proSx (ProRSx) gene (green), GT4 (gray), WbqC (blue), and lmbE (brown). In some organisms, tRNAProT is encoded in an operon composed of either only tRNAProT and murG or a truncated proSx gene with a flanking methionine tRNA gene (tRNAMet, orange). tRNAProN is only found in operons with the tRNAMet and truncated proSx configuration. The complete list of operons from each organism is shown in Fig. S2. B, phylogeny of bacterial-encoded ProRS enzymes. The branches corresponding to the Bacterial-type and Archaeal/Eukaryotic-type ProRS isoforms (canonical isoforms in bacteria) are shown in the purple and light blue clusters, respectively. ProRSx sequences form a distinct cluster from the canonical ProRS enzymes. The tRNAProA-associated ProRSx sequences (green) diverge from the ProRSx encoded with tRNAProT or tRNAProN (red). Branches with bootstrap support higher than 80% are indicated by black triangles.
Phylogenetic distribution and evolution of ProRSx and tRNAProX
The 46 organisms now known to encode ProRSx and tRNAProX are found in bacteria from the Actinomycetes class, primarily in Streptomyces species and a few Kitasatospora species (Table S1 and Fig. S1). These organisms represent a small subset of the more than 3000 Streptomycetaceae genomes that are publicly available. Notably, these genes are encoded in 4 of the more than 12 species known to cause plant diseases, including strains of Streptomyces ipomoeae (34), S. turgidiscabies, Streptomyces reticuliscabiei (35) (same genomospecies), Streptomyces griseiscabiei (36), and Streptomyces caniscabiei (37, 38). However, in several cases, ProRSx/tRNAProX is only present in one of the several available strains of these pathogens, suggesting either a recent acquisition or ancestral loss with maintenance in only one known lineage.
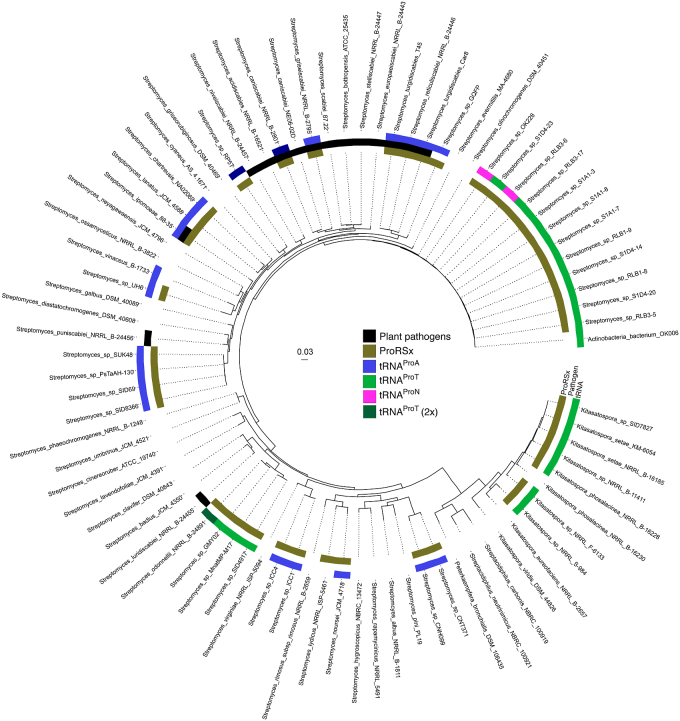
To better contextualize the phylogenetic distribution and evolution of ProRSx and tRNAProX, we constructed a phylogenetic tree to represent the relationship between select strains of Streptomyces, Kitasatospora, and Streptacidiphilus using multilocus sequence analysis (39). In addition to the 46 genomes encoding ProRSx and tRNAProX, we included 38 strains to represent the large diversity of Streptomyces (39), the three major clades of Kitasatospora (40), and Streptacidiphilus (41). While no available genomes of Streptacidiphilus contain ProRSx, the genus was included because it is sister to the Kitasatospora genus (40). Whether the ancestor of Streptacidiphilus lost ProRSx or if ancestral lineages of Kitasatospora and Streptomyces independently gained ProRSx remains unknown. The analysis revealed that ProRSx and all three tRNAProX families are distributed sporadically within the Streptomyces genus, while ProRSx and tRNAProT are present in two of the three defined clades of Kitasatospora (Fig. 3). Based on its presence across multiple genera, we hypothesize that tRNAProT is the most ancestral tRNAProX, even though tRNAProA is more prevalent in Streptomyces. Moreover, the presence of tRNAProN in only two closely related Streptomyces strains indicates that it emerged more recently. Interestingly, Streptomyces odonnellii clustered with other strains that encode the tRNAProT, suggesting that the duplication of the tRNAProT gene is a recent event. Whether the polyphyletic distribution of ProRSx and the three members of tRNAProX results from horizontal gene transfer of the genomic locus or ancestral loss of the genes in most lineages of Streptomycetaceae is unknown.
Figure 3.
Phylogenetic distribution of ProRSx and tRNAProX. Maximum Likelihood phylogenetic tree constructed using multilocus sequence analysis from protein sequences of Streptomyces housekeeping genes atpD, gyrB, recA, rpoB, and trpB. Legend indicates the presence of ProRSx (brown, inner ring), pathogenicity (black, middle ring), and anticodon sequence for the corresponding tRNAProX (outer ring). The scale bar represents the phylogenetic distance of 0.03 nucleotide substitutions per site. Strains NRRL S-984 and NRRL F-6133 were renamed from Streptomyces to Kitasatospora based on this multilocus sequence analysis (MLSA).
tRNAProT translates Thr codons as Pro
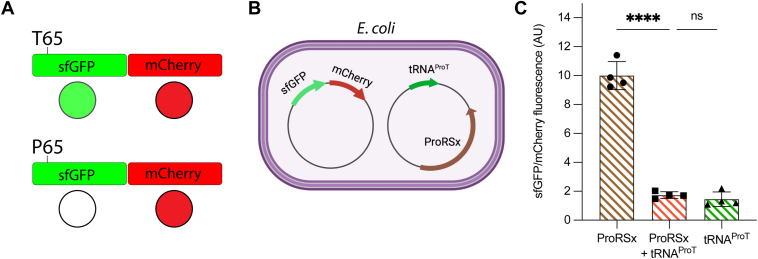
To investigate whether tRNAProT mistranslates Thr codons with Pro, we adapted our previously developed dual fluorescence reporter consisting of a fusion of superfolder green fluorescent protein (sfGFP) and the red fluorescent mCherry in E. coli (Fig. 4A) (31). The reporter is based on the critical role of the residue Thr65 in forming the sfGFP fluorophore. Pro at this position prevents proper fluorophore formation, causing a complete loss of the sfGFP fluorescence (31, 42). mCherry is used to normalize sfGFP expression. Thus, we expected that cells expressing tRNAProT and sfGFP-mCherry would display a lower sfGFP/mCherry emission ratio than cells without tRNAProT (Fig. 4B). Indeed, we observed an ∼7-fold reduction in sfGFP/mCherry fluorescence emission in E. coli cells expressing tRNAProT (Fig. 4C), indicating that tRNAProT can incorporate Pro in response to Thr codons. Interestingly, the coexpression of tRNAProT with its cognate ProRSx only marginally increased sfGFP/mCherry fluorescence, suggesting that endogenous E. coli ProRS aminoacylates tRNAProT.
Figure 4.
Detection of Thr-to-Pro mistranslation by tRNAProT.A, schematic representation of the sfGFP/mCherry fusion reporter used for monitoring in vivo mistranslation of Thr (ACU) codons with Pro. T65 in sfGFP is essential for fluorophore formation and fluorescence (green circle), whereas Pro at this position impairs sfGFP fluorescence resulting in nonfluorescent protein (white circle). In cells expressing tRNAProT, Thr65 can be ambiguously translated as Thr and Pro, reducing the sfGFP fluorescence relative to cells without tRNAProT. mCherry fluorescence is used to normalize the expression of sfGFP. B, E. coli MG1655 was transformed with plasmids encoding the sfGFP/mCherry reporter and tRNAProT (with or without ProRSx), respectively. C, the ratio of sfGFP and mCherry fluorescence at mid-exponential growth was measured in the presence of tRNAProT only (green bar), ProRSx only (brown bar), or both (orange bar). Bars represent the average of four biological replicates with the SD indicated. ns, not significant; ∗∗∗∗p < 0.0001 by t test.
