Abstract
Endoplasmic reticulum (ER) membrane protein complex (EMC) is required for the co-translational insertion of newly synthesized multi-transmembrane proteins. Compromised EMC function in different cell types has been implicated in multiple diseases. Using inducible genetic mouse models, we revealed defects in retinal vascularization upon endothelial cell (EC) specific deletion of Emc1, the largest subunit of EMC. Loss of Emc1 in ECs led to reduced vascular progression and vascular density, diminished tip cell sprouts, and vascular leakage. We then performed an unbiased transcriptomic analysis on human retinal microvascular endothelial cells (HRECs) and revealed a pivotal role of EMC1 in the β-catenin signaling pathway. Further in-vitro and in-vivo experiments proved that loss of EMC1 led to compromised β-catenin signaling activity through reduced expression of Wnt receptor FZD4, which could be restored by lithium chloride (LiCl) treatment. Driven by these findings, we screened genomic DNA samples from familial exudative vitreoretinopathy (FEVR) patients and identified one heterozygous variant in EMC1 that co-segregated with FEVR phenotype in the family. In-vitro expression experiments revealed that this variant allele failed to facilitate the expression of FZD4 on the plasma membrane and activate the β-catenin signaling pathway, which might be a main cause of FEVR. In conclusion, our findings reveal that variants in EMC1 gene cause compromised β-catenin signaling activity, which may be associated with the pathogenesis of FEVR.
Keywords: Angiogenesis, β-catenin signaling, EMC1, Familial exudative vitreoretinopathy, LiCl
Introduction
Blood vessels form the biggest network and provide nutrients and oxygen for organs.1, 2, 3, 4 Abnormal vascular development is related to numerous inherited diseases, including cavernous hemangioma,5 familial exudative vitreoretinopathy (FEVR),6 and primary pulmonary hypertension.7 FEVR is a severe ocular disorder characterized by defective retinal vascular development.8 A survey of early ocular examinations among more than 64,000 Chinese newborns revealed an estimated FEVR incidence of approximately 4.4‰.9,10 Retinal vascularization requires precise regulation of multiple signaling pathways, among which the canonical Wnt signaling is one of the most important signaling axis regulating vascular development.11, 12, 13, 14 Generally, Wnt or Norrin ligands bind to frizzled (FZD) family receptors and lipoprotein receptor-related proteins (LRP) co-receptors, to activate intracellular transduction through phosphorylation of GSK3β and subsequentially accumulation of β-catenin.15, 16, 17 With the assistance of LRP5/LRP6, frizzled 4 (FZD4) acts as the exclusive receptor of Norrin to promote retinal vascular development.18, 19, 20
Endoplasmic reticulum (ER) membrane protein complex (EMC) was first identified as a cluster of six subunits in yeast, with pivotal roles in optimal ER homeostasis.21 Being widely conserved among species, the mammalian EMC complex consists of a combined cluster of ten EMC subunits (EMC1–EMC10) and depleting any one of EMC1, EMC2, EMC3, EMC5, or EMC6 completely disrupted the EMC complex.22,23 Recent studies have shown that EMC in vertebrates not only functions as an insertase for C-terminal transmembrane domains (TMDs) of tail-anchored (TA) membrane proteins,24,25 but also mediates the expression of non-TA membrane proteins like G protein-coupled receptors (GPCRs), which are essential for vessel myogenic tone and vascular permeabilities.26, 27, 28
During the past years, several potential pathogenic EMC1 (ER membrane protein complex subunit 1) variants have been identified by phenotype–genotype correlation analysis. The first EMC1 variant was reported to be associated with retinitis pigmentosa (RP).29 Since then, a total of eight monoallelic or biallelic variants were identified in patients affected with various phenotypes, manifesting congenital cardiac pathologies, developmental delay, visual impairment, hypotonia, scoliosis, and cerebellar atrophy.30, 31, 32 Recently, a Xenopus model was established to illuminate the pathogenesis of various phenotypes upon Emc1 depletion, with the Frizzled protein and Wnt signaling levels diminished in genetically modified embryos.33 However, the role of EMC1 in mammalian vascularization remains unelucidated.
We generated an endothelial cell-specific knockout model for Emc1 and assessed its role in retinal angiogenesis. Emc1 deletion in endothelial cells led to aberrant retinal vascularization, manifesting reduced vascular progression and vascular density, diminished tip cell sprouts, and vascular leakage. Using an unbiased RNA transcriptomics analysis and in-vitro experiments, we revealed a pivotal role of EMC1 in Wnt signaling. By screening FEVR samples with high-throughput whole-exome sequencing, we identified a heterozygous missense variant in EMC1 gene that is associated with FEVR. The mutant EMC1 protein led to compromised β-catenin signaling activity via diminished expression of FZD4 receptor. Our study uncovers the essential role of EMC1 in retinal vascularization and sheds light on the importance of EMC in the regulation of β-catenin signaling during retinal angiogenesis.
Material and methods
Generation of Emc1 knockout mice and mice breeding
All animal experiments were conducted in accordance with the relevant guidelines and were approved by the Animal Care and Use Committee of Sichuan Provincial People's Hospital. All mice were raised in a specific pathogen-free barrier facility with standard rodent water and diet.
Mice carrying loxp sites flanking exons 5 and 6 of Emc1 (Cyagen Biosciences, Suzhou, China; https://www.cyagen.com/cn/zh-cn/sperm-bank-cko/230866) generated using the CRISPR/Cas9 system in C57BL/6J genetic background were bred into Pdgfb-iCre-ER transgenic mice line,34,35 The offspring animals were then bred with Emc1loxp/loxp homozygous mice to produce Emc1loxp/loxp Pdgfb-iCre-ER mice (hereafter named Emc1iECKO/iECKO). The Emc1iECKO/iECKO mice were subjected to intraperitoneal tamoxifen injections to knockout Emc1 in vascular endothelial cells.36 Tamoxifen-treated littermates carrying wild-type alleles, or without Pdgfb-iCre-ER allele were used as the controls. Mice were genotyped by polymerase chain reaction (PCR) using genomic DNA templates obtained from mouse tails using the primers listed in Table S2. All amplification was performed using an EasyTaq® DNA Polymerase (AS111-11, TransGen Biotech, CN). The PCR protocol of Emc1 floxed mice consisted of 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 60 °C for 20 s, and 72 °C for 30 s, and 1 cycle of 72 °C for 10 min. For Pdgfb-CreER mice, the first cycle consisted of 94 °C for 3 min, followed by 35 cycles of 95 °C for 30 s, 60 °C for 32 s, and 72 °C for 45 s, and 1 cycle of 72 °C for 5 min.
Immunohistochemistry and EdU labeling of mice retinae
To evaluate the development of retinal vessels, retinae were dissected from the eyes and fixed in 4% PFA at 4 °C overnight, followed by a rinse in phosphate-buffered saline (PBS). After being blocked in blocking buffer containing 1% fetal bovine serum with 0.25% Triton X-100 for 1 h at room temperature, the retinae were incubated overnight with Isolectin B4 (I21413, Thermo Fisher Scientific, USA), and primary antibodies including rat anti-mouse Ter-119 (553670, BD Biosciences, USA), rat anti-mouse plasmalemma vesicle-associated protein (PLVAP)/MECA-32 (553849, BD Biosciences, USA), rabbit anti-LEF1 (76010, Cell Signaling Technologies, USA), rabbit anti-ERG (ab92513, Abcam, USA), and Alexa Fluor 488-conjugated Claudin-5 monoclonal antibody (352588, Thermo Fisher Scientific, USA). The retinae were washed 3 times in PBS followed by incubation with appropriate Alexa Fluor-conjugated secondary antibodies (A-11006, A32721, or A27018, Thermo Fisher Scientific, USA) for 4 h. The Click-iT EdU Alexa Fluor-488 Imaging Kit (C10337, Thermo Fisher Scientific, USA) was used in combination with ERG antibody to detect EdU positive endothelial cells in retinae of pups that were sacrificed 3 h after intraperitoneal EdU injection.
