Abstract
Mitochondrial DNA (mtDNA) encodes proteins and RNAs that are essential for mitochondrial function and cellular homeostasis, and participates in important processes of cellular bioenergetics and metabolism. Alterations in mtDNA are associated with various diseases, especially cancers, and are considered as biomarkers for some types of tumors. Moreover, mtDNA alterations have been found to affect the proliferation, progression and metastasis of cancer cells, as well as their interactions with the immune system and the tumor microenvironment (TME). The important role of mtDNA in cancer development makes it a significant target for cancer treatment. In recent years, many novel therapeutic methods targeting mtDNA have emerged. In this study, we first discussed how cancerogenesis is triggered by mtDNA mutations, including alterations in gene copy number, aberrant gene expression and epigenetic modifications. Then, we described in detail the mechanisms underlying the interactions between mtDNA and the extramitochondrial environment, which are crucial for understanding the efficacy and safety of mtDNA-targeted therapy. Next, we provided a comprehensive overview of the recent progress in cancer therapy strategies that target mtDNA. We classified them into two categories based on their mechanisms of action: indirect and direct targeting strategies. Indirect targeting strategies aimed to induce mtDNA damage and dysfunction by modulating pathways that are involved in mtDNA stability and integrity, while direct targeting strategies utilized molecules that can selectively bind to or cleave mtDNA to achieve the therapeutic efficacy. This study highlights the importance of mtDNA-targeted therapy in cancer treatment, and will provide insights for future research and development of targeted drugs and therapeutic strategies.
Keywords: Mitochondria, mtDNA, Cancer, Cancer therapy
Graphical abstract
1. Introduction
Mitochondria play critical organelles roles in various aspects of biological processes such as energy generation, immune responses, programmed cell death and metabolism of various bioactive molecules including calcium, iron-sulfur cluster, one-carbon units, nucleotide, amino acid and lipid metabolism (Huertas et al., 2017; Li et al., 2018; Lu et al., 2018; Spinelli & Haigis, 2018; Zhang et al., 2020). Various factors such as free radical production, mtDNA mutation, etc., can cause mitochondrial dysfunction. Mitochondria with abnormal function has also been implicated in the development of various diseases, and have long been proposed as driving forces for malignant transformation (Bennett et al., 2022; Iske et al., 2020; Kim et al., 2017; Ma et al., 2018). Recent research highlighted that mitochondria can be regard as an important target for cancer therapy.
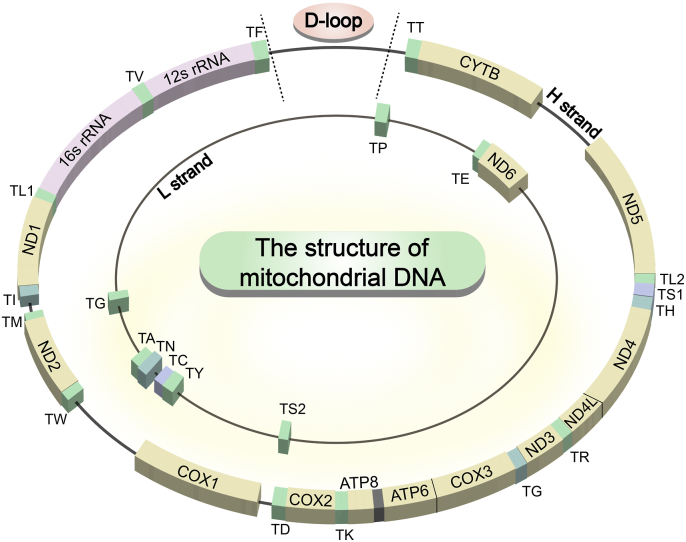
Mitochondrial DNA is a fundamental indicator of mitochondrial function. It encodes 37 coding genes, including 13 essential subunits of oxidative phosphorylation (OXPHOS) complex proteins and RNAs (22 tRNAs and 2 rRNAs) required for protein synthesis (Fig. 1). (Guerra et al., 2017) Mitochondrial DNA plays an irreplaceable role in the maintenance of metabolic activity and cellular function. Mitochondrial DNA mutations and copy number alterations have been found to be closely related to the acceleration of tumor development (Hu et al., 2016; Smith et al., 2022b). For example, Tid1 knockdown in gastric cancer cells leads to a decrease in mtDNA copy number, which regulates cell migration and invasion (Wang et al., 2020). Reduced mtDNA content has been found to be associated with the induction of a progressive phenotype of breast cancer and the promotion of tumor formation (Aminuddin et al., 2020; Singh et al., 2009). In addition, a 4977-bp deletion from 8470 to 13,459 bp in mtDNA has been proved as a major inducement in the carcinogenesis of hepatocellular carcinoma and colorectal cancer (Aminuddin et al., 2020). Mutation and microsatellite instability in the non-coding D-loop region of mtDNA would contribute to laryngeal cancer (Wang et al., 2021). These observations highlight the significant association between mtDNA and cancer progression.
Fig. 1.
The detailed structure of mtDNA. The whole mtDNA sequence included a total of 37 coding genes and a D-loop region. And the coding genes including 13 essential proteins (ND1, ND2, ND3, ND4, ND4L, ND5, ND6, CYTB, COX1, COX2, COX3, ATP6 and ATP8), 22 tRNAs (TA, TR, TN, TD, TC, TE, TQ, TG, TH, TI, TL1, TL2, TK, TM, TF, TP, TS1, TS2, TT, TW, TY and TV) and 2 rRNAs (12s rRNA and 16s rRNA).
Compared to the nuclear genome (nDNA), the lack of histone protection, the void of introns and a weak DNA repair ability, together makes mtDNA more vulnerable to damages, thereby leading to single-strand breaks, abasic sites, oxidative base damage and DNA crosslinking (Hou et al., 2018; Pustylnikov et al., 2018). Several endogenous factors such as oxidative stress (Nadalutti et al., 2022), enzyme-induced damage (Geden et al., 2021), and the absence of repair mechanisms (Boguszewska et al., 2020), as well as some exogenous factors such as industrial byproducts including alkylating agents, aldehydes and polycyclic aromatic hydrocarbons, therapeutic drugs including nucleoside analogues, and radiation (Cline, 2012), may introduce mutations in mtDNA. On the other hand, changes in key genes that are associated with the stability of the mtDNA genome are the main endogenous factors that cause mtDNA mutation. It has been demonstrated that numerous genes participate in the generation of mtDNA mutations, such as SLC25A4 (Finsterer & Zarrouk-Mahjoub, 2018), FBXL4 (Alsina et al., 2020), thymidine kinase 2 (Laine-Menéndez et al., 2021) and so on. P53 is an important regulator of normal mitochondrial and cellular physiology, and its aberrant expression affects mitochondrial function, mitochondrial biogenesis, mtDNA abundance and tumor suppressor activity (Bergeaud et al., 2013; Koczor et al., 2012; Yadav et al., 2015). It has been reported that P53 could bind to glycogen synthase kinase 3β, mitochondrial transcription factor A (TFAM) and mtDNA regulatory D-loop region and participate in stimulating mtDNA repair to maintain mtDNA content in HepG2 cells (Vadrot et al., 2012). Loss of P53 can lead to increased susceptibility of mtDNA to mutations (Wu et al., 2018). Furthermore, ANKLE1 is an endonuclease involved in response to DNA damage stimulus, and its ectopic expression could cause mtDNA instability and cleavage in breast epithelial-derived cells (Przanowski et al., 2023). These risk factors may result in the accumulation of mtDNA mutations and significantly increase the likelihood of tumor progression.
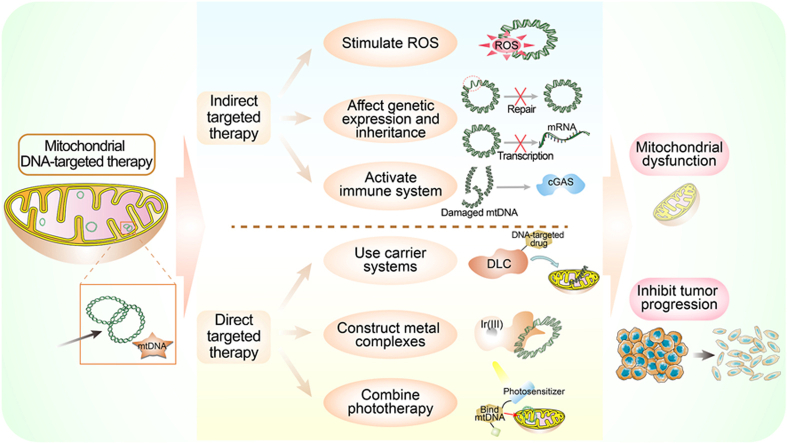
Considering its important role in cancer development, mtDNA-targeted therapies have emerged as promising strategies in cancer treatment recently. The application of immunotherapy by inducing mtDNA release, targeted therapy by constructing mitochondria-targeted complexes and combination of chemotherapy and photodynamic therapy (PDT) have greatly promoted the progress of anticancer targeting mtDNA. Here, we reviewed the mtDNA-targeted cancer treatment strategies and summarized the significant progress. Initially, we provided an overview of the association between mtDNA mutations and tumorigenesis. Then we elucidated the vital messenger role of mtDNA in biological processes, which is conducive to a deeper understanding of the mechanisms, efficacy, physiological response and prognosis of targeted therapeutic strategies. Finally, we screened relevant studies in recent years based on the criterion of causing mtDNA damage and leading to cancer cell death. Based on the pathways of mtDNA damage caused by the therapeutic strategies mentioned, we classified the included studies into two categories, namely the indirect targeted therapy and direct targeted therapy. Meanwhile, we summarized the main success of these studies in two tables (Table 1, Table 2). We hope this study will provide useful insights for researchers interested in mtDNA-targeted treatment.
Table 1.
Summary of indirect targeting therapies in recent years.
| Strategies for targeted therapy | Drugs, compounds or key genes used | The core of therapeutic strategy (Characteristics of mtDNA targeting) | Cancer | year | |
|---|---|---|---|---|---|
| Wang et al. (Wang et al., 2023) | Influence genetic functions of mtDNA, increasing ROS | TFAM and gemcitabine | Inhibition of TFAM results in mitochondrial dysfunction, amplifies gemcitabin-induced oxidative stress, and augments cytotoxicity. | Pancreatic cancer | 2023 |
| Yin et al. (Yin et al., 2023) | Activate immunity | Raddeanin A | It could bind to TDP-43, induce mtDNA leakage, and ultimately enhance DC-mediated antigen cross-presentation and T cell activation. | Melanoma and colon cancer | 2023 |
| Xiao et al. (Xiao et al., 2022) | Activate immunity | PLGA@Icaritin NPs | Resulted in overproduction of oxidative-mitochondrial DNA, which activates the release of DAMPs | Gastric cancer | 2022 |
| Li et al. (Li et al., 2022) | Activate immunity | (BPA + CPI)@PLGA NPs | The release of mtDNA caused by mitochondrial metabolism disorder further activated the cGAS/STING signal pathway | Liver cancer | 2022 |
| Witkowska et al. (Witkowska et al., 2022) | Activate immunity | Ethyl 3-((tert-butoxycarbonyl)amino)-2-hydroxy-9H-carbazole-1-carboxylate; 3-((tert-butoxycarbonyl) amino)-2-hydroxy-9H-carbazole-1-carboxylic acid | It induced the release of mtDNA into the cytosol by increasing the permeabilization of the mitochondrial IM, which could lead to cell death-associated inflammation | Colon cancer and osteosarcoma | 2022 |
| Zhao et al. (Zhao et al., 2022) | Activate immunity | A herpesvirus-mimicking nanoparticle (named Vir-ZM@TD) | Vir-ZM@TD evaded rapid clearance in the blood circulation and mimicked the serial infection processes of herpesvirus, including TFAM deficiency-triggered mtDNA stress, as well as the release of Mn2+ from organelles into the cytosol, priming cGAS-STING pathway-mediated innate immunity. | Breast carcinoma | 2022 |
| Yan et al. (Yan et al., 2022) | Activate immunity | RocA | RocA promoted NK cell infiltration by activating cGAS-STING signaling via targeting mtDNA. | NSCLC | 2022 |
| Benedetti et al. (Benedetti et al., 2022) | Increase ROS | Acyclovir | The continuous generation of ROS caused dose-dependent damage to mtDNA | NSCLC | 2022 |
| Somuncu et al. (Somuncu et al., 2022) | Increase ROS, influence genetic functions of mtDNA | 3,3'-[(1,1′-Biphenyl)-4′,4′-diyl)bis (azo)]bis [4-amino-1-naphthalenesulfonic acid](CR) | CR was a high-affinity binder to the Pol γ protein, causing mitochondrial dysfunction by inhibiting Pol γ activity and oxidative mtDNA damage repair. | Colonic carcinoma | 2022 |
| Wang et al. (Wang et al., 2022a) | Influence genetic functions of mtDNA | CircRNAs | miR-1182/TFAM axis was inhibited, resulting in transcriptional repression of ND1 and ATP6 | NSCLC | 2022 |
| Kong et al. (Kong et al., 2022) | Influence genetic functions of mtDNA | The combination of epoxomicin and cisplatin | Inhibition of TFAM and POLRMT function, affecting mitochondrial genome transcription | ovarian carcinoma | 2022 |
| Guo et al. (Guo et al., 2022) | Influence genetic functions of mtDNA, increase ROS | VB12- sericin-PBLG-IR780 | VB12- sericin-PBLG-IR780 could significantly inhibit the expression of ATP synthase and lead to ROS generation. | Gastric cancer | 2022 |
| Hu et al. (Hu et al., 2021) | Activate immunity | ATM protein inhibition | ATM inhibition potently activated the cGAS/STING pathway and enhanced lymphocyte infiltration into the TME by TFAM, leading to mtDNA leakage into the cytoplasm. | Breast carcinoma and melanoma | 2021 |
| Jiang et al. (Jiang, Guo, et al., 2021) | Activate immunity, metal complexes | MSN-Ru2+/Fe2+ | MSN-Ru2+/Fe2+ could enter mitochondria to bind with mtDNA due to the lipophilic and DNA affinity of Ru2+ complex. Oxidative mtDNA is able to escape from the tumor cells and results in the reactivated immunoresponse of macrophages against cancer cells. | Pancreatic cancer | 2021 |
| Panchangam et al. (Panchangam et al., 2021) | Increase ROS | Novel C–N-cyclometalated 2H-indazole-Ir(III) complex | Increased ROS damage mtDNA. And mtDNA may also be damaged as a target during metal delivery. | Triple negative mammary gland cancer | 2021 |
| Koshikawa et al. (Koshikawa et al., 2021) | Increase ROS, carrier system | A five-ring PIP-TPP | It localized in the mitochondria in HeLamtA3243G cells and induced mitochondrial ROS production, mitophagy and apoptosis in a mutation-specific fashion | Cervical carcinoma | 2021 |
| Li et al. (Li, Zhang, et al., 2021) | Influence genetic functions of mtDNA, Activate immunity | Zalcitabine | Zalcitabine-induced TFAM degradation could trigger oxidative DNA damage, mtDNA release to the cytosol, and subsequent activation of the CGAS-STING1 pathway | Pancreatic cancer | 2021 |
| Cheng et al. (Cheng et al., 2020) | Activate immunity | Overexpression of Lon | Upregulation of Lon induced the secretion of extracellular vehicles, which carry mtDNA and PD-L1. | Oral squamous cell carcinoma | 2020 |
| Pandey et al. (Pandey & Verma, 2020) | Influence genetic functions of mtDNA | Violacein and silver nanoparticles | Dyad drug system could structurally bind and inhibit TFAM at the interface of TFAM-DNA complex during replication and thus can hinder majority of pathways contributing to cancer proliferation. | Pan-carcinoma | 2020 |
| Bonekamp et al. (Bonekamp et al., 2020) | Influence genetic functions of mtDNA | IMTs | IMT1 and IMT1B significantly reduced the levels of mtDNA transcriptions and respiratory chain subunits in tumor cells by inhibiting POLRMT. | NSCLC, cervical carcinoma and ovarian carcinoma | 2020 |
| Inamura et al. (Inamura et al., 2019) | Increase ROS | Gemcitabine | Gemcitabine depleted the cellular pool of deoxyribonucleotides and inhibits the synthesis of mtDNA,resulting in the acceleration of ROS generation in mitochondria. | Insulinoma | 2019 |
| Fan et al. (Fan et al., 2017) | Influence genetic functions of mtDNA | miR-199a-3p | The up-regulation of miR-199a-3p expression could inhibit mitochondria by inhibiting TFAM transcription | Breast carcinoma | 2017 |
Table 2.
