Abstract
Human cytomegalovirus (HCMV) DNA quantitation in whole blood (WB) by real-time or quantitative polymerase chain reaction (qPCR) is a highly sensitive and reproducible diagnostic procedure for monitoring HCMV DNAemia (DNAemia is the detection of DNA in samples of plasma, whole blood, isolated peripheral blood leukocytes or in buffy-coat specimens) in patients. We provided a comparative analysis of HCMV DNA extraction performance by two different techniques, one performed by an automated extractor and the other by a manual method. We observed that the automated extraction method allowed HCMV DNA detection in the presence of weak viremia while no differences are observed when the viral load is greater. Therefore, automated DNA extraction is a suitable and recommended protocol not only for early detection of HCMV infection but also for more accurate monitoring of HCMV DNAemia during post-therapy follow-up.
Keywords: HCMV, DNA extraction, Hematological diseases
1. Introduction
Human cytomegalovirus (HCMV) belongs to the Orthoherpesviridae family and that is widespread globally, indeed it affects 40–95% of the population worldwide, usually without symptoms. After the infection, the virus remains in a latent stage due to the host immune response, although an inflammatory context could contribute to HCMV reactivation. The HCMV reactivation could play a oncogenic role in which virus will promote the development and spread of cancerous cells [1,2]. Acute HCMV infections are associated with poor prognosis in particular for immunocompromised and immunodeficient patients. The management of HCMV infection is based on antiviral drugs (e.g., ganciclovir, valganciclovir, cidofovir, foscarnet) somministration but their efficacy is limited by side effects, toxicity, resistance issues, and other problems. In recent years, novel drugs (e.g., letermovir and maribavir) have been developed characterized by less toxicity [3]. In this scenario, HCMV is the most relevant opportunistic pathogen in immunosuppressed patients, particularly in solid organs or bone marrow transplant recipients [4]. Detection and monitoring of HCMV infection is fundamental for the efficient management of transplant recipients. The detection method should be: (i) easy to perform; (ii) rapid; (iii) sufficiently sensitive to facilitate early detection of patients at high risk of HCMV disease; (iv) specific for the diagnosis of HCMV infection or reactivation. Wide varieties of techniques for HCMV DNAemia evaluation have been developed, including both non-molecular and molecular tests. Because of its high sensitivity, qualitative PCR was the first method used for the diagnosis of HCMV infection in transplant recipients [5]. However, the introduction of qPCR technology provided a quantitative as well as highly sensitive and reproducible approach for HCMV DNA measurement, allowing monitoring of HCMV kinetics in infected patients from the beginning of HCMV infection and during the antiviral therapy. Notably, by using quantitative PCR methods, it has been demonstrated that immunosuppressed patients with a high virus load have a major risk of developing HCMV disease [[6], [7], [8]]. Indeed, qPCR technology combines a higher sensitivity with a wider dynamic range as compared to end-point PCR [9] Literature data suggest that assessment of HCMV DNA levels in whole blood (WB), plasma, peripheral blood mononuclear cells, and peripheral blood leukocytes, analyzed independently or comparatively with various assays, yields variable results [[10], [11], [12]]. Remarkably, WB appears to be the matrix of choice for the quantification of HCMV DNAemia, since it allows the quantification of viral load in both cells and the extra-cellular compartment [[13], [14], [15]] and, for this reason, it is crucial to a successful clinical application of HCMV DNA detection. Thus, the purpose of this study was to compare HCMV DNAemia assessed by qPCR on WB DNA isolated by an automated extraction method and on DNA samples extracted using a manual method. This analysis was performed on WB specimens derived from a cohort of immunosuppressed and transplanted patients with suspected or proven HCMV disease at the first diagnosis and during the post-antiviral therapy follow-up.
2. Materials and methods
2.1. Patients and samples
The comparative study was conducted on 25 subjects affected by different types of hematological malignancies, enrolled at the IRCCS-CROB, Referral Cancer Center of Basilicata (Table 1). Patients selected for this study based on tumor type included 4 acute myeloid leukemia (AML), 8 non-Hodgkin lymphomas (NHL), 1 Hodgkin lymphoma (HL), 6 multiple myeloma (MM), 2 acute lymphocytic leukemia (ALL), 2 myeloproliferative neoplasm (MPNs), 2 chronic lymphocytic leukemia (CLL). Five of these patients, (3 with NHL, 1 with MM and, 1 with ALL) underwent autologous stem cell transplant. All patients analyzed followed combination therapy for their condition. At the time of sampling, all patients had a fever; they were negative for other infections (TORCH and monotest) and 30% of them displayed an increase in liver enzymes. All patients gave their informed consent to participate in the study.
