Abstract
De novo nucleotide biosynthetic pathway is a highly conserved and essential biochemical pathway in almost all organisms. Both purine nucleotides and pyrimidine nucleotides are necessary for cell metabolism and proliferation. Thus, the dysregulation of the de novo nucleotide biosynthetic pathway contributes to the development of many human diseases, such as cancer. It has been shown that many enzymes in this pathway are overactivated in different cancers. In this review, we summarize and update the current knowledge on the de novo nucleotide biosynthetic pathway, regulatory mechanisms, its role in tumorigenesis, and potential targeting opportunities.
Keywords: Biosynthetic pathway, Cancer, de novo synthesis, Metabolism, Nucleotide
Introduction
DNA and RNA are two main types of nucleic acids in cells. Both DNA and RNA are made of nucleotides, each containing a five-carbon sugar backbone, a phosphate group, and a nitrogen base. There are two major pathways to synthesize nucleotides in cells: the de novo nucleotide biosynthetic pathway and the salvage synthesis pathway.1, 2, 3 The salvage synthesis pathway uses exogenous nucleic acid raw materials or endogenous nucleic acid degradation intermediates as substrates. Most proliferating cells synthesize nucleotides de novo. So, we mainly focus on de novo nucleotide biosynthetic pathway here. De novo synthesis of both purine and pyrimidine nucleotides occurs from readily available components. Purine de novo synthesis uses amino acids, carbon dioxide and formyl as raw materials; Pyrimidine de novo synthesis uses glutamine, carbon dioxide, one carbon unit and aspartic acid as raw materials. Liver is the main organ for nucleoside synthesis in vivo.4
To meet the requirement of cell doubling, dividing cells need to replenish nucleotides at the same rate as cell division. Thus, cells need to take nutrients, generate metabolic energy and to drive anabolism, including nucleotide biosynthesis.5 For cancer cells, metabolic adaptations leading to increased synthesis of nucleotides by de novo biosynthesis pathways are emerging as key alterations driving cancer growth. Genes involved in de novo nucleotide biosynthetic pathway are up-regulated in high-speed proliferation cells, such as dividing cells and tumor cells6 (Table 1). Targeting the key enzymes of these pathways has become a novel therapeutic strategy for cancers.7 In this review, we give an overview of de novo nucleotide biosynthetic pathway and how this may lead to potential new approaches to drug development in cancer.
Table 1.
Enzymes of de novo nucleotide biosynthetic pathway overactivated in cancers.
| Gene | Protein | Cancer |
|---|---|---|
| PRPS1/2 | Phosphoribosyl Pyrophosphate Synthetase |
Acute myeloid leukemia Colorectal cancer Burkitt's lymphoma |
| GMPS | GMP synthetase | Esophageal squamous cell carcinoma |
| PPAT | phosphoribosyl pyrophosphate amidotransferase |
Lung cancer |
| CAD | Carbamoyl-Phosphate Synthetase 2 Aspartate Transcarbamylase Dihydroorotase |
Liver cancer Triple-negative breast cancer Glioblastoma |
| DHODH | Dihydroorotate Dehydrogenase | Liver cancer |
| HK-II | Hexokinase | Gallbladder cancer |
| TKT | Transketolase | multiple cancers |
| RPIA | Ribose-5-phosphate isomerase-A | Colorectal cancer |
| MTHFD2 | Methylene-THF dehydrogenase/cyclohydrolase | Breast cancer |
De novo purine and pyrimidine biosynthesis in cells
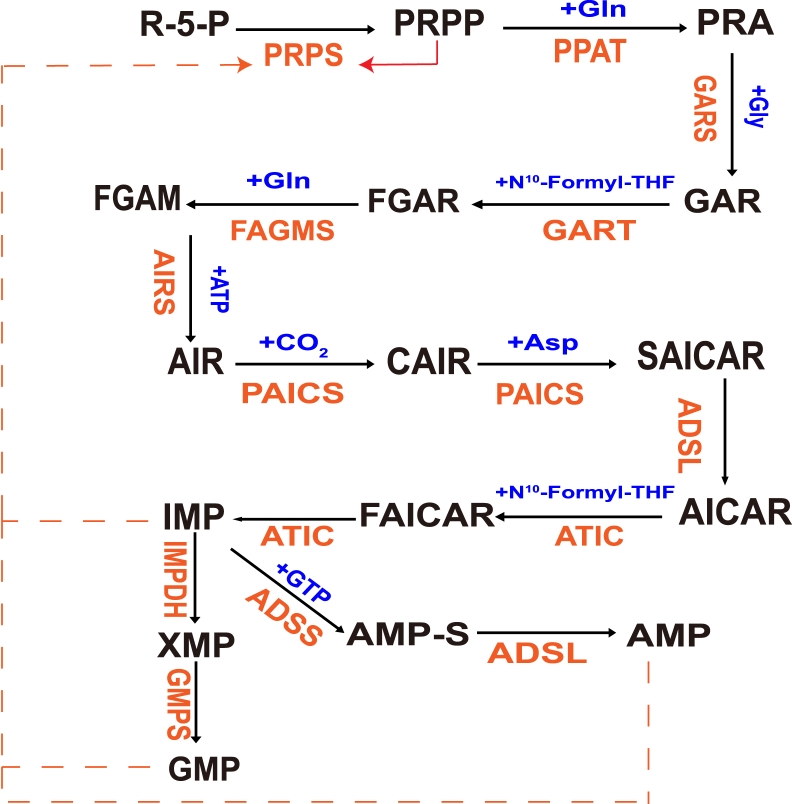
De novo purine biosynthesis (DNPB) is characterized by high conservation and energy intensive. Numerous amino acid substrates and one-carbon units, such as glutamine, ATP and formate, contribute to the 10-step enzymatic process, which keeps the homeostasis of purine to meet the needs of cell proliferation, survival and metabolism in different environments and states. In all higher organisms, de novo purine biosynthesis is catalyzed by six enzymes, which is converted from phosphoribosyl pyrophosphate (PRPP) to inosine monophosphate (IMP) in 10 consecutive steps. Five molecules of ATP, two molecules of glutamine and formate, and one molecule of glycine, aspartate, and carbon dioxide are necessary for generation of one molecule of IMP. IMP is converted into adenosine 5 ′- monophosphate (AMP) or guanosine 5' - monophosphate (GMP) under the action of four enzymes. Therefore, IMP plays an intermediate role in the de novo synthesis pathway. On the other hand, PRPP can bind free purine bases hypoxanthine, adenine and guanine and be transformed into their respective single nucleotides by the rescue enzymes hypoxanthine guanine phosphoribosyltransferase (HGPRT) and adenine phosphoribosyltransferase (APRT).8 DNPB pathway is regulated by metabolites composed of at least 9 enzymes. The synergistic effect of various enzymes increases the pathway flux by about 7 times and gives priority to the distribution of IMP to AMP, which makes the yield of AMP 3–4 times higher than that of GMP. In recent years, some studies have also proposed the concept of purine corpuscles. Purine corpuscles are located at the proximal end of mitochondria and are regarded as “active” DNPB metabolites (Fig. 1). Purine body metabolism is not only of great significance to human health, but also plays an important role in invasive cancer with high purine demand.9
Figure 1.
