Abstract
The TFIIF-like Rpc53/Rpc37 heterodimer of RNA polymerase (pol) III is involved in various stages of transcription. The C-terminal region of Rpc53 dimerizes with Rpc37 to anchor on the lobe domain of the pol III cleft. However, structural and functional features of the Rpc53 N-terminal region had not been characterized previously. Here, we conducted site-directed alanine replacement mutagenesis on the Rpc53 N-terminus, generating yeast strains that exhibited a cold-sensitive growth defect and severely compromised pol III transcriptional activity. Circular dichroism and NMR spectroscopy revealed a highly disordered 57-amino acid polypeptide in the Rpc53 N-terminus. This polypeptide is a versatile protein-binding module displaying nanomolar-level binding affinities for Rpc37 and the Tfc4 subunit of the transcription initiation factor TFIIIC. Accordingly, we denote this Rpc53 N-terminus polypeptide as the TFIIIC-binding region or CBR. Alanine replacements in the CBR significantly reduced its binding affinity for Tfc4, highlighting its functional importance to cell growth and transcription in vitro. Our study reveals the functional basis for Rpc53’s CBR in assembly of the pol III transcription initiation complex.
Keywords: transcription, RNA polymerase III, Rpc53, Rpc25, Rpc17, TFIIIC, Tfc4, microscale thermophoresis
Genetic information stored as DNA is transcribed into RNA by evolutionarily conserved multi-subunit RNA polymerases (pol) (1). Transcription in eukaryotic cells is performed by three related multi-subunit RNA polymerases, namely, pol I, II, and III. RNA pol I and pol II are responsible for rRNA and mRNA transcription, respectively, whereas pol III transcribes short untranslated RNAs such as tRNA, 5S rRNA, U6 snRNA, and 7SL RNA (2). The process of transcription can be divided into three steps starting with “initiation,” proceeding with “elongation,” and finally ending with “termination.” To initiate transcription, pol III relies on a set of transcription factors (TFs) to recognize the DNA promoter and subsequently assemble the pre-initiation complex (PIC). For example, to initiate tRNA transcription, the initiation factor TFIIIC interacts with the DNA element A and B boxes within the tRNA gene (3). TFIIIC comprises six subunits that form two individual subcomplexes called τA and τB (3). The τA subcomplex is composed of the subunits τ131 (Tfc4), τ95 (Tfc1) and τ55 (Tfc7), and it binds the A box (3). The τB subcomplex composed of τ60 (Tfc8), τ91 (Tfc6) and τ138 (Tfc3) binds to the B box (3). TFIIIC subsequently recruits TFIIIB composed of Brf1, Bdp1, and TBP to form the TFIIIB–TFIIIC–DNA complex (3, 4). Pol III is finally recruited to the pre-assembled protein-DNA complex through interactions with Brf1, Bdp1, and τ131 (Tfc4) to form the PIC (5, 6, 7, 8, 9, 10, 11, 12).
Amongst the three polymerases, pol III is the largest, consisting of 17 subunits with a total molecular weight of ∼0.7 MDa (2). Apart from a 12-subunit core, pol III has five specific subunits that form two subcomplexes Rpc82/34/31 and Rpc53/37, respectively, involved in distinct transcriptional functions. Cryo-electron microscopy (cryo-EM) studies have shown that these two pol III-specific subcomplexes are positioned adjacent to the pol III active site cleft formed by the two largest subunits, Rpc160 and Rpc128 (Fig. 1A) (13, 14, 15, 16, 17). The heterotrimeric Rpc82/34/31 complex participates in promoter opening and transcription initiation and elongation (18, 19, 20, 21), whereas the heterodimeric Rpc53/37 complex is involved in promoter opening, transcription termination, and re-initiation (6, 22, 23, 24, 25, 26). The Rpc53/37 heterodimer is also known as the TFIIF-like subcomplex as it displays regions homologous to TFIIFα (RAP74) and TFIIFβ (RAP30) subunits, and it has a conserved dimerization module localized in the lobe domain of the pol III cleft, similar to that of TFIIF in the pol II PIC (16, 17, 27, 28, 29, 30, 31, 32).
Figure 1.
Pol III PIC model and genetic analysis of the Rpc53 N-terminus.A, two orientations of a pol III PIC model consisting of pol III, TFIIIB, and DNA (PDB code: 6eu0). The 12-subunit pol III core is colored gray and the template and non-template DNA strands are dark blue. The pol III-specific subunits are colored as indicated. TFIIIB subunits: Brf1, TBP, Bdp1. The lysine residues on the pol III surface involved in cross-linking with the Rpc53 N-terminus are highlighted in red. In the right panel, lysine (K) residues that cross-link to the Rpc53 N-terminus are indicated. The structural illustration was generated by the Chimera program (52). B, schematic of Rpc53. The N-terminal 1 to 57 amino acid region, which we term the CBR (TFIIIC binding region), is colored gray. C, multiple sequence alignment of the Rpc53 N-terminal region. Alanine replacement mutations in S. cerevisiae Rpc53 are as indicated. Residues involved in cross-linking analyses are colored as in Fig. S1, A and B. Amino acids within the orange box in human Rpc4 are structurally resolved in the human pol III structure by Wang et al. (36). Species and accession numbers for amino acid sequences (Uniprot): Saccharomyces cerevisiae, P25441; Kluyveromyces lactis, A0A5P2U400; Homo sapiens, P05423; Mus musculus, Q91WD1. The alignment was generated by the ClustalW and ESPript programs (53, 54). D, cell growth of yeast strains harboring alanine replacement mutations. Image panels show serially diluted spotted cells at the indicated temperatures.
In the yeast Saccharomyces cerevisiae, Rpc53 protein is composed of 422 amino acids, with the C-terminal dimerization domains (amino acids (aa.) 283–317 and 356–421) forming the Rpc53/37 dimerization module (Fig. 1B). Although cryo-EM analyses of yeast pol III in the elongating complex or the initiation complex with TFIIIB have revealed the Rpc53 dimerization domain, the entire N-terminal peptide segment has not been resolved (Fig. 1, A and B). The lack of a resolved structure raises the question whether the N-terminal region of Rpc53 is required for pol III transcription. Studies have also shown that this region is subjected to phosphorylation and SUMOylation (Fig. S1A), with these post-translational modifications negatively regulating pol III transcriptional activity (33, 34). However, a previous mutational analysis indicated that the N-terminal region is essential for optimal cell growth, and a site-directed photo-cross-linking analysis using the photoreactive amino acid p-Benzoyl-L-Phenylalanine (BPA) revealed that the Rpc53 N-terminal region is involved in multiple intermolecular interactions (6). In particular, the peptide segment encompassing the first 58 aa. interacts with multiple proteins within the PIC, including the pol III subunits Rpc25, Rpc17, Rpc82, and Rpc34, as well as the TFIIIC subunit Tfc4 (Fig. S1, B and C). Supporting the importance of these protein interactions, deletion of aa. 11 to 40 of Rpc53 resulted in defective cell growth and reduced transcriptional activity in vitro (6). Consistent with that BPA cross-linking study, a lysine–lysine cross-linking analysis coupled with mass spectrometry on purified pol III revealed that the Rpc53 N-terminus interacts with Rpc17, Rpc25, Rpc82, ABC23 (Rpb6), Rpc128, and the Rpc53 dimerization domain (Fig. 1A, residues marked in red; Fig. S1C) (35). Thus, the Rpc53 N-terminus is localized either near the Rpc25/17 dimer or the Rpc53/37 dimerization module (Fig. 1A).
Recent cryo-EM studies on human pol III have provided structural information on the Rpc53 N-terminal region. In the human pol III structure generated by Wang et al. (36), aa. 29 to 46 of Rpc4 (an Rpc53 homolog) were resolved as localizing near the Rpc4/5 (Rpc53/37 in yeast) dimerization module (Fig. S1, A and D). In another human pol III structure by Girbig et al. (37), aa. 99 to 133 of Rpc4 were resolved as an elongated structure wrapping around the exterior of the pol III jaw (Fig. S1, A and E). In addition, aa. 134 to 157 of Rpc4 folds into an extension of the Rpc4/5 dimerization module (Fig. S1E). Consistent with the localization of the Rpc4 dimerization domain extension, a site-directed photo-cross-linking analysis revealed that the homologous Rpc53 peptide region interacts with Rpc37 (Rpc5 in human) (Fig. S1, A, B, and E) (6). However, comparing the positions of the aa. 29 to 46 peptide and the dimerization domain extension respectively in the Wang et al. and Girbig et al. structures (36, 37) (Fig. S1D versus Fig. S1E), these two different Rpc4 N-terminal regions coincidently interact with the same surface of the Rpc5 dimerization domain. Therefore, it is likely that the entire N-terminal region of human Rpc4 is highly adaptable for protein interactions to permit multiple protein interfaces on the pol III surface. We believe this specific feature is shared among all Rpc53 homologs, as our previous cross-linking studies on yeast also support multiple intermolecular interactions for its N-terminal region (Fig. S1, B and C) (6, 35).
