Abstract
Poultry is one of the most commonly farmed species and the most widespread meat industries. However, numerous poultry flocks have been long threatened by pathogenic bacterial infections, especially antimicrobial resistant pathogens. Here the prevalence and the antimicrobial resistance (AMR) profiles of bacterial pathogens isolated from poultry in Jiangxi Province, China were investigated. From 2020 to 2022, 283 tissue and liquid samples were collected from clinically diseased poultry, including duck, chicken, and goose, with an overall positive isolation rate of 62.90%. Among all the 219 bacterial isolates, 29 strains were gram-positive and 190 strains were gram-negative. Major bacteria species involved were avian pathogenic Escherichia coli (APEC; 57.53%; 126/219), followed by Salmonella spp. (11.87%, 26/219), Pasteurella multocida (6.39%, 14/219), and Staphylococcus spp. (1.22%, 11/219). Antimicrobial susceptibility testing showed the APEC isolates displayed considerably higher levels of AMR than the Salmonella and P. multocida isolates. The APEC isolates showed high resistance rate to amoxicillin (89.68%), ampicillin (89.68%), and florfenicol (83.33%), followed by streptomycin (75.40%), cefradine (65.87%), and enrofloxacin (64.29%). Multidrug-resistant isolates were observed in APEC (99.21%), Salmonella spp. (96.16%), and P. multocida (85.71%), and nearly 3 quarters of the APEC strains were resistant to 7 or more categories of antimicrobial drugs. Moreover, blaNDM genes associated with carbapenemase resistance and mcr-1 associated with colisitin resistance were detected in the APEC isolates. Our findings could provide evidence-based guidance for veterinarians to prevent and control bacterial diseases, and be helpful for monitoring the emerging and development of AMR in poultry bacterial pathogens.
Key words: poultry, bacterial pathogen, antimicrobial resistance, avian pathogenic Escherichia coli, Jiangxi Province
INTRODUCTION
Poultry is one of the most widespread types of meat food industries worldwide because of its relatively low production costs and the absence of cultural and religious restrictions for its consumption (Nhung et al., 2017). Poultry flocks are often raised under intensive conditions, and the consequent threat of bacterial infection leads to huge economical losses and decreased animal welfare, such as avian pathogenic Escherichia coli (APEC) infections (Christensen et al., 2021). Bacterial infections have traditionally and preferentially been controlled by the use of a large diversity of antimicrobials. However, the emergence of antimicrobial resistance (AMR) in bacteria from global animal production results in severe risks of ineffective antimicrobials and veterinary treatment failure (Christensen et al., 2021). Additionally, AMR in poultry pathogens is likely to threaten human health because of poultry is an important source of zoonoses transmissible to humans (Munang'andu et al., 2012). Nowadays, many concerns have been raised about the horizontal transmission transfer of AMR bacteria and genes within and between poultry flocks and farms, and even zoonotic transfer via the food chain (Christensen et al., 2021). Thus, monitoring the AMR profiles of pathogenic bacteria is necessary for optimizing effective antimicrobial treatments in poultry and following up the development of bacterial drug resistance.
Jiangxi Province is one of the largest provinces in poultry breeding of China and possesses many excellent native chicken breeds (Li et al., 2021). By the end of 2022, the poultry stock of Jiangxi Province was 241.674 million, up by 4.2% year over year; 592.635 million poultry were brought to market, up by 2.7% from a year earlier. However, the bacterial infection investigation and the relative antimicrobial susceptibility information of poultry industry in Jiangxi Province have not been well reported in the last decades. This study aims to identify the prevalence of bacterial pathogens associated with poultry morbidity and mortality, and obtain baseline information of AMR in clinical bacterial pathogens. The results could provide evidence-based guidance for veterinarians to prevent and control bacterial diseases and help reduce the development of AMR in Jiangxi Province, China.
MATERIALS AND METHODS
Clinical Sample Collection
During a 2-yr period from April 2020 to December 2022, 283 clinical samples were collected from sick or dead poultry in 11 different cities within Jiangxi Province, China. Samples of duck (n = 192), chicken (n = 68), and goose (n = 23) were mainly livers, followed by brains, lungs, ascites, and semen. The samples were stored in a low-temperature environment and transferred to the laboratory. Among the collected samples, the breeds of ducks were mainly Sheldrake and White duck; the breeds of chickens were mainly white-feathered chickens and local breeds, such as Taihe Silky Fowl and Ningdu Yellow Chicken; the breeds of geese are Xingguo Grey Goose. The age of the sick or dead poultry ranged from 10 d to 350 d.
Bacterial Isolation
Bacterial isolation was performed under sterile conditions. The surface of organ or tissue samples were sterilized with 75% alcohol. Then, a small piece of meat (weight approx. 0.2 g) was cut and homogenized in 1 mL of sterile physiological saline. Each 50 μL of the sample homogenate and the liquid samples were cultured aerobically on tryptic soy agar (Hopebio, Qingdao, China) with 5% (v/v) newborn bovine serum (NBS; Sijiqing, Hangzhou, China) at 37°C for 24 h. The samples were simultaneously cultured on MacConkey agar (Hopebio) for rapid preliminary identification of bacterial species.