Summary of direct targeting strategies in recent years.
| Strategies for targeted therapy | Drugs or compounds used | The core of therapeutic strategy (Characteristics of mtDNA targeting) | Cancer | year | |
|---|---|---|---|---|---|
| Mukerabigwi et al. (Mukerabigwi et al., 2023) | Carrier system | TF@CNM + DOX | Design tumor and mitochondrial dual targeting multiprodrug to improve intracellular ros level and solve multi-drug resistance in cancers. | Breast carcinoma | 2023 |
| Yao et al. (Yao et al., 2023) | Carrier system | PIP-TPP (CCC-h1005) | CCC-h1005 can be used to treat many cervical cancers harboring high copies of the target variant | Cervical carcinoma | 2023 |
| Faria et al. (Faria et al., 2022) | Carrier system | PEI–DQA/TAT/pDNA, MTS-CPP/pDNA | Polymer and peptide delivery systems increased mitochondrial localization of targeted therapy. | Cervical carcinoma and lung cancer | 2022 |
| Luo et al. (Luo et al., 2022) | Carrier system | NP(Pt)@AL | The activity of thioredoxin reductase 1 inhibited by AL and the adducts of Pt (II) with mtDNA can costimulate ROS and reactivate the mitochondrial pathway of apoptosis. | Ovarian carcinoma | 2022 |
| Tsuji et al. (Tsuji et al., 2022) | Carrier system | novel PIP-TPP (CCC-021-TPP) | CCC-021-TPP caused cell senescence, accompanied by significant induction of anti-apoptotic BCL-XL | NSCLC | 2022 |
| Mondal et al. (Mondal et al., 2022) | Direct bonding | Ym155 | Ym155 binds mtDNA leading to mitochondrial dysfunction, including a decrease in OXPHOS and TCA cycle intermediates, and an increase in mitochondrial permeability. | Lung cancer | 2022 |
| Wang et al. (Wang et al., 2022c) | Metal complex | Zn (II)–cryptolepine–cyclen | It showed efficient plasmid DNA intercalation, and has a high binding affinity to mtDNA to cleave DNA, further causing mitochondrial damage, and can be used for cisplatin resistance | Lung cancer | 2022 |
| Echevarría et al. (Echevarría et al., 2022) | Metal complex, PDT | A family of Ir (III) complexes | Complexes [1a] Cl and [3a] Cl were able to cause severe cleavage on mtDNA, resulting in the inhibition of the expression of mitochondrial genes. | Prostate cancer and melanoma | 2022 |
| Bajpai et al. (Bajpai et al., 2022b) | Metal complex | Cholesterol-based chimeric nanoparticles consisting of cisplatin, camptothecin, and tigecycline | Particles localized efficiently into the mitochondria of cancer cells within 6 h, simultaneously impairing mtDNA, mt-Top1, and mitochondrial ribosomes. | Breast carcinoma, cervical carcinoma and lung cancer | 2022 |
| Jiang et al. (Jiang, Guo, et al., 2021) | Activate immunity, metal complexes | MSN-Ru2+/Fe2+ | MSN-Ru2+/Fe2+ could enter mitochondria to bind with mtDNA due to the lipophilic and DNA affinity of Ru2+ complex. Oxidative mtDNA is able to escape from the tumor cells and results in the reactivated immunoresponse of macrophages against cancer cells. | Pancreatic cancer | 2021 |
| Muhammad et al. (Muhammad et al., 2021) | Carrier system | c,c,t -[Pt-(NH3)2Cl2 (TPP) (Dox)] (PPD) | Enhanced mitochondrial localization and overcoming cisplatin resistance | Breast carcinoma | 2021 |
| Koshikawa et al. (Koshikawa et al., 2021) | Increase ROS, carrier system | A five-ring PIP-TPP | It localized in the mitochondria in HeLamtA3243G cells and induced mitochondrial ROS production, mitophagy and apoptosis in a mutation-specific fashion | Cervical carcinoma | 2021 |
| Zhang et al. (Zhang et al., 2021) | PTT, PDT | IR780@Pt NPs | Massive ROS generation and photothermal effects under 808 nm laser irradiation resulted in MMP loss, significantly reduced cellular ATP production, decreased cellular GSH levels, mtDNA damage, and mitochondrial dysfunction. | Osteosarcoma and cervical carcinoma | 2021 |
| Nair et al. (Nair et al., 2020) | Carrier system | A folic acid anchored p-sulfo-calix [4] arene capped hollow gold nanoparticles was meticulously loaded with Dox and Mt-Dox. | Overcomed off-target effects and eradicated both nDNA and mtDNA | Cervical carcinoma and lung cancer | 2020 |
| Chen et al. (Chen et al., 2020) | Carrier system | Choil-TPP | It could inhibit the transcription of mtDNA and damage mtDNA. | pancreatic cancer | 2020 |
| Yang et al. (Yang et al., 2020) | Carrier system, PTT | Pt (IV)-NPs contain IR780 | Pt (IV)-NPs could markedly facilitate cancer-specific mitochondrial targeting, inducing mitochondrial dysfunction and mtDNA damage, thus greatly increasing the Pt accumulation in cisplatin resistant cancer cells. | Lung cancer | 2020 |
| Liu et al. (Liu et al., 2020) | Direct bonding | Pentamidine | Pentamidine targets AT sequences in mtDNA, resulting in decreased transcription levels of mitochondrial coding genes, decreased mtDNA, and changes in mitochondrial morphology and function | Prostate cancer | 2020 |
| Qin et al. (Qin et al., 2020) | Metal complex | Cyclometalated iridium (III) complexes | Ir complexes induced an increase in intracellular ROS levels, a reduction in ATP production, mtDNA damage, an increase in lipid eroxidation levels, and proteasomal activity inhibition | Lung cancer | 2020 |
| Li et al. (Li, Wu, et al., 2020) | Metal complex, PDT | Dinuclear Ir(III)-containing luminescent metallohelices | It had stronger mtDNA binding affinity and better PDT effect. And mtDNA were cleaved by the generated intracellular 1O2. | Breast carcinoma and lung cancer | 2020 |
| Cao et al. (Cao et al., 2019) | Metal complex | Ir3: [Ir (dfppy)2 (dppz)](PF6); Ir4: [Ir (ptz)2 (dppz)](PF6) | Complexes Ir3 and Ir4 bound to mtDNA, intercalated to mtDNA in situ and induced mtDNA damage, resulting in decline of mitochondrial membrane potential, disability of adenosine triphosphate generation, disruption of mitochondrial energetic and metabolic status | Lung cancer | 2019 |
| Chen et al. (Chen et al., 2018) | Carrier system | peptide nucleic acids coupled with TPP | It targeted the D-loop regulatory region of mtDNA.TPP is a DLC for mitochondrial targeting, and PNA can bond to DNA and RNA targets efficiently | Lung cancer | 2018 |
| Wang et al. (Wang et al., 2018) | Carrier system, PTT | Nanoparticles: a core–shell –SS–shell architecture are composed of a core of Fe3O4 colloidal nanocrystal clusters, an inner shell of PDA functionalized with TPP, and an outer shell of methoxy poly (ethylene glycol) linked to the PDA by disulfide bonds. | The magnetic core can increase the accumulation of nanoparticles at the tumor site. A photothermal effect is generated from the PDA photosensitizer using NIR, leading to a dramatic decrease in mitochondrial membrane potential. Simultaneously, the loaded Dox can enter the mitochondria and subsequently damage the mtDNA. | Melanoma | 2018 |
| Hu et al. (Hu et al., 2017) | Carrier system, PCT | UCNP core combined with mesoporous silica and Ru2+ complex. | Under NIR irradiation, the accumulated H2O2 in intratumoral mitochondrion will react with Fe2+ to efficiently generate localized –OH radicals which cause mtDNA damage. | Liver cancer | 2017 |
| Yang et al. (Yang et al., 2017) | Direct bonding, PDT | D112 | D112 bound to mtDNA, and induced mtDNA damage, ROS production and complex I inhibition. | Breast carcinoma | 2017 |
| Pokrzywinski et al. (Pokrzywinski et al., 2016) | Carrier system | mitoTEMPOL, mitoquinone and mitochromanol-acetate conjugated to TPP+ | Reduced mitochondrial function by affecting mtDNA integrity and oxidative respiration. | Breast carcinoma and lung cancer | 2016 |
2. mtDNA mutations: a trigger for cancerogenesis
Mutations in the mitochondrial genome constitute a significant component of the cancer mutant genome (Ju et al., 2014). mtDNA dysfunction and gene mutations are closely related to tumorigenesis and can confer different degrees of advantages to cancer cells depending on the mutation types, locations, and heteroplasmy level. The composition of this network involves alterations in gene copy number, aberrant gene expression, and mtDNA epigenetic modifications that frequently impact cancer occurrence and malignant transformation through metabolic regulation, reactive oxygen species (ROS) generation, intercellular interaction, and other mechanisms. Here we provide a concise overview of the intricate relationship between mtDNA damage, gene mutations, and cancer progression, to clarify the status of mtDNA as a biomarker in tumors and its value as a therapeutic target.
Abnormal gene expression of mtDNA is the most common manifestation of mitochondrial genome changes, which often causes imbalance of ROS level, changes of cellular metabolism, and the transformation of tumor energy form. Research findings have demonstrated that mutations in mtDNA can impact the constituents of electron transport chain, thereby resulting in elevated levels of ROS (Smith et al., 2022a). In most cases, Complex I is preferentially affected, accompanied by silencing of complex III and complex V (Kim et al., 2022). Abnormal ROS levels can promote tumor initiation and growth by inducing mtDNA mutations, genomic instability, metabolic reprogramming, and activation of oncogenic signaling pathways (Kuo et al., 2022). Furthermore, metabolic alterations serve as a crucial mechanism in regulating tumorigenesis following mtDNA mutations. Schöpf et al. (Schöpf et al., 2020) studied the alterations in mitochondrial metabolism and mtDNA in prostate cancer tissue samples. They found that malignant tissue samples have lower “NADH-pathway” capacity and stronger succinate oxidation than benign tissue samples, especially in high-grade tumors. The accumulation of harmful mtDNA mutations, particularly in mitochondrial complex I genes, and the presence of mtDNA heteroplasmy causes it and contributes to the development and aggressiveness of prostate cancer. This study has emphasized that mtDNA mutations can induce metabolic reprogramming in tumor tissues, leading to high-risk transformation of tumors. In addition, deregulation of enzymes caused by mtDNA mutations can lead to the accumulation of intermediate products in cancer cells, accelerating tumorigenesis (Badrinath & Yoo, 2018). From the perspective of energy generation, mtDNA mutations may affect the cellular energy capacity, increase oxidative stress, indirectly lead to cancer aggravation, and also have an energy programming effect on cancer metabolism that depends on OXPHOS (Carew & Huang, 2002; Davis et al., 2014).
Epigenetic changes in mtDNA are modifications that affect the expression or function of mtDNA without altering its sequence, including mtDNA methylation, RNA methylation, and histone modifications (Wagner et al., 2022). Some studies have shown that epigenetic changes in mtDNA can promote tumor progression by affecting various aspects of mitochondrial function and communication. In colorectal cancer, Feng et al. (Feng et al., 2012) observed a significant correlation between the expression change of ND2 and clinical pathological stage, and they identified that demethylation of D-loop plays an important regulatory role in this process. In addition, 8 mtDNA aberrantly methylated sites were found to be associated with enhanced invasion and metastasis of breast cancer (Han et al., 2017). And this potentially malignant methylation in D-loop region can be maternally inherited. The progression of bone metastatic tumors of renal cell carcinoma was also shown to be related to the methylation status of mtDNA D-loop region, and could be induced by hypoxic microenvironment (Liu, Tian, et al., 2022). However, even though mtDNA methylation may be used as a marker to predict cancer progression, its relationship with cancer remains to be further elucidated. Mitochondria retain their own replication, transcription, and translation systems, and abnormal methylation modifications of their own mitochondrial RNA may also affect mtDNA and mitochondrial function by regulating the stability and structure of mitochondrial transcription. rRNA methylation is mainly affected by mitochondrial rRNA methyltransferases, including mitochondrial large subunit methyltransferases (MRM1, MRM2, MRM3, TRMT61B) and mitochondrial small subunit methyltransferases (TFB1M, TRMT2B, NSUN4, METTL15) (Lopez Sanchez et al., 2020). Abnormalities of mitochondrial rRNA methyltransferases have been found to be associated with diabetes, Alzheimer's disease, and breast cancer (Couch et al., 2016; Koeck et al., 2011). mtDNA lacks histones, but mtDNA binding proteins can serve as targets for post-translational modifications, mainly involving TFAM, mitochondrial single-stranded binding protein and DNA polymerase γ (POL γ), which participates in mitochondrial transcription (Chatterjee et al., 2022). Histone modifications can affect the accessibility, replication, and transcription of mtDNA, and may promote cancer progression by regulating OXPHOS, mitochondrial biogenesis, apoptosis, and retrograde signaling in cancer cells.
The change in mtDNA copy number level is also important for cancer progression. So far, an increase or decrease in mtDNA copy number has been observed in various cancers, which may represent the potential role of mtDNA copy number in cancer progression (Li, Sundquist, et al., 2021, p. 13). In recent years, researchers have tried to utilize it as a biomarker for cancer diagnosis and efficacy prediction. Zhang et al. (Zhang et al., 2023) investigated the correlation between leukocyte mtDNA copy number and clinical outcomes in breast cancer patients, revealing for the first time that high mtDNA copy numbers are associated with worse prognosis in hormone receptor-positive cases. And they proposed using mtDNA copy number as a biomarker to predict prognosis. In addition, Sun et al. (Sun et al., 2018) attempted to demonstrate that increased mtDNA copy number can promote the metastasis of microsatellite-stable colorectal cancer cells by increasing mitochondrial OXPHOS level. Similarly, Reznik et al. (Ed et al., 2015) found that a large number of tumor tissues exhibit alterations in the content of mtDNA, manifested as depletion (bladder cancer, breast cancer, esophageal cancer, head and neck squamous cell carcinoma, kidney cancer) or accumulation (lung cancer), which is closely related to the regulation of tumor pathology and patient survival. The association between mtDNA copy number and tumor progression is undoubtedly authentic. On the contrary, some studies have shown that the change in mtDNA copy number was not sufficient to serve as a clinical marker for cancer assessment (Haja Mohideen et al., 2015). Therefore, the clinical value of mtDNA copy number remains to be further clarified.
In the process of mtDNA mutation and disease induction, the level of mtDNA heteroplasmy significantly impacts the clinical progression of the disease. Generally speaking, only when the mutant heteroplasmy of mtDNA reaches a certain proportion can it induce disease. In recent years, scholars have conducted a more in-depth analysis of this dynamic evolutionary process. Wallace et al. (Picard et al., 2014) conducted a study in 2014 to assess the impact of varying level of mtDNA tRNALeu (UUR) 3243 A > G mutation on gene expression profile, signaling pathway and epigenetic changes in cells. Their findings suggested that the degree of heteroplasmy has differential effects on cell phenotype. This variability of heteroplasmy negated the concept of “single mutation-single disease”, providing an explanation for clinical variability in mtDNA diseases. In 2019, Wallace et al. (Kopinski et al., 2019) further delved into a more comprehensive examination of the impact of heteroplasmy level on the nDNA. They incorporated nuclear NAD+/NADH levels, changes in histone modifications, mitochondrial intermediates, and alterations in redox states into the intricate network of “mtDNA-nDNA”. This perspective enhances our understanding of the process by which mtDNA mutations cause diseases and plays a crucial role in guiding the appropriate utilization of mtDNA as a biomarker.
The alterations in mtDNA can affect various aspects of mitochondrial function and cellular homeostasis, including energy generation, ROS production, metabolic reprogramming, epigenetic regulation and nuclear gene expression. Moreover, the level of mtDNA heteroplasmy can modulate the phenotypic variability and severity of mtDNA diseases. Therefore, mtDNA damage and gene mutations are important factors that contribute to cancer development and progression. Besides, the interactions between mtDNA and other cellular components, such as nDNA, immune system, and neighboring cells, can also influence the tumorigenicity, metastasis, chemoresistance, and immunogenicity of cancer cells.
3. The messenger role of mtDNA: basis and concerns of targeted therapy
As an important intermediate medium, mtDNA can extensively communicate with extramitochondrial biological systems and trigger a series of biological effects, especially when its stable state is disrupted. Understanding how mtDNA interacts with other components will help reveal the mechanism, effect prediction and prognosis evaluation of mtDNA-targeted therapies. Here, we discuss the important role of mtDNA in intercellular and intracellular communication.
3.1. The interactions between mtDNA and nDNA
The extensive interactions between mtDNA and nDNA include the expression regulation of related nuclear genes and the maintenance of mtDNA stability under nuclear regulation (Kosmider et al., 2019; Yamazoe et al., 2021; Zhou et al., 2010). mtDNA can influence nDNA expression through various pathways. These pathways include activating related signaling pathways during damage, directly acting on nDNA through ROS generated by mitochondrial dysfunction, or integrating mtDNA into nDNA to form nuclear mitochondrial sequences (NUMTs). The influences on the expression nuclear genes may confer both advantages and disadvantages for the vital functions of cells. Han et al. (Han et al., 2021) found an increase in stemness-associated genes, such as OCT4, NANOG, and SOX2, after the depletion of mtDNA in Hep3B/rho0 cells, causing the obtaining and maintaining of cancer stem-like properties. And Lee et al. (Lee et al., 2015) discovered the pivotal roles that the activation of retrograde NUPR1-granulin pathway after mitochondrial dysfunction played in human liver cancer. Moreover, the process of epithelial-mesenchymal transition has also been shown to be associated with the activation of the TGF-β-c-Jun/AP-1 signaling pathway caused by mitochondrial dysfunction (Yi et al., 2015). NUMTs are fragments of mtDNA that have been transferred and integrated into nDNA. When exogenous or endogenous factors cause DNA double-strand break (DSB), mtDNA can integrate into nDNA through microhomology-mediated end joining, retro-transposition, endocytosis of mitochondria-derived vesicles, encapsulation of mtDNA in the nucleus, and other pathways, affecting the structure and regulation of nDNA genetic information (Gaziev & Shaikhaev, 2010). Some studies have shown that these integrated sequences are involved in processes such as carcinogenesis and aging (Wei et al., 2022). Therapeutically, we hope that mtDNA damage caused by mtDNA-targeted therapy can induce the expression of genes related to cell death. It has been reported the alteration of mtDNA copy number can initiate retrograde signaling, which causes the impairment of the expression profile of nuclear genes and modifies cell physiology and morphology (Abd Radzak et al., 2022). In addition, the immune- and stress-related genes have been found the most highly differentially expressed in mtDNA-deleted 4T1/rho0 cells, which might be the basis of immune activation (Grasso et al., 2020).
In addition to affecting the gene expression of nDNA, mtDNA damage can also activate DNA damage response (DDR) to promote mtDNA repair and regulate cellular senescence and apoptosis activities. mtDNA damage can activate the DDR of nDNA to repair mtDNA and maintain mtDNA stability. mtDNA damage can be sensed by different molecules and pathways that translocate to the nucleus and modulate the expression of nuclear genes that are involved in mtDNA replication, repair, and turnover, such as p53, ataxia telangiectasia mutation (ATM) protein, and NF-κB (Lee & Paull, 2020; Shin et al., 2022). Moreover, the nuclear DDR can also regulate the availability of nucleotides for mtDNA synthesis by balancing the nucleotide pool (Rai, 2010). The DDR can regulate the nucleotide pool by activating enzymes that synthesize or salvage nucleotides, such as RNR (Zuo et al., 2022), and/or by inhibiting enzymes that degrade or consume nucleotides, such as SAMHD1, DUT, and PARN (Murphy & Kleiman, 2020; St Gelais et al., 2012). In addition, the nuclear DDR can also influence the communication between the nucleus and the mitochondria by modulating the expression of proteins that mediate mitochondrial-nuclear crosstalk, or altering the localization of proteins that shuttle between the two organelles (Fairbrother-Browne et al., 2021). Therefore, the nuclear DDR is an important mechanism for maintaining mitochondrial function and integrity after mtDNA damage. As another important double-stranded DNA in cells, the interactions between mtDNA and nDNA merit more research. And it is an aspect worth paying attention to after the disruption of mtDNA homeostasis caused by either therapeutic reasons or natural factors.
