Summary
Platelets play an important role in the pathogenesis of sepsis and platelet transfusion is a therapeutic option for sepsis patients, although the exact mechanisms have not been elucidated so far. ITGA2B encodes the αIIb protein in platelets, and its upregulation in sepsis is associated with increased mortality rate. Here, we generated a Itga2b (Q887X) knockin mouse, which significantly reduced ITGA2B expression of platelet and megakaryocyte. The decrease of ITGA2B level aggravated the death of septic mice. We analyzed the transcriptomic profiles of the platelets using RNA sequencing. Our findings suggest that ITGA2B upregulates PTPN6 in megakaryocytes via the transcription factors Nfkb1 and Rel. Furthermore, PTPN6 inhibits platelet apoptosis and necroptosis during sepsis by targeting the Ripk1/Ripk3/Mlkl and caspase-8 pathways. This prevents Kupffer cells from rapidly clearing activated platelets, and eventually maintains vascular integrity during sepsis. Our findings indicate a new function of ITGA2B in the regulation of platelet death during sepsis.
Subject areas: Natural sciences, Biological sciences, Physiology, Pathophysiology, Immunology
Graphical abstract

Highlights
-
•
ITGA2B upregulates PTPN6 in the megakaryocytes via Nfkb1 and Rel
-
•
The high level of PTPN6 protects platelets from apoptosis and necroptosis during sepsis
-
•
ITGA2B prevents Kupffer cells from clearing platelets during sepsis
Natural sciences; Biological sciences; Physiology; Pathophysiology ; Immunology ;
Introduction
Invasion of pathogenic bacteria in the bloodstream and the ensuing bacteremia increases the levels of bacterial toxins and other metabolites, which can progress to sepsis if not treated promptly. It is clinically manifested by the systemic inflammatory response syndrome, which comprises of fever, tachycardia, decreased blood pressure, tachypnea, and leukocytosis.1,2 Sepsis is one of the major causes of infection-related deaths and hospitalizations, and about 40 million sepsis patients are treated worldwide every year.3 In addition, the high treatment costs of bacterial septicemia result in considerable economic burden to the patients.4 The high incidence and mortality rates sepsis can be attributed to the lack of knowledge regarding the pathogenesis of sepsis, which translates to a lack of effective targeted therapeutic drugs.5 Therefore, it is crucial to elucidate the pathological basis of sepsis, particularly the interaction between the bacteria and immune cells, in order to develop new targeted interventions and improve patient survival.
Thrombocytopenia is very common during sepsis and is partly the result of disseminated intravascular coagulation.6 Platelets are nuclear-free blood cells involved in hemostasis and thrombosis,7 and play various roles in the inflammatory response.8 The chemokines released by activated platelets enhance the migration of neutrophils and other leukocytes to inflammatory tissues by stimulating endothelial cells.9,10 Platelet infusion has proved to be effective in patients with non-hemorrhagic sepsis, and can reduce the death of mice caused by Escherichia coli infection.8,11,12 It remains to be clarified whether platelets play a protective role in the inflammatory response.
The integrin α2B (Itga2b) gene is upregulated in circulating platelets and is associated with higher mortality rates of septic patients and mice.13 It encodes integrin αIIb (CD41), which forms a complex with integrin 3, a marker of the megakaryocyte/platelet lineage. Integrin αIIbβ3 is necessary for normal platelet hemostatic function,14 and ITGA2B mRNA is actively converted into new proteins in platelets during sepsis, accompanied by increased activation of integrin αIIbβ3.13 However, the mechanism of platelet ITGA2B protecting sepsis patients or animals is still unclear.
Research shows that Itga2b p.Q891X mutation in platelets of GT patients produces truncated pro-αIIb protein, forms truncated pro-αIIbβ3 complex with β3, remains in the endoplasmic reticulum, and is eventually degraded in large quantities.15 In the previous study, we studied the family of three Itga2b p.Q891X homozygote, and the results showed that the ITGA2B protein in platelets of patients and carriers was significantly reduced.16 We created a mouse model with point mutation (Q887X) at mouse Itga2b locus by CRISPR-Cas-mediated genome engineering, which was the same sequence loci as human Itga2b p.Q891X. Mutations in Itga2b can affect a series of signaling pathways in platelets, thereby affecting platelet function. Here, we employed parallel techniques of RNA sequencing (RNA-seq) to interrogate the transcriptional and translational landscape in platelets from murine platelets of wild type (WT) and Itga2b (Q887X) knockin mice, and found that the inflammation-related factor PTPN6 was differentially expressed. Protein tyrosine phosphatase non-receptor type 6 (PTPN6) is a key regulator of bone marrow hematopoiesis, and relays signals via the immune receptor tyrosine group inhibitory motif.17 PTPN6 controls the activity of about 60 cell surface receptors and cytoplasmic signaling proteins.18 PTPN6 negatively regulates activation of p38 mitogen-activated protein kinase to control IL-1α and tumor necrosis factor dual function in expression and maintenance of Ripk1 function to prevent Ripk3-Mlkl and caspase-8-dependent cell death and the associated IL-1 α/β dual role of release.19,20 However, the mechanism by which platelet apoptosis and necroptosis is controlled by ITGA2B in sepsis is not well known.
In this study, we found that platelet ITGA2B prevents platelet death during sepsis by upregulating PTPN6, which in turn inhibits the pro-apoptotic caspase-8 and Ripk3/Mlkl pathways. Our findings suggest a potentially novel mechanism by which ITGA2B protects platelet death to drive pathological inflammation during sepsis.
Results
ITGA2B plays an important role in inflammatory response
Integrins play critical roles in platelet functions, and αIIbβ3 is a megakaryocyte and platelet-specific integrin that regulates the adhesion of inflammatory endothelial cells and promotes thrombotic response.21 Itga2b encodes the αIIb subunit of integrin IIbβ3. We generated an Itga2b (Q887X) knockin mouse model with c.2659C > T substitution in the gene sequence (Figure 1A). Itga2b mRNA levels were significantly decreased in the Itga2b (Q887X) knockin mice compared to the WT littermates (Figure 1B). No clear spontaneous bleeding phenotypes were observed in Itga2b (Q887X) knockin mice. However, Itga2b (Q887X) knockin mice have platelet dysfunction,16 but the potential impact of platelet dysfunction on the death of mice caused by sepsis will be excluded through the following study.
Figure 1.
ITGA2B plays an important role in the inflammatory response
(A) Platelet Itga2b (Q887X) knockin mice were genotyped by PCR and subsequent sequence analysis.
(B) Itga2b mRNA expression was detected by RT-qPCR. n = 5 mice/group. Data were mean ± SD of at least 3 independent experiments and compared between groups using unpaired t test. Statistical significance was represented by asterisks, ∗∗∗∗p < 0.001.
(C) Scatterplot showing enriched pathways of the downregulated genes. The size of each circle indicates the number of significantly downregulated genes in each corresponding pathway. The enrichment factor was calculated using the number of enriched genes divided by the number of all background genes in each pathway. p < 0.05 indicates statistical significance.
(D) Heatmap showing upregulated (red) and downregulated (green) genes. E-F. GSEA using MSigDB identified differential gene enrichment between Itga2b (Q887X) knockin mice and WT mice. NES, Normal p and FDR q values for each gene set are shown. The results revealed differential expression of genes involved in inflammatory responses.
We profiled the transcriptomes of the platelets from Itga2b (Q887X) knockin mice and WT littermates by RNA-seq, and compared the gene expression patterns by principal components analysis. As shown in Figure S1A, the Itga2b (Q887X) knockin platelets were grouped separately from the WT control group of principal component 1, which accounted for 98% of the transcriptional variation. Compared to the WT group, 559 genes were significantly upregulated and 1772 were significantly downregulated in the platelets from Itga2b (Q887X) knockin mice (Figures S1B and S1C). Gene Ontology analysis of the upregulated genes revealed significant enrichment of pathways related to inflammation (Figure 1C). We established a gene set with all the genes in the inflammation-related pathway, and constructed a heatmap to confirm that most of these genes are upregulated after Itga2b inactivation (Figure 1D). Gene set enrichment analysis confirmed significant enrichment of upregulated genes involved in inflammatory responses and cytokine-mediated signaling pathways (Figures 1E and 1F). Taken together, Itga2b downregulation may significantly increase the risk of inflammation-related diseases.