To further characterize the functional importance of Rpc53 in yeast, we introduced point mutations into the N-terminal 57-aa. segment that was previously shown to interact with multiple pol III subunits and the Tfc4 subunit of TFIIIC (Figs. 1B and S1, B and C). We observed both cell growth and in vitro transcriptional defects associated with those Rpc53 N-terminal mutations. We further characterized pair-wise protein interactions and found that a Rpc53 N-terminal peptide binds mildly to the Rpc25/17 dimer and the Rpc53/37 dimerization module and strongly to Rpc37. Subsequent analysis revealed a strong binding affinity between the Rpc53 N-terminal peptide and the N-terminal tetratricopeptide repeats (TPR) of Tfc4. Consistent with the defective cell growth and in vitro transcription phenotypes, mutations in the Rpc53 N-terminal peptide severely compromised the Rpc53-Tfc4 interaction in a microscale thermophoresis (MST) analysis. Thus, our study characterizes the Rpc53 N-terminus as a versatile protein-binding module that interacts with multiple subunits of the pol III complex and further contributes to PIC-specific binding with the TFIIIC to support cell growth and pol III transcription initiation.
Results
Mutational analysis of the Rpc53 N-terminal region reveals amino acids essential for cell growth
In order to structurally characterize the N-terminal region of Rpc53, we purified two Rpc53 peptide fragments containing the first 57 or 122 amino acids for circular dichroism (CD) analysis (Fig. S2, A and B). The resulting CD spectra indicate that these two Rpc53 N-terminal peptides have low numbers of secondary structures, with the α-helix percentage ranging from 4.99% to 13.53% and the β-strand percentage ranging from 16.13% to 17.8%. Moreover, for the Rpc53 (1–57) peptide, 1D proton nuclear magnetic resonance (NMR) data support the lack of β-strand structures (Fig. S2C). Furthermore, a small shift in the chemical range of backbone amide protons (∼7.4–8.6 ppm) in a 2D 1H,15N-HMQC spectrum (Fig. S2D) indicates that the Rpc53 (1–57) peptide may adopt a random coil or a highly helical structure. Therefore, our combined CD and NMR data (Fig. S2, B–D) indicate that the Rpc53 (1–57) peptide adopts a random coil structure. We postulate that the Rpc53 N-terminal region lacks a globular fold, and so the structure can flexibly interact with a large surface area of the pol III complex and TFIIIC, as evidenced by previous cross-linking analyses (Figs. 1A and S1B) (6, 35).
We have previously reported that deletion of the peptide spanning Pro11 to Lys40 (ΔP11- K40) from S. cerevisiae Rpc53 leads to a cold-sensitive cell growth phenotype and decreased transcriptional activity in vitro (6). In order to identify functionally important amino acids within this peptide, we conducted a mutational analysis by replacing selected amino acids with alanine. As shown in Figure 1C, sequence alignment of Rpc53 homologs reveals that their N-terminal regions harbor a number of positively charged residues (Lys or Arg) flanked by hydrophobic amino acids, as well as high numbers of serine, glycine and proline residues. We introduced our alanine replacement mutants into yeast by plasmid shuffling and then assayed cell growth at 16 °C, 25 °C, 30 °C, and 35 °C. Yeast cells containing multiple alanine replacement mutations at Ser23-Leu28, Phe30-Pro32, and Arg37-Ser39 showed a cold-sensitive growth defect at 16 °C (Fig. 1D). Given that a “cold-sensitive” (cs) growth phenotype is often associated with defects in multi-subunit complexes (38, 39), our mutational and CD data indicate that the non-globular Rpc53 N-terminal region likely functions as an important interface for the large protein assembly in pol III transcription.
Mutations in the Rpc53 N-terminal region lead to reduced transcriptional activity
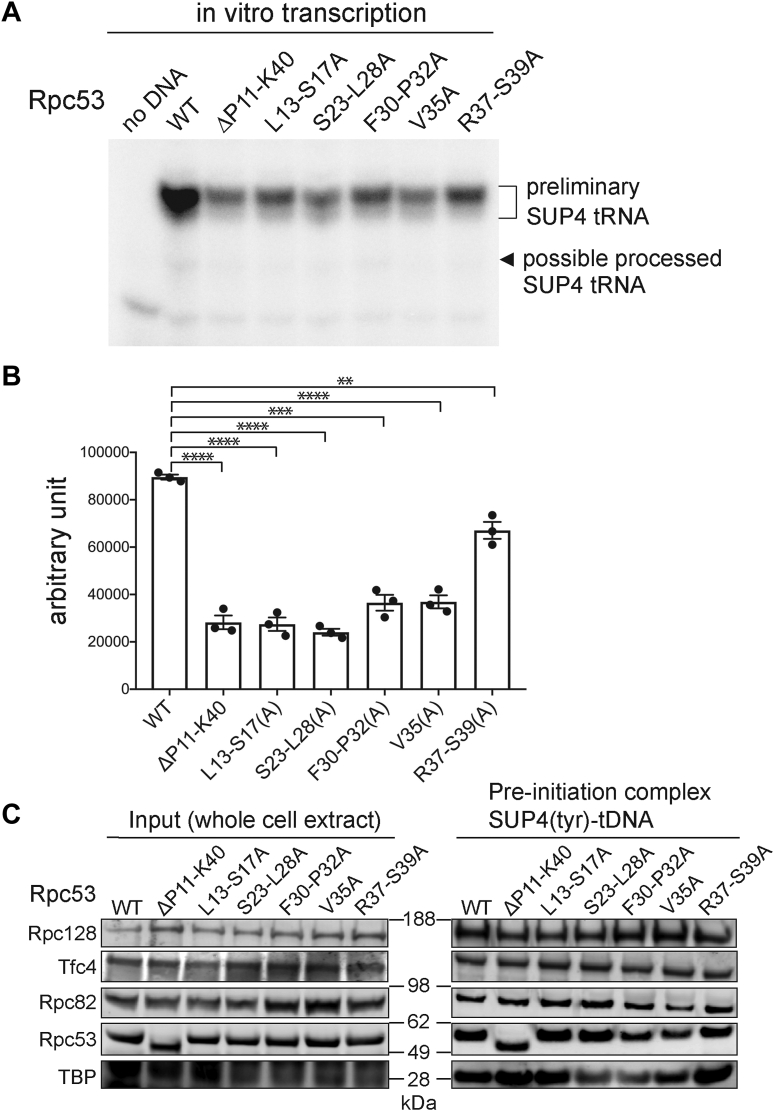
To assay pol III transcriptional activity, we carried out an in vitro transcription assay using SUP4(tyr)-tDNA template and whole cell extracts (WCE) prepared from yeast containing either wild-type (WT) or mutagenized Rpc53 characterized by the cell growth assay (Fig. 1, C and D). The radiolabeled pol III-transcribed RNA products from the in vitro assay were then separated by denaturing polyacrylamide gel electrophoresis, which revealed that the Rpc53 N-terminal mutations generally compromised pol III transcription activity (Fig. 2, A and B). We notice that the L13-S17(A) and V35A mutations, which did not cause any defects in cell growth (Fig. 1D), also compromised in vitro transcription activity. We speculate that additional cellular pathways other than the pol III transcription are involved in regulating cell growth in the assays. Nevertheless, our data show that the Rpc53 N-terminal region encompassing aa. 10 to 40 contains important residues for the proper functioning of pol III in transcription.
Figure 2.
In vitro transcription and PIC formation analyses.A, a representative autoradiograph showing SUP4 tRNA transcripts from a multi-round transcription assay using whole cell extracts (WCE) from indicated yeast strains. Mutations of Rpc53 are indicated. B, scatter plot reporting the reproducibility of independent biological replicates shown with filled circles (n = 3). Error bars represent the mean ± SD values from three independent experiments using separately prepared WCEs. Unpaired t test was used to detect significant differences compared with each sample to the WT (Wild Type) (∗∗∗∗ denotes p < 0.0001, ∗∗∗ denotes p = 0.0001, and ∗∗ denotes p = 0.003). C, western analysis of proteins in the isolated pol III PICs from the immobilized template assay. The DNA template contained the SUP4 tRNA gene. The positions of the molecular weight markers (kDA) are shown between the Western blotting panels. The gel bands corresponding to each indicated protein subunit were validated by specific antisera for individual polypeptides.