De novo purine biosynthetic pathway. The de novo purine biosynthetic pathway uses six enzymes to catalyze the transformation of phosphoribosylpyrophosphate (PRPP) into inosine 5′-monophosphate (IMP) via 10 highly conserved step reactions. In the first step, pyrophosphate was replaced by the amino group of glutamine, and the imidazole ring was constructed on phosphoribosyl. After that, glycine, methylchuanyl and amino groups were connected in turn to form 5-aminoimidazole nucleotide (AIR). Then carboxylate to obtain the amino group of aspartic acid, formylate, and finally dehydrate to form IMP. When IMP gets amino group, it is converted into AMP. The amino group comes from aspartic acid, which forms adenylate succinic acid, and then splits to liberate fumaric acid (fumaric acid). The first reaction is catalyzed by adenylate succinate synthase (ADSS) and GTP provides energy. The second reaction is catalyzed by adenylate succinate lyase (ADSL). IMP can be oxidized by hypoxanthine nucleotide dehydrogenase (IMPDH) to produce xanthine, and then catalyzed by guanylate synthase (GMPs) to accept the amino group of glutamine to produce GMP. The remedial pathway of purine nucleotides mainly involves three enzymes: adenosine kinase (ADK), adenine phosphoribosyltransferase (APRT) and hypoxanthine guanine phosphoribosyltransferase (HGPRT). All three enzymes catalyze the production of NMP, but the substrate of ADK is adenosine, which consumes ATP; The substrate of the other two enzymes is the corresponding purine and PRPP is consumed.
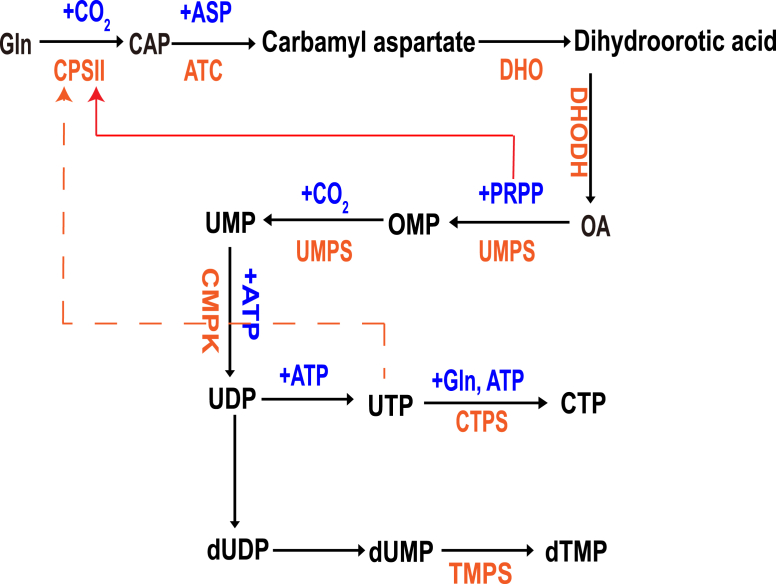
The de novo pyrimidine biosynthesis pathway consists of 6 enzymatic reactions, which are performed by 3 key enzymes: 1-Carbamoyl-Phosphate Synthetase 2, 2-Aspartate Transcarbamylase, and 3-Dihydroorotase (CAD); Dihydroorotate dehydrogenase (quinone) (DHODH); and 1-Orotate Phosphoribosyl Transferase and 2-Orotidine-5′-Decarboxylase/Uridine Monophosphate Synthetase (UMPS).10 Carbamoyl-phosphate synthetase II (CPS II) is the first enzyme in de novo pyrimidine biosynthesis and catalyzes the formation of carbamoyl phosphate from l-glutamine, bicarbonate, and 2 mol of MgATP.11 PRPP boosts CPS II activity while UTP inhibits its activity (Fig. 2). CAD is the rate limiting enzyme which mediates first three enzymatic reactions where glutamine and bicarbonate are sequentially processed into carbamoyl-phosphate, carbonyl-l-aspartate and dihydroorotate.12 DHODH, which localizes in the inner mitochondrial membrane, mediates the oxidation of dihydroorotate to orotate by reducing ubiquinone. In the mitochondria, DHODH also interacts with respiratory complexes II and III to regulate mitochondrial electron transport chain and the oxidative phosphorylation (OXPHOS).13 UMPS is the main enzyme for the 2 last steps of the de novo pyrimidine biosynthetic pathway. UMPS produces orotidine monophosphate (OMP) from orotate and the pentose phosphate phosphoribosyl pyrophosphate (PRPP) through its orotate phosphoribosyl transferase activity (Fig. 2). Uridine monophosphate (UMP) is the product of decarboxylation of OMP. UMP, which is used to synthesize UDP, UTP, dTTP, CTP, and dCTP, is the common metabolite between the de novo and the salvage pyrimidine synthesis pathway.14
Figure 2.