Bacterial Identification
Bacterial colonies were expanded at 37°C in tryptone soya broth (Hopebio) with 5% NBS or lysogeny broth (Hopebio). Bacterial morphology was identified using gram staining. The bacterial genome was extracted using Bacterial DNA Kit (Omega Bio-Tek, Norcross, Georgia) according to the manufacturer's instructions. PCR targeting the gene coding for 16S rRNA was performed with the primer pair 27F/1492R (Table S1). Amplification products were then sent to Sangon Biotech (Shanghai) Co., Ltd. for sequencing. Bacterial sequences were further matched with the database in NCBI using Nucleotide BLAST. All bacteria were stored with glycerin at a final concentration of 25% at −80°C.
Scanning Electron Microscope
Bacteria were cultured at 37°C in tryptone soya broth with 5% NBS or lysogeny broth to the mid-log phase. Cells were harvested and fixed with 2.5% glutaraldehyde at 4°C overnight. The subsequent dehydration steps with ethanol and metal plating were performed in accordance with previously described methods (Tan et al., 2017). Electron microscope observation was performed by using a JSM-6390LV SEM (NTC, Tokyo, Japan).
Antibiotic Drugs
A total of 17 commercially available antimicrobial drugs for veterinary and human use, namely, penicillins (amoxicillin [AMX; 10 μg per tablet] and ampicillin [AMP; 10 μg per tablet]), cephalosporins (cefuroxime [CXM; 30 μg per tablet], cefradine [CED; 30 μg per tablet], and ceftazidime [CAZ; 30 μg per tablet]), tetracycline (doxycycline [DOX; 30 μg per tablet]), chloramphenicol (florfenicol [FON; 30 μg per tablet]), macrolide (erythromycin [ERM; 15 μg per tablet]), peptide antibiotic (polymyxin B [PMB; 300 IU per tablet]), lincosamide (lincomycin [LCM; 2 μg per tablet]), quinolones (enrofloxacin [ENR; 10 μg per tablet] and ciprofloxacin [CIP; 5 μg per tablet]), aminoglycosides (neomycin [NEO; 30 μg per tablet], spectinomycin [SPT; 100 μg per tablet], gentamicin [GEN; 10 μg per tablet], streptomycin [STR; 10 μg per tablet], and kanamycin [KAN; 30 μg per tablet]), were prepared for antimicrobial susceptibility assays. Antibiotic disks were purchased from Hangzhou Microbial Reagent Co., Ltd. (Hangzhou, China).
Antimicrobial Susceptibility Assay
Antimicrobial susceptibility assays of all the bacterial isolates were performed using the disk diffusion method on Mueller-Hinton agar (Hopebio) in accordance with the standardized protocol suggested by the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2022). For bacteria with high nutritional requirements, 5% NBS was also added into the plates. Briefly, bacterial cultures were adjusted to be a 0.5 McFarland standard with sterile physiological saline and evenly spread on Mueller-Hinton agar plates using aseptic cotton swabs. The specific antibiotic disks were placed on plates with a disk dispenser. Subsequently, the plates were incubated at 37°C for 18 to 24 h. E. coli ATCC 25922 was used as the quality control strain. The diameter of the inhibition zone was measured with a vernier caliper. The resistance breakpoints for the zone diameter were defined according to the CLSI guidelines (CLSI, 2022). The results were identified as susceptible, intermediate, or resistant. Histograms were drawn on GraphPad prism 8.0 software (San Diego, CA).
Identification of Antibiotic Resistance Genes
General PCR amplification was conducted for the identification of acquired carbapenemase gene (blaNDM) (Zafer et al., 2015) and colistin resistance genes (mcr-1 to mcr-8) (Rebelo et al., 2018; Yang et al., 2019). The 9 sets of primers, the expected fragment sizes, and the references are listed in Table S1. The addition of ddH2O instead of bacterial DNA was served as the negative control during PCR assays. Aliquots were analyzed using electrophoresis on a 0.8% (w/v) agarose gel.
Ethics Statement
When samples needed to be obtained from living sick poultry, the experiment protocol was approved by the Animal Ethics Committee of the Institute of Animal Husbandry and Veterinary, Jiangxi Academy of Agricultural Science (2010-JXAAS-XM-01). All efforts were made to minimize suffering.
RESULTS
Prevalence of Bacteria Isolated From Clinical Samples
In this study, a total of 283 tissues and liquid samples from clinically sick or dead poultry were collected in Jiangxi Province from April 2020 to December 2022 for bacteria isolation. Through morphological observation and PCR identification, 219 bacterial strains were recovered from 178 (62.90%; n = 283) of all the samples (Table 1). The prevalence of bacteria in ducks, chickens, and geese was 64.58% (124/192), 64.71% (44/68), and 43.48% (10/23), respectively. Especially, mixed infections were observed in the 3 kinds of poultry, including 29 (16.29%; n = 178) dual-infected cases and 5 (1.77%; n = 178) multiple-infected cases.
Table 1.