Next, we analyzed PIC formation by means of an immobilized template (IMT) assay on WCEs from our mutant strains. The IMT assay utilizes an immobilized DNA containing a pol III-transcribed gene to be incubated with the WCE for PIC formation, followed by subsequent Western analysis to reveal proteins on the DNA template. As shown in Figure 2C, protein levels of pol III subunits (Rpc128, Rpc82, and Rpc53) and Tfc4 (a subunit of TFIIIC), as well as TBP (a DNA-binding subunit of TFIIIB), on the SUP4-tDNA template were not changed significantly by Rpc53 mutations. In addition, similar results were obtained from analyses using DNA templates with Leu3-tRNA and 5S-rRNA genes (Fig. S3). Thus, our IMT analyses suggest that despite its role in supporting pol III transcription, the N-terminal region of Rpc53 does not play a major role in PIC stability.
The Rpc53 N-terminal peptide interacts with the Rpc25/Rpc17 heterodimer
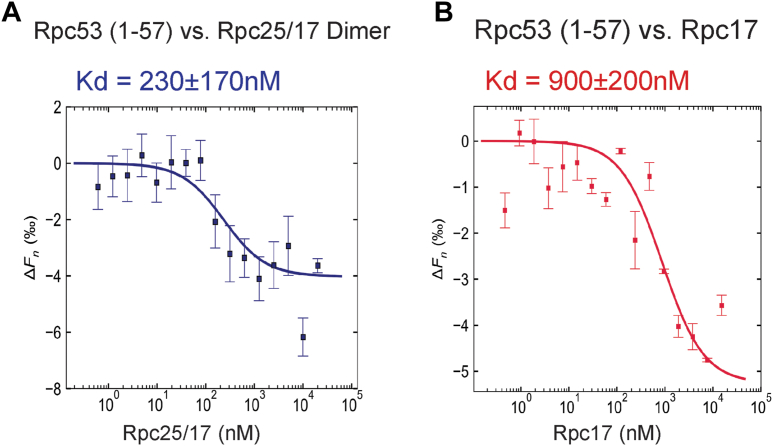
Based on our cross-linking analysis (Figs. 1A and S1, B and C) (6), we observed that the Rpc53 N-terminal region likely localizes adjacent to the Rpc25/17 heterodimer of the pol III complex. We recombinantly expressed this Rpc25/17 heterodimer and purified it to homogeneity (Fig. S4, A and B) for a binding analysis with Rpc53. To quantitate the binding affinity between the Rpc25/17 heterodimer and the Rpc53 N-ter (1–57) fragment, we carried out a pairwise interaction analysis by microscale thermophoresis (MST). The MST data showed that Rpc25/17 and Rpc53 N-ter (1–57) interact with one another with a dissociation constant (Kd) of 230 nM (Fig. 3A). We also purified the Rpc17 subunit alone (Fig. S4B) and found that it interacted with the Rpc53 N-ter (1–57) fragment with a Kd of 900 nM (Fig. 3B). Hence, our MST analysis demonstrates that the Rpc53 N-terminal region interacts mildly with the heterodimeric form of the Rpc25/17 subcomplex of pol III, an outcome consistent with our previous cross-linking study revealing an interaction between the Rpc25/17 heterodimer and the Rpc53 N-terminus (6).
Figure 3.
Determination of the dissociation constant between Rpc53 (1–57) peptide and Rpc25/17 dimer.A, MST binding curve for the interaction between Rpc53 (1–57) and Rpc25/17 dimer. For this MST experiment, the concentration of the Alexa 647-labeled Rpc53 (1–57) peptide was kept constant (5 nM), whereas the concentration of the non-labeled binding partner (Rpc25/17 dimer) varied from 20 μM to 0.00061 μM. An MST-on time of 5 s was used for this analysis, giving a Kd of 230 ± 170 nM (n = 3 independent measurements, error bars represent the standard deviation). B, MST binding curve for the interaction between Rpc53 (1–57) and Rpc17. MST assay conditions were as in (A), with the concentration of the Alexa 647-labeled Rpc53 (1–57) peptide kept constant (5 nM), whereas the concentration of the non-labeled binding partner Rpc17 varied from 15.3 μM to 0.000467 μM. An MST-on time of 5 s was used for this analysis, giving a Kd of 900 ± 200 nM.
The Rpc53 N-terminal region binds to Rpc37 with a nanomolar-level binding affinity
Recent cryo-EM structural analysis of human pol III has revealed the binding of an N-terminal peptide of Rpc4 (Rpc53 in yeast) to the region adjacent to the Rpc4/5 dimerization module (36). As illustrated in the model of the human elongation complex by Wang et al. (Fig. S1D) (36), the Rpc4 N-terminal peptide displays close contacts with the Rpc4/5 dimerization module, Rpc10 (Rpc11 in yeast), and Rpc2 (Rpc128 in yeast). Based on sequence homology (Figs. 1C and S1A), the Rpc53 N-terminus could potentially interact with the Rpc53/37 dimerization module, Rpc11, and Rpc128. To characterize pairwise interactions, we recombinantly expressed and purified the dimeric protein complex containing the Rpc53/37 dimerization module (Figs. 4A and S4C; Rpc53 (274–422)/Rpc37 (71–181)) for binding analysis with the Rpc53 N-ter (1–57) fragment. The MST analysis shown in Figure 4B indicates a binding affinity of 1.62 μM (Kd) between the Rpc53/37 dimerization module and the Rpc53 N-ter (1–57) fragment. Next, we purified another Rpc53/37 dimer comprising the dimerization module and an additional Rpc37 C-terminus (Rpc53 (274–422)/Rpc37 (71–282); Figs. 4A and S4C). The Rpc37 C-terminus contains a peptide termed the “termination loop” that regulates selection of an optimal terminator sequence by pol III (Fig. 4A). Our MST data revealed a binding affinity of 691 nM between the Rpc53 N-ter (1–57) fragment and the Rpc53/37 dimerization module with the Rpc37 C-terminal extension (Fig. 4C). Furthermore, we purified full-length Rpc37 and analyzed by MST its interaction with the Rpc53 N-ter (1–57) fragment, revealing a binding affinity of 8.2 nM (Figs. 4D and S4C). Therefore, our MST data indicate that the Rpc53 N-terminus is involved in intra-subcomplex interactions within the Rpc53/37 dimer and, notably, this region interacts with Rpc37 alone with a nanomolar-level binding affinity.
Figure 4.
Determination of the dissociation constant between Rpc53 (1–57) peptide and Rpc53/37 dimer.A, structural regions in Rpc53 and Rpc37. Structurally resolved regions in Rpc53 and Rpc37 are colored cyan and purple, respectively. Gray colored regions are unstructured. B, MST binding curve for the interaction between Rpc53 (1–57) and the Rpc53/37 dimerization module (dm). Dimerization module: Rpc53 aa. 274 to 422 and Rpc37 aa. 71 to 181. For this MST experiment, the concentration of the Alexa 647-labeled Rpc53 (1–57) peptide was kept constant (5 nM), whereas the concentration of the non-labeled binding partner (Rpc53/37 dimerization module) varied from 35.2 μM to 0.00107 μM, giving a Kd of 1.62 ± 0.65 μM. C, MST binding curve for the interaction between Rpc53 (1–57) and the Rpc53/37 dimerization module with the Rpc37 C-terminal extension. For this MST experiment, the concentration of the Alexa 647-labeled Rpc53 (1–57) peptide was kept constant (5 nM), whereas the concentration of the non-labeled binding partner (Rpc53 (274–422)/Rpc37 (71–282)) varied from 35.2 μM to 0.00107 μM, giving a Kd of 691 ± 266 nM. D, MST binding curve for the interaction between Rpc53 (1–57) and Rpc37. For this MST experiment, the concentration of the Alexa 647-labeled Rpc53 (1–57) peptide was kept constant (5 nM), whereas the concentration of the non-labeled Rpc37 binding partner varied from 20.1 μM to 0.000613 μM, giving a Kd of 8.2 ± 1.7 nM.