3.2. The role of mtDNA in intercellular transfer
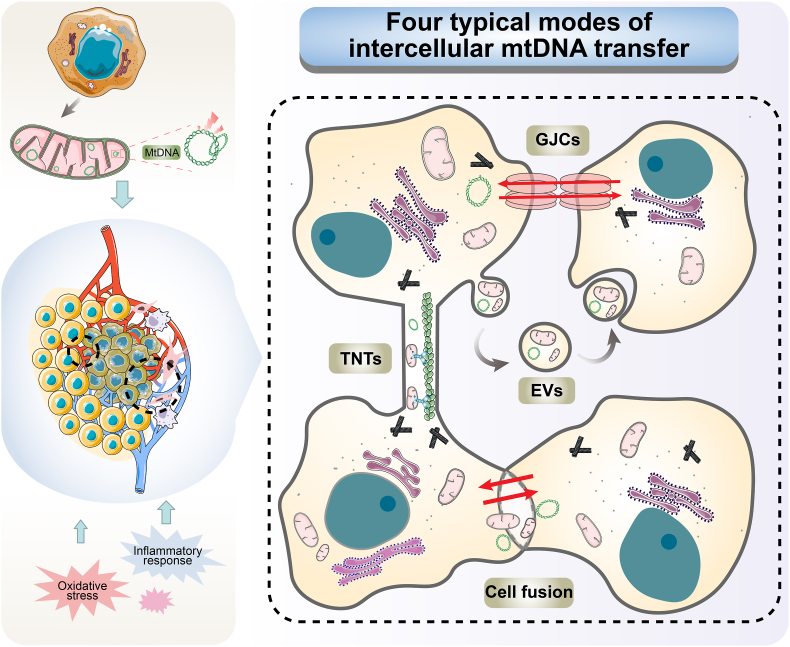
Similarly, mtDNA is crucial for cell-to-cell communications. Cancer cells with mtDNA depletion can obtain functional mtDNA from adjacent cells to repair mitochondrial function, including stromal cells and tumor cells. At the same time, some mtDNA carrying certain pathogenic mutations can be transferred to other cancer cells. These mechanisms of intercellular transfer include tunneling nanotubes (TNTs), cell fusion, gap junction channels (GJCs) and extracellular vesicles (EVs), and occur under physiological and pathological conditions (Fig. 2). (Liu, Sun, et al., 2022; Valenti et al., 2021a) TNTs are slender, membrane-enclosed tubular connections that enable the intercellular transfer of various molecules and organelles, including mtDNA, between distant cells (Dubois et al., 2020). The generation of TNTs primarily involves two mechanisms, namely filopodial interplay and cell dislodgement, which additionally necessitate the modulation of cortical actin dynamics, plasma membrane deformation and curvature (Turos-Korgul et al., 2022). The movement of mtDNA and mitochondria within TNTs is contingent on the cytoskeleton elements, predominantly F-actin and microtubules, and can be influenced by cytoskeletal composition, membrane dynamics, molecular motors, oxidative stress and extracellular signals (Dagar et al., 2021). TNTs-mediated mtDNA transmission can modulate the metabolic and genetic diversity of tumor cells and influence their proliferation, metastasis and adaptation (Marlein et al., 2019). For example, cancer-associated fibroblasts can donate functional mitochondria and mtDNA to prostate cancer cells via TNTs and increase their glycolytic activity and chemoresistance, promoting their malignant transformation (Ippolito et al., 2019). Furthermore, another study reported that glioblastoma (GBM) cells can transmit mutated mitochondria and mtDNA to neighboring primary astrocytes via TNTs, modulating metabolism, facilitating cancer progression and tumor metabolism adaptation to the microenvironment (Valdebenito et al., 2021). As another important pattern, GJCs are composed of connexin (Cx) proteins that form hexameric hemichannels on each cell membrane (Valenti et al., 2021b). However, owing to the pore size restriction, they preclude the passage of organelles and usually assist in TNT-mediated mitochondrial transfer (Liu, Sun, et al., 2022). Cx43 is an important regulator of TNT generation, which has been reported to act in the mechanism of mitochondrial transfer between mesenchymal stem cells and epithelial cells in asthma inflammation (Yao et al., 2018).
Fig. 2.
Schematic diagram of four typical modes of intercellular mtDNA transfer. Stimulation of TME tissues by endogenous factors, such as damaged mtDNA in tumor cells, or exogenous factors like oxidative stress, inflammatory response and pathological stimuli can trigger intercellular transfer of mtDNA. mtDNA transfer mainly includes four typical forms: tunneling nanotubes (TNTs), cell fusion, gap junction channels (GJCs) and extracellular vesicles (EVs). Multiple mechanisms frequently co-exist and synergistically interact in TME.
EVs-mediated pattern can transfer mtDNA or mitochondria-related components by fusing with the plasma membrane or endocytosis, affecting the functions of the recipient cells. Takenage et al. (Takenaga et al., 2021) have revealed that metastasis-enhancing pathogenic mtDNA derived from metastatic tumor cells is transferred to low-metastatic tumor cells via EV to promote the metastatic potential of tumors. And in the treatment of breast cancer, EVs can carry intact mtDNA and act as infectious mediators between cancer-associated fibroblasts and cancer cells, rescuing breast cancer cells with OXPHOS defects induced by hormone therapy (Sansone et al., 2017). The findings suggest a strong correlation between EVS-mediated mtDNA transfer and the development of tumors as well as the acquisition of drug resistance. Moreover, EVs that remain in the extracellular environment may have a close association with mtDNA-mediated immune and inflammatory responses in TME. TNTs, EVs and GJCs represent the main pathways of mtDNA intercellular transfer in tumor tissues, but there are often multiple patterns co-existing and synergizing in reality (Yang et al., 2023). In addition, mtDNA can also be transferred between cells through cell fusion. It has been demonstrated that the formation of hybrid cells through spontaneous fusion between pre-malignant (IMR90 E6E7) and malignant (IMR90 E6E7 RST) mesenchymal cells facilitate the metastatic dissemination of tumors (Brito et al., 2021). Nevertheless, it is important to note that cell fusion involves the sharing of a large number of organelles and cellular structures, and cannot be solely attributed to mtDNA or mitochondrial exchange.
In the recent studies, intercellular transmission of mtDNA is usually associated with functional repair and remodeling of cells with mitochondrial dysfunction. In cancer cells, intercellular mitochondrial transfer can modulate the bioenergetics, metabolism, oxidative stress, gene expression and signaling pathways of tumor cells and non-tumor cells in the TME, affecting the tumorigenicity, metastasis, chemoresistance and immunogenicity of tumor cells and other phenotypes (Berridge et al., 2015). Intercellular mtDNA transfer is an emerging phenomenon that has important implications for understanding and treating mitochondrial diseases. As a dynamic and complex process, it has beneficial or detrimental effects on both donor and recipient cells depending on the context and direction of transfer. Also, it offers new insights into the molecular mechanisms and consequences of mitochondrial dysfunction and communication in different cell types and tissues, penning new avenues for developing novel therapeutic strategies based on manipulating intercellular mtDNA transfer to enhance or inhibit mitochondrial function in target cells (Valenti et al., 2021a).
3.3. The role of mtDNA in immune activation
As a damage-associated molecular pattern (DAMP), mtDNA release and immune activation are also frequently associated with each other. The release of mtDNA is the first step in triggering immune responses and boosting immunosurveillance (Bai & Liu, 2019; Choi et al., 2020). Two mechanisms have been analyzed directed at mtDNA release, including releasing through the formation of BAX and BAK in the outer membrane and subsequent inner membrane (IM) permeabilization in apoptotic cells, and involving the oligomerization of voltage-dependent anion channel (VDAC) in the outer membrane of non-apoptotic cells (Bahat et al., 2021b). During apoptosis, apoptosis factors can inhibit the anti-apoptotic members of BCL-2 family including BCL-2 and BCL-xL, thereby unleashing BAX and BAK to cause mitochondrial pore formation in outer mitochondrial membrane, leading to release of oxidized mtDNA and activating the NLRP3 inflammasome (Paik et al., 2021; Singh et al., 2022). The activation of BAX and BAK can be induced through direct binding to a subset of BH3-only proteins named BIM, BID, PUMA and possibly NOXA (Ludwig et al., 2020). In non-apoptotic macrophages, the mechanism that ox-mtDNA could exit mitochondria via mitochondrial permeability transition pores- and VDAC-dependent channels when it was cleaved by the endonuclease FEN1 to 500–650 bp fragments has been found (Xian et al., 2022). VDAC can oligomerize under oxidative stress conditions and form large mitochondrial outer membrane pores which mediate mtDNA release in live cells (Kim et al., 2019). The release of damaged-mtDNA into the cytosol can stimulate pattern recognition receptors including Toll-like receptors and NOD-like receptors as a DAMP originated from cell itself (Bahat et al., 2021a). The cyclic GMP-AMP synthase (cGAS)-STING pathway plays an important role in leaked-mtDNA induced immune responses (Li, Zhang, et al., 2021; Zhang, Tang, et al., 2020). The cGAS is identified as a DNA sensors and can be activated by dsDNA including mtDNA, leading to the synthesis of dinucleotide second messenger 2′,3′-cyclic GMP-AMP (cGAMP) from ATP and GTP (Morimoto et al., 2021). cGAMP can finally stimulate innate immune by activating an endoplasmic reticulum-localized stimulator of interferon genes (STING), which increase production of inflammatory cytokines (Pokatayev et al., 2020; Wang et al., 2020). Besides, extracellular cGAMP can act as a soluble immune mediator in the TME to promote immune cell activation (Skopelja-Gardner et al., 2022). Extracellular release of mtDNA also plays a significant role in immune activation and inflammatory response, and is associated with the polarization and functional regulation of macrophages, dendritic cells, and T cells (Amari & Germain, 2021). The predominant pathway for mtDNA release in the TME is EV-mediated, including exosomes, microvesicles and apoptotic bodies. mtDNA can be encapsulated into EVs either through direct contact or via mitochondria-derived vesicles, resulting in extracellular release. Additionally, during apoptosis, mtDNA can also be passively released through cell membrane rupture caused by mechanical trauma (De Gaetano et al., 2021). It can be speculated that it is theoretically feasible to achieve therapy by inducing mtDNA activated immune response through targeting mtDNA. The understanding of the subsequent immune response to mtDNA-targeted therapy is of great significance for evaluating the therapeutic process.
In summary, mtDNA is not only essential for mitochondrial function, but also plays important roles in various biological processes that involve communication and interaction with other cellular components. mtDNA can regulate the expression of nuclear genes, be transferred to other cells to repair or enhance mitochondrial function, and activate immune responses by acting as a DAMP. These roles of mtDNA have implications for the development and progression of various diseases, especially cancers. As a result, understanding the mechanisms and consequences of mtDNA-mediated communication and immune activation is crucial for developing novel mtDNA-targeted therapeutic strategies.
4. Cancer therapies targeting mtDNA
4.1. Indirect targeted therapy
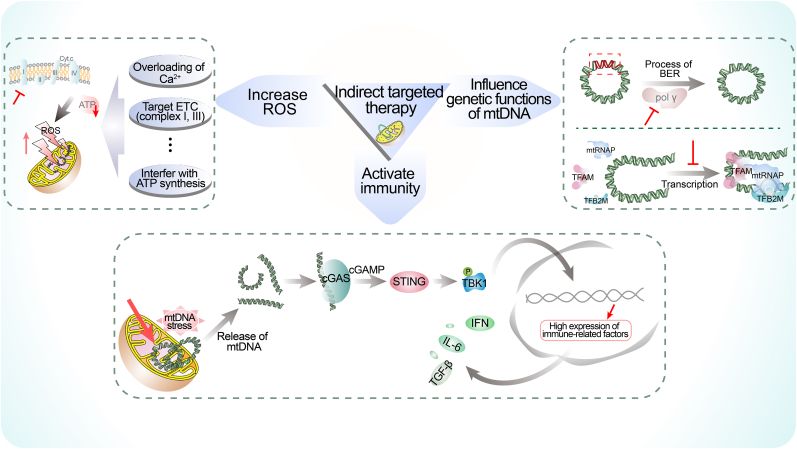
In this part, we discussed how to indirectly target mtDNA for cancer therapy by stimulating ROS, inhibiting POL γ and TFAM, and activating cGAS-STING pathway. These strategies exert their effects by influencing pathways that are involved in mtDNA stability and integrity instead of binding to mtDNA, thereby inducing mtDNA damage and dysfunction. The main contents of indirect targeted therapy are summarized in Fig. 3. Also, more details concerning recent indirect mtDNA-targeted researches were gathered and reported in Table 1.
Fig. 3.
The main aspects of indirect targeted therapy. We summarize the related studies of indirect targeted therapy into three categories, including increasing ROS, influencing genetic functions of mtDNA and activating immunity. Treatments that disrupt the normal states of these aspects will further cause damage of mtDNA, resulting in tumor suppression mediated by mitochondrial dysfunction. BER, base excision repair; IFN, interferon; IL-6, interleukin- 6; mtRNAP, mitochondrial RNA polymerase; TBK1, TANK Binding Kinase 1; TFAM, mitochondrial transcription factor A; TFB2M, mitochondrial transcription factor B2; TGF-β, transforming growth factor beta.
4.1.1. Stimulation of ROS
ROS can be produced in mitochondria. The excessive accumulation of ROS results in mtDNA damage and mitochondrial dysfunction that may lead to severe cellular damage and apoptosis (Urbina-Varela et al., 2020; Zhang, Guo, et al., 2020). Inducing intracellular ROS accumulation to cause mtDNA damage is a viable strategy that has been developed in various studies exploring cancer therapy. Several studies have demonstrated the anti-cancer effects of different agents, such as the new Ir(III)-2-indazole complex with an isopropyl substituent which could induce mtDNA target inhibition and mitochondrion-mediated apoptosis by stimulating ROS in triple negative breast cancer (Panchangam et al., 2021). The novel VB12-sericin-PBLG-IR780 nanomicelles synthesized by Guo et al. have been proved to have anti-cancer effects by significantly inhibiting the expression of ATP synthase, inducing oxidative stress in mitochondria, increasing the generation of ROS and ultimately leading to the oxidative damage of mtDNA in the presence of near-infrared (NIR) (Guo et al., 2022). In addition to that, the antiviral drug Acyclovir may inhibit the progression of cancers through mitochondrial mechanism by causing ROS-mediated mtDNA damage (Benedetti et al., 2022). The pathways that can cause increasing production of ROS can be potential targets for indirect mtDNA therapy. Several conditions including overloading of Ca2+, affecting the electron transport chain (mainly referring to complexes I and III) and interfering with the process of ATP synthesis can contribute to aberrant ROS generation, becoming possible targeting strategies (Karam et al., 2017; Sun et al., 2014; Vo et al., 2019).
4.1.2. Targeting the genetic expression and inheritance of mtDNA
The mtDNA POL γ is required for the replication and damage repair of the mitochondrial genome, and plays important roles in the biogenesis of mitochondria (De & Campbell, 2007; Siibak et al., 2017). Base excision repair is the prominent pathway for the repair of oxidative DNA lesions by excising and replacing damaged bases like 8-oxodeoxyguanine in mtDNA, using various proteins encoded in the nucleus and imported into the mitochondria (Fu et al., 2019; Zheng et al., 2020). Therefore, diminished repair capacity can be one of the factors responsible for high mutation frequencies of mtDNA which is closely related to the anti-cancer treatment (Singh et al., 2015). 3,3'-[(1,1′-Biphenyl)-4′,4′-(diyl)bis (azo)]bis [4-amino-1-naphthalenesulfonic acid] (CR) has been identified with a high binding affinity to the POL γ protein and can cause mitochondrial dysfunction and cell apoptosis by inhibiting POL γ activity and oxidative mtDNA damage repair in MLH1 deficient human colon cancer (Somuncu et al., 2022). As one of the most significant polymerases in mtDNA synthesis and damage repair, it can become an important target of tumor therapy.
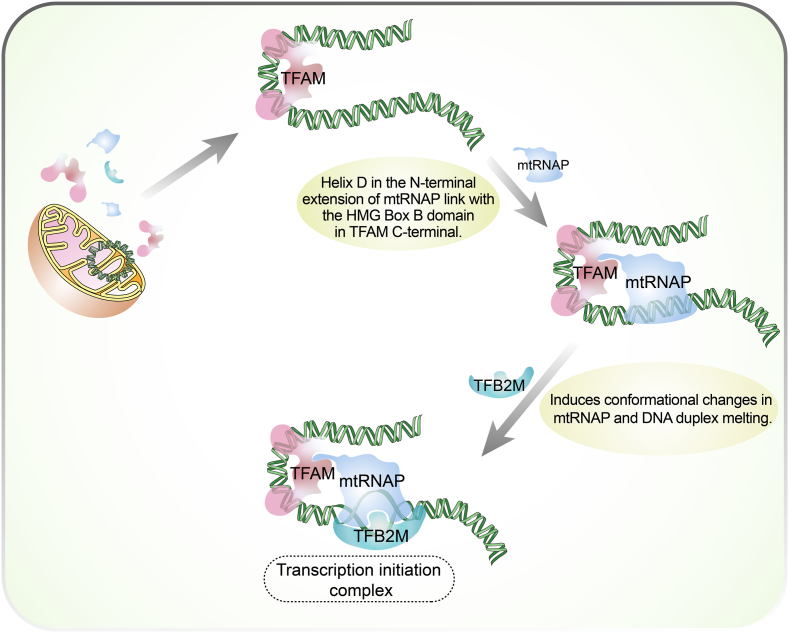
Besides POL γ, nuclear-encoded TFAM is a high mobility group (HMG) protein and acts as a key regulator in the replication, transcription, and inheritance of mtDNA (Wen et al., 2014). It supports the maintenance and stability of mitochondrial genome and the stability of TFAM is essential for biological function in cancer progression (Lan et al., 2017). As a product of the nucleus, TFAM contains five different domains including a cleavable matrix targeting sequence, two HMG homology domains connected by a linker, a tail, and short leader sequence that is located between matrix targeting sequence and HMG1 in human and murine (Kim et al., 2018; Kozhukhar & Alexeyev, 2022). TFAM is a protein that attaches to mtDNA and regulate mitochondrial transcription process (Fu et al., 2021). The completion of the process requires the formation of a transcription initiation complex composed of TFAM, mitochondrial transcription factor B2 (TFB2M), mitochondrial RNA polymerase (mtRNAP) and promoter DNA (Fig. 4). (Uchida et al., 2017) Besides, the interaction of TFAM with 8-oxodG-containing DNA substrates has shown the important role of TFAM in regulating DNA repair, while mtDNA packaging is possibly affected by the binding between TFAM and different non-canonical DNA structures (Chew & Zhao, 2021). As TFAM plays an important role in cellular activities and is indispensable in cancer progression, its aberrant expression in colorectal cancer cells can be a helpful marker for tumor progression in colorectal cancer patients (Zhao et al., 2021). Silencing its expression can inhibit mitochondrial function and significantly reduce tumor initiation potential in colon cancer cells and KrasG12D-driven lung cancer (Wen et al., 2019). TFAM can also increase the expression of angiogenesis and invasion-related genes and play a pro-tumorigenic role (Araujo et al., 2018). Moreover, TFAM affects the sensitivity of tumor treatment. Cellular sensitivity to cisplatin treatment is correlated with microRNA-199a-3p-mediated attenuation of TFAM expression in MDA-MB-231 cells (Zhang & Wang, 2018). Similarly, TFAM modulates the response of tumor cells to ionizing radiation via P53/TIGAR signaling in various cancers (Jiang & Wang, 2019). The irreplaceable position of TFAM in tumor growth and progression highlights its potential as a promising avenue for cancer treatment.
Fig. 4.
