Abstract
Objective
This study aimed to update the genetic diversity of Rotavirus (RV) infections in children under five years old in Beijing, China.
Methods
A 5-year active hospital-based surveillance for sporadic acute gastroenteritis (AGE) from January 2018 to December 2022 in the capital of China was performed. A total of 748 fecal samples from AGE patients were collected for followed by RV antigen detection by ELSIA, RNA detection by reverse transcription PCR, G/P genotyping and phylogenetic analyzing.
Results
RV antigen was detected in 11.0% of the collected samples, with 54 samples confirmed to be RV RNA positive. G9 and G8 genotypes were identified in 43 (79.6%) and 7 (13.0%) samples, respectively, all of which were allocated to P[8]. The predominant G/P combination was G9P[8] (79.6%), following by G8P[8] (13.0%), G4P[8] (5.6%) and G3P[8] (1.9%). A significant change in G/P-type distribution was observed, with the G9P[8] being predominant from 2018 to 2021, followed by the emergence of an uncommon G8P[8] genotype, which was first reported in 2021 and became predominant in 2022. Blast analysis showed that one G1 isolate had a high similarity of 99.66% on nucleotide acid with RotaTeq vaccine strain with only one amino acid difference L150V. Additionally, one P[8] isolate was clustered into a branch together with RotaTeq vaccine strain G6P[8].
Conclusions
The study reveals that G8P[8] has become the predominant genotype in pediatric outpatients in China for the first time, indicating a significant change in the composition of RV genetic diversity. The importance of RVA genotyping in surveillance is emphasized, as it provides the basis for new vaccine application and future vaccine efficacy evaluation.
Keywords: Rotavirus, Surveillance, China, G8P[8], G9P[8]
1. Introduction
Group A rotaviruses (RVAs) are a significant cause of severe acute diarrhea among children under 5 years of age worldwide [1]. The WHO prequalification and introduction of oral RVA vaccines (ORV) in many countries over the last decade have significantly reduced the global burden of the disease [2]. However, the effectiveness of RVA vaccines has been higher in high-income countries (80%–90%) than in low-to-middle income countries (40%–70%) [3], and there is still much work to be done in reducing the disease's global impact. Furthermore, a long-term impact of RVA vaccines on genetic diversity of circulating RVAs has been observed in many neighboring countries of China, including Korea, Japan, Thailand, and Russia [[4], [5], [6], [7], [8]].
In 2001, the Lanzhou Lamb RV vaccine (LLR) became the only domestic RV vaccine licensed in China [9,10] with more than 83 million doses of LLR lot-released as of October 2019 [11]. At the end of 2018, the MSD RotaTeq vaccine (G1-G4, G9, P [5]) was also licensed in China. In addition, two novel RVA vaccines, LLR3 (G2-G4) and pentavalent (G1-G4, G8-G9), finished phase III clinical trials in China [[11], [12], [13]]. Coincidently, in the past two decades, the predominant G genotype of RVAs have undergone two major changes in China, including G1 to G3 around 2001 and G3 to G9 after 2012 [14,15], while the P genotype remains unchanged as P [8,16]. Unusual G8 genotype have emerged and rapidly spread in several countries [17], but it has been reported only in a limited number of cities in the south of China, including Kunming, Shanghai, Suzhou, Guangzhou and Fuzhou [[18], [19], [20], [21]]. As of our study, there were no reports of G8P [8] RVs in northern cities of China.
In Beijing, the capital of China, we established a hospital-based surveillance network for sporadic AGE since 2011. The common diarrhea-related viral pathogens including RV, Sapovirus (SaV), Norovirus (NoV), Astrovirus (AstV) and Enteric Adenovirus (AdV) were routinely monitored. The previous surveillance data in our lab from 2011 to 2017 showed that RVA was the main pathogen causing viral diarrhea in infants and young children in Beijing (25.8%, 374/1451) [22], but unfortunately there was no details about genetic diversity. The other recent surveillance demonstrated that G9P[8] was still most predominant till to 2019, followed by G3P[8], G2P[4], G1P[8], G2P[4], and G4P[6] [[23], [24], [25]].