The Rpc53 N-terminal region binds to the N-terminal TPR of Tfc4
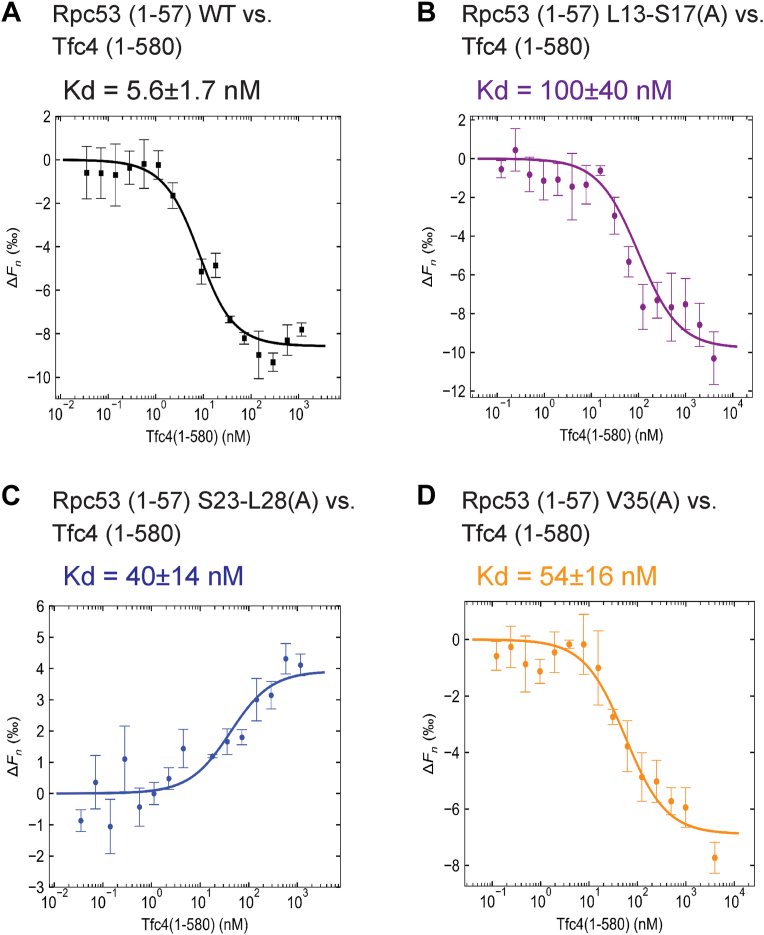
Our previous site-directed cross-linking study revealed that the Rpc53 N-terminal region interacts with the Tfc4 subunit of TFIIIC within the PIC (6). Tfc4 has also been found to interact with the Brf1 and Bdp1 subunits of TFIIIB, as well as the ABC10α subunit of pol III (8, 40, 41, 42, 43). In addition, the region of Tfc4 involved in interactions with TFIIIB and ABC10α has been mapped to its N-terminal TPR. Accordingly, we wondered if the Rp53 N-terminal region also interacts with the N-terminal TPR of Tfc4. To assay for interaction between Rpc53 and Tfc4, we recombinantly expressed and purified the Tfc4 N-terminal region (aa. 1–580, Fig. S5A), which harbors the first 10 TPR repeats (aa. 123–570) and additional flanking sequences (8). Then, we subjected the Rpc53 N-ter (1–57) and Tfc4 N-ter (1–580) peptides to MST analysis, revealing a Kd of 5.6 nM (Fig. 5A). Thus, our MST data indicate that the Rpc53 N-terminal region interacts strongly with the N-terminal region of Tfc4.
Figure 5.
Determination of the dissociation constant between Rpc53 (1–57) peptide and the Tfc4 N-terminal domain.A, MST binding curve for the interaction between wild-type (WT) Rpc53 (1–57) and Tfc4 N-terminal domain. For the MST experiment, the concentration of the Alexa 647-labeled Rpc53 (1–57) was kept constant (5 nM), whereas the concentration of the non-labeled binding partner, i.e., Tfc4 (1–580), varied from 1.16 μM to 0.000035 μM. An MST-on time of 5 s was used for this analysis, giving a Kd of 5.6 ± 1.7 nM for this interaction (n = 3 independent measurements, error bars represent the standard deviation). B–D, MST binding curves for interactions between various Rpc53 (1–57) alanine replacement mutant peptides and the Tfc4 N-terminal domain. MST assay conditions were as in (A), and respective Kd values are presented above the plots. Notice that the sigmoidal curve in C is different from the others. The shape of the sigmoidal binding curve can depend on the fluorescent dye-labeled protein or the ligand. However, Kd values are calculated irrespective of curve shape.
Mutations in the Rpc53 N-terminal region compromise its interaction with Tfc4
Our mutational analysis generated a number of Rpc53 N-terminal mutants displaying a cold-sensitive yeast growth phenotype and reduced in vitro pol III transcriptional activity. Since we found that the Rpc53 N-ter (1–57) peptide interacts strongly with Tfc4, we wondered if the Rpc53 N-terminal mutations affect binding with Tfc4. Accordingly, we purified Rpc53 N-ter (1–57) peptides (Fig. S5B) hosting alanine replacement mutations that yielded defective cell growth and in vitro transcription phenotypes. These Rpc53 N-ter mutant peptides were then used in MST binding assays to quantitate pairwise binding affinities with the Tfc4 N-ter (1–580) peptide. As demonstrated in Figure 5, B–D, Kd values for binding of three Rpc53 N-ter mutant peptides with Tfc4 increased, ranging from 40 to 100 nM. Therefore, given the Kd value of 5.6 nM between respective WT Rpc53 N-ter and Tfc4 peptides, the Rpc53 N-ter mutations appear to compromise pairwise interactions. Accordingly, we conclude that the first 57 residues of Rpc53 interact specifically with Tfc4, consistent with the observed functional importance of this region for cell growth and pol III transcriptional activity.
Discussion
The TFIIF-related Rpc53/37 heterodimer is positioned at the lobe domain of Rpc128 to regulate promoter opening, transcription termination, and re-initiation (Fig. 1A). The structure and function of Rpc37 is known in great detail, including its dimerization domain (residues 67–181) for heterodimer formation and lobe binding and its C-terminal termination/initiation loop (residues 197–224) that interacts with the TFIIE-related subunit Rpc34 and the TFIIIB subunit Bdp1 (6, 9, 13, 16, 17, 23). In contrast, other than its C-terminal dimerization domain (Fig. 1B), structural and functional information on most of the Rpc53 N-terminal region is limited. In this work, we adopted genetic, biochemical and biophysical approaches to study Rpc53’s N-terminal 1 to 57 amino acid region, which was previously shown to integrate a multi-protein network responsible for PIC formation and proper transcription (6). Based on our CD analysis, the Rpc53 N-ter (1–57) peptide is likely unstructured, lacking any globular fold (Fig. S2). We carried out an alanine replacement mutational study of Rpc53 N-ter (1–57) to generate a series of mutant yeast strains, which displayed a cold-sensitive growth defect (Fig. 1, C and D). By means of an in vitro transcription assay (Fig. 2, A and B), we observed that the alanine replacement mutations in Rpc53 resulted in significantly reduced tRNA synthesis. We further used an MST protein-binding assay to reveal that the Rpc53 N-ter (1–57) peptide demonstrates nanomolar-level binding affinities for Rpc37 and Tfc4 (Figs. 4D and 5A). Further MST analysis revealed a strong dependency on Rpc53 N-terminal amino acids for Tfc4 binding (Fig. 5, B–D), supporting a role for the Rpc53 N-terminus in assembly of the PIC. Given that Tfc4 is a subunit of the transcription factor TFIIIC, we denote this Rpc53 N-terminal region as the CBR (for the TFIIIC binding region) (Fig. 1B).
The strong interaction between the CBR and Rpc37 revealed from our MST binding assay supports the localization of a Rpc4 (the Rpc53 homolog) N-terminal peptide adjacent to the Rpc4/5 (Rpc53/37 in yeast) dimerization module in the cryo-EM structure of the human pol III complex (Fig. S1D) (36). In addition to Rpc37, our MST binding assays indicate that the CBR also interacts with the Rpc25/17 heterodimer, albeit with a moderate binding affinity in the sub-micromolar range (Fig. 3A). The Rpc25/17 binding data support previous cross-linking analyses showing that the CBR cross-links with both constitutive subunits in purified pol III and the PIC (6, 35). Cross-linking analyses have also identified other pol III subunits that are binding targets of the CBR, including Rpc128, Rpc82, Rpc34, and ABC23 (Figs. 1A and S1, B and C). Therefore, it is likely that the large flexible region of the Rpc53 N-terminus enables the CBR to interact with a large surface area of pol III and with an array of its subunits. However, it is apparent that except for its binding with Rpc37, the CBR does not interact strongly with other pol III subunits, as those interactions are not resolved in cryo-EM structures of the yeast and human pol III complexes (16, 17, 37). Furthermore, the CBR potentially functions in DNA interaction, as a similar region in the N-terminus of human Rpc4 (Rpc53) has been predicted to possess DNA-binding properties (37). However, we did not observe any pairwise interaction between the CBR and SUP4 tDNA in our MST binding analysis.
