Abstract
Background
Immunotherapy plus tyrosine kinase inhibitor (IO-TKI) has become the first-line management for metastatic renal cell carcinoma (RCC), despite the absence of biomarkers. Recently, pyrroline-5-carboxylate reductase 1 (PYCR1) and proline metabolism have been reported regulatory roles in the anti-tumor response.
Methods
There were three cohorts enrolled: two from our institution (ZS-MRCC and ZS-HRRCC) and one from a clinical trial (JAVELIN-101). The PYCR1expression in each sample was evaluated by RNA sequencing. Flow cytometry and immunohistochemistry were performed to assess immune infiltration. Single-cell RNA-seq (scRNA-seq) data was used for cluster analysis of T cells and macrophages. Primary endpoints were set as response and progression-free survival (PFS).
Results
Patients in the low-PYCR1 group had greater objective response rate (52.2% vs 18.2%) and longer PFS in both cohorts (ZS-MRCC cohort, P=0.01, HR=2.80; JAVELIN-101 cohort, P<0.001, HR=1.85). In responders, PYCR1 expression was decreased (P<0.05). In the high PYCR1 group, CD8+ T cells exhibited an exhausted phenotype with decreased GZMB (Spearman's ρ=-0.36, P=0.02). scRNA-seq revealed tissue-resident memory T (Trm) (P<0.05) and tissue-resident macrophage (P<0.01) were decreased in samples with high PYCR1 expression. A machine learning score was further built by random forest, involving PYCR1 and Trm markers. Only in the subgroup with the lower RFscore did IO+TKI show a favorable outcome, compared to TKI monotherapy.
Conclusions
Immunosuppression and IO+TKI resistance were correlated with high PYCR1 expression. T cell exhaustion and dysfunction were also related with the expression of PYCR1. PYCR1 has the potential to be employed as a biomarker to discriminate between IO+TKI and TKI monotherapy as the optimal patient treatment strategy.
Keywords: Renal cell carcinoma, PYCR1, Immune checkpoint inhibition plus tyrosine kinase inhibition, T cell dysfunction
Introduction
RCC is a physiologically heterogeneous tumor with various tumorigenesis drivers, and >75% of RCC tumors are clear cell type in histology. The high-dose interleukin-2 (HDIL-2) and interferon-alpha (IFN-α) used to be the cornerstones of treatment for advanced diseases with generally poor outcomes in the majority of patients. Over the past two decades, we have witnessed a paradigm shift in the treatment of metastatic renal cell carcinoma (mRCC), with the advent of innovative therapeutic methods that have drastically improved survival. With the emergence of immune checkpoint inhibitors (ICIs) and anti-VEGF tyrosine kinase inhibitors (TKI), the therapy landscape for ccRCC has undergone a radical transformation. The current standard first-line treatment for advanced mRCC is the combination therapy of immunotherapy (IO) plus TKI [1]. There are four FDA-approved combinations therapy: pembrolizumab+axitinib (KEYNOTE 426) [2], avelumab+axitinib (JAVELIN 101) [3], nivolumab+cabozantinib (CheckMate 9ER) [4], and pembrolizumab+lenvatinib (CLEAR) [5]. In mRCC, the IO+TKI improved overall survival and progression-free survival (PFS), and induced an exceptional durable response. However, a substantial proportion of patients continue to exhibit intrinsic resistance to IO+TKI or acquire acquired resistance, ultimately succumbing to the progression of mRCC. Consequently, there is an urgent need for the development of novel biomarkers for clinical decision-making to predict and improve the prognosis of mRCC.
In order to satisfy the higher energy and anabolic demands of persistent cell proliferation, cancer cells undergo metabolic reprogramming [6]. The reprogramming of metabolic pathways has shed light on the intricacy of cancer metabolism. Proline is a unique non-essential amino acid that plays important roles in protein structure and cellular stress response [7]. Pyrroline 5-carboxylate reductase 1 (PYCR1) is an essential enzyme for proline synthesis that is abundantly expressed in cancer stroma and CAFs. Previous studies have demonstrated that the activity of the proline dehydrogenase (PRODH) and PYCR1 enzymes might work as a cycle between the mitochondria and the cytosol, transferring proline as intermediates and redox equivalents [8]. It is likely that the activity of each of these enzymes depends on the cellular context and microenvironment. Mitochondrial proline synthesis, specifically via PYCR1 activity, is crucial for hypoxia metabolism and survival [9]. Reducing PYCR1 levels in tumor-associated fibroblasts (CAFs) is sufficient to inhibit tumor collagen synthesis, tumor growth, and metastatic spread resulting in ineffective drug delivery and immune cell recruitment [10].