Cell line culture and transfection
HEK 293T cells and those stably transfected with SuperTopflash plasmids (HEK 293STF) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 mg/mL penicillin, and 100 mg/mL streptomycin. Cells were seeded onto appropriate dishes and were transfected with plasmids using lipofectamine 3000 reagent (L3000001, Thermo Fisher Scientific, USA) according to the manufacturer's instructions. Luciferase assays, real-time quantitative PCR (RT-qPCR) or Western blot analysis were conducted 48 h after transfection.
Cell culture and lentivirus-mediated knockdown
Human retinal microvascular endothelial cells (HRECs) were purchased from Cell Systems (ACBRI-181, Cell Systems, USA) and maintained in EGM™-2 medium (CC-3162, Lonza, CH). Lentivirus carrying shEMC1 (5′- AGAGAACTACTGCTC-ATTT -3′) or negative control shRNA (5′- TTCTCCGAACGTG-TCACGT -3′) were used to infect HRECs at 15% confluence. Cells were harvested 120 h after infection and used for RT-qPCR and Western blot analysis.
Cloning of plasmids and luciferase assays
The cDNA of EMC1 was cloned into pcDNA3.1 vector, and the variant was introduced using the Q5 Site-Directed Mutagenesis Kit (E0554, New England Biolabs, UK). Luciferase assays were performed as previously described.18 Briefly, EMC1 knockdown (KD) HEK 293STF cells in 24-well plates were co-transfected with EMC1 (100 ng WT, 50 ng WT + 50 ng I762V, 100 ng I762V, or Vector) and 200 ng pGL4-Renilla for 48 h before detection. The pGL4-Renilla plasmid was transfected as an internal control.
RT-qPCR and RT-PCR (reverse transcription PCR)
Total RNAs were extracted from cultured cells and lung tissue of postnatal day-26 mice using TRIzol® reagent (TransGen Biotech, CN). EasyScript One-Step RT-PCR SuperMix (TransGen Biotech, CN) was used to synthesize cDNA, which was amplified using TransStart Tip Green qPCR SuperMix (TransGen Biotech, CN) and in a 7500 Fast Real-Time PCR System (Thermo Fisher Scientific, USA). Three biological replicates were performed using ΔΔCT methods and normalized to reference gene β-actin. RT-PCR was carried out by PCR-based methods using the TransStart Tip Green qPCR SuperMix (TransGen Biotech, CN). The primers for RT-qPCR or RT-PCR are listed in Table S3.
Western blotting
HEK 293T, HRECs or mouse lung tissue were sonicated three times in lysis buffer with the addition of protease and phosphatase inhibitor cocktail (Med Chem Express, USA). The lysates were mixed with 4 × sodium dodecyl sulfate (SDS) sample buffer (Solarbio, CN) and then loaded on 12% polyacrylamide gel. Western blotting was conducted, and signals were detected by iBright™ CL1500 Imaging System (Thermo Fisher Scientific, USA). The following antibodies were used: rabbit anti-CyclinD1 (MA5-14512, Thermo Fisher Scientific, USA), rabbit anti-c-Myc (T55150, Abmart, CN), rabbit anti-FZD4 (A8161, ABclonal Technology, CN), rabbit anti-p-GSK3β-Ser-9 (5558, Cell Signaling Technologies, USA), rabbit anti-GSK3β (12456, Cell Signaling Technologies, USA), rabbit anti-β-actin (3700, Cell Signaling Technologies, USA), mouse anti-FLAG (F3165, Merck, GER), rabbit anti-LRP5 (5731, Cell Signaling Technologies, USA), rabbit anti-FZD6 (A10503, ABclonal Technology, CN), HRP-conjugated goat anti-mouse secondary antibody (7076, Cell Signaling Technologies, USA), and HRP-conjugated goat anti-rabbit secondary antibody (7074, Cell Signaling Technologies, USA).
Transcriptome sequencing and bioinformatic analysis
A high-quality total RNA sample (1 μg) extracted from HRECs was used for library construction according to the manufacturer's instructions for the TruSeq RNA sample preparation kit (Illumina, USA). Briefly, mRNA was selected by oligo(dT) beads and fragmented. The following steps were completed before sequencing: cDNA synthesis, end repair, A-base addition, and ligation. NovaSeq 6000 sequencing system (Illumina) was used to obtain raw reads, which were trimmed to get clean reads before being aligned to the reference genome by using hisat2 software (https://ccb.jhu.edu/software/hisat2/index.shtml).37 The quality evaluation was conducted using qualimap_v2.2.1 (http://qualimap.bioinfo.cipf.es/).38 The htseq was used to count reads of each gene (https://htseq.readthedocs.io/en/release_0.11.1/).39
To unravel differential expression genes (DEGs) between the control group and the EMC1-null group, we calculated the gene expression levels via the fragments per kilobase of exon per million mapped reads (FPKM) method (Table S1). R package edgeR (https://bioconductor.org/packages/edgeR) was used to perform differential expression analysis on protein-coding genes with the following criteria40,41: (i) the threshold of |log2FC| should be over 0.585; (ii) the p-value was less than 0.05; (iii) the gene abundance in control or EMC1-null group should be over 0. The filtered DEGs (Table S1) were further analyzed by gene pathway classification using PANTHER (www.pantherdb.org).42 The volcano plot and heat map were generated using the OmicShare tools (www.omicshare.com/tools).
LiCl treatment
After being serum-starved overnight, HRECs were cultured in a serum-null medium with a final concentration of 60 mM of lithium chloride (LiCl) for 24 h. Then the cells were harvested for further analysis.
DNA sequencing of FEVR patients
This study was approved by the Institutional Research Committee of Sichuan Provincial People's Hospital and Xin Hua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine (China). FEVR-affected patients without variants in known FEVR-associated genes were recruited from XinHua Hospital. According to the Declaration of Helsinki, informed consent was obtained from each participant involved in this study. Firstly, genomic DNA was extracted from peripheral blood samples using a DNA extraction kit (TianGen, China). Whole-exome sequencing (WES) was performed on DNA samples to identify candidate pathogenic genes for FEVR. Briefly, genomic DNA was fragmented to an average length of ∼150 bp using an S220 Focused-ultrasonicator (Covaris, USA). The standard Illumina libraries were prepared using an Agilent SureSelect Human All Exon V5 kit (Agilent Technologies, USA), including the processes of end repair, adapter ligation, and PCR amplification. Afterward, the libraries were sequenced by a HiSeq2500 sequencer (Illumina). The BWA software package (http://bio-bwa.sourceforge.net/)43 was utilized for the alignment of raw reads to UCSC h19 (http://genome.ucsc.edu/). We further used SAMTOOLS (http://samtools.sourceforge.net/) to identify SNPs, insertions, and deletions. Sanger sequencing was applied for validation and co-segregation analysis. Primers for Sanger sequencing were listed in Table S2.
Image acquisition and statistical analysis
The immunofluorescent signal was collected by LSM 800 confocal scanning microscope (Zeiss, GER). Retinal vascular density was calculated using Angiotool 0.5 beta. Signals of Western blot were analyzed using ImageJ software version 1.48. Statistical analysis was performed by GraphPad Prism 8.0 software. Student's t-tests with Welch's correction or one-way ANOVA with Tukey's or Dunnett's multiple comparison tests were used to evaluate statistical significance.