Our binding study, combined with data from previous cross-linking analyses, supports CBR-Tfc4 interaction. This interaction plays an important role in pol III transcription, as evidenced by our mutational analyses of the CBR resulting in reduced tRNA synthesis and a cold-sensitive cell growth phenotype. According to the current stepwise model of pol III PIC formation, pol III is recruited to the pre-assembled TFIIIC–TFIIIB–DNA complex mainly through interactions with the Brf1 and Bdp1 subunits of TFIIIB. Consequently, our study adds the CBR as an additional protein domain contributing to PIC formation. Our binding analysis further mapped the CBR interaction site to the N-terminal TPR of Tfc4. As previous studies also indicated that the Tfc4 N-ter TPR serves as the binding site for Brf1 and Bdp1, the CBR likely strengthens the network of protein interactions in the PIC (8, 12). However, as revealed in the in vitro PIC formation assay, mutations of CBR appear not to compromise the stability of PIC formation (Figs. 2C and S3). We speculate that Brf1 and Bdp1 likely compensate the loss of CBR interaction in the protein network of PIC. Alternatively, the CBR displaces the TFIIIB-TFIIIC interaction to prime initiation of RNA synthesis and subsequent elongation. However, it remains speculative if the CBR interacts with proteins, including subunits of pol III and Tfc4, in a stage-dependent manner to regulate the transcription cycle.
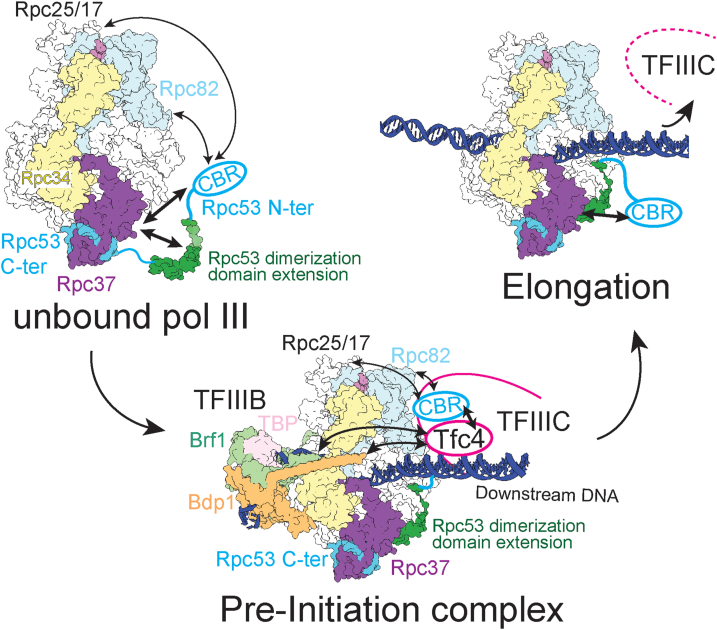
Since the CBR lies N-terminal to Rpc53’s jaw-binding domain, a model for how the CBR functions in transcription initiation can be derived from our MST binding analysis and the available cryo-EM structures of yeast and human pol IIIs (36, 37). As depicted in our model shown in Figure 6, the CBR (cyan oval), which extends from the jaw-binding domain, initially competes with the dimerization domain extension of Rpc53 (green) to interact with Rpc37 (purple). Upon PIC formation, the CBR interacts with the N-terminal TPR of Tfc4 of the TFIIIC complex. Subsequently, pol III initiates RNA synthesis to form the elongation complex, and the CBR releases its PIC-specific binding of Tfc4. In the elongation complex, the CBR again interacts with Rpc37 for a yet-to-be-defined functional role. Alternatively, the CBR displaces the interaction between Tfc4 and Tfc3 (τ138) of the τB subcomplex to promote the disassembly of TFIIIC. The interaction between the N-ter TPR of Tfc4 and the τIR domain of Tfc3 was observed based on the previous cross-linking and binding analyses by the Muller group (8), and this interaction links the τA and τB subcomplexes within TFIIIC. In that study and subsequent works (44), the Muller group also characterized interactions among Brf1, Bdp1, TBP, and Tfc4 and proposed that the TFIIIB–TFc4 interaction displaces the τA-τB link. It is likely that by binding to Tfc4, the CBR and TFIIIB function to disassemble TFIIIC to pave the way for pol III elongation. As revealed from several studies using in vivo ChIP assays (45, 46), dissociation of TFIIIC from pol III-transcribed genes represents an important step leading to productive RNA synthesis for cell growth.
Figure 6.
A model for the TFIIIC binding region (CBR) of the Rpc53 N-terminus in transcription. The CBR is shown as the cyan oval linked to the Rpc53 dimerization domain extension (green). Rpc37 and the C-terminal dimerization domain of Rpc53 are colored purple and cyan, respectively. As indicated, both the CBR and dimerization domain extension of Rpc53 interacts with the dimerization domain of Rpc37 in the pol III complex. In the pre-initiation complex (PIC), the dimerization domain extension is stably bound to Rpc37 and the CBR interacts with Tfc4. The CBR can also interact with Rpc82 and the Rpc25/17 dimer. Brf1 and Bdp1 of the transcription factor TFIIIB have been shown previously to interact with Tfc4. During elongation, the CBR is released from its interaction with Tfc4, and TFIIIC dissociates from the DNA. Thus, the CBR can again compete for binding with Rpc37.
Experimental procedures
Plasmids, yeast strains, and cell growth
For genetic analysis on S. cerevisiae Rpc53, we used the reagents generated and described in detail in our previous publication (6). In short, the gene sequence for Rpc53 with a V5 epitope tag at the C-terminus was cloned into the pRS315 (cen ars Leu2+) vector to generate the plasmid pRS315 Rpc53V5. Plasmids for pRS315 Rpc53V5_ΔP11toK40 and multiple alanine replacement mutants were prepared by introducing the respective number of alanine into the Rpc53 coding sequence of pRS315 Rpc53V5 by in vitro mutagenesis. The plasmids containing the WT and mutant Rpc53 genes were transformed into an Rpc53 strain containing pRS316 Rpc53 (Ura3+) plasmid, Rpc53::KanMx. Rpc53 WT and alanine replacement mutant yeast strains were then generated through plasmid shuffling on 5-fluoroorotic acid plates, replacing the WT pRS316 Rpc53 plasmid (Ura3+) with the mutant ones (Leu2+). For cell growth analysis, the Rpc53 WT and mutant strains were grown in yeast extract, peptone-dextrose (YPD) medium, and harvested at an optical density at 600 nm of 1.0. The cell cultures were then diluted to concentrations of 10−1, 10−3, 10−5, and 10−7. Each diluted cell culture (5 μl) was spotted onto either YPD plates or synthetic Glucose Complete plates (without leucine). Plates were incubated at temperatures of 16, 25, 30 and 37 °C. The incubation time for cell growth at 30 and 37 °C was 3 days, at 25 °C it was 5 days, and at 16 °C it was 7 days.
In vitro transcription and immobilized template assays
The in vitro transcription assay was conducted according to a previously established protocol (6). In brief, 30 μg of WCE was pre-incubated with 150 ng plasmid DNA containing the SUP4 tRNA gene in a final volume of 17 μl transcription buffer containing 200 ng α-amanitin, 4 units of RNase inhibitor (Roche), and 1 mM dithiothreitol (DTT). Transcription was started by adding 3 μl NTPs (ATP [500 μM], UTP [500 μM], CTP [500 μM], GTP [50 μM], and α-32PGTP [0.5 μl]) to reach a final volume of 20 μl. After reaction at 30 °C for 30 min, RNA products were extracted and analyzed by denaturing polyacrylamide (6%) gel electrophoresis. The SUP4 tRNA transcripts were visualized by autoradiography.
To isolate the pol III PIC via IMT assay, yeast WCEs from 1 l cell culture were incubated with the 5′-end biotin-conjugated DNA fragment containing the S. cerevisiae SUP4 tRNA, LEU3 tRNA, or 5S rRNA gene sequences, as described in detail previously (6). Briefly, the biotinylated DNA was amplified by PCR and subsequently immobilized on Streptavidin magnetic beads (Dynal) in a transcription buffer containing 20 mM HEPES (pH 7.9), 80 mM KCl, 5 mM MgCl2, 1 mM EDTA, 2% glycerol, 0.01% Tween 20, and 1 mM DTT. Each WCE (800 μg) was mixed with 2 μg of immobilized DNA in a final volume of 100 μl transcription buffer for 30 min incubation at 30 °C. The isolated PICs were washed three times with transcription buffer before undergoing Western blot analysis.