Table 1.
Clinical information of HM patients.
| Title 1 | Clinical features |
|---|---|
| sex | M (15) |
| F (10) | |
| Mean age and range (years) | 58 M (21–28) |
| 61 F (29–83) | |
| Malignancy | AML (4), NHL (8), ALL (2), HL (1), MPNs (2), CLL (2), MM (6) |
| Transplantation | Stem cell transplant LNH (3), ALL (1), MM (1) |
2.2. DNA extraction
Whole blood is collected from the patients in a tube containing EDTA as an anticoagulant. DNA was automatically isolated from 400 μL of whole blood by Magna Pure instrument. The Magna Pure LC DNA isolation kit I was used according to the manufacturer's recommendations (Roche) with a elution volume of 100 μL. DNA was extracted manually from 200 μL of whole blood with High Pure Viral Nucleic Acid Kit (Roche) with a elution volume of 50 μL, according to the manufacturer's instructions. HCMV DNA was detected and quantified with the LightCycler system [[16], [17], [18]].
To assess the quantity and quality of the extracted viral DNA, the absorbance at 260 nm of all samples was measured by a NanoDrop Lite spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The absorbance ratio 260/280 for pure DNA free of protein was reported. Each sample was measured 3 times and average values are reported in this study.
2.3. PCR
qPCR was carried out with the Fast Start DNA master hybridization probes (Roche Molecul Biochemicals). Extracted DNA (5 μl) was added to the PCR mixture containing MgCl2, primers, and probes according to the LightMix kit Human Cytomegalovirus (TIB MolBiol GmbH, Berlin, Germany) recommendations (sequences of primers and probe are summarized in Table 2) [19]. The conditions were as follows: initial denaturation of 1 cycle of 10 min at 95 °C, followed by 50 cycles of 5 s at 95 °C, 5 s at 62 °C and 15 s at 62 °C. The reaction, data acquisition, and analysis were all performed by using the LightCycler instrument. Each sample was analyzed in triplicate and, to monitor contamination, a HCMV DNA negative sample and a DNase- and RNase-free sterile water control were included in each PCR run. Plasmids of known concentration (provided by the kit) were used as a reference standard. Plasmids were diluted to generate a standard curve of 1–6 log10 genome copies per reaction volume. The qPCR in terms of sensitivity can to detect 10 copies of HCMV DNA. The concentration of target DNA was calculated by plotting the crossing point of each sample on the standard curves by using the Light Cycler software. HCMV DNA was expressed as log10 genome copy numbers/mL x 200. The quantitative values expressed the number of copies/quantity of templates. These need to be multiplied by 200 (conversion factor) to obtain the number of copies per mL. The negative control (NTC) showed no signal. An internal control (IC) was included in each PCR reaction to monitor extraction and demonstrate the ability to run an amplification reaction. Amplification after CT 32 (less than 100 copies) is assumed to be low positive. In each run, samples have been tested in three technical triplicates, and mean CT values were used to estimate the DNA viral titer. If there was >30% variation among the three CT values, the result was considered unacceptable and the sample was tested again. The samples showed an amplification curve with a CT between 28 and 32. A result less than 10 copies was considered negative.
Table 2.
List of sequences of primers and probe used for qPCR analysis.
| gene | HCMV |
|---|---|
| Primer forward | GAGGACAACGAAATCCTGTTGGGCA |
| Primer reverse (t* LDC640 labeled) | GTCGACGGTGGAGATACTGCT*GAGG |
| Probe (fluorescein-labeled) | GGACTACCTCTTCAAACGCATGATTGAC |
2.4. Statistical data analysis
All statistical analyses were performed using by chi-square test. Mean CT values of each sample and standard deviation were calculated using Grubbs tests on the samples used and extracted 3 times for each protocol. Each sample has been tested in triplicates, and mean CT values were used to determine the viral load. A p-value of less than 0.01 was considered statistically significant.