Mistranslation of Asn codons by tRNAProN
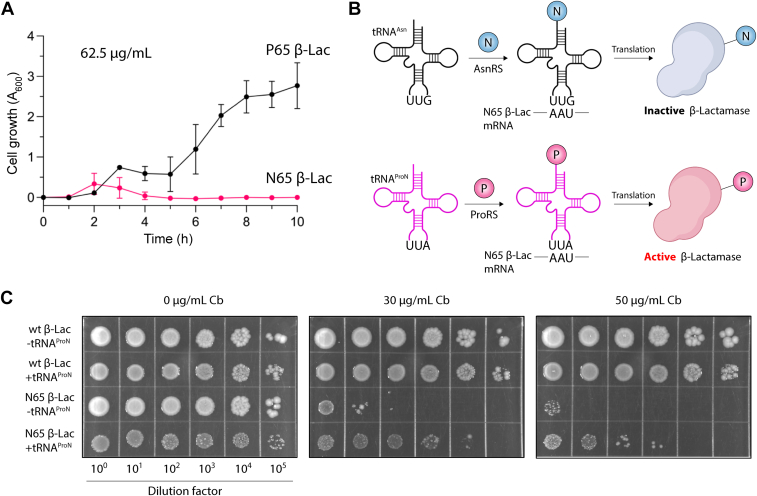
We next sought to test whether tRNAProN can mistranslate Asn codons with Pro. Because Asn at position 65 of sfGFP also impairs chromophore formation (42), this reporter was unsuitable for studying Asn-to-Pro mistranslation. Instead, we adopted a gain-of-function reporter based on β-lactamase activity. We previously reported that a Pro residue at position 65 of β-lactamase is essential for its catalytic activity. Consequently, cells encoding the P65A β-lactamase mutant are sensitive to the antibiotics ampicillin and carbenicillin (31). To evaluate whether Asn has a similar effect, we mutated the Pro65 codon to AAU (Asn) in β-lactamase and compared its activity with wildtype (wt) β-lactamase in E. coli. We observed that cells expressing the P65N β-lactamase did not grow in media containing ampicillin, whereas cells expressing the wt enzyme displayed robust growth (Fig. 5A). This result further supports that Pro65 is essential for β-lactamase and enabled us to repurpose its activity to investigate Asn-to-Pro mistranslation. We expected that mistranslation of Asn at position 65 with Pro would yield a sufficient fraction of active β-lactamase that should increase antibiotic tolerance (Fig. 5B). Thus, we expressed P65N β-lactamase in E. coli in the absence or presence of tRNAProN and tested cell growth on agar plates containing varying concentrations of carbenicillin. Because tRNAProN is found next to a truncated proSx gene, we tested the tRNA alone. As expected, E. coli expressing the P65N β-lactamase variant alone showed significantly impaired growth in the presence of the antibiotic (Fig. 5C). In contrast, cells expressing tRNAProN displayed a substantial tolerance to carbenicillin, even at the highest concentration tested (Fig. 5C).
Figure 5.
In vivo translation activity of tRNAProN.A, an N65 β-lactamase mutant is inactive and does not protect E. coli from β-lactam antibiotics. Cell growth (A600) was monitored for E. coli expressing wt (P65) or N65 β-lactamase in LB medium containing ampicillin (62.5 μg/ml). Each time point is the average of three biological replicates with the SD indicated by the error bars. B, schematic representation of the β-lactamase reporter designed to monitor the translation of Asn AAU codons by tRNAProN based on the critical functional role of the P65 residue. Mistranslation of Asn AAU at position 65 with Pro by tRNAProN produces a mixed population of P65 and N65 β-lactamase, which can afford antibiotic resistance to cells. C, serial dilutions of E. coli cultures expressing either wt β-lactamase (wt β-Lac) or N65 β-lactamase (65N β-Lac) in the presence or absence of tRNAProN were spotted on LB-agar plates containing 0 μg/ml, 30 μg/ml, and 50 μg/ml carbenicillin (Cb).
To confirm the incorporation of Pro in response to Asn codons, we used an sfGFP reporter with an N-terminal extension previously developed to facilitate the detection of mistranslation events using liquid chromatography–tandem mass spectrometry (LC-MS/MS) (43). The 26-residue extension contains a hydrophobic peptide sequence that provides reliable ionization. The sfGFP reporter was recombinantly expressed and purified from E. coli cells with and without tRNAProN coexpression and analyzed by LC-MS/MS. Interestingly, while no Pro misincorporation was identified at the targeted position of the N-terminal extension, Asn-to-Pro mistranslation was detected at three different positions within sfGFP corresponding to N115, N144, and N185 (Fig. S4). These substitutions were not observed in the sfGFP purified in the absence of tRNAProN. These results demonstrate that tRNAProN can mistranslate Asn codons with Pro.
Expression of tRNAProX is toxic in E. coli
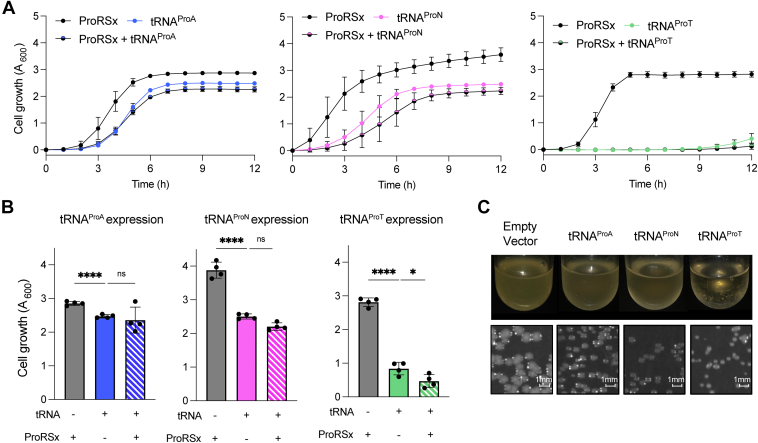
To investigate the effect of mistranslation on cell fitness, we constitutively expressed S. turgidiscabies tRNAProA, Streptomyces sp. OK228 tRNAProN, and Kitasatospora setae tRNAProT in E. coli with or without their cognate ProRSx, except for tRNAProN, which is only encoded with a truncated ProRSx (Figs. 2A and S2). Consequently, we chose K. setae ProRSx (KsProRSx) for coexpression with tRNAProN, given the phylogenetic relationship between KsProRSx and the truncated Streptomyces sp. OK228 ProRSx (Fig. 2B). Notably, individual expression of the three tRNAs was toxic in E. coli cells, albeit with different degrees of severity (Fig. 6). tRNAProA caused a minor growth defect, whereas tRNAProN robustly impaired growth. In contrast, tRNAProT expression almost completely inhibited cell growth. Interestingly, cell cultures expressing tRNAProT formed white precipitated clumps, suggesting that Thr-to-Pro mistranslation causes cell wall disruption and lysis (Fig. 6C). Lastly, coexpression of the cognate proSx genes slightly augmented the growth defect of tRNAProT but not of tRNAProA and tRNAProN. The severity of the growth phenotypes by tRNAProX was also visible in the colony sizes of cells expressing the tRNAs after plasmid transformation. (Fig. 6C). These data suggest that the constitutive mistranslation of Ala, Asn, and Thr codons with Pro is toxic for E. coli.
Figure 6.
Toxicity of tRNAProXgenes in E. coli. tRNAProA, tRNAProN, and tRNAProT were constitutively expressed in E. coli MG1655 cells in the presence and absence of cognate ProRSx. KsProRSx was coexpressed with tRNAProN because tRNAProN is found with a truncated ProRSx gene. Cell growth (A600) was monitored for 12 h (A) or measured after 20 h (B). The plotted results represent the average of four biological replicates with the corresponding SD indicated by the error bars ns, not significant; ∗p< 0.05; ∗∗∗∗p< 0.0001 by t test. C, the density of overnight E. coli cultures expressing empty pCDF plasmid (control) or tRNAProX genes alone is shown. Distinct colony sizes of E. coli cells were also observed on LB-agar plates (bottom panel).