Purification of Rpc53 (1–57) peptide
The gene sequence for Rpc53 aa. 1 to 57 with a C-terminal V5 epitope was cloned between the NdeI and XhoI sites in the multiple cloning site of a pET21a vector (Stratagene). A TEV protease cleavage site was inserted by means of in vitro mutagenesis between the 57th amino acid and the V5-6xHis tag, resulting in the pET21a Rpc53 (1–57) (TEV) V5His6 plasmid. The plasmid was transformed into Escherichia coli strain BL21 (DE3) RIL (Stratagene). The protein was overexpressed upon addition of 0.4 mM isopropyl-β-D-thiogalactoside (IPTG) at 20 °C for 5 h. Cells from the culture were harvested and washed using lysis buffer (20 mM Tris-Cl (pH 7.5), 1 M NaCl, 30 mM Imidazole, 10% Glycerol). After lysis using a microfluidizer (Microfluidics), the cell extract was clarified by ultracentrifugation. The Rpc53 (1–57) V5His6 peptide was purified using Ni-Sepharose beads (GE Healthcare), and eluted in elution buffer (20 mM Tris-Cl (pH7.5), 150 mM NaCl, 250 mM Imidazole, 10% Glycerol). The eluates were subsequently dialyzed against dialysis buffer (20 mM Tris-Cl (pH 7.5), 50 mM KCl, 10% Glycerol). All buffers were supplemented with 2 mM β-mercaptoethanol and phenylmethylsulphonyl fluoride (PMSF). Further purification of the Rpc53(1–57) peptide was achieved using a Source 15S column on an AKTA Purifier (GE) with a 10-column volume linear gradient from 160 mM to 1 M KCl in a buffer containing 20 mM Tris-Cl (pH7.5) and 10% Glycerol. The Rpc53(1–57) peptide eluted at ∼550 mM KCl. The peak fractions were pooled, concentrated, and stored at −80 °C until further use.
Purification of recombinant Rpc25/17 dimer
The gene sequences for Rpc25 and Rpc17 were cloned into a pETDuetHis6SUMO vector. The Rpc25/17 co-expression plasmid was transformed into E. coli. The dimeric protein corresponding to His6SUMO-Rpc25•Rpc17-Cter-Flag was overexpressed upon the addition of 0.4 mM IPTG at 18 °C for 12 h (overnight). Cells from the culture were harvested and then lysed in lysis buffer (50 mM Tris-Cl (pH 8), 1 M NaCl, 10 mM Imidazole, 10% Glycerol). After lysis by microfluidizer (Microfluidics), the cell extract was clarified by centrifugation, and the fusion peptide was purified using Ni-Sepharose beads (GE Healthcare). The SUMO fusion Rpc25•Rpc17-Cter-Flag polypeptide was eluted in elution buffer (50 mM Tris-Cl (pH 8), 1 M NaCl, 150 mM Imidazole, 10% Glycerol), and the eluates were subsequently dialyzed against dialysis buffer (20 mM Tris-Cl (pH7.5), 50 mM KCl, 10% Glycerol). All buffers were supplemented with 2 mM β-mercaptoethanol and PMSF. The dialyzed eluate was subjected to SUMO protease digestion by adding purified yeast Ulp1 (SUMO protease) to the eluate at a concentration of 2 μg/ml for 1 h at room temperature. SUMO protease, SUMO tag, and uncleaved fusion proteins were removed by means of a second passage through Ni-Sepharose beads to aid the separation of His6SUMO from the Rpc25/Rpc17 dimer complex. Further purification was conducted via a HiTrap Heparin 5 ml column (GE) using a 10-column volume linear gradient from 100 mM to 1 M KCl, with the protein eluting at ∼500 mM KCl. The peak fractions containing Rpc25/Rpc17-Flag were pooled and further purified using a MonoQ column on an AKTA Purifier (GE) using a 10-column volume linear gradient from 100 mM to 1 M KCl, with the proteins eluting at ∼640 mM KCl. The fractions containing Rpc25/Rpc17-Flag dimer were pooled and stored at −80 °C until further use.
Purification of the recombinant Rpc53/37 dimerization module (dm) and the Rpc53/37 dm with the Rpc37 C-terminal extension
The Rpc53/37 dimerization module (dm) contains Rpc53 aa. 274 to 422 and Rpc37 aa. 71 to 181 (Fig. 4A). The gene sequence encoding Rpc53 aa. 274 to 422 (dimerization domain; dd) along with the gene sequences for Rpc37 aa. 71 to 181 (dimerization domain; dd) or Rpc37 aa. 71 to 282 (dimerization domain with the C-terminal extension; Rpc37dd+C-term) with a C-terminal V5 epitope tag were cloned between the BamHI and XhoI sites in the multiple cloning site of a pET21aHis6SUMO vector (Novagen), resulting in the plasmids pET21aHisSUMOC53dd•C37ddV5 and pET21aHisSUMOC53dd•C37dd+CtermV5, respectively. The plasmids were transformed individually into E. coli strain BL21 (DE3) RIL (Stratagene) for protein overexpression. The proteins were overexpressed upon addition of 0.4 mM IPTG at 18 °C for 12 h (overnight). Cells from the culture were harvested, lysed using lysis buffer (20 mM Tris-Cl (pH 7.5), 1 M NaCl, 30 mM Imidazole, 10% Glycerol). After lysis by microfluidizer (Microfluidics), the cell extract was clarified by centrifugation. The Rpc53dd/Rpc37ddV5 (Rpc53/37 dm) and Rpc53dd/Rpc37dd+CtermV5 dimeric proteins were separately purified using Ni-Sepharose beads (GE Healthcare) and eluted in elution buffer (20 mM Tris-Cl (pH7.5), 150 mM NaCl, 250 mM Imidazole, 10% Glycerol). The eluates were subsequently dialyzed against dialysis buffer (20 mM Tris-Cl (pH 7.5), 50 mM KCl, 10% Glycerol) to remove imidazole. All buffers were supplemented with 2 mM β-mercaptoethanol and PMSF. The dialyzed eluates were subjected to SUMO protease digestion by adding purified yeast Ulp1 (SUMO protease) to the eluate at a concentration of 2 μg/ml for 2 h at 4 °C. SUMO protease, SUMO tag, and uncleaved fusion proteins were removed by a second passage through Ni-Sepharose beads. For the Rpc53dd/Rpc37ddV5 dimer, further purification post SUMO protease digestion and SUMO tag removal was conducted by using a HiTrap Heparin column (GE) with a 10-column volume linear gradient from 200 mM to 1 M KCl, with the protein eluting at ∼450 mM KCl. The fractions containing Rpc53dd/Rpc37ddV5 dimer were pooled and stored at −80 °C until further use. The Rpc53dd/Rpc37ddV5 dimer is also referred to as the Rpc53 (274–422)/Rpc37 (71–181) dimeric protein.
For the Rpc53dd/Rpc37dd+CtermV5 dimer, further purification was conducted by using a HiTrap Heparin column (GE) with a 10-column volume linear gradient from 160 mM to 1 M KCl, with the protein eluting at ∼490 mM KCl. The peak fractions containing Rpc53dd/Rpc37dd+CtermV5 were pooled and further purified using a Source15S column using a 10-column volume linear gradient from 160 mM to 1 M KCl, with the proteins eluting at 400 mM KCl. The fractions containing Rpc53dd/Rpc37dd+CtermV5 dimer were pooled and stored at −80 °C until further use. The Rpc53dd/Rpc37dd+CtermV5 dimer is also referred to as the Rpc53 (274–422)/Rpc37 (71–282) dimeric protein.
Purification of recombinant Rpc37
The gene sequence for Rpc37 aa. 1 to 282 (full-length) with a C-terminal Flag epitope was cloned between the XbaI and XhoI sites in the multiple cloning site of a pET21a (Novagen) vector, resulting in pET21aC37Flag plasmid. The Rpc37 protein was overexpressed upon addition of 0.4 mM IPTG at 25 °C for 5 h. Cells from the culture were harvested and then lysed using lysis buffer (50 mM Tris-Cl (pH 7.5), 1 M NaCl, 20 mM Imidazole, 10% Glycerol). After lysis by microfluidizer (Microfluidics), the cell extract was clarified by centrifugation, and the Rpc37 (Full length)_Flag protein was purified using Ni-Sepharose beads (GE Healthcare). The protein was eluted in elution buffer (50 mM Tris-Cl (pH7.5), 150 mM NaCl, 250 mM Imidazole, 10% Glycerol), and the eluates were subsequently dialyzed against dialysis buffer (50 mM Tris-Cl (pH7.5), 50 mM KCl, 10% Glycerol). All buffers were supplemented with 2 mM β-mercaptoethanol and PMSF. The Rpc37_Flag protein was further purified using a Source15Q column using a 10-column volume linear gradient from 160 mM to 1 M KCl, with Rpc37_Flag eluting at ∼490 mM KCl. The fractions containing Rpc37_Flag were pooled and stored at −80 °C until further use.