3. Results
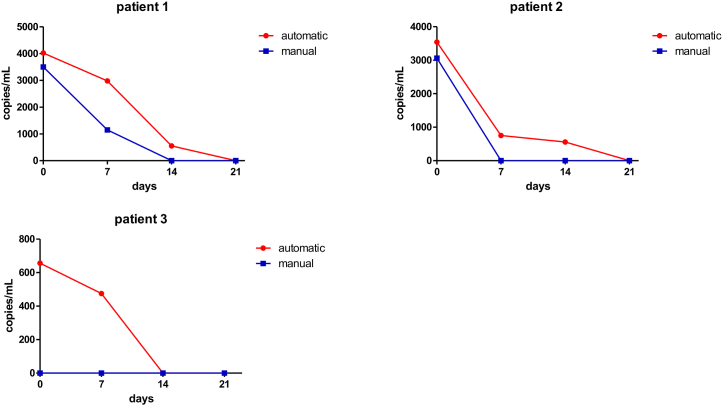
Quantitative qPCR for HCMV DNA was performed on WB specimens derived from patients enrolled in this study [20]. The analysis was performed utilizing a filter combination 498–660 quantification mode (Second Derivative Maximum) for the control reaction, and filter combination 498–640 quantification mode (Second Derivative Maximum) obtained from the standard row and samples for HCMV detection. Each analysis performed the amplification curves of a standard of known HCMV concentration (101–106), the absolute quantification of a positive HCMV PCR result, and on the baseline, curves obtained from negative samples that were used as negative controls in subsequent analyses. Quantitative qPCR analyses for HCMV carried out on DNA samples extracted by using the manual method, allowed the detection of HCMV DNA in WB specimens of 2 of 25 patients (8%), while a weak positivity was detected in 3 of 25 (12%), and a negative result in 20 of 25 (80%). Interestingly, quantitative analyses performed on DNA samples extracted by using the automated Magna Pure Compact (Roche) not only confirmed the HCMV positivity in the same 2 of 25 patients (12%), but also allowed the detection of a weak positivity in 11 of 25 patients (44%), while 12 of 25 (48%) patients resulted negative. Differences between manual and automated extraction were significant (p = 0.01) when analyzed by chi-square test. Viral load was found to be lower in HCMV -positive DNA samples extracted using the manual method when compared to the same samples obtained by using the automated method. A substantial concordance among the results obtained from quantification of HCMV DNA extracted with the two different procedures was observed for the two positive samples. qPCR assessment of HCMV DNAemia was also repeated in seven patients of the same cohort, during the post-antiviral therapy follow-up (after 1, 2, and 3 weeks of therapy) (Table 3). Among the seven patients analyzed, two samples were found strongly positive by using both methods; in one case, viral loads of 4020 copies/mL and 3500 copies/mL, at the first sampling, were detected in DNA samples extracted by using the automated method and the manual protocol, respectively. After 1 week of ganciclovir treatment (GVC), viral loads of 2977 copies/mL and 1146 copies/mL were detected in the automatically and in the manually extracted DNA samples, respectively. Sampling was repeated after a further week and a weak positive (553 copies/mL) was observed only in the automatically isolated DNA sample. After a further week, in the WB sample collected at the end of the follow-up period, no viral load was detectable even using the automated extraction method. In the second patient viral loads of 3540 copies/mL and 3060 copies/mL, at the first sampling, were detected in DNA samples extracted by using automated method and manual protocol, respectively. After 1 week of ganciclovir treatment (GVC), a viral load of 750 copies/mL was detected automatically extracted DNA sample, while HCMV DNA was undetectable in the manually extracted DNA sample derived from the same patient. The sample was repeated after a further week and a weak positive (558 copies/mL) was observed only in the automatically isolated DNA sample. After a further week, in the WB sample collected at the end of the follow-up period, no viral load was detectable even using the automated extraction method.
Table 3.
Monitoring of HCMV DNAemia during the follow-up.
| Patients | DNA extraction method | DNAemia (copies/mL) | |||
|---|---|---|---|---|---|
| Initial | After 1 week | After 2 week | After 3 week | ||
| 1 |
|
|
|
|
|
| 2 |
|
|
|
|
|
| 3 |
|
|
|
|
|
| 4 |
|
|
|
||
| 5 |
|
|
|
||
| 6 |
|
|
|
||
| 7 |
|
|
|
||
In the third patient, a weak positivity, with 656 copies/mL, was observed after the automated DNA isolation, while no HCMV DNA copies were detectable using the manual method. Following 7 days of anti-viral therapy, the patient still resulted weakly positive (475 copies/mL), as revealed by the quantitative analysis carried out on the automatically isolated DNA sample. On the contrary, the same test had negative results when DNA was isolated from blood using the manual method. In DNA samples isolated from blood specimens of the same patient, after a further three weeks, no viral load was found, even when DNA extraction was automatically performed. A graphical representation of the time trend of HCMV DNA concentrations measured in WB samples extracted by using the automated and manual protocols at the diagnosis and during the follow-up of three patients with different viral loads is reported in Fig. 1.