De novo pyrimidine biosynthetic pathway. The first step of pyrimidine synthesis is to produce carbamoyl phosphate. Carbamoyl phosphate synthase Ⅱ (CPS Ⅱ) catalyzes the condensation of CO2 and glutamine. Aspartate carbamoylase (ATCase) catalyzes the condensation of aspartic acid and carbamoyl phosphate to produce carbamoyl aspartate. This reaction is the rate limiting step of pyrimidine synthesis. ATCase is a rate limiting enzyme, which is inhibited by the feedback of the product. It does not consume ATP and is powered by carbamoyl phosphate hydrolysis. Dihydro whey acid with pyrimidine ring was formed by dehydration and intramolecular rearrangement of carbamoyl aspartic acid catalyzed by dihydro whey enzyme. Catalyzed by dihydroorotate reductase, dihydroorotate is oxidized to orotate. This enzyme requires FMN and non heme Fe2+, which is located on the outer side of the mitochondrial inner membrane. Quinones provide oxidation ability. The other five enzymes in pyrimidine synthesis exist in the cytosol. The reaction of whey acid with PRPP was catalyzed by whey acid phosphoribosyltransferase to produce orotidine-5 ′- monophosphate (OMP). It is powered by PRPP hydrolysis. OMP decarboxylase catalyzes OMP decarboxylation to produce UMP. The synthesis of uridine triphosphate (UTP) is similar to that of purine nucleoside triphosphate. Cytidine triphosphate (CTP) is produced by ammonia addition of UTP catalyzed by CTP synthase. In animals, the amino group is provided by glutamine, while in bacteria, it is directly provided by NH3. This reaction consumes 1 ATP.
The regulatory mechanisms of nucleotide de novo synthesis
To maintain homeostasis, dividing cells need to replenish nucleotides at the same rate as cell division. So, the de novo synthesis of purine and pyrimidine is strictly regulated at multiple levels in cells. Here, we will introduce the regulatory mechanisms of de novo nucleotide biosynthetic pathway from the following three aspects: regulation of precursors; regulation of the key enzymes at transcription level; regulation of the rate limiting enzymes at post-translational level.
Regulation of precursors
Regulation of glutamine synthesis
Glutamine is the core nutrient that maintains cell growth and metabolism in both normal cells and tumor cells, and its metabolites can also be used as precursors for nucleotide de novo synthesis.15 Metabolic reprogramming of cancer cells is regulated by multiple oncogenic factors and tumor suppressors.16 MYC plays an important role in glutamine catabolism and glutamine-dependent nucleotide de novo synthesis. MYC regulates glutamine metabolism through multiple mechanisms. MYC promotes glutamine uptake by activating glutamine transporters SLC1A5 and SLC38A5.17 MYC transcriptionally inhibits miR-23a/b to enhance the translation of GLS1 (encodes glutaminase 1, also called GLS), leading to elevated glutaminolysis in cells.18 Moreover, MYC induces active demethylation of the Glutamine synthetase (GS, also termed glutamate-ammonia ligase) promoter and its increased expression through transcriptional upregulation of thymine DNA glycosylase, promoting glutamine synthesis and nucleotide biosynthesis.19 Roles of MYC in glutamine metabolism strictly depend on tumor context. In different tumor types and environments, MYC has different effects on glutamine metabolism.
In addition to MYC, other oncogenes, such as K-RAS, also affects glutamine metabolism and nucleotide de novo synthesis. Overexpression of K-RAS in tumor cells increases the expression of enzymes related to glutamine metabolism, and the absorption of glutamine is significantly enhanced.20 The above studies suggest that oncogenes promote the metabolism of glutamine in tumor cells and provide more raw materials for the synthesis of nucleotides, thereby satisfying the rapid proliferation of tumor cells.
Regulation of nucleotide synthesis by affecting glucose metabolism
The Warburg effect is a hallmark of cancer. Cancer cells prefer to convert most glucose to lactate regardless of whether oxygen is present (aerobic glycolysis).21 Cancer cells use glucose 6-phosphate produced in the glycolysis pathway as raw materials, and use the pentose phosphate pathway (PPP) to produce 5-phosphate ribose for nucleotide synthesis, or use other metabolic intermediates as the raw material for nucleotide synthesis.22 Glycolysis is regulated by many oncogenes, the most common ones are MYC, nuclear factor-κB (NF-κB), protein kinase B (protein kinase B), PKB, also known as AKT and RAS etc.