Aminoacylation of tRNAProX by E. coli ProRS
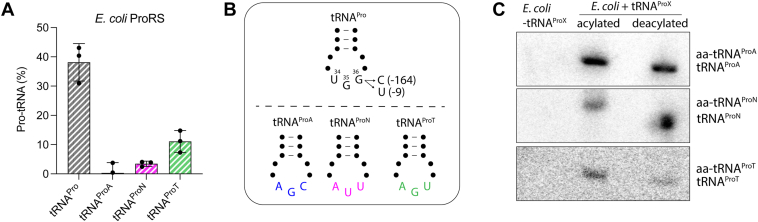
An intriguing observation from our results is that expression of ProRSx was not necessary for tRNAProN- and tRNAProT-mediated mistranslation in E. coli (Figs. 4 and 6), suggesting that endogenous E. coli ProRS aminoacylates both tRNAs. Efficient aminoacylation by E. coli ProRS relies on the recognition of conserved bases in the tRNAPro anticodon (G35 and G36) and acceptor stem (A73 and G72) (44, 45). All members of the tRNAProX family have A73 and G72 but contain either U or C at position 36 (Figs. 1 and S1). Moreover, tRNAProT and tRNAProA have a G35, whereas tRNAProN has a U35. We posited that the presence of these elements in tRNAProX might enable their recognition and aminoacylation by E. coli ProRS. To test this hypothesis, we performed in vitro aminoacylation assays using E. coli ProRS and in vitro tRNA transcripts. The results showed that E. coli ProRS has a lower aminoacylation efficiency toward all three tRNAProX relative to its cognate tRNAPro substrate (Figs. 7A and S5). However, the enzyme displayed different efficiencies toward each tRNAProX. Although tRNAProT was aminoacylated almost 4 times less than tRNAPro, it was charged ∼3 and 36 times better than tRNAProN and tRNAProA, respectively. The higher aminoacylation of E. coli ProRS for tRNAProT may be due to the recognition of U36, as E. coli ProRS was shown to aminoacylate a G36U tRNAPro mutant 19-fold more efficiently than a G36C mutant (Fig. 7B) (45). Next, we tested the aminoacylation level of tRNAProX in E. coli. Total RNA was extracted from E. coli expressing a tRNAProX under acidic conditions to retain the amino acid bound to tRNAs. A fraction of each tRNAProX sample was treated with a basic solution to cleave the amino acid, which allows distinguishing between aminoacylated and deacylated tRNA using specific probes for each tRNA and Northern blotting. Total RNA from E. coli without tRNAProX was used as a negative control. Interestingly, these assays showed that the three tRNAs are mostly aminoacylated in vivo (Fig. 7C). While this experiment does not reveal the amino acid attached to the tRNAs, aminoacylation by E. coli ProRS is likely possible, given the Pro identity elements of tRNAProX.
Figure 7.
Aminoacylation of tRNAProXby E. coli ProRS.A, in vitro aminoacylation of tRNAProA, tRNAProN, and tRNAProT by E. coli ProRS. The reactions were carried out with 1 μM ProRS, 7 μM tRNA, and 1 mM proline at 37 °C for 30 min. Bars represent the average of three technical replicates with the SD indicated. Aminoacylation curves are shown in Fig. S5. B, anticodon stem–loop representations of tRNAPro, tRNAProA, tRNAProN, and tRNAProT are shown to highlight the difference in their anticodon sequence. Substitution of G36 to C or U in E. coli tRNAPro is known to decrease aminoacylation efficiency by E. coli ProRS by 164- and 9-fold, respectively (45). C, the steady-state aminoacylation levels of tRNAProX expressed in E. coli were determined by Northern blotting. Total RNA was extracted from cells expressing plasmid-borne tRNAProN, tRNAProA, or tRNAProT under acidic conditions. A fraction of the sample was then treated with NaOH to remove the amino acids from tRNAs (“deacylated”). The RNA was separated in an acid-urea gel, and tRNAs were detected by hybridization with specific radiolabeled DNA probes.
Pro mistranslation of Ala and Asn codons benefits E. coli under antibiotic stress
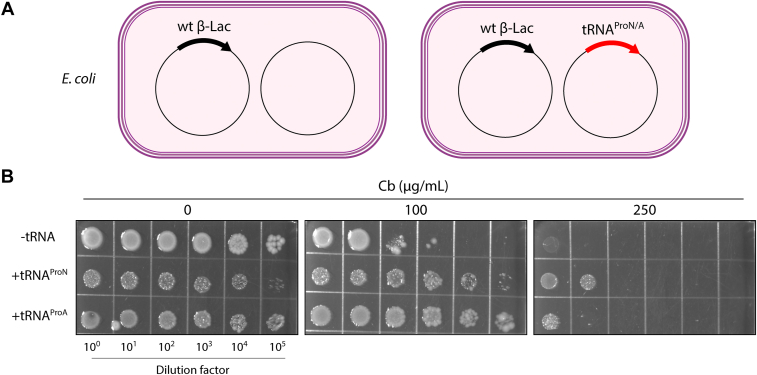
As shown above, mistranslation by tRNAProN and tRNAProA negatively affects cell growth under normal growth conditions (Fig. 6). However, mistranslation may offer an adaptive mechanism for survival under specific stress conditions, such as in the presence of antibiotics (28, 46, 47). To assess whether mistranslation by tRNAProA or tRNAProN is beneficial, we grew E. coli expressing the tRNAs with β-lactamase on LB-agar plates containing different concentrations of carbenicillin (Fig. 8A). We reasoned that, if mistranslation offers an advantage, cells expressing the mistranslating tRNA could tolerate higher antibiotic concentrations. Notably, we found that, while mistranslating cells showed a growth defect on plates without carbenicillin, they grew on plates with higher antibiotic concentrations than cells with an empty plasmid (Fig. 8B). Mistranslating cells also showed higher tolerance to ampicillin (Fig. S6). The decreased sensitivity to these β-lactam antibiotics was only observed when β-lactamase was coexpressed with the tRNAs (Fig. S6). These results provide new evidence supporting the positive consequences of mistranslation.
Figure 8.
Increased antibiotic tolerance of E. coli cells expressing tRNAProNor tRNAProA.A, E. coli MG1655 cells harboring the plasmid encoding wt β-lactamase were transformed with a plasmid containing the tRNAProN or tRNAProA gene. B, serial dilutions of overnight cultures were spotted on LB-agar plates containing 0 μg/ml, 100 μg/ml, and 250 μg/ml carbenicillin (Cb). The plates were incubated at 37 °C for 24 h.
Discussion
Here we described the identification and characterization of tRNAProN and tRNAProT, a new class of bacterial tRNAs, sparsely present throughout two genera of Actinomycetota, that emerged to mistranslate the genetic code. The discovery of tRNAProN and tRNAProT adds to the growing number of naturally occurring tRNA mistranslators reported in recent years, including several bacterial and human tRNAs (9, 48, 49, 50, 51, 52). The discovery of mistranslating tRNAs has been possible by re-examining sequenced genomes and reclassifying the annotated tRNA genes using a holistic analysis of each tRNA sequence and the predicted secondary structure. This approach allows the recognition of small nuances passed over by automated, high-throughput tRNA search algorithms. With the vast number of sequenced genomes, we predict that more examples of mistranslating tRNAs may be uncovered by carefully reanalyzing tRNA genes detected in these genomes.