Fig. 1.
Viral load in DNA samples extracted by using the automated method and the manual protocol at the diagnosis and during the follow-up.
In the remaining cases, specimens collected at the time of the first sampling, resulted weakly positive when the automated method was used; they were, instead, negative when the manual method was carried out. Following three weeks of the anti-viral therapy, patients showed a total absence of viral load in DNA samples isolated by both methods (Table 3).
4. Discussion
Human health is affected by HCMV-associated diseases since they represent a challenge for the management of chemotherapy side effects. In this scenario, exploring alternative strategies and drugs to mitigate HCMV acute and/or latent infections is of great importance, despite current guidelines provide that antiviral drugs should be administered to treat HCMV infections clinically [3]. Indeed, the mortality rate associated with HCMV disease among transplant recipients and immunosuppressed patients remains relatively high and one of the best strategies to control the infection is, undoubtedly, prevention [[21], [22], [23]]. There are two main approaches to prevent the clinical syndrome associated with HCMV infection: antiviral prophylaxis and early therapy. However, serious hematological adverse effects associated with antiviral prophylaxis, based on the administration of GVC and other antiviral drugs, is not negligible. Therefore, patients that receive GVC treatment must be monitored for potential side effects of the drugs, as well as for their clinical response. Among GVC side effects, ganciclovir-related neutropenia, which increases the risk for fungal and bacterial infections as well as for the generation of resistant strains of HCMV is frequently reported [16]. The pre-emptive therapy, on the contrary, consists in the administration of antiviral drugs only when the viral load in the blood exceeds a threshold value indicative of progression to HCMV -related disease (10,000 copies/ml of WB in bone marrow transplant recipients [[24], [25], [26]]. Therefore, to be an efficient means of prevention, pre-emptive therapy requires constant monitoring of viral load over time [[27], [28], [29]].
Moreover, some authors indicated the comparative evaluation of different automated systems for DNA extraction associated with various commercially available qPCR assays for the quantitation of plasma HCMV DNAemia. In particular, it was demonstrated that they are not equally efficient for HCMV DNA extraction from plasma specimens and that, consequently, they can result in significantly different HCMV DNA quantitation [30].
Although literature data consistently indicate the difference between the two methods, research has poorly investigated the identification of an efficient method of DNA extraction from blood, which is a critical step for an accurate HCMV viral load quantitation [9,31,32].
Based on this rationale, we evaluated the hypothesis that is a significant difference in the efficiency of two different DNA extraction methods from WB of patients with suspected HCMV disease for a subsequent HCMV DNA quantitation by qPCR [33,34].
First, in one patient characterized by high viral load, during the follow-up, the automated method and the manual protocol detected viral DNA after 7 days of antiviral treatment but after a further week of treatment (14 days), no viral DNA was detectable even using the manual extraction method.
Then, in the second patient after 1 week of ganciclovir treatment (GVC), a viral DNA was detected in the automatically extracted DNA sample, which was undetectable in the manually extracted DNA sample derived from the same patient.
Lastly, in the third patient characterized by weak positivity, only automated DNA isolation led to detect viral DNA. Moreover, after 7 days of anti-viral therapy, the patient still resulted weakly positive to HCMV DNA. On the contrary, no viral DNA was detected using the manual DNA isolation method (Fig. 1).
Overall, our data suggested that the automated DNA extraction protocol carried out with the MagNA Pure Instrument (Roche) is a more efficient method for the detection of low load DNA, if compared to the manual one, performed with the High Pure Viral Nucleic Acid kit (Roche). In particular, we reported that a strong HCMV positivity is detectable with both extraction systems. By contrast, in weakly positive patients, the manual protocol allowed us to identify as positive only 12% of patients, while the automated DNA extraction method allowed us to identify HCMV positivity in 44% of cases. Differences between the two extraction protocols emerged in the detection of HCMV DNA in WB of patients whose viral load was monitored during follow-up. As shown in Fig. 1 automatic method ensured a more sensitive detection of a weak HCMV positivity. Our previous experimental evidence showed that other extraction methods checked were affected by poor reliability of the results and false results. Our results (data not shown) suggested that other extraction and amplification methods were very susceptible to change in eluent volume, poor yield and/or specificity depending on the sample type. In this perspective, our works suggests further research using and verifying it on different sample types to confirm the benefit of this alternative method.