With the emergence of uncommon genotypes found around the world post the vaccination era, we continued the active surveillance along with genotyping analysis in Beijing from 2018 to 2022, in order to update the genetic diversity of RVA infections in children under five years old in Beijing, China, and provide the basis for measuring vaccine efficacy in the near future.
2. Methods
2.1. Subjects enrollments
Briefly, from January 2018 to December 2022, outpatient children younger than 5 years, who met the case definition of diarrhea according to the National Surveillance Protocol for Viral Diarrhea (2007 version), were enrolled in Beijing Pediatric Research Institute. The inclusion criteria for the study subjects are as follows: patients with gastrointestinal symptoms such as diarrhea and vomiting as their primary complaint, diagnosed within 5 days of onset, without considering clinical diagnosis. Patients who have had ≥3 episodes of diarrhea within 24 h and abnormal stool consistency (such as watery stool, mucous stool, or bloody stool, etc.) are included, while cases of diarrhea caused by the use of antibiotics or inappropriate intake of chemicals, etc. are excluded. The study is part of the hospital-based surveillance network for sporadic AGE in Beijing since 2011. It complies with the Declaration of Helsinki and was performed in accordance with Good Laboratory Practice (GLP). Prior to study initiation, the protocol, informed consent form from the guardians of the enrolled outpatient children and other study-related documents were approved by the ethics committee of Chaoyang District Center for Disease Control and Prevention, Beijing.
2.2. Sample collection, preparation and viral pathogen detection
About 15 specimens were collected every month, except for 2020 owing to lack of outpatients in the circumvent of COVID-19 pandemic. Stool samples were collected three days after the onset of diarrhea. The samples were suspended with 10% phosphate salt buffer (Hyclone), and centrifuged at 5000 g for 5 min. Viral RNA was extracted from 200 μl of the supernatant by a QIAamp Viral RNA Mini Kit (Qiagen), in accordance with the manufacturer's instructions. All samples were tested for RV by using ProSpecT Rotavirus Kit (OXOID, UK), for NoV, SaV, Enteric AdV, AstV by commercial real-time RT-PCR kits (DAAN GENE, China) in accordance with the manufacturers' instructions respectively.
2.3. RV G/P genotyping and genetic analysis
For RV-positive samples, partial G and P genes were amplified by multiplex semi-nested reverse transcription PCR (RT-PCR) (the primers listed in Table 1) according to the previous report [15]. The complete coding genes of VP7 were amplified by using the primer pair VP7-F: GGCTTTAAAAGAGAGAATTTCCGTCTGG and VP7-R: GGTCACATCATACAATTCTAATCTAAG. The sequences obtained were assembled by Vector NTI Advance 11.0. Edited sequences were analyzed and genotyped via the online tool - Rotavirus A Genotype Determination (https://www.viprbrc.org/brc/rvaGenotyper.spg). The similarity analysis was performed by nucleotide and protein BLAST (Basic Local Alignment Search Tool) which provided by the National Center for Biotechnology Information (National Institutes of Health, USA). Phylogenetic trees of the viral genome were constructed by using the neighbor-joining method with bootstrap analysis of 1000 replicates in MEGA 6.0. Bootstrap values estimated with 1000 replicate data sets were indicated at each node. The scale bar indicates the number of nucleotide substitutions per site. The reference sequences was downloaded from GenBank (National Institutes of Health, USA) and listed in Supplementary Table 1.
Table 1.