Results
EC-specific knockout of Emc1 causes defects in vessels
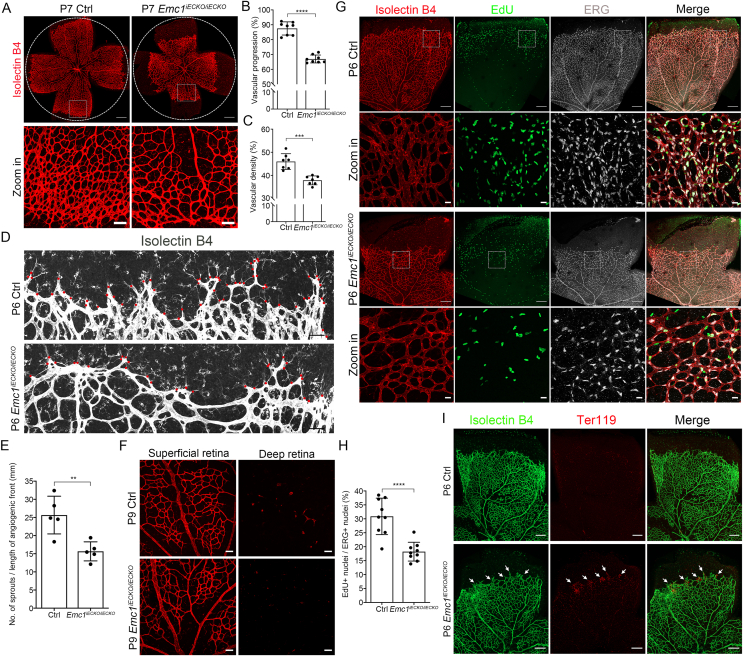
To uncover the role of EMC1 in postnatal retinal vascularization, Emc1iECKO/iECKO mice and control littermates were treated with tamoxifen from postnatal day 1 (P1) to P3. The superficial plexus and vertical growth of retinal vessels were analyzed on P6∼P9 (Fig. 1), respectively. Immunostaining of whole-mounted retinae revealed that the vascular progression and density of superficial vessels were reduced in P7 Emc1iECKO/iECKO retinae in contrast to controls (Fig. 1A–C). The sprouts at the angiogenic front were also diminished in the mutant vessels (Fig. 1D, E). After reaching the periphery retina approximately on P8, the vessels sprout vertically into the deeper retina.44 As expected, vessels sprouted vertically into the deep retina of wild-type mice on P9, whereas they were absent in the deep retinae of P9 Emc1iECKO/iECKO mice (Fig. 1F). Consistent with the reduced outgrowth and decreased sprouts of Emc1-null retinal vessels, retinal endothelial cells of P6 Emc1iECKO/iECKO mice exhibited prominently decreased proliferation compared to that of controls (Fig. 1G, H). In addition to the developmental defects of the Emc1iECKO/iECKO vasculature, marked hemorrhage in retinae was also observed in Emc1iECKO/iECKO mice (Fig. 1I). Thus, inactivation of endothelial Emc1 not only results in defective vascularization but also disturbs the integrity of the blood-retina barrier.
Figure 1.
Loss of Emc1 in mouse endothelial cells results in retinal vascular defects. (A) Representative overview (top panels) and high-magnification images (bottom panels) of P7 control (Ctrl) and Emc1iECKO/iECKO mouse retinae stained with Isolectin B4 (IB4). The circles indicate vessel outgrowth of Ctrl retinae. Scale bars, 500 μm and 100 μm. (B, C) Quantification of vascular progression (B) and vascular density (C) of P7 control (Ctrl) and Emc1iECKO/iECKO mouse retinae. Error bars, standard deviations (SDs). Student's t-test (n ≥ 7), ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. (D) Representative immunofluorescence images of P6 Ctrl and Emc1iECKO/iECKO mouse retinae stained with IB4. Red dots indicate sprouts at the angiogenic front. Scale bars, 50 μm. (E) Quantification of the numbers of sprouts per unit of front length (mm). Error bars, SDs. Student's t-test (n = 5), ∗∗P < 0.01. (F) Representative immunofluorescence image of superficial and deep IB4-staining retinae of P9 Ctrl and Emc1iECKO/iECKO mice. Scale bars, 50 μm. (G) Representative overview and high-magnification images of P6 Ctrl and Emc1iECKO/iECKO mouse retinae co-stained with IB4, EdU, and ERG. Dotted boxes indicate magnified areas. Scale bars, 20 μm and 200 μm. (H) Quantification of the percentage of EdU+/ERG + cells to total ERG + cells per field. Error bars, SDs. Student's t-test (n = 9), ∗∗∗∗P < 0.0001. (I) Representative immunofluorescence images of P6 Ctrl and Emc1iECKO/iECKO mouse retinae co-stained with IB4 and Ter119. White arrows indicate leakage of erythrocytes. Scale bars, 200 μm.
EMC1 depletion compromised Wnt signaling
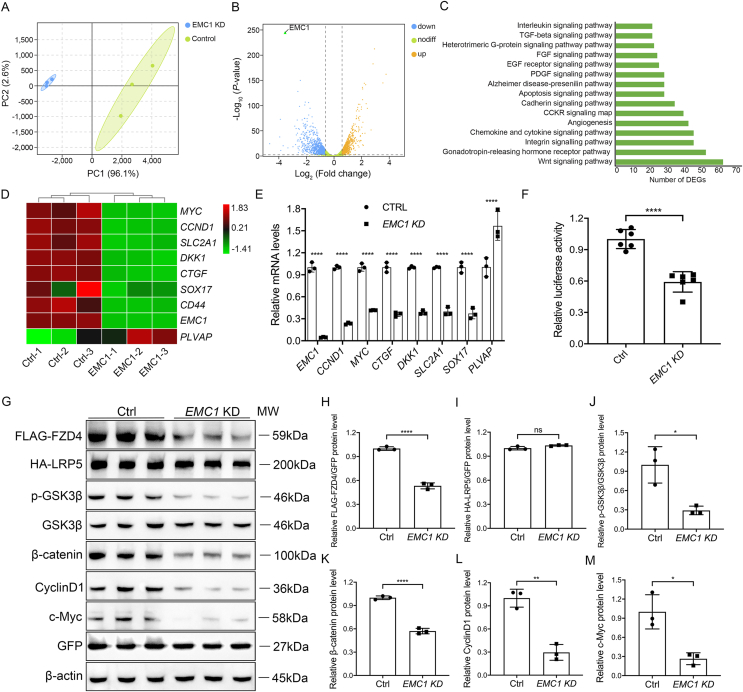
To unravel the insights into the mechanism of how EMC1 regulates retinal angiogenesis, we conducted an unbiased transcriptomic analysis in controls and EMC1-null HRECs. The principal component analysis revealed an apparent separation between controls and EMC1-null groups (Fig. 2A). Volcano plot revealed numerous changes in mRNA levels due to EMC1 depletion (Fig. 2B). Using the PANTHER pathway classification, the DEGs mostly clustered in the Wnt pathway, gonadotropin-releasing hormone receptor pathway, and integrin signaling pathway (Fig. 2C). Interfered Wnt signaling has been linked to deficient vascularization, and variants in components of the Wnt pathway genes are associated with FEVR.45 To date, several genetically modified mouse models, which conditionally or globally depleted the core genes of Wnt signaling (Fzd4−/−, Lrp5−/−, or Tspan12−/−), have been established to reproduce clinical phenotypes of FEVR.6,45, 46, 47, 48 Interestingly, the defective retinal vascularization due to EC-specific Emc1 ablation also mimics retinal vascular defects in Wnt-deficient mouse models (Fig. 1). We conjectured that EMC1 functions in retinal vascularization via the Wnt signaling pathway.
Figure 2.
EMC1 regulates Wnt signaling through FZD4 receptor. (A) Principal component analysis for Ctrl and EMC1 KD (knockdown) HRECs. (B) Volcano plot of differentially expressed genes (DEGs) in Ctrl and EMC1 KD HRECs. The dash lines indicate the threshold of P = 0.05 and |log2FC| = 0.585. The significantly down-regulated and up-regulated genes are shown as blue and yellow dots, respectively. Genes with no significant alterations are shown as green dots. (C) Pathway classification of DEGs using the PANTHER classification system online. (D) Heatmap of the expression fold changes of several Wnt-targeted genes in Ctrl and EMC1 KD HRECs as determined by RNA-seq analysis. (E) RT-qPCR quantification of the relative mRNA levels of EMC1, MYC, CCND1, SLC2A1, DKK1, CTGF, SOX17, CD44, and PLVAP in Ctrl and EMC1 KD HRECs. Error bars, SDs. Student's t-test (n = 3), ∗∗∗∗P < 0.0001. (F) Relative luciferase activity in Ctrl and EMC1 KD HEK 293STF cells transfected with pGL4-Renilla as an internal control. Error bars, SDs. Student's t-test (n = 6), ∗∗∗∗P < 0.0001. (G–M) Western blot and quantification analysis of the expression levels of FLAG-FZD4, HA-LRP5, p-GSK3β, β-catenin, CyclinD1, and c-Myc in Ctrl and EMC1 KD HEK 293T cells co-transfected with FLAG-tagged FZD4, HA-tagged LRP5, and GFP. GFP and β-actin were used as internal controls for over-expressed proteins and endogenous proteins (unless otherwise noted), respectively. GSK3β was used as an internal control for p-GSK3β (unless otherwise noted). Error bars, SDs. Student's t-test (n = 3), ∗P < 0.05, ∗∗P < 0.01, ∗∗∗∗P < 0.0001. ns, not significant.