In this study, we identified that elevated PYCR1 expression could become a potential biomarker for IO+TKI treatment response and PFS. We also found the correlation between elevated PYCR1 expression, vessels, and fibroblasts, as well as CD8+ T cells dysfunction, tissue-resident memory T cells, and resistant macrophages. Furthermore, we developed a random-forest model based on PYCR1 and Trm markers in RCC to predict the efficacy of IO+TKI versus TKI monotherapy. Our findings revealed the predictive function and immunologic correlations of PYCR1 and suggest that inhibiting PYCR1 may boost the efficacy of IO/TKI in RCC patients.
Materials and methods
Study cohorts and data collection
ZS-HRRCC cohort: 43 patients with high-risk localized RCC were enrolled who underwent radical nephrectomy at Zhongshan Hospital, Fudan University from Jan 2020 to Dec 2021. Inclusion Criteria and exclusion Criteria were previously descrbied [11]. Three patients were excluded because of the unavailability of tissue samples or failure to meet sample quality control standards, clinical.
ZS-MRCC cohort: Totally, 51 MRCC patients with TKI+IO combination therapy were enrolled from January 2017 to December 2020. Inclusion Criteria and exclusion Criteria were previously descrbied [11]. Six patients were excluded due to the unavailability of tissue samples or loss of follow-up. Therapeutic response and disease progression were defined by the RECIST 1.1 criteria [12].
The JAVELIN-101 cohort was from a clinical trial that enrolled 726 metastatic advanced RCC participants as previously reported by Robert J. Motzer et al [3,13].
The Cancer Genome Atlas (TCGA) project enrolled 530 clear cell RCC patients in the TCGA-KIRC cohort (https://xena.ucsc.edu/) [14].
RNA-seq and data processing
The RNA of samples was extracted from resected primary RCC tissues. The sample preprocessing and library construction were previously described [11]. Further normalization of sequencing data was performed to read count and FPKM values.
Hematoxylin & eosin (H&E) staining and immunohistochemistry
Immunohistochemistry procedure and antibodies were performed as described before [11,15]. Slides were scanned with PANNORAMIC® 250 Flash III DX (3DHISTECH Ltd.). The densities of targeted cells were calculated as the mean number of cells/mm2. The micro vessels were counted as the mean number of MVD/mm2. Three researchers were asked to quantify positive cells on six randomly selected fields.
Flow cytometry
The protocol and antibodies of flow cytometry were described previously [11]. Briefly, after blocking Fc receptors, single-cell suspensions and white blood cells were stained for 30 min at 4 °C with fluorescently labeled membrane marker antibodies. Proteins were stained intracellularly using antibodies Intracellular Fixation & Permeabilization Buffer (Thermo Fisher Scientific). Flowjo v10.0 was used to analyze BD LSRFortessaTM X-20 (BD Biosciences) FACS data (Tree Star).
In silico approaches
The R program (https://www.r-project.org) was utilized for all bioinformatic analyses. The "forestplot" R package created the forest plots. The waterfall plot and heatmaps were calculated and plotted by the R packages "ComplexHeatmap" and "ggplot2." The random forest model was constructed with the "randomForestSRC" and "ggRandomForests" packages.
scRNA-seq data analysis
The scRNA-seq data was obtained from dataset GSE178481 in Gene Expression Omnibus (GEO). The analysis was carried out by "Seurat" R package [16]. nFeature_RNA > 200 & percent.mt < 12.5 & percent.rp < 60 & nFeature_RNA < 7500 & log10GenesPerUMI > 0.80 & nCount_RNA < 20000 were set as the quality control parameters. The subcluster analysis of T cells and macrophages was performed according to the previous study [17].
Statistical analysis
The Kruskal-Wallis H test was used to compare continuous variables between groups. The categorical variables were used for the chi-square test. Spearman's correlation analysis was utilized for quantitative correlation analysis. Kaplan-Meier analysis with log-rank regression was performed for survival analysis. Cox proportional hazard models were utilized for prognostic analysis.