Molecular weight of amplification products and primer sequences for rotavirus VP4 (P) and VP7 (G) genotyping by semi-nested RT-PCR.
| Genotype | Primer | Squence (5’→3′) | Molecular weight (bp) |
|---|---|---|---|
| VP7 (G) 1st round | VP7F | ATGAATGTATTGAATATCCAC | |
| VP7 (G) 1st round | VP7R | AACATGGCACCATTTTTCC | |
| VP7(G) | VP7R | AACATGGCACCATTTTTCC | |
| G1 | aBT1 | CAAAGTACTCAATCAATATGG | 618 |
| G2 | aCT2 | CATGATATTAACCATTTACTGTG | 521 |
| G3 | G3 | ACCAACTCACACTAGAGG | 682 |
| G4 | aDT4 | CGATTCTGGTGACGAGTTG | 452 |
| G8 | aAT8 | GTCCCACCATATGTAATTCG | 754 |
| G9 | G9 | CTTATGTGCTATAATTTAC | 179 |
| VP4 (G) 1st round | VP4F | TATGCTCCAGTAAATTGG | |
| VP4 (G) 1st round | VP4R | ATTGCAATTCTTTGCATATG | |
| VP4(G) | VP4F | TATGCTCCAGTAAATTGG | |
| P[4] | 2T-1 | CTTTGTTAGAAGGTAGAGTC | 362 |
| P[6] | 3T-1 | TGTGGATTAGTTGGCTCAA | 146 |
| P[8] | 1T-1D | TCCACTGGRTAAACCTGC | 224 |
| P[9] | 4T-1 | TGACACATGCAATAGGAC | 270 |
| P[10] | 5T-1 | ATCAAAGTTAGTCGTCGG | 462 |
| P[11] | P[11] | GTAACATCCCAGACTGTG | 191 |
3. Results
3.1. AGE-related viral pathogens detected among children under 5 years in the capital of China (2018–2022)
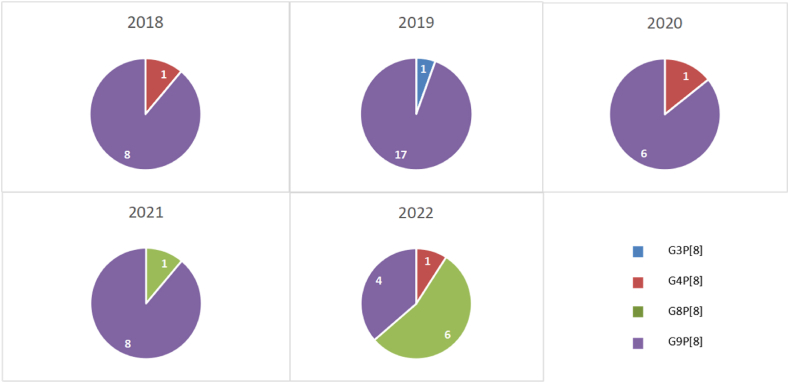
A total of 748 fecal samples from AGE patients were collected. RVA antigen was detected in 11.0% of the collected samples, while NoV, Enteric AdV, AstV and SaV RNAs were detected in 15.6%, 10.4%, 3.3%, and 2.8%, respectively. Furthermore, 54 samples confirmed to be RV RNA positive. Overall, forty-three (79.6%) and 7 (13.0%) allocated to G9 and G8 genotypes respectively, all of which allocated to P [8] genotype. The predominant G/P combination was G9P[8] (79.6%), following byG8P[8] (13.0%), G1P [8] (3.7%), G4P [8] (1.9%) and G3P [8] (1.9%). From the view of time, G9P[8] was still the predominant genotype from 2018 to 2021, but the uncommonG8P[8] was first reported with a detection rate of 11.1% (1/10) in 2021 and increased rapidly to be the predominant genotype in 2022 with a detection rate of 54.5% (6/11) (Fig. 1).
Fig. 1.
Annual distribution of RVAs with different genotypes among children under five years old in Beijing, China from 2018 to 2022.
3.2. Genetic diversity of RV among children under 5 years in the capital of China (2018–2022)
In generally, several RVA isolates showed the highest homology with the current used RotaTeq vaccine, while the others were highly similar to domestic isolates or isolates from neighboring countries in the same period. The detail information of referenced sequences was supplied in the Supplementary Table 1.