ITGA2B deficiency exacerbated systemic inflammation and organ injury in mice during sepsis
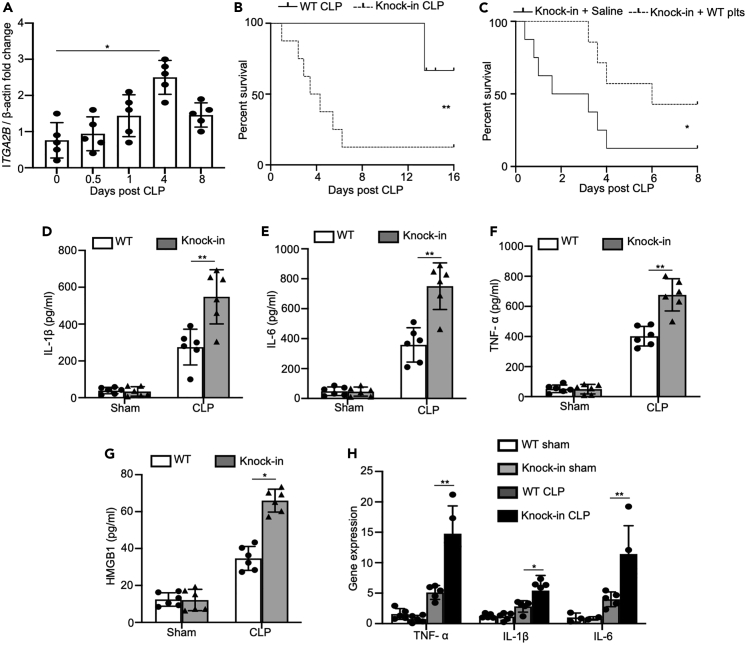
ITGA2B expression increased significantly in the platelets of mice within 8 days following cecal ligation and puncture (CLP), and peaked after 4 days (Figure 2A). Thus, ITGA2B is upregulated in platelets during sepsis. To determine its potential role in the pathogenesis of sepsis, we challenged WT and Itga2b (Q887X) knockin mice with CLP model. About 40% of WT mice and 90% of Itga2b (Q887X) knockin mice were dead 16 days following CLP (Figure 2B). We also conducted survival rate tests on mice of different ages, such as 1 month, 6 months, and 1 year. Although the mortality rate of sepsis varies among mice of different ages, the mortality rate of Itga2b (Q887X) knockin mice is significantly higher than that of WT mice of the same age (Figure S2). We injected WT platelets (4×108 platelets/mice) intravenously into Itga2b (Q887X) knockin mice 4 h after CLP. The control group was given physiological saline. 95% Itga2b (Q887X) knockin mice given physiological saline died within 8 days, while 45% of mice infused with WT platelets survived (Figure 2C). This strongly suggested that platelet ITGA2B plays a protective role in sepsis, which may be related to systemic inflammation. To this end, we analyzed the levels of inflammatory cytokines in the plasma of WT and Itga2b (Q887X) knockin mice. Indeed, plasma levels of IL-1β, TNF-a, IL-6, and HMGB-1 were significantly higher in the Itga2b (Q887X) knockin mice compared to the WT mice 24 h after sepsis induction (Figures 2D–2G), indicating that platelet ITGA2B relieves sepsis-induced systemic inflammatory response. We also analyzed the expression of TNF-a, IL-6, and IL-1β mRNAs in the peripheral blood mononuclear cells from WT and Itga2b (Q887X) knockin mice, and observed similar results (Figure 2H), indicating that ITGA2B regulates the biosynthesis of inflammatory cytokines.
Figure 2.
ITGA2B is upregulated in murine platelets during sepsis and inhibits CLP-induced cytokine production in mice
(A) Itga2b mRNA levels in platelets isolated from sham or CLP mice after 0.5, 1, 4, and 8 days post perform CLP, 8 days was considered a recovery time point (n = 5/group).
(B) The WT and Itga2b (Q887X) knockin mice were performed CLP (n = 15 mice/group). The survival rate was monitored and analyzed using log rank test. Statistical significance was represented by asterisks, ∗∗, p < 0.01.
(C) The Itga2b (Q887X) knockin mice were performed CLP (n = 8 mice/group). 108 WT platelets or saline were injected intravenously into Itga2b (Q887X) knockin mice 4 h after CLP. The survival rate was monitored and analyzed using log rank test. Statistical significance was represented by asterisks, ∗, p < 0.05.
(D–G) Itga2b (Q887X) knockin mice and WT littermates were performed sham or CLP for 24 h. TNF-α, IL-1β, IL-6, and HMGB-1 levels in the plasma were measured by ELISA. n = 6. H. TNF-α, IL-1β, and IL-6 mRNA levels in PBMCs. n = 5. Data were mean ± SD of at least 3 independent experiments and were analyzed using 1-way ANOVA followed by Tukey’s test for multiple groups and were representative of 3 independent experiments. Statistical significance was represented by asterisks, ∗, p < 0.05; ∗∗, p < 0.01.
Since multiple organ dysfunction is a frequent complication of sepsis, we also compared the histological changes in the major organs of WT and Itga2b (Q887X) knockin mice following CLP. The lungs of Itga2b (Q887X) knockin mice had alveolar cavity atrophy, inflammatory injury, alveolar wall edema, and interstitial thickening compared to the WT mice (Figure 3A). Scanning electron microscopy further revealed severe hemorrhage and alveolar swelling, as well as excessive fibrin deposition in the lungs of Itga2b (Q887X) knockin mice compared to that of WT mice subjected to CLP (Figure 3B). Sepsis induces an acute inflammatory reaction in the lungs, damages the pulmonary vascular endothelial cells, activates white blood cells, and increases production of free radicals that cause lung injury.22 Consistent with this, CLP mice significantly increased the number of inflammatory cells and protein content in the BALF compared to that in the sham group (Figures 3C and 3D). Furthermore, these inflammatory changes were exacerbated in Itga2b (Q887X) knockin mice compared to the WT counterparts following CLP (Figures 3C and 3D). Myeloperoxidase (MPO) is a peroxidase enzyme that contains prosthetic heme group and is predominantly released by activated neutrophils. Not surprisingly, the level of MPO in the lungs was significantly higher in the Itga2b (Q887X) knockin mice compared to the WT mice (Figure 3E).
Figure 3.
ITGA2B is upregulated in murine platelets and ameliorates multiple organ injury and severe hemorrhage during sepsis
(A) Itga2b (Q887X) knockin mice and WT littermates were performed sham or CLP for 24 h, followed by organ harvest. Representative images of H&E-stained liver sections 24 h after CLP. Scale bars: 20 μm.
(B) Representative images of scanning electron micrographs of lung tissues are shown. Scale bars: 50 μm.
(C–E) Total protein concentration and white blood cells in BALF, and MPO activity in lung homogenates were measured 24 h after CLP.
(F) Representative images of H&E-stained liver sections and quantification of injuries 24 h after CLP. Scale bars: 20 μm.
(G) Representative images of the intestines from sham or CLP mice 24 h after CLP.
(H and I) Vascular permeability index (Evans blue concentration/weight of the tissue) 24 h after CLP.
(J) Representative images of BALF from WT mice and Itga2b (Q887X) knockin mice 24 h after CLP.
(K) Presence of RBCs in the lavage of septic mice. (n = 5. Data were mean ± SD of at least 3 independent experiments and were analyzed using 1-way ANOVA followed by Tukey’s test for multiple groups and were representative of 3 independent experiments. Statistical significance was represented by asterisks, ∗, p < 0.05; ∗∗, p < 0.01.)
The liver tissues of Itga2b (Q887X) knockin mice showed unclear sinuses, damaged lobular structure, swollen peripheral cells, edema, and connective tissue hyperplasia compared to the WT mice 24 h following CLP, indicating considerable liver damage. In addition, ballooning degeneration, a form of apoptosis, was also significantly higher in the Itga2b (Q887X) knockin mice compared to the WT mice (Figure 3F). These results suggest that platelet ITGA2B can alleviate the multiple organ failure caused by sepsis.
Meanwhile, in the process of dissecting mice, we found that the Itga2b (Q887X) knockin mice also exhibited massive bleeding in the mesenteric lymph nodes and small intestines after CLP (Figure 3G), which suggested that ITGA2B may regulate vascular wall permeability during the systemic inflammatory response. To test this hypothesis, we injected the mice intravenously with Evans Blue, and analyzed the amount of extravasated dye in the lungs, livers, and other tissues to determine vascular leakage. Compared to the WT mice, the Itga2b (Q887X) knockin mice showed a significant increase in vascular permeability in multiple organs following CLP (Figures 3H and 3I). And we found massive pulmonary bleeding exclusively in Itga2b (Q887X) knockin mice 24 h after CLP (Figures 3J and 3K). Therefore, we can surmise that platelet ITGA2B plays an essential role in maintaining the integrity of vascular wall during sepsis.
Platelets underwent apoptosis and necroptosis in Itga2b (Q887X) knockin mice during sepsis
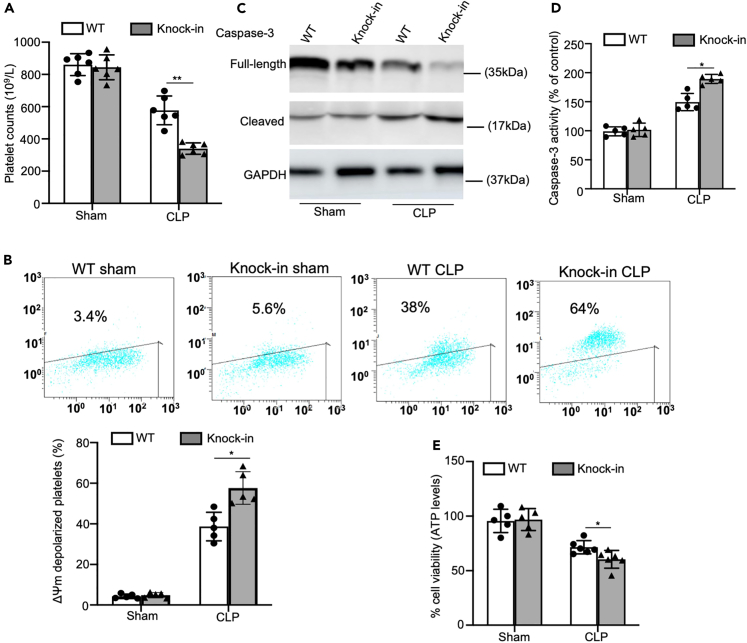
Platelets can promote the activation of endothelial cells and the recruitment of inflammatory cells, and are indispensable for maintaining vascular integrity in inflammatory tissues.23 Given that inflammation induces thrombocytopenia, we next measured the platelet counts of the mice during sepsis. At 24 h after inducing CLP, the peripheral platelet count of the Itga2b (Q887X) knockin mice was considerably lower than that of WT controls (Figure 4A). There is also evidence of platelet death during sepsis. Since mitochondrial membrane depolarization is a marker of early apoptosis, we analyzed the changes in mitochondrial membrane potential (ΔΨm) in the platelets of WT and Itga2b (Q887X) knockin mice following CLP. The platelets from CLP mice emitted an intense green fluorescence compared to that in the control group (Figure 4B). Furthermore, the number of JC-1 monomer-positive cells (green fluorescence) increased significantly in the Itga2b (Q887X) knockin mice compared to the WT mice after inducing CLP. These findings indicated that platelet Itga2b (Q887X) knockin reduced ΔΨm, thereby initiating the mitochondrial apoptosis pathway during sepsis. Caspases combined with other apoptogenic enzymes disrupt plasma membrane integrity leading to phosphatidylserine externalization during apoptosis. Consistent with this, sepsis significantly elevated caspase-3 activity in platelets of Itga2b (Q887X) knockin mice 24 h after inducing CLP, as indicated by the appearance of 17-kDa fragments (Figure 4C), and total caspase-3 activity (Figure 4D).