Assembly of transcription initiation complexes. TFAM and three other components (TFB2M, mtRNAP and promoter DNA) form a complex on the mtDNA to start the expression process. At the beginning of the transcription, TFAM firstly binds to DNA 16–39 nt upstream of the transcription start site and induces a ∼180° bend into DNA (Hillen et al., 2017; Rubio-Cosials et al., 2011). And then the helix D in the N-terminal extension of mtRNAP will link with the HMG Box B domain of TFAM, resulting in anchoring to the promoter (Hillen et al., 2017). The formed pre-IC lacks DNA specificity and transcriptional activity, unless the N-terminus of TFB2M bound to the active site of mtRNAP and induces conformational changes in mtRNAP (Morozov et al., 2015). mtRNAP, mitochondrial RNA polymerase; TFAM, mitochondrial transcription factor A; TFB2M, mitochondrial transcription factor B2.
An antiviral drug for a human immunodeficiency virus infection, zalcitabine, has recently been found by causing TFAM degradation through a LONP1-dependent pathway in pancreatic cancer, resulting in triggering mtDNA stress, injury, cytosolic release and finally trigger an antiviral innate immune response via the activation of cGAS-STING1 pathway (Li, Zhang, et al., 2021). Consistent with this, Wang et al. demonstrated the pivotal role of TFAM, which exhibits high expression levels in pancreatic cancer, preserving stability of mitochondrial function and promoting biogenesis (Wang et al., 2023). Targeting TFAM inhibition may offer a potential solution to overcome resistance to gemcitabine in pancreatic cancer cells. Moreover, melatonin and the combination of epoxomicin and cisplatin can also inhibit mitochondrial functions, and ultimately promote cellular apoptosis by inhibiting TFAM (Franco et al., 2018, p. 23; Kong et al., 2022). The interactions between the expression of TFAM and other genetic activities can also be breakthrough points for targeted therapy. Up-regulation of Mir-199a-3p expression can induce mitochondrial dysfunction by inhibiting TFAM transcription in breast cancer MDA-MB-231 (Fan et al., 2017). The knockdown of Circ_0002476 caused mtDNA alterations by inhibiting Mir-1182/TFAM axis, leading to the transcriptional inhibition of ND1 (encoding the subunit of NADH dehydrogenase) and ATP6 (encoding the subunits of ATP synthase) (Wang et al., 2022a). Computer-based virtual screening have been widely used in novel drug discovery and capability prediction (Arévalo & Amorim, 2022; Song et al., 2021). The combination of violacein and silver nanoparticles that was predicted to have an anticancer activity by inhibiting TFAM as a dyad drug system (Pandey & Verma, 2020). The application of this technology will provide new strategies for the development of new TFAM-targeted drugs and solving the resistance of traditional drugs through mitochondrial pathway.
mt-RNAP, also known as POLRMT, is another important enzyme that regulates mtDNA transcription and replication. POLRMT is essential for the expression of mitochondrial genes and the maintenance of mtDNA integrity and copy number. Consequently, POLRMT is a potential target for anti-cancer therapy. Bonekamp et al. (Bonekamp et al., 2020) developed mitochondrial transcription inhibitors (IMTs), namely IMT1 and IMT1B. These compounds inhibited POLRMT non-competitively, altering its conformation, stability and activity. They could cause a significant reduction in the levels of mtDNA transcriptions and respiratory chain subunits in tumor cells, leading to dose-dependent inhibition of OXPHOS. The anti-tumor efficacy of IMTs has also been confirmed in ovarian cancer, cervical cancer and lung cancer cells.
4.1.3. Activation of the immune system
The interaction between mtDNA and immune system has attracted considerable attention in the recent discussion (Cushen et al., 2020; Yumnamcha et al., 2020). The manipulation of mtDNA to activate immune process represents a significant therapeutic strategy for enhancing anti-tumor effects (Mohanty et al., 2019). Two novel substituted carbazole derivatives (Ethyl 3-((tert-butoxycarbonyl) amino)-2-hydroxy-9H-carbazole-1-carboxylate and 3-((tert-butoxycarbonyl) amino)-2-hydroxy-9H-carbazole-1-carboxylic acid) synthesized by Witkowska et al. have displayed a better anticancer activity by inducing massive accumulation of mtDNA DSBs that were oxidatively damaged, increasing the permeabilization of the mitochondrial IM and stimulating the immune responses in colon and osteosarcoma (Witkowska et al., 2022).
One of the most common methods is employing some nano-carrier systems, that could activate innate immune response pathways while eliciting mtDNA damage and release. The BCP nanoparticles ((BPA + CPI) @ PLGA NPs), which combined 3-BPA and CPI-613 via a nano-drug delivery system, could not only promote the release of mtDNA by causing mitochondrial metabolism disorder, but also enhance the dimerization of STING and activation of immune (Li et al., 2022). And icaritin loading poly (lactic-co-glycolic acid) nanoparticles (PLGA@Icaritin NPs) could increase production of oxidative-mitochondrial DNA which followed by immunogenic cell death, and recruit infiltrating CD4+ cells, CD8+ T cells and cytokine to activate the anti-tumor immunity (Xiao et al., 2022). Virus-based immunotherapy, a promising method for cancer treatment, has also been used in this approach (Thomas et al., 2019). Recently, a herpesvirus-mimicking nanoparticle (named Vir-ZM@TD) is designed, in which the combinations of DNAzyme-loaded Mn-doped zeolitic imidazolate framework-90 nanoparticles, erythrocyte membrane, RGD and HA2 peptides were used to mimic virus structure (Zhao et al., 2022). As a result, mtDNA stress was triggered by Vir-ZM@TD, releasing it into cytosol, and the sensitivity of cGAS to mtDNA was increase by Mn2+. Moreover, the natural product rocaglamide (RocA) has been shown to dramatically activate the cGAS-STING pathway and promote the cytoplasmic release of damaged mtDNA induced by itself (Yan et al., 2022).
In addition to that, using endogenous proteins to regulate mtDNA and immune system is also a potential strategy in cancer treatment. For example, overexpression of mitochondrial Lon that can accumulate ROS, increase oxidative damage of mtDNA and decrease mtDNA repair efficiency, promote mtDNA release into the cytosol and induce the secretion of extracellular vehicles carrying mtDNA, ultimately affecting the immune response in TME (Cheng et al., 2020). Besides, ATM protein has been identified as a prospective target in cancer therapy for its inhibition can enhance immune checkpoint blockade therapeutic effects by promoting cytoplasmic leakage of mtDNA, activation of cGAS-STING pathway and lymphocyte infiltration into TME (Hu et al., 2021). And Yin et al. (Yin et al., 2023) discovered the special effect of Raddeanin A in tumor immunotherapy, which is an oleanane class triterpenoid saponin isolated from Anemone raddeana Regel. It has the ability to interact with the transactive responsive DNA-binding protein 43 (TDP-43), trigger the release of mtDNA from mitochondria, and consequently enhance dendritic cells (DCs) mediated antigen cross-presentation and T cell activation. A specific protein, vaccinia virus-associated kinase 2 (VRK2), has been identified as a facilitator of mtDNA binding to VDAC, leading to the oligomerization of VDAC1 and subsequent release of mtDNA (He et al., 2021). This finding suggested that VRK2 could be a promising therapeutic target for viral infectious diseases associated with mtDNA release. Moreover, recent studies have identified VRK2 as a crucial target for enhancing tumor immunotherapy and overcoming tumor drug resistance (Chen et al., 2022; Moore et al., 2022; Peled et al., 2021; Zhu et al., 2021). However, it remains unclear whether targeting VRK2 exerts its effects via the mtDNA release pathway in cancer therapy. In conclusion, identifying and utilizing the connection between mtDNA and immune response in order to devise innovative and efficacious cancer treatment approaches is an area worth exploring.
4.2. Direct targeted therapy
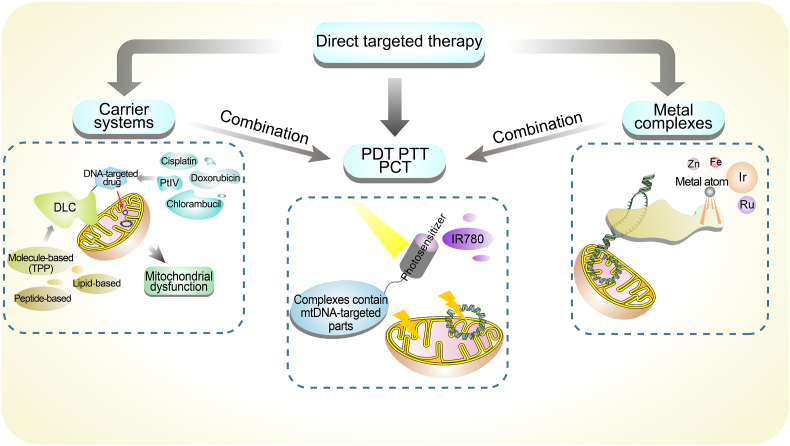
The field of directly mtDNA-targeted therapy faces the challenge of preventing the impact on nuclear genes due to their genetic orthology. Nevertheless, some recent methods have overcome this problem and applied mtDNA-targeted therapy to clinical settings (Yang et al., 2022). These methods utilize mitochondrial localization molecules that can modulate the function of mtDNA by directly targeting it. This part summarized several specific strategies in the field of direct targeted therapy to mtDNA that may be extensively applied and implemented (Fig. 5). And more details of related studies about direct targeting recently were summarized in Table 2.
Fig. 5.
The main aspects of direct targeted therapy. We summarize the relevant research into three aspects: carrier systems, metal complexes and application of phototherapy. And among the studies we included, carrier systems, metal complexes, and phototherapy frequently overlapped in their research techniques, drug construction, therapeutic effects and so on. We suspect that future research may need to enhance the integration of the three methods to develop more effective strategies. DLC, delocalized lipophilic cation; PCT, photochemotherapy; PDT, photodynamic therapy; PTT, photothermal therapy; TPP, triphenylphosphonium.
4.2.1. The application of carrier systems
How to prevent the impact on nuclear genes while targeting mtDNA has always been a problem needs to be solved. To address the challenges of targeting mtDNA specially, the use of nano-carrier systems to deliver agents specifically targeting mtDNA has gained widespread adoption in recent years (Tsuji et al., 2022). Delocalized lipophilic cations are commonly used as mitochondrion-targeted agents to construct molecule-based nano-carriers that selectively accumulate in mitochondria with a high membrane potential (Δ Ψ m) and deliver the bioactive cargo into mitochondria (Hall et al., 2021; Jiang, Zhou, et al., 2021). D112 is a cyanine molecule that was identified by the Eastman Kodak Company as a potential anti-cancer drug with high cytotoxic activity and selectivity (Yang et al., 2015). As a delocalized lipophilic cation (DLC), D112 shows promise as it can enter mitochondria preferentially, damage mtDNA, inhibit complex I and induce ROS (Yang et al., 2017). Besides, triphenylphosphonium (TPP) is the most frequently used carrier as a DLC due to its non-toxicity, easy synthesis and purification, and high stability in a biological system (Muhammad et al., 2021; Valdez-Alegría et al., 2020). It exhibits great mitochondrial aggregation by taking advantage of hydrophobicity and positive charge, with widely explored for guiding mtDNA-targeted therapy (Wang, Xiang, et al., 2020). TPP that is loaded with traditional DNA targeting chemotherapy drugs can achieve satisfactory therapeutic effects (Koshikawa et al., 2021, 2021b; Pokrzywinski et al., 2016). To overcome CDDP resistance in triple negative breast cancer cells, a mitochondrial targeting prodrug c,c,t -[Pt-(NH3)2Cl2 (TPP) (Dox)] (PPD), which conjugated PtIV to doxorubicin (Dox) and TPP cation, was synthesized by Muhammad et al. (Muhammad et al., 2021) The novel compound exhibited a superior mitochondrial localization compared to the one that is devoid of TPP and triggered mtDNA impairment, bioenergetics disruption, ROS production and necrosis induction. Moreover, nitrogen mustard chlorambucil (a DNA alkylating drug), cisplatin and other agents were also used to synthesize TPP-conjugated drugs that target mitochondria and mtDNA (Chen et al., 2020). A TPP-loaded organoarsenic prodrug system, which incorporated cisplatin and Dox (targets DNA) with t-As (interrupts the functioning of the thioredoxin system in mitochondria) via cleavable linkers, could induce mitochondria-mediated apoptosis in HepG2 (Luo et al., 2021). Pyrrole-imidazole polyamides, which are cell-permeable minor groove binders that show sequence-specific binding to double-stranded DNA and inhibit the transcription, always conjugate with TPP to target mtDNA and work well in a variety of cancers (Koshikawa, Yasui, et al., 2021). Chen et al. (Chen et al., 2018) designed a peptide nucleic acids which can bind efficiently to DNA and RNA targets to form Watson-Crick type double helix and result in interruption of transcription or RNA degradation. And the loading of TPP improves its mtDNA targeting capability, which is crucial for the mtDNA-targeted therapy.
At the same time, the novel polymer-based, lipid-based and peptide-based carrier systems for mitochondrial targeting have been developed. Faria et al. (Faria et al., 2022) designed a polyethylenimine–dequalinium polymer to load and complex a mitochondrial-gene-based plasmid, affecting gene delivery and protein expression in this organelle, and they also developed a cell-penetrating peptides based system added mitochondrial targeting sequences to achieve the efficacy of mitochondrial gene therapy. A cholesterol-based chimeric nanoparticle consisting of cisplatin, camptothecin and tigecycline was created with abilities of impairing mt-DNA, mitochondrial topoisomerase I and mitochondrial ribosomes in lung, cervical and breast cancer cells (Bajpai et al., 2022a). These kinds of delivery systems that target mitochondria have been widely developed and have great prospects in the treatment of cancers targeting mtDNA.
4.2.2. The construction of metal complexes
Complexes comprising metallic elements including Ir, Zn, Fe and Ru also play important roles in building nano-systems that target mtDNA (Nair et al., 2020; Wang et al., 2022b). Some of the metallic elements involve in chemical responses in mitochondria, while others bind DNA specifically and stabilize the complexes. Jiang et al. (Jiang, Guo, et al., 2021) have constructed a nanocatalytic medicine that induce oxidative damage in the mtDNA of tumor cells. The medicine is a Fe2+-Ru2+-loaded mesoporous silica nanoparticle named as MSN-Ru2+/Fe2+, where Ru2+ complex can bind with mtDNA and Fe2+-Fenton reagents can accelerate the intramitochondrial ROS generation by the Fenton reaction. Ir may be the most common applicational one among various metal elements because of its notable ROS generation efficiency, lifetime sensitivity to microviscosity, straightforward ligand tuning, cell permeability and photostability (Lee, Nam, et al., 2021; Qin et al., 2020). A family of Ir(III) complexes of general formula [Ir(C^N)2(N^N′)]Cl (N^N′ = thiabendazole-based ligands; C^N = ppy (2-phenylpyridinate) (Series A), or dfppy (2-(2,4-difluorophenyl)pyridinate) (Series B)) has been explored and two of them have shown anticancer activity under irradiation by inducing apoptosis through a multimodal mechanism of action that involves photocatalytic oxidation of mtDNA and mitochondrial membrane depolarization (Echevarría et al., 2022). Li et al. (Li, Wu, et al., 2020) developed metallohelices that were fabricated using the dynamic imine-coupling chemistry between aldehyde end-capped fac-Ir (ppy)3 handles and linear alkanediamine spacers, showing stronger DNA-binding affinities in a minor groove manner and better ability of inducing cell apoptosis under white light irradiation. These complexes also show their potential as PDT agents for cancer treatment. Additionally, cyclometalated Ir(III) complexes Ir3 and Ir4 with dipyrido [3,2- a:2′,3′- c]phenazine (dppz) ligands have shown their affinity to mtDNA and effects to damage mtDNA (Cao et al., 2019).
4.2.3. Combined with phototherapy
Phototherapy is an emerging strategy for cancer treatment that uses light to activate drugs or materials that can kill cancer cells by targeting mtDNA, which is essential for their survival and proliferation. PDT, photothermal therapy (PTT) and photochemotherapy (PCT) are three types of phototherapeutic methods that have shown promising results in recent studies. Metal complexes and DLC-based systems often intersected with the application of Phototherapy due to their unique optical and chemical properties (Hu et al., 2017; Li, Wu, et al., 2020; Wang et al., 2018). Using photosensitizers that produce ROS when exposed to light, PDT is a non-invasive modality that causes oxidative damage to the cancer cells (Shi & Sadler, 2020). And PDT offers several benefits, such as light-controlled treatment area, minimal invasiveness, low side effects and negligible drug resistance (Liu, Wang, et al., 2022; Shen et al., 2022). The mitochondria-targeted NIR photosensitizer, IR780, are widely used in the phototherapy. A multifunctional nanomedicine IR780@Pt NPs that was constructed by Zhang et al. through a supramolecular self-assembly strategy (Zhang et al., 2021). It can sufficiently induce mitochondrial dysfunction under NIR light irradiation through both photothermal and photodynamic effects, inhibiting the nucleotide excision repair pathway for the repair of DNA-Pt caused by Pt. Besides, the possibility of PDT as an adjunct to DLC-based methods has been demonstrated by Yang et al. (Yang et al., 2017) PTT that uses NIR absorbents for facilitating efficient heat production and induces cancer cell death with NIR irradiation (Nomura et al., 2020). Wang et al. (Wang et al., 2018) constructed a complex nanoparticle which contained a core-shell-SS-shell architecture with a core of Fe–O colloidal nanocrystal clusters, an inner shell of polydopamine (PDA) functionalized with TPP and an outer shell of methoxy poly (ethylene glycol) linked to the PDA by disulfide bonds. This nanoparticle can generate photothermal effect from the PDA under NIR light, ending in dramatic decreasing in MMP and Dox-mediated damage of mtDNA. And Yang et al. (Yang et al., 2020) precisely assembled IR780 with a biotin-labeled Pt (IV) prodrug derivative at a 1:1 molecular ratio, achieving mitochondria-targeted chemo-photothermal synergistic therapy under NIR irradiation in A549R tumors and almost completely eradicating the tumors. In addition, the integration of PDT and PTT has been recognized as a potential approach for achieving complete tumor ablation therapy, showing excellent tumor treatment effect (Chen et al., 2021; Li et al., 2019). PCT is a type of therapy that uses light and a photosensitizer to produce cytotoxic oxygen species within the tumors (Bossu et al., 1999). A PCT strategy of cancer therapy using NIR-assisted tumor-specific Fenton reactions has been proposed. In this study, the PCT agent, which consisted of three key components including upconversion nanoparticles core based on lanthanide-doped nanocrystals (convert NIR light to UV or visible photons to catalyze photo-Fenton reaction), mesoporous silica shell coat (load and deliver Fe2+) and Ru2+ complex on the surface of mesoporous silica shell (bind mtDNA), showed much enhanced and tumor-specific therapeutic efficacy in HegG2 cells (Hu et al., 2017). As we can see, phototherapy techniques have various forms and applications that can produce different therapeutic effects, and are highly effective in conjunction with mtDNA-targeted chemotherapy in cancer treatment.