The characterization of tRNAProN and tRNAProT in E. coli showed that the impact of Pro mistranslation on cell fitness depends greatly on the identity of the mistranslated codon. Thr-to-Pro mistranslation was significantly more toxic than Asn-to-Pro mistranslation. In contrast, Ala-to-Pro mistranslation was more tolerated. This difference in toxicity is remarkable because replacing residues with Pro is expected to severely affect protein structure and function (53). However, our results suggest that the substitution of Thr and Asn with Pro is more disruptive to proteins than Ala substitutions with Pro. This may be explained by the expanded catalytic and structural roles of Thr and Asn in proteins relative to the more inert function of Ala. The tRNAProX aminoacylation efficiency of E. coli ProRS may also contribute to the distinct cellular toxicities. The harmful effect of each tRNA correlates with the in vitro aminoacylation activity of E. coli ProRS. Thus, the ability of E. coli ProRS to aminoacylate tRNAProT more efficiently could result in higher levels of mistranslation relative to the other two tRNAs. tRNAProT could also influence Pro-tRNAPro synthesis by competing for ProRS aminoacylation with endogenous tRNAPro. We also considered the possible contribution of codon usage to the differences in toxicity. In E. coli, Ala codons represent 9.46% of predicted codons, whereas Thr and Asn codons comprise 5.38% and 3.92%, respectively (54). The higher Ala codon usage is inversely correlated to the severity of the observed phenotype for mistranslation of Ala codons, thus reinforcing the notion that proteins can endure Ala-to-Pro substitutions more effectively than Thr-to-Pro and Asn-to-Pro mutations. On the other hand, the mild toxicity of Asn-to-Pro relative to Thr-to-Pro may be due to the lower number of Asn codons. Different patterns of cellular toxicity have also been observed for other forms of mistranslation. Notably, E. coli was shown to withstand a variety of mistranslation forms such as Cys and Glu mistranslation at Pro and Gln codons, respectively (55). Recently, Gly-to-Ala mistranslation in human cells was shown to be less harmful than Ser misincorporation at Phe codons (52). In yeast, Ala mistranslation revealed codon-specific growth defects (56). Therefore, it is becoming clear that not all forms of mistranslation are equal and that the nature of mistranslated codons determines the biological impact of mistranslation, while additional factors may contribute to the distinct growth phenotypes. Lastly, the differences in tRNAProX-induced toxicity may provide clues about the phylogenetic distribution and evolution of the tRNAProX family. tRNAProA, the least toxic of the three tRNAProX, is present in more species relative to tRNAProT and tRNAProN, and it is always encoded with a full-length ProRSx (except for S. vinaceus). Thus, selection may act against the more toxic tRNAProT and tRNAProN, resulting in the partial deletion or inactivation of ProRSx and a more limited phylogenetic distribution.
The specific mechanism for how Pro mistranslation of Asn and Ala codons increases carbenicillin tolerance in E. coli has yet to be elucidated. Interestingly, other forms of mistranslation have also been shown to decrease antibiotic sensitivity. In E. coli, indiscriminate mistranslation with Met protects cells from chloramphenicol (46), whereas Asp-to-Asn and Glu-to-Gln mistranslation endows Mycobacteria with increased resistance to rifampicin (47). The chemical diversity of these mistranslation events suggests that the randomization of the cellular protein pool may be more important than the identity of the misincorporated amino acid to decrease antibiotic susceptibility. Testing the effect of other forms of mistranslation on antibiotic resistance could provide more clues into this possibility and ultimately establish mistranslation as a bona fide mechanism of antibiotic resistance (57).
The biological function of the tRNAProX family in their host organisms is yet to be investigated. However, the increased carbenicillin tolerance of E. coli cells expressing tRNAProN suggests that Pro mistranslation may help tRNAProX-encoding organisms endure antibiotic exposure. It may also be possible that mistranslation offers an advantage in other environmental conditions, including in the adaptation of plant pathogenicity. However, ProRSx/tRNAProX are not collocated with known plant virulence genes. Finally, despite the tRNAs' capacity to mistranslate the genetic code in E. coli, their biological function in the host species may lie outside protein synthesis. tRNAs are known to have diverse functions beyond translation (58). In some cases, a particular tRNA isoacceptor can be partitioned for nonribosomal activities (59, 60). Thus, an alternative function of tRNAProX regulated by cellular factors such as posttranslational modifications or elongation factors is possible. Further investigations will define the biological role of tRNAProX and establish the environmental conditions that promote their expression.
Experimental procedures
Identification of tRNAProA and tRNAProA-like genes
tRNAProA and tRNAProA-like genes were identified using the basic local alignment search tool (BLASTN) (61) hosted by the National Center for Biotechnology Information. The Streptomyces turgidiscabies tRNAProA sequence (STRTUCAR8_03649) was used as the query. Additional tRNAProA and tRNAProA-like genes were found using the “Compare Region Viewer” tool in the Bacterial and Viral Bioinformatics Resource Center (BR-BRC) (62, 63). The secondary structure and identity of the putative tRNAProA genes were predicted using tRNAscan-SE 2.0 (64). The manually aligned tRNA sequences are shown in Fig. S1.
Phylogenetic analysis and distribution of ProRSx and tRNAProX
A total of 121 sequences corresponding to ProRSx (45), Bacterial-type ProRS (42), and Archaeal/Eukaryotic-type ProRS (34) were aligned using Clustal Omega (65). The Bacterial- and Archaeal/Eukaryotic-type ProRS sequences are from the organisms encoding ProRSx (Table S1). The sequence alignment was used to build the phylogenetic tree using MEGAX (66) with the Maximum Likelihood method with 100 bootstraps. The tree was visualized with iTOL (67). Multilocus sequence analysis alignments were generated with protein sequences of housekeeping genes from Streptomyces scabiei 87.22 (strepdb.streptomyces.org.uk) for atpD (SCO5373), gyrB (SCO3874), recA (SCO5769), rpoB (SCO4654), and trpB (SCO2037) against a local database of 84 genomes using automlsa2 (68, 69). The 46 genomes of strains that encode ProRSx or tRNAProX were complemented with 38 additional genomes selected to represent the diversity of Streptomyces, Streptacidiphilus, and Kitasatospora (39, 40, 41, 70). The S. turgidiscabies gene sequence (STRTUCAR8_03650) was used as the query against the local sequence database with a cutoff threshold of 1e−10 to confirm that ProRSx was not present in the other genomes. The ML tree was constructed using default parameters for iqtree2.2 and RAxML embedded in automlsa2 with 1000 bootstrap iterations. The R package ggtree was used to plot the phylogeny (71) and covariates.
Bacterial strains and plasmids
E. coli Stellar cells (Takara) were used for cloning, and E. coli MG1655 was used for in vivo experiments. The HiFi DNA assembly kit (New England Biolabs) was used for molecular cloning. DNA sequencing was performed by Keck Biotechnology Resource Laboratory at Yale University to confirm plasmid sequences. The KsProRSx sequence was codon-optimized for expression in E. coli using the codon optimization tool from Integrated DNA Technologies. The corresponding DNA was purchased from Twist Bioscience. The S. turgidiscabies ProRSx (StProRSx) gene was cloned into the pCDF vector. The KsProRSx was cloned into pCDF and pEVOL vectors (72). The tRNAProA, tRNAProN, and tRNAProT genes were cloned into either pCDF or pEVOL vectors under the E. coli constitutive promoter proK. A previously developed sfGFP-mCherry fusion reporter (31) was used to investigate in vivo mistranslation by tRNAProT. The ACC (Thr) codon at position 65 in sfGFP was mutated to ACU (Thr).
Growth conditions
E. coli strains were grown in LB medium at 37 °C and constant shaking. For selection, appropriate antibiotics carbenicillin (Cb, 100 μg/ml), spectinomycin (Spe, 100 μg/ml), kanamycin (Kan, 50 μg/ml), and chloramphenicol (Chl, 35 μg/ml) were supplemented to the medium, unless otherwise indicated.
Growth assays
Chemically competent E. coli MG1655 cells were transformed with the pCDF plasmid encoding a tRNAProX gene with or without a proSx gene (K. setae or S. turgidiscabies). After transformation, cells were recovered in S.O.C. medium (Invitrogen) at 37 °C for 1 h with constant shaking. Following recovery, transformants were plated on LB-agar with respective antibiotics and grown overnight at 37 °C. Single colonies were used to inoculate 150 μl of fresh LB medium with the corresponding antibiotics in black-sided 96-well plates with clear flat bottoms (Corning). Cell growth (absorbance, A) was measured at 600 nm (A600) for 20 h of incubation at 37 °C and constant shaking using a BioTek synergy HTX plate reader.
Fluorescence-based assays
The pEVOL plasmid encoding tRNAProT and the sfGFP-mCherry reporter plasmid were transformed into E. coli MG1655 cells. Single colonies were used to inoculate LB medium containing Kan, Chl, and 0.2 mM IPTG to induce the expression of sfGFP-mCherry in a 96-well microplate. Cells were incubated for 24 h with constant shaking at 37 °C using a BioTek HTX plate reader. sfGFP (excitation 485 nm, emission 510 nm) and mCherry (excitation 587 nm, emission 610 nm) fluorescence and A600 were measured. For consistency, we chose the individual mid-exponential growth phase of E. coli for determining sfGFP-mCherry fluorescence and mistranslation frequency. This was accomplished by determining the time at which E. coli cells reached the mid-exponential growth phase using the preset function “Sigmoidal, 4P, X is concentration” in Prism 9 (GraphPad). The sfGFP and mCherry fluorescence proportion was calculated at the mid-exponential growth phase.