Results
PYCR1 expression associated with response and prognosis of IO+TKI therapy
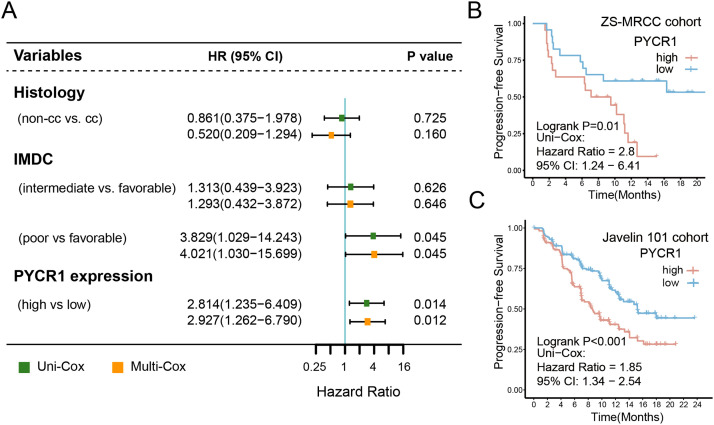
The recommendations of EAU RCC guideline suggest IO+TKI combination therapy as the standard first-line treatment for mRCC. In clinical practice, however, our ZS-MRCC cohort demonstrated that mRCC patients had a variety of therapeutic responses (Fig. 1E–G). The expression of PYCR1 was elevated in RCC tissues compared to normal tissues (P<0.001, Fig. 1A), and was enhanced significantly in TNM stage IV and ISUP grade G4 (Fi. 1B and C). Although no significant difference in PFS was observed between PYCR1-low and PYCR1-high TxNxM1 RCC patients in the TCGA cohort (Fig. S1A), responders to IO+TKI exhibited significantly reduced PYCR1 expression (P<0.05, Fig. 1D). The therapy response was classified as complete response (CR), partial response (PR), stable disease (SD), or progressing disease (PD) based on RECIST 1.1 criteria. Meanwhile, the proportion of responders (PR/CR) in the low-PYCR1 expression subgroup increased (52.2% vs 18.2%, Fig. 1F and G). This may be due to that the TCGA cohort comprise a large number of patients with earlier stages and the treatment was totally different at that time. Patients in our ZS-MRCC cohort with lower PYCR1 expression had longer PFS (P=0.01, Fig. 2B), which was verified in another cohort, Javelin 101 (P<0.001, Fig. 2C). The cutoff was set at 50%. Subsequently, clinical and pathological variables, including age, histology, and IMDC group, along with the expression of PYCR1 were included in the Cox regression model. PYCR1 indicated poor prognosis independently for PFS (univariate: hazard ratio (HR) = 2.814, 95% confidence interval (CI) = 1.235-6.409, P = 0.014; multivariate: HR = 2.927, 95% CI = 1.262–6.790, P = 0.012; Fig. 2A). Collectively, our data suggested that the expression of PYCR1 could serve as an independent negative prognosticator for patients receiving IO+TKI therapy.
Fig. 1.
PYCR1 related with resistance to tyrosine kinase inhibitor (TKI) plus immunotherapy (IO) combination therapy in renal cell carcinoma (RCC). (A) Expression of PYCR1 in RCC and peritumor tissues. P values, Kruskal-Wallis H test. (B) and (C) Association between PYCR1 and TNM stage/ ISUO grade in RCC. P values, Kruskal-Wallis H test. (D) Expression of PYCR1 between responders and non-responders of TKI+IO combination therapy in the ZS-MRCC cohort. P values, Kruskal-Wallis H test. (E&F) Therapeutic response (E) and representative chest computed tomography (F) according to PYCR1 in the ZS-MRCC cohort under TKI+IO combination therapy. (G) Tumor best percentage change from baseline and PYCR1 expression in our ZS-MRCC cohort of TKI+IO combination therapy. *, P<0.05; **, P<0.01; ***, P<0.001; ns, not significant.
Fig. 2.
PYCR1 related with prognosis of tyrosine kinase inhibitor (TKI) plus immunotherapy (IO) combination therapy in renal cell carcinoma (RCC). (A) Univariate and multivariate Cox regression model was used to calculate HR and 95% CI. HR < 1 indicates better survival. The cutoff of PYCR1 expression was the median value. (B) and (C) Progression-free survival (PFS) after TKI+IO therapy according to PYCR1 in the ZS-MRCC cohort (B) and Javelin 101 cohort (C) of TKI+IO combination therapy. P value, Kaplan-Meier analysis, and log-rank test.
PYCR1 was not associated with T cell infiltration
As previously indicated, PYCR1 may influence the IO-TKI sensitivity. By H&E and immunohistochemistry, we sought to assess the TME elements of mRCC samples (Fig. 3A). Fig. 3A shows the features ranked by the expression of PYCR1. TILs showed no significant difference between high and low PYCR1 samples (Figure S1B), and neither CD8+ T cells (Fig. 3B) nor CD4+ T cells (Fig. 3C) displayed a trend of difference. Subsequently, flow cytometry was applied to resected nephrectomy samples from our ZS-HRRCC cohort in order to verify the tumor-infiltrating T cells. RNA-seq was also used to determine the expression levels of PYCR1. In consistent with the results of IHC, the quantity of TILs, CD8+ T cells or CD4+ T cells did not differ with PYCR1 expression levels (Fig. 3D).
Fig. 3.
Relationship between PYCR1 and tumor microenvironment in RCC. (A) Heatmap displaying tumor microenvironment components ranked by PYCR1 in the ZS-HRRCC cohort. (B) and (C) Representative images and quantification of tumor-infiltrating CD8+ T cells (B), and CD4+ T cells (C) sorted by PYCR1 level. P values, Kruskal-Wallis H test. (D) Representative images of flow cytometry and the association between CD8+ T cells or CD4+ T cells, and PYCR1 expression in the ZS-HRRCC cohort. (E–H) Representative images of flow cytometry and the association between GZMB+ CD8+ T cells (E), GZMB+ CD4+ T cells (F), PD1+ CD8+ T cells (G), or PD1+ CD4+ T cells (H), and the expression of PYCR1. ρ and P values, Spearman's rank-order correlation.