Characteristics of clinical poultry samples.
| Poultry species | Sample number | Positive samples (rates) | No. of mono-infected samples | No. of dual-infected samples | No. of multiple-infected samples | No. of isolated bacteria |
|---|---|---|---|---|---|---|
| Duck | 192 | 124 (64.58%) | 101 | 21 | 2 | 149 |
| Chicken | 68 | 44 (64.71%) | 36 | 6 | 2 | 55 |
| Goose | 23 | 10 (43.48%) | 7 | 2 | 1 | 15 |
| Total | 283 | 178 (62.90%) | 144 | 29 | 5 | 219 |
Isolate Distribution
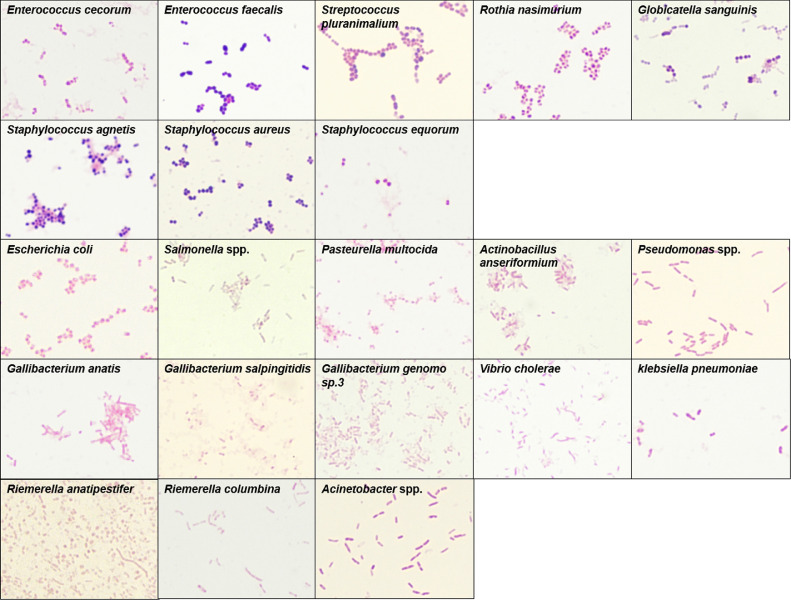
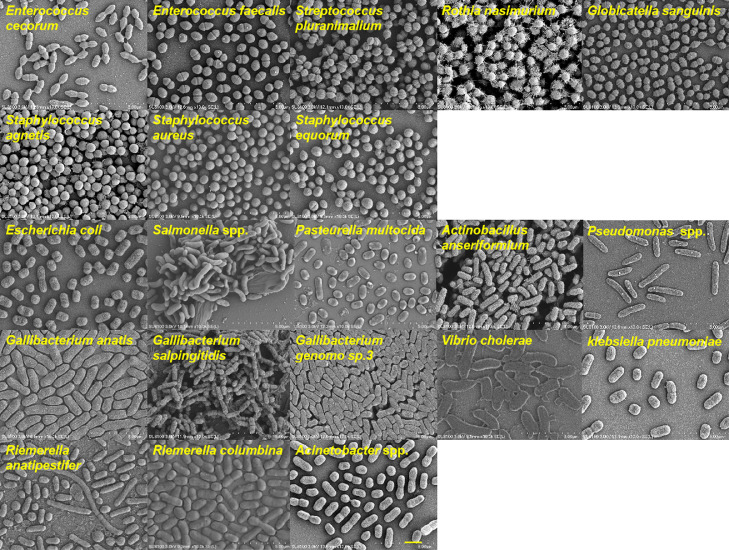
Among the 219 bacterial isolates, 29 strains were gram-positive, and 190 strains were gram-negative (Table S2). The detailed information of host, sample, and collection time and city of each strain are shown in Table S2. The gram staining micrographs (Figure 1) and scanning electron micrographs (Figure 2) of 8 strains of gram-positive bacteria and 13 strains of gram-negative bacteria were organized and provided.
Figure 1.
Gram staining of pathogenic bacteria (1,000×).
Figure 2.
Morphological micrographs of pathogenic bacteria. Bar, 2 μm.
The bacterial isolates could be divided into 16 genera (Table 2). Our data indicated that APEC was the predominant pathogenic bacteria, with a proportion of 57.53% (126/219), followed by Salmonella spp. (11.87%, 26/219), Pasteurella multocida (6.39%, 14/219), and Staphylococcus spp. (1.22%, 11/219). Further analysis showed the proportion of APEC in duck and chicken source isolates was the highest, and the proportion of Salmonella spp. in goose source isolates was the highest. Eight strains of Riemerella anatipestifer were isolated from duck livers, whereas 1 strain of Riemerella columbina was isolated from chicken ascites (Table S2). Additionally, Streptococcus spp. (5 strains), Globicatella sanguinis (3 strains), Vibro cholerae (1 strain), Pseudomonas spp. (1 strain), Acinetobacter spp. (1 strain), and Actinobacillus anseriformium (1 strain) were identified only in duck source samples (Table 2).
Table 2.