Purification of recombinant Tfc4(1–580)
The gene sequence for Tfc4 aa. 1 to 580 with a C-terminal Flag epitope was cloned between the BamHI and XhoI sites in the pET21aHis6SUMO vector. The confirmed pET21aHisSUMOTfc4(1–580)Flag plasmid was transformed individually into E. coli strain BL21 (DE3) RIL (Stratagene) for protein overexpression. The protein was overexpressed upon addition of 0.4 mM IPTG at 18 °C for 12 h (overnight). Cells from the culture were harvested and then lysed using lysis buffer (50 mM Tris-Cl (pH 7.5), 500 mM NaCl, 10 mM Imidazole, 4 mM MgCl2, 5% Glycerol). After lysis by microfluidizer (Microfluidics), the cell extract was clarified by centrifugation and the Tfc4(1–580) peptide was purified using Ni-Sepharose beads (GE Healthcare). The protein was eluted in elution buffer (50 mM Tris-Cl (pH7.5), 300 mM NaCl, 250 mM Imidazole, 5% Glycerol), and the eluates were subsequently dialyzed against dialysis buffer (20 mM Tris-Cl (pH7.5), 50 mM KCl, 10% Glycerol). All buffers were supplemented with 2 mM β-mercaptoethanol and PMSF. The dialyzed eluate of His6SUMOTfc4(1–580)_Flag was subjected to SUMO protease digestion by adding purified yeast Ulp1 (SUMO protease) to the eluate at a concentration of 2 μg/ml for 2 h at 4 °C. SUMO protease, SUMO tag and uncleaved fusion proteins were removed by a second passage through Ni-Sepharose beads. The Tfc4(1–580)_Flag protein was further purified using a HiTrap Heparin 5 ml column (GE) with a 10-column volume linear gradient from 200 mM to 1 M KCl, with Tfc4(1–580)_Flag eluting at ∼400 mM KCl. Pooled fractions of Tfc4(1–580)_Flag were subjected to gel filtration-based chromatographic purification via a Superdex 200 column (GE) containing 50 mM Tris-Cl (pH7.5), 450 mM KCl, and 5% Glycerol. The fractions containing Tfc4(1–580) Flag were pooled and stored at −80 °C until further use.
Circular dichroism spectroscopy
A Chirascan-plus CD spectrometer (Applied Photophysics) was used to assess the secondary structures of the Rpc53 N-terminal peptide fragments Rpc53 (1–57) and Rpc53 (1–122). Protein samples at a concentration of 0.2 μg/μl in 5 mM phosphate (pH 7.5) buffer were loaded into the quartz cell with 1-mm path length. The samples were scanned from 280 to 190 nm at 25 °C to obtain far-UV CD spectra. The CD-derived secondary structures were predicted using the DICROWEB and K2D3 programs (47, 48).
NMR spectroscopy
The NMR sample contained ∼2.2 mM Rpc53 (1–57) in pH 5.5 buffer (90% H2O/10% D2O) containing 5 mM phosphate and 100 mM KCl, with a total volume of 300 μl in a 5 mm optical density. Shigemi tube. A trace amount (33 μM) of 4,4-dimethyl-4-silapentane-1-sulfonic acid was added to the solution as an internal chemical shift standard. All NMR data were collected at 298 K (25 °C) on a Bruker 800 MHz NMR spectrometer (AV800) equipped with a TXI cryogenic probe. The two-dimensional (2D) 1H,15N-SoFast HMQC spectrum (49) was collected using the following parameters: 1600 scans per FID, 100 increments in the 15N-dimension, 0.1 s interscan delay, and the center of the N-H proton selective pulses was set to 8.0 PPM.
Microscale thermophoresis
The Rpc53 (1–57) peptide was labeled using Alexa Fluor 647 NHS Ester (Thermo Fischer). The labeling reaction was performed by applying a concentration of 20 μM protein (molar ratio of dye/protein = 3) at room temperature for 30 min in the dark. Unreacted dye in the labeling reaction was removed by passing it through a PD Midi Trap G-25 (GE) desalting column, and the labeled protein was eluted with 500 μl of 5 mM sodium phosphate buffer. Labeling efficiency was determined as ∼0.8 according to UV/VIS spectrophotometry at 650 and 280 nm. The concentration of Alexa647-labeled Rpc53 (1–57) protein samples was adjusted to 5 nM by means of 5 mM sodium phosphate pH 8.0 supplemented with 0.05% Tween 20 (MST buffer). The respective ligands—Rpc25/17 dimer, Rpc17, Rpc53/37 dm, Rpc53/37 dm with the Rpc37 C-terminal extension, Rpc37, and Tfc4 (1–580)—were also dissolved in MST buffer. The ligand protein samples were subjected to serial dilution to prepare sample concentrations starting from μM to pM level. Each ligand dilution sample was mixed with dye-labeled Rpc53 (1–57) peptide in a 1:1 volume ratio. After 10-min incubation, the samples were loaded into Monolith NT.115 MST Premium Coated capillaries for MST measurements in a Monolith NT.115pico instrument (NanoTemper Technologies) at 25 °C. Instrument parameters were adjusted to 40% LED power and medium MST power. Data from three independently pipetted measurements were analyzed and the sigmoidal binding curves were obtained by plotting the change in normalized fluorescence (ΔFnorm in [‰]) against the concentration of the ligand. The binding curves were fitted using the PALMIST analysis software, and the binding curves were plotted using GUSSI (50, 51) to derive binding constants (Kd). Error bars = sd.
Data availability
All data are included in the article and supporting information.
Supporting information
This article contains supporting information (6, 35, 36, 37).
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Seok-Kooi Khoo, Yi-Yu-Wei, Yu-Chun Lin, and Chih-Syuan Liu for technical assistance throughout the study. We thank Dr John O'Brien for English editing. We thank Dr Shu-Chuan Jao in the Biophysics Core Facility, funded by Academia Sinica Core Facility and Innovative Instrument Project (AS-CFII108-111), for providing technical assistance and useful discussion on the MST experiments. We thank Academia Sinica High-Field NMR Center (HFNMRC) for technical support; HFNMRC is funded by Academia Sinica Core Facility and Innovative Instrument Project (AS-CFII-108-112).
Author contributions
A. C. S. and H.-T. C. conceptualization; A. C. S. and W.-J. W. methodology; A. C. S. and W.-J. W. formal analysis; A. C. S. and W.-J. W. investigation; A. C. S. writing-original draft; W.-J. W. and H.-T. C. supervision; W.-J. W. and H.-T. C. writing-review and editing.
Funding and additional information
This work was supported by the grants 105-2627-M-001-009, 106-2627-M-001-008, 107-2627-M-001-004, and 108-2311-B-001-010 from the Ministry of Science and Technology, Taiwan, R.O.C. The work was also supported in part from the Taiwan Protein Project (Grant No. AS-KPQ-109-TPP2).
Reviewed by members of the JBC Editorial Board. Edited by Craig Cameron
Supporting information
References
- 1.Werner F., Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat. Rev. Microbiol. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 2.Vannini A., Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol. Cell. 2012;45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 3.Schramm L., Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 4.Geiduschek E.P., Kassavetis G.A. The RNA polymerase III transcription apparatus. J. Mol. Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- 5.Khoo S.K., Wu C.C., Lin Y.C., Lee J.C., Chen H.T. Mapping the protein interaction network for TFIIB-related factor Brf1 in the RNA polymerase III preinitiation complex. Mol. Cell. Biol. 2014;34:551–559. doi: 10.1128/MCB.00910-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu C.C., Lin Y.C., Chen H.T. The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Mol. Cell. Biol. 2011;31:2715–2728. doi: 10.1128/MCB.05151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferri M.L., Peyroche G., Siaut M., Lefebvre O., Carles C., Conesa C., et al. A novel subunit of yeast RNA polymerase III interacts with the TFIIB-related domain of TFIIIB70. Mol. Cell. Biol. 2000;20:488–495. doi: 10.1128/mcb.20.2.488-495.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Male G., von Appen A., Glatt S., Taylor N.M., Cristovao M., Groetsch H., et al. Architecture of TFIIIC and its role in RNA polymerase III pre-initiation complex assembly. Nat. Commun. 2015;6:7387. doi: 10.1038/ncomms8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu H.L., Wu C.C., Lee J.C., Chen H.T. A region of Bdp1 necessary for transcription initiation that is located within the RNA polymerase III active site cleft. Mol. Cell. Biol. 2015;35:2831–2840. doi: 10.1128/MCB.00263-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khoo S.K., Wu C.C., Lin Y.C., Chen H.T. The TFIIE-related Rpc82 subunit of RNA polymerase III interacts with the TFIIB-related transcription factor Brf1 and the polymerase cleft for transcription initiation. Nucleic Acids Res. 2018;46:1157–1166. doi: 10.1093/nar/gkx1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrau J.C., Sentenac A., Werner M. Mutagenesis of yeast TFIIIB70 reveals C-terminal residues critical for interaction with TBP and C34. J. Mol. Biol. 1999;288:511–520. doi: 10.1006/jmbi.1999.2724. [DOI] [PubMed] [Google Scholar]