In accordance with our speculation, reduced transcriptional levels of Wnt-targeted genes, including MYC, CCND1, SLC2A1, DKK1, CTGF, SOX17, and CD44 were identified by transcriptional analysis (Fig. 2D). Interestingly, we noticed up-regulation of PLVAP, which was reported to be a readout of impaired β-catenin signaling,6 in EMC1-deficient HRECs (Fig. 2D). Most of the Wnt-targeted DEGs were further confirmed by RT-qPCR (Fig. 2E). To validate the role of EMC1 in Norrin/β-catenin signaling, we conducted luciferase assays in HEK 293STF cells. The results showed compromised β-catenin signaling activity in the absence of EMC1, which was consistent with transcriptomics data (Fig. 2F).
EMC1 depletion affects Wnt signal transduction via FZD4
Given the regulatory role of EMC1 in the synthesis of multipass transmembrane proteins,26, 27, 28 we first explored whether EMC1 regulates endothelial Wnt signaling through Frizzled (FZDs) receptors. RT-PCR of HRECs revealed that the mRNA abundance of FZD4 and FZD6 was much higher than other FZDs (Fig. S1A), indicating that FZD4 and FZD6, rather than other FZDs, might mainly participate in the retinal vascularization. In the absence of EMC1, the protein levels of FZD4 and FZD6 were further proved to be decreased in HRECs (Fig. S1B, C). Since Golan et al49 have previously reported that FZD6 could not activate Wnt-induced canonical β-catenin signaling, and that FZD4 is widely recognized as a specific receptor of the Norrin/β-catenin signaling pathway, which plays a pivotal role in retinal vascular development,6,45, 46, 47, 48 we speculate that EMC1 might regulate Wnt signaling activity mainly through FZD4 receptor rather than FZD6 receptor. Generally, Norrin binds to receptor FZD4 and co-receptor LRP5 in the cell membrane of retinal ECs, which leads to phosphorylation of GSK3β at Ser9, thereby stabilizing the β-catenin for Wnt signal transduction.50 Thus, we over-expressed FLAG-tagged FZD4 and HA-tagged LRP5 in HEK 293T cells to investigate whether EMC1 regulates the expression of exogenous over-expressed receptor FZD4 or co-receptor LRP5. Interestingly, we detected an obvious reduction of FLAG signal rather than HA signal upon EMC1 depletion (Fig. 2G–I), indicating that EMC1 might more specifically regulate multipass transmembrane protein FZD4 rather than single-transmembrane protein LRP5. Meanwhile, the mRNA level of FLAG-tagged FZD4 was unaffected in the absence of EMC1 (Fig. S1D), indicating that EMC1 regulates FZD4 at the post-transcriptional level. Additionally, the protein levels of phosphorylated GSK3β (p-GSK3β-Ser9, the inactivated state of GSK3β that protects β-catenin from degradation) as well as β-catenin and its downstream targets (CyclinD1 and c-Myc) were also significantly declined (Fig. 2G–M).
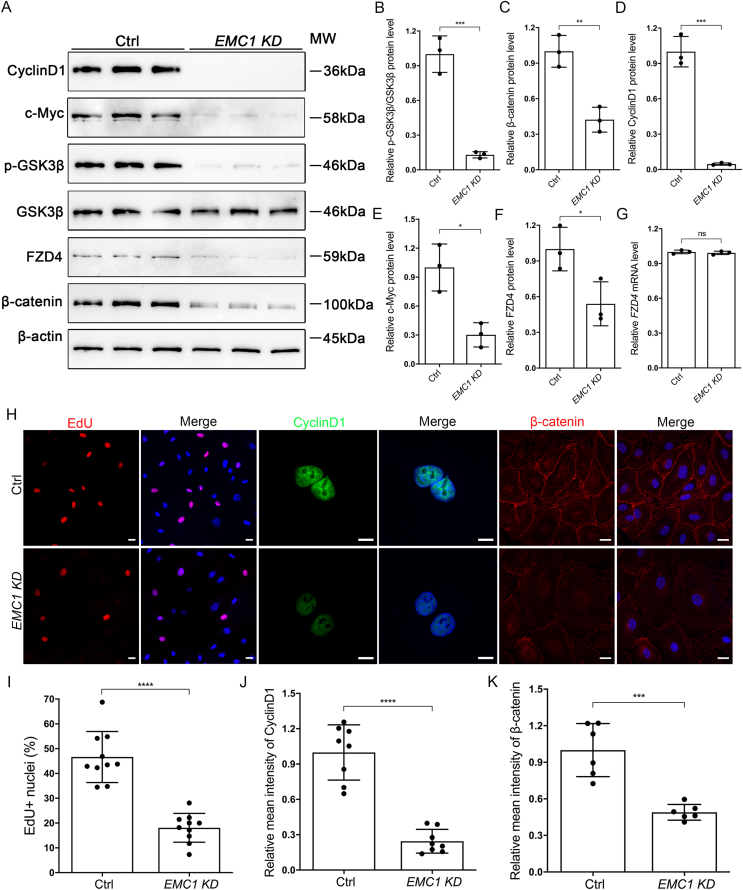
To further confirm the effect of EMC1 on the Wnt/FZD4 axis in HRECs, we evaluated FZD4 and downstream targets of the Norrin/β-catenin signaling pathway via Western blot analysis in controls and EMC1-depleted HRECs. Consistently, the immunoblotting analysis revealed markedly attenuated Wnt signaling in the absence of EMC1, manifested by reduced protein levels of p-GSK3β-Ser9, β-catenin, CyclinD1 and c-Myc (Fig. 3A–E). As expected, FZD4 was decreased at the protein level rather than the mRNA level in EMC1-depleted HRECs (Fig. 3A, F, G).
Figure 3.
EMC1-depletion in HRECs results in compromised Wnt signaling and cell proliferation. (A–F) Western blot and quantification analysis of the expression levels of CyclinD1, c-Myc, p-GSK3β, FZD4, and β-catenin in Ctrl and EMC1 KD HRECs. Error bars, SDs. Student's t-test (n = 3), ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. (G) RT-qPCR quantification of the relative FZD4 mRNA level in Ctrl and EMC1 KD HRECs. Error bars, SDs. Student's t-test (n = 3). ns, not significant. (H) Representative immunofluorescence images of Ctrl and EMC1 KD HRECs labeled with EdU, β-catenin, or CyclinD1 and DAPI (blue). Scale bars, 20 μm (EdU and β-catenin) and 10 μm (CyclinD1). (I–K) Quantification of the percentage of EdU + nuclei (I), and relative levels of CyclinD1 (J) and β-catenin (K) in Ctrl and EMC1 KD HRECs. Error bars, SDs. Student's t-test (n ≥ 6), ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
Given that decreased EC proliferation and compromised Wnt signaling was observed in Emc1iECKO/iECKO mice (Fig. 1G, H) and EMC1-depleted HRECs (Fig. 2), respectively, we further explored whether loss of EMC1 could cause decreased cell proliferation in vitro. We performed an EdU-dependent proliferation assay in controls and EMC1-depleted HRECs and observed less percentage of EdU-positive cells upon EMC1 depletion (Fig. 3H, I). Additionally, interfered cell cycle and decreased Wnt activity were also confirmed in EMC1-KD HRECs by drastic reduction of the immunofluorescent signal of CyclinD1 and β-catenin (Fig. 3H–K). Taken together, our data demonstrated that EMC1 contributes to the Wnt signaling via FZD4 receptor that mediates the proliferation of endothelial cells.