3.2.1. G1 genotype
One strain of G1 RVA isolated in 2022 (in-house No.: 22051027) had a 99.66% nucleotide similarity with RotaTeq-WI79-9/1992/G1P7[5] VP7 gene (GenBank Accession No.: GU565057) on VP7 gene through Blast, with only one amino acid difference (L150V) between them. The other G1 RVA isolated in 2018 (in-house No.: 18051012) had the highest similarity 99.66% with VP7 gene of JL18-1009 G1P [8] (GenBank Accession No.: OM920777) which was also isolated in 2018 in China from human stool sample.
3.2.2 G3 genotype
The only G3 RVA isolated in 2019 (in-house No.: 19051030) had the 100% similarity with VP7 gene of G3P8 (GenBank Accession No.: MW331238) which was isolated in 2018 in Thailand, while it had a similarity of 93.19% with RotaTeq-WI78-8/1992/G3P7 [5] VP7 gene (GenBank Accession No.: GU565079).
3.2.3. G4 genotype
The only G4 RVA isolated in 2020 (in-house No.: 20051063) had the highest similarity (91.49%) with RotaTeq-BrB-9/1996/G4P7[5] VP7 gene (GenBank Accession No.: GU565090), and no more than 90% similarity with the other isolates recorded in GenBank.
3.2.4. G8 genotype
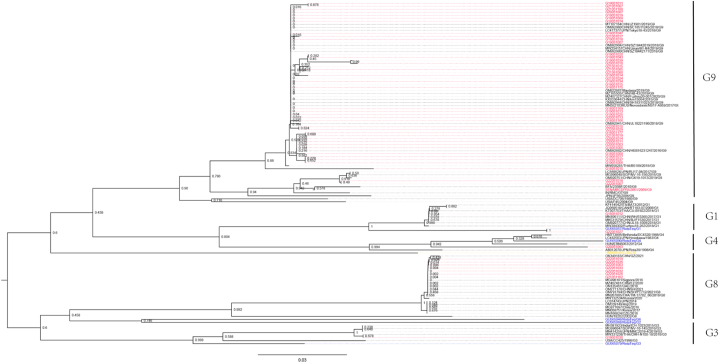
The seven G8 RVA isolated in this study showed a wide nucleotide similarity range from 67.6% to 99.8%, which can be classified into three groups. Group 1 contains five isolates (in-house No.: G22051026, G22051033, G22051019, G22051036, G21051162), and their homology is between 93.1% and 99.8%. The groups 2 and 3 contain only one virus respectively (in-house No.: G22051032 for group 2 and G22051003 for group 3). The homology between group 2 and group 1 comes from 80.5% to 86.5%, while the homology between group 3 and 1 was as low as 76.0%–82.9%. To be noted, the homology of group 2 and group 3 was only 67.6%. Seven G8 isolates from Beijing has only 46.9%–79.9% homology with isolates from southern China during 2020 and 2021 (Guangzhou, Fuzhou and Shanghai) (GenBank Accession Nos.: OK349183, MZ407481, OM777170). Among Beijing isolates, group 1 has 94.3%–98.8% homology with Thailand isolate in 2018 (GenBank Accession No.: MN207885), group 2 has 94.9% homology with Russia isolate in 2020 (GenBank Accession No.: MW132534), and group 3 has 81.7% homology with Thailand isolate in 2018 (GenBank Accession No.: MN207885). Phylogenetically, all the G8 isolates clustered to a branch together with domestic Guangzhou isolates, rather than Fuzhou and Shanghai isolates (Fig. 2).
Fig. 2.
