Abstract
The nucleocapsid (NC) domain of the retrovirus Gag protein plays several important roles in the viral life cycle, including virus assembly, viral genomic RNA encapsidation, primer tRNA placement, and enhancement of viral reverse transcription. In this study, deletion of NC domain of human immunodeficiency virus type 1 (HIV-1) Gag was found to drastically reduce virus particle production in CD4+ T cells. Cellular fractionation experiments showed that although most of the uncleaved wild-type HIV-1 Gag, unmyristylated Gag, and p6Gag domain-truncated Gag molecules copurified with the host cell cytoskeleton, most of the mutant Gag molecules lacking both the NC and p6Gag domains failed to cofractionate with cytoskeleton. In wild-type virus-infected cells, in which the viral protease was active, the cleaved NCp7 copurified with the cytoskeleton, whereas most of the MAp17 and CAp24 did not. Monoclonal antibody against actin coimmunoprecipitated full-length Gag and p6Gag domain-truncated Gag molecules from cell lysates but failed to precipitate the truncated mutant Gag molecules lacking NC plus p6Gag. Purified recombinant NCp7, but not CAp24, was able to bind F-actin in cosedimentation experiments. Furthermore, wild-type NCp7 and a zinc finger mutant NCp7(F16A), like a cellular actin-binding protein (the villin headpiece), bound F-actin in a dose-dependent fashion in vitro. Taken together, these results suggest that HIV-1 NCp7 can bind F-actin directly and that interaction between HIV-1 Gag and the actin cytoskeleton through the NC domain may play an important role in HIV-1 assembly and/or other steps of the viral life cycle.
In the case of most retroviruses, with the possible exception of foamy viruses (3, 23), the Gag molecule can act alone to direct the assembly and release of immature virus-like particles (32, 60, 62). Several functional virus assembly domains in Gag, including a membrane binding domain (M), an interaction domain (I), and a late virus budding domain (L) (32, 60, 62), have been proposed to exist. However, with the exception of membrane targeting and binding, the functions of virus assembly domains I and L remain to be defined.
The Gag molecule of human immunodeficiency virus type 1 (HIV-1) is initially synthesized as a 55-kDa polyprotein precursor (Pr55Gag) and subsequently cleaved by the viral protease encoded in the pol gene region to yield the matrix (MAp17), capsid (CAp24), nucleocapsid (NCp7), p6Gag, and two spacer peptides P2 and P1 (30). MAp17 contains a major determinant for plasma membrane targeting and binding (35), the M domain; both the N-terminal myristylation (8, 28, 58, 66, 69) and internal (20, 24, 58, 66, 69) amino acid sequences of MAp17 are important for plasma membrane targeting and binding. NCp7 can function as an I domain when fused with the Rous sarcoma virus Gag molecule (5) and is known to be critical for HIV-1 assembly (15, 19, 54, 67, 68). The L domain of HIV-1 Gag maps to the p6Gag region, which is required for efficient virus release (27, 31, 47, 57, 65).
Retroviral Gag molecules are first synthesized in the cytoplasm and then subsequently transported to the site of virus budding on the plasma membrane. During transport, Gag molecules directly or indirectly encounter the host cell cytoskeleton, which comprises microtubule filaments, intermediate filaments, and actin microfilaments. Increasing evidence is accumulating to indicate that cytoskeleton components, especially actin microfilaments, play an important role in HIV-1 assembly. In HIV-1-infected T cells and macrophages, actin and HIV-1 Gag proteins are colocalized in the pseudopod structures, where virus budding is concentrated (48, 50), and interaction between HIV-1 Gag precursor molecules and actin has been reported (52). Cytochalasin D, which alters intracellular actin structures (13), also affects the intracellular distribution of HIV-1 Gag (52) and partially inhibits HIV-1 production (55). Finally, purified HIV-1 virions have been shown to contain actin and several actin-binding proteins (46).
In this study we have analyzed and compared the effects of the three assembly domains of HIV-1 Gag on virus assembly in CD4+ T cells, and we have investigated the possible interactions of these assembly domains with the host cell cytoskeleton. We found that the NC domain and myristylation of HIV-1 Gag were essential for virus production from CD4+ T cells, whereas the L domain in p6Gag enhanced virus production. Mutations that destroyed myristylation of HIV-1 Gag or the p6Gag domain did not affect cofractionation of the mutant Gag with the host cell cytoskeleton, whereas deletion of the NC domain significantly reduced the cofractionation of the mutant Gag with the cytoskeleton. We found that purified NC protein was able to bind F-actin directly in vitro. Furthermore, interaction between full-length HIV-1 Gag and truncated Gag containing NC but not NC truncated Gag molecules and actin in H9 cells was detected by coimmunoprecipitation experiments, suggesting a possible link between actin binding and HIV-1 replication.
MATERIALS AND METHODS
DNA constructs and cells.
The wild-type infectious proviral plasmid HXB2Neo, the myristylation-minus mutant proviral plasmid (Myr-), and the pol region deletion mutant HXB2NeoΔPol (Pr55) proviral plasmid have been previously described (40, 41). The Pr48 construct is isogenic to HXB2Neo, except for the presence of a premature stop codon at the beginning of p6Gag coding region (64). The Pr41 construct is also isogenic to HXB2Neo, except for a premature stop codon at the end of CAp24 coding region (15). H9 cell lines expressing each of the viral constructs were generated and maintained as previously described (40, 41).
Virus production analysis by immunoblotting.
Virus produced from H9 cells over a period of 24 h was harvested. Cells were washed twice with phosphate-buffered saline (PBS) to remove released virions and then resuspended in fresh RPMI 1640 with 10% fetal bovine serum and antibiotics. After 24 h supernatants were cleared of cell debris by centrifugation for 10 min at 3,000 rpm in a Sorvall RT 6000B centrifuge, filtered through 0.2-μm-pore-size filters (Millipore, Bedford, Mass.), and then centrifuged at 100,000 × g over a 20% sucrose cushion in a Sorvall ultracentrifuge with an AH-629 rotor. Viral pellets were resuspended in radioimmunoprecipitation assay buffer (0.05 M Tris [pH 7.2], 0.15 M NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) at approximately 1/1,000 of the starting supernatant volume, and the samples were mixed with loading buffer for SDS-polyacrylamide gel electrophoresis (PAGE) (12% gel). Cell pellets were collected and lysed in a small volume of radioimmunoprecipitation assay buffer (approximately 1/100 of the starting supernatant volume). Cell lysates were cleared by centrifugation at 12,000 × g in Eppendorf tubes for 30 min in a tabletop microcentrifuge; the supernatant from this spin was removed to a fresh tube and mixed with sample loading buffer for SDS-PAGE (12% gel). Immunoblotting of cell lysates and viral lysates was performed as previously described (40, 41).
Cell fractionation by detergent extraction.