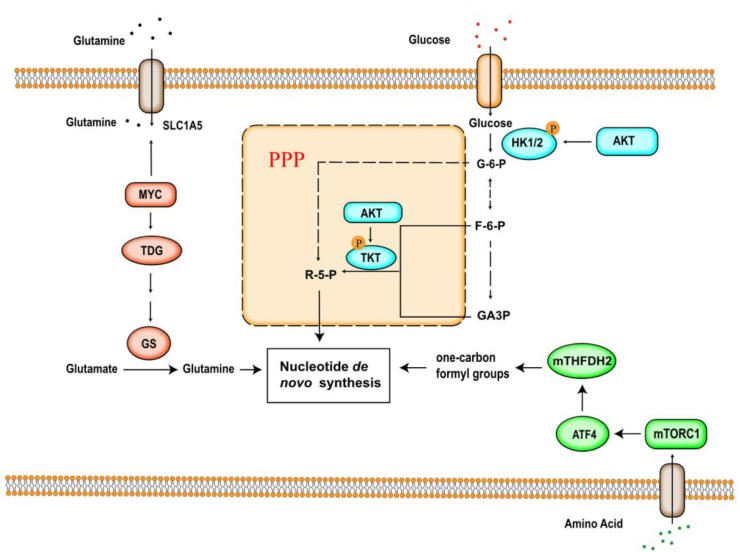
AKT is a serine/threonine protein kinase, which controls a variety of cell activities such as proliferation, autophagy, and metabolism. Among them, AKT plays an extremely important role in the glycolysis. In addition to affecting the expression of GLUT1, AKT also increases the activities of phosphofructokinase-1 (PFK-1) and hexokinase (HK II).23, 24, 25 Both PFK-1 and HK-II are the rate limiting enzymes of glycolysis. The increased activity of PFK-1 and HK-II promotes the efficiency of glycolysis. The intermediate product 3-phosphoglyceric acid is converted into serine after a series of reactions, which further generates glycine, which provides one carbon unit for the nucleotide de novo synthesis(Fig. 3). In addition, AKT activates transketolase (TKT) through phosphorylation of TKT in the non-oxidative pathway of PPP, promotes the synthesis of 5-phosphoribose and PRPP, and increases purine ribonucleotides synthesis.26
Figure 3.
Regulation of precursors in de novo nucleotide biosynthetic pathway. One carbon metabolism is regulated by upregulation of MTHFD2; Glutamine metabolism is regulated by MYC; Glucose metabolism is regulated by AKT through phosphorylating HK1/2 and TKT. All three mechanisms control de novo nucleotide biosynthesis.
Regulation of one carbon metabolism
One carbon metabolism produces one carbon unit, which is important for cellular biosynthesis, methylation, regulation of redox status and amino-acid homeostasis.27 Moreover, one carbon unit is the precursor of the de novo synthesis of nucleotides.28 MTHFD2(mitochondrial tetrahydrofolate cycle enzyme methylenetetrahydrofolate dehydrogenase 2) is the most highly overexpressed metabolic enzyme in human cancers, and one carbon metabolism is regulated by MTHFD2.29 mTOR is a serine/threonine kinase protein kinase which controls cell growth.30 mTORC1 has transcriptional effects on multiple enzymes contributing to purine synthesis, with expression of MTHFD2 being closely associated with mTORC1 signaling in both normal and cancer cells. MTHFD2 expression and purine synthesis were stimulated by activating transcription factor 4 (ATF4), which was activated by mTORC1.31
Regulation of the key enzymes at transcription level
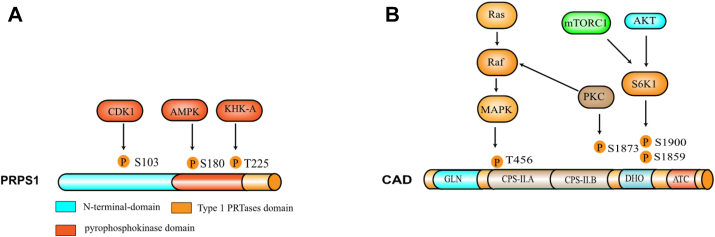
As we described above, MYC regulates nucleotide de novo synthesis by affecting glutamine synthesis and glucose metabolism. In addition, MYC also directly affects the expression of some key enzymes in de novo nucleotide biosynthetic pathway. For example, MYC directly binds the promoter regions of TS, UMPS, IMPDH2 and PRPS2 genes, and regulates these gene expression.32,33 Cunningham et al also reported another mechanism by which MYC regulates PRPS2. Overexpressing MYC in cancer cells enhanced the activity of eukaryotic initiation factor 4E (eIF4E), which binds the pyrimidine-rich translational element (PRTE) on PRPS2 mRNA to promote the translation of PRPS2. In the nucleotide de novo synthesis pathway, PRPS1/2 are the rate limiting enzymes for phosphoric acid pyrophosphate (PRPP) production (Fig. 4A), and IMP, AMP and GMP form the negative feedback loop to inhibit its activity, while PRPP activates PRPS1/2 (Fig. 1). Therefore, the up-regulation of PRPS2 protein expression promotes the synthesis of nucleotides and tumor cell rapid proliferation.34
Figure 4.
Modification sites and structure of CAD and PRPS1. (A) PRPS1 is phosphorylated by the kinases CDK1, AMPK and KHK-A to regulate its activity in cells. CDK1 and KHK-A mediated PRPS1 phosphorylation promotes its activity, and AMPK mediated PRPS1 phosphorylation inhibits its activity. (B) CAD is phosphorylated by the kinases MAPK, S6K1 and PKC to promote its activity in cells.
CAD is another rate-limiting enzyme for the initial three steps in the de novo synthesis of pyrimidines, which is overexpressed in many cancers (Table 1). MYC also directly regulates the expression of CAD. MYC binds MAX to form heterodimers, which controls cell proliferation through activating the downstream target genes. MYC-MAX binds to the E-box sequence of CAD promoter to initiate CAD transcription, thereby increasing the de novo synthesis of pyrimidines35(Fig. 4B).