Figure 4.
Platelets undergo apoptosis and necroptosis in Itga2b (Q887X) knockin mice during sepsis
(A) Peripheral platelet counts in Itga2b (Q887X) knockin mice and WT littermates at 24 h after CLP.
(B) Representative flow cytometric figures showing platelet Δψm depolarization at 24 h after CLP. The monomeric form of JC-1 accumulates in the cytosol after mitochondrial membrane depolarization, and the JC-1 aggregates are present in the intact mitochondria.
(C) Western blot analysis of caspase-3 with anti-caspase-3 antibody at 24 h after CLP.
(D) Analysis of caspase-3 activity with ELISA at 24 h after CLP, n = 5.
(E) Viability of platelets from WT and Itga2b (Q887X) knockin mice at 24 h after CLP. n = 6. (Data were mean ± SD of at least 3 independent experiments and were analyzed using 1-way ANOVA followed by Tukey’s test for multiple groups and were representative of 3 independent experiments. Statistical significance was represented by asterisks, ∗p < 0.05. ∗∗p < 0.01. ∗∗∗p < 0.001.)
We next assessed if sepsis could initiate necroptosis in platelets of Itga2b (Q887X) knockin mice. Platelet viability was assessed by the Cell Titer Glo assay, measuring ATP abundance 24 h post CLP. Itga2b (Q887X) knockin mice led to extensive platelet loss compared to the WT mice following CLP, which was consistent with a predominantly necrotic form of death (Figure 4E).
Taken together, Itga2b (Q887X) knockin enhanced apoptosis and necroptosis in platelets during sepsis.
Itga2b (Q887X) knockin increased platelet clearance in the liver
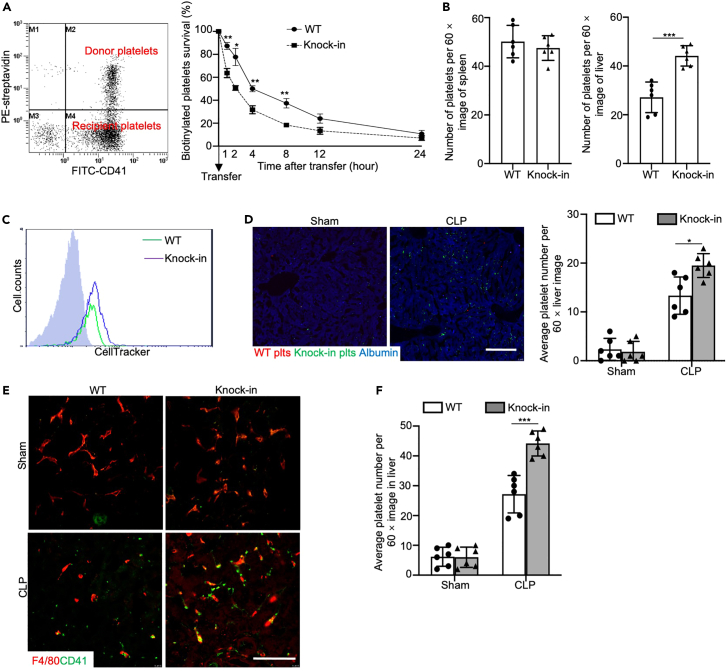
Compared to the WT mice, the Itga2b (Q887X) knockin mice had a reduced number of platelets during sepsis (Figure 4A). To determine whether accelerated platelet clearance plays an essential role in the pathogenesis of sepsis in the Itga2b (Q887X) knockin mice, we transfused 108 biotin-labeled septic WT or septic Itga2b (Q887X) knockin platelets into WT recipient mice. According to the proportion (%) of platelets transfused at each time point, the Itga2b (Q887X) knockin mice platelets were cleared faster than WT platelets, and a rapid decrease in the transfused Itga2b (Q887X) knockin platelets occurred within the first 4 h (Figure 5A). Thus, Itga2b (Q887X) knockin increased clearance of platelets in the septic mice, resulting in a significant reduction in the number of peripheral platelets.
Figure 5.
Itga2b (Q887X) knockin increased platelet clearance in the liver
(A) Representative dot plot showing transfusion of biotinylated platelets from septic WT mice or septic Itga2b (Q887X) knockin mice into WT recipients. Peripheral blood samples were taken from recipient mice at 0, 1, 2, 4, 8, 12, and 24 h after transfusion. Data represent means ± SD at each time point, n = 4 experiments.
(B) Bar graphs are quantification of platelets per 60× image. Data represent mean ± SD. n = 6 images/genotype.
(C) Flow cytometry analysis of labeling efficiency of WT and Itga2b (Q887X) knockin platelets by CellTracker CMRA (red) and CMFDA (green), respectively.
(D) Representative fluorescence microscopy images of liver sections of WT recipients 4 h after the competitive transfusion of labeled control or septic WT and control or septic Itga2b (Q887X) knockin platelets with additional staining of F4/80 (blue). Scale bar, 50 μm. Bar graphs show number of platelets per 60× field, n = 6 experiments.
(E) Typical confocal images of frozen sections of liver from WT and Itga2b (Q887X) knockin septic mice stained with anti-CD41 (green) and anti-F4/80 (red) antibodies. Scale bar, 10 μm.
(F) Number of platelets in the liver sections per image (60×). n = 6. (Data were mean ± SD of at least 3 independent experiment. Data from multiple groups were compared by a one-way ANOVA followed by a Tukey's test and two groups were compared by two-tailed student’s t-test and were representative of 3 independent experiments. Statistical significance was represented by asterisks, ∗p < 0.05. ∗∗p < 0.01. ∗∗∗p < 0.001.)
To further explore the site of platelet clearance after apoptosis and necroptosis, we analyzed the number of platelets in the liver and spleen of the WT and Itga2b (Q887X) knockin mice following CLP. There was no difference in the number of platelets in the spleen of both groups, whereas the number of platelets in the liver sections was almost 2-fold higher in the Itga2b (Q887X) knockin group compared to the WT group at 24 h after inducing CLP (Figure 5B). To confirm that the liver is the main site of platelet clearance in the Itga2b (Q887X) knockin mice during sepsis, we simultaneously transfused the same number of CMRA-labeled CLP WT and CMFDA-labeled CLP Itga2b (Q887X) knockin platelets into WT recipient mice (Figure 5C). The number of Itga2b (Q887X) knockin platelets (green) in the liver was significantly higher than that of the WT platelets (red) (Figures 5D and S3). Furthermore, immunofluorescence staining preliminarily showed the involvement of Kupffer cells in platelet clearance (Figures 5E, 5F, and S4). Taken together, the Itga2b (Q887X) knockin platelets were rapidly cleared via the liver, which indicates that ITGA2B prevents platelet clearance during sepsis.
PTPN6 is a negative regulator of platelet death during sepsis
PTPN6, a cytoplasmic phosphatase with anti-inflammatory action, was among the top 20 downregulated genes in the platelets from Itga2b (Q887X) knockin mice (Table S1; Figure 6A). Furthermore, PTPN6 was significantly downregulated in the whole lysates as well as on the surface of Itga2b (Q887X) knockin platelets compared to the WT (Figures 6B and 6C). The protein-protein interaction network of the DEGs further confirmed the important role played by PTPN6 in the pathology of inflammation (Figure 6D). Studies show that PTPN6 inhibits Ripk3/Mlkl and caspase-8-dependent inflammation.20 Therefore, the aggravated disease phenotype in the Itga2b (Q887X) knockin mice suggests that inactivation of PTPN6 may partially affect the ability of Ripk1/Ripk3/Mlkl and caspase-8 pathways to inhibit inflammatory damage. We detected increased expression of RIPk1, p-RIPk1, Ripk3, p-RIP3, MLKL, and p-MLKL in the platelets from Itga2b (Q887X) knockin mice, indicating that the Ripk1/Ripk3/MLKL pathway was hyperactivated in the platelets both before and after sepsis (Figures 7A–7C and S5A–S5C). Furthermore, cleaved caspase-8 was detected in Itga2b (Q887X) knockin but not in WT platelets during sepsis (Figures 7D and S5D). Therefore, the sepsis process is driven by the downregulation of PTPN6, and the consequent induction of caspase-8-dependent apoptosis and Ripk3-Mlkl-dependent platelet death.
Figure 6.
Platelet PTPN6 was decreased in Itga2b (Q887X) knockin mice
(A) Volcano plot of significantly (FDR,0.05) upregulated (log2 fold-change 0.1.5, red) and downregulated (log2 fold change, 21.5, green) genes in the platelets from WT and Itga2b (Q887X) knockin mice. PTPN6 is highlighted.
(B) Immunoblot showing PTPN6 protein levels in the lysates of WT and Itga2b (Q887X) knockin platelets. n = 6.