Phylogenetic analysis based on partial VP7 genes. The tree was constructed by using the neighbor-joining method with bootstrap analysis of 1000 replicates in MEGA 6.0. Bootstrap values estimated with 1000 replicate data sets were indicated at each node. The scale bar indicates the number of nucleotide substitutions per site. The sequences displayed in red, blue, yellow and black fonts are from this study, RotaTeq vaccine (G1P[5], G2P[5], G3P[5], G4P[5] and G6P[8]), LLR vaccine (G10P[15]), and other reference sequences downloaded from GenBank, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2.5. G9 genotype
The forty-three G9 RVA isolated in this study also showed a wide nucleotide similarity range from 65.1% to 100%, which can be classified into four groups. Thirty-eight isolates located in group 1 and their homology is between 88.5% and 100%. The group 2 contains only one isolate (G18051015). The group 3 contains two isolates (G22051010 and G22051029), and the group 4 also contains two isolates (G22051018 and G22051007). The similarities between the two are 83.2%–88.9% (group 1 and 2), 64.7%–71.9% (group 1 and 3), 70.4%–77.2% (group 1 and 4), 58.7% (group 2 and 3), 65.6% (group 2 and 4), and 73.0% (group 3 and 4). Through blast, isolates in group 1, 2, 3, and 4 showed nearly 100%, ∼90%, ∼95% and ∼98% homology respectively with those isolated in Japan and different cities of China. The above similarity analysis was further confirmed by the phylogenetic tree based on partial VP7 (Fig. 2). Although G9 isolates can be divided into three groups based on genetic similarity which also supported by the phylogenetic tree, all G9 isolates located in the large branch together with domestic isolates or isolates from neighboring countries including Russia, Japan, Malaysia, Thailand, Singapore, and India, instead of the USA isolates.
3.2.6. P genotype
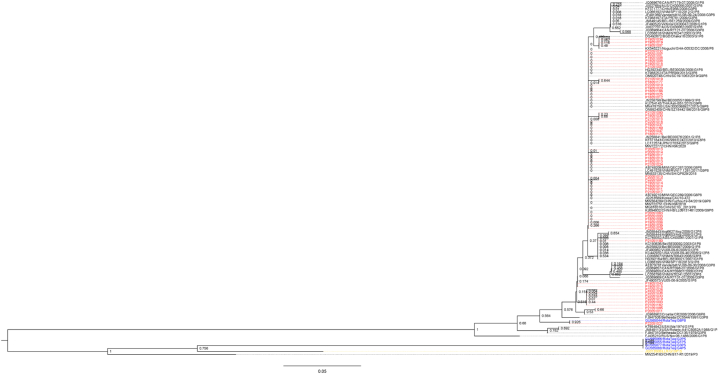
The VP4 gene sequences of 54 P [8] RVA isolated in this study were very conservative, with more than 95% homology between each other. In addition, they all showed more than 99% homology with isolates in domestic and neighbor countries through blast. To be noted, one isolate (in-house No.: P22051027) showed 99.47% similarity with RotaTeq-WI79-4/1992/G6P1A[8] VP4 gene (GenBank Accession No.: GU565044). Phylogenetically, all the other VP4 sequences of P[8] strains were clustered into lineage III referenced to the genetic analysis of Ghanaian G1P[8] and G9P[8] RVA strains [26], except for one isolate was clustered into a branch together with RotaTeq vaccine G6P[8] (Fig. 3).
Fig. 3.
Phylogenetic analysis based on partial VP4 genes. The tree was constructed by using the neighbor-joining method with bootstrap analysis of 1000 replicates in MEGA 6.0. Bootstrap values estimated with 1000 replicate data sets were indicated at each node. The scale bar indicates the number of nucleotide substitutions per site. The sequences displayed in red, blue, yellow and black fonts are from this study, RotaTeq vaccine (G1P[5], G2P[5], G3P[5], G4P[5] and G6P[8]), LLR vaccine (G10P[15]), and other reference sequences downloaded from GenBank, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
This is the first report of G8 RVA in northern China, which has been reported in southern China since 2015 [18]. This is also the first time that the G/P combination G8P[8] has taken the lead in China, indicating that the composition of RVA genetic diversity is undergoing a significant change, following G1 to G3 around 2001 and G3 to G9 after 2012 [14,25]. Therefore, in order to investigate the changes of RVAs genetic diversity and to provide the basis for new vaccine application as well as a baseline for future vaccine efficacy evaluation, it is very necessary to carry out extensive surveillance in other regions of China.