In addition to MYC, other oncogenes, such as E2F7 and RAS also directly regulate the expression of key enzymes in the nucleotide de novo synthesis pathway. E2F7 can bind to the promoter of ribonucleotide reductase M2 (RRM2) to inhibit the transcription of RRM2, which in turn leads to a decrease in dNTPs in cells, promoting DNA damage and cell senescence.36 RAS can also induce cell senescence by down-regulating the expression of certain enzymes during nucleotide synthesis, such as thymidylate synthase (TS), RRM1 and RRM2.37
Regulation of the rate limiting enzyme by post-translational modification
In addition to transcriptional level regulation, posttranslational modification of key enzymes is also very important for de novo nucleotide biosynthetic pathway. As described above, PRPS1 is a rate-limiting enzyme for purine biosynthesis. The PRPS1 active unit is a hexamer, consisting of three homodimers arranged in a propeller-like shape. Each of the homodimers has an active site and two regulatory allosteric sites. The active site allows for ATP and R-5-P to bind at the junction of two domains within one homodimer. PRPS1 can be powered by ATP, which catalyzes the conversion of R-5-P to phosphoribosyl pyrophosphate (PRPP) and produces the metabolic by-product adenosine monophosphate (AMP).38 PRPS1 is phosphorylated and regulated by several kinases in cells.39 The Ser103 site of PRPS1 is phosphorylated by CDK1(cell cycle-dependent kinase 1), which increases the catalytic activity of PRPS1,40 nucleotide de novo synthesis, and cancer cell proliferation. The Thr225 site of PRPS1 is phosphorylated by fructokinase A, which weakens the inhibition of PRPS1 activity by ADP/GDP and promotes the de novo synthesis of DNA and RNA.41 Moreover, the energy senor in cells, AMPK (AMP-activated protein kinase), directly phosphorylates PRPS1 on Ser180 upon lack of nutrition in cells.42 AMPK-mediated phosphorylation of Ser180 promotes the conversion of PRPS1 from a hexamer to a monomer, decreasing PRPS1 activity and thus attenuating purine synthesis.43
mTOR is a genetically conserved serine/threonine protein kinase, which controls cell growth, proliferation, and survival.44 S6 kinase 1 (S6K1) is a downstream ribosomal protein target of mTORC1 and directly phosphorylates Ser1859 on CAD. mTOR-S6K1 mediated CAD phosphorylation stimulates the first three steps of the de novo pyrimidine synthesis and thus helps to advance the cells overall progression through S phase of the cell cycle.45 In resting cells, CAD is localized mainly in the cytoplasm where it carries out pyrimidine synthesis. As proliferating cells enter S phase, MAP Kinase (Erk1/2) phosphorylates CAD at Thr456, resulting in CAD translocation to the nucleus.46 MAPK mediated CAD phosphorylation attenuates the inhibition of CAD activity by UTP and increases the sensitivity of CAD to substrates.47 In addition, protein kinase C phosphorylates CAD at Ser1873, boosts its activity, and promotes pyrimidine synthesis, cell growth and proliferation.48
De novo nucleotide synthesis and cancer
In cancer cells, nucleotide production greatly increases because of the huge demand of ribosomal RNA, which synthesizes DNA and maintains the transcriptome during the rapid cell proliferation.49 Nucleotide also functions as an important downstream of major nutrient pathways and chemotherapeutic target of almost all types of cancer.50 Many enzymes of the de novo nucleotide biosynthetic pathway are upregulated or overactivated in cancers (Table 1).51, 52, 53, 54, 55 For example, PRPS1 exerts a pro-tumorigenic role in colorectal cancer. During the S phase of the cell cycle, the Ser103 site of PRPS1 is phosphorylated by CDK1, which increases PRPS1 activity and enhances the de novo synthesis of purines. This mechanism eventually promotes the colon carcinogenesis. In relapsed childhood acute lymphoblastic leukemia, investigators have performed whole-exome sequencing of samples obtained at diagnosis, remission and relapse and identified multiple relapse-specific mutations in PRPS1, especially the mutation of A190T, T303S, K176N and N144S. These recurrence-specific mutations in PRPS1 enhance drug resistance in relapsed childhood acute lymphoblastic leukemia. The underlying mechanism is that these sites are located at the dimeric interface of PRPS1, predominantly near the allosteric sites of PRPS1. PRPS1 mutants cause defects in nucleotide feedback inhibition of their activities, which will enhance PRPS1 enzyme activity, the de novo synthesis of purines and eventually lead to drug resistance and recurrence of tumors. Several inhibitors of purine synthesis have been used to conduct clinical trials, including the inhibitor of GART-Lometrexol. Treatment of cancers expressing PRPS1 A190T and S103T with Lometrexol attenuated purine synthesis in tumor cells, and may relief PRPS1 mutation-driven chemotherapeutic drugs resistance in relapsed ALL.56
Overactivation of de novo synthesis of pyrimidine is a very common occurrence in cancers (Table 1). For example, the increased de novo synthesis of pyrimidine promotes glioblastoma progression. EGFR-MAPK/ERK axis mediates phosphorylation of CAD by ERK, enhances de novo synthesis of pyrimidines and promotes glioblastoma giant cell growth.57 More and more evidence suggests that CAD or DHODH inhibitor may serve as a potential cancer treatment strategy.58 Triple-negative breast cancer (TNBC) is considered an aggressive cancer because it grows quickly, is poorly treated with chemotherapy and prone to recurrence.59 TNBC is susceptible to resistance to adriamycin, a kind of chemotherapeutic drug. The main mechanism is that adriamycin promotes the phosphorylation of CAD at Thr456, which enhances intracellular de novo synthesis of pyrimidine.60 Treatment of TNBC with PALA, an inhibitor of CAD, significantly increased cellular sensitivity to adriamycin, and administration of an inhibitor of DHODH also has the same effect. On the other hand, DHODH inhibitors also inhibit the oxidative phosphorylation (OXPHOS) and mitochondrial function, because DHODH is localized in the mitochondria, linked to the respiratory chain via the coenzyme Q pool.61 But current DHODH inhibitors lack specificity and show mixed results in cancer trials. Specific DHODH inhibitors and new combination treatments might improve efficacy.