(C) PTPN6 expression on the surface of platelets as detected by flow cytometry. n = 6.
(D) PPI network based on significantly downregulated genes. The size of each circle indicates the node degree of the corresponding gene, and the gray lines represent interacting partners. (Data were mean ± SD of at least 3 independent experiments and compared between groups using unpaired t test. Statistical significance was represented by asterisks, ∗p < 0.05. ∗∗p < 0.01. ∗∗∗p < 0.001.)
Figure 7.
PTPN6 drives inflammation through caspase-8 and Ripk3/MLKl pathway in Itga2b (Q887X) knockin mice during sepsis
(A–C) Immunoblots showing levels of p-RIP1, RIP1(A), p-RIP3, RIP3(B); and p-MLKL, p-MLKL(C) in platelets from the sham or CLP mice at 24 h after CLP.
(D) Immunoblot showing cleaved caspase-8 (cl.Casp8) levels in freshly isolated WT and Itga2b (Q887X) knockin platelets.
(E) Meg-01 cells transfected with control siRNA or Ptpn6 siRNA, and knockdown efficiency was determined by western blotting.
(F) Immunoblot showing cleaved caspase-8 (cl.Casp8) in Meg-01 cell treated with control siRNA or PTPN6 siRNA in the presence and absence of LPS.
(G–I) Immunoblots showing levels of p-RIP1, RIP1(G), p-RIP3, RIP3(H); and p-MLKL, p-MLKL(I) in Meg-01 cell treated with control siRNA or PTPN6 siRNA in the presence and absence of LPS. Luminescence was quantified using a ChemiDoc Gel Imaging System and Image Lab software. n = 6. Data were normalized to the amount of the loading control protein. (Data were presented as the mean ± SD of six independent experiments and compared between groups using unpaired t test. Statistical significance was represented by asterisks, ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001.)
To further verify whether platelet death signals are regulated by PTPN6 during sepsis, we knocked down the genes in Meg-01 cells (Figure 7E). The inhibition of PTPN6 increased LPS-induced tyrosine phosphorylation of the Ripk1/Ripk3/MLKL and caspase-8 pathways (Figures 7F–7I). Taken together, PTPN6 is a negative regulator of caspase-8 and Ripk3-Mlkl-dependent platelet death during sepsis.
ITGA2B regulates PTPN6 expression via the transcription factors NFkb1 and Rel
Platelets are produced by bone marrow megakaryocytes.24 To further explore the mechanisms regulating PTPN6 in the megakaryocytes of Itga2b (Q887X) knockin mice, we screened for the downregulated transcription factors in order to identify the potential regulators of PTPN6 (Figure 8A). We identified Ebf1, Hes1, Ikzf1, Nfkb1, Nr3c1, Pax5, Rel, and Usf1 (Figure 8B; Tables S2 and S3), of which Hes1, Nfkb1, and Rel reportedly are involved in the inflammatory response, and Nfkb1 and Rel are subunits of the pro-inflammatory nuclear factor κB (NF-κB),25 so we focus on Nfkb1 and Rel. Furthermore, a marked decrease in Nfkb1 and Rel protein and mRNA was observed in the bone marrow megakaryocytes of Itga2b (Q887X) knockin mice (Figures 8C, 8D and S6). To ascertain whether PTPN6 is the target gene of Nfkb1 and Rel, we performed a luciferase reporter assay, which confirmed the binding of Nfkb1 and Rel to the PTPN6 promoter. The luciferase activity of the reporter gene placed under the PTPN6 promoter increased following co-transfection with Nfkb1 and Rel-expressing vectors (Figures 8E and 8F). Taken together, our results suggest that ITGA2B upregulates PTPN6 in murine megakaryocytes via NF-κB, which directly binds to its promoter region.
Figure 8.
ITGA2B influences the expression of PTPN6 by regulating the transcription factor NFkb1 and Rel
(A) Transcription factors (TFs) that target PTPN6 were predicted using the PROMO ALGGEN database within a dissimilarity margin less than or equal to 5%.
(B) Venn diagram showing downregulated TFs and predicated TFs of PTPN6 in the platelets. The number of congruently downregulated genes is indicated in the middle, and their gene symbols are listed at the bottom.
(C) Nfkb1 and Rel mRNA levels in the WT mice and Itga2b (Q887X) knockin mice. n = 6.
(D) Representative confocal microscopic images of WT and Itga2b (Q887X) knockin mice bone marrow megakaryocytes. Arrows point to the megakaryocytes. n = 6. Scale bar, 10 μm.
(E and F) Luciferase activity levels in HEK293T cells co-transfected with the PTPN6 promoter-Luc reporter and GV230-Nfkb1, GV230-Rel, or empty vector. Luciferase activity levels in HEK293T cells co-transfected with the PTPN6 promoter-Luc reporter and GV230-vector containing Nfkb1, and Rel or an empty vector as the control. n = 9 mice/group. (Data were presented as the mean ± SD of three independent experiments and were analyzed using 1-way ANOVA followed by Tukey’s test for multiple groups and were representative of 3 independent experiments. Statistical significance was represented by asterisks, ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001.)
Discussion
We have shown that platelet/megakaryocytes ITGA2B plays a crucial role in the pathogenesis of sepsis. ITGA2B upregulates PTPN6 in the megakaryocytes via Nfkb1 and Rel, and the high levels of PTPN6 protect the platelets from apoptosis and necroptosis during sepsis by inhibiting the Ripk1/Ripk3/MLKL and caspase-8 pathways. This prevents the rapid clearance of platelets by Kupffer cells, and helps maintain vascular integrity during sepsis.
In fact, platelets are increasingly being considered a potential therapeutic target for reducing thrombosis and the excessive inflammatory response during sepsis.21,26 The acute changes in platelet function that accompany septic progression are often related to adverse clinical results.27,28 The expression of ITGA2B in the platelets of sepsis patients is about 9-fold higher than that in healthy controls, and eventually normalizes in the survivors.13 High ITGA2B expression in platelets during sepsis is the result of its increased transport from the megakaryocytes to platelets.13 However, it is unclear how sepsis upregulation of ITGA2B could relieve sepsis-associated systemic inflammatory response. We generated an Itga2b c.2659C > T (p.Q887X) knockin mouse model using CRISPR-Cas9 technology.16 The levels of Itga2b mRNA and protein were very low in platelets of Itga2b c.2659C > T (p.Q887X) knockin mouse. Thus, we used the Itga2b c.2659C > T (p.Q887X) knockin mouse to study the role of platelet ITGA2B in sepsis and analyzed the transcriptional changes in the platelets. However, Itga2b (Q887X) knockin mice have platelet dysfunction,16 but the potential impact of platelet dysfunction on the death of mice caused by sepsis will be excluded through the following study. We found that the lack of ITGA2B exacerbated the sepsis-induced inflammatory response compared to that in WT mice, indicating that platelet ITGA2B plays an important role in inflammatory response during sepsis. Under physiological conditions, they do not cause platelet apoptosis or necroptosis, nor do they show significant organ bleeding. During sepsis, there was a significant increase in abnormal platelet death and severe organ bleeding in Itga2b c.2659C > T (p.Q887X) knockin mouse, indicating that their phenotype during sepsis was not caused by abnormal platelet function caused by ITGA2B mutations.
The interaction among inflammation, coagulation, and platelet activation determines the degree of organ dysfunction at the different stages of sepsis.29 ITGA2B is significantly upregulated in the platelets and megakaryocytes during sepsis, and its sequences are conserved between human and mouse.13 We found that CLP mice significantly increased the expression of platelet ITGA2B, and CLP-induced tissue damage and mortality were aggravated in the Itga2b (Q887X) knockin mice. The mRNA level of inflammatory factors in leukocytes of Itga2b (Q887X) knockin mice is significantly increased, suggesting that platelet ITGA2B regulates the expression of inflammatory factors from the mRNA level, indicating that ITGA2B is an important regulatory medium for sepsis. Sepsis usually leads to thrombocytopenia, thus triggering massive production of platelets to avoid bleeding.30,31 In the Itga2b (Q887X) knockin mice, organ bleeding was only visible following sepsis, which is consistent with previous data showing that the Itga2b mutation itself is not enough to induce bleeding in mice.32 It is possible that platelet ITGA2B is crucial to preventing inflammatory bleeding, and platelets may play a role in a paracrine manner.33,34,35 Although it is unclear how bacteria induce thrombocytopenia, there is evidence that LPS injection in WT mice can promote the formation of platelet aggregates in the pulmonary and hepatic microvascular circulation, leading to severe thrombocytopenia.36 We observed significant thrombocytopenia in the Itga2b (Q887X) knockin mice during sepsis, suggesting that ITGA2B is crucial to the survival of platelets. Furthermore, the platelets in Itga2b (Q887X) knockin mice showed high rates of apoptosis and necrosis were rapidly cleared by the Kupffer cells in the liver, which also contributed to a decrease in the number of peripheral platelets.
Preclinical models of sepsis are essential to elucidate the mechanisms underlying sepsis and identify novel therapeutic targets.37 The tyrosine phosphatase PTPN6 was among the most significantly downregulated genes in the platelets of Itga2b (Q887X) knockin mice. It is mainly expressed in the hematopoietic cells, and regulated multiple signaling pathways38 involved in inflammatory diseases.39,40 Furthermore, PTPN6 inhibits IL-1R-mediated inflammation.17,20 However, little is known about the regulatory mechanism of PTPN6 expression in platelets/megakaryocytes. Studies show that PTPN6 maintains the function of Ripk1, and blocks the Ripk3-Mlkl and caspase-8 apoptotic pathways.41 Mlkl regulates platelet function and inflammation. The necrosis signal of megakaryocytes is dispensable for platelet production.42 Consistent with this, the Ripk3-MLKl and caspase-8 pathways were upregulated in the Itga2b (Q887X) knockin mice during sepsis.