The procedure for detergent extraction was modified from that of Rey et al. (52). Cells (5 × 105) were washed once with 1 ml of PBS by centrifugation at 1,500 rpm for 10 min in a Sorvall RT 6000B centrifuge and then twice with 50 μl of cytoskeleton stabilizing buffer [CSB; 100 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.9) with 1 mM EGTA, 3% polyethylene glycol 8000, and 66 μg of phalloidin/ml]. Cell pellets were collected by centrifugation at 800 × g for 2 min and lysed with 50 μl of CSB containing 1% Triton X-100, 2 mM GTP, aprotinin (2 μg/ml) phenylmethylsulfonyl fluoride (50 μg/ml), and leupeptin (2 μg/ml) by gentle pipetting 10 times. Cell lysates were centrifuged at 800 × g for 5 min. The supernatants (enriched for cytoplasmic and detergent-soluble membrane fractions) were carefully separated from the pellets and clarified for 5 min at 1,500 × g, and the resulting supernatants were termed soluble fractions. The detergent-insoluble pellets were washed twice with 50 μl of CSB (insoluble fraction). The soluble and insoluble fractions were dissolved in equal volumes of 1× sample buffer (0.08 M Tris-HCl [pH 6.8] with 2.0% SDS, 10% glycerol, 0.1 M dithiothreitol, and 0.2% bromophenol blue) and analyzed by SDS-PAGE and immunoblotting with a rabbit polyclonal anti-CAp24 serum (AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health), a sheep polyclonal anti-MAp17 serum (AIDS Research and Reference Reagent Program), a monoclonal anti-NCp7 antibody (a gift from Larry Arthur), an HIV-1-positive human antiserum, or a monoclonal antiactin antibody (Sigma, St. Louis, Mo.).
Coimmunoprecipitation experiments.
Cells (5 × 105) were lysed by incubation at room temperature for 10 min in 100 μl of PBS containing 1% Triton X-100, and the cell lysates were clarified by centrifugation at 8,000 × g for 20 min. The supernatants were incubated overnight at 4°C with protein A-Sepharose beads that had been preincubated with a monoclonal antiactin antibody (Sigma) or a monoclonal antitubulin antibody (Sigma). The unbound fractions were separated from the Sepharose beads by centrifugation at 1,000 × g for 2 min. Pellet samples were washed five times with 500 μl of PBS containing 1% Triton X-100. To release the immunoprecipitates (bound fraction), the Sepharose beads were boiled in 1× sample buffer for 5 min and then briefly centrifuged at 12,000 × g. The unbound and bound fractions were adjusted to equal volumes with 1× sample buffer and separated by SDS-PAGE, and the viral proteins were visualized by immunoblotting with an HIV-1-positive human serum.
F-actin cosedimentation.
Actin polymerization and F-actin cosedimentation were performed according to the traditional methods (43). Chicken muscle actin (Sigma) was stored in monomeric form in buffer G (5 mM Tris-HCl [pH 7.5] with 0.1 mM CaCl, 0.1 mM ATP, and 10 mM 2-mercaptoethanol). For measuring the dose response of wild-type NCp7 (AIDS Research and Reference Reagent Program), F16A mutant NCp7 [NCp7(F16A)], or villin headpiece (a gift from James McKnight) binding to actin, G-actin at concentrations of 0, 1.25, 2.5, 5, 10, and 20 μM was subjected to polymerizing conditions (addition of 50 mM KCl, 1 mM MgCl2, and 1 mM ATP) and incubated for 1 h at room temperature. Purified NCp7 (2.5 μM) or villin headpiece (2.5 μM) in buffer G was then added to each actin sample, and the mixtures were incubated for 30 min at room temperature and then centrifuged at 150,000 × g for 30 min. The supernatants and pellets were separated for subsequent analysis by 15% tricine SDS-PAGE, and the proteins were visualized by Coomassie blue staining. To compare the actin-binding capacities of CAp24 and NCp7, 50 μM G-actin was subjected to polymerization and then mixed with 1 μg of CAp24 or NCp7 before ultracentrifugation.
Preparation of NCp7(F16A) protein.
Mutant NCp7 protein was prepared from a thioredoxin-NCp7 fusion protein as follows. The coding region for the NCp7(F16A) protein was obtained by PCR amplification of an HIV-1 MN proviral clone, using the primers F16A-sense (5′-ATG CCG GGG TAC CGA CGA CGA CAA GAT GCA GAG AGG CAA TTT TAG GAA TCA AAG AAA GAT TAT CAA GTG Cgc CAA TTG TGG CAA AGA AGG-3′) and F16A-antisense (5′-GGG TAC GGT CGA CCT AAT TAG CCT GTC TCT CAG TAC AAT CTT TCA TTT GG-3′), obtained from Operon Technologies, Inc., Alameda, Calif. Oligonucleotide F16A-sense contains a KpnI site (underlined) and the sequence that codes for the enterokinase cleavage site (Asp-Asp-Asp-Asp-Lys); the remaining 63 nucleotides (3′ end) code for the 21 N-terminal residues of the HIV-1 NCp7(F16A) protein from the MN proviral clone (NC protein starts with a Met residue). The location of the F16A mutation is indicated in lowercase. Oligonucleotide F16A-antisense contains a SalI site (underlined), the complement of a TAG stop codon (lowercase), and the complement of the sequence that codes for the 11 C-terminal residues of NCp7 ending with an Asn residue. The fragment that codes for the 55-amino-acid NCp7 protein with the enterokinase cleavage site at the N terminus and the flanking KpnI and SalI sites was prepared by PCR as follows. The HIV-1 MN proviral plasmid (∼50 ng) was amplified by using AmpliTaq core reagents (Perkin-Elmer, Roche Molecular Systems, Inc., Branchburg, N.J.), 4 mM MgCl2, 2 μM each oligonucleotides F16A-sense and F16A-antisense, and AmpliTaq Gold according to the manufacturer’s procedures. Amplification was performed in a Perkin-Elmer PE9600 thermocycler as follows: incubation at 94 for 9 min; 35 cycles of 94°C for 10 s, 65°C for 15 s, and 70°C for 30 s; and incubation at 70°C for 8 min. The primers and nucleotides were removed from the PCR products by dilution to 200 μl of 10 mM Tris–1.0 mM EDTA (pH 8.0) and centrifugation through a Microcon 50 column until the volume was ∼0.1 that of the starting volume. This wash procedure was repeated. The retentate was then digested with KpnI and SalI. The fragment containing the NCp7 coding region was then ligated into the homologous sites of the pET32a vector (Novagen, Inc., Madison, Wis.). Plasmids were screened, and sequences were verified by nucleotide sequence analysis.
Competent Escherichia coli BL21(DE3) was transformed with plasmid pET32a containing the NCp7(F16A) gene. In a 4-liter Erlenmeyer flask, 500 ml of LB broth containing 100 μg of ampicillin per ml was inoculated with the transformed bacteria and grown at 37°C with shaking to an optical density at 600 nm of 0.6. Cultures were stored at 4°C without shaking overnight. The next day, bacteria were collected by centrifugation at 3,600 × g in a Beckman (Fullerton, Calif.) JS-4.2 rotor for 20 min at 4°C. The cell pellet was suspended in 1 liter of LB broth containing 100 μg of ampicillin per ml and grown at 37°C with shaking until an optical density at 600 nm of 0.6 was reached. Isopropylthio-β-galactoside was added to a final concentration of 0.4 mM, and the culture was incubated with shaking for an additional 3 h. Bacteria were pelleted at 3,600 × g in a Beckman JS-4.2 rotor for 30 min at 4°C. The pellet was suspended in 40 ml of 100 mM CAPS buffer (pH 10) and sonicated for ∼1.5 h in a model SC 101TH sonicating bath (Sonicor Instrument Corp., Copiague, N.Y.) in two 50-ml polypropylene conical tubes. The sonicated material was centrifuged 3,600 × g in a Beckman JS-4.2 rotor for 20 min at 4°C, and the pellet was discarded.
To 10 ml of the supernatant containing the fusion protein, 106 μl of 4 M Tris buffer (pH 8.5), 213 μl of 5 M NaCl, 0.7 μl of 30 mM zinc acetate, and 21 μl of 1 M CaCl2 were added. The solution was slowly stirred on a magnetic stirrer, 7 U of EKMax enterokinase (Invitrogen Corporation, Carlsbad, Calif.) was added, and the solution was digested for 3 h at room temperature. Treatment of the fusion protein with enterokinase liberates NCp7(F16A) mutant protein of the correct amino acid sequence for the 55-amino-acid form (30). NCp7(F16A) was purified by reverse-phase high-pressure liquid chromatography using a C18 reverse-phase column as described previously (30a). The purified NCp7(F16A) was analyzed for the correct molecular mass by matrix-assisted laser description ionization time-of-flight mass spectrometry using a Shimadzu Kompact Maldi-II laser desorption mass spectrometer. Quantitation of the purified protein was performed by amino acid analysis on a Beckman System 6300 amino acid analyzer (Beckman Coulter, Inc., Fullerton, Calif.). Purified protein was aliquoted, one equivalent of zinc acetate per Zn2+ finger was added, and the sample was lyophilized. The dried NCp7(F16A) sample was stored at −70°C. The mutant protein was dissolved in buffer G.
RESULTS
Mutant constructs and virus production from CD4+ T cells.
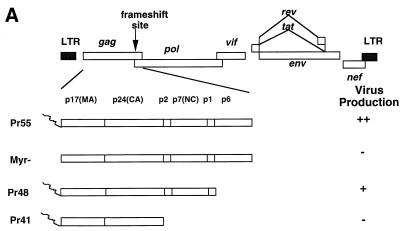
We established CD4+ T cells (H9) containing various mutant constructs as a means of evaluating the effect of specific assembly domains of HIV-1 Gag on virus production (Fig. 1A). The full-length HIV-1 Gag containing all three assembly domains was expressed from the Pr55 construct (Fig. 1). This construct contains a point mutation that destroyed the activity of viral protease (41). The M-domain mutant was represented by the Myr- construct. Pr48 contained a truncation of the p6Gag, the L domain, and the Pr41 contained a truncation of both the I domain (NCp7) and the L domain.
FIG. 1.
(A) Diagram of truncation mutants of HIV-1 Gag. For details on construction of mutants, see Materials and Methods. Construct Pr55 expresses full-length HIV-1 Gag. Myr-contains a mutation that destroyed the signal for myristylation. Construct Pr48 contains a stop codon at the beginning of the p6Gag, while Pr41 has a stop codon at the end of the CAp24. LTR, long terminal repeat. (B) Virus production from H9 cells. Cell and virus lysates from H9 cells expressing Pr55 and mutant Gag constructs were separated by SDS-PAGE, transferred to nitrocellulose filters, and analyzed by immunoblotting with an HIV-1-positive human serum. (C) Cellular fractionation of viral proteins. Triton X-100-insoluble (I) and -soluble (S) materials from uninfected H9 cells (H9), and from H9 cells expressing Pr55 and mutant Gag constructs were prepared as described in Materials and Methods. Lysates were separated by SDS-PAGE, transferred to nitrocellulose filters, and analyzed by immunoblotting with a rabbit polyclonal anti-CAp24 serum.
H9 cell lines carrying each of the DNA constructs were generated, and virus production was evaluated. Consistent with previous observations in a variety of target cells other than CD4+ T cells, virus production in cells expressing mutant forms of Myr- (Fig. 1B, lane 3) or Pr41 (Fig. 1B, lane 4) Gag molecules was much lower than that in cells expressing wild-type Gag molecules (Fig. 1B, lane 5). The virus yield from the cells expressing the mutant Pr48 (Fig. 1B, lane 2) Gag molecules was slightly less than that from cells expressing wild-type Gag molecules. It is expected that there is no Gag processing in H9 cells expressing Pr55, Myr-, and Pr41 constructs. The Pr55 construct contains a point mutation that has destroyed the activity of viral protease (41). Our previous observations have indicated that the Myr- construct has a defect in Gag processing in H9 cells (39). The Pr41 construct does not express Pol because of a premature stop codon before the frameshifting signal. Our group and others have previously reported that truncation of p6Gag (Pr48 construct) results in reduced Gag processing in COS and HeLa cells (31, 64). It seems that this construct also has a defect in Gag processing in H9 cells (Fig. 1B).
Cofractionation of various forms of HIV-1 Gag molecules with the host cell cytoskeleton.
It has been reported that HIV-1 Gag cofractionates with host cell cytoskeleton (52). To address the question of whether mutant Gag molecules could influence cofractionation with the host cell cytoskeleton, we extracted H9 cells expressing various forms of the Gag molecules with nonionic detergent and separated the cell extracts into detergent-insoluble (cytoskeleton-enriched) and detergent-soluble (cytosolic and membrane) fractions. Most of the wild-type Gag molecules (Pr55) fractionated with the detergent-insoluble cytoskeleton fraction (Fig. 1C, lane 3); a small fraction of the wild-type Gag molecules (Pr55) was associated with the detergent-soluble fraction (Fig. 1C, lane 4). A similar distribution was observed for the mutant Myr- (Fig. 1C, lanes 5 and 6) and Pr48 (Fig. 1C, lanes 7 and 8) Gag molecules. In contrast, most of the mutant Pr41 Gag molecules cosedimented with the detergent-soluble fraction (Fig. 1C, lane 10), although some still remained associated with the detergent-insoluble fraction (Fig. 1C, lane 9). The cytoskeleton fraction purified in this manner contains both cytoskeleton, cytoskeleton-associated complexes, and unbroken nuclei. We have examined H9 cells expressing various Gag constructs by immunofluorescence and have not detected significant accumulation of wild-type or mutant Gag molecules in the nuclei (data not shown). Therefore, cofractionation of HIV-1 Gag molecules with the cytoskeleton fraction is not the result of a nuclear localization of HIV-1 Gag molecules. However, cofractionation experiments cannot address whether HIV-1 Gag molecules bind directly or indirectly to the cytoskeleton or associate with noncytoskeleton complexes which copelleted with the cytoskeleton.
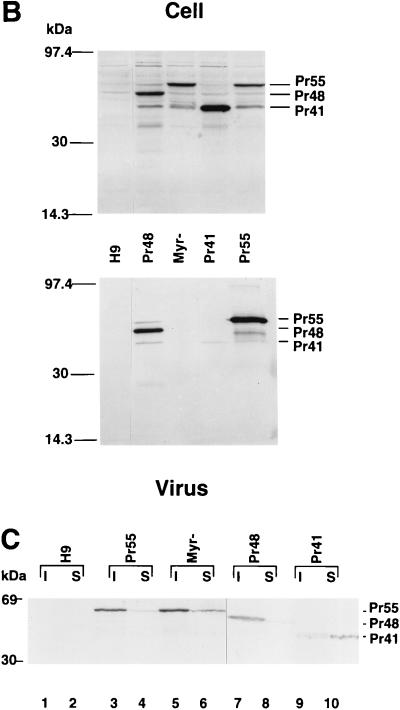
Cofractionation of various cleaved HIV-1 Gag molecules with the host cell cytoskeleton.
To identify which domain(s) of the HIV-1 Gag molecule might contribute to cofractionation with cytoskeleton, we also fractionated HIV-1-infected H9 cells as described above. In HIV-1-infected H9 cells that expressed active viral protease, the uncleaved Gag precursor Pr55 as well as the cleaved Gag proteins MAp17, CAp24, and NCp7 could be detected. Although most of the uncleaved Gag precursor Pr55 cosedimented with the cytoskeletal proteins as described above, most of the CAp24 cosedimented with the detergent-soluble fraction (Fig. 2A, upper panel, lane 4). MAp17 was largely detected in the detergent-soluble fraction (Fig. 2A, middle panel, lane 4), whereas NCp7 was detected only in the cytoskeleton fraction (Fig. 2A, lower panel, lane 3). When we analyzed the distribution of actins in uninfected and HIV-1-infected H9 cells, we observed that most of the intracellular actins cosedimented with the detergent-insoluble cytoskeleton (Fig. 2B). HIV-1 infection did not drastically change the distribution of intracellular actin. These data suggest that NCp7 is a major determinant for cofractionation of HIV-1 Gag molecule with the host cell cytoskeleton.
FIG. 2.
Cellular fractionation of cleaved HIV-1 Gag proteins. Triton X-100-insoluble (I) and -soluble (S) materials from uninfected H9 cells (H9) or from wild-type HIV-1-infected H9 cells (H9/HIV-1) were prepared as described in Materials and Methods. (A) Lysates were separated by SDS-PAGE, transferred to nitrocellulose filters, and analyzed by immunoblotting with an HIV-1-positive human serum (top panel), a sheep anti-MAp17 antiserum (middle panel), or a monoclonal anti-NCp7 antibody (lower panel). (B) Lysates were separated by SDS-PAGE, transferred to nitrocellulose filters, and analyzed by immunoblotting with a monoclonal antiactin antibody.
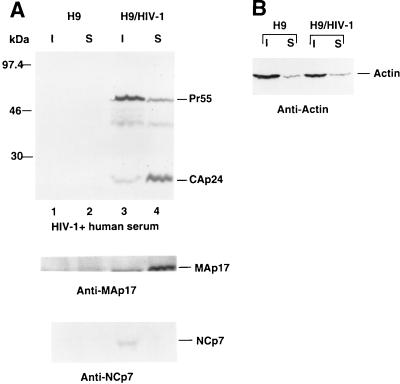
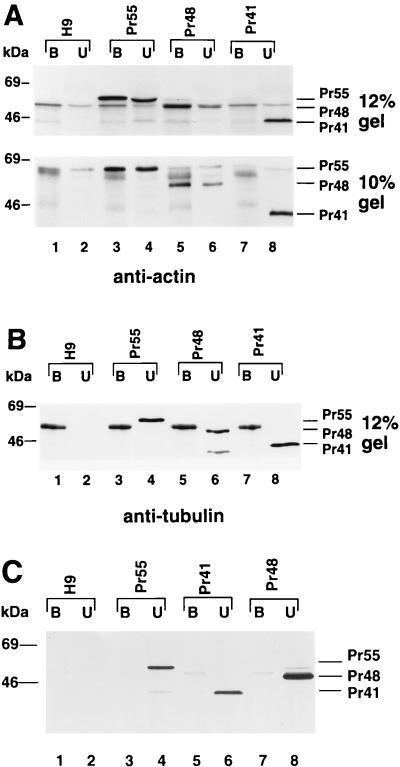
Coimmunoprecipitation of HIV-1 Gag and actin from infected cells with a monoclonal antibody against actin.
To determine whether HIV-1 Gag molecules could bind actin in infected cells, we performed coimmunoprecipitation experiments. Cells were lysed with 1% Triton X-100 in PBS, and the detergent-soluble cell lysates were immunoprecipitated with a monoclonal antiactin antibody that had been preadsorbed onto protein A-Sepharose beads. The bound and unbound fractions were dissolved in equal volumes of sample buffer, separated by SDS-PAGE, and analyzed by immunoblotting with an HIV-1-positive human serum. The wild-type Gag molecules (Pr55) from H9 cells were precipitated with the antiactin antibody (Fig. 3A, upper panel, lane 3); some wild-type Gag molecules (Pr55), however, were not (Fig. 3A, upper panel, lane 4). Similarly, the mutant Pr48 Gag molecules from H9 cells were precipitated with the antiactin antibody (Fig. 3A, lower panel, lane 5). Since Pr48 comigrated with a protein from mock-infected H9 cells on SDS–12% polyacrylamide gels (Fig. 3A, upper panel), the original samples were rerun on SDS–10% polyacrylamide gels (Fig. 3A, lower panel). On 10% gels, both Pr48 and Pr41 could be clearly separated from background protein bands. In contrast to the results obtained for Pr55 and Pr48, none of the mutant Pr41 Gag molecules were precipitated with the antiactin antibody (Fig. 3A, lower panel, lane 7). In control experiments, neither wild-type Gag molecules (Pr55), mutant Pr48 Gag molecules, nor mutant Pr41 Gag molecules were immunoprecipitated with a monoclonal antitubulin antibody (Fig. 3B). Furthermore, when no antibody was added to the protein A-Sepharose beads, neither wild-type Gag molecules (Pr55), mutant Pr48 Gag molecules, nor mutant Pr41 Gag molecules were detected in the pellet fractions (Fig. 3C). A 50-kDa protein was detected in all of the protein A-Sepharose beads pellet fractions (even that from the uninfected H9 cells) after incubation with antitubulin antibody (Fig. 3B). The precise nature of this protein band is unknown; it was not detected in the absence of antibody (Fig. 3C) and thus may represent the heavy chain of the mouse immunoglobulin precipitated by the protein A-Sepharose beads. The presence of this protein in the immunoblot did not alter the conclusion drawn from these experiments: that some Pr55 and Pr48 molecules were associated with actin in H9 cells, while the majority of the Pr41 molecules were not associated with actin in H9 cells.
FIG. 3.
Coimmunoprecipitation analysis. Cell lysates from uninfected H9 cells (lanes 1 and 2) and H9 cells expressing Pr55 (lanes 3 and 4), Pr48 (lanes 5 and 6), and Pr41 (lanes 7 and 8) were prepared by lysing the cells in PBS containing 1% Triton X-100, followed by immunoprecipitation with a monoclonal antiactin antibody (A), a monoclonal antitubulin antibody (B), or no antibody (C). The immunoprecipitated (bound [B]) and unprecipitated (unbound [U]) materials were separated by SDS-PAGE on a 12% (A, upper panel) or 10% (A, lower panel) gel and transferred onto two nitrocellulose filters. Viral proteins on the filters were visualized by immunoblotting with an HIV-1-positive human serum.
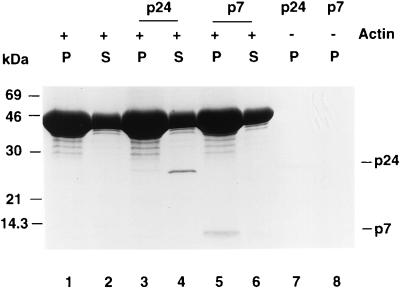
Direct binding of NCp7, but not CAp24, to F-actin.
Since NCp7 and, to a lesser extent, CAp24 copurified with the cytoskeletal components, we asked whether purified CAp24 and NCp7 could bind actin directly. Binding of CAp24 and NCp7 to actin was assayed by measuring cosedimention with in vitro-polymerized F-actin. Most of the CAp24 did not cosediment with F-actin after ultracentrifugation (Fig. 4, lane 3) and remained in the supernatant (Fig. 4, lane 4). At the same time, most of the NCp7 cosedimented with the F-actin pellet (Fig. 4, lane 5), and very little remained in the supernatant (Fig. 4, lane 6). When CAp24 or NCp7 was incubated in the same buffer in the absence of actin, no significant amount of CAp24 (Fig. 4, lane 7) or NCp7 (Fig. 4, lane 8) could be pelleted, suggesting that under these experimental conditions there was no significant aggregation of CAp24 or NCp7.
FIG. 4.
F-actin cosedimentation. Chicken muscle actin (50 μM) was polymerized as described in Materials and Methods and then mixed with buffer alone, 1 μg of CAp24, or 1 μg of NCp7 before ultracentrifugation. The same amount of CAp24 or NCp7 was also incubated in the actin polymerization buffer in the absence of actin. The supernatants (S) and pellets (P) were separated after ultracentrifugation and analyzed by 15% tricine SDS-PAGE; the proteins were visualized by Coomassie blue staining.
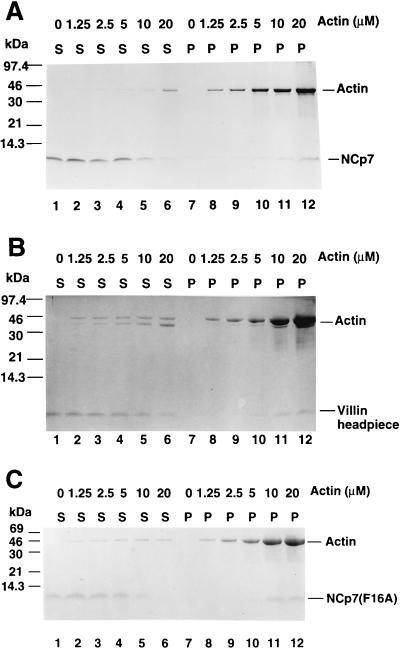
A more quantitative analysis of NCp7 binding to actin was accomplished by cosedimentation with various concentrations of F-actin. With increasing concentrations of actin, NCp7 was progressively depleted from the residual supernatant following ultracentrifugation (Fig. 5A). Although some binding of NCp7 (2.5 μM) to F-actin in the ultracentrifugation pellet was detected when 1.25 and 2.5 μM G-actin was used for polymerization (Fig. 5A, lanes 8 and 9), increased binding of NCp7 to F-actin was detected when 5 μM or more G-actin was used for polymerization (Fig. 5A, lanes 10 to 12). Approximately 40% of the NCp7 bound to F-actin when 10 μM G-actin was polymerized to F-actin (Fig. 5A, lane 11). When 20 μM G-actin was used to polymerize F-actin, more than 70% of the NCp7 was fractionated with the F-actin pellet (Fig. 5A, lane 12). The dose-dependent F-actin binding by NCp7 was very similar to that of a known cellular F-actin-binding protein, villin headpiece (Fig. 5B). Although the actin binding experiments performed here represent a traditional method for testing and confirming putative actin-binding proteins (44, 51), it remains to be demonstrated that NCp7 and actin binding are relevant in HIV-1-infected cells.
FIG. 5.
Dose-dependent F-actin binding determined by cosedimentation. Actin at concentrations of 0, 1.25, 2.25, 5, 10, and 20 μM was subjected to polymerizing conditions and incubated with 2.5 μM purified wild-type NCp7 (A), villin headpiece (B), or mutant NCp7 (C) before ultracentrifugation. The supernatants (S) and pellets (P) were separated after ultracentrifugation and analyzed by 15% tricine SDS-PAGE; the proteins were visualized by Coomassie blue staining.
We also studied whether certain mutations in NCp7 could affect actin binding. Zinc fingers of HIV-1 NC have been shown to be critical for viral genomic RNA packaging but not for virus assembly and release (32, 60, 62). We tested the F-actin binding of one such mutant form of NCp7F16A (19). The dose-dependent F-actin binding by the mutant NCp7(F16A) was very similar to that of the wild-type NCp7 (Fig. 5A). These results indicate that certain mutations in NC that inhibited viral genomic RNA packaging did not affect F-actin binding (Fig. 5C), suggesting that there may not be a direct link between actin binding and viral RNA packaging. Further study is under way to identify structural features of NCp7 that are critical for F-actin binding.
DISCUSSION
In this study, we demonstrated that the NC domain of HIV-1 Gag is a major determinant in the process of viral protein association with the host cell cytoskeleton. Mutations in the other two virus assembly domains, M (myristylation) and L (p6Gag), had little effect on the association of mutant Gag molecules with cytoskeletal components. Cellular fractionation experiments using wild-type virus-infected cells which express active viral protease indicated that cleaved NCp7 almost exclusively fractionated with the cytoskeleton. In contrast, the most of the mature MAp17 and CAp24 molecules were not associated with cytoskeleton.
It is possible that the association of HIV-1 Gag molecule with the cytoskeleton is mediated by the NC domain through actin binding. This argument is supported by our observation that HIV-1 Gag could be coprecipitated with actin in cell lysates and that purified NCp7 could bind directly to F-actin in vitro. The observation that HIV-1 Gag and actin colocalize to the pseudopod structures of virus-infected T cells and macrophages (48, 50) is also consistent with the concept that HIV-1 Gag binds to actin filaments in vivo (52). It is worth noting that small amounts of MAp17 plus CAp24 (Fig. 1, Pr41) and mature CAp24 (Fig. 2A) cofractionated with cytoskeletal components. The crystal structure of the carboxy-terminal region of CAp24 (amino acids 151 to 231) resembles the putative actin-binding subdomain of myosin (26). Although we did not observe direct binding of CAp24 to F-actin in vitro (Fig. 4), it is still possible that CAp24 or MAp17 plus CAp24 can bind to actin or actin-binding proteins in vivo and become associated with the cytoskeleton of the host cell. Alternatively, CAp24 might bind to other components of the cytoskeleton, such as microtubules or intermediate filaments, either directly or indirectly.
Previous studies have indicated that the retroviral NC domain plays several important roles in the life cycle of the virus. However, the functional significance of the interaction between HIV-1 NC and actin remains to be characterized. Binding of the NCp7 domain to actin filaments could be involved in intracellular transport of HIV-1 Gag molecules; this idea would be consistent with the observation that HIV-1 Gag molecules are colocalized with actin to the pseudopods in virus-infected T cells and macrophages (48, 50). Truncation of NCp7 also resulted in a lower degree of association of mutant Gag molecules with the plasma membrane than was observed for the full-length HIV-1 Gag molecules (54).
Actin binding through the NCp7 domain of HIV-1 Gag molecules could also play a role in virus assembly and budding. One proposed role for the retroviral Gag assembly domain I (NC) is to stimulate intermolecular interaction (5, 62); deletion of the NC domain of HIV-1 Gag has been shown to drastically reduce virus production (15, 34, 54, 67, 68). Substitution of the HIV-1 NC domain with protein modules known to interact fully or partially restored virus assembly and release (68). Interaction among HIV-1 Gag molecules during virus assembly might be enhanced by binding to actin polymers. Actin and associated actin-binding proteins might also serve as nucleation sites for the assembly of Gag molecules. Consistent with this idea, it has been shown that cytochalasin D, a drug that interferes with cellular actin structures (13), partially reduces virus production (55).
It is also possible that actin may serve as a structural component of HIV-1 particles. Actin has been detected in purified HIV-1 virions at approximately 10% of the molar concentration of Gag (46). The virus-like structures that are released from cells expressing NC deletion mutant Gag molecules have a density lower than those of viruses containing the NC domain (5, 15, 34, 54, 67, 68). Interaction of the NC domain with actin may influence the packing density (amount of viral protein per unit of lipid) of the HIV-1 Gag molecules during assembly (5, 62). The interaction among Gag molecules that is mediated through the MA and CA domains alone might be less tight than that mediated by MA and CA plus NC and actin. It is worth mentioning that interaction of NC with RNA has also been proposed to play a role in virus assembly and in controlling particle density (5, 7, 10, 15, 34). Interactions between cleaved NCp7 and actin may also play a structural role in the formation of HIV-1 core, since NCp7 mutants frequently display abnormal core structures (1, 6, 12, 19).
Extensive mutational analyses have indicated that the retroviral NC protein is necessary for encapsidation of viral genomic RNA (56), annealing of the primer tRNA to the viral genomic primer binding site (4, 17), and formation of the viral genomic RNA dimer (14, 22). In addition to their structural roles in virus assembly and maturation, nucleocapsids from several viruses have been shown to be important during early stages of viral replication, including minus-strand DNA transfer during viral DNA synthesis (2, 29, 53), prevention of nonspecific reverse transcription (29, 37, 42), and enhancement of the processivity of reverse transcriptase activity (33, 49, 53, 61, 63). As a component of the viral nucleocapsid, NC protein, by interacting with actin and cellular actin-binding proteins, may enhance the penetration of the viral nucleocapsid into the cytoplasm after virus fusion and uncoating. NC is also a member of the viral reverse transcription complex (59). Therefore, it is also intriguing to consider that the transport of the complex from the site of virus entry to the nucleus could be enhanced by interaction between NC and actin filaments, a strategy used also by other viruses such as baculoviruses (11, 36).
The complete and partial structures of several cellular F-actin-binding proteins including the C-terminal 36 amino acids of villin headpiece (45), yeast cofilin (21), and gelsolin (9), have been determined by X-ray crystallography or nuclear magnetic resonance spectroscopy. Among them, no common structural motif that is critical for F-actin binding has been identified (44, 51). However, the C-terminal α-helix of the villin headpiece (18, 25), cofilin (38), and the F-actin binding S2 subdomain of gelsolin (44, 51) have been found to be important in the interaction with F-actin, as demonstrated by mutagenesis studies (18) and synthetic peptide binding experiments (25). In the case of the villin headpiece, both positively charged amino acids in the C-terminal α-helix and positively charged amino acids in the N-terminal region are critical for F-actin binding (18). The nuclear magnetic resonance structure of the HIV-1 NC-RNA complex reveals beta sheets representing the zinc knuckle domains and an N-terminal 3-10 helix (16). NCp7 exhibits very low homology to all known F-actin-binding proteins and did not reveal any significant structural similarities with other known F-actin-binding proteins. However, there are several positively charged amino acids in the 3-10 helix as well as in the linker region between the two zinc fingers. By analogy to villin headpiece, some of these positively charged residues in NCp7 could be involved in F-actin binding. Further study is required to map the NCp7 recognition site for F-actin.
ACKNOWLEDGMENTS
We thank Susan Craig and Richard McCann for helpful suggestions. We gratefully acknowledge the generous gifts of anti-p7 antibodies from Larry Arthur and purified recombinant villin headpiece from James McKnight. We also thank Alan Rein for the NCp7(F13A)-pET32a expression clone and Bradley P. Kane and Donald G. Johnson for assistance in the production and purification of the NCp7(F16A) protein. The following reagents were obtained through the AIDS Research and Reference Reagent Program, NIAID, NIH: HIV-1MN p7 from Louis Henderson; HIV-1SF2 p25/24 Gag from Kathelyn Steimer; and HIV-1 p24 antiserum from Julia Hurwitz.
This work was supported in part by Public Health Service grant AI-35525 from the National Institutes of Health and was sponsored in part by the National Cancer Institute, Department of Health and Human Services under contract NO1-CO-56000 with SAIC Frederick.
REFERENCES
- 1.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allain B, Lapadat-Tapolsky M, Berlioz C, Darlix J L. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994;13:973–981. doi: 10.1002/j.1460-2075.1994.tb06342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldwin D N, Linial M L. The roles of Pol and Env in the assembly pathway of human foamy virus. J Virol. 1998;72:3658–3665. doi: 10.1128/jvi.72.5.3658-3665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barat C, Schatz O, Le Grice S, Darlix J L. Analysis of the interactions of HIV1 replication primer tRNA(Lys,3) with nucleocapsid protein and reverse transcriptase. J Mol Biol. 1993;231:185–190. doi: 10.1006/jmbi.1993.1273. [DOI] [PubMed] [Google Scholar]
- 5.Bennett R P, Nelle T D, Wills J W. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1993;67:6487–6498. doi: 10.1128/jvi.67.11.6487-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berthoux L, Pechoux C, Ottmann M, Morel G, Darlix J L. Mutations in the N-terminal domain of human immunodeficiency virus type 1 nucleocapsid protein affect virion core structure and proviral DNA synthesis. J Virol. 1997;71:6973–6981. doi: 10.1128/jvi.71.9.6973-6981.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowzard J B, Bennett R B, Krishna N K, Ernst S M, Rein A, Wills J W. Importance of basic residues in nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J Virol. 1998;72:9034–9044. doi: 10.1128/jvi.72.11.9034-9044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burtnick L D, Koepf E K, Grimes J, Jones E Y, Stuart D I, McLaughlin P J, Robinson R C. The crystal structure of plasma gelsolin: implications for actin severing, capping, and nucleation. Cell. 1997;90:661–670. doi: 10.1016/s0092-8674(00)80527-9. [DOI] [PubMed] [Google Scholar]
- 10.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charlton C A, Volkman L E. Penetration of Autographa californica nuclear polyhedrosis virus nucleocapsids into IPLB Sf 21 cells induces actin cable formation. Virology. 1993;197:245–254. doi: 10.1006/viro.1993.1585. [DOI] [PubMed] [Google Scholar]
- 12.Clavel F, Orenstein J M. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J Virol. 1990;64:5230–5234. doi: 10.1128/jvi.64.10.5230-5234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper J A. Effects of cytochalasin and phalloidin on actin. J Cell Biol. 1987;105:1473–1478. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darlix J L, Gabus C, Nugeyre M T, Clavel F, Barre-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J Mol Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 15.Dawson L, Yu X F. The role of nucleocapsid of HIV-1 in virus assembly. Virology. 1998;251:141–157. doi: 10.1006/viro.1998.9374. [DOI] [PubMed] [Google Scholar]
- 16.De Guzman R N, Wu Z R, Stalling C C, Pappalardo L, Borer P N, Summers M F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 17.De Rocquigny H, Gabus C, Vincent A, Fournie-Zaluski M C, Roques B, Darlix J L. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc Natl Acad Sci USA. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doering D S, Matsudaira P. Cysteine scanning mutagenesis at 40 of 76 positions in villin headpiece maps the F-actin binding site and structural features of the domain. Biochemistry. 1996;35:12677–12685. doi: 10.1021/bi9615699. [DOI] [PubMed] [Google Scholar]
- 19.Dorfman T, Luban J, Goff S P, Haseltine W A, Gottlinger H G. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Facke M, Janetzko A, Shoeman R L, Krausslich H G. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J Virol. 1993;67:4972–4980. doi: 10.1128/jvi.67.8.4972-4980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fedorov A A, Lappalainen P, Fedorov E V, Drubin D G, Almo S C. Structure determination of yeast cofilin. Nat Struct Biol. 1997;4:366–369. doi: 10.1038/nsb0597-366. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 22.Feng Y X, Copeland T D, Henderson L E, Gorelick R J, Bosche W J, Levin J G, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer N, Heinkelein M, Lindemann D, Enssle J, Baum C, Werder E, Zentgraf H, Muller J G, Rethwilm A. Foamy virus particle formation. J Virol. 1998;72:1610–1615. doi: 10.1128/jvi.72.2.1610-1615.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freed E O, Orenstein J M, Buckler-White A J, Martin M A. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friederich E, Vancompernolle K, Huet C, Goethals M, Finidori J, Vandekerckhove J, Louvard D. An actin-binding site containing a conserved motif of charged amino acid residues is essential for the morphogenic effect of villin. Cell. 1992;70:81–92. doi: 10.1016/0092-8674(92)90535-k. [DOI] [PubMed] [Google Scholar]
- 26.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sundquist W I, Hill C P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 27.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Göttlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5787. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo J, Henderson L E, Bess J, Kane B, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henderson L E, Bowers M A, Sowder R C N, Serabyn S A, Johnson D G, Bess J W, Jr, Arthur L O, Bryant D K, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Henderson L E, Sowder R, Copeland T D, Smythers G, Oroszlan S. Quantitative separation of murine leukemia virus proteins by reversed-phase high-pressure liquid chromatography reveals newly described gag and env cleavage products. J Virol. 1984;52:492–500. doi: 10.1128/jvi.52.2.492-500.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang M, Orenstein J M, Martin M A, Freed E O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunter E. Macromolecular interations in the assembly of HIV and other retroviruses. Semin Virol. 1994;5:71–83. [Google Scholar]
- 33.Ji X, Klarmann G J, Preston B D. Effect of human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein on HIV-1 reverse transcriptase activity in vitro. Biochemistry. 1996;35:132–143. doi: 10.1021/bi951707e. [DOI] [PubMed] [Google Scholar]
- 34.Jowett J B, Hockley D J, Nermut M V, Jones I M. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J Gen Virol. 1992;73:3079–3086. doi: 10.1099/0022-1317-73-12-3079. . (Erratum, 74:943, 1993.) [DOI] [PubMed] [Google Scholar]
- 35.Krausslich H G, Welker R. Intracellular transport of retroviral capsid components. Curr Top Microbiol Immunol. 1996;214:25–63. doi: 10.1007/978-3-642-80145-7_2. [DOI] [PubMed] [Google Scholar]
- 36.Lanier L M, Volkman L E. Actin binding and nucleation by Autographa california M nucleopolyhedrovirus. Virology. 1998;243:167–177. doi: 10.1006/viro.1998.9065. [DOI] [PubMed] [Google Scholar]
- 37.Lapadat-Tapolsky M, Gabus C, Rau M, Darlix J L. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J Mol Biol. 1997;268:250–260. doi: 10.1006/jmbi.1997.0978. [DOI] [PubMed] [Google Scholar]
- 38.Lappalainen P, Fedorov E V, Fedorov A A, Almo S C, Drubin D G. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee Y-M, Tian C-J, Yu X-F. A bipartite membrane-binding signal in the human immunodeficiency virus type-1 matrix protein is required for the proteolytic processing of Gag precursors in a cell type-dependent manner. J Virol. 1998;72:9061–9068. doi: 10.1128/jvi.72.11.9061-9068.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee Y M, Tang X B, Cimakasky L M, Hildreth J E, Yu X-F. Mutations in the matrix protein of human immunodeficiency virus type 1 inhibit surface expression and virion incorporation of viral envelope glycoproteins in CD4+ T lymphocytes. J Virol. 1997;71:1443–1452. doi: 10.1128/jvi.71.2.1443-1452.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee Y M, Yu X-F. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology. 1998;243:78–93. doi: 10.1006/viro.1998.9064. [DOI] [PubMed] [Google Scholar]
- 42.Li X, Quan Y, Arts E J, Li Z, Preston B D, de Rocquigny H, Roques B P, Darlix J L, Kleiman L, Parniak M A, Wainberg M A. Human immunodeficiency virus type 1 nucleocapsid protein (NCp7) directs specific initiation of minus-strand DNA synthesis primed by human tRNALys3 in vitro: studies of viral RNA molecules mutated in regions that flank the primer binding site. J Virol. 1996;70:4996–5004. doi: 10.1128/jvi.70.8.4996-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCann R O, Craig S W. The I/LWEQ module: a conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc Natl Acad Sci USA. 1997;94:5679–5684. doi: 10.1073/pnas.94.11.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGough A. F-actin-binding proteins. Curr Opin Struct Biol. 1998;8:166–176. doi: 10.1016/s0959-440x(98)80034-1. [DOI] [PubMed] [Google Scholar]
- 45.McKnight C J, Matsudaira P T, Kim P S. NMR structure of the 35-residue villin headpiece subdomain. Nat Struct Biol. 1997;4:180–184. doi: 10.1038/nsb0397-180. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 46.Ott D E, Coren L V, Kane B P, Busch L K, Johnson D G, Sowder R C N, Chertova E N, Arthur L O, Henderson L E. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J Virol. 1996;70:7734–7743. doi: 10.1128/jvi.70.11.7734-7743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearce-Pratt R, Malamud D, Phillips D M. Role of the cytoskeleton in cell-to-cell transmission of human immunodeficiency virus. J Virol. 1994;68:2898–2905. doi: 10.1128/jvi.68.5.2898-2905.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peliska J A, Balasubramanian S, Giedroc D P, Benkovic S J. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 50.Perotti M E, Tan X, Phillips D M. Directional budding of human immunodeficiency virus from monocytes. J Virol. 1996;70:5916–5921. doi: 10.1128/jvi.70.9.5916-5921.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Puius Y A, Mahoney N M, Almo S C. The modular structure of actin-regulatory proteins. Curr Opin Cell Biol. 1998;10:23–34. doi: 10.1016/s0955-0674(98)80083-5. [DOI] [PubMed] [Google Scholar]
- 52.Rey O, Canon J, Krogstad P. HIV-1 Gag protein associates with F-actin present in microfilaments. Virology. 1996;220:530–534. doi: 10.1006/viro.1996.0343. [DOI] [PubMed] [Google Scholar]
- 53.Rodriguez-Rodriguez L, Tsuchihashi Z, Fuentes G M, Bambara R A, Fay P J. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J Biol Chem. 1995;270:15005–15011. doi: 10.1074/jbc.270.25.15005. [DOI] [PubMed] [Google Scholar]
- 54.Sandefur S, Varthakavi V, Spearman P. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J Virol. 1998;72:2723–2732. doi: 10.1128/jvi.72.4.2723-2732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sasaki H, Nakamura M, Ohno T, Matsuda Y, Yuda Y, Nonomura Y. Myosin-actin interaction plays an important role in human immunodeficiency virus type 1 release from host cells. Proc Natl Acad Sci USA. 1995;92:2026–2030. doi: 10.1073/pnas.92.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlesinger S, Makino S, Linial M L. cis-acting genomic elements and trans-acting proteins involved in the assembly of RNA viruses. Semin Virol. 1994;5:39–49. doi: 10.1006/smvy.1994.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwartz M D, Geraghty R J, Panganiban A T. HIV-1 particle release mediated by Vpu is distinct from that mediated by p6. Virology. 1996;224:302–309. doi: 10.1006/viro.1996.0532. [DOI] [PubMed] [Google Scholar]
- 58.Spearman P, Wang J J, Vander Heyden N, Ratner L. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J Virol. 1994;68:3232–3242. doi: 10.1128/jvi.68.5.3232-3242.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stevenson M. Portals of entry: uncovering HIV nuclear transport pathways. Trends Cell Biol. 1996;6:9–15. doi: 10.1016/0962-8924(96)81032-4. [DOI] [PubMed] [Google Scholar]
- 60.Swanstrom R, Wills J. Synthesis, assembly, and processing of viral proteins. In: Coffin J, Hughes S, Varmus H, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1998. pp. 263–334. [PubMed] [Google Scholar]
- 61.Tanchou V, Gabus C, Rogemond V, Darlix J L. Formation of stable and functional HIV-1 nucleoprotein complexes in vitro. J Mol Biol. 1995;252:563–571. doi: 10.1006/jmbi.1995.0520. [DOI] [PubMed] [Google Scholar]
- 62.Wills J W, Craven R C. Form, function, and use of retroviral gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. . (Editorial.) [DOI] [PubMed] [Google Scholar]
- 63.Wu W, Henderson L E, Copeland T D, Gorelick R J, Bosche W J, Rein A, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J Virol. 1996;70:7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu X-F, Dawson L, Tian C J, Flexner C, Dettenhofer M. Mutations of the human immunodeficiency virus type 1 p6Gag domain result in reduced retention of Pol proteins during virus assembly. J Virol. 1998;72:3412–3417. doi: 10.1128/jvi.72.4.3412-3417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu X-F, Matsuda Z, Yu Q C, Lee T H, Essex M. Role of the C terminus Gag protein in human immunodeficiency virus type 1 virion assembly and maturation. J Gen Virol. 1995;76:3171–3179. doi: 10.1099/0022-1317-76-12-3171. [DOI] [PubMed] [Google Scholar]
- 66.Yuan X, Yu X, Lee T H, Essex M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol. 1993;67:6387–6394. doi: 10.1128/jvi.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, Barklis E. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J Virol. 1997;71:6765–6776. doi: 10.1128/jvi.71.9.6765-6776.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y, Qian H, Love Z, Barklis E. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J Virol. 1998;72:1782–1789. doi: 10.1128/jvi.72.3.1782-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]