Our findings are consistent with other domestic reports. In China, the most common RVA strains in children under 5-years old were G1P [8] and G2P [4] during the period of 2000–2011 [27]. In the neighbor countries, the most prevalent G-P combination was G9P[8] from 2011 to 2018 [28,29], exceptG8P[8] in Korea [6]. Other studies also showed that the proportion of G9P[8] was reported to increase remarkably from 3.4% in 2009 to 60.9% in 2015 [30], even to up to 76.61%in Shandong province, China from July 2017 to June 2018 [31]. The limitation of our studies is that it is not yet clear whether this change occurred in the vaccinated, unvaccinated or mixed populations. Therefore, the clinical information and vaccination status of infected patients should be collected and further analyzed to evaluate the efficacy of RVA vaccines and their effects on the changes of RVA genotype as previous studies [6,29]. In addition, although the COVID-19 pandemic led to hesitation in routine infant vaccination uptake [32,33], the cases of RVGE have sharply decreased since early 2020 attributed to the physical distancing and mask wearing measures compared to our study performed in the pre-COVID-19 era [34]. Similarly, a nation-wide observational study also showed a profound change of the activity of enteric pathogens and a great reduction of the positive rates for almost all enteric viruses among acute diarrhea patients [35]. Therefore, how the RV genotype changes in the post-COVID-19 era is still an unknown event, which needs continuous monitoring.
The change of RVA genotypes may be part of the reason for the lack cross-protection against G9P[8] andG8P[8], which prompts development of novel RVA vaccines which cover newly emerged RVA genotypes. RVA vaccine was introduced into the second-category list of the national immunization program in China, which is not compulsory. Currently two RVA vaccines are available. The first one is the live monovalent oral Lanzhou lamb RVA vaccine (LLR) (G10P [12]) manufactured by the Lanzhou Institute of Biological Products and licensed early in 2000 in China [36]. The second is pentavalent RotaTeq (RV5; MSD) containing five reassortant RVAs (G1, G2, G3, G4, and P [8]) derived from human and bovine parent strains, which completed phase III clinical trial in Guangxi province, China [37] and has been approved in April 2018. Therefore, in the RVA vaccine development pipeline candidate vaccines including additional G9 strain or providing cross-protection against G9 strain should be developed to ensure protection coverage on circulating G9 strains. These candidate vaccines were a combination of the US National Institutes of Health (NIH) UK-Compton bovine RV vaccine (UK-BRV) and reassortants including components for G8 (strain 1290) and/or G9 (strain AU32). The former UK-BRV is a multivalent bovine-human RV reassortant vaccine comprised of the G6P [5] bovine rotavirus backbone with the VP7 genes from the common human RV strains incorporated as reassortants into the vaccine strains [38]. In China, non-exclusive licenses for the development and production of the UK-BRV vaccine were granted to Chengdu Institute of Biological Products and Wuhan Institute of Biological Products [39]. Searched from Chinese Clinical Trial Registry website, the hexavalent RVA vaccine manufactured by Wuhan Institute of Biological Products started the phase I clinical trial from July 2016, and the phase III clinical trial was expected to be finished in December 2021. The results in phase I clinical trial showed that the novel oral hexavalent RVA vaccine was generally well-tolerated in all adults, toddlers and infants, and the vaccine was immunogenic in infants, with higher anti-RV IgA antibody geometric mean concentrations (GMC) and seroconversion rate in the vaccine group than in the placebo group post-3rd dose immunization [12]. In addition, another novel human-lamb trivalent live vaccine LLR3 (containing G2, G3 and G3 strains) also showed dramatic cross-protection against severe RVGE induced by genotype G9 with a 70.3% (95%CI, 59.9–