Perspectives and conclusions
In addition to being the basic unit of RNA and DNA, nucleotide is also involved in all major aspects of metabolism, and the importance of this is reflected in the strict regulation of their intracellular levels. ATP is the primary carrier of energy in cells, which is used for the transfer of energy from energy-yielding reactions to energy-requiring processes,62 and GTP is of essential for protein synthesis. Nucleotides can react as activated intermediates, which are involved in the synthesis of carbohydrates and lipids and are the components of major coenzymes.63 Nucleotides can also act as metabolic signal molecules, such as AMP. Maintenance of nucleotide levels is therefore fundamental to cellular functions. Unsurprisingly, dysregulation of de novo nucleotide biosynthetic pathway is linked to cancer and other human diseases.64
Although the basic de novo nucleotide biosynthetic pathway is well established, the detailed regulatory network for modulating nucleotide biosynthesis in physiological and pathological conditions remain unclear, particularly in terms of dependence on cell type and cancer context. Existing regulatory mechanisms are just the tip of the iceberg. This is in part due to the lack of powerful tools for investigating the actual paths from nutrient precursors through various feeder pathways to de novo synthesized nucleotides. Characterization of protein expression and modification in the pathway can provide more insights, but this need to be further investigated by animal model in vivo and clinical samples. Stable isotope tracers in conjunction with the state-of-the-art metabolomics and clinical proteomics will provide strong support for such functional studies.
Although there is still a long way to apply abnormal nucleic acid metabolism as a drug target in the clinical treatment of malignant tumors, some drugs, such as DHODH inhibitors, especially leflunomide, as an immunosuppressant, has been widely used in the clinical treatment of rheumatic diseases with good safety. Teriflunomide, the active metabolite of leflunomide, is used to treat adriamycin resistant breast cancer. Although it can significantly increase the sensitivity of breast cancer cells to adriamycin, current DHODH inhibitors lack specificity and show mixed results in cancer trials. Specific DHODH inhibitors and new combination treatments might improve efficacy. The change of metabolic pathway is often accompanied by the change of the whole cell metabolism, the side effect, especially the effects on other metabolic pathways and other cell signal pathways are not clear. The relationship between targeted drugs for abnormal nucleic acid metabolism and other antitumor drugs needs to be further studied, so as to provide a new scheme for the treatment of multi-target tumors. Due to the metabolic heterogeneity in cancer, the nucleic acid metabolism in different tumors is different, and the adaptability of tumor cells to metabolic changes is also needed to be considered. In the future we expect the systematic research about dysregulations of nucleotide metabolism in cancer pathogenesis, such understanding is expected to have important diagnostic and therapeutic benefits in relevant human diseases such as cancer.
Conflict of interests
Authors declare no competing interests.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Yi Yao, Email: yaoyi2018@whu.edu.cn.
Huadong Pei, Email: huadong.pei@georgetown.edu.
References
- 1.Thierry D., Timothy L.B., Yuen Y.H., et al. De novo purine synthesis inhibition and antileukemic effects of mercaptopurine alone or in combination with methotrexate in vivo. Blood. 2002;100(4):1240–1247. doi: 10.1182/blood-2002-02-0495. [DOI] [PubMed] [Google Scholar]
- 2.Siddiqui A., Ceppi P. A non-proliferative role of pyrimidine metabolism in cancer. Mol Metabol. 2020;35(7):100962. doi: 10.1016/j.molmet.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincenzetti S., Polzonetti V., Micozzi D., Pucciarelli S. Enzymology of pyrimidine metabolism and neurodegeneration. Curr Med Chem. 2016;23(14):1408–1431. doi: 10.2174/0929867323666160411125803. [DOI] [PubMed] [Google Scholar]
- 4.Pareek V., Tian H., Winograd N., Benkovic S.J. Metabolomics and mass spectrometry imaging reveal channeled de novo purine synthesis in cells. Science. 2020;368(29):283–287. doi: 10.1126/science.aaz6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J., Thompson C.B. Metabolic regulation of cell growth and proliferation. Nat Rev Mol Cell Biol. 2019;20(7):436–450. doi: 10.1038/s41580-019-0123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin J., Ren W., Huang X., Deng J., Li T., Yin Y. Potential mechanisms connecting purine metabolism and cancer therapy. Front Immunol. 2018;9(30):1697. doi: 10.3389/fimmu.2018.01697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villa E., Ali E.S., Sahu U., Ben-Sahra I. Cancer cells tune the signaling pathways to empower de novo synthesis of nucleotides. Cancers (Basel) 2019;11(5):688. doi: 10.3390/cancers11050688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allsop J., Watts R. Purine de novo synthesis and salvage during testicular development in the rat. Pediatr Res. 1985;19(3):744. doi: 10.1007/978-1-4684-1248-2_52. [DOI] [PubMed] [Google Scholar]
- 9.Robinson A.D., Eich M.L., Varambally S. Dysregulation of de novo nucleotide biosynthetic pathway enzymes in cancer and targeting opportunities. Cancer Lett. 2020;470(3):134–140. doi: 10.1016/j.canlet.2019.11.013. [DOI] [PubMed] [Google Scholar]
- 10.Lane A.N., Fan T.W. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015;43(4):2466–2485. doi: 10.1093/nar/gkv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simmer J.P., Kelly R.E., Rinker A., et al. Mammalian carbamyl phosphate synthetase (CPS). DNA sequence and evolution of the CPS domain of the Syrian hamster multifunctional protein CAD. J Biol Chem. 1990;265(18):10395–10402. [PubMed] [Google Scholar]