PYCR1 correlated with dysfunction of T cells
With the ability to recognize and respond to tumor-specific antigens, tumor-infiltrating lymphocytes are critical for antitumor immunity. However, the number of lymphocytes infiltrating the tumor is not always correlated with the efficacy and resistance of immunotherapy or IO+TKI therapy, which may be attributed to the function level or exhausted state of T cells. Flow cytometry was used to further assess the functional state of tumor-infiltrating lymphocytes. The proportion of GZMB+CD8+ T cells was found to be inversely linked with PYCR1 expression (Spearman's = -0.36, P = 0.02 Fig. 3E), rather than CD4+ T cells (Fig. 3F). Moreover, both PD1+CD8+ T cells and PD1+CD4+ T cells were found strongly associated with PYCR1 expression (Spearman's ρ=0.44, P<0.001, Fig. 3G & Spearman's ρ=0.22, P=0.17, Fig. 3H). The results suggested the expression of PYCR1 was related with the function of T cells.
Suppressive TME in RCC with elevated PYCR1 expression
Suppressive TME results in malfunctioning T cells. In the current study, flow cytometry and immunohistochemistry were used to examine regulatory cells in the TME. Flow cytometry (P<0.05, Fig. 4A) and immunohistochemistry (P<0.05, Fig. 4A) demonstrated that the infiltration of regulatory T cells was increased in samples with high PYCR1 expression. Furthermore, CD34 (P<0.05, Fig. 4B) and αSMA (P<0.01, Fig. 4C) indicated that the micro vessels and fibroblasts were enriched in high PYCR1 samples. Additionally, KI67 was found to be positively correlated with PYCR1(Spearman's =0.38, P=0.01, Fig. 4D). These results suggested a probable association between PYCR1 expression and suppressive TME. The association between PYCR1 and suppressive molecules in RCC was investigated. PYCR1 and CXCL8 have a positive correlation (Spearman's =0.29, P<0.001, Fig. 4E). Moreover, the expression of PYCR1 was significantly linked with extracellular matrix remodeling, MMP9 (Spearman's =0.4, P<0.001, Fig. 4F). Besides, through GSEA analysis, we found that negative regulation of lymphocyte activation, regulatory T cell differentiation, negative regulation of IL−2 production, negative regulation of immune effector process, and negative regulation of immune response were enriched in high-PYCR1 samples (Fig. 4G).
Fig. 4.
Suppressive microenvironment in RCC with elevated PYCR1 expression. (A) Flow cytometric gating strategy and Immunohistochemical quantification of regulatory T cells and its increase in high PYCR1 expression samples. P value, Wilcoxon signed-rank test. (B) Immunohistochemical quantification of CD34+, α-SMA+ and KI67+ cells association with PYCR1 expression. P value, Wilcoxon signed-rank test. ρ and P values, Spearman's rank-order correlation. (E) and (F) Correlation between CXCL8 or MMP9 and PYCR1 expression. ρ and P values, Spearman's rank-order correlation. (G) Gene-set enrichment analysis of high-PYCR1 vs. low-PYCR1 samples. Red dots represent pathways enriched in high- PYCR1 samples. *, P<0.05; **, P<0.01.
Correlation between PYCR1 and somatic mutations in RCC
A summary of the JAVELIN-101 cohort's somatic mutations classified by PYCR1 expression was performed. VHL (55%), PBRM1 (32%), SETD2 (25%), and BAP1 (16%) mutations were identified as prevalent mutations in mRCC. None of these was significantly related with PYCR1 expression (Fig. S2A and S2B), suggesting that the poor prognosis of IO+TKI in high-PYCR1 patients may not be a result of these well-known mutations.