5. Discussion
mtDNA has emerged as a target of cancer therapy. The alteration of mtDNA is frequently found in a variety of cancers and acts as inducement, biomarker or subsequent phenomenon in the process of tumor development, which underscores its irreplaceable position in cancer treatment. Further studying the functions of genes and encoded proteins in mtDNA is the key to elucidating mtDNA-related diseases. Mok et al. (Mok et al., 2020) have developed a new tool to manipulate mtDNA using RNA-free DddA (a double-stranded DNA deaminase) derived cytidine base editors (DdCBEs) to introduce targeted base conversions, C•G to T•A. This tool employed CRISPR-free mitochondrial base editing to precisely manipulate mtDNA and mimic disease-related mtDNA mutations. It has also been validated in five mtDNA genes (MT-ND1, MT-ND2, MT-ND4, MT-ND5 and MT-ATP8), which were stable without mtDNA deletions or copy number loss. In addition, by combining DdCBE with Cre/loxP system, Tan et al. (Tan et al., 2023) established a cell and rat resource library of mtDNA-encoded mitochondrial proteins (mt-Proteins) depletion, which can be used to study the biological functions and disease mechanisms of mt-Proteins. It provided a powerful and versatile platform for advancing mitochondrial research and therapy.
mtDNA plays a crucial role as a messenger in biological reactions. Understanding the response after mtDNA damage has important implications for drug efficacy appraisal, adverse reaction evaluation, and patient prognosis assessment. Rho0 cancer cells, which lacking mtDNA, have been frequently used in mtDNA-related studies (Crider et al., 2012; Guerrero-Castillo et al., 2021; Kuwahara et al., 2016). Rho0 cells can be obtained by treating with low concentration of ethidium bromide, which doesn't affect nDNA synthesis (Kuwahara et al., 2016). Related researches involve cancer treatment mechanisms, cancer drug resistance, factors affecting tumor progression, TME, etc., with a wide range, showing its important role in mitochondrial researches (Gong et al., 2018; Takenaga et al., 2021; Yi et al., 2015). More information about the rho0-related studies in recent years were summarized in Table S1. Accurately describing the mechanisms of mtDNA in intracellular environment, cell communication and tumor progression is decisive in the development of mtDNA-targeted therapy, which require further investigation in future studies.
The stability and integrity of mtDNA are important for cancer targeted therapy. One of the main mechanisms of achieving favorable therapeutic outcomes by mtDNA targeting is to disrupt the stability and integrity of mtDNA. Various targeting strategies can be employed to achieve this goal, depending on the initiating or rescuing factors involved. For the initiating factors, the integrity of mtDNA can be compromised by enhancing mitochondrial oxidative stress, or by utilizing mitochondrial-targeted drug delivery systems that enhance the accumulation of DNA-damaging agents in mitochondria. For instance, nanocarriers have emerged as a promising tool for mtDNA targeting, as they offer an optimal platform for drug delivery and facilitate mitochondrial accumulation. They can physically encapsulate insoluble drugs to achieve controlled drug release, ultimately improving drug metabolism and reducing systemic drug toxicity (Luo et al., 2022). Moreover, other molecule-based, polymer-based, lipid-based and peptide-based nano-carrier systems have also been applied in this field, contributing to the advancement of cancer therapy. The development of effective mtDNA targeting delivery strategies remains a research hotspot for mtDNA-targeted cancer therapy. For the rescuing factors, cells have evolved several mtDNA repair pathways to prevent or repair mtDNA damage, including direct reversal, base excision repair, mismatch repair, translesion synthesis, and DSB repair (Li, Guan, et al., 2020). These pathways can restore the integrity and stability of mtDNA by removing the lesions or rejoining the broken strands. Inhibiting mtDNA repair pathways can increase the sensitivity of cancer cells to toxic agents, leading to an accumulation of more mtDNA mutations and a loss of mitochondrial functions (Xie et al., 2015). This mechanism has been explored by some existing studies we discussed above.
Depending on the treatment approaches, we can categorize the anti-cancer therapeutic strategies targeting mtDNA into “elimination strategies” (which disrupt the stability and integrity of mtDNA) and “rescue strategies” (which restore mitochondrial function through gene editing, such as providing normal mtDNA or repairing mutated mtDNA). However, most of the current mtDNA-targeted anti-cancer research focused on the “elimination strategies”, while the development of the “rescue strategies” has not been extensively explored. Gene therapy requires a nucleic acid targeting delivery system, which has been proven effective in treating some diseases (Yu et al., 2013). Yoshinaga and Numata conducted a comprehensive review of the rapidly evolving mitochondrial-targeted nucleic acid delivery vectors in recent years, categorizing them into four groups: mitochondrial targeting signal (MTS) peptides, systems based on lipophilic cations, systems based on mitochondrial fusion lipids, and physical delivery methods (Yoshinaga & Numata, 2022). Among them, the system based on MTS peptides is one of the most powerful tools. MTS peptide is a short amino acid sequence that is usually located at the N-terminus of the protein and consists of alternating hydrophobic and positively charged amino acids (Bykov et al., 2022). It has been widely employed by researchers for plasmid DNA delivery and viral gene delivery targeting mitochondria (Yoshinaga & Numata, 2022). In mitochondrial gene therapy of cancer cells, Faria et al. (Faria et al., 2022) constructed MTS-CPP/pDNA complexes and achieved promising outcomes, highlighting the potential of MTS peptide system in targeted treatment of cancer through gene therapy. In addition to the MTS peptide system, Yamada et al. (Yamada et al., 2012) designed a liposome-based carrier DF-MITO-Porter, which can deliver macromolecules more efficiently to mitochondria in cancer cells. Nevertheless, the progress towards developing mtDNA “rescue strategies” that have practical therapeutic value and clinical significance remains relatively sluggish. In the future, multi-functional carriers should be developed to enhance their capabilities in terms of mitochondrial localization, mitochondrial penetration, gene-specific editing and damage repair. And more nucleic acid delivery tools should be utilized in practical mitochondrial gene therapy experiments.
Therapeutic strategies that target mtDNA have considerable significance for anti-cancer drug research and clinical treatment. Targeting mtDNA can help overcome tumor drug resistance and complete elimination of tumors. This is because the acquisition of some biological characteristics of tumors are related to mtDNA. For example, mutations in a gene called MT-ND4 can make tumor cells resistant to cisplatin, a drug that damages DNA by forming bonds with it (Abedi et al., 2020; Alam et al., 2017; van Gisbergen et al., 2015). Cisplatin bonds are harder to removal from mtDNA than from nDNA, because mtDNA lacking a repair system called nucleotide excision repair. Therefore, it would be effective to direct cisplatin or similar compounds (Pt complexes) to the mitochondria using a carrier molecule such as TPP (Muhammad et al., 2021). At the same time, IR780 overcomes cisplatin resistance to improve the therapeutic efficacy via increasing the Pt accumulation in mitochondria and combining with phototherapy as a mitochondria-targeting phototherapy agent (Ma et al., 2019; Yang et al., 2020). Another considerable significance of targeting mtDNA lies in the close involvement of mtDNA in the disease process in clinical diseases. The presence of mtDNA has been observed to have a significant impact on a variety of clinical diseases. Mutations in any of the 37 genes encoded by mtDNA can cause mitochondrial dysfunction and disease, including cancer (Möllers et al., 2005). And the balance between mutant and WT mtDNA determines the process of mitochondrial diseases with clinical phenotypes (Lee, Lee, et al., 2021). Therefore, targeting mtDNA can be a promising candidate in drug-resistant cancer treatment and provide a wider range of strategies for clinical treatment.
However, the development and application of mtDNA-targeting drugs for anti-cancer therapy face many challenges. (i) The association between mtDNA mutations and clinical characteristics of tumors requires further elucidation. It is essential to identify the causal relationship between mtDNA alterations (including mtDNA damage, mtDNA mutation and mtDNA heteroplasmy levels) and tumor progression (including tumor occurrence, malignant transformation, tumor metastasis, tumor resistance and clinical heterogeneity of cancer patients), rather than solely simple correlations. This is of utmost importance in utilizing mtDNA as a biomarker for clinical evaluation of tumors and as a preferred target for tumor therapy. (ii) Most of the studies are still limited to validation in animal models. Only a few mtDNA-targeting drugs are in clinical trials for anti-cancer therapy, although there are already many drugs for mitochondrial therapy (Singh et al., 2021). A clinical trial of mitochondria-targeted tamoxifen (MitoTam) has been reported. MitoTam is a TPP-conjugated drug that can localize to mitochondria and induce ROS accumulation, mtDNA damage and cell injury in cancer treatment. In a clinical trial sponsored by the University of Pisa, Italy, MitoTam has achieved satisfactory results in a phase I trial and is currently being extended to a phase II trial (Dong et al., 2020). However, most of the existing mtDNA-targeting drugs were originally designed for other purposes, such as neurodegenerative diseases, viral infections, or genetic disorders (Rowe et al., 2001; Xu et al., 2022). Phototherapy may be able to achieve a faster translation to clinical application, as there are many photosensitizers for PDT that have entered clinical use or clinical trials, which have been summarized by Shi et al. (Shi & Sadler, 2020) How to promote the validation of mtDNA-targeting anticancer drugs in clinical trials is the key to mtDNA-targeted therapy. (iii) Most therapeutic strategies that activate immunity by targeting mtDNA rely on the cGAS-STING pathway to sense dsDNA. The inhibition of cGAS activity would severely influence the efficacy of these strategies. Several factors can influence the dsDNA recognition by cGAS, including its transcriptional and post-transcriptional regulation, post-translational modifications, genetic mutations of cGAS-binding proteins and drug effects in patients with autoimmune diseases (Skopelja-Gardner et al., 2022; Yu & Liu, 2021). The combination of multiple therapies may overcome this challenge. (iv) As a promising option for combination therapy, phototherapy faces many challenges that need to be addressed. The widespread application of phototherapy requires overcoming some difficulties, such as the low ability of light to reach deep tumors, the abnormal antioxidant mechanisms that may confer resistance of tumor cells to light damage and the safety of the drugs that enable phototherapy (Shi & Sadler, 2020).
6. Conclusion
mtDNA is a key factor in various clinical pathologies, especially cancers. As a trigger, there is a causal relationship between mtDNA alterations and tumor development. Targeting mtDNA has become a promising strategy for cancer therapy. The mechanisms of the responses after mtDNA damage are crucial for developing mtDNA-targeted drugs, predicting drug efficacy, and analyzing potential drug resistance. In recent years, mtDNA-targeted therapy has rapidly developed, including direct targeting of mtDNA and indirect targeting of mtDNA through intermediate mediators. These strategies have their own characteristics in tumor treatment. Despite the challenges and limitations of mtDNA-targeted research, such as the scarcity of clinical studies and the possible resistance, we have revealed the huge potential of mtDNA as a therapeutic target for cancer treatment. We hoped this study will offer insights for future research and development of novel mtDNA-targeting drugs and strategies.
Author contributions
Yumeng Lin: Conceptualization, Investigation, Methodology, Project administration and Roles/Writing - original draft; Bowen Yang: Conceptualization and Writing - review & editing; Yibo Huang: Conceptualization and Writing - review & editing; You Zhang: Conceptualization and Writing - review & editing; Yu Jiang: Investigation and Writing - review & editing; Longyun Ma: Writing - review & editing; Ying-Qiang Shen: Conceptualization, Funding acquisition, Project administration and Writing - review & editing.
Conflict of interest
The authors declare no competing interests.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
☐ The authors declare the following financial interests/personal relationships which may be considered as potential competing interests.
Acknowledgements:
This work was supported by the Natural Science Foundation of China (81902784) and the fund of the Sichuan Provincial Department of science and technology (2022YFSY0058).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cellin.2023.100113.
Contributor Information
Yumeng Lin, Email: 1460193021@qq.com.
Bowen Yang, Email: 516084218@qq.com.
Yibo Huang, Email: refined_fish@foxmail.com.
You Zhang, Email: 2015181641028@stu.scu.edu.cn.
Yu Jiang, Email: 2399818927@qq.com.
Longyun Ma, Email: 1014510106@qq.com.
Ying-Qiang Shen, Email: shen@scu.edu.cn.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- Abd Radzak S.M., Mohd Khair S.Z.N., Ahmad F., Patar A., Idris Z., Mohamed Yusoff A.A. Insights regarding mitochondrial DNA copy number alterations in human cancer. International Journal of Molecular Medicine. 2022;50 doi: 10.3892/ijmm.2022.5160. (Review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abedi S., Yung G., Atilano S.R., Thaker K., Chang S., Chwa M.…Kenney M.C. Differential effects of cisplatin on cybrid cells with varying mitochondrial DNA haplogroups. PeerJ. 2020;8 doi: 10.7717/peerj.9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S., Wallrabe H., Svindrych Z., Chaudhary A., Christopher K., Chandra D., Periasamy A. Investigation of mitochondrial metabolic response to doxorubicin in prostate cancer cells: An NADH, FAD and tryptophan FLIM assay. Scientific Reports. 2017;7 doi: 10.1038/s41598-017-10856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsina D., Lytovchenko O., Schab A., Atanassov I., Schober F.A., Jiang M.…Larsson N.G. FBXL4 deficiency increases mitochondrial removal by autophagy. EMBO Molecular Medicine. 2020;12 doi: 10.15252/emmm.201911659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amari L., Germain M. Mitochondrial extracellular vesicles - origins and roles. Frontiers in Molecular Neuroscience. 2021;14 doi: 10.3389/fnmol.2021.767219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminuddin A., Ng P., Leong C., Chua E. Mitochondrial DNA alterations may influence the cisplatin responsiveness of oral squamous cell carcinoma. Scientific Reports. 2020;10:7885. doi: 10.1038/s41598-020-64664-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo L.F., Siena A.D.D., Plaça J.R., Brotto D.B., Barros, Muys B.R.…Silva W.A., Jr. Mitochondrial transcription factor A (TFAM) shapes metabolic and invasion gene signatures in melanoma. Scientific Reports. 2018;8:14190. doi: 10.1038/s41598-018-31170-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arévalo J., Amorim J. Virtual screening, optimization and molecular dynamics analyses highlighting a pyrrolo[1,2-a]quinazoline derivative as a potential inhibitor of DNA gyrase B of Mycobacterium tuberculosis. Scientific Reports. 2022;12:4742. doi: 10.1038/s41598-022-08359-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrinath N., Yoo S.Y. Mitochondria in cancer: In the aspects of tumorigenesis and targeted therapy. Carcinogenesis. 2018;39:1419–1430. doi: 10.1093/carcin/bgy148. [DOI] [PubMed] [Google Scholar]
- Bahat A., MacVicar T., Langer T. Metabolism and innate immunity meet at the mitochondria. Frontiers in Cell and Developmental Biology. 2021;9 doi: 10.3389/fcell.2021.720490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahat A., MacVicar T., Langer T. Metabolism and innate immunity meet at the mitochondria. Frontiers in Cell and Developmental Biology. 2021;9 doi: 10.3389/fcell.2021.720490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J., Liu F. The cGAS-cGAMP-STING pathway: A molecular link between immunity and metabolism. Diabetes. 2019;68:1099–1108. doi: 10.2337/dbi18-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajpai A., Desai N., Pandey S., Shukla C., Datta B., Basu S. Chimeric nanoparticles for targeting mitochondria in cancer cells. Nanoscale Advances. 2022;4:1112–1118. doi: 10.1039/d1na00644d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajpai A., Desai N.N., Pandey S., Shukla C., Datta B., Basu S. Chimeric nanoparticles for targeting mitochondria in cancer cells. Nanoscale Advances. 2022;4:1112–1118. doi: 10.1039/d1na00644d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti S., Catalani S., Canonico B., Nasoni M., Luchetti F., Papa S., Potenza L., Palma F. The effects of Acyclovir administration to NCI-H1975 non-small cell lung cancer cells. Toxicology in Vitro : An International Journal Published in Association with BIBRA. 2022;79 doi: 10.1016/j.tiv.2021.105301. [DOI] [PubMed] [Google Scholar]
- Bennett C., Latorre-Muro P., Puigserver P. Mechanisms of mitochondrial respiratory adaptation. Nature Reviews Molecular Cell Biology. 2022;23:817–835. doi: 10.1038/s41580-022-00506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeaud M., Mathieu L., Guillaume A., Moll U.M., Mignotte B., Le Floch N., Vayssière J.L., Rincheval V. Mitochondrial p53 mediates a transcription-independent regulation of cell respiration and interacts with the mitochondrial F₁F0-ATP synthase. Cell Cycle. 2013;12:2781–2793. doi: 10.4161/cc.25870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge M.V., Dong L., Neuzil J. Mitochondrial DNA in tumor initiation, progression, and metastasis: Role of horizontal mtDNA transfer. Cancer Research. 2015;75:3203–3208. doi: 10.1158/0008-5472.CAN-15-0859. [DOI] [PubMed] [Google Scholar]
- Boguszewska K., Szewczuk M., Kaźmierczak-Barańska J., Karwowski B. The similarities between human mitochondria and bacteria in the context of structure, genome, and base excision repair system. Molecules. 2020;Vol. 25 doi: 10.3390/molecules25122857. Basel, Switzerland) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonekamp N.A., Peter B., Hillen H.S., Felser A., Bergbrede T., Choidas A.…Nussbaumer P., et al. Small-molecule inhibitors of human mitochondrial DNA transcription. Nature. 2020;588:712–716. doi: 10.1038/s41586-020-03048-z. [DOI] [PubMed] [Google Scholar]