β-Lactamase-based assays
The mistranslation capacity of tRNAProN was tested using a previously designed β-lactamase reporter (a gift from Dr Ahmed Badran, Scripps) (31). The essential Pro residue at position 65 of β-lactamase was mutated to the Asn AAU codon. The activity of the resulting mutant was tested in E. coli S6020 cells. Cultures were prepared by inoculating LB medium containing Kan with single colonies and grown overnight at 37 °C. A volume of 99 μl of fresh LB medium with Kan and 62.5 μg/ml ampicillin was mixed with 1 μl of overnight culture. A600 was monitored for 10 h at 37 °C with constant shaking using a BioTek synergy plate reader. Wildtype (wt) β-lactamase was used as a control. To test tRNAProN, the pCDF plasmid harboring tRNAProN was cotransformed with the β-lactamase (P65N or wt) reporter plasmid into E. coli MG1655 cells. After transformation, cells were recovered for 1 h in S.O.C. medium with shaking at 37 °C. Subsequently, cells were plated on a selective LB-agar medium and incubated overnight at 37 °C. Cell cultures were obtained by inoculating 5 ml LB medium with a single colony and incubating overnight with constant shaking at 37 °C. The overnight cultures were diluted to A600 of 1, which corresponded to the 100 dilutions, and then serially diluted to 101, 102, 103, 104, and 105. A volume of 4 μl of diluted cells was spotted on LB-agar plates with varying concentrations of carbenicillin (0 μg/ml, 20 μg/ml, 30 μg/ml, 40 μg/ml, and 50 μg/ml). The plates were imaged after 18 to 20 h of incubation at 37 °C.
GFP purification and mass spectrometry analysis
The peptide sequence MSKGPGKVPGAGVPGXGVPGVGKGGGT (43) was fused to the N terminus of sfGFP containing a C-terminal 6xHis-tag in pBAD using HiFi DNA assembly. Position X indicates the Asn AAU codon. The resulting sfGFP variant was cotransformed with pCDF with or without the tRNAProN gene into E. coli MG1655. Single colonies were used to inoculate LB medium containing the antibiotics Cb and Spe. Cells were grown overnight at 37 °C. The overnight culture was used to inoculate fresh LB medium with the corresponding antibiotics, and cells were grown to an A600 of 0.4 to 0.6. sfGFP expression was induced with 0.15% arabinose for 5 h. The suspension cultures were harvested and lysed with Bugbuster (Merck Millipore) and roll-incubated at room temperature for 20 min in a buffer containing 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 10 mM Imidazole, and cOmplete EDTA-free protease inhibitor mixture tablets (Roche), and 1 μl Benzonase (Sigma-Aldrich). Lysed cells were centrifuged at 17,000g for 45 min at 4 °C. The lysate was passed through a TALON Metal Affinity Resin (Takara Bio), and the proteins were eluted with 300 mM imidazole. Subsequently, the buffer was exchanged with 50 mM Tris-HCl (pH 8.0) and 100 mM NaCl using Amicon Ultra-4 centrifugal filter units (10.000 k, Merck Millipore) following protein concentration using the same filter units.
Samples were reduced with DTT and alkylated with iodoacetamide before digestion with trypsin and LysC. LC-MS/MS analysis was performed on a Thermo Scientific Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher), equipped with a nanospray ionization source in positive ion mode with a Thermo Fisher Ultimate 3000 RSLCnano HPLC System (Thermo Fisher). Peptide mixtures were loaded into a PEPMAP100 C18 5uM trap column (Thermo Fisher) at a constant flow of 30 uL/min, holding at 60 °C. Peptides were eluted at a rate of 0.25 μl/min and separated using a PepSep Reprosil c18, 1.9 μm, 15 cm ID 75 μM analytical column (Bruker) throughout a 60-min gradient: 0 to 50 min, 4% - 32% acetonitrile + 0.1% formic acid; 50 to 55 min, 95% acetonitrile + 0.1% formic acid; and 55 to 60 min, 4% acetonitrile + 0.1% formic acid.
Mass spectrometry data were acquired using the data-dependent mode with a cycle time of 3 s. MS1 scan was performed in orbitrap within a range of 400 to 1600 m/z, at resolution 120,000 and autoinjection time with a standard AGC target. RF lens was set to 30%. Isolation for MS2 scans was performed in the quadrupole, with an isolation window of 0.7. MS2 scans were done in the linear ion trap at turbo scan rate, with an auto maximum injection time and a standard AGC target level. HCD was used for generating the MS2 spectrum as fixed normalized collision energy of 30%. MS2 spectra were acquired at a 15,000 resolution. All LC-MS/MS was performed at Bioinformatics Solutions Inc. in Waterloo, Ontario, Canada.
MS Raw Files were processed using PEAKS XPro (v10.6, Bioinformatics Solutions Inc). The data were searched against a custom database containing the sfGFP sequence in the E. coli K12 UniProt-reviewed database. Precursor ion mass error tolerance was set to 10 ppm, and fragment ion mass error tolerance was set to 0.02 Da. Semispecific cleavage with trypsin or chymotrypsin was selected with a maximum of three missed cleavages. A fixed modification of carbamidomethylation (+57.02 Da) on cysteine residues was specified. Variable modifications of deamidation (+0.98 Da) on asparagine and glutamine, as well as oxidation (+15.99 Da) on methionine, were specified. A maximum of two variable modifications per peptide was allowed. The peptide false discovery rate was set to 1% for the database search. Only confident mutations with an A-Score of 20 or higher and minimum mutation ion intensities of 1% were considered. To ensure the accuracy in identifying mutations, we applied a false discovery rate filter of <1% for peptide identification and minimum fragment ion intensity of 1% for each mutation.
Radiolabeling of oligonucleotide probes
A 50-μl reaction mix containing 5 μl of 10 μM DNA oligonucleotide probes, 2.5 μl T4-polynucleotide kinase buffer (New England Biolabs), 37.5 μl RNase-free water, 5 μl γ-32P-ATP (PerkinElmer), and 2.5 μl T4-polynucleotide kinase (New England Biolabs) was prepared and incubated for 30 min at 37 °C. Excess γ-32P-ATP was removed with Sephadex G-25 spin columns (Cytiva). Radioactivity of the oligonucleotides was measured with a scintillation counter (Tri-Carb 3110 TR, PerkinElmer).
Northern blot analysis
E. coli MG1655 cells expressing tRNAProA, tRNAProN, or tRNAProT from a pCDF plasmid were grown overnight to inoculate fresh 20 ml LB medium with appropriate antibiotics. Cells were grown to an A600 of 0.4 and harvested by centrifugation at 4000g at 4 °C. The cell pellet was resuspended in 0.5 ml extraction buffer (0.3 M sodium acetate pH 4.5, 10 mM EDTA) and lysed with 0.5 ml acidic phenol (pH 4.5). Samples were vortexed for 10 s and incubated on ice for 3 min. This step was repeated 5 times. Samples were then centrifuged for 12 min at 12,000g at 4 °C. The aqueous phase containing total RNA was collected, and the sample was re-extracted with 0.25 ml extraction buffer. RNA in the aqueous supernatant was precipitated by adding 2.5 × V absolute ice-cold ethanol and incubated at −80 °C for 1 h. Samples were centrifuged for 20 min at 17,000g and 4 °C. The supernatant was discarded, and the RNA pellet was washed with 1 ml of 70% ice-cold ethanol. Centrifugation was repeated for 12 min. The supernatant was discarded, and the RNA pellet was air dried. “Deacylated tRNA” samples were resuspended in 30 μl 200 μM Tris-HCl pH 8.0 and incubated for 30 min at 37 °C, while the “acylated tRNA” samples were resuspended in 30 μl 10 mM sodium acetate pH 4.5. RNA concentrations were measured with a spectrophotometer (NanoDrop C 2000, Thermo Scientific). Total RNA from E. coli MG1655 was isolated as described above. One microgram of total RNA (15 μg for samples containing tRNAProT) was mixed with 2× loading buffer (0.1 M sodium acetate pH 5.0, 8 M Urea, 0.05% bromophenol blue, and 0.05% xylene cyanole). The samples were separated on a 6.5% PAGE/7 M Urea gel (Outer Plate 22.3 × 20 cm [Bio-Rad], Inner Plate 20.0 × 20.0 cm [Bio-Rad], Spacer 0.5 mm [Bio-Rad]) in 0.1 M sodium acetate pH 5.0 at 150 V for 30 min and then 250 V for 7 h at 4 °C with a PROTEAN II xi electrophoresis cell (Bio-Rad). After separation, the gel region between bromophenol blue and xylene cyanole was excised and transferred to a wet Nylon membrane (GE Healthcare) in 0.5× TBE using a Trans-Blot SD cell (Bio-Rad) at constant 25 V for 32 min. The RNA was cross-linked to the membrane in a UVC 500 Crosslinker (Amersham) and then hybridized with 32P-labeled DNA probes targeting tRNAProA (5′-CAGGCTCCTCGGCAACG-3′), tRNAProN (5′-CGGCTCCTCGGCAGCAA-3′), and tRNAProT (5′-CAGCTCCTTGGCAACAC-3′) overnight at 42 °C. The membrane was exposed against a phosphor screen for 30 min for tRNAProA and tRNAProN and overnight for tRNAProT. The screen was imaged using a phosphoimager (Typhoon, Amersham).