Distribution of bacteria isolates.
| Bacteria | Duck | Chicken | Goose | Total | Proportion of bacteria (n = 219) |
|---|---|---|---|---|---|
| Avian pathogenic Escherichia coli | 105 | 19 | 2 | 126 | 57.53% |
| Salmonella spp. | 0 | 17 | 9 | 26 | 11.87% |
| Pasteurella multocida | 9 | 5 | 0 | 14 | 6.39% |
| Staphylococcus spp. | 6 | 5 | 0 | 11 | 1.22% |
| Riemerella spp. | 8 | 1 | 0 | 9 | 4.11% |
| Gallibacterium spp. | 4 | 4 | 0 | 8 | 3.65% |
| Enterococcus spp. | 4 | 1 | 1 | 6 | 2.74% |
| Streptococcus spp. | 5 | 0 | 0 | 5 | 2.28% |
| Rothia nasimurium | 1 | 1 | 2 | 4 | 1.83% |
| Globicatella sanguinis | 3 | 0 | 0 | 3 | 1.37% |
| Klebsiella pneumoniae | 0 | 1 | 1 | 2 | 0.91% |
| Vibro cholerae | 1 | 0 | 0 | 1 | 0.46% |
| Pseudomonas spp. | 1 | 0 | 0 | 1 | 0.46% |
| Acinetobacter spp. | 1 | 0 | 0 | 1 | 0.46% |
| Actinobacillus anseriformium | 1 | 0 | 0 | 1 | 0.46% |
| Avibacterium paragallinarum | 0 | 1 | 0 | 1 | 0.46% |
| Total | 149 | 55 | 15 | 219 | − |
Antimicrobial Resistance Analysis
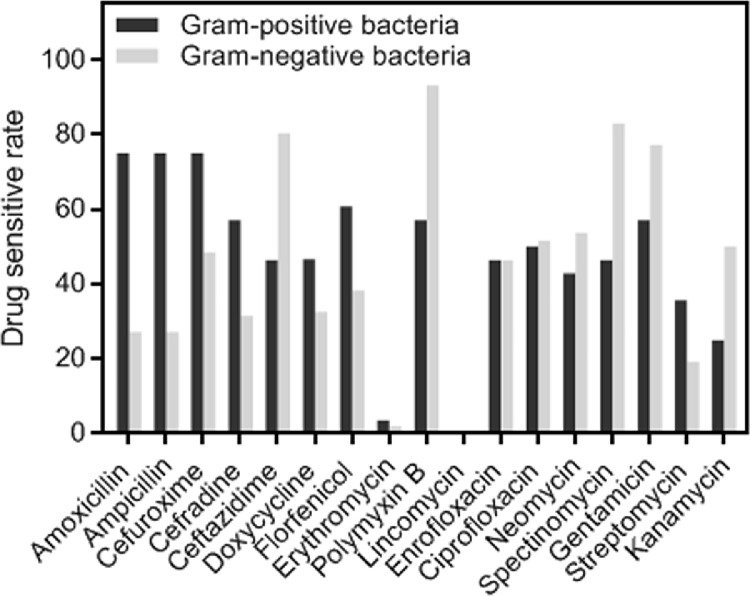
The antimicrobial susceptibility of bacteria strains was determined using the disk diffusion method with 17 antimicrobial drugs as belonging to 9 categories. The diameters of inhibition zone of bacteria strains are shown in Table S2. The antimicrobial susceptibility profiles between gram-positive bacteria and gram-negative bacteria were compared. As shown in Figure 3, gram-positive bacteria were the most sensitive to AMX, AMP, and CXM, and the sensitivity rates of the 3 drugs were 75%. Gram-negative bacteria were the most sensitive to PMB (93.09%), followed by SPT (82.98%), CAZ (80.32%), and GEN (77.13%).
Figure 3.
Comparison of antimicrobial susceptibility profiles between gram-positive and gram-negative bacteria.
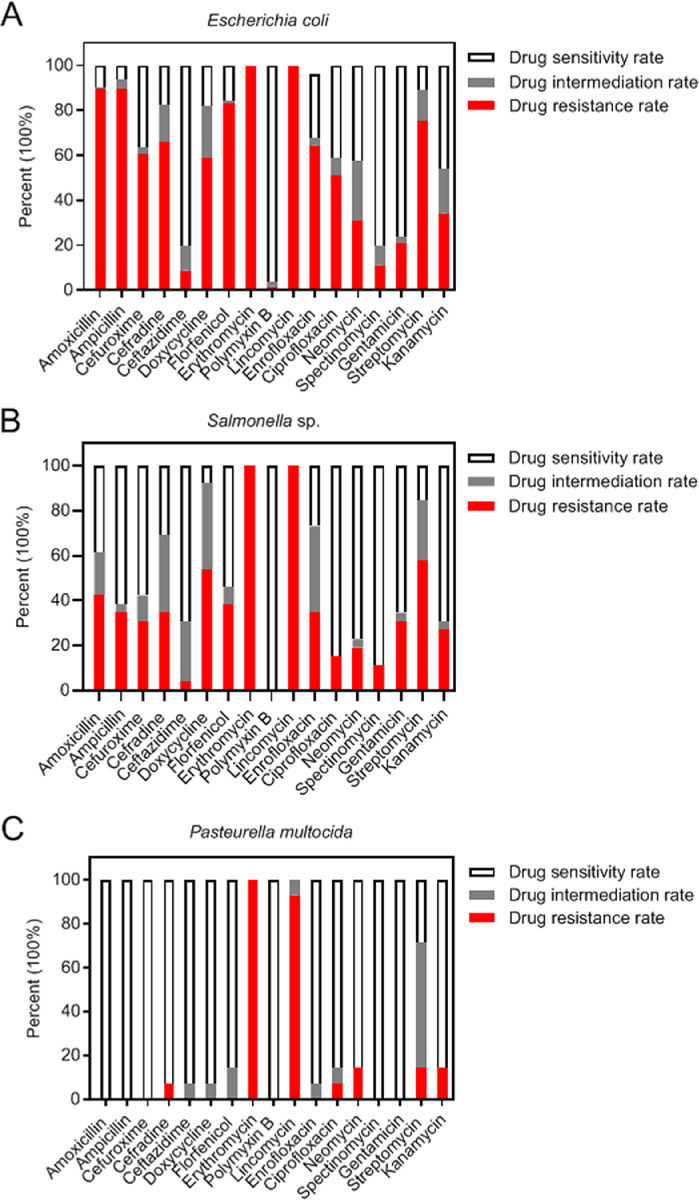
The AMR profiles of APEC, Salmonella spp., and P. multocida were analyzed because of high isolation rates of the 3 kinds of bacteria (Figure 4). The results showed APEC, Salmonella spp., and P. multocida were nearly 100% resistant to ERM and LCM. The APEC strains showed high resistance rate to AMX (89.68%), AMP (89.68%), and FON (83.33%), followed by STR (75.40%), CED (65.87%), and ENR (64.29%). Conversely, the APEC strains showed high sensitivity rate to PMB (96.03%), SPT and CAZ (80.16%), and GEN (76.19%). In addition, more than half of Salmonella strains were resistant to STR (57.69%) and DOX (53.85%), whereas most Salmonella strains were sensitive to PMB (100.00%), SPT (88.46%), CIP (84.62%), and NEO (76.92%). Moreover, all the P. multocida strains were completely sensitive to a variety of antimicrobial drugs, such as AMP, AMP, CXM, PMB, SPT, and GEN. The P. multocida strains also had high sensitivity rates to the other drugs (>85%) except for STR (28.57%).