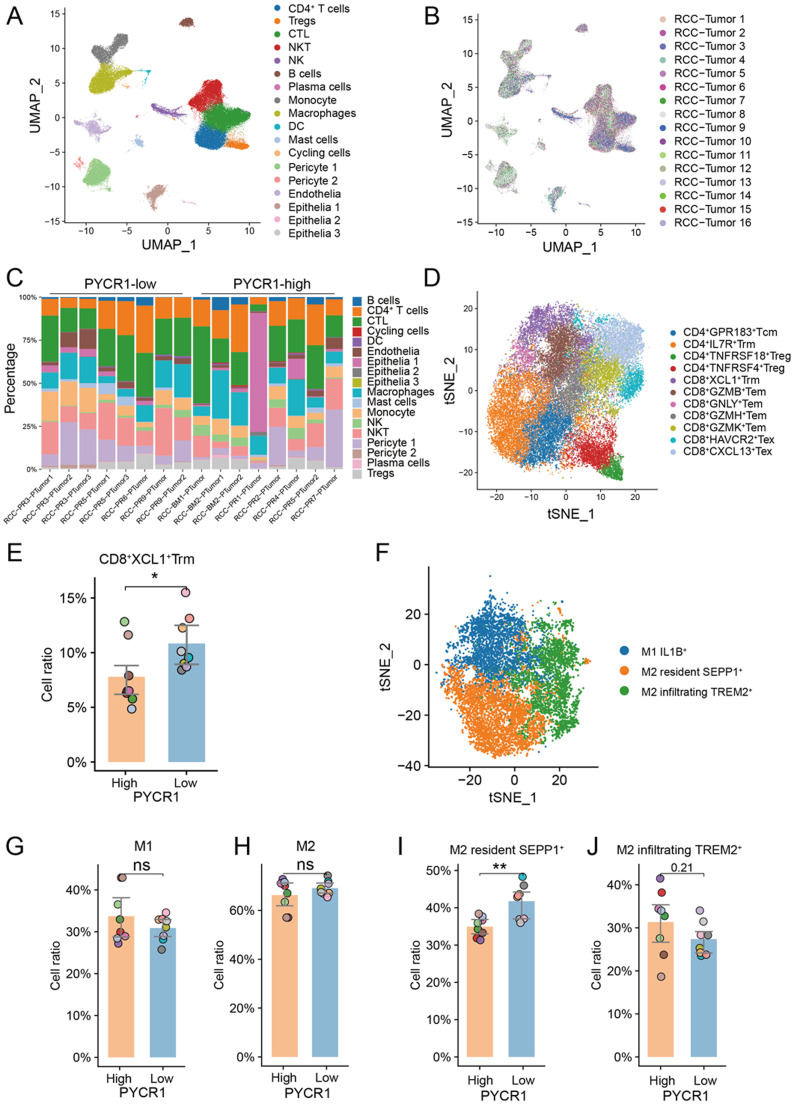
Single-cell RNA-seq (scRNA-seq) showed that high PYCR1 tumors establish an immunosuppressive tumor microenvironment
The scRNA-seq profiling (10x Genomics) of 16 samples from freshly resected primary ccRCC samples [18], in the GSE178481 cohort. After quality control, we obtained 118,957 cells, and samples were integrated using joint analysis of the heterogenous samples (Fig. 5A and 5B). Unsupervised clustering identified 18 distinct clusters, including immune cells, pericyte cells, endothelial cells, and epithelial cells (Fig. 5C). Overall, in line with the results of flow cytometry, high-PYCR1 samples did not show different immune infiltration modes compared to low-PYCR1 samples, with no significant change in cytotoxic T lymphocytes, CD4+ T cells, Tregs, and macrophages (Fig. S3). Subsequently, subcluster analysis was performed according to previous sutdy [17], identifying four clusters of CD4+ T cells, nine clusters of CTLs (Fig. 5D and S4), and three clusters of macrophages (Fig. 5F). We observed that tissue-resident memory T (Trm) (highly expressing XCL1) (P<0.05, Fig. 5E) and tissue-resident macrophage (expressing SEPP1) (P<0.01, Fig. 5I) were decreased in samples with high PYCR1 expression, rather than M1 macrophages (Fig. 5G), M2 macrophages (Fig. 5H) and another cluster of M2, M2 infiltrating TREM2+ cells (Fig. 5J).
Fig. 5.
Single-cell RNA-sequencing analysis of RCC samples. (A) and (B) Integrative analysis of scRNA-seq samples from 16 RCC samples, visualized using a common UMAP identified by 18 cell clusters (A) or by tissues (B). (C) Summary of tumor cell clusters, in each sample, sorted by PYCR expression level. (D) Subcluster analysis for T cells. (G) CD8+ XCL1+ tissue-resident memory T cells (Trm) were decreased in high-PYCR1 samples. (F) Subcluster analysis for macrophages. (G) and (H) The quantity of subtypes of macrophages in high- and low-PYCR1 samples. *, P<0.05; **, P<0.01.