tRNA in vitro transcription and labeling
tRNAs for in vitro aminoacylation assays were transcribed in vitro. The DNA template for in vitro transcription was generated via PCR from a plasmid containing a T7 promotor. After the PCR, DNA fragments were purified with a NucleoSpin kit (Macherey-Nagel). For in vitro transcription, a 1-ml reaction mix of 1× transcription buffer (0.1 M Tris-HCl pH 8.0, 0.1 M MgCl2, 2.3 M spermidine, 10% Triton, 10 mg/ml bovine serum albumin, 50 mM DTT), 4 mM ATP, 4 mM GTP, 4 mM CTP, 4 mM UTP, 10 μg DNA template, 20 mM MgCl2, DTT 5 mM, 10 μl pyrophosphatase (Roche), and T7 RNA polymerase was incubated at 37 °C for 16 h. Subsequently, the reaction was mixed with a 0.05% bromophenol blue, 0.05% xylene cyanole solution and 50% formamide and separated with a 12% PAGE/8 M urea gel at 120 V overnight. tRNA bands were visualized under UV light and excised from the gel. The tRNA was extracted from the gel by grinding the excised gel piece and adding 1:2 extraction buffer (0.5 M ammonium acetate and 1 mM EDTA, pH 8.0). The gel solution was incubated at 37 °C with constant shaking overnight. The gel debris was sedimented by centrifugation at 4000g for 10 min at 4 °C. The supernatant was filtered and concentrated using an Amicon Ultra (Merck) 10K filter. To precipitate the tRNA, 1 × V absolute ice-cold ethanol was added and incubated at −80 °C for 1 h. The tRNAs were precipitated by centrifugation at 13,000g for 30 min at 4 °C. The supernatant was discarded, and the tRNA pellet was washed with ice-cold 70% ethanol and centrifuged at 13,000g for 10 min at 4 °C. The supernatant was discarded, and the pellet was air dried and resuspended in 150 μl diethyl pyrocarbonate water. Subsequently, the tRNAs were labeled in a 100-μl reaction containing 50 mM glycine pH 9.0, 10 mM MgCl2, 1 μM tRNA, 0.82 μM α-32P-ATP (PerkinElmer), 50 μM sodium pyrophosphate, and 8.77 μg E. coli tRNA nucleotidyltransferase (73). The reaction was incubated for 5 min at 37 °C. A volume of 1 μl 100 μM CTP and 5 μl pyrophosphatase (Roche) were immediately added to the reaction mixture and incubated for another 2 min at 37 °C. The reaction was placed on ice, and the tRNA was extracted with 1 × V acidic phenol pH 4.6 and centrifuged at 13,000g for 2 min at 4 °C. The aqueous phase was collected and re-extracted with acidic phenol. Excess α-32P-ATP was removed with Sephadex G-25 spin columns (Cytiva). tRNA was precipitated by adding 2 × V absolute ice-cold ethanol and 0.1 × V ammonium acetate and incubated at −20 °C overnight. The tRNA solution was centrifuged at 13,000g for 45 min at 4 °C, and the supernatant was discarded. Subsequently, the tRNA pellet was washed with 300 μl 70% ice-cold ethanol and centrifuged at 13,000g for 10 min at 4 °C. The supernatant was discarded, and the 32P-tRNA was resuspended in 30 μl of RNase-free water.
In vitro aminoacylation assays
Aminoacylation of tRNAProA, tRNAProN, tRNAProT, and E. coli tRNAPro by E. coli ProRS was carried out in 50 mM Hepes pH 7.3, 8 mM ATP, 50 mM MgCl2, 0.2 mg/ml bovine serum albumin, 40 mM KCl, 40 mM β-mercaptoethanol, 1 mM proline, 7 μM tRNA, 3 μl γ-32P-tRNA, and 1 μM ProRS in a final volume of 15 μl. Reactions were incubated at 37 °C and quenched at the indicated time points by mixing 1 μl of the reaction mix with 5 μl quenching solution (200 mM sodium acetate pH 5.0 and 0.1 U/μl nuclease P1 [Sigma Aldrich]). The quenched solutions were incubated for 1 h at room temperature. A volume of 1 μl was spotted on a cellulose matrix thin layer chromatography plate (Merck), prewashed with water. The chromatography plate was run in a buffer containing 0.1 M sodium acetate pH 5.0 and 0.5% glacial acetic acid at room temperature. The plate was air dried and exposed to a phosphor screen overnight. The screen was scanned using a phosphoimager, and the AMP and Pro-AMP spots were quantified using ImageJ 1.53 (NIH).
Statistical analysis
All assays were conducted independently, and the number of biological and technical replicates is specified in the figure legends. Statistical analyses were carried out using GraphPad Prism version 9 (GraphPad), with a two-tailed t test used to assess statistical significance. A p-value of ≤0.05 was considered statistically significant. The results are presented as the mean ± standard deviation (SD). SDs are indicated as error bars.
Data availability
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE (74) partner repository with the dataset identifier PXD041335 and 10.6019/PXD041335. All the other data are contained in the article. Requests for plasmids and other materials should be sent to oscar.vargas@yale.edu.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Dr Natalie Krahn for enlightening discussions and technical support and Drs Noah Reynolds and Jeffery Tharp for critically reading the manuscript.
Author contributions
D. S. and O. V.-R. conceptualization; D. B. S., K. S. H., C. R. C., and O. V.-R. methodology; D. B. S., K. S. H., and O. V.-R. validation; D. B. S. and O. V.-R. formal analysis; D. B. S., J. T. F., J. B., S. A. G., B. A. S., A. A., and O. V.-R. investigation; M. J., D. S., C. R. C., and O. V.-R. resources; D. B. S., J. T. F., S. A. G., and O. V.-R. data curation; D. B. S. and O. V.-R. writing – original draft; D. B. S., J. T. F., B. A. S., M. J., K. S. H., E. W., D. S., C. R. C., and O. V.-R. writing – review and editing; D. B. S., B. A. S., K. S. H., E. W., and O. V.-R. visualization; D. S. and O. V.-R. supervision; D. S. and O. V.-R. project administration; D. S. and O. V.-R. funding acquisition.
Funding and additional information
J. B. was, in part, supported by an educational gift from Dr Cecil B. Pickett. This work was supported by grants from the National Science Foundation (IOS-2151063 to O. V-R.) and NIGMS, National Institutes of Health (R35GM122560-05S1 to D. S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Ronald Wek
Footnotes
Present address for Oscar Vargas-Rodriguez: Department of Molecular Biology and Biophysics, University of Connecticut Health Center, Farmington, Connecticut 06030, USA.
Contributor Information
Dieter Söll, Email: dieter.soll@yale.edu.
Oscar Vargas-Rodriguez, Email: oscar.vargas@yale.edu.