To further explore the mechanisms regulating PTPN6 expression in the platelets, we screened the RNA-seq data of the Itga2b (Q887X) knockin platelet/megakaryocytes for the downregulated transcription factors, and identified Hes1, Nfkb1, and Rel. Nfkb1 and Rel are subunits of the transcription factor NF-κB.25 The phenotype induced by Nfkb1 or Rel deletion is observed in some tissues but not in others,43,44 thus possibly reflecting the redundancy of pathways involving NF-κB signaling proteins. A dysregulated NF-κB pathway may increase the risk of inflammation during sepsis.45 We found that Nfkb1 and Rel directly bind to the PTPN6 promoter to increase its expression in the megakaryocytes. Therefore, we hypothesized that Nfkb1 and Rel are critical transcriptional regulators of PTPN6 during sepsis.
Studies on murine and baboon models of sepsis have shown that pharmaceutical inhibition of αIIbβ3 improved the parameters of organ failure, endothelial function, and mortality,46 which is consistent with abnormal thrombosis that frequently accompanies sepsis. We found that increased ITGA2B expression correlated with higher mortality rates in infected patients and mice. A clear understanding of the functional significance of increased Itga2b mRNA and the translation of integrin subunit αIIb should be the focus of future research. In conclusion, we have shown for the time that platelet ITGA2B inhibits inflammatory platelet death during sepsis by downregulating Ptpn6, and this pathway is a potential therapeutic target that warrants further investigation.
Limitations of the study
Our data provide the novel insights into the mechanisms of platelet death during sepsis. They also suggest potential therapeutic avenues for septic patients. We suggest a potentially novel mechanism underlying the protection of platelet function during sepsis. However, although ITGA2B is predominantly expressed in the MK/platelet lineage, inflammatory stimuli and conditions can be expressed by T cells and monocytic lineage. We have bypassed that in our study in the in vivo assays during the experiment. In additional, CRISPR-Cas system by itself has off-target effects that may underline the described genetic variations between WT mice and CRISPR-Cas9 Itga2b knockin mice. In order for any of the data to be reliable, the control mice should be derived from the heterozygote of the knockin mice.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Antibodies against caspase-3 | Cell Signaling Technology | Cat#9662; RRID:AB_841281 |
| PE anti-CD41 antibody | BD Biosciences | Cat#558040; RRID: AB_397004 |
| FITC anti-CD41 antibody | eBioscience | Cat#11-0411 |
| Chicken polyclonal anti-albumin antibody | Abcam | Cat#ab106582; RRID: AB_10888110 |
| Rat monoclonal anti-F4/80 | Abcam | Cat#ab6640; RRID: AB_1140040 |
| Rabbit monoclonal anti-CD41 | Abcam | Cat#ab134131; RRID: AB_2732852 |
| Alexa Fluor 405–conjugated goat anti-chicken IgY | Abcam | Cat#ab175674; RRID: AB_2890171 |
| Alexa Fluor 594–conjugated goat anti-rat IgG | Abcam | Cat#ab150160; RRID: AB_2756445 |
| Alexa Fluor 647–conjugated donkey anti-rabbit IgG | Abcam | Cat#ab150075; RRID: AB_2752244 |
| PE-anti-SHP1 antibody | Abcam | Cat#ab209002 |
| SHP1 Rabbit mAb | SAB | Cat#48636 |
| Rabbit polyclonal anti-p-RIP1 | CST | Cat#53,286 |
| Rabbit polyclonal anti-RIP1 | Abcam | Cat#ab56815; RRID: AB_945152 |
| Rabbit polyclonal anti-RIP3 | Abcam | Cat#ab62344; RRID: AB_956268 |
| Rabbit polyclonal anti-p-RIP3 | Abcam | Cat#ab195117; RRID: AB_2768156 |
| Rabbit polyclonal anti-Mlkl | Abcam | Cat#ab71399; RRID: AB_2145458 |
| Rabbit monoclonal anti-p-Mlkl | Abcam | Cat#ab196436; RRID: AB_2687465 |
| Rabbit monoclonal anti-caspase8 | CST | Cat#: 4790 |
| Rabbit polyclonal anti-GAPDH | Abcam | Cat#ab2251-1; RRID: AB_1267174 |
| Rabbit polyclonal anti-Nfkb1 | SAB | Cat#41225 |
| Rabbit polyclonal anti-Rel | SAB | Cat#21020 |
| HRP conjugated goat anti-rabbit IgG secondary antibody | SAB | Cat#3012 |
| HRP conjugated goat anti-rat IgG secondary antibody | SAB | Cat#L35017 |
| Chemicals, peptides, and recombinant proteins | ||
| CellTracker Orange CMRA (red fluorescence) | Life Technologies | Cat# C34551 |
| CMFDA (green fluorescence) | Life Technologies | Cat# C2925 |
| Anti-PE microbeads | Invitrogen | Cat# 130-048-801 |
| DreamTaq Green PCR Master Mix | Thermo Fisher | Cat#: K1082 |
| DAPI | Invitrogen | Cat#D1306 |
| Polybead microspheres 3.00 μm | Polysciences | Cat#17134-15 |
| Evans Blue dye | Merck | Cat#E2129 |
| SYBR Green Master Mix | Yeasen | Cat#11203ES03 |
| TRIzol™ Reagent | Invitrogrn | Cat#15596026CN |
| H&E staining | Beyotime | Cat#C0105S |
| Lipofectamine 3000 | Invitrogrn | Cat#L3000008 |
| RPMI 1640 | Gibco | Cat#61870036 |
| Critical commercial assays | ||
| Evo M-MLV RT Kit with gDNA clean for qPCR | AG | Cat#: AG11711 |
| Ribo-ZeroTM Magnetic Kit | Epicentre | Cat#: MRZSR116 |
| QiaQuick PCR extraction kit | Qiagen | Cat#: 28104 |
| RNA Extraction Kit | AG | Cat#: AG21017 |
| TNF-a ELISA kit | R&D Systems | Cat#: MTA00B |
| IL-1 beta ELISA kit | R&D Systems | Cat#: MTB00B |
| IL-6 ELISA kit | R&D Systems | Cat#: M6000B |
| HMGB1 ELISA kit | SAB | Cat#: EK1715 |
| MPO ELISA kit | SAB | Cat#: EK15531 |
| Luciferase Assay System | Promega | Cat#: E1500 |
| Deposited data | ||
| RAN-seq | BIG Data Center | CRA011409 |
| Experimental models: Cell lines | ||
| None | ||
| Experimental models: Organisms/strains | ||
| Mouse: Itga2b (Q887X) knock-in | Cyagen corporation | N/A |
| Mouse: C57BL/6J | Cyagen corporation | N/A |
| Oligonucleotides | ||
| qPCR primers see Table S4 | This study | N/A |
| siRNA primers see Table S5 | This study | N/A |
| Software and algorithms | ||
| GraphPad Prism 9 | This study | N/A |
| FlowJo | ACEA NovoCyte | N/A |
| Adobe Illustrator 2022 | This study | N/A |
| ImageJ software | This study | N/A |
| Quan Studio5 system | Applied Biosystems | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Zhanli Xie (zhanlixie1989@126.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Generation of Itga2b (Q887X) knock-in mice
Itga2b (Q887X) knock-in mice were constructed by Cyagen corporation. The Cas9 mRNA, gRNA produced by in vitro transcription and template oligonucleotide introduction of mouse ES cells into C57BL/6 mice by homologous recombination codon 2659 of αIIb contains substitution of C to T to obtain heterozygous (heterologous) knock-in mice. Homozygous knock-in mice were obtained by mating heterozygous mice.
CLP model of sepsis in mice
A CLP model was used to induce sepsis.47 In this study, heterozygous knockout mice were mated, and their homozygous knockout offspring and their WT littermates were used in the experiments at 8–10 weeks of age. Female mice are less susceptible to sepsis and male mice have been the choice in the vast majority of the studies of sepsis reported in the literature.48,49 Thus, only male mice were used in in vivo studies. Briefly, mice were anesthetized, and then the skin and peritoneum were opened to expose the cecum. The caecum was ligated with 2-0 silk thread at the distal end of the ileocecal valve. Two consecutive punctures were performed with the 21G needle, and a small amount of stool was squeezed out. Then, the cecum was returned to the abdominal cavity, and the abdominal incision was closed in two layers with 2-0 silk thread. After surgery, mice received a subcutaneous injection of normal saline solution (1 mL/mouse). Mice were sacrificed at 1, 2, 4 and 8d after CLP surgery and the blood and liver samples were collected for subsequent experiments.
Method details
Whole transcriptome library preparation and RNA-sequencing
Total platelet RNA of 6 male WT mice and 6 male Itga2b (Q887X) knock-in mice was extracted using Trizol reagent kit according to the manufacturer’s protocol, and eukaryotic mRNA was enriched using oligo(dT) beads. Ribo-ZeroTM Magnetic Kit was used to remove rRNA and enrich for prokaryotic mRNA. The enriched mRNA was cut into short fragments using fragmentation buffer, diluted to 100 ng/μl, and reverse transcribed to cDNA using random primers. Second-strand cDNA was synthesized using dNTP, DNAse I, RNase H and buffer solution, and purified with the QiaQuick PCR extraction kit. After end-repairing and addition of add poly (A) tail, the fragments were connected to Illumina sequencing adapter and selected by agarose gel electrophoresis. The sequences were amplified by PCR and sequenced on the Illumina HiSeq2500 platform by Gene Denovo Biotechnology Co. The clean reads were filtered and aligned with ribosome RNA (rRNA). The number of genes/transcripts with FDR parameter lower than 0.05 and absolute multiple change ≥ 2 were considered as differentially expressed genes (DEGs). After correcting the gene length deviation, the DEGs were functionally annotated by the gene ontology (GO) analysis for GO-seq R package. The Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was performed using the KOBAS software to determine the pathways that were significantly associated with the DEGs.