Defects in Wnt signaling of Emc1-depleted mouse ECs
To assess the Wnt signaling activity in vivo, we performed RT-qPCR and Western blot analysis on lung lysates of P26 Emc1iECKO/iECKO mice and littermate controls. The efficiency of inducible Emc1 knockout in mouse lung tissue was verified by RT-qPCR (Fig. S1E). As expected, the mRNA levels of Wnt-targeted genes, including Ccnd1, Myc, Cldn5, Sox17, and Axin2, were prominently decreased in Emc1iECKO/iECKO mice lung tissues (Fig. S1F–J). Consistently, the protein levels of CyclinD1, c-Myc, p-GSK3β-Ser9, and β-catenin were largely decreased due to the loss of endothelial-Emc1 in mice lung lysates (Fig. S1K–O). The protein expression of FZD4 was decreased with an unaltered mRNA level (Fig. S1K, P, Q), which confirmed that EMC1 regulated Wnt signaling through its effect on the post-transcription of FZD4. We further performed immunostaining on the retinal flat mount to explore whether the Wnt signaling activity was compromised in the Emc1-deficient retinae. Compromised Norrin/β-catenin signaling has been reported to cause reduced tight junction protein Claudin-5, accompanied by abnormally increased expression of plasmalemma vesicle-associated protein (PLVAP) in mice retinae.6,10 The vascular permeability was also increased in Emc1iECKO/iECKO mice retinae, manifested with attenuated Claudin-5 (Fig. S2A, B) and accumulated PLVAP signals in veins (Fig. S2C, D). Additionally, the lymphoid enhancer-binding factor 1 (LEF1), a transcription Wnt/β-catenin signaling in endothelial cells, is markedly decreased in Emc1iECKO/iECKO retinal veins (Fig. S3).
LiCl restores proliferation and Wnt signaling in EMC1-depleted HRECs
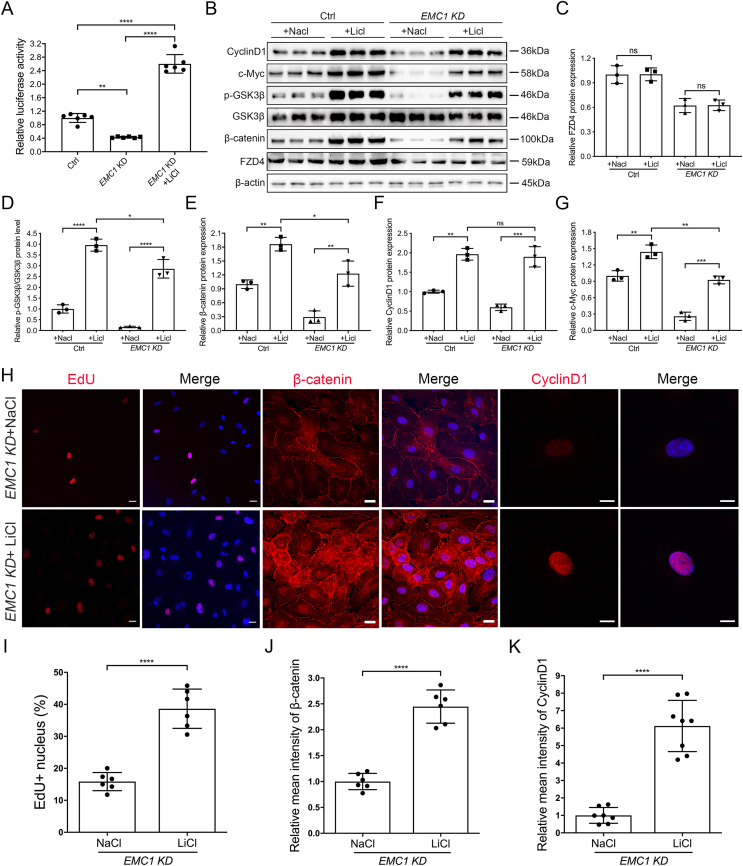
Given the reduction of Wnt signaling in the absence of EMC1, we considered that activation of Wnt by GSK3 inhibitor LiCl might rescue the endothelial defects in EMC1-depleted HRECs. We first tested whether LiCl treatment could enhance the luciferase activity of HRECs in the absence of EMC1. The results showed an approximately six-fold enhancement of Wnt luciferase activity in the EMC1-depleted HEK 293STF compared to that treated with vehicle (Fig. 4A). Additionally, we identified prominently increased protein levels of β-catenin, p-GSK3β-Ser9 as well as Wnt downstream targets CyclinD1 and c-Myc in LiCl-treated cells (Fig. 4B–G). Interestingly, the protein level of FZD4 was unchanged upon LiCl treatment, indicating that LiCl treatment could restore the compromised Wnt signaling activity by bypassing FZD4 (Fig. 4B, C). We next applied immunocytochemistry to investigate whether enhancement of Wnt signaling by LiCl could restore the CyclinD1 and β-catenin levels as well as hypo-proliferation in EMC1-depleted HRECs. As expected, LiCl treatment successfully restored the expression of CyclinD1 and β-catenin, which further stimulated cell proliferation in EMC1-depleted HRECs (Fig. 4H–K).
Figure 4.
LiCl treatment restores Wnt signaling activity and proliferation of EMC1 KD HRECs. (A) Relative luciferase activity in Ctrl, EMC1 KD, and EMC1 KD HEK 293STF cells treated with LiCl. pGL4-Renilla plasmid was transfected as an internal control. Error bars, SDs. The P-values are from multiple comparisons in one-way ANOVA with Tukey's multiple comparison test (n = 6); ∗∗P < 0.01, ∗∗∗∗P < 0.0001. (B–G) Western blot and quantification analysis of the expression levels of CyclinD1, c-Myc, p-GSK3β, β-catenin, and FZD4 in Ctrl and EMC1 KD HRECs treated with LiCl or NaCl. Error bars, SDs. The P-values are from multiple comparisons in two-way ANOVA with Sidak's multiple comparison test (n = 3); ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. ns, not significant. (H) Representative immunofluorescence images of EMC1 KD HRECs treated with NaCl or LiCl and labeled with EdU, β-catenin, or CyclinD1 and DAPI (blue). Scale bars, 20 μm (EdU and β-catenin) and 10 μm (CyclinD1). (I–K) Quantification of the percentage of EdU + nuclei (I), and relative levels of β-catenin (J) and CyclinD1 (K) in EMC1 KD HRECs treated with NaCl or LiCl. Error bars, SDs. Student's t-test (n ≥ 6), ∗∗∗∗P < 0.0001.
The pathogenesis of EMC1 variants in FEVR patients
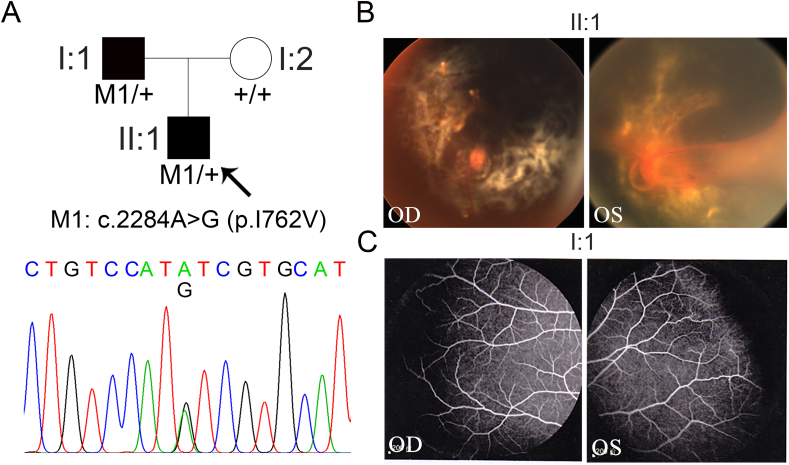
In view of the FEVR-like phenotypes and compromised Wnt signaling activity in the retinae of Emc1iECKO/iECKO mice, we applied WES on the genomic DNA samples from FEVR patients to identify candidate EMC1 variants for FEVR. These approaches identified one missense heterozygous variant in the EMC1 gene (NM_001271429: c.2284A > G; p. I762V, rs540813358) in two patients from one family, which was validated by Sanger sequencing and co-segregation analysis (Fig. 5A). The frequency of this variant was less than 0.001 in ExAC, 1000 Genomes, gnomAD, and TOPMed databases. The proband was firstly diagnosed with FEVR at 3 years old, manifested with exudation in the right eye and falciform retinal fold in the left eye (Fig. 5B). His mother had normal vision with wild-type EMC1 allele, whilst his heterozygote father showed incomplete peripheral vessels and intersections between capillaries in bilateral retinae (Fig. 5C).