Supporting information
References
- 1.Ribas de Pouplana L., Santos M.A.S., Zhu J.-H., Farabaugh P.J., Javid B. Protein mistranslation: friend or foe? Trends Biochem. Sci. 2014;39:355–362. doi: 10.1016/j.tibs.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Kapur M., Ackerman S.L. mRNA translation gone awry: translation fidelity and neurological disease. Trends Genet. 2018;34:218–231. doi: 10.1016/j.tig.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H., Lyu Z., Fan Y., Evans C.R., Barber K.W., Banerjee K., et al. Metabolic stress promotes stop-codon readthrough and phenotypic heterogeneity. Proc. Natl. Acad. Sci. U. S. A. 2020;117:22167–22172. doi: 10.1073/pnas.2013543117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bullwinkle T.J., Reynolds N.M., Raina M., Moghal A., Matsa E., Rajkovic A., et al. Oxidation of cellular amino acid pools leads to cytotoxic mistranslation of the genetic code. Elife. 2014;3 doi: 10.7554/eLife.02501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raina M., Moghal A., Kano A., Jerums M., Schnier P.D., Luo S., et al. Reduced amino acid specificity of mammalian tyrosyl-tRNA synthetase is associated with elevated mistranslation of Tyr codons. J. Biol. Chem. 2014;289:17780–17790. doi: 10.1074/jbc.M114.564609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ling J., Söll D. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4028–4033. doi: 10.1073/pnas.1000315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brilkova M., Nigri M., Kumar H.S., Moore J., Mantovani M., Keller C., et al. Error-prone protein synthesis recapitulates early symptoms of Alzheimer disease in aging mice. Cell Rep. 2022;40 doi: 10.1016/j.celrep.2022.111433. [DOI] [PubMed] [Google Scholar]
- 8.Shcherbakov D., Nigri M., Akbergenov R., Brilkova M., Mantovani M., Petit P.I., et al. Premature aging in mice with error-prone protein synthesis. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abl9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lant J.T., Berg M.D., Heinemann I.U., Brandl C.J., O'Donoghue P. Pathways to disease from natural variations in human cytoplasmic tRNAs. J. Biol. Chem. 2019;294:5294–5308. doi: 10.1074/jbc.REV118.002982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bacher J.M., de Crécy-Lagard V., Schimmel P.R. Inhibited cell growth and protein functional changes from an editing-defective tRNA synthetase. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1697–1701. doi: 10.1073/pnas.0409064102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nangle L.A., Motta C.M., Schimmel P. Global effects of mistranslation from an editing defect in mammalian cells. Chem. Biol. 2006;13:1091–1100. doi: 10.1016/j.chembiol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Lee J.W., Beebe K., Nangle L.A., Jang J., Longo-Guess C.M., Cook S.A., et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 13.Pataskar A., Champagne J., Nagel R., Kenski J., Laos M., Michaux J., et al. Tryptophan depletion results in tryptophan-to-phenylalanine substitutants. Nature. 2022;603:721–727. doi: 10.1038/s41586-022-04499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Netzer N., Goodenbour J.M., David A., Dittmar K.A., Jones R.B., Schneider J.R., et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462:522–526. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan Y., Wu J., Ung M.H., De Lay N., Cheng C., Ling J. Protein mistranslation protects bacteria against oxidative stress. Nucl. Acids Res. 2015;43:1740–1748. doi: 10.1093/nar/gku1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuite M.F., Santos M.A.S. Codon reassignment in Candida species: an evolutionary conundrum. Biochimie. 1996;78:993–999. doi: 10.1016/s0300-9084(97)86722-3. [DOI] [PubMed] [Google Scholar]
- 17.Miranda I., Rocha R., Santos M.C., Mateus D.D., Moura G.R., Carreto L., et al. A genetic code alteration is a phenotype diversity generator in the human pathogen Candida albicans. PLoS One. 2007;2:e996. doi: 10.1371/journal.pone.0000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki T., Ueda T., Watanabe K. The ‘polysemous’ codon - a codon with multiple amino acid assignment caused by dual specificity of tRNA identity. EMBO J. 1997;16:1122–1134. doi: 10.1093/emboj/16.5.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rocha R., Pereira P.J.B., Santos M.A.S., Macedo-Ribeiro S. Unveiling the structural basis for translational ambiguity tolerance in a human fungal pathogen. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14091–14096. doi: 10.1073/pnas.1102835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones T.E., Alexander R.W., Pan T. Misacylation of specific nonmethionyl tRNAs by a bacterial methionyl-tRNA synthetase. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6933–6938. doi: 10.1073/pnas.1019033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohler K., Ibba M. Translational fidelity and mistranslation in the cellular response to stress. Nat. Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samhita L., Raval P.K., Agashe D. Global mistranslation increases cell survival under stress in Escherichia coli. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1008654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan T. Adaptive translation as a mechanism of stress response and adaptation. Annu. Rev. Genet. 2013;47:121–137. doi: 10.1146/annurev-genet-111212-133522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchner S., Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015;16:98–112. doi: 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- 25.Bacher J.M., Waas W.F., Metzgar D., de Crécy-Lagard V., Schimmel P. Genetic code ambiguity confers a selective advantage on Acinetobacter baylyi. J. Bacteriol. 2007;189:6494–6496. doi: 10.1128/JB.00622-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ling J., O'Donoghue P., Söll D. Genetic code flexibility in microorganisms: novel mechanisms and impact on physiology. Nat. Rev. Microbiol. 2015;13:707–721. doi: 10.1038/nrmicro3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman K.S., O'Donoghue P., Brandl C.J. Mistranslation: from adaptations to applications. Biochim. Biophys. Acta Gen. Subj. 2017;1861:3070–3080. doi: 10.1016/j.bbagen.2017.01.031. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz M.H., Pan T. Function and origin of mistranslation in distinct cellular contexts. Crit. Rev. Biochem. Mol. Biol. 2017;52:205–219. doi: 10.1080/10409238.2016.1274284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyu Z., Wilson C., Ling J. Translational fidelity during bacterial stresses and host interactions. Pathogens. 2023;12:383. doi: 10.3390/pathogens12030383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garofalo R., Wohlgemuth I., Pearson M., Lenz C., Urlaub H., Rodnina M.V. Broad range of missense error frequencies in cellular proteins. Nucl. Acids Res. 2019;47:2932–2945. doi: 10.1093/nar/gky1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vargas-Rodriguez O., Badran A.H., Hoffman K.S., Chen M., Crnković A., Ding Y., et al. Bacterial translation machinery for deliberate mistranslation of the genetic code. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2110797118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giegé R., Sissler M., Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucl. Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ibba M., Söll D. Quality control mechanisms during translation. Science. 1999;286:1893–1897. doi: 10.1126/science.286.5446.1893. [DOI] [PubMed] [Google Scholar]
- 34.Guan D., Grau B.L., Clark C.A., Taylor C.M., Loria R., Pettis G.S. Evidence that thaxtomin C is a pathogenicity determinant of Streptomyces ipomoeae, the causative agent of Streptomyces soil rot disease of sweet potato. Mol. Plant Microbe Interact. 2012;25:393–401. doi: 10.1094/MPMI-03-11-0073. [DOI] [PubMed] [Google Scholar]
- 35.Bouchek-Mechiche K., Gardan L., Andrivon D., Normand P. Streptomyces turgidiscabies and Streptomyces reticuliscabiei: one genomic species, two pathogenic groups. Int. J. Syst. Evol. Microbiol. 2006;56:2771–2776. doi: 10.1099/ijs.0.63161-0. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen H.P., Shelley B.A., Mowery J., Clarke C.R. Description of Streptomyces griseiscabiei sp. nov. and reassignment of Streptomyces sp. strain NRRL B-16521 to Streptomyces acidiscabies. Int. J. Syst. Evol. Microbiol. 2022;72 doi: 10.1099/ijsem.0.005574. [DOI] [PubMed] [Google Scholar]
- 37.Nguyen H.P., Weisberg A.J., Chang J.H., Clarke C.R. Streptomyces caniscabiei sp. nov., which causes potato common scab and is distributed across the world. Int. J. Syst. Evol. Microbiol. 2022;72 doi: 10.1099/ijsem.0.005225. [DOI] [PubMed] [Google Scholar]