Correlation between clinical symptoms and patient history is an important factor to verify and resulting viral amplification by real-time PCR in patient follow-up and outcome.
Our data highlighted that, the automated method of DNA extraction compared to manual protocol, could be a more helpful laboratory tool for the early detection of HCMV infection and, consequently, from a clinical perspective for a more successful pre-emptive anti- HCMV therapy ensuring excellent performance in assessing viral load.
Future studies will be aimed to increase the number of samples and protocols to apply to different biological fluids and sample volumes.
Author contribution statement
Gabriella Bianchino, Vitina Grieco: Conceived and designed the experiments; Performed the experiments; Wrote the paper.
Pietrantuono Giuseppe, Geppino Falco: Contributed reagents, materials, analysis tools or data; materials-methods.
Sabino Russi: Analyzed and interpreted the data.
Del Vecchio Luigi: Conceived and designed the experiments.
Tiziana Notarangelo: Analyzed and interpreted the data; Wrote the paper.
Data availability statement
Data included in article/supp. material/referenced in article.
Funding
This work was supported by the Italian Ministry of Health -Ricerca Corrente 2022.
Ethics statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the individual for the publication of any potentially identifiable images or data included in this article.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the patient and his family for giving consent to this case report.
References
- 1.Herbein G. Tumors and cytomegalovirus: an intimate interplay. Viruses. 2022;14 doi: 10.3390/v14040812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herbein G. High-risk oncogenic human cytomegalovirus. Viruses. 2022;14 doi: 10.3390/v14112462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen S.-J., Wang S.-C., Chen Y.-C. Challenges, recent advances and perspectives in the treatment of human cytomegalovirus infections. Trav. Med. Infect. Dis. 2022;7 doi: 10.3390/tropicalmed7120439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mengelle C., Sandres-Sauné K., Pasquier C., Rostaing L., Mansuy J.M., Marty M., Da Silva I., Attal M., Massip P., Izopet J. Automated extraction and quantification of human cytomegalovirus DNA in whole blood by real-time PCR assay. J. Clin. Microbiol. 2003;41:3840–3845. doi: 10.1128/JCM.41.8.3840-3845.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halwachs-Baumann G., Wilders-Truschnig M., Enzinger G., Eibl M., Linkesch W., Dornbusch H.J., Santner B.I., Marth E., Kessler H.H. Cytomegalovirus diagnosis in renal and bone marrow transplant recipients: the impact of molecular assays. J. Clin. Virol. 2001;20:49–57. doi: 10.1016/s1386-6532(00)00155-4. [DOI] [PubMed] [Google Scholar]
- 6.Caliendo A.M., Schuurman R., Yen-Lieberman B., Spector S.A., Andersen J., Manjiry R., Crumpacker C., Lurain N.S., Erice A. CMV working group of the complications of HIV disease RAC, AIDS clinical trials group, comparison of quantitative and qualitative PCR assays for cytomegalovirus DNA in plasma. J. Clin. Microbiol. 2001;39:1334–1338. doi: 10.1128/JCM.39.4.1334-1338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinberg T.N., Hodges S., Li G., Cai M., Zamora R. Comparison of PCR, antigenemia assay, and rapid blood culture for detection and prevention of cytomegalovirus disease after lung transplantation. J. Clin. Microbiol. 2000;38:768–772. doi: 10.1128/JCM.38.2.768-772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tong C.Y., Cuevas L.E., Williams H., Bakran A. Comparison of two commercial methods for measurement of cytomegalovirus load in blood samples after renal transplantation. J. Clin. Microbiol. 2000;38:1209–1213. doi: 10.1128/JCM.38.3.1209-1213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon S., Jung B.K., Ko S.-Y., Lee C.K., Cho Y. Comparison of quantitation of cytomegalovirus DNA by real-time PCR in whole blood with the cytomegalovirus antigenemia assay. Ann. Lab. Med. 2015;35:99–104. doi: 10.3343/alm.2015.35.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emery V.C., Sabin C.A., Cope A.V., Gor D., Hassan-Walker A.F., Griffiths P.D. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. Lancet. 2000;355:2032–2036. doi: 10.1016/S0140-6736(00)02350-3. [DOI] [PubMed] [Google Scholar]