Quantitative RT-PCR
Total RNA was isolated from murine and human platelets using TRIzol, and 2μg was used for reverse-transcription. Quantitative PCR was performed using SYBR Green Master Mix in 20μL reaction volumes. The samples were analyzed in duplicates with β-actin as the internal control.
Cytokine mRNA expression in leukocyte was quantified using the Quan Studio5 system.50
Histological analysis
The WT and Itga2b (Q887X) knock-in mice were performed sham or CLP, and the mice were anesthetized 24 hours later. Mouse lung and liver tissues were fixed, embedded in paraffin, cut into 5μm-thick sections, and stained with H&E. The slides were imaged with Leica DM2000 microscope.
Scanning electron microscopy
The WT and Itga2b (Q887X) knock-in mice were performed sham or CLP, and the mice were anesthetized 24 hours later. The lung tissues were resected and fixed in glutaraldehyde. Followed by dehydration through a tert-butanol gradient, the tissue samples were vacuum pumped, sprayed with gold, and observed under a scanning electron microscope.
Measurement of cytokine levels in plasma
Blood was collected from the orbital vein plexus of mice 24 hours after CLP. The plasma levels of IL-6, TNF-a, IL-1β and HMGB-1 were measured by specific ELISA kits according the manufacturer’s instructions.
Quantification of inflammatory cells and proteins in BALF
The WT and Itga2b (Q887X) knock-in mice were performed sham or CLP, and anesthetized 24 hours later with 5% chloral hydrate. The skin of the neck was cut, and the bronchi were exposed and separated. One milliliter cold PBS was injected into the lungs, and aspirated twice to obtain about 900 mL of the bronchoalveolar lavage fluid (BALF). The samples were centrifuged at 800 g for 10 minutes, and the clarified supernatant was aspirated. After measuring the protein content of BALF using the BCA kit, the samples were diluted with 0.5 mL PBS and the total number of cells were determined using whole blood analyzer. The lung tissues were also extracted, weighed, and macerated in cold PBS. The homogenates were centrifuged at 500 g for 5 minutes, and the supernatants were diluted 50 times. The MPO concentration in the supernatants were measured using the MPO ELISA kit.
Assessment of vascular permeability in tissues
Vascular permeability in the tissues was quantitatively assessed by the extravasation of Evans Blue dye (EBD).51 Briefly, the mice were performed sham or CLP, and Evans blue dye was injected 24 hours later via the intravenous route. The mice were euthanized 10 minutes after injection, and immediately perfused with the buffer solution through the left ventricle to remove the dye from systemic and pulmonary circulation via the right atrial stoma. Vascular leakage in the tissues was evaluated by measuring the intensity of Evans blue dye.
Platelet count analysis
Two microliters whole mouse blood was diluted 1:50 in FACS buffer, and incubated with PE anti-CD41 antibody and Polybead® 3μM microspheres. The number of peripheral blood platelets was determined according to the standard concentration of microsphere.
Platelet preparation and platelet survival analysis
Platelets were isolated and prepared based on previous publications.52 In brief, whole blood was obtained from the inferior vena cava and collected into test-tubes containing sodium citrate as an anticoagulant, diluted 1:1 with Tyrode buffer, and centrifuged at 100 g for 10 min at room temperature. The supernatant was collected and centrifuged at 800 g for 8 mins to obtain the platelet pellet, followed by two subsequent washes with Tyrode buffer. The WT and Itga2b (Q887X) knock-in mice were performed sham or CLP, and the mice were anesthetized 24 hours later. Platelets were isolated from donor WT or Itga2b (Q887X) knock-in mice and labeled with 1 mg/ml Sulfo-NHS-biotin for 30 min at room temperature. The recipient mice were transfused with 108 biotin-labeled platelets via the retroorbital vascular plexus. Four hours after transfusion, 2μL blood drawn through the tail vein at different time points, diluted 1:50 with FACS solution, and stained with PE-streptavidin and FITC anti-CD41 antibody. The percentage of viable donor platelets in the peripheral blood of recipient mice was analyzed by flow cytometry.
Apoptosis assay
To detect apoptosis in the mouse platelets, the mitochondrial inner transmembrane potential (ΔΨm) was evaluated by staining with JC-1 (2 μg/mL). The cells were analyzed by flow cytometr to determine mitochondrial depolarization.
Competitive platelet transfusion experiment
The WT and Itga2b (Q887X) knock-in mice were performed sham or CLP, and the mice were anesthetized 24 hours later. Platelets were isolated from donor WT or Itga2b (Q887X) knock-in mice and suspended in Tyrode’s buffer (without BSA). The WT and Itga2b knock-in platelets were respectively stained with 2.5μM CMRA (red) and CMFDA (green) at room temperature for 20 min in the dark. The labeled cells were washed once with 1 ml Tyrode’s buffer (without BSA), centrifuged at 850 g for 8 min, and suspended in Tyrode’s buffer. The WT recipient mice were transfused simultaneously with 5 × 107 CMRA-labeled WT platelets and 5 × 107 CMFDA-labeled Itga2b knock-in platelets. 24 hours later, the mice were euthanized and the liver tissues were extracted. The tissue samples were stained with rat anti-rat Albumin to observe hepatocyte.
Caspase-3 activity assay
The WT and Itga2b (Q887X) knock-in mice were performed sham or CLP, and anesthetized 24 hours later with 5% chloral hydrate. Washed mouse platelets (6 × 108/mL) were lysed with lysis buffer on ice for 30 min. The caspase-3 activity assay was performed according to the manufacturer’s protocol. Briefly, 10 μL of platelet lysate per sample was mixed with 80 μL of reaction buffer and 10 μL of caspase-3 substrate (Ac-DEVD-pNA, 2 μM). Samples were further incubated at 37°C for 4 h and activity was determined by an ELISA reader at an absorbance of 450 nm. The specific activity of caspase 3 was normalized for the total protein of sample.
Western blotting
The WT and Itga2b (Q887X) knock-in mice were performed sham or CLP, and anesthetized 24 hours later with 5% chloral hydrate. Freshly isolated platelets were washed with phosphate-buffered saline, and the pellet was re-suspended in cell lysis buffer containing protease inhibitor and phosphatase inhibitors (100 ng/mL). Further details are given in the Online Supplementary Data. The lysates were centrifuged at 14,000 rpm for 20 min at 4°C, and the supernatants were collected. Fifty microgram total protein per sample was fractionated by SDS-PAGE and then transferred onto PVDF membrane. After blocking with 3% BSA in TBS at room temperature for one hour, the blots were incubated overnight with primary antibodies targeting p-RIP1 (Ser166, 1:1000), RIP1 (1:1000), p-RIP3 (Ser232, 1:500), RIP3 (1:1000), p-MLKL(Ser 473, 1:500), MLKL (1:1000) and Caspase-3 at 4°C. Subsequently, the membranes were incubated with HRP-conjugated goat anti-rabbit or anti-mouse secondary antibody at room temperature for 1h in the dark. After washing with TBST, the positive protein bands were visualized by the ECL system and quantified using ImageJ software.
Transient transfection
PTPN6-specific siRNAs and negative controls (NC) were synthesized by GenePharma. The cells were transfected with the respective constructs using Lipofectamine 3000 according to the manufacturer’s instructions.
Luciferase reporter assay
The PTPN6 gene sequence was cloned into the Sac I and Xhol I sites of the luciferase reporter vector (Luc). HEK293T cells were seeded into 24-well dishes and cultured in 10% FBS-supplemented DMEM under 5% CO2 till 70% confluent. The cells were transiently transfected with 1μg PTPN6 pro-Luc or empty-Luc plasmid (empty-Luc) and 0.2μg Rel-pGL4 and Nfkb1-pGL4 plasmids using Lipofectamine 3000. The cells were harvested 24 hours later, and dual luciferase activity was monitored using a specific kit as per the manufacturer’s instructions.
Isolation of megakaryocytes
The bone marrow was extracted from the femora and tibia of 10-weeks-old mice, and the red blood cells were lysed with RBC lysis buffer. After washing with PBS, the cells were labeled with anti-CD41 microbeads and then loaded onto a MACS column in the MACS separato. The magnetically-retained megakaryocytes were eluted using the attached plungers.53
Study approval
Animal studies were conducted with protocols approved by the Institutional Animal Care and Use Committee of the Soochow University.
Quantification and statistical analysis
Statistical analysis
Data are expressed as the mean ± standard deviation (SD) of at least 3 independent experiments unless otherwise mentioned. Sigma Plot and GraphPad Prism 9 were used for statistical analysis. The survival curves of animal experiment were generated with a log rank (Mantel-Cox) test. Data from multiple groups were compared by a one-way ANOVA followed by a Tukey’s test and two groups were compared by two-tailed Student’s t test, and p < 0.05 was considered statistically significant (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
Acknowledgments
This study was supported by Suzhou People’s Livelihood Science and Technology Project (grant no. SKY2022155 and SKJYD2021089), National Natural Science Foundation of China (grant no. 81800128), Suzhou New District Science and Technology Project (grant no. 2020Z002), the Natural Science Foundation of Jiangsu Province (grant no. BK20170361), Core Medical Science Subjects in Suzhou (grant no. szxk202131) and Research project of Suzhou College of Nanjing Medical University (GSKY20210241).