- 12.Li G., Li D., Wang T., He S. Pyrimidine biosynthetic enzyme CAD: its function, regulation, and diagnostic potential. Int J Mol Sci. 2021;22(19):10253. doi: 10.3390/ijms221910253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bajzikova M., Kovarova J., Coelho A.R., et al. Reactivation of dihydroorotate dehydrogenase-driven pyrimidine biosynthesis restores tumor growth of respiration-deficient cancer cells. Cell Metabol. 2019;29(2):399–416. doi: 10.1016/j.cmet.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loffler M., Fairbanks L.D., Zameitat E., Marinaki A.M., Simmonds H.A. Pyrimidine pathways in health and disease. Trends Mol Med. 2005;11(5):430–437. doi: 10.1016/j.molmed.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Aaron M.H., Vivian C.H., Laura V.D., et al. Amino acids rather than glucose account for the majority of cell mass in proliferating mammalian cells. Dev Cell. 2016;36(5):540–549. doi: 10.1016/j.devcel.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boroughs L.K., DeBerardinis R.J. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17(4):351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao X.A., Petrashen A.P., Sanders J.A., Peterson A.L., Sedivy J.M. SLC1A5 glutamine transporter is a target of MYC and mediates reduced mTORC1 signaling and increased fatty acid oxidation in long-lived Myc hypomorphic mice. Aging Cell. 2019;18(3) doi: 10.1111/acel.12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao P., Irina T., Chang T.C., et al. c-Myc suppression of miR-23 enhances mitochondrial glutaminase and glutamine metabolism. Cancer Res. 2009;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alex J.B., Peng I.C., Fan Y.J., et al. Oncogenic myc induces expression of glutamine synthetase through promoter demethylation. Cell Metabol. 2015;22(6):1068–1077. doi: 10.1016/j.cmet.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernfeld E., Foster D.A. Glutamine as an essential amino acid for KRas-driven cancer cells. Trends Endocrin Met. 2019;30(16):357–368. doi: 10.1016/j.tem.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pei Y.Z., Qian Y.J., Wang H., Tan L. Epigenetic regulation of ferroptosis-associated genes and its implication in cancer therapy. Front Oncol. 2022;12(23):112–117. doi: 10.3389/fonc.2022.771870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng J., Li J.J., Wu L.W., et al. Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39(1):126. doi: 10.1186/s13046-020-01629-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robey R.B., Hay N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene. 2006;25(7):4683–4696. doi: 10.1038/sj.onc.1209595. [DOI] [PubMed] [Google Scholar]
- 25.Wiernan H.L., Wofford J.A., Rathmell J.C. Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Mol Biol Cell. 2007;18(4):1437–1446. doi: 10.1091/mbc.E06-07-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saha A., Connelly S., Jiang J.J., et al. Akt phosphorylation and regulation of transketolase is a nodal point for amino acid control of purine synthesis. Mol Cell. 2014;55(2):264–276. doi: 10.1016/j.molcel.2014.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luciano-Mateo F., Hernandez-Aguilera A., Cabre N., et al. Nutrients in energy and one-carbon metabolism: learning from metformin users. Nutrients. 2017;9(2):121. doi: 10.3390/nu9020121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrea R., Alessandra N., Francesca R., Antonella T., Marina B., Delia M. One-carbon metabolism: biological players in epithelial ovarian cancer. Int J Mol Sci. 2018;19(7):2092. doi: 10.3390/ijms19072092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nilsson R., Jain M., Madhusudhan N., et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014;5(17):3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169(2):361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 31.Issam B.S., Gerta H., Stephane J.H.R., John M.A., Brendan D.M. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351(6274):728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsieh A.L., Walton Z.E., Altman B.J., Stine Z.E., Dang C.V. MYC and metabolism on the path to cancer. Semin Cell Dev Biol. 2015;43(18):11–21. doi: 10.1016/j.semcdb.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mannava S., Grachtchouk V., Wheeler L.J., et al. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle. 2008;7(15):2392–2400. doi: 10.4161/cc.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cunningham J.T., Moreno M.V., Lodi A., Ronen S.M., Ruggero D. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell. 2014;157(18):1088–1103. doi: 10.1016/j.cell.2014.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyd K.E., Farnham P.J. Myc versus USF: discrimination at the cad gene is determined by core promoter elements. Mol Cell Biol. 1997;17(5):2529–2537. doi: 10.1128/mcb.17.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carvajal L.A., Hamard P.J., Tonnessen C., Manfredi J.J. E2F7, a novel target, is up-regulated by p53 and mediates DNA damage-dependent transcriptional repression. Gene Dev. 2012;26(14):1533–1545. doi: 10.1101/gad.184911.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang P., Du W.J., Wu M.A. Regulation of the pentose phosphate pathway in cancer. Protein Cell. 2014;5(18):592–602. doi: 10.1007/s13238-014-0082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S., Lu Y., Peng B., Ding J. Crystal structure of human phosphoribosylpyrophosphate synthetase 1 reveals a novel allosteric site. Biochem J. 2007;401(1):39–47. doi: 10.1042/BJ20061066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian X., Li X., Tan L., et al. Conversion of PRPS hexamer to monomer by AMPK-mediated phosphorylation inhibits nucleotide synthesis in response to energy stress. Cancer Discov. 2018;8(1):94–107. doi: 10.1158/2159-8290.CD-17-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jing X., Wang X.J., Zhang T., et al. Cell-cycle-dependent phosphorylation of PRPS1 fuels nucleotide synthesis and promotes tumorigenesis. Cancer Res. 2019;79(28):4650–4664. doi: 10.1158/0008-5472.CAN-18-2486. [DOI] [PubMed] [Google Scholar]