Risk model construction and contribution of components
The therapeutic efficacy of TKI+IO varies between patients. TKI monotherapy is still an alternative first-line therapy for mRCC patients. Therefore, it is required to develop a model capable of identifying whether a subgroup responds better or worse to IO-TKI therapy. Random forest, a well-known machine learning approach, was performed to conduct a predictive model. The expression levels of PYCR1 and Trm markers, which are CD6, XCL1, XCL2, MYADM, CAPG, RORA, NR4A1, NR4A2, NR4A3, CD69, and ITGAE, were used as model parameters. The variable importance of each parameter to the final random forest model was illustrated (Fig. 6A). Kaplan-Meier corroborated the prognostic significance of RFscore in patients with avelumab+axitinib in the Javelin 101 cohort (P<0.001, Fig. 6C and D), rather than patients in the sunitinib arm. Univariate Cox regression was performed, showing that the low RFscore subgroup in the avelumab+axitinib arm exhibited a trend toward longer PFS than the sunitinib arm (HR 0.393, 95% CI 0.278–0.556; p<0.001, p-value for interaction<0.001, Fig. 6B), whereas high RFscore did not show prognosis value in therapy selection (HR 0.995, 95% CI 0.762–1.299, p=0.0.970, Fig. 6B). These results indicated that the RFscore could help to select low RFscore subgroup of mRCC patients with prolonged PFS by treated IO-TKI (6 months AUC=0.73, 12 months AUC=0.77, 18 months AUC=0.74, Fig. 6E).
Fig. 6.
An integrated risk score for IO+TKI benefit prediction(A) Construction and variables’ importance of the random forest score (RFscore) involving the expression of PYCR1 and Trm markers, including CAPG, CD6, MYADM, XCL2, ITGAE, NR4A1, XCL1, NR4A2, RORA, NR4A3, and CD69. (B) The benefit of IO+TKI vs. TKI monotherapy, according to the expression of the parameters of random forest model and the RFscore. The Cox regression model was used to calculate HR and 95% CI. HR < 1 indicates a favorable outcome of IO+TKI. HR >1 indicates better survival of the TKI monotherapy. The cutoffs of the gene expression were median values. (B) Progression-free survival according to high- and low-RFscore in RCC treated by IO+TKI or TKI monotherapy. (D) Progression-free survival of high- or low-RFscore in patients with IO+TKI combination therapy. (E) The RFscore receiver operating characteristic (ROC) curve analyze for the PFS of IO+TKI. ***, P<0.001.
Discussion
The PYCR1, which catalyzes the last step in proline synthesis, was determined to be essential for carcinogenesis by genomic screening. PYCR1 is one of the top five metabolic enzymes that are continuously overexpressed in 19 types of cancer, whereas it is significantly underexpressed or absent in normal tissues [19]. In silico analysis confirmed that PYCR1 was widely overexpressed in 22 cancer types from TCGA database, suggesting that disordered proline metabolism may be a general metabolic signature for most cancer types [20]. PYCR1 also promoted tumor cell development, and metastasis, and might be a biomarker for the prognosis of breast cancer, prostate cancer, hepatocellular carcinoma, renal cell carcinoma, gastric cancer, and non-small cell lung cancer [20], [21], [22], [23], [24], [25]. However, in the current study, we focused on the systemic management of advanced RCC. There was no statistically significant trend for patients with low PYCR1 expression to have a longer OS than those with high PYCR1 expression. It did not match the result of the previous report [23]. The reason may be that most cases enrolled in the TCGA cohort were at an early stage without effective treatment at the time, although PYCR1 expression was associated with stage and grade (Fig. 1B and C). Importantly, PYCR1 was associated with IO+TKI therapeutic outcome and poor prognosis in ZS-MRCC and Javelin 101 cohorts undergoing IO+TKI combination therapy (Fig. 2B and C).
In the past decade, the use of VEGFR-TKI has led to significant advancements in the treatment of mRCC. Subsequently, the EAU revised its recommendations for first-line therapy to IO+TKI combinations. However, the response rate to IO+TKI treatment varies from patient to patient and is generally low (Fig. 1E–G). The PYCR1 expression has the potential to predict the prognosis of IO+TKI therapy. Furthermore, the RFscore, calculated by our model, which was based on PYCR1 and Trm markers, demonstrated predictive value for treatment selection between IO-TKI and TKI monotherapy, since only patients with a low-RFscore might benefit from IO-TKI (Fig. 6C–E).
Cancer cells have a strong ability to survive under harsh microenvironments, evade regulatory systems such as apoptosis, and withstand the immune system's defenses. Cancer cells change their metabolism to sustain unrestrained proliferation and metastasis. Increasing number of reports suggest that the metabolism of non-essential amino acids plays a crucial role in TME [26]. In particular, the availability of proline regulates collagen production and maturation, as well as the development of cancer cell plasticity and heterogeneity [27]. In a recent investigation on lung cancer, an intriguing interaction between Proline metabolism/collagen production and microenvironment was revealed [28]. PYCR1 is an essential enzyme for proline synthesis and is abundantly expressed in stroma and CAFs [10]. These results indicated that the expression of PYCR1 may influence TME. In the present study, expression of PYCR1 also correlated with CD8+ T cell dysfunction and exhaustion, as well as immune-suppressive factors (Figs. 3E–H and 4A–C).