Figure 5.
Identification of a missense EMC1 variant in a FEVR-associated family. (A) FEVR pedigrees, Sanger sequencing analysis, and amino acids alignment in a Chinese FEVR-associated family. Patients are denoted in black. Black arrows indicate the proband of the family. (B) Fundus image of the proband's bilateral eyes. (C) Fundus fluorescein angiography of the proband's affected father.
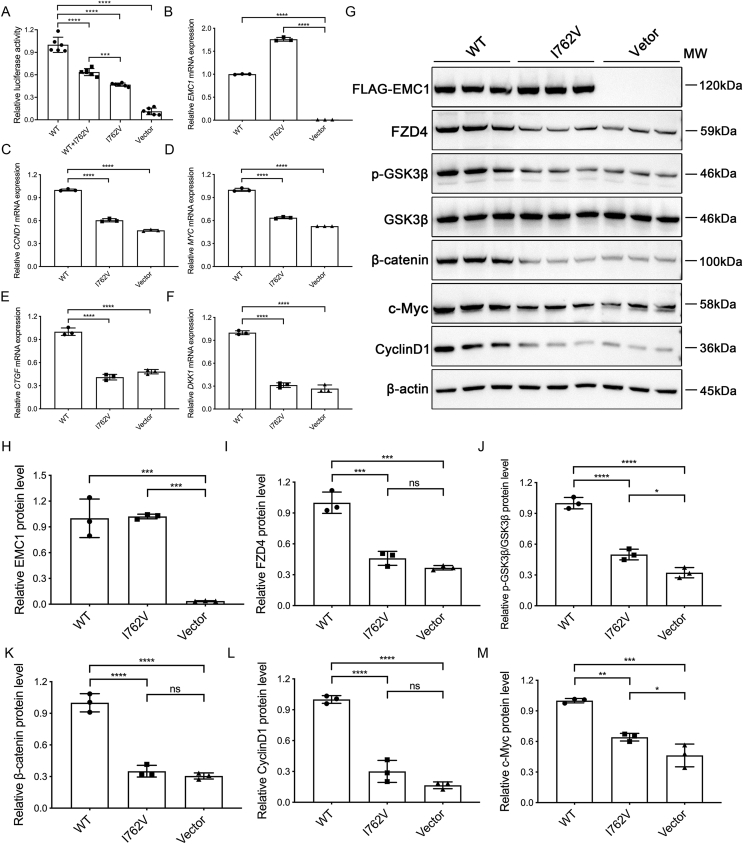
We then constructed a FLAG-tagged human EMC1 (wild-type or mutant) over-expression plasmid to investigate the pathogenicity of this identified variant in heterozygous or homozygous states. We first applied luciferase assay and found that wild-type EMC1 remarkably promoted Wnt signaling activity compared to that of the vehicle (Fig. 6A). Although the heterozygous I762V variant exhibited a milder inhibitory effect than the homozygous form, both of them showed compromised Wnt signaling activity compared to that of wild-type EMC1 (Fig. 6A). As expected, the mRNA levels of Wnt targeted genes, including CCND1, MYC, CTGF and DKK1 were decreased in the I762V-transfected HEK 293T cells compared to that of wild-type EMC1 (Fig. 6B–F). In addition, Western blot analysis on HEK 293T cells transfected with FLAG-tagged EMC1 (wild-type, variant, or empty vector) revealed that I762V variant did not affect the expression of EMC1 (Fig. 6G, H). I762V hardly promoted the Wnt activity, manifested with almost comparable protein levels of FZD4, p-GSK3β-Ser9, β-catenin, CyclinD1, and c-Myc with that of vector plasmid, which was drastically decreased compared to that of wild-type EMC1 (Fig. 6G–M). These data indicated that the I762V variant disrupts the function of EMC1 on Wnt signaling.
Figure 6.
EMC1 I762V variant results in compromised Wnt signaling activity via reduced FZD4 expression. (A) Relative luciferase activity in HEK 293STF cells co-transfected with EMC1 (wild-type, wild-type/I762V, I762V variant, or vector) and pGL4-Renilla. pGL4-Renilla plasmid was transfected as an internal control. Error bars, SDs. The P-values are from multiple comparisons in one-way ANOVA with Tukey's multiple comparison test (n = 4); ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. (B–F) RT-qPCR quantification of the relative mRNA levels of EMC1, CCND1, MYC, CTGF, and DKK1 in HEK 293T cells transfected with EMC1 (wild-type, I762V variant, or vector). Error bars, SDs. The P-values are from multiple comparisons in one-way ANOVA with Dunnett's multiple comparison test (n = 3); ∗∗∗∗P < 0.0001. (G–M) Western blot and quantification analysis of the expression levels of FLAG-EMC1, FZD4, p-GSK3β, β-catenin, c-Myc, and CyclinD1 in Ctrl and EMC1 KD HRECs. Error bars, SDs. The P-values are from multiple comparisons in one-way ANOVA with Tukey's multiple comparison test (n = 3); ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. ns, not significant.
Discussion
Given the regulatory role of EMC complex in the expression of multipass transmembrane proteins, the variants in EMC1 gene have been identified in various tissue-specific congenital diseases such as retinal dystrophy, developmental delay, hypotonia, scoliosis, and cerebellar atrophy.30, 31, 32 A previous research has figured out the mechanism that decreased Wnt signaling contributes to the defective development of Emc1-depleted neural crest cells.33 However, the role of EMC1 in vascular development remains unclear.
In this study, we constructed an EC-specific Emc1-knockout mouse model that exhibited FEVR-like phenotypes, providing insights into the regulatory roles of EMC1 in postnatal retinal vascularization. Since EMC1 is critical for basic biological processes in almost all cell types, it was not surprising to observe deficient retinal vessel development in Emc1iECKO/iECKO mice, including decreased vascular sprouts, compromised EC proliferation, delayed vascular progression, and vascular leakage (Fig. 1). The retinal vascular defects of Emc1iECKO/iECKO retinal vessels closely resembled that of Fzd4−/−, Lrp5−/−, Ndp-, and Tspan12−/− mice, which shared a common mechanism of down-regulated Wnt signaling pathway.6,16,45 An unbiased transcriptomic analysis of DEGs between controls and EMC1-depleted HRECs revealed enrichment of Wnt signaling related genes with remarkable down-regulation of several Wnt-targeted genes. We further applied in-vivo and in-vitro experiments and concluded that EMC1 mediates Wnt signaling through FZD4 receptor, which can be restored by downstream LiCl treatment.