- 41.Li X., Qian X., Peng L.X., et al. A splicing switch from ketohexokinase-C to ketohexokinase-A drives hepatocellular carcinoma formation. Nat Cell Biol. 2016;18(5):561–571. doi: 10.1038/ncb3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X.J., Qian X., Lu Z.M. Fructokinase A acts as a protein kinase to promote nucleotide synthesis. Cell Cycle. 2016;15(20):2689–2690. doi: 10.1080/15384101.2016.1204861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aoki T., Weber G. Carbamoyl phosphate synthetase (Glutamine-Hydrolyzing) - increased activity in cancer-cells. Science. 1981;212(13):463–465. doi: 10.1126/science.7209543. [DOI] [PubMed] [Google Scholar]
- 44.Wullschleger S., Loewith R., Hall M.N. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 45.Issam B.S., Jessica J.H., John M.A., Brendan D.M. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339(6125):1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sigoillot F.D., Kotsis D.H., Serre V., Sigoillot S.M., Evans D.R., Guy H.I. Nuclear localization and mitogen-activated protein kinase phosphorylation of the multifunctional protein CAD. J Biol Chem. 2005;280(10):25611–25620. doi: 10.1074/jbc.M504581200. [DOI] [PubMed] [Google Scholar]
- 47.Sigoillot F.D., Berkowski J.A., Sigoillot S.M., Kotsis D.H., Guy H.I. Cell cycle-dependent regulation of pyrimidine biosynthesis. J Biol Chem. 2003;278(34):3403–3409. doi: 10.1074/jbc.M211078200. [DOI] [PubMed] [Google Scholar]
- 48.Sigoillot F.D., Kotsis D.H., Masko E.M., Bame M., Evans D.R., Evans H.I. Protein kinase C modulates the up-regulation of the pyrimidine biosynthetic complex, CAD, by MAP kinase. Front Biosci. 2007;12(4):3892–3898. doi: 10.2741/2358. [DOI] [PubMed] [Google Scholar]
- 49.Katherine M.A., Zhang G., Li H., et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Rep. 2013;3(25):1252–1265. doi: 10.1016/j.celrep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chon J., Stover P.J., Field M.S. Targeting nuclear thymidylate biosynthesis. Mol Aspect Med. 2017;53(16):48–56. doi: 10.1016/j.mam.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moloy T.G., Chen G.A., Balabhadrapatruni V.S.K.C., et al. Role and regulation of coordinately expressed de novo purine biosynthetic enzymes PPAT and PAICS in lung cancer. Oncotarget. 2015;6(27):23445–23461. doi: 10.18632/oncotarget.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ridder D., Schindeldecker M., Berndt K., et al. Key enzymes in pyrimidine synthesis, CAD and CPS1, predict prognosis in hepatocellular carcinoma. Cancers. 2021;59(1):1722–1727. doi: 10.3390/cancers13040744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen J.N., Yu Y., Li H., et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol Cancer. 2019;18(33):1–16. doi: 10.1186/s12943-019-0947-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Qiu Z.P., Guo W.J., Wang Q.F., et al. MicroRNA-124 reduces the pentose phosphate pathway and proliferation by targeting PRPS1 and RPIA mRNAs in human colorectal cancer cells. Gastroenterol. 2015;14(10):1587–1598. doi: 10.1053/j.gastro.2015.07.050. [DOI] [PubMed] [Google Scholar]
- 55.Zhang X., Li F., Zhou Y., et al. Long non-coding RNA AFAP1-AS1 promotes tumor progression and invasion by regulating the miR-2110/Sp1 axis in triple negative breast cancer. Oncol. 2020;11(10):101–132. doi: 10.1038/s41419-021-03917-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li B., Li H., Bai Y., et al. Negative feedback-defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL. Nat Med. 2015;21(6):563–571. doi: 10.1038/nm.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joseph T.M., Armand B., Christine R.C., Hollis D.S., Nouri N. Revisiting the role of dihydroorotate dehydrogenase as a therapeutic target for cancer. Pharmacol Ther. 2019;195(6):111–131. doi: 10.1016/j.pharmthera.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 58.Wang X., Yang K., Wu Q., et al. Targeting pyrimidine synthesis accentuates molecular therapy response in glioblastoma stem cells. Sci Transl Med. 2019;11(504):4972. doi: 10.1126/scitranslmed.aau4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patricia C., Zhang L., Michael U., et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384(9938):164–172. doi: 10.1016/S0140-6736(13)62422-8. [DOI] [PubMed] [Google Scholar]
- 60.Brown K.K., Spinelli J.B., Asara J.M., Toker A. Adaptive reprogramming of de novo pyrimidine synthesis is a metabolic vulnerability in triple-negative breast cancer. Cancer Discov. 2017;7(4):391–399. doi: 10.1158/2159-8290.CD-16-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He Y.C., Cao J., Chen J.W., Pan D.Y., Zhou Y.K. Influence of methionine/valine-depleted enteral nutrition on nucleic acid and protein metabolism in tumor-bearing rats. World J Gastroenterol. 2003;9(4):145–148. doi: 10.3748/wjg.v9.i4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vultaggio-Poma V., Sarti A.C., Di Virgilio F. Extracellular ATP: a feasible target for cancer therapy. Cells. 2020;9(11):2496. doi: 10.3390/cells9112496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Naffouje R., Grover P., Yu H., et al. Anti-tumor potential of IMP dehydrogenase inhibitors: a century-long story. Cancers (Basel) 2019;11(9):1346. doi: 10.3390/cancers11091346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koch J., Mayr J.A., Alhaddad B., et al. CAD mutations and uridine-responsive epileptic encephalopathy. Brain. 2017;140(2):279–286. doi: 10.1093/brain/aww300. [DOI] [PubMed] [Google Scholar]