- 38.Weisberg A.J., Kramer C.G., Kotha R.R., Luthria D.L., Chang J.H., Clarke C.R. A novel species-level group of Streptomyces exhibits variation in phytopathogenicity despite conservation of virulence loci. Mol. Plant Microbe Interact. 2021;34:39–48. doi: 10.1094/MPMI-06-20-0164-R. [DOI] [PubMed] [Google Scholar]
- 39.Labeda D.P., Dunlap C.A., Rong X., Huang Y., Doroghazi J.R., Ju K.S., et al. Phylogenetic relationships in the family Streptomycetaceae using multi-locus sequence analysis. Antonie Van Leeuwenhoek. 2017;110:563–583. doi: 10.1007/s10482-016-0824-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y., Wang M., Sun Z.Z., Xie B.B. Comparative genomic insights into the taxonomic classification, diversity, and secondary metabolic potentials of Kitasatospora, a genus closely related to Streptomyces. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.683814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malik A., Kim Y.R., Kim S.B. Genome mining of the genus Streptacidiphilus for biosynthetic and biodegradation potential. Genes (Basel) 2020;11:1166. doi: 10.3390/genes11101166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakano H., Okumura R., Goto C., Yamane T. In vitro combinatorial mutagenesis of the 65th and 222nd positions of the green fluorescent protein of Aequarea victoria. Biotechnol. Bioproc. Eng. 2002;7:311–315. [Google Scholar]
- 43.Mohler K., Aerni H.R., Gassaway B., Ling J., Ibba M., Rinehart J. MS-READ: quantitative measurement of amino acid incorporation. Biochim. Biophys. Acta Gen. Subj. 2017;1861:3081–3088. doi: 10.1016/j.bbagen.2017.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McClain W.H., Schneider J., Gabriel K. Distinctive acceptor-end structure and other determinants of Escherichia coli tRNAPro identity. Nucl. Acids Res. 1994;22:522–529. doi: 10.1093/nar/22.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H., Peterson R., Kessler J., Musier-Forsyth K. Molecular recognition of tRNAPro by Escherichia coli proline tRNA synthetase in vitro. Nucl. Acids Res. 1995;23:165–169. doi: 10.1093/nar/23.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz M.H., Waldbauer J.R., Zhang L., Pan T. Global tRNA misacylation induced by anaerobiosis and antibiotic exposure broadly increases stress resistance in Escherichia coli. Nucl. Acids Res. 2016;44:10292–10303. doi: 10.1093/nar/gkw856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Javid B., Sorrentino F., Toosky M., Zheng W., Pinkham J.T., Jain N., et al. Mycobacterial mistranslation is necessary and sufficient for rifampicin phenotypic resistance. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1132–1137. doi: 10.1073/pnas.1317580111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mukai T., Vargas-Rodriguez O., Englert M., Tripp H.J., Ivanova N.N., Rubin E.M., et al. Transfer RNAs with novel cloverleaf structures. Nucl. Acids Res. 2017;45:2776–2785. doi: 10.1093/nar/gkw898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun L., Gomes A.C., He W., Zhou H., Wang X., Pan D.W., et al. Evolutionary gain of alanine mischarging to noncognate tRNAs with a G4:U69 base pair. J. Am. Chem. Soc. 2016;138:12948–12955. doi: 10.1021/jacs.6b07121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lant J.T., Kiri R., Duennwald M.L., O'Donoghue P. Formation and persistence of polyglutamine aggregates in mistranslating cells. Nucl. Acids Res. 2021;49:11883–11899. doi: 10.1093/nar/gkab898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berg M.D., Giguere D.J., Dron J.S., Lant J.T., Genereaux J., Liao C., et al. Targeted sequencing reveals expanded genetic diversity of human transfer RNAs. RNA Biol. 2019;16:1574–1585. doi: 10.1080/15476286.2019.1646079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hasan F., Lant J.T., O'Donoghue P. Perseverance of protein homeostasis despite mistranslation of glycine codons with alanine. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2023;378 doi: 10.1098/rstb.2022.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.MacArthur M.W., Thornton J.M. Influence of proline residues on protein conformation. J. Mol. Biol. 1991;218:397–412. doi: 10.1016/0022-2836(91)90721-h. [DOI] [PubMed] [Google Scholar]
- 54.Chan P.P., Lowe T.M. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucl. Acids Res. 2016;44:D184–D189. doi: 10.1093/nar/gkv1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruan B., Palioura S., Sabina J., Marvin-Guy L., Kochhar S., Larossa R.A., et al. Quality control despite mistranslation caused by an ambiguous genetic code. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16502–16507. doi: 10.1073/pnas.0809179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cozma E., Rao M., Dusick M., Genereaux J., Rodriguez-Mias R.A., Villén J., et al. Probing the genetic code and impacts of mistranslation using tRNAAla anticodon variants. bioRxiv. 2022 doi: 10.1101/2022.11.23.517754. [preprint] [DOI] [Google Scholar]
- 57.Witzky A., Tollerson R., 2nd, Ibba M. Translational control of antibiotic resistance. Open Biol. 2019;9 doi: 10.1098/rsob.190051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katz A., Elgamal S., Rajkovic A., Ibba M. Non-canonical roles of tRNAs and tRNA mimics in bacterial cell biology. Mol. Microbiol. 2016;101:545–558. doi: 10.1111/mmi.13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giannouli S., Kyritsis A., Malissovas N., Becker H.D., Stathopoulos C. On the role of an unusual tRNAGly isoacceptor in Staphylococcus aureus. Biochimie. 2009;91:344–351. doi: 10.1016/j.biochi.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 60.Rietmeyer L., Fix-Boulier N., Le Fournis C., Iannazzo L., Kitoun C., Patin D., et al. Partition of tRNAGly isoacceptors between protein and cell-wall peptidoglycan synthesis in Staphylococcus aureus. Nucl. Acids Res. 2021;49:684–699. doi: 10.1093/nar/gkaa1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 62.Olson R.D., Assaf R., Brettin T., Conrad N., Cucinell C., Davis J.J., et al. Introducing the bacterial and viral bioinformatics resource center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucl. Acids Res. 2023;51:D678–D689. doi: 10.1093/nar/gkac1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wattam A.R., Davis J.J., Assaf R., Boisvert S., Brettin T., Bun C., et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucl. Acids Res. 2017;45:D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chan P.P., Lin B.Y., Mak A.J., Lowe T.M. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucl. Acids Res. 2021;49:9077–9096. doi: 10.1093/nar/gkab688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stecher G., Tamura K., Kumar S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 2020;37:1237–1239. doi: 10.1093/molbev/msz312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Letunic I., Bork P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucl. Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davis E.W., 2nd, Okrent R.A., Manning V.A., Trippe K.M. Unexpected distribution of the 4-formylaminooxyvinylglycine (FVG) biosynthetic pathway in Pseudomonas and beyond. PLoS One. 2021;16 doi: 10.1371/journal.pone.0247348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davis Ii E.W., Weisberg A.J., Tabima J.F., Grunwald N.J., Chang J.H. Gall-ID: tools for genotyping gall-causing phytopathogenic bacteria. PeerJ. 2016;4:e2222. doi: 10.7717/peerj.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Labeda D.P. Taxonomic evaluation of putative Streptomyces scabiei strains held in the ARS Culture Collection (NRRL) using multi-locus sequence analysis. Antonie Van Leeuwenhoek. 2016;109:349–356. doi: 10.1007/s10482-015-0637-6. [DOI] [PubMed] [Google Scholar]
- 71.Yu G., Smith D.K., Zhu H., Guan Y., Lam T.T.-Y. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Met. Ecol. Evol. 2017;8:28–36. [Google Scholar]
- 72.Young T.S., Ahmad I., Yin J.A., Schultz P.G. An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 73.Ledoux S., Uhlenbeck O.C. [3'-32P]-labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods. 2008;44:74–80. doi: 10.1016/j.ymeth.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perez-Riverol Y., Bai J., Bandla C., Garcia-Seisdedos D., Hewapathirana S., Kamatchinathan S., et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucl. Acids Res. 2022;50:D543–D552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE (74) partner repository with the dataset identifier PXD041335 and 10.6019/PXD041335. All the other data are contained in the article. Requests for plasmids and other materials should be sent to oscar.vargas@yale.edu.