Figure 4.
Antimicrobial resistance profiles of Escherichia coli (A), Salmonella (B), and Pasteurella multocida (C).
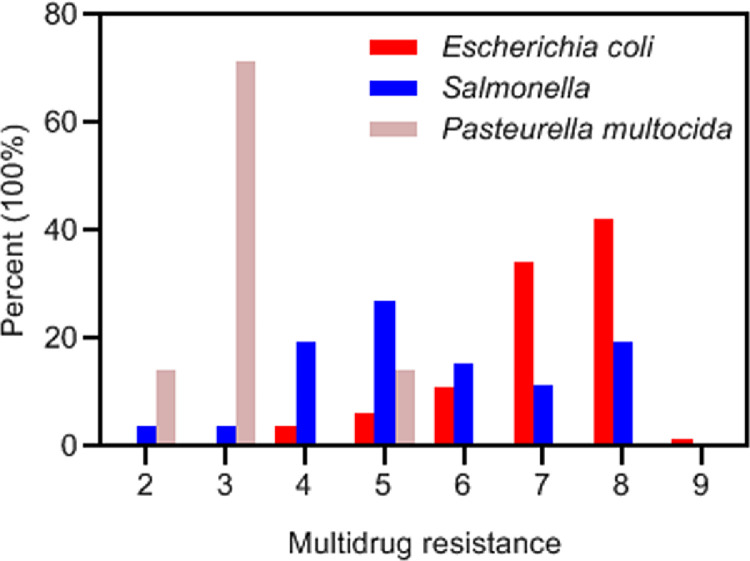
Multidrug Resistance
The standardized definition for multidrug resistance is presented as a bacteria isolate that is not susceptible to at least 3 antimicrobial categories (Sweeney et al., 2018). Multidrug resistance of APEC, Salmonella spp., and P. multocida was counted and showed in Figure 5. Multidrug-resistant isolates were observed in APEC (99.21%), Salmonella spp. (96.16%), and P. multocida (85.71%). Among all the APEC isolates, 42.00% were resistant to 8 categories of antimicrobial drugs, and 34.13% were resistant to 7 categories of antimicrobial drugs, accounting for the highest portion. Notably, one of the APEC isolates exhibited drug insensitivity to the 9 categories; 47.62% of the APEC isolates exhibited drug insensitivity to 12 or more antibacterial drugs, and nearly 3 quarters of the APEC strains were resistant to 7 or more categories of antimicrobial drugs. For the Salmonella isolates, 26.92% were resistant to 5 categories of antimicrobial drugs with the highest portion. In addition, 71.43% of the P. multocida isolates were resistant to 3 categories of antimicrobial drugs.
Figure 5.
Analysis of multidrug resistance. X-axis indicates resistance of the isolates to 2 to 9 categories of antimicrobial drugs.
Distribution of Antimicrobial Resistance Genes
The blaNDM genes encoding metallo-β-lactamases are associated with carbapenemase resistance (Poirel et al., 2011). The mcr genes encoding phosphoetanolamine transferases are associated with colisitin resistance (Barbieri et al., 2021). Previous studies show that blaNDM and mcr-1-positive bacteria disseminate from poultry farms to supermarkets along the poultry production chain in China and contaminate the environment (Wang et al., 2017). Therefore, the prevalence of blaNDM and mcr in all bacteria isolates was investigated through PCR assays.