Norrin/LRP5/FZD4 complex, which triggers activation of canonical Wnt signaling pathway, plays a crucial role in retinal vascularization.13 Of note, variants in FZD4 gene account for the second proportion of Chinese FEVR cases and the largest proportion of Korean FEVR cases.51, 52, 53 Thus, the defects in retinal vascular development in Emc1iECKO/iECKO mice might be largely attributable to the decreased FZD4 protein. Yet, we cannot exclude the possibility that other multipass membrane proteins contribute partially to Emc1iECKO/iECKO retinal vascular phenotypes, since the vascular defects were not completely the same as that of Wnt-down-regulated mouse models.6,16,45
Interestingly, we identified a monoallelic variant I762V in EMC1 from a family with FEVR and considered it as a candidate pathogenic variant due to the compromised activation effect of I762V variant on the Norrin/β-catenin signaling. Except for the novel variant in the present study, a total of eight disease-causing variants have been reported previously, including five homozygous variants, one compound heterozygous variant, and one de novo heterozygous variant,30, 31, 32 indicating that disease-causing EMC1 variants could be inherited as both autosomal-recessive and -dominant manners. Notably, clinical features of patients who carried EMC1 variants were considerably overlapped in multi-organ defects, including retrognathia, development delay, intellectual disability, scoliosis, truncal hypotonia, diminished deep tendon reflexes, and heterogeneous ophthalmological impairments.32 However, two individuals with homozygous A144T variants showed only retinal dystrophy without other EMC1-related phenotypes,29 suggesting that EMC1 variants might cause non-syndromic diseases.
In the present study, EMC1-I762V heterozygotes were first found to be associated with defective retinal vascular development, which expanded the spectrum of EMC1-associated clinical features. It is reasonable to assume that some missense variants in EMC1 could affect the normal expression of specific multipass membrane proteins like rhodopsin and FZD4, which results in local lesions other than multi-organ pathologies. However, given the fact that four of nine previously reported variants were not located on the functional domains of EMC1 protein, it seems that there is no correlation between the location of variants and phenotypes. Further studies to identify more causative EMC1 variants are needed for the investigation of insights into the genotype–phenotype correlations between EMC1 variants and FEVR, which are essential for the perspicuous pathogenic mechanisms and precise pathogenic prediction of EMC1 variants.
Conclusions
In summary, we demonstrate the regulatory role of EMC1 in retinal vascularization via FZD4/β-catenin axis. Additionally, we identify one heterozygous variant in EMC1 gene that is causative for FEVR and illustrate the mechanism of the variant with down-regulated β-catenin signaling activity. Thus, our study reports a novel candidate gene for FEVR, which expands the spectrum of EMC1 variants and EMC1-related phenotypes, providing evidence for the prenatal diagnosis of candidate disease-causing EMC1 variants in FEVR.
Author contributions
X.J.Z. and Z.L.Y. conceived the research. S.J.L., M.Y., and R.L.Z. performed the animal model study, cell biology, immunohistochemistry, and gene expression studies. L.P., W.J.L., X.Y.J., Y.Q.H., and E.K.D. performed the construction of plasmids. L.Z. and Y.M.Y. performed the breeding of animals. P.Q.Z. and R.L.Z. recruited the participants and made clinical diagnoses for the FEVR cases. S.J.L., M.Y., Y.S., and X.J.Z. analyzed the data. S.J.L., M.Y., and X.J.Z wrote the manuscript, with input from other authors.
Conflict of interests
The authors have declared that no conflict of interests exists.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82101153, 82000913, 81970841, 82121003, and 82071009); the Sichuan Science and Technology Program, China (No. 2022YFS0598, 2021YFS0386, 2021YFS0369, and 2021JDGD0036); the CAMS Innovation Fund for Medical Sciences, China (No. 2019–12M-5-032); the Department of Science and Technology of Qinghai Province, China (No. 2022-HZ-814); the fund for Sichuan Provincial People's Hospital, China (No. 2021QN01); the Department of Chengdu Science and Technology, China (No. 2021-YF05-01316-SN); the Huanhua Outstanding Scholar Program for Sichuan Provincial People's Hospital (China) to Xianjun Zhu. The funders had no role in the study design, data collection, analysis, or preparation of the manuscript.
Data availability
All the data and materials are available within the article and the supplementary materials.
Acknowledgements
The authors thank all patients and their family members for their helpful participation in this study.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.10.003.
Contributor Information
Zhenglin Yang, Email: yangzhenglin@cashq.ac.cn.
Xianjun Zhu, Email: xjzhu@uestc.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Carmeliet P. Angiogenesis in health and disease. Nat Med. 2003;9(6):653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 2.Xue B., Wang P., Yu W., et al. CD146 as a promising therapeutic target for retinal and choroidal neovascularization diseases. Sci China Life Sci. 2022;65(6):1157–1170. doi: 10.1007/s11427-021-2020-0. [DOI] [PubMed] [Google Scholar]
- 3.Liu W., Jiang X., Li X., et al. LMBR1L regulates the proliferation and migration of endothelial cells through Norrin/β-catenin signaling. J Cell Sci. 2022;135(6):jcs259468. doi: 10.1242/jcs.259468. [DOI] [PubMed] [Google Scholar]
- 4.Qian Y., Wang Z., Lin H., et al. TRIM47 is a novel endothelial activation factor that aggravates lipopolysaccharide-induced acute lung injury in mice via K63-linked ubiquitination of TRAF2. Signal Transduct Targeted Ther. 2022;7(1):148. doi: 10.1038/s41392-022-00953-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon A.M., McWhorter A.R. Vascular abnormalities in mice lacking the endothelial gap junction proteins connexin37 and connexin40. Dev Biol. 2002;251(2):206–220. doi: 10.1006/dbio.2002.0826. [DOI] [PubMed] [Google Scholar]
- 6.Junge H.J., Yang S., Burton J.B., et al. TSPAN12 regulates retinal vascular development by promoting norrin- but not wnt-induced FZD4/β-catenin signaling. Cell. 2009;139(2):299–311. doi: 10.1016/j.cell.2009.07.048. [DOI] [PubMed] [Google Scholar]
- 7.Yeager M.E., Halley G.R., Golpon H.A., et al. Microsatellite instability of endothelial cell growth and apoptosis genes within plexiform lesions in primary pulmonary hypertension. Circ Res. 2001;88(1):E2–E11. doi: 10.1161/01.res.88.1.e2. [DOI] [PubMed] [Google Scholar]
- 8.Criswick V.G., Schepens C.L. Familial exudative vitreoretinopathy. Am J Ophthalmol. 1969;68(4):578–594. doi: 10.1016/0002-9394(69)91237-9. [DOI] [PubMed] [Google Scholar]
- 9.Fei P., Liu Z., He L., et al. Early detection of ocular abnormalities in a Chinese multicentre neonatal eye screening programme-1-year result. Acta Ophthalmol. 2021;99(3):e415–e422. doi: 10.1111/aos.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang M., Li S., Huang L., et al. CTNND1 variants cause familial exudative vitreoretinopathy through the Wnt/cadherin axis. JCI Insight. 2022;7(14) doi: 10.1172/jci.insight.158428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franco C.A., Liebner S., Gerhardt H. Vascular morphogenesis: a Wnt for every vessel? Curr Opin Genet Dev. 2009;19(5):476–483. doi: 10.1016/j.gde.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 12.van de Schans V.A.M., Smits J.F., Blankesteijn W.M. The Wnt/frizzled pathway in cardiovascular development and disease: friend or foe? Eur J Pharmacol. 2008;585(2–3):338–345. doi: 10.1016/j.ejphar.2008.02.093. [DOI] [PubMed] [Google Scholar]
- 13.Zerlin M., Julius M.A., Kitajewski J. Wnt/frizzled signaling in angiogenesis. Angiogenesis. 2008;11(1):63–69. doi: 10.1007/s10456-008-9095-3. [DOI] [PubMed] [Google Scholar]
- 14.Yan X., Yang Z., Chen Y., et al. Endothelial cells-targeted soluble human Delta-like 4 suppresses both physiological and pathological ocular angiogenesis. Sci China Life Sci. 2015;58(5):425–431. doi: 10.1007/s11427-015-4834-3. [DOI] [PubMed] [Google Scholar]