- Bossu E., Padilla J.J., A'Amar O., Parache R.M., Notter D., Vigneron C., Guillemin F. Determination of endogenous porphyrins and the maximal HpD tumor/normal skin ratio in SKH-1 hairless mice by light induced fluorescence spectroscopy. Artificial Cells Blood Substitutes and Immobilization Biotechnology. 1999;27:109–117. doi: 10.3109/10731199909117686. [DOI] [PubMed] [Google Scholar]
- Brito A., Merle C., Lagarde P., Faustin B., Devin A., Lartigue L., Chibon F. Cell fusion enhances energy metabolism of mesenchymal tumor hybrid cells to sustain their proliferation and invasion. BMC Cancer. 2021;21:863. doi: 10.1186/s12885-021-08561-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykov Y.S., Flohr T., Boos F., Zung N., Herrmann J.M., Schuldiner M. Widespread use of unconventional targeting signals in mitochondrial ribosome proteins. EMBO Journal. 2022;41 doi: 10.15252/embj.2021109519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Zheng Y., Wu X., Tan C., Chen M., Wu N., Ji L., Mao Z. Anticancer cyclometalated iridium(III) complexes with planar ligands: Mitochondrial DNA damage and metabolism disturbance. Journal of Medicinal Chemistry. 2019;62:3311–3322. doi: 10.1021/acs.jmedchem.8b01704. [DOI] [PubMed] [Google Scholar]
- Carew J.S., Huang P. Mitochondrial defects in cancer. Molecular Cancer. 2002;1:9. doi: 10.1186/1476-4598-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D., Das P., Chakrabarti O. Mitochondrial epigenetics regulating inflammation in cancer and aging. Frontiers in Cell and Developmental Biology. 2022;10 doi: 10.3389/fcell.2022.929708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Cao J., Zhang K., Zhang Y.N., Lu J., Zubair Iqbal M., Zhang Q., Kong X. Synergistic photodynamic and photothermal therapy of BODIPY-conjugated hyaluronic acid nanoparticles. Journal of Biomaterials Science, Polymer Edition. 2021;32:2028–2045. doi: 10.1080/09205063.2021.1954138. [DOI] [PubMed] [Google Scholar]
- Chen J., Qiao K., Zhang C., Zhou X., Du Q., Deng Y., Cao L. VRK2 activates TNFα/NF-κB signaling by phosphorylating IKKβ in pancreatic cancer. International Journal of Biological Sciences. 2022;18:1288–1302. doi: 10.7150/ijbs.66313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Tu X., Xie L., Xiong L., Song J., Ye X. Peptide nucleic acids targeting mitochondria enhances sensitivity of lung cancer cells to chemotherapy. American Journal of Tourism Research. 2018;10:2940–2948. [PMC free article] [PubMed] [Google Scholar]
- Chen W., Hu S., Mao S., Xu Y., Guo H., Li H.…Neamati N. Discovery of mitochondrial transcription inhibitors active in pancreatic cancer cells. ChemMedChem. 2020;15:2029–2039. doi: 10.1002/cmdc.202000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A., Cheng L., Kuo C., Lo Y., Chou H., Chen C., Wang Y., Chuang T., Cheng S., Lee A. Mitochondrial Lon-induced mtDNA leakage contributes to PD-L1-mediated immunoescape via STING-IFN signaling and extracellular vesicles. Journal for immunotherapy of cancer. 2020;8 doi: 10.1136/jitc-2020-001372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew K., Zhao L. Interactions of mitochondrial transcription factor A with DNA damage: Mechanistic insights and functional implications. Genes. 2021;12 doi: 10.3390/genes12081246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Kim H., Lee S., Lee S., Kim B. viaA telomerase-derived peptide exerts an anti-hepatitis B virus effect mitochondrial DNA stress-dependent type I interferon production. Frontiers in Immunology. 2020;11:652. doi: 10.3389/fimmu.2020.00652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline S.D. Mitochondrial DNA damage and its consequences for mitochondrial gene expression. Biochimica et Biophysica Acta. 2012;1819:979–991. doi: 10.1016/j.bbagrm.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch F.J., Kuchenbaecker K.B., Michailidou K., Mendoza-Fandino G.A., Nord S., Lilyquist J.…Barkardottir R.B., et al. Identification of four novel susceptibility loci for oestrogen receptor negative breast cancer. Nature Communications. 2016;7:11375. doi: 10.1038/ncomms11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crider D.G., Garcia-Rodriguez L.J., Srivastava P., Peraza-Reyes L., Upadhyaya K., Boldogh I.R., Pon L.A. Rad53 is essential for a mitochondrial DNA inheritance checkpoint regulating G1 to S progression. The Journal of Cell Biology. 2012;198:793–798. doi: 10.1083/jcb.201205193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushen S., Sprouse M., Blessing A., Sun J., Jarvis S., Okada Y.…Goulopoulou S. Cell-free mitochondrial DNA increases in maternal circulation during healthy pregnancy: A prospective, longitudinal study. American Journal of Physiology - Regulatory, Integrative and Comparative Physiology. 2020;318:R445–R452. doi: 10.1152/ajpregu.00324.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagar S., Pathak D., Oza H.V., Mylavarapu S.V.S. Tunneling nanotubes and related structures: Molecular mechanisms of formation and function. Biochemical Journal. 2021;478:3977–3998. doi: 10.1042/BCJ20210077. [DOI] [PubMed] [Google Scholar]
- De Gaetano A., Solodka K., Zanini G., Selleri V., Mattioli A.V., Nasi M., Pinti M. Molecular mechanisms of mtDNA-mediated inflammation. Cells. 2021:10. doi: 10.3390/cells10112898. [Online] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C.F., Ricketts C.J., Wang M., Yang L., Cherniack A.D., Shen H.…Bristow C.A., et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26:319–330. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De A., Campbell C. A novel interaction between DNA ligase III and DNA polymerase gamma plays an essential role in mitochondrial DNA stability. Biochemical Journal. 2007;402:175–186. doi: 10.1042/BJ20061004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Gopalan V., Holland O., Neuzil J. Mitocans revisited: Mitochondrial targeting as efficient anti-cancer therapy. International Journal of Molecular Sciences. 2020;21 doi: 10.3390/ijms21217941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois F., Bénard M., Jean-Jacques B., Schapman D., Roberge H., Lebon A.…Galas L. Investigating tunneling nanotubes in cancer cells: Guidelines for structural and functional studies through cell imaging. BioMed Research International. 2020;2020 doi: 10.1155/2020/2701345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echevarría I., Zafon E., Barrabés S., Martínez M., Ramos-Gómez S., Ortega N.…Massaguer A. Rational design of mitochondria targeted thiabendazole-based Ir(III) biscyclometalated complexes for a multimodal photodynamic therapy of cancer. Journal of Inorganic Biochemistry. 2022;231 doi: 10.1016/j.jinorgbio.2022.111790. [DOI] [PubMed] [Google Scholar]
- Ed R., Martin L.M., Yasin S., Nadeem R., William L., Chris S. Mitochondrial DNA copy number variation across human cancers. bioRxiv. 2015 [Google Scholar]
- Fairbrother-Browne A., Ali A.T., Reynolds R.H., Garcia-Ruiz S., Zhang D., Chen Z., Ryten M., Hodgkinson A. Mitochondrial-nuclear cross-talk in the human brain is modulated by cell type and perturbed in neurodegenerative disease. Communications Biology. 2021;4:1262. doi: 10.1038/s42003-021-02792-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Zhou S., Zheng M., Deng X., Yi Y., Huang T. MiR-199a-3p enhances breast cancer cell sensitivity to cisplatin by downregulating TFAM (TFAM) Biomedicine & Pharmacotherapy = Biomedecine & pharmacotherapie. 2017;88:507–514. doi: 10.1016/j.biopha.2017.01.058. [DOI] [PubMed] [Google Scholar]
- Faria R., Paul M., Biswas S., Vivès E., Boisguérin P., Sousa Â., Costa D. Peptides vs. Polymers: Searching for the most efficient delivery system for mitochondrial gene therapy. Pharmaceutics. 2022;14 doi: 10.3390/pharmaceutics14040757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Xiong L., Ji Z., Cheng W., Yang H. Correlation between increased ND2 expression and demethylated displacement loop of mtDNA in colorectal cancer. Molecular Medicine Reports. 2012;6:125–130. doi: 10.3892/mmr.2012.870. [DOI] [PubMed] [Google Scholar]
- Finsterer J., Zarrouk-Mahjoub S. Phenotypic spectrum of SLC25A4 mutations. Biomed Rep. 2018;9:119–122. doi: 10.3892/br.2018.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco D., Moretti I., Marie S. Mitochondria transcription factor A: A putative target for the effect of melatonin on U87MG malignant glioma cell line. Molecules (Basel, Switzerland) 2018 doi: 10.3390/molecules23051129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M.H., Chen I.C., Lee C.H., Wu C.W., Lee Y.C., Kung Y.C., Hung C.Y., Wu K.L.H. Anti-neuroinflammation ameliorates systemic inflammation-induced mitochondrial DNA impairment in the nucleus of the solitary tract and cardiovascular reflex dysfunction. Journal of Neuroinflammation. 2019;16:224. doi: 10.1186/s12974-019-1623-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z., Dean J.W., Xiong L., Dougherty M.W., Oliff K.N., Chen Z.E., Jobin C., Garrett T.J., Zhou L. Mitochondrial transcription factor A in RORγt(+) lymphocytes regulate small intestine homeostasis and metabolism. Nature Communications. 2021;12:4462. doi: 10.1038/s41467-021-24755-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaziev A.I., Shaikhaev G.O. Nuclear mitochondrial pseudogenes. Molecular Biology. 2010;44:358–368. [PubMed] [Google Scholar]
- Geden M.J., Romero S.E., Deshmukh M. p53 is required for nuclear but not mitochondrial DNA damage-induced degeneration. Cell Death & Disease. 2021;12:104. doi: 10.1038/s41419-020-03373-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X., Dan J., Li F., Wang L. Suppression of mitochondrial respiration with local anesthetic ropivacaine targets breast cancer cells. Journal of Thoracic Disease. 2018;10:2804–2812. doi: 10.21037/jtd.2018.05.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso C., Eccles D.A., Boukalova S., Fabre M.S., Dawson R.H., Neuzil J., Herst P.M., Berridge M.V. Mitochondrial DNA affects the expression of nuclear genes involved in immune and stress responses in a breast cancer model. Frontiers in Physiology. 2020;11 doi: 10.3389/fphys.2020.543962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra F., Arbini A.A., Moro L. Mitochondria and cancer chemoresistance. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2017;1858:686–699. doi: 10.1016/j.bbabio.2017.01.012. [DOI] [PubMed] [Google Scholar]
- Guerrero-Castillo S., van Strien J., Brandt U., Arnold S. Ablation of mitochondrial DNA results in widespread remodeling of the mitochondrial complexome. The EMBO Journal. 2021;40 doi: 10.15252/embj.2021108648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Li Z., Huang H., Xu Z., Chen Z., Shen G.…Hu Y. VB12-Sericin-PBLG-IR780 nanomicelles for programming cell pyroptosis via photothermal (PTT)/Photodynamic (PDT) effect-induced mitochondrial DNA (mitoDNA) oxidative damage. ACS Applied Materials and Interfaces. 2022;14:17008–17021. doi: 10.1021/acsami.1c22804. [DOI] [PubMed] [Google Scholar]
- Haja Mohideen A.M.S., Dicks E., Parfrey P., Green R., Savas S. Mitochondrial DNA polymorphisms, its copy number change and outcome in colorectal cancer. BMC Research Notes. 2015;8:272. doi: 10.1186/s13104-015-1250-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Robertson A., Hill L., Rendina L. Synthesis and tumour cell uptake studies of gadolinium(III)-phosphonium complexes. Scientific Reports. 2021;11:598. doi: 10.1038/s41598-020-79893-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Zhao Z., Zhang M., Li G., Yang C., Du F.…Sun Y. Maternal trans-general analysis of the human mitochondrial DNA pattern. Biochemical and Biophysical Research Communications. 2017;493:643–649. doi: 10.1016/j.bbrc.2017.08.138. [DOI] [PubMed] [Google Scholar]
- Han Y.S., Yi E.Y., Jegal M.E., Kim Y.J. Cancer stem-like phenotype of mitochondria dysfunctional Hep3B hepatocellular carcinoma cell line. Cells. 2021:10. doi: 10.3390/cells10071608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W.R., Cao L.B., Yang Y.L., Hua D., Hu M.M., Shu H.B. VRK2 is involved in the innate antiviral response by promoting mitostress-induced mtDNA release. Cell Molecular Immunology. 2021;18:1186–1196. doi: 10.1038/s41423-021-00673-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen H.S., Morozov Y.I., Sarfallah A., Temiakov D., Cramer P. Structural basis of mitochondrial transcription initiation. Cell. 2017;171:1072–1081.e10. doi: 10.1016/j.cell.2017.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X.S., Wang H.S., Mugaka B.P., Yang G.J., Ding Y. Mitochondria: Promising organelle targets for cancer diagnosis and treatment. Biomaterials Science. 2018;6:2786–2797. doi: 10.1039/c8bm00673c. [DOI] [PubMed] [Google Scholar]
- Huertas J., Al Fazazi S., Hidalgo-Gutierrez A., López L., Casuso R. Antioxidant effect of exercise: Exploring the role of the mitochondrial complex I superassembly. Redox Biology. 2017;13:477–481. doi: 10.1016/j.redox.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P., Wu T., Fan W., Chen L., Liu Y., Ni D., Bu W., Shi J. Near infrared-assisted Fenton reaction for tumor-specific and mitochondrial DNA-targeted photochemotherapy. Biomaterials. 2017;141:86–95. doi: 10.1016/j.biomaterials.2017.06.035. [DOI] [PubMed] [Google Scholar]
- Hu L., Yao X., Shen Y. Altered mitochondrial DNA copy number contributes to human cancer risk: Evidence from an updated meta-analysis. Scientific Reports. 2016;6 doi: 10.1038/srep35859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Zhou M., Bao X., Pan D., Jiao M., Liu X., Li F., Li C. ATM inhibition enhances cancer immunotherapy by promoting mtDNA leakage and cGAS/STING activation. The Journal of Clinical Investigation. 2021:131. doi: 10.1172/JCI139333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamura A., Muraoka-Hirayama S., Sakurai K. Loss of mitochondrial DNA by gemcitabine triggers mitophagy and cell death. Biological and Pharmaceutical Bulletin. 2019;42:1977–1987. doi: 10.1248/bpb.b19-00312. [DOI] [PubMed] [Google Scholar]
- Ippolito L., Morandi A., Taddei M.L., Parri M., Comito G., Iscaro A.…Giannoni E. Cancer-associated fibroblasts promote prostate cancer malignancy via metabolic rewiring and mitochondrial transfer. Oncogene. 2019;38:5339–5355. doi: 10.1038/s41388-019-0805-7. [DOI] [PubMed] [Google Scholar]
- Iske J., Seyda M., Heinbokel T., Maenosono R., Minami K., Nian Y.…Tullius S. Senolytics prevent mt-DNA-induced inflammation and promote the survival of aged organs following transplantation. Nature Communications. 2020;11:4289. doi: 10.1038/s41467-020-18039-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Guo Y., Wei C., Hu P., Shi J. Nanocatalytic innate immunity activation by mitochondrial DNA oxidative damage for tumor-specific therapy. Advanced Materials. 2021;33 doi: 10.1002/adma.202008065. [DOI] [PubMed] [Google Scholar]
- Jiang X., Wang J. Down-regulation of TFAM increases the sensitivity of tumour cells to radiation via p53/TIGAR signalling pathway. Journal of Cellular and Molecular Medicine. 2019;23:4545–4558. doi: 10.1111/jcmm.14350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Zhou S., Zhang X., Li C., Ji S., Mao H., Jiang X. Mitochondrion-specific dendritic lipopeptide liposomes for targeted sub-cellular delivery. Nature Communications. 2021;12:2390. doi: 10.1038/s41467-021-22594-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Y.S., Alexandrov L.B., Gerstung M., Martincorena I., Nik-Zainal S., Ramakrishna M.…Green A.R., et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. Elife. 2014;3 doi: 10.7554/eLife.02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Gupta R., Blanco L., Yang S., Shteinfer-Kuzmine A., Wang K., Zhu J., Yoon H., Wang X., Kerkhofs M., Kang H., Brown A., Park S., Xu X., Zandee van Rilland E., Kim M., Cohen J., Kaplan M., Shoshan-Barmatz V., Chung J. VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science (New York, N.Y.) 2019;366:1531–1536. doi: 10.1126/science.aav4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.S., Chuenchor W., Chen X., Cui Y., Zhang X., Zhou Z.H., Gellert M., Yang W. Cracking the DNA code for V(D)J recombination. Molecules and Cells. 2018;70:358–370.e4. doi: 10.1016/j.molcel.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Mahmood M., Reznik E., Gammage P. Mitochondrial DNA is a major source of driver mutations in cancer. Trends in cancer. 2022;8:1046–1059. doi: 10.1016/j.trecan.2022.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam C., Yi J., Xiao Y., Dhakal K., Zhang L., Li X.…Zhou J. Absence of physiological Ca transients is an initial trigger for mitochondrial dysfunction in skeletal muscle following denervation. Skeletal Muscle. 2017;7:6. doi: 10.1186/s13395-017-0123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.K., Noh Y.H., Nilius B., Ko K.S., Rhee B.D., Kim N., Han J. Current and upcoming mitochondrial targets for cancer therapy. Seminars in Cancer Biology. 2017;47:154–167. doi: 10.1016/j.semcancer.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Koczor C.A., White R.C., Zhao P., Zhu L., Fields E., Lewis W. p53 and mitochondrial DNA: their role in mitochondrial homeostasis and toxicity of antiretrovirals. American Journal Of Pathology. 2012;180:2276–2283. doi: 10.1016/j.ajpath.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeck T., Olsson A.H., Nitert M.D., Sharoyko V.V., Ladenvall C., Kotova O.…Poulsen P., et al. A common variant in TFB1M is associated with reduced insulin secretion and increased future risk of type 2 diabetes. Cell Metab. 2011;13:80–91. doi: 10.1016/j.cmet.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Kong Q., Yan X., Cheng M., Jiang X., Xu L., Shen L., Yu H., Sun L. p62 promotes the mitochondrial localization of p53 through its UBA domain and participates in regulating the sensitivity of ovarian cancer cells to cisplatin. International Journal of Molecular Sciences. 2022;23 doi: 10.3390/ijms23063290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopinski P., Janssen K., Schaefer P., Trefely S., Perry C., Potluri P.…Wallace D. Regulation of nuclear epigenome by mitochondrial DNA heteroplasmy. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:16028–16035. doi: 10.1073/pnas.1906896116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshikawa N., Kida Y., Yasui N., Shinozaki Y., Tsuji K., Watanabe T.…Nagase H. A linear five-ring pyrrole-imidazole polyamide-triphenylphosphonium conjugate targeting a mitochondrial DNA mutation efficiently induces apoptosis of HeLa cybrid cells carrying the mutation. Biochemical and Biophysical Research Communications. 2021;576:93–99. doi: 10.1016/j.bbrc.2021.08.088. [DOI] [PubMed] [Google Scholar]