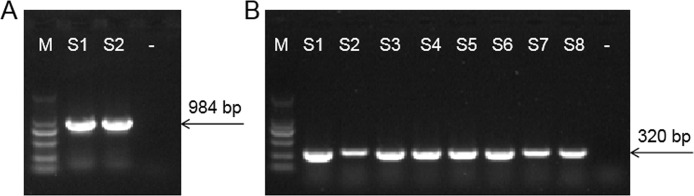
Two blaNDM-positive APEC (Figure 6A) and 8 mcr-1-positive APEC (Figure 6B) were identified as a consequence, whereas mcr-2 to mcr-8 were not detected (Table S2). None of the strains possesses the 2 drug resistance genes. The 2 blaNDM-positive APEC isolates exhibited a broad spectrum of AMR to all antimicrobial drugs except for PMB. However, the phenotypes of 5 PMB-insensitive APEC isolates were not fully consistent with the genotypes, which indicated that further genome sequencing and analysis and particular characterization of colistin resistance is needed.
Figure 6.
PCR amplification of blaNDM (A) and mcr-1 (B). M represents the DNA marker (100–2,000 bp). S1–S2 in the left figure refers to the 2 blaNDM-positive APEC strains. S1–S8 in the right figure the 8 mcr-1-positive APEC strains. - represents the negative control.
DISCUSSION
To the knowledge of the authors, well-directed studies have been rarely carried out on poultry bacterial diseases in Jiangxi Province despite its major, expanding poultry industry. Thus, 283 clinical tissue and liquid samples were collected from poultry flocks for the period of 2020 to 2022. The bacteriology results showed a high bacterial detection rate of samples, up to 62.90%. In particular, the isolation rate of gram-negative bacteria (86.76%; 190/219) far exceeds that of gram-positive bacteria (13.24%, 29/219). The results indicated that despite available diverse categories of antibacterial drugs nowadays, bacterial infections still require more attentions, especially the threat from gram-negative bacteria.
Acting as primary and secondary pathogen, APEC cause colibacillosis in different types of poultry of all ages and is associated with a syndrome of severe extraintestinal disease (Aleksandrowicz et al., 2021). APEC infections constitute a major threat to the poultry industry worldwide due to high morbidity and mortality, reduced productivity within poultry flocks, extensive economic losses, and contamination of the food chain (Alber et al., 2021). The results of bacteria isolations from broiler and layer chicks in Zambia showed the main pathogens were APEC, Salmonella gallinarum, and Proteus species (Munang'andu et al., 2012). In the United States, APEC and Clostridium perfringens are 2 important pathogenic bacteria readily found in the broiler environment (Fancher et al., 2020). In this study, major bacteria species involved were APEC (57.53%), Salmonella spp. (11.87%), and P. multocida (6.39%), which showed APEC is the primary bacterial pathogen in poultry breeding industry in Jiangxi Province, China.
In addition to the high detection rate, the AMR of APEC isolated in Jiangxi Province is severe. A previous review summarized phenotypic resistance in thousands of APEC isolates from diseased chickens from Asia, Africa, the United States, Spain, and Brazil with disk diffusion assays (Nhung et al., 2017). Among these APEC isolates, 243 were isolated from diseased chickens, ducks, and geese during the period from 2007 to 2014 in eastern China and tested for the AMR profiles (Dou et al., 2016). Drug resistance rates of the APEC isolates in this study were AMX (89.68%), AMP (89.68%), STR (75.40%), KAN (34.13%), FON (83.33%), and ENR (64.29%), which were higher than the data from the average resistance levels of these above strains (Nhung et al., 2017). APEC showed high insensitive rate to most of the tested antibacterial drugs and strong multidrug resistance. Moreover, AMR-specific genes, blaNDM and mcr-1, were observed in APEC isolates.
Salmonella spp. is one of the most important foodborne pathogens worldwide and causes severe public health problems. The authors used to investigate the presence of Salmonella spp. in chicken and duck samples from retail markets in different provinces of China from 2010 to 2014; the overall prevalence of Salmonella spp. in chicken and duck meat were 14.3 and 11.6%, respectively (Zeng et al., 2019). The results in this study showed Salmonella spp. were only recovered from chickens and geese, with the prevalence of 25% (17/68) and 39.13% (9/23), respectively. Considering the risk of Salmonella infections in human beings, further investigations should be conducted and the AMR of foodborne pathogens from extensive sources should be continuously monitored.
One of the main concerns of this study is the emergence of some newly developing bacteria and infection cases. Rothia nasimurium is a facultative anaerobic gram-positive coccus and part of the commensal flora of humans and other animals. Until recently, R. nasimurium was reported to be an opportunistic pathogen to ducks (Wang et al., 2021) and chickens (Zhang et al., 2022). Here, R. nasimurium strains were recovered from not only ducks and chickens but also geese. Globicatella sanguinis is a rare gram-positive cocci known to affect the bloodstream, urinary tract, and central nervous system in human beings (Gupta et al., 2022). Streptococcus pluranimalium, a gram-positive aerobic coccus, has been isolated primarily from human beings and several farm animals, such as horses, cattles, and pigs (Ahmed et al., 2018). Here, G. sanguinis and S. pluranimalium strains were isolated from duck brain and liver samples, which predicted that the 2 bacteria are potential zoonotic pathogens. A case of Streptococcus uberis-infection in ducks was also observed. Interestingly, S. uberis is the leading pathogen in dairy herds, causing clinical and subclinical infections (Tabashiri et al., 2022). Moreover, 1 strain of A. anseriformium was isolated from duck liver, and the bacterium has once been isolated from a case of conjunctivitis in duck in 2012 (Bisgaard and Christensen, 2012).