Author contributions
Z.X., J.J., W.L., L.Z., and D.L. conceived the project, designed experiments, analyzed the data, and wrote the manuscript. Z.X., J.J., W.L., L.Z., D.L., and Y.W. performed experiments. J.A. and S.Q. provided key recommendations and analyze data. All data were generated in-house, and no paper mill was used. All authors agree to be accountable for all aspects of work ensuring integrity and accuracy.
Declaration of interests
The authors have no relevant financial or non-financial interests to disclose.
Published: July 19, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107414.
Supplemental information
Data and code availability
-
•
The RNA-seq data from this study have been deposited in the Genome Sequence Archive in BIG Data Center (https://bigd.big.ac.cn), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences under the accession number: CRA011409 (https://bigd.big.ac.cn/gsa/browse/CRA011409).
-
•
All scripts used for analyses, differential expression results were performed using the OmicShare tools, a free online platform for data analysis (https://www.omicshare.com/tools).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Walter K.L., Seymour C.W. Hydrocortisone, Vitamin C, and Thiamine for Treatment of Sepsis: Making Evidence Matter. JAMA. 2021;325:730–731. doi: 10.1001/jama.2020.26029. [DOI] [PubMed] [Google Scholar]
- 2.Seymour C.W., Liu V.X., Iwashyna T.J., Brunkhorst F.M., Rea T.D., Scherag A., Rubenfeld G., Kahn J.M., Shankar-Hari M., Singer M., et al. Assessment of Clinical Criteria for Sepsis: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315:762–774. doi: 10.1001/jama.2016.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angus D.C., Linde-Zwirble W.T., Lidicker J., Clermont G., Carcillo J., Pinsky M.R. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit. Care Med. 2001;29:1303–1310. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Angus D.C., Bindman A.B. Achieving Diagnostic Excellence for Sepsis. JAMA. 2022;327:117–118. doi: 10.1001/jama.2021.23916. [DOI] [PubMed] [Google Scholar]
- 5.Klimiankou M., Skokowa J. Old drug revisited: disulfiram, NETs, and sepsis. Blood. 2021;138:2604–2605. doi: 10.1182/blood.2021013438. [DOI] [PubMed] [Google Scholar]
- 6.Weiss L.J., Manukjan G., Pflug A., Winter N., Weigel M., Nagler N., Kredel M., Lâm T.T., Nieswandt B., Weismann D., Schulze H. Acquired platelet GPVI receptor dysfunction in critically ill patients with sepsis. Blood. 2021;137:3105–3115. doi: 10.1182/blood.2020009774. [DOI] [PubMed] [Google Scholar]
- 7.Celikel R., McClintock R.A., Roberts J.R., Mendolicchio G.L., Ware J., Varughese K.I., Ruggeri Z.M. Modulation of alpha-thrombin function by distinct interactions with platelet glycoprotein Ibalpha. Science. 2003;301:218–221. doi: 10.1126/science.1084183. [DOI] [PubMed] [Google Scholar]
- 8.Xiang B., Zhang G., Guo L., Li X.A., Morris A.J., Daugherty A., Whiteheart S.W., Smyth S.S., Li Z. Platelets protect from septic shock by inhibiting macrophage-dependent inflammation via the cyclooxygenase 1 signalling pathway. Nat. Commun. 2013;4:2657. doi: 10.1038/ncomms3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claushuis T.A.M., de Stoppelaar S.F., Stroo I., Roelofs J.J.T.H., Ottenhoff R., van der Poll T., Van't Veer C. Thrombin contributes to protective immunity in pneumonia-derived sepsis via fibrin polymerization and platelet-neutrophil interactions. J. Thromb. Haemost. 2017;15:744–757. doi: 10.1111/jth.13625. [DOI] [PubMed] [Google Scholar]
- 10.Bai M., Grieshaber-Bouyer R., Wang J., Schmider A.B., Wilson Z.S., Zeng L., Halyabar O., Godin M.D., Nguyen H.N., Levescot A., et al. CD177 modulates human neutrophil migration through activation-mediated integrin and chemoreceptor regulation. Blood. 2017;130:2092–2100. doi: 10.1182/blood-2017-03-768507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Provan D., Arnold D.M., Bussel J.B., Chong B.H., Cooper N., Gernsheimer T., Ghanima W., Godeau B., González-López T.J., Grainger J., et al. Updated international consensus report on the investigation and management of primary immune thrombocytopenia. Blood Adv. 2019;3:3780–3817. doi: 10.1182/bloodadvances.2019000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieberman L., Bercovitz R.S., Sholapur N.S., Heddle N.M., Stanworth S.J., Arnold D.M. Platelet transfusions for critically ill patients with thrombocytopenia. Blood. 2014;123:1146–1151. doi: 10.1182/blood-2013-02-435693. quiz 1280; quiz 1280. [DOI] [PubMed] [Google Scholar]
- 13.Middleton E.A., Rowley J.W., Campbell R.A., Grissom C.K., Brown S.M., Beesley S.J., Schwertz H., Kosaka Y., Manne B.K., Krauel K., et al. Sepsis alters the transcriptional and translational landscape of human and murine platelets. Blood. 2019;134:911–923. doi: 10.1182/blood.2019000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emambokus N.R., Frampton J. The glycoprotein IIb molecule is expressed on early murine hematopoietic progenitors and regulates their numbers in sites of hematopoiesis. Immunity. 2003;19:33–45. doi: 10.1016/s1074-7613(03)00173-0. [DOI] [PubMed] [Google Scholar]
- 15.Shen W.Z., Jin P.P., Ding Q.L., Wang X.F., Li S.M., Jiang Y.Z., Wang H.L. [Molecular mechanisms of Glanzmann thrombasthenia caused by alpha II b L721R and Q860X compound heterozygous mutation] Zhonghua Xue Ye Xue Za Zhi. 2008;29:577–582. [PubMed] [Google Scholar]
- 16.Xie Z., Jiang J., Cao L., Jiang M., Yang F., Ma Z., Wang Z., Ruan C., Liu H., Zhou L. Nonsense-mediated mRNA decay efficiency influences bleeding severity in ITGA2B c.2659C > T (p.Q887X) knock-in mice. Clin. Genet. 2021;100:213–218. doi: 10.1111/cge.13975. [DOI] [PubMed] [Google Scholar]
- 17.Tartey S., Gurung P., Karki R., Burton A., Hertzog P., Kanneganti T.D. Ets-2 deletion in myeloid cells attenuates IL-1alpha-mediated inflammatory disease caused by a Ptpn6 point mutation. Cell. Mol. Immunol. 2021;18:1798–1808. doi: 10.1038/s41423-020-0398-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson H.A., Dolton G., Ohme J., Ladell K., Vigar M., Wehenkel S., Hindley J., Mohammed R.N., Miners K., Luckwell R.A., et al. Purity of transferred CD8(+) T cells is crucial for safety and efficacy of combinatorial tumor immunotherapy in the absence of SHP-1. Immunol. Cell Biol. 2016;94:802–808. doi: 10.1038/icb.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wicki S., Gurzeler U., Wei-Lynn Wong W., Jost P.J., Bachmann D., Kaufmann T. Loss of XIAP facilitates switch to TNFalpha-induced necroptosis in mouse neutrophils. Cell Death Dis. 2016;7:e2422. doi: 10.1038/cddis.2016.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Speir M., Nowell C.J., Chen A.A., O'Donnell J.A., Shamie I.S., Lakin P.R., D'Cruz A.A., Braun R.O., Babon J.J., Lewis R.S., et al. Ptpn6 inhibits caspase-8- and Ripk3/Mlkl-dependent inflammation. Nat. Immunol. 2020;21:54–64. doi: 10.1038/s41590-019-0550-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pu Q., Wiel E., Corseaux D., Bordet R., Azrin M.A., Ezekowitz M.D., Lund N., Jude B., Vallet B. Beneficial effect of glycoprotein IIb/IIIa inhibitor (AZ-1) on endothelium in Escherichia coli endotoxin-induced shock. Crit. Care Med. 2001;29:1181–1188. doi: 10.1097/00003246-200106000-00019. [DOI] [PubMed] [Google Scholar]
- 22.Zhu J., Feng B., Xu Y., Chen W., Sheng X., Feng X., Shi X., Liu J., Pan Q., Yu J., et al. Mesenchymal stem cells alleviate LPS-induced acute lung injury by inhibiting the proinflammatory function of Ly6C(+) CD8(+) T cells. Cell Death Dis. 2020;11:829. doi: 10.1038/s41419-020-03036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goerge T., Ho-Tin-Noe B., Carbo C., Benarafa C., Remold-O'Donnell E., Zhao B.Q., Cifuni S.M., Wagner D.D. Inflammation induces hemorrhage in thrombocytopenia. Blood. 2008;111:4958–4964. doi: 10.1182/blood-2007-11-123620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macaulay I.C., Thon J.N., Tijssen M.R., Steele B.M., MacDonald B.T., Meade G., Burns P., Rendon A., Salunkhe V., Murphy R.P., et al. Canonical Wnt signaling in megakaryocytes regulates proplatelet formation. Blood. 2013;121:188–196. doi: 10.1182/blood-2012-03-416875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ni Y., Yap T., Silke N., Silke J., McCullough M., Celentano A., O'Reilly L.A. Loss of NF-kB1 and c-Rel accelerates oral carcinogenesis in mice. Oral Dis. 2021;27:168–172. doi: 10.1111/odi.13508. [DOI] [PubMed] [Google Scholar]
- 26.O'Connell K.E., Mikkola A.M., Stepanek A.M., Vernet A., Hall C.D., Sun C.C., Yildirim E., Staropoli J.F., Lee J.T., Brown D.E. Practical murine hematopathology: a comparative review and implications for research. Comp. Med. 2015;65:96–113. [PMC free article] [PubMed] [Google Scholar]