- Koshikawa N., Yasui N., Kida Y., Shinozaki Y., Tsuji K., Watanabe T., Takenaga K., Nagase H. A PI polyamide-TPP conjugate targeting a mtDNA mutation induces cell death of cancer cells with the mutation. Cancer Science. 2021;112:2504–2512. doi: 10.1111/cas.14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmider B., Lin C.R., Karim L., Tomar D., Vlasenko L., Marchetti N.…Bahmed K. Mitochondrial dysfunction in human primary alveolar type II cells in emphysema. EBioMedicine. 2019;46:305–316. doi: 10.1016/j.ebiom.2019.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozhukhar N., Alexeyev M.F. TFAM's contributions to mtDNA replication and OXPHOS biogenesis are genetically separable. Cells. 2022:11. doi: 10.3390/cells11233754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.-L., Ponneri Babuharisankar A., Lin Y.-C., Lien H.-W., Lo Y.K., Chou H.-Y.…Lee A. Y.-L. Mitochondrial oxidative stress in the tumor microenvironment and cancer immunoescape: Foe or friend? Journal of Biomedical Science. 2022;29:74. doi: 10.1186/s12929-022-00859-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara Y., Roudkenar M., Suzuki M., Urushihara Y., Fukumoto M., Saito Y., Fukumoto M. The involvement of mitochondrial membrane potential in cross-resistance between radiation and docetaxel. International Journal of Radiation Oncology, Biology, Physics. 2016;96:556–565. doi: 10.1016/j.ijrobp.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Laine-Menéndez S., Domínguez-González C., Blázquez A., Delmiro A., García-Consuegra I., Fernández-de la Torre M.…Morán M. Preferent diaphragmatic involvement in TK2 deficiency: An autopsy case study. International Journal of Molecular Sciences. 2021;22 doi: 10.3390/ijms22115598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Lee S., Baek G., Kim A., Kang B., Seo H., Kim J. Mitochondrial DNA editing in mice with DddA-TALE fusion deaminases. Nature Communications. 2021;12:1190. doi: 10.1038/s41467-021-21464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L., Guo M., Ai Y., Chen F., Zhang Y., Xia L.…Lu B. Tetramethylpyrazine blocks TFAM degradation and up-regulates mitochondrial DNA copy number by interacting with TFAM. Bioscience Reports. 2017:37. doi: 10.1042/BSR20170319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Nam J., Lee C., Park M., Yoo C., Rhee H., Seo J., Kwon T. Analysing the mechanism of mitochondrial oxidation-induced cell death using a multifunctional iridium(III) photosensitiser. Nature Communications. 2021;12:26. doi: 10.1038/s41467-020-20210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Paull T.T. Mitochondria at the crossroads of ATM-mediated stress signaling and regulation of reactive oxygen species. Redox Biology. 2020;32 doi: 10.1016/j.redox.2020.101511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang S., Wang B., Wei H., Liu X., Hao J.…Yan X. High-fat-diet impaired mitochondrial function of cumulus cells but improved the efficiency of parthenogenetic embryonic quality in mice. Animal Cells and Systems. 2018;22:243–252. doi: 10.1080/19768354.2018.1497707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.Y., Guan Y.D., Chen X.S., Yang J.M., Cheng Y. DNA repair pathways in cancer therapy and resistance. Frontiers in Pharmacology. 2020;11 doi: 10.3389/fphar.2020.629266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Sundquist K., Wang X., Zhang N., Hedelius A., Sundquist J., Memon A.A. Cancers [Online]; 2021. Association of mitochondrial DNA copy number and telomere length with prevalent and incident cancer and cancer mortality in women: A prospective Swedish population-based study; p. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Sun Y., Qi Z., Cao L., Ding S. Mitochondrial transfer/transplantation: An emerging therapeutic approach for multiple diseases. Cell & Bioscience. 2022;12:66. doi: 10.1186/s13578-022-00805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Tian J., Peng F., Wang J. Hypermethylation of mitochondrial DNA facilitates bone metastasis of renal cell carcinoma. Journal of Cancer. 2022;13:304–312. doi: 10.7150/jca.62278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Wang Q., Qiu W., Lyu Y., Zhu Z., Zhao X., Zhu W. AIE-active luminogens as highly efficient free-radical ROS photogenerator for image-guided photodynamic therapy. Chemical Science. 2022;13:3599–3608. doi: 10.1039/d2sc00067a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang F., Tong Y., Li L., Liu Y., Gao W. Pentamidine inhibits prostate cancer progression via selectively inducing mitochondrial DNA depletion and dysfunction. Cell Proliferation. 2020;53 doi: 10.1111/cpr.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Yang J., Luo L., Jiang M., Qin B., Yin H.…You J. Targeting photodynamic and photothermal therapy to the endoplasmic reticulum enhances immunogenic cancer cell death. Nature Communications. 2019;10:3349. doi: 10.1038/s41467-019-11269-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wu J., Wang L., He C., Chen L., Jiao Y., Duan C. Mitochondrial-DNA-targeted Ir -containing metallohelices with tunable photodynamic therapy efficacy in cancer cells. Angewandte Chemie (International ed. in English) 2020;59:6420–6427. doi: 10.1002/anie.201915281. [DOI] [PubMed] [Google Scholar]
- Li Q., Chen Q., Yang X., Zhang Y., Lv L., Zhang Z.…Fu B. Cocktail strategy based on a dual function nanoparticle and immune activator for effective tumor suppressive. Journal of Nanobiotechnology. 2022;20:84. doi: 10.1186/s12951-022-01241-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Jee B., Kwon S., Yoon Y., Xu W., Wang H.…Yoon G. Identification of a mitochondrial defect gene signature reveals NUPR1 as a key regulator of liver cancer progression. Hepatology (Baltimore, Md. 2015;62:1174–1189. doi: 10.1002/hep.27976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Zhang Y., Liu J., Kang R., Klionsky D., Tang D. Mitochondrial DNA stress triggers autophagy-dependent ferroptotic death. Autophagy. 2021;17:948–960. doi: 10.1080/15548627.2020.1739447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Sanchez M., Cipullo M., Gopalakrishna S., Khawaja A., Rorbach J. Methylation of ribosomal RNA: A mitochondrial perspective. Frontiers in Genetics. 2020;11:761. doi: 10.3389/fgene.2020.00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Chen X., Wang Q., Xu X., Xu B. TNFα promotes glioblastoma A172 cell mitochondrial apoptosis via augmenting mitochondrial fission and repression of MAPK-ERK-YAP signaling pathways. OncoTargets and Therapy. 2018;11:7213–7227. doi: 10.2147/OTT.S184337. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ludwig L., Roach L., Katz S., LaBelle J. Loss of BIM in T cells results in BCL-2 family BH3-member compensation but incomplete cell death sensitivity normalization. Apoptosis : An International Journal on Programmed Cell Death. 2020;25:247–260. doi: 10.1007/s10495-020-01593-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Gong X., Su L., Lin H., Yang Z., Yan X., Gao J. Activatable mitochondria-targeting organoarsenic prodrugs for bioenergetic cancer therapy. Angewandte Chemie. 2021;60:1403–1410. doi: 10.1002/anie.202012237. [DOI] [PubMed] [Google Scholar]
- Ma H., Lee Y., Hayama T., Van Dyken C., Marti-Gutierrez N., Li Y.…Mitalipov S. Germline and somatic mtDNA mutations in mouse aging. PLoS One. 2018;13 doi: 10.1371/journal.pone.0201304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Luo Y., Liang X., Lyu Q., Meng F., Chen X.…Zhou D. Alantolactone-loaded pegylated prodrug nanocarriers for synergistic treatment of cisplatin-resistant ovarian cancer via reactivating mitochondrial apoptotic pathway. ACS Biomaterials Science & Engineering. 2022;8:2526–2536. doi: 10.1021/acsbiomaterials.2c00316. [DOI] [PubMed] [Google Scholar]
- Ma B., Sheng J., Wang P., Jiang Z., Borrathybay E. Combinational phototherapy and hypoxia-activated chemotherapy favoring antitumor immune responses. International Journal of Nanomedicine. 2019;14:4541–4558. doi: 10.2147/IJN.S203383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlein C.R., Piddock R.E., Mistry J.J., Zaitseva L., Hellmich C., Horton R.H.…Rushworth S.A. CD38-Driven mitochondrial trafficking promotes bioenergetic plasticity in multiple myeloma. Cancer Research. 2019;79:2285–2297. doi: 10.1158/0008-5472.CAN-18-0773. [DOI] [PubMed] [Google Scholar]
- Mohanty A., Tiwari-Pandey R., Pandey N. Mitochondria: The indispensable players in innate immunity and guardians of the inflammatory response. Journal of cell communication and signaling. 2019;13:303–318. doi: 10.1007/s12079-019-00507-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok B.Y., de Moraes, Zeng J., Bosch D.E., Kotrys A.V.…Liu D.R. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature. 2020;583:631–637. doi: 10.1038/s41586-020-2477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möllers M., Maniura-Weber K., Kiseljakovic E., Bust M., Hayrapetyan A., Jaksch M., Helm M., Wiesner R., von Kleist-Retzow J. A new mechanism for mtDNA pathogenesis: Impairment of post-transcriptional maturation leads to severe depletion of mitochondrial tRNASer(UCN) caused by T7512C and G7497A point mutations. Nucleic Acids Research. 2005;33:5647–5658. doi: 10.1093/nar/gki876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal A., Jia D., Bhatt V., Akel M., Roberge J., Guo J.Y., Langenfeld J. Ym155 localizes to the mitochondria leading to mitochondria dysfunction and activation of AMPK that inhibits BMP signaling in lung cancer cells. Scientific Reports. 2022;12 doi: 10.1038/s41598-022-17446-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore E., Strazza M., Mor A. Combination approaches to target PD-1 signaling in cancer. Frontiers in Immunology. 2022;13 doi: 10.3389/fimmu.2022.927265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto D., Matsumura S., Bustos-Villalobos I., Sibal P., Ichinose T., Naoe Y.…Kasuya H. C-REV retains high infectivity regardless of the expression levels of cGAS and STING in cultured pancreatic cancer cells. Cells. 2021;10 doi: 10.3390/cells10061502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov Y.I., Parshin A.V., Agaronyan K., Cheung A.C., Anikin M., Cramer P., Temiakov D. A model for transcription initiation in human mitochondria. Nucleic Acids Research. 2015;43:3726–3735. doi: 10.1093/nar/gkv235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad N., Cai-Ping T., Nasreen S., Mao Z. Redirecting cisplatin and doxorubicin to mitochondria affords highly effective platinum prodrug against triple negative breast cancer. Chemistry - An Asian Journal. 2021;16:2276–2279. doi: 10.1002/asia.202100593. [DOI] [PubMed] [Google Scholar]
- Mukerabigwi J., Tang R., Cao Y., Mohammed F., Zhou Q., Zhou M., Ge Z. Mitochondria-targeting polyprodrugs to overcome the drug resistance of cancer cells by self-amplified oxidation-triggered drug release. Bioconjugate Chemistry. 2023;34:377–391. doi: 10.1021/acs.bioconjchem.2c00559. [DOI] [PubMed] [Google Scholar]
- Murphy M.R., Kleiman F.E. Vol. 11. Wiley Interdiscip Rev RNA; 2020. (Connections between 3' end processing and DNA damage response: Ten years later). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadalutti C.A., Ayala-Peña S., Santos J.H. Mitochondrial DNA damage as driver of cellular outcomes. American Journal of Physiology - Cell Physiology. 2022;322:C136–c150. doi: 10.1152/ajpcell.00389.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair J., Joseph M., Arya J., Sreedevi P., Sujai P., Maiti K. Elucidating a thermoresponsive multimodal photo-chemotherapeutic nanodelivery vehicle to overcome the barriers of doxorubicin therapy. ACS Applied Materials and Interfaces. 2020;12:43365–43379. doi: 10.1021/acsami.0c08762. [DOI] [PubMed] [Google Scholar]
- Nomura S., Morimoto Y., Tsujimoto H., Arake M., Harada M., Saitoh D.…Ueno H. Highly reliable, targeted photothermal cancer therapy combined with thermal dosimetry using a near-infrared absorbent. Scientific Reports. 2020;10:9765. doi: 10.1038/s41598-020-66646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik S., Kim J., Silwal P., Sasakawa C., Jo E. An update on the regulatory mechanisms of NLRP3 inflammasome activation. Cellular and Molecular Immunology. 2021;18:1141–1160. doi: 10.1038/s41423-021-00670-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchangam R., Rao R., Balamurali M., Hingamire T., Shanmugam D., Manickam V., Chanda K. HAntitumor effects of Ir(III)-2-Indazole complexes for triple negative breast cancer. Inorganic Chemistry. 2021;60:17593–17607. doi: 10.1021/acs.inorgchem.1c02193. [DOI] [PubMed] [Google Scholar]
- Pandey A., Verma S. Combination drug therapy for multimodal treatment of cancer by targeting mitochondrial transcriptional pathway: An in-silico approach. Medical Hypotheses. 2020;143 doi: 10.1016/j.mehy.2020.110075. [DOI] [PubMed] [Google Scholar]
- Peled M., Tocheva A., Adam K., Mor A. VRK2 inhibition synergizes with PD-1 blockade to improve T cell responses. Immunology Letters. 2021;233:42–47. doi: 10.1016/j.imlet.2021.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M., Zhang J., Hancock S., Derbeneva O., Golhar R., Golik P.…Wallace D. Progressive increase in mtDNA 3243A>G heteroplasmy causes abrupt transcriptional reprogramming. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E4033–E4042. doi: 10.1073/pnas.1414028111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokatayev V., Yang K., Tu X., Dobbs N., Wu J., Kalb R., Yan N. Homeostatic regulation of STING protein at the resting state by stabilizer TOLLIP. Nature Immunology. 2020;21:158–167. doi: 10.1038/s41590-019-0569-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokrzywinski K., Biel T., Kryndushkin D., Rao V. Therapeutic targeting of the mitochondria initiates excessive superoxide production and mitochondrial depolarization causing decreased mtDNA integrity. PLoS One. 2016;11 doi: 10.1371/journal.pone.0168283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przanowski P., Przanowska R., Guertin M. ANKLE1 cleaves mitochondrial DNA and contributes to cancer risk by promoting apoptosis resistance and metabolic dysregulation. Communications Biology. 2023;6:231. doi: 10.1038/s42003-023-04611-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pustylnikov S., Costabile F., Beghi S., Facciabene A. Targeting mitochondria in cancer: Current concepts and immunotherapy approaches. Translational Research. 2018;202:35–51. doi: 10.1016/j.trsl.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W., Pan Z., Cai D., Li Y., He L. Cyclometalated iridium(iii) complexes for mitochondria-targeted combined chemo-photodynamic therapy. Dalton Transactions (Cambridge, England : 2003) 2020;49:3562–3569. doi: 10.1039/d0dt00180e. [DOI] [PubMed] [Google Scholar]
- Rai P. Oxidation in the nucleotide pool, the DNA damage response and cellular senescence: Defective bricks build a defective house. Mutation Research, Genetic Toxicology and Environmental Mutagenesis. 2010;703:71–81. doi: 10.1016/j.mrgentox.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Rowe T., Weissig V., Lawrence J. Mitochondrial DNA metabolism targeting drugs. Advanced Drug Delivery Reviews. 2001;49:175–187. doi: 10.1016/s0169-409x(01)00133-8. [DOI] [PubMed] [Google Scholar]
- Rubio-Cosials A., Sidow J.F., Jiménez-Menéndez N., Fernández-Millán P., Montoya J., Jacobs H.T., Coll M., Bernadó P., Solà M. Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nature Structural & Molecular Biology. 2011;18:1281–1289. doi: 10.1038/nsmb.2160. [DOI] [PubMed] [Google Scholar]
- Sansone P., Savini C., Kurelac I., Chang Q., Amato L.B., Strillacci A.…Gasparre G., et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E9066–e9075. doi: 10.1073/pnas.1704862114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöpf B., Weissensteiner H., Schäfer G., Fazzini F., Charoentong P., Naschberger A.…Klocker H. OXPHOS remodeling in high-grade prostate cancer involves mtDNA mutations and increased succinate oxidation. Nature Communications. 2020;11:1487. doi: 10.1038/s41467-020-15237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Hu T., Liu X., Zha J., Meng F., Wu Z.…Tan C. Defect engineering of layered double hydroxide nanosheets as inorganic photosensitizers for NIR-III photodynamic cancer therapy. Nature Communications. 2022;13:3384. doi: 10.1038/s41467-022-31106-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Sadler P.J. How promising is phototherapy for cancer? British Journal of Cancer. 2020;123:871–873. doi: 10.1038/s41416-020-0926-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Hong S.G., Choi S.Y., Rath M.E., Saredy J., Jovin D.G.…Park J.Y. Flow-induced endothelial mitochondrial remodeling mitigates mitochondrial reactive oxygen species production and promotes mitochondrial DNA integrity in a p53-dependent manner. Redox Biology. 2022;50:102252. doi: 10.1016/j.redox.2022.102252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K., Ayyasamy V., Owens K., Koul M., Vujcic M. Mutations in mitochondrial DNA polymerase-gamma promote breast tumorigenesis. Journal of Human Genetics. 2009;54:516–524. doi: 10.1038/jhg.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siibak T., Clemente P., Bratic A., Bruhn H., Kauppila T.E.S., Macao B.…Wredenberg A. A multi-systemic mitochondrial disorder due to a dominant p.Y955H disease variant in DNA polymerase gamma. Human Molecular Genetics. 2017;26:2515–2525. doi: 10.1093/hmg/ddx146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Faccenda D., Campanella M. Pharmacological advances in mitochondrial therapy. EBioMedicine. 2021;65 doi: 10.1016/j.ebiom.2021.103244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G., Guibao C., Seetharaman J., Aggarwal A., Grace C., McNamara D., Vaithiyalingam S., Waddell M., Moldoveanu T. Structural basis of BAK activation in mitochondrial apoptosis initiation. Nature Communications. 2022;13:250. doi: 10.1038/s41467-021-27851-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G., Pachouri U.C., Khaidem D.C., Kundu A., Chopra C., Singh P. Mitochondrial DNA Damage and Diseases. 2015;4:176. doi: 10.12688/f1000research.6665.1. F1000Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopelja-Gardner S., An J., Elkon K.B. Role of the cGAS-STING pathway in systemic and organ-specific diseases. Nature Reviews Nephrology. 2022;18:558–572. doi: 10.1038/s41581-022-00589-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A., Whitehall J., Greaves L. Mitochondrial DNA mutations in ageing and cancer. Molecular Oncology. 2022;16:3276–3294. doi: 10.1002/1878-0261.13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.L.M., Whitehall J.C., Greaves L.C. Mitochondrial DNA mutations in ageing and cancer. Molecular Oncology. 2022;16:3276–3294. doi: 10.1002/1878-0261.13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somuncu B., Ekmekcioglu A., Antmen F., Ertuzun T., Deniz E., Keskin N.…Muftuoglu M. Targeting mitochondrial DNA polymerase gamma for selective inhibition of MLH1 deficient colon cancer growth. PLoS One. 2022;17 doi: 10.1371/journal.pone.0268391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Wu C., Wu K., Han Q., Miao X., Ma D., Leung C. Ubiquitination regulators discovered by virtual screening for the treatment of cancer. Frontiers in Cell and Developmental Biology. 2021;9 doi: 10.3389/fcell.2021.665646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli J.B., Haigis M.C. The multifaceted contributions of mitochondria to cellular metabolism. Nature Cell Biology. 