- 11.Sia I.G., Wilson J.A., Groettum C.M., Espy M.J., Smith T.F., Paya C.V. Cytomegalovirus (CMV) DNA load predicts relapsing CMV infection after solid organ transplantation. J. Infect. Dis. 2000;181:717–720. doi: 10.1086/315242. [DOI] [PubMed] [Google Scholar]
- 12.Sia I.G., Wilson J.A., Espy M.J., Paya C.V., Smith T.F. Evaluation of the COBAS AMPLICOR CMV MONITOR test for detection of viral DNA in specimens taken from patients after liver transplantation. J. Clin. Microbiol. 2000;38:600–606. doi: 10.1128/JCM.38.2.600-606.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allice T., Cerutti F., Pittaluga F., Varetto S., Franchello A., Salizzoni M., Ghisetti V. Evaluation of a novel real-time PCR system for cytomegalovirus DNA quantitation on whole blood and correlation with pp65-antigen test in guiding pre-emptive antiviral treatment. J. Virol Methods. 2008;148:9–16. doi: 10.1016/j.jviromet.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Allice T., Enrietto M., Pittaluga F., Varetto S., Franchello A., Marchiaro G., Ghisetti V. Quantitation of cytomegalovirus DNA by real-time polymerase chain reaction in peripheral blood specimens of patients with solid organ transplants: comparison with end-point PCR and pp65 antigen test. J. Med. Virol. 2006;78:915–922. doi: 10.1002/jmv.20641. [DOI] [PubMed] [Google Scholar]
- 15.Razonable R.R., Brown R.A., Wilson J., Groettum C., Kremers W., Espy M., Smith T.F., Paya C.V. The clinical use of various blood compartments for cytomegalovirus (CMV) DNA quantitation in transplant recipients with CMV disease. Transplantation. 2002;73:968–973. doi: 10.1097/00007890-200203270-00025. [DOI] [PubMed] [Google Scholar]
- 16.Gault E., Michel Y., Dehée A., Belabani C., Nicolas J.C., Garbarg-Chenon A. Quantification of human cytomegalovirus DNA by real-time PCR. J. Clin. Microbiol. 2001;39:772–775. doi: 10.1128/JCM.39.2.772-775.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gouarin S., Vabret A., Scieux C., Agbalika F., Cherot J., Mengelle C., Deback C., Petitjean J., Dina J., Freymuth F. Multicentric evaluation of a new commercial cytomegalovirus real-time PCR quantitation assay. J. Virol Methods. 2007;146:147–154. doi: 10.1016/j.jviromet.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Boaretti M., Sorrentino A., Zantedeschi C., Forni A., Boschiero L., Fontana R. Quantification of cytomegalovirus DNA by a fully automated real-time PCR for early diagnosis and monitoring of active viral infection in solid organ transplant recipients. J. Clin. Virol. 2013;56:124–128. doi: 10.1016/j.jcv.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 19.Laurino S., Brancaccio M., Angrisano T., Calice G., Russi S., Mazzone P., Di Paola G., Aieta M., Grieco V., Bianchino G., Falco G., Notarangelo T. Role of IL-6/STAT3 Axis in resistance to cisplatin in gastric cancers. Biomedicines. 2023;11 doi: 10.3390/biomedicines11030694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.La Rocca F., Grieco V., Ruggieri V., Zifarone E., Villani O., Zoppoli P., Russi S., Laurino S., Falco G., Calice G., Marinaccio A., Natalicchio M.I., Albano F., Musto P. Superiority of droplet digital PCR over real-time quantitative PCR for JAK2V617F allele mutational burden assessment in myeloproliferative neoplasms: a retrospective study. Diagnostics. 2020;10 doi: 10.3390/diagnostics10030143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lizaola-Mayo B.C., Rodriguez E.A. Cytomegalovirus infection after liver transplantation. World J. Transplant. 2020;10:183–190. doi: 10.5500/wjt.v10.i7.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Limaye A.P., Babu T.M., Boeckh M. Progress and challenges in the prevention, diagnosis, and management of cytomegalovirus infection in transplantation. Clin. Microbiol. Rev. 2020;34 doi: 10.1128/CMR.00043-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenks C.M., Hoff S.R., Mithal L.B. Congenital cytomegalovirus infection: epidemiology, timely diagnosis, and management. NeoReviews. 2021;22 doi: 10.1542/neo.22-9-e606. [DOI] [PubMed] [Google Scholar]