It has been a common understanding that high usage levels of antibacterial drugs would give advantage to the development and transmission of organisms with higher levels of resistance (Nhung et al., 2017). Studies on larger collection of E. coli strains from humans and food animals have conclusively demonstrated increases in resistance over time against most antimicrobials (Tadesse et al., 2012). In recent times, many strategies have been proposed to control poultry bacterial diseases, such as using plant extracts (Abiala et al., 2016), improvement of host genetic resistance (Monson and Lamont, 2021), vaccination and use of competitive exclusion (Christensen et al., 2021), and alternative therapies, especially virulence inhibitors (Kathayat et al., 2021). At present, various vaccine candidates, mostly live-attenuated and recombinant vaccines, have been investigated to protect poultry against APEC and Salmonella infections (Kathayat et al., 2021). However, effective bacteria vaccines that can provide protection against diverse serotypes and heterogenous infections are still urgently needed.
In conclusion, the present study investigated the current information about bacterial infections, AMR profiles, and specific AMR genes throughout the poultry industry in Jiangxi Province. Our findings showed a high bacterial isolation rate from clinical poultry flocks and infections from gram-negative bacteria were more usual, especially APEC infections. The results of antimicrobial susceptibility testing indicated that multidrug resistance has widely emerged in avian pathogenic bacteria in Jiangxi Province. In addition, the presence of specific AMR genes is increasing the risk of the intra- and interspecies horizontal transfers. Thus, a continuous clinical surveillance of bacterial AMR is urgently needed. Taking advantage of stringent measures is also important to reduce the development of AMR and treatment failure of poultry diseases, such as improving management protocols, encouraging responsible antimicrobial usage, and establishing a knowledge base on the AMR profile of poultry pathogens. Moreover, further research focusing on the understanding of AMR-associated mechanisms would be helpful and necessary for developing effective preventive measures of bacterial infections.
ACKNOWLEDGMENTS
This work was supported by the Jiangxi Collaborative Innovation Fund of Modern Agricultural Scientific Research (JXXTCXQN202201), the Earmarked Fund for Jiangxi Agriculture Research System (JXARS-09), the Jiangxi Joint Research Project for Improved Variety (2022JXCQZY04), and the Jiangxi Collaborative Innovation Fund of Modern Agricultural Scientific Research (JXXTCX202106).
DISCLOSURES
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in the present study.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2023.102830.
Appendix. Supplementary materials
REFERENCES
- Abiala M., Olayiwola J., Babatunde O., Aiyelaagbe O., Akinyemi S. Evaluation of therapeutic potentials of plant extracts against poultry bacteria threatening public health. BMC Complement. Altern. Med. 2016;16:417. doi: 10.1186/s12906-016-1399-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A., Zaman G., Gardezi A., Satti L., Sabir N., Haleem A. Cerebral abscess caused by novel species: Streptococcus pluranimalium. J. Coll. Phys. Surg. Pak. 2018;28:S181–S183. doi: 10.29271/jcpsp.2018.09.S181. [DOI] [PubMed] [Google Scholar]
- Alber A., Stevens M.P., Vervelde L. The bird's immune response to avian pathogenic Escherichia coli. Avian Pathol. 2021;50:382–391. doi: 10.1080/03079457.2021.1873246. [DOI] [PubMed] [Google Scholar]
- Aleksandrowicz A., Khan M.M., Sidorczuk K., Noszka M., Kolenda R. Whatever makes them stick - adhesins of avian pathogenic Escherichia coli. Vet. Microbiol. 2021;257 doi: 10.1016/j.vetmic.2021.109095. [DOI] [PubMed] [Google Scholar]
- Barbieri N.L., Pimenta R.L., de Melo D.A., Nolan L.K., de Souza M.M.S., Logue C.M. mcr-1 identified in fecal Escherichia coli and avian pathogenic E. coli (APEC) from Brazil. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.659613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgaard M., Christensen H. Classification of avian haemolytic Actinobacillus-like organisms (Bisgaard taxon 26) associated with anseriforme birds as Actinobacillus anseriformium sp. nov. Int. J. Syst. Evol. Microbiol. 2012;62:352–358. doi: 10.1099/ijs.0.028746-0. [DOI] [PubMed] [Google Scholar]
- Christensen H., Bachmeier J., Bisgaard M. New strategies to prevent and control avian pathogenic Escherichia coli (APEC) Avian Pathol. 2021;50:370–381. doi: 10.1080/03079457.2020.1845300. [DOI] [PubMed] [Google Scholar]
- CLSI . 32nd ed. Clinical and Laboratory Standards Institute; Wayne, PA: 2022. Performance Standards for Antimicrobial Susceptibility Testing, M100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou X., Gong J., Han X., Xu M., Shen H., Zhang D., Zhuang L., Liu J., Zou J. Characterization of avian pathogenic Escherichia coli isolated in eastern China. Gene. 2016;576:244–248. doi: 10.1016/j.gene.2015.10.012. [DOI] [PubMed] [Google Scholar]