- 12.Moir R.D., Sethy-Coraci I., Puglia K., Librizzi M.D., Willis I.M. A tetratricopeptide repeat mutation in yeast transcription factor IIIC131 (TFIIIC131) facilitates recruitment of TFIIB-related factor TFIIIB70. Mol. Cell. Biol. 1997;17:7119–7125. doi: 10.1128/mcb.17.12.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann N.A., Jakobi A.J., Moreno-Morcillo M., Glatt S., Kosinski J., Hagen W.J., et al. Molecular structures of unbound and transcribing RNA polymerase III. Nature. 2015;528:231–236. doi: 10.1038/nature16143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vannini A., Ringel R., Kusser A.G., Berninghausen O., Kassavetis G.A., Cramer P. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell. 2010;143:59–70. doi: 10.1016/j.cell.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez-Tornero C., Bottcher B., Rashid U.J., Steuerwald U., Florchinger B., Devos D.P., et al. Conformational flexibility of RNA polymerase III during transcriptional elongation. EMBO J. 2010;29:3762–3772. doi: 10.1038/emboj.2010.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vorlander M.K., Khatter H., Wetzel R., Hagen W.J.H., Muller C.W. Molecular mechanism of promoter opening by RNA polymerase III. Nature. 2018;553:295–300. doi: 10.1038/nature25440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abascal-Palacios G., Ramsay E.P., Beuron F., Morris E., Vannini A. Structural basis of RNA polymerase III transcription initiation. Nature. 2018;553:301–306. doi: 10.1038/nature25441. [DOI] [PubMed] [Google Scholar]
- 18.Brun I., Sentenac A., Werner M. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 1997;16:5730–5741. doi: 10.1093/emboj/16.18.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thuillier V., Stettler S., Sentenac A., Thuriaux P., Werner M. A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J. 1995;14:351–359. doi: 10.1002/j.1460-2075.1995.tb07009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werner M., Chaussivert N., Willis I.M., Sentenac A. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J. Biol. Chem. 1993;268:20721–20724. [PubMed] [Google Scholar]
- 21.Wei Y.Y., Chen H.T. Functions of the TFIIE-related tandem winged-helix domain of Rpc34 in RNA polymerase III initiation and elongation. Mol. Cell. Biol. 2018;38 doi: 10.1128/MCB.00105-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kassavetis G.A., Prakash P., Shim E. The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening. J. Biol. Chem. 2010;285:2695–2706. doi: 10.1074/jbc.M109.074013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arimbasseri A.G., Maraia R.J. Mechanism of transcription termination by RNA polymerase III utilizes a non-template strand sequence-specific signal element. Mol. Cell. 2015;58:1124–1132. doi: 10.1016/j.molcel.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arimbasseri A.G., Maraia R.J. Distinguishing core and holoenzyme mechanisms of transcription termination by RNA polymerase III. Mol. Cell. Biol. 2013;33:1571–1581. doi: 10.1128/MCB.01733-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dieci G., Sentenac A. Facilitated recycling pathway for RNA polymerase III. Cell. 1996;84:245–252. doi: 10.1016/s0092-8674(00)80979-4. [DOI] [PubMed] [Google Scholar]
- 26.Landrieux E., Alic N., Ducrot C., Acker J., Riva M., Carles C. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J. 2006;25:118–128. doi: 10.1038/sj.emboj.7600915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carter R., Drouin G. The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol. Biol. Evol. 2010;27:1035–1043. doi: 10.1093/molbev/msp316. [DOI] [PubMed] [Google Scholar]
- 28.Chen H.T., Warfield L., Hahn S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat. Struct. Mol. Biol. 2007;14:696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schilbach S., Hantsche M., Tegunov D., Dienemann C., Wigge C., Urlaub H., et al. Structures of transcription pre-initiation complex with TFIIH and mediator. Nature. 2017;551:204–209. doi: 10.1038/nature24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murakami K., Tsai K.L., Kalisman N., Bushnell D.A., Asturias F.J., Kornberg R.D. Structure of an RNA polymerase II preinitiation complex. Proc. Natl. Acad. Sci. U. S. A. 2015;112:13543–13548. doi: 10.1073/pnas.1518255112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He Y., Yan C., Fang J., Inouye C., Tjian R., Ivanov I., et al. Near-atomic resolution visualization of human transcription promoter opening. Nature. 2016;533:359–365. doi: 10.1038/nature17970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plaschka C., Hantsche M., Dienemann C., Burzinski C., Plitzko J., Cramer P. Transcription initiation complex structures elucidate DNA opening. Nature. 2016;533:353–358. doi: 10.1038/nature17990. [DOI] [PubMed] [Google Scholar]
- 33.Lee J., Moir R.D., McIntosh K.B., Willis I.M. TOR signaling regulates ribosome and tRNA synthesis via LAMMER/Clk and GSK-3 family kinases. Mol. Cell. 2012;45:836–843. doi: 10.1016/j.molcel.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z., Wu C., Aslanian A., Yates J.R., 3rd, Hunter T. Defective RNA polymerase III is negatively regulated by the SUMO-ubiquitin-Cdc48 pathway. Elife. 2018;7 doi: 10.7554/eLife.35447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu C.C., Herzog F., Jennebach S., Lin Y.C., Pai C.Y., Aebersold R., et al. RNA polymerase III subunit architecture and implications for open promoter complex formation. Proc. Natl. Acad. Sci. U. S. A. 2012;109:19232–19237. doi: 10.1073/pnas.1211665109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q., Li S., Wan F., Xu Y., Wu Z., Cao M., et al. Structural insights into transcriptional regulation of human RNA polymerase III. Nat. Struct. Mol. Biol. 2021;28:220–227. doi: 10.1038/s41594-021-00557-x. [DOI] [PubMed] [Google Scholar]
- 37.Girbig M., Misiaszek A.D., Vorländer M.K., Lafita A., Grötsch H., Baudin F., et al. Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states. Nat. Struct. Mol. Biol. 2021;28:210–219. doi: 10.1038/s41594-020-00555-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 39.Scheraga H.A., Nemethy G., Steinberg I.Z. The contribution of hydrophobic bonds to the thermal stability of protein conformations. J. Biol. Chem. 1962;237:2506–2508. [PubMed] [Google Scholar]
- 40.Dumay H., Rubbi L., Sentenac A., Marck C. Interaction between yeast RNA polymerase III and transcription factor TFIIIC via ABC10alpha and tau131 subunits. J. Biol. Chem. 1999;274:33462–33468. doi: 10.1074/jbc.274.47.33462. [DOI] [PubMed] [Google Scholar]
- 41.Dumay-Odelot H., Acker J., Arrebola R., Sentenac A., Marck C. Multiple roles of the tau131 subunit of yeast transcription factor IIIC (TFIIIC) in TFIIIB assembly. Mol. Cell. Biol. 2002;22:298–308. doi: 10.1128/MCB.22.1.298-308.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao Y., Moir R.D., Willis I.M. Interactions of Brf1 peptides with the tetratricopeptide repeat-containing subunit of TFIIIC inhibit and promote preinitiation complex assembly. Mol. Cell. Biol. 2006;26:5946–5956. doi: 10.1128/MCB.00689-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao Y., Willis I.M., Moir R.D. The Brf1 and Bdp1 subunits of transcription factor TFIIIB bind to overlapping sites in the tetratricopeptide repeats of Tfc4. J. Biol. Chem. 2003;278:44467–44474. doi: 10.1074/jbc.M308354200. [DOI] [PubMed] [Google Scholar]
- 44.Vorländer M.K., Jungblut A., Karius K., Baudin F., Grötsch H., Kosinski J., et al. Structure of the TFIIIC subcomplex τA provides insights into RNA polymerase III pre-initiation complex formation. Nat. Commun. 2020;11:4905. doi: 10.1038/s41467-020-18707-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ciesla M., Skowronek E., Boguta M. Function of TFIIIC, RNA polymerase III initiation factor, in activation and repression of tRNA gene transcription. Nucleic Acids Res. 2018;46:9444–9455. doi: 10.1093/nar/gky656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts D.N., Wilson B., Huff J.T., Stewart A.J., Cairns B.R. Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol. Cell. 2006;22:633–644. doi: 10.1016/j.molcel.2006.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andrade M.A., Chacón P., Merelo J.J., Morán F. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 1993;6:383–390. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- 48.Whitmore L., Wallace B.A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schanda P., Brutscher B. Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J. Am. Chem. Soc. 2005;127:8014–8015. doi: 10.1021/ja051306e. [DOI] [PubMed] [Google Scholar]
- 50.Tso S.C., Chen Q., Vishnivetskiy S.A., Gurevich V.V., Iverson T.M., Brautigam C.A. Using two-site binding models to analyze microscale thermophoresis data. Anal. Biochem. 2018;540-541:64–75. doi: 10.1016/j.ab.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheuermann T.H., Padrick S.B., Gardner K.H., Brautigam C.A. On the acquisition and analysis of microscale thermophoresis data. Anal. Biochem. 2016;496:79–93. doi: 10.1016/j.ab.2015.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 53.Gouet P., Robert X., Courcelle E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the article and supporting information.