The role of T cells in tumor immunity has been well-established. Trm can actively participate in cancer immunosurveillance and antitumor immunity due to their specific functions, which include tissue retention and a quick response to rechallenge [29]. Emerging evidence presented a correlation between responses to ICIs and Trm. [30] Trm showed early proliferation during anti–PD-1 treatment in the responder group and produced high IFN-γ in response to anti–PD-1 therapy [31,32]. This finding expands the potential clinical relevance of Trm detection as a biomarker of favorable therapeutic response to PD-1–blocking immunotherapy. In the current study, we demonstrated that Trm was decreased in high PYCR1 samples, indicating the unfavorable outcome of IO+TKI (Fig. 5E).
The current study has several limitations. The retrospective design might cause enrollment biases. Secondly, the sample size was limited, preventing further subgroup analysis. Future prospective validation studies with larger cohorts will be performed. We would like to investigate the potential of PYCR1 as a target for therapy in mRCC in future animal models and clinical studies.
Conclusions
PYCR1 has the potential to serve as an indication of poor survival and IO-TKI response rate. PYCR1 expression correlated with CD8+ T cell dysfunction and exhaustion, as well as the reduced quantity of Trm. The machine-learning RFscore conducted by PYCR1 and Trm markers could suggest drug selection between IO-TKI versus TKI monotherapy in m RCC.
Declarations
Ethics approval and consent to participate
The study followed the Declaration of Helsinki and was approved by the Clinical Research Ethics Committee of Zhongshan Hospital, Fudan University (B2021-119). Informed consent was obtained from each participate.
Consent for publication
Not applicable.
Funding
This study was funded by grants from National Natural Science Foundation of China (81700660, 81902898, 81772696, 81974393), Shanghai Sailing Program (19YF1407900), and Experimental Animal Project of Shanghai Science and Technology Commission (19140905200). All these study sponsors have no roles in the study design, in the collection, analysis, and in the interpretation of data.
Availability of data and materials
The datasets in the current study are open to the public at the TCGA (https://xena.ucsc.edu/) and Javelin 101 clinical trial (https://www.nature.com/articles/s41591-020-1044-8). Further inquiries can be directed to the corresponding authors. The data of our cohorts that support the results of this study are available from the corresponding author upon reasonable request.
CRediT authorship contribution statement
Xianglai Xu: Conceptualization, Methodology, Software, Writing – review & editing. Ying Wang: Data curation, Writing – original draft, Methodology, Visualization, Writing – review & editing. Xinyu Hu: Visualization, Investigation, Writing – review & editing. Yanjun Zhu: Supervision, Writing – review & editing. Jiajun Wang: Software, Methodology, Writing – original draft, Writing – review & editing. Jianming Guo: Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank those authors who released and shared their datasets on the TCGA databases and Javelin 101 clinical trial.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2023.100919.
Contributor Information
Yanjun Zhu, Email: zhu.yanjun@zs-hospital.sh.cn.
Jiajun Wang, Email: w.jiajun@hotmail.com.
Jianming Guo, Email: guo.jianming@zs-hospital.sh.cn.
Appendix. Supplementary materials
References
- 1.Ljungberg B., et al. European association of urology guidelines on renal cell carcinoma: the 2022 update. Eur. Urol. 2022;82:399–410. doi: 10.1016/j.eururo.2022.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Rini B.I., et al. Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N. Engl. J. Med. 2019;380:1116–1127. doi: 10.1056/NEJMoa1816714. [DOI] [PubMed] [Google Scholar]
- 3.Motzer R.J., et al. Avelumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N. Engl. J. Med. 2019;380:1103–1115. doi: 10.1056/NEJMoa1816047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choueiri T.K., et al. Nivolumab plus cabozantinib versus sunitinib for advanced renal-cell carcinoma. N. Engl. J. Med. 2021;384:829–841. doi: 10.1056/NEJMoa2026982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Motzer R., et al. Lenvatinib plus pembrolizumab or everolimus for advanced renal cell carcinoma. N. Engl. J. Med. 2021;384:1289–1300. doi: 10.1056/NEJMoa2035716. [DOI] [PubMed] [Google Scholar]