- Fancher C.A., Zhang L., Kiess A.S., Adhikari P.A., Dinh T.T.N., Sukumaran A.T. Avian pathogenic Escherichia coli and Clostridium perfringens: challenges in no antibiotics ever broiler production and potential solutions. Microorganisms. 2020;8:1533. doi: 10.3390/microorganisms8101533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta B., Jain A.K., Saini M., Sardana M., Soni R., Angrup A. Globicatella sanguinis corneal abscess with endophthalmitis. J. AAPOS. 2022;26:46–48. doi: 10.1016/j.jaapos.2021.08.305. [DOI] [PubMed] [Google Scholar]
- Kathayat D., Lokesh D., Ranjit S., Rajashekara G. Avian pathogenic Escherichia coli (APEC): an overview of virulence and pathogenesis factors, zoonotic potential, and control strategies. Pathogens. 2021;10:467. doi: 10.3390/pathogens10040467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Tan M., Zhang F., Ji H., Zeng Y., Yang Q., Tan J., Huang J., Su Q., Huang Y., Kang Z. Diversity of avian leukosis virus subgroup J in local chickens, Jiangxi, China. Sci. Rep. 2021;11:4797. doi: 10.1038/s41598-021-84189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monson M.S., Lamont S.J. Genetic resistance to avian pathogenic Escherichia coli (APEC): current status and opportunities. Avian Pathol. 2021;50:392–401. doi: 10.1080/03079457.2021.1879990. [DOI] [PubMed] [Google Scholar]
- Munang'andu H.M., Kabilika S.H., Chibomba O., Munyeme M., Muuka G.M. Bacteria isolations from broiler and layer chicks in Zambia. J. Pathog. 2012;2012 doi: 10.1155/2012/520564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nhung N.T., Chansiripornchai N., Carrique-Mas J.J. Antimicrobial resistance in bacterial poultry pathogens: a review. Front. Vet. Sci. 2017;4:126. doi: 10.3389/fvets.2017.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Dortet L., Bernabeu S., Nordmann P. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 2011;55:5403–5407. doi: 10.1128/AAC.00585-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo A.R., Bortolaia V., Kjeldgaard J.S., Pedersen S.K., Leekitcharoenphon P., Hansen I.M., Guerra B., Malorny B., Borowiak M., Hammerl J.A., Battisti A., Franco A., Alba P., Perrin-Guyomard A., Granier S.A., De Frutos Escobar C., Malhotra-Kumar S., Villa L., Carattoli A., Hendriksen R.S. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eur. Surveill. 2018;23:17–00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M.T., Lubbers B.V., Schwarz S., Watts J.L. Applying definitions for multidrug resistance, extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J. Antimicrob. Chemother. 2018;73:1460–1463. doi: 10.1093/jac/dky043. [DOI] [PubMed] [Google Scholar]
- Tabashiri R., Sharifi S., Pakdel A., Bakhtiarizadeh M.R., Pakdel M.H., Tahmasebi A., Hercus C. Genome-wide post-transcriptional regulation of bovine mammary gland response to Streptococcus uberis. J. Appl. Genet. 2022;63:771–782. doi: 10.1007/s13353-022-00722-y. [DOI] [PubMed] [Google Scholar]
- Tadesse D.A., Zhao S., Tong E., Ayers S., Singh A., Bartholomew M.J., McDermott P.F. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950-2002. Emerg. Infect. Dis. 2012;18:741–749. doi: 10.3201/eid1805.111153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M.F., Liu W.Q., Zhang C.Y., Gao T., Zheng L.L., Qiu D.X., Li L., Zhou R. The involvement of MsmK in pathogenesis of the Streptococcus suis serotype 2. MicrobiologyOpen. 2017;6:e00433. doi: 10.1002/mbo3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Li Y., Lin X., Xu H., Li Y., Xue R., Wang G., Sun S., Li J., Lan Z., Chen J. Genetic characterization, mechanisms and dissemination risk of antibiotic resistance of multidrug-resistant Rothia nasimurium. Infect. Genet. Evol. 2021;90 doi: 10.1016/j.meegid.2021.104770. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang R., Li J., Wu Z., Yin W., Schwarz S., Tyrrell J.M., Zheng Y., Wang S., Shen Z., Liu Z., Liu J., Lei L., Li M., Zhang Q., Wu C., Zhang Q., Wu Y., Walsh T.R., Shen J. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2017;2:16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- Yang F., Shen C., Zheng X., Liu Y., El-Sayed Ahmed M.A.E., Zhao Z., Liao K., Shi Y., Guo X., Zhong R., Xu Z., Tian G.B. Plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli and Klebsiella pneumoniae isolated from market retail fruits in Guangzhou, China. Infect. Drug Resist. 2019;12:385–389. doi: 10.2147/IDR.S194635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafer M.M., Al-Agamy M.H., El-Mahallawy H.A., Amin M.A., El Din Ashour S. Dissemination of VIM-2 producing Pseudomonas aeruginosa ST233 at tertiary care hospitals in Egypt. BMC Infect. Dis. 2015;15:122. doi: 10.1186/s12879-015-0861-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y.B., Xiong L.G., Tan M.F., Li H.Q., Yan H., Zhang L., Yin D.F., Kang Z.F., Wei Q.P., Luo L.G. Prevalence and antimicrobial resistance of Salmonella in pork, chicken, and duck from retail markets of China. Foodborne Pathog. Dis. 2019;16:339–345. doi: 10.1089/fpd.2018.2510. [DOI] [PubMed] [Google Scholar]
- Zhang J., Mo S., Li H., Yang R., Liu X., Xing X., Hu Y., Li L. Rothia nasimurium as a cause of disease: first isolation from farmed chickens. Vet. Sci. 2022;9:653. doi: 10.3390/vetsci9120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.