2018;20:745–754. doi: 10.1038/s41556-018-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Gelais C., de Silva S., Amie S.M., Coleman C.M., Hoy H., Hollenbaugh J.A., Kim B., Wu L. SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology. 2012;9:105. doi: 10.1186/1742-4690-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Zhan L., Chen Y., Wang G., He L., Wang Q.…Huang Q. Increased mtDNA copy number promotes cancer progression by enhancing mitochondrial oxidative phosphorylation in microsatellite-stable colorectal cancer. Signal Transduction and Targeted Therapy. 2018;3:8. doi: 10.1038/s41392-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Cheng B., Dong Y., Li T., Xie Z., Zhang Y. Time-dependent effects of anesthetic isoflurane on reactive oxygen species levels in HEK-293 cells. Brain Sciences. 2014;4:311–320. doi: 10.3390/brainsci4020311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaga K., Koshikawa N., Nagase H. Intercellular transfer of mitochondrial DNA carrying metastasis-enhancing pathogenic mutations from high- to low-metastatic tumor cells and stromal cells via extracellular vesicles. BMC molecular and cell biology. 2021;22:52. doi: 10.1186/s12860-021-00391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Qi X., Kong W., Jin J., Lu D., Zhang X.…Wang C., et al. A conditional knockout rat resource of mitochondrial protein-coding genes via a DdCBE-induced premature stop codon. Science Advances. 2023;9 doi: 10.1126/sciadv.adf2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S., Kuncheria L., Roulstone V., Kyula J., Mansfield D., Bommareddy P.…Coffin R. Development of a new fusion-enhanced oncolytic immunotherapy platform based on herpes simplex virus type 1. Journal for immunotherapy of cancer. 2019;7:214. doi: 10.1186/s40425-019-0682-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji K., Kida Y., Koshikawa N., Yamamoto S., Shinozaki Y., Watanabe T., Lin J., Nagase H., Takenaga K. Suppression of non-small-cell lung cancer A549 tumor growth by an mtDNA mutation-targeting pyrrole-imidazole polyamide-triphenylphosphonium and a senolytic drug. Cancer Science. 2022;113:1321–1337. doi: 10.1111/cas.15290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turos-Korgul L., Kolba M., Chroscicki P., Zieminska A., Piwocka K. Tunneling nanotubes facilitate intercellular protein transfer and cell networks function. Frontiers in Cell and Developmental Biology. 2022;10 doi: 10.3389/fcell.2022.915117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida A., Murugesapillai D., Kastner M., Wang Y., Lodeiro M.F., Prabhakar S.…Cameron C.E. Unexpected sequences and structures of mtDNA required for efficient transcription from the first heavy-strand promoter. Elife. 2017;6 doi: 10.7554/eLife.27283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina-Varela R., Castillo N., Videla L., Del Campo A. Impact of mitophagy and mitochondrial unfolded protein response as new adaptive mechanisms underlying old pathologies: Sarcopenia and non-alcoholic fatty liver disease. International Journal of Molecular Sciences. 2020;21 doi: 10.3390/ijms21207704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadrot N., Ghanem S., Braut F., Gavrilescu L., Pilard N., Mansouri A., Moreau R., Reyl-Desmars F. Mitochondrial DNA maintenance is regulated in human hepatoma cells by glycogen synthase kinase 3β and p53 in response to tumor necrosis factor α. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdebenito S., Malik S., Luu R., Loudig O., Mitchell M., Okafo G., Bhat K., Prideaux B., Eugenin E.A. Tunneling nanotubes, TNT, communicate glioblastoma with surrounding non-tumor astrocytes to adapt them to hypoxic and metabolic tumor conditions. Scientific Reports. 2021;11 doi: 10.1038/s41598-021-93775-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez-Alegría C., Fuentes-Rivas R., García-Rivas J., Zavala Arce R., Jiménez Núñez M., García-Gaitán B. Synthesis of chitosan-polyvinyl alcohol biopolymers to eliminate fluorides from water. Biomolecules. 2020;10 doi: 10.3390/biom10010156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenti D., Vacca R.A., Moro L., Atlante A. Mitochondria can cross cell boundaries: An overview of the biological relevance, pathophysiological implications and therapeutic perspectives of intercellular mitochondrial transfer. International Journal of Molecular Sciences. 2021;22 doi: 10.3390/ijms22158312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenti D., Vacca R.A., Moro L., Atlante A. Mitochondria can cross cell boundaries: An overview of the biological relevance, pathophysiological implications and therapeutic perspectives of intercellular mitochondrial transfer. International Journal of Molecular Sciences. 2021:22. doi: 10.3390/ijms22158312. [Online] [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gisbergen M., Voets A., Starmans M., de Coo I., Yadak R., Hoffmann R., Boutros P., Smeets H., Dubois L., Lambin P. How do changes in the mtDNA and mitochondrial dysfunction influence cancer and cancer therapy? Challenges, opportunities and models. Mutation Research, Reviews in Mutation Research. 2015;764:16–30. doi: 10.1016/j.mrrev.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Vo M., Smith B., Nicholas J., Choi Y. Activation of NIX-mediated mitophagy by an interferon regulatory factor homologue of human herpesvirus. Nature Communications. 2019;10:3203. doi: 10.1038/s41467-019-11164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A., Kosnacova H., Chovanec M., Jurkovicova D. Mitochondrial genetic and epigenetic regulations in cancer: Therapeutic potential. International Journal of Molecular Sciences. 2022:23. doi: 10.3390/ijms23147897. [Online] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Cheng H., Zhou Y., Ma M. Clinical significance of the D-loop gene mutation in mitochondrial DNA in laryngeal cancer. OncoTargets and Therapy. 2021;14:3461–3466. doi: 10.2147/OTT.S304836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Huang K., Tseng W., Lo J., Li A., Fang W.…Lee H. DNAJA3/Tid1 is required for mitochondrial DNA maintenance and regulates migration and invasion of human gastric cancer cells. Cancers. 2020;12 doi: 10.3390/cancers12113463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Jiang C., Yin H., Gao S., Yu B. Hepatobiliary & pancreatic diseases international : HBPD INT. 2023. Targeting mitochondrial transcription factor A sensitizes pancreatic cancer cell to gemcitabine. [DOI] [PubMed] [Google Scholar]
- Wang W., Sun H., Ma X., Zhu T., Zhang H. Circ_0002476 regulates cell growth, invasion, and mtDNA damage in non-small cell lung cancer by targeting miR-1182/TFAM axis. Thoracic Cancer. 2022 doi: 10.1111/1759-7714.14631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wei G., Zhang X., Huang X., Zhao J., Guo X., Zhou S. Multistage targeting strategy using magnetic composite nanoparticles for synergism of photothermal therapy and chemotherapy. Small. 2018;14 doi: 10.1002/smll.201702994. [DOI] [PubMed] [Google Scholar]
- Wang H., Zang C., Ren M., Shang M., Wang Z., Peng X.…Zhou C. Cellular uptake of extracellular nucleosomes induces innate immune responses by binding and activating cGMP-AMP synthase (cGAS) Scientific Reports. 2020;10:15385. doi: 10.1038/s41598-020-72393-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Xiang Y., Pan W., Wang H., Li N., Tang B. Dual-targeted photothermal agents for enhanced cancer therapy. Chemical Science. 2020;11:8055–8072. doi: 10.1039/d0sc03173a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhou X., Wei Q., Qin Q., Li J., Tan M., Zhang S. Novel bifluorescent Zn(II)-cryptolepine-cyclen complexes trigger apoptosis induced by nuclear and mitochondrial DNA damage in cisplatin-resistant lung tumor cells. European Journal of Medicinal Chemistry. 2022;238 doi: 10.1016/j.ejmech.2022.114418. [DOI] [PubMed] [Google Scholar]
- Wang Z.F., Zhou X.F., Wei Q.C., Qin Q.P., Li J.X., Tan M.X., Zhang S.H. Novel bifluorescent Zn(II)-cryptolepine-cyclen complexes trigger apoptosis induced by nuclear and mitochondrial DNA damage in cisplatin-resistant lung tumor cells. European Journal of Medicinal Chemistry. 2022;238 doi: 10.1016/j.ejmech.2022.114418. [DOI] [PubMed] [Google Scholar]
- Wei W., Schon K.R., Elgar G., Orioli A., Tanguy M., Giess A., Tischkowitz M., Caulfield M.J., Chinnery P.F. Nuclear-embedded mitochondrial DNA sequences in 66,083 human genomes. Nature. 2022;611:105–114. doi: 10.1038/s41586-022-05288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y.A., Xiong X., Scott T., Li A.T., Wang C., Weiss H.L.…Gao T. The mitochondrial retrograde signaling regulates Wnt signaling to promote tumorigenesis in colon cancer. Cell Death & Differentiation. 2019;26:1955–1969. doi: 10.1038/s41418-018-0265-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z., Lei Z., Jin-An M., Xue-Zhen L., Xing-Nan Z., Xiu-Wen D. The inhibitory role of miR-214 in cervical cancer cells through directly targeting mitochondrial transcription factor A (TFAM) European Journal of Gynaecological Oncology. 2014;35:676–682. [PubMed] [Google Scholar]
- Witkowska M., Maciejewska N., Ryczkowska M., Olszewski M., Bagiński M., Makowiec S. From tryptophan to novel mitochondria-disruptive agent, synthesis and biological evaluation of 1,2,3,6-tetrasubstituted carbazoles. European Journal of Medicinal Chemistry. 2022;238 doi: 10.1016/j.ejmech.2022.114453. [DOI] [PubMed] [Google Scholar]
- Wu S., Akhtari M., Alachkar H. Characterization of mutations in the mitochondrial encoded electron transport chain complexes in acute myeloid leukemia. Scientific Reports. 2018;8 doi: 10.1038/s41598-018-31489-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian H., Watari K., Sanchez-Lopez E., Offenberger J., Onyuru J., Sampath H.…Karin M. Oxidized DNA fragments exit mitochondria via mPTP- and VDAC-dependent channels to activate NLRP3 inflammasome and interferon signaling. Immunity. 2022;55:1370–1385.e8. doi: 10.1016/j.immuni.2022.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Yao W., Lin M., Huang W., Li B., Peng B., Ma Q., Zhou X., Liang M. Icaritin-loaded PLGA nanoparticles activate immunogenic cell death and facilitate tumor recruitment in mice with gastric cancer. Drug Delivery. 2022;29:1712–1725. doi: 10.1080/10717544.2022.2079769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M., Doetsch P.W., Deng X. Bcl2 inhibition of mitochondrial DNA repair. BMC Cancer. 2015;15:586. doi: 10.1186/s12885-015-1594-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Du W., Zhao Y., Lim K., Lu L., Zhang C., Li L. Mitochondria targeting drugs for neurodegenerative diseases-Design, mechanism and application. Acta Pharmaceutica Sinica B. 2022;12:2778–2789. doi: 10.1016/j.apsb.2022.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav N., Kumar S., Marlowe T., Chaudhary A.K., Kumar R., Wang J.…Chandra D. Oxidative phosphorylation-dependent regulation of cancer cell apoptosis in response to anticancer agents. Cell Death & Disease. 2015;6:e1969. doi: 10.1038/cddis.2015.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Kawamura E., Harashima H. Mitochondrial-targeted DNA delivery using a DF-MITO-Porter, an innovative nano carrier with cytoplasmic and mitochondrial fusogenic envelopes. Journal of Nanoparticle Research. 2012;14:1013. [Google Scholar]
- Yamazoe M., Sasano T., Ihara K., Takahashi K., Nakamura W., Takahashi N., Komuro H., Hamada S., Furukawa T. Sparsely methylated mitochondrial cell free DNA released from cardiomyocytes contributes to systemic inflammatory response accompanied by atrial fibrillation. Scientific Reports. 2021;11:5837. doi: 10.1038/s41598-021-85204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Gilman P., Mirzayans R., Sun X., Touret N., Weinfeld M., Goping I.S. Characterization of the apoptotic response induced by the cyanine dye D112: A potentially selective anti-cancer compound. PLoS One. 2015;10 doi: 10.1371/journal.pone.0125381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Yao C., Fang C., Han M., Gong C., Hu D.…Zhu S. Rocaglamide promotes the infiltration and antitumor immunity of NK cells by activating cGAS-STING signaling in non-small cell lung cancer. International Journal of Biological Sciences. 2022;18:585–598. doi: 10.7150/ijbs.65019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Pan Z., Zhang D., Cao Q., Ji L., Mao Z. Precisely assembled nanoparticles against cisplatin resistance via cancer-specific targeting of mitochondria and imaging-guided chemo-photothermal therapy. ACS Applied Materials and Interfaces. 2020;12:43444–43455. doi: 10.1021/acsami.0c12814. [DOI] [PubMed] [Google Scholar]
- Yang J., Liu L., Oda Y., Wada K., Ago M., Matsuda S., Hattori M., Goto T., Ishibashi S., Kawashima-Sonoyama Y., Matsuzaki Y., Taketani T. Extracellular vesicles and cx43-gap junction channels are the main routes for mitochondrial transfer from ultra-purified mesenchymal stem cells, RECs. International Journal of Molecular Sciences. 2023:24. doi: 10.3390/ijms241210294. [Online] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Shen X., Sun H., Zhu W., Zhang J., Lu L. Neutrophil extracellular traps in hepatocellular carcinoma are enriched in oxidized mitochondrial DNA which is highly pro-inflammatory and pro-metastatic. Journal of Cancer. 2022;13:1261–1271. doi: 10.7150/jca.64170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Weinfeld M., Lemieux H., Montpetit B., Goping I. Photo-activation of the delocalized lipophilic cation D112 potentiates cancer selective ROS production and apoptosis. Cell Death & Disease. 2017;8 doi: 10.1038/cddis.2017.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Takenaga K., Koshikawa N., Kida Y., Lin J., Watanabe T.…Nagase H. Anticancer effect of a pyrrole-imidazole polyamide-triphenylphosphonium conjugate selectively targeting a common mitochondrial DNA cancer risk variant in cervical cancer cells. International Journal of Cancer. 2023;152:962–976. doi: 10.1002/ijc.34319. [DOI] [PubMed] [Google Scholar]
- Yao Y., Fan X., Jiang D., Zhang Y., Li X., Xu Z.…Fu Q. Connexin 43-mediated mitochondrial transfer of iPSC-MSCs alleviates asthma inflammation. Stem Cell Reports. 2018;11:1120–1135. doi: 10.1016/j.stemcr.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi E., Park S., Jung S., Jang W., Kim Y. Mitochondrial dysfunction induces EMT through the TGF-β/Smad/Snail signaling pathway in Hep3B hepatocellular carcinoma cells. International Journal of Oncology. 2015;47:1845–1853. doi: 10.3892/ijo.2015.3154. [DOI] [PubMed] [Google Scholar]
- Yin M., Dong J., Sun C., Liu X., Liu Z., Liu L.…Deng H. Raddeanin A enhances mitochondrial DNA-cGAS/STING axis-mediated antitumor immunity by targeting transactive responsive DNA-binding protein 43. Advanced Science. 2023;2206737 doi: 10.1002/advs.202206737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga N., Numata K. Rational designs at the forefront of mitochondria-targeted gene delivery: Recent progress and future perspectives. ACS Biomaterials Science & Engineering. 2022;8:348–359. doi: 10.1021/acsbiomaterials.1c01114. [DOI] [PubMed] [Google Scholar]
- Yu L., Liu P. Cytosolic DNA sensing by cGAS: Regulation, function, and human diseases. Signal Transduction and Targeted Therapy. 2021;6:170. doi: 10.1038/s41392-021-00554-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Mehta A., Wang G., Hauswirth W.W., Chiodo V., Boye S.L., Guy J. Next-generation sequencing of mitochondrial targeted AAV transfer of human ND4 in mice. Molecular Vision. 2013;19:1482–1491. [PMC free article] [PubMed] [Google Scholar]
- Yumnamcha T., Guerra M., Singh L., Ibrahim A. Vol. 9. Antioxidants; Basel, Switzerland): 2020. (Metabolic dysregulation and neurovascular dysfunction in diabetic retinopathy). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Guo S., Zhu X., Qiu J., Deng G., Qiu C. Alpinetin inhibits breast cancer growth by ROS/NF-κB/HIF-1α axis. Journal of Cellular and Molecular Medicine. 2020;24:8430–8440. doi: 10.1111/jcmm.15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du X., Liu B., Li C., Long J., Zhao M.…Lai Y. Engineering supramolecular nanomedicine for targeted near infrared-triggered mitochondrial dysfunction to potentiate cisplatin for efficient chemophototherapy. ACS Nano. 2021;16:1421–1435. doi: 10.1021/acsnano.1c09555. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Tang Z., An R., Ye L., Zhong B. USP29 maintains the stability of cGAS and promotes cellular antiviral responses and autoimmunity. Cell Research. 2020;30:914–927. doi: 10.1038/s41422-020-0341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang J. HuR stabilizes TFAM mRNA in an ATM/p38-dependent manner in ionizing irradiated cancer cells. Cancer Science. 2018;109:2446–2457. doi: 10.1111/cas.13657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Lin S., Zeng B., Chen X., Chen L., Chen M.…He P., et al. High leukocyte mitochondrial DNA copy number contributes to poor prognosis in breast cancer patients. BMC Cancer. 2023;23:377. doi: 10.1186/s12885-023-10838-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang R., Hu D., Sun X., Fujioka H., Lundberg K.…Qi X. Oligodendroglial glycolytic stress triggers inflammasome activation and neuropathology in Alzheimer’s disease. Science Advances. 2020;6 doi: 10.1126/sciadv.abb8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Wang Y., Zhao J., Zhang Z., Jin M., Zhou F.…Ren T. PDE2 inhibits PKA-mediated phosphorylation of TFAM to promote mitochondrial Ca(2+)-induced colorectal cancer growth. Frontiers Oncology. 2021;11:663778. doi: 10.3389/fonc.2021.663778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Wang Y., Jiang W., Wang Q., Li J., Wen Z., Li A., Zhang K., Zhang Z., Shi J., Liu J. Herpesvirus-mimicking DNAzyme-loaded nanoparticles as a mitochondrial DNA stress inducer to activate innate immunity for tumor therapy. Advanced Materials. 2022;34 doi: 10.1002/adma.202204585. [DOI] [PubMed] [Google Scholar]
- Zheng J., Akbari M., Schirmer C., Reynaert M.L., Loyens A., Lefebvre B.…Bohr V.A. Hippocampal tau oligomerization early in tau pathology coincides with a transient alteration of mitochondrial homeostasis and DNA repair in a mouse model of tauopathy. Acta Neuropathol Commun. 2020;8:25. doi: 10.1186/s40478-020-00896-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Liu L., Chen J. Mitochondrial DNA heteroplasmy in Candida glabrata after mitochondrial transformation. Eukaryotic Cell. 2010;9:806–814. doi: 10.1128/EC.00349-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Li Q., Zhao Y., Peng H., Guo L., Zhu J.…Chen S. Vaccinia-related kinase 2 drives pancreatic cancer progression by protecting Plk1 from Chfr-mediated degradation. Oncogene. 2021;40:4663–4674. doi: 10.1038/s41388-021-01893-4. [DOI] [PubMed] [Google Scholar]
- Zuo Z., Zhou Z., Chang Y., Liu Y., Shen Y., Li Q., Zhang L. Genes & Diseases; 2022. Ribonucleotide reductase M2 (RRM2): Regulation, function and targeting strategy in human cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.