- 6.Boroughs L.K., DeBerardinis R.J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 2015;17:351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phang J.M., Pandhare J., Liu Y. The metabolism of proline as microenvironmental stress substrate. J. Nutr. 2008;138:2008S–2015S. doi: 10.1093/jn/138.10.2008S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phang J.M., Liu W., Hancock C., Christian K.J. The proline regulatory axis and cancer. Front. Oncol. 2012;2:60. doi: 10.3389/fonc.2012.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westbrook R.L., et al. Proline synthesis through PYCR1 is required to support cancer cell proliferation and survival in oxygen-limiting conditions. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2022.110320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kay E.J., et al. Cancer-associated fibroblasts require proline synthesis by PYCR1 for the deposition of pro-tumorigenic extracellular matrix. Nat. Metab. 2022;4:693–710. doi: 10.1038/s42255-022-00582-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J., et al. Alternative complement pathway signature determines immunosuppression and resistance to immunotherapy plus tyrosine kinase inhibitor combinations in renal cell carcinoma. Urol. Oncol. 2022 doi: 10.1016/j.urolonc.2022.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Eisenhauer E.A., et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur. J. Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 13.Motzer R.J., et al. Avelumab plus axitinib versus sunitinib in advanced renal cell carcinoma: biomarker analysis of the phase 3 JAVELIN Renal 101 trial. Nat. Med. 2020;26:1733–1741. doi: 10.1038/s41591-020-1044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldman M.J., et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J., et al. Tumor-infiltrating neutrophils predict therapeutic benefit of tyrosine kinase inhibitors in metastatic renal cell carcinoma. Oncoimmunology. 2019;8 doi: 10.1080/2162402X.2018.1515611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hao Y., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(3573-3587):e3529. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L., et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564:268–272. doi: 10.1038/s41586-018-0694-x. [DOI] [PubMed] [Google Scholar]
- 18.Alchahin A.M., et al. A transcriptional metastatic signature predicts survival in clear cell renal cell carcinoma. Nat. Commun. 2022;13:5747. doi: 10.1038/s41467-022-33375-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilsson R., et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao S., Li S., Yuan Z., Zhou L. Pyrroline-5-carboxylate reductase 1 (PYCR1) upregulation contributes to gastric cancer progression and indicates poor survival outcome. Ann. Transl. Med. 2020;8:937. doi: 10.21037/atm-19-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai F., et al. Pyrroline-5-carboxylate reductase 1 promotes proliferation and inhibits apoptosis in non-small cell lung cancer. Oncol. Lett. 2018;15:731–740. doi: 10.3892/ol.2017.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng T., et al. Knockdown of PYCR1 inhibits cell proliferation and colony formation via cell cycle arrest and apoptosis in prostate cancer. Med. Oncol. 2017;34:27. doi: 10.1007/s12032-016-0870-5. [DOI] [PubMed] [Google Scholar]
- 23.Weijin F., et al. The clinical significance of PYCR1 expression in renal cell carcinoma. Medicine. 2019;98:e16384. doi: 10.1097/MD.0000000000016384. (Baltimore) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo C.L., et al. Mitochondrial oxidative stress by Lon-PYCR1 maintains an immunosuppressive tumor microenvironment that promotes cancer progression and metastasis. Cancer Lett. 2020;474:138–150. doi: 10.1016/j.canlet.2020.01.019. [DOI] [PubMed] [Google Scholar]
- 25.Ding J., et al. Human mitochondrial pyrroline-5-carboxylate reductase 1 promotes invasiveness and impacts survival in breast cancers. Carcinogenesis. 2017;38:519–531. doi: 10.1093/carcin/bgx022. [DOI] [PubMed] [Google Scholar]
- 26.Choi B.H., Coloff J.L. The diverse functions of non-essential amino acids in cancer. Cancers. 2019:11. doi: 10.3390/cancers11050675. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Aniello C., Patriarca E.J., Phang J.M., Minchiotti G. Proline metabolism in tumor growth and metastatic progression. Front. Oncol. 2020;10:776. doi: 10.3389/fonc.2020.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo L., et al. Kindlin-2 links mechano-environment to proline synthesis and tumor growth. Nat. Commun. 2019;10:845. doi: 10.1038/s41467-019-08772-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yenyuwadee S., Sanchez-Trincado Lopez J.L., Shah R., Rosato P.C., Boussiotis V.A. The evolving role of tissue-resident memory T cells in infections and cancer. Sci. Adv. 2022;8:eabo5871. doi: 10.1126/sciadv.abo5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caushi J.X., et al. Transcriptional programs of neoantigen-specific TIL in anti-PD-1-treated lung cancers. Nature. 2021;596:126–132. doi: 10.1038/s41586-021-03752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edwards J., et al. CD103(+) Tumor-resident CD8(+) T cells are associated with improved survival in immunotherapy-naive melanoma patients and expand significantly during anti-PD-1 treatment. Clin. Cancer Res. 2018;24:3036–3045. doi: 10.1158/1078-0432.CCR-17-2257. [DOI] [PubMed] [Google Scholar]
- 32.Liikanen I., et al. Hypoxia-inducible factor activity promotes antitumor effector function and tissue residency by CD8+ T cells. J. Clin. Invest. 2021;131 doi: 10.1172/JCI143729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets in the current study are open to the public at the TCGA (https://xena.ucsc.edu/) and Javelin 101 clinical trial (https://www.nature.com/articles/s41591-020-1044-8). Further inquiries can be directed to the corresponding authors. The data of our cohorts that support the results of this study are available from the corresponding author upon reasonable request.