- 24.Gerna G., Lilleri D., Furione M., Baldanti F. Management of human cytomegalovirus infection in transplantation: validation of virologic cut-offs for preemptive therapy and immunological cut-offs for protection. New Microbiol. 2011;34:229–254. [PubMed] [Google Scholar]
- 25.Lilleri D., Gerna G., Furione M., Bernardo M.E., Giorgiani G., Telli S., Baldanti F., Locatelli F. Use of a DNAemia cut-off for monitoring human cytomegalovirus infection reduces the number of preemptively treated children and young adults receiving hematopoietic stem-cell transplantation compared with qualitative pp65 antigenemia. Blood. 2007;110:2757–2760. doi: 10.1182/blood-2007-03-080820. [DOI] [PubMed] [Google Scholar]
- 26.Salzberger B., Bowden R.A., Hackman R.C., Davis C., Boeckh M. Neutropenia in allogeneic marrow transplant recipients receiving ganciclovir for prevention of cytomegalovirus disease: risk factors and outcome. Blood. 1997;90:2502–2508. doi: 10.1182/blood.V90.6.2502. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka Y., Kanda Y., Kami M., Mori S., Hamaki T., Kusumi E., Miyakoshi S., Nannya Y., Chiba S., Arai Y., Mitani K., Hirai H., Mutou Y. Japan Hematology and Oncology Clinical Study Group (J-HOCS), Monitoring cytomegalovirus infection by antigenemia assay and two distinct plasma real-time PCR methods after hematopoietic stem cell transplantation. Bone Marrow Transplant. 2002;30:315–319. doi: 10.1038/sj.bmt.1703661. [DOI] [PubMed] [Google Scholar]
- 28.Khawaja F., Spallone A., Kotton C.N., Chemaly R.F. Cytomegalovirus infection in transplant recipients: newly approved additions to our armamentarium. Clin. Microbiol. Infect. 2023;29:44–50. doi: 10.1016/j.cmi.2022.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Petrisli E., Chiereghin A., Gabrielli L., Zanfi C., Lauro A., Piccirilli G., Baccolini F., Altimari A., Bagni A., Cescon M., Pinna A.D., Landini M.P., Lazzarotto T. Early and late virological monitoring of cytomegalovirus, Epstein-Barr virus, and human herpes virus 6 infections in small bowel/multivisceral transplant recipients. Transplant. Proc. 2010;42:74–78. doi: 10.1016/j.transproceed.2009.12.032. [DOI] [PubMed] [Google Scholar]
- 30.Bravo D., Clari M.Á., Costa E., Muñoz-Cobo B., Solano C., José Remigia M., Navarro D. Comparative evaluation of three automated systems for DNA extraction in conjunction with three commercially available real-time PCR assays for quantitation of plasma Cytomegalovirus DNAemia in allogeneic stem cell transplant recipients. J. Clin. Microbiol. 2011;49:2899–2904. doi: 10.1128/JCM.00785-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennett D., Bergantini L., Ferrara P., Cusi M.G., Scolletta S., Montagnani F., Paladini P., Sestini P., Refini R.M., Luzzi L., Fossi A., Bargagli E. Cytomegalovirus infection is associated with development of chronic lung allograft dysfunction. Lung. 2022;200:513–522. doi: 10.1007/s00408-022-00551-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zangger N., Oxenius A. T cell immunity to cytomegalovirus infection. Curr. Opin. Immunol. 2022;77 doi: 10.1016/j.coi.2022.102185. [DOI] [PubMed] [Google Scholar]
- 33.Costa C., Mantovani S., Balloco C., Sidoti F., Fop F., Cavallo R. Comparison of two nucleic acid extraction and testing systems for HCMV-DNA detection and quantitation on whole blood specimens from transplant patients. J. Virol Methods. 2013;193:579–582. doi: 10.1016/j.jviromet.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 34.Uwiringiyeyezu T., El Khalfi B., Saile R., Belhachmi J., Soukri A. Comparability of CMV DNA extraction methods and validation of viral load. Methods Protoc. 2022;5 doi: 10.3390/mps5010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supp. material/referenced in article.