- 27.Gawaz M., Dickfeld T., Bogner C., Fateh-Moghadam S., Neumann F.J. Platelet function in septic multiple organ dysfunction syndrome. Intensive Care Med. 1997;23:379–385. doi: 10.1007/s001340050344. [DOI] [PubMed] [Google Scholar]
- 28.Russwurm S., Vickers J., Meier-Hellmann A., Spangenberg P., Bredle D., Reinhart K., Lösche W. Platelet and leukocyte activation correlate with the severity of septic organ dysfunction. Shock. 2002;17:263–268. doi: 10.1097/00024382-200204000-00004. [DOI] [PubMed] [Google Scholar]
- 29.Xie Z., Shao B., Hoover C., McDaniel M., Song J., Jiang M., Ma Z., Yang F., Han J., Bai X., et al. Monocyte upregulation of podoplanin during early sepsis induces complement inhibitor release to protect liver function. JCI Insight. 2020;5 doi: 10.1172/jci.insight.134749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haas S., Hansson J., Klimmeck D., Loeffler D., Velten L., Uckelmann H., Wurzer S., Prendergast Á.M., Schnell A., Hexel K., et al. Inflammation-Induced Emergency Megakaryopoiesis Driven by Hematopoietic Stem Cell-like Megakaryocyte Progenitors. Cell Stem Cell. 2015;17:422–434. doi: 10.1016/j.stem.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Valet C., Magnen M., Qiu L., Cleary S.J., Wang K.M., Ranucci S., Grockowiak E., Boudra R., Conrad C., Seo Y., et al. Sepsis promotes splenic production of a protective platelet pool with high CD40 ligand expression. J. Clin. Invest. 2022;132 doi: 10.1172/JCI153920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunishima S., Kashiwagi H., Otsu M., Takayama N., Eto K., Onodera M., Miyajima Y., Takamatsu Y., Suzumiya J., Matsubara K., et al. Heterozygous ITGA2B R995W mutation inducing constitutive activation of the alphaIIbbeta3 receptor affects proplatelet formation and causes congenital macrothrombocytopenia. Blood. 2011;117:5479–5484. doi: 10.1182/blood-2010-12-323691. [DOI] [PubMed] [Google Scholar]
- 33.McVerry B.J., Peng X., Hassoun P.M., Sammani S., Simon B.A., Garcia J.G.N. Sphingosine 1-phosphate reduces vascular leak in murine and canine models of acute lung injury. Am. J. Respir. Crit. Care Med. 2004;170:987–993. doi: 10.1164/rccm.200405-684OC. [DOI] [PubMed] [Google Scholar]
- 34.Jeewandara C., Gomes L., Wickramasinghe N., Gutowska-Owsiak D., Waithe D., Paranavitane S.A., Shyamali N.L.A., Ogg G.S., Malavige G.N. Platelet activating factor contributes to vascular leak in acute dengue infection. PLoS Negl. Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flaumenhaft R., Tanaka E., Graham G.J., De Grand A.M., Laurence R.G., Hoshino K., Hajjar R.J., Frangioni J.V. Localization and quantification of platelet-rich thrombi in large blood vessels with near-infrared fluorescence imaging. Circulation. 2007;115:84–93. doi: 10.1161/CIRCULATIONAHA.106.643908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montrucchio G., Bosco O., Del Sorbo L., Fascio Pecetto P., Lupia E., Goffi A., Omedè P., Emanuelli G., Camussi G. Mechanisms of the priming effect of low doses of lipopoly-saccharides on leukocyte-dependent platelet aggregation in whole blood. Thromb. Haemost. 2003;90:872–881. doi: 10.1160/TH03-02-0085. [DOI] [PubMed] [Google Scholar]
- 37.Claushuis T.A.M., van Vught L.A., Scicluna B.P., Wiewel M.A., Klein Klouwenberg P.M.C., Hoogendijk A.J., Ong D.S.Y., Cremer O.L., Horn J., Franitza M., et al. Thrombocytopenia is associated with a dysregulated host response in critically ill sepsis patients. Blood. 2016;127:3062–3072. doi: 10.1182/blood-2015-11-680744. [DOI] [PubMed] [Google Scholar]
- 38.Croker B.A., Lawson B.R., Rutschmann S., Berger M., Eidenschenk C., Blasius A.L., Moresco E.M.Y., Sovath S., Cengia L., Shultz L.D., et al. Inflammation and autoimmunity caused by a SHP1 mutation depend on IL-1, MyD88, and a microbial trigger. Proc. Natl. Acad. Sci. USA. 2008;105:15028–15033. doi: 10.1073/pnas.0806619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gurung P., Fan G., Lukens J.R., Vogel P., Tonks N.K., Kanneganti T.D. Tyrosine Kinase SYK Licenses MyD88 Adaptor Protein to Instigate IL-1alpha-Mediated Inflammatory Disease. Immunity. 2017;46:635–648. doi: 10.1016/j.immuni.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lukens J.R., Vogel P., Johnson G.R., Kelliher M.A., Iwakura Y., Lamkanfi M., Kanneganti T.D. RIP1-driven autoinflammation targets IL-1alpha independently of inflammasomes and RIP3. Nature. 2013;498:224–227. doi: 10.1038/nature12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo M., Xu X., Liu X., Shen W., Yang L., Zhu Z., Weng S., He J., Zuo H. The Non-Receptor Protein Tyrosine Phosphatase PTPN6 Mediates a Positive Regulatory Approach From the Interferon Regulatory Factor to the JAK/STAT Pathway in Litopenaeus vannamei. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.913955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moujalled D., Gangatirkar P., Kauppi M., Corbin J., Lebois M., Murphy J.M., Lalaoui N., Hildebrand J.M., Silke J., Alexander W.S., Josefsson E.C. The necroptotic cell death pathway operates in megakaryocytes, but not in platelet synthesis. Cell Death Dis. 2021;12:133. doi: 10.1038/s41419-021-03418-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Loo G., De Lorenzi R., Schmidt H., Huth M., Mildner A., Schmidt-Supprian M., Lassmann H., Prinz M.R., Pasparakis M. Inhibition of transcription factor NF-kappaB in the central nervous system ameliorates autoimmune encephalomyelitis in mice. Nat. Immunol. 2006;7:954–961. doi: 10.1038/ni1372. [DOI] [PubMed] [Google Scholar]
- 44.Franke A., McGovern D.P.B., Barrett J.C., Wang K., Radford-Smith G.L., Ahmad T., Lees C.W., Balschun T., Lee J., Roberts R., et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Reilly L.A., Hughes P., Lin A., Waring P., Siebenlist U., Jain R., Gray D.H.D., Gerondakis S., Strasser A. Loss of c-REL but not NF-kappaB2 prevents autoimmune disease driven by FasL mutation. Cell Death Differ. 2015;22:767–778. doi: 10.1038/cdd.2014.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matan M., King D., Peled E., Ackerman S., Bar-Lavi Y., Brenner B., Nadir Y. Heparanase level and procoagulant activity are reduced in severe sepsis. Eur. J. Haematol. 2018;100:182–188. doi: 10.1111/ejh.12997. [DOI] [PubMed] [Google Scholar]
- 47.Rittirsch D., Huber-Lang M.S., Flierl M.A., Ward P.A. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat. Protoc. 2009;4:31–36. doi: 10.1038/nprot.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Angele M.K., Pratschke S., Hubbard W.J., Chaudry I.H. Gender differences in sepsis: cardiovascular and immunological aspects. Virulence. 2014;5:12–19. doi: 10.4161/viru.26982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacMillan-Crow L.A., Mayeux P.R. Female mice exhibit less renal mitochondrial injury but greater mortality using a comorbid model of experimental sepsis. Intern. Med. Rev. 2018;4 doi: 10.18103/imr.v4i10.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang A., Xie Z., Wang B., Colman R.W., Dai J., Wu Y. Correction: An essential role of high-molecular-weight kininogen in endotoxemia. J. Exp. Med. 2019;216:244. doi: 10.1084/jem.2016190012072018c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsugawa-Shimizu Y., Fujishima Y., Kita S., Minami S., Sakaue T.A., Nakamura Y., Okita T., Kawachi Y., Fukada S., Namba-Hamano T., et al. Increased vascular permeability and severe renal tubular damage after ischemia-reperfusion injury in mice lacking adiponectin or T-cadherin. Am. J. Physiol. Endocrinol. Metab. 2021;320:E179–E190. doi: 10.1152/ajpendo.00393.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan Y., Yago T., Fu J., Herzog B., McDaniel J.M., Mehta-D'Souza P., Cai X., Ruan C., McEver R.P., West C., et al. Podoplanin requires sialylated O-glycans for stable expression on lymphatic endothelial cells and for interaction with platelets. Blood. 2014;124:3656–3665. doi: 10.1182/blood-2014-04-572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma X., Li Y., Kondo Y., Shi H., Han J., Jiang Y., Bai X., Archer-Hartmann S.A., Azadi P., Ruan C., et al. Slc35a1 deficiency causes thrombocytopenia due to impaired megakaryocytopoiesis and excessive platelet clearance in the liver. Haematologica. 2021;106:759–769. doi: 10.3324/haematol.2019.225987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The RNA-seq data from this study have been deposited in the Genome Sequence Archive in BIG Data Center (https://bigd.big.ac.cn), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences under the accession number: CRA011409 (https://bigd.big.ac.cn/gsa/browse/CRA011409).
-
•
All scripts used for analyses, differential expression results were performed using the OmicShare tools, a free online platform for data analysis (https://www.omicshare.com/tools).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.