- 15.MacDonald B.T., Tamai K., He X. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang M., Li S., Liu W., et al. The ER membrane protein complex subunit Emc3 controls angiogenesis via the FZD4/WNT signaling axis. Sci China Life Sci. 2021;64(11):1868–1883. doi: 10.1007/s11427-021-1941-7. [DOI] [PubMed] [Google Scholar]
- 17.He Y., Yang M., Zhao R., et al. Novel truncating variants in CTNNB1 cause familial exudative vitreoretinopathy. J Med Genet. 2023;60(2):174–182. doi: 10.1136/jmedgenet-2021-108259. [DOI] [PubMed] [Google Scholar]
- 18.Xu Q., Wang Y., Dabdoub A., et al. Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell. 2004;116(6):883–895. doi: 10.1016/s0092-8674(04)00216-8. [DOI] [PubMed] [Google Scholar]
- 19.Li S., Yang M., He Y., et al. Variants in the Wnt co-receptor LRP6 are associated with familial exudative vitreoretinopathy. J Genet Genomics. 2022;49(6):590–594. doi: 10.1016/j.jgg.2021.11.010. [DOI] [PubMed] [Google Scholar]
- 20.Zhao R., Wang S., Zhao P., et al. Heterozygote loss-of-function variants in the LRP5 gene cause familial exudative vitreoretinopathy. Clin Exp Ophthalmol. 2022;50(4):441–448. doi: 10.1111/ceo.14037. [DOI] [PubMed] [Google Scholar]
- 21.Jonikas M.C., Collins S.R., Denic V., et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323(5922):1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volkmar N., Thezenas M.L., Louie S.M., et al. The ER membrane protein complex promotes biogenesis of sterol-related enzymes maintaining cholesterol homeostasis. J Cell Sci. 2019;132(2):jcs223453. doi: 10.1242/jcs.223453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chitwood P.J., Hegde R.S. The role of EMC during membrane protein biogenesis. Trends Cell Biol. 2019;29(5):371–384. doi: 10.1016/j.tcb.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 24.Guna A., Volkmar N., Christianson J.C., et al. The ER membrane protein complex is a transmembrane domain insertase. Science. 2018;359(6374):470–473. doi: 10.1126/science.aao3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shurtleff M.J., Itzhak D.N., Hussmann J.A., et al. The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins. Elife. 2018;7 doi: 10.7554/eLife.37018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chitwood P.J., Juszkiewicz S., Guna A., et al. EMC is required to initiate accurate membrane protein topogenesis. Cell. 2018;175(6):1507–1519. doi: 10.1016/j.cell.2018.10.009. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kauffenstein G., Laher I., Matrougui K., et al. Emerging role of G protein-coupled receptors in microvascular myogenic tone. Cardiovasc Res. 2012;95(2):223–232. doi: 10.1093/cvr/cvs152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehta D., Malik A.B. Signaling mechanisms regulating endothelial permeability. Physiol Rev. 2006;86(1):279–367. doi: 10.1152/physrev.00012.2005. [DOI] [PubMed] [Google Scholar]
- 29.Abu-Safieh L., Alrashed M., Anazi S., et al. Autozygome-guided exome sequencing in retinal dystrophy patients reveals pathogenetic mutations and novel candidate disease genes. Genome Res. 2013;23(2):236–247. doi: 10.1101/gr.144105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harel T., Yesil G., Bayram Y., et al. Monoallelic and biallelic variants in EMC1 identified in individuals with global developmental delay, hypotonia, scoliosis, and cerebellar atrophy. Am J Hum Genet. 2016;98(3):562–570. doi: 10.1016/j.ajhg.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geetha T.S., Lingappa L., Jain A.R., et al. A novel splice variant in EMC1 is associated with cerebellar atrophy, visual impairment, psychomotor retardation with epilepsy. Mol Genet Genomic Med. 2018;6(2):282–287. doi: 10.1002/mgg3.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cabet S., Lesca G., Labalme A., et al. Novel truncating and missense variants extending the spectrum of EMC1-related phenotypes, causing autism spectrum disorder, severe global development delay and visual impairment. Eur J Med Genet. 2020;63(6) doi: 10.1016/j.ejmg.2020.103897. [DOI] [PubMed] [Google Scholar]
- 33.Marquez J., Criscione J., Charney R.M., et al. Disrupted ER membrane protein complex-mediated topogenesis drives congenital neural crest defects. J Clin Invest. 2020;130(2):813–826. doi: 10.1172/JCI129308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraccaroli A., Pitter B., Taha A.A., et al. Endothelial alpha-parvin controls integrity of developing vasculature and is required for maintenance of cell-cell junctions. Circ Res. 2015;117(1):29–40. doi: 10.1161/CIRCRESAHA.117.305818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L., Zhang S.S., Wang K.F., et al. Overexpression of Twist1 in vascular endothelial cells promotes pathological retinal angiogenesis in mice. Zool Res. 2022;43(1):64–74. doi: 10.24272/j.issn.2095-8137.2021.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun K.X., Jiang X.Y., Li X., et al. Deletion of phosphatidylserine flippase β-subunit Tmem30a in satellite cells leads to delayed skeletal muscle regeneration. Zool Res. 2021;42(5):650–659. doi: 10.24272/j.issn.2095-8137.2021.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Contreras-López O., Moyano T.C., Soto D.C., et al. Step-by-step construction of gene Co-expression networks from high-throughput Arabidopsis RNA sequencing data. Methods Mol Biol. 2018;1761:275–301. doi: 10.1007/978-1-4939-7747-5_21. [DOI] [PubMed] [Google Scholar]
- 38.García-Alcalde F., Okonechnikov K., Carbonell J., et al. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics. 2012;28(20):2678–2679. doi: 10.1093/bioinformatics/bts503. [DOI] [PubMed] [Google Scholar]
- 39.Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mi H., Muruganujan A., Ebert D., et al. PANTHER version 14:more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2018;47(D1):D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selvam S., Kumar T., Fruttiger M. Retinal vasculature development in health and disease. Prog Retin Eye Res. 2018;63:1–19. doi: 10.1016/j.preteyeres.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 45.Ye X., Wang Y., Cahill H., et al. Norrin, frizzled-4, and Lrp5 signaling in endothelial cells controls a genetic program for retinal vascularization. Cell. 2009;139(2):285–298. doi: 10.1016/j.cell.2009.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu X., Yang M., Zhao P., et al. Catenin α 1 mutations cause familial exudative vitreoretinopathy by overactivating Norrin/β-catenin signaling. J Clin Invest. 2021;131(6) doi: 10.1172/JCI139869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park H., Yamamoto H., Mohn L., et al. Integrin-linked kinase controls retinal angiogenesis and is linked to Wnt signaling and exudative vitreoretinopathy. Nat Commun. 2019;10(1):5243. doi: 10.1038/s41467-019-13220-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y., Smallwood P.M., Williams J., et al. A mouse model for kinesin family member 11 (Kif11)-associated familial exudative vitreoretinopathy. Hum Mol Genet. 2020;29(7):1121–1131. doi: 10.1093/hmg/ddaa018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Golan T., Yaniv A., Bafico A., et al. The human Frizzled 6 (HFz6) acts as a negative regulator of the canonical Wnt. beta-catenin signaling cascade. J Biol Chem. 2004;279(15):14879–14888. doi: 10.1074/jbc.M306421200. [DOI] [PubMed] [Google Scholar]
- 50.Nusse R., Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169(6):985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 51.Seo S.H., Yu Y.S., Park S.W., et al. Molecular characterization of FZD4, LRP5, and TSPAN12 in familial exudative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2015;56(9):5143–5151. doi: 10.1167/iovs.14-15680. [DOI] [PubMed] [Google Scholar]
- 52.Chen C., Wang Z., Sun L., et al. Next-generation sequencing in the familial exudative vitreoretinopathy-associated rhegmatogenous retinal detachment. Invest Ophthalmol Vis Sci. 2019;60(7):2659–2666. doi: 10.1167/iovs.19-26619. [DOI] [PubMed] [Google Scholar]
- 53.Tian T., Chen C., Zhang X., et al. Clinical and genetic features of familial exudative vitreoretinopathy with only-unilateral abnormalities in a Chinese cohort. JAMA Ophthalmol. 2019;137(9):1054–1058. doi: 10.1001/jamaophthalmol.2019.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data and materials are available within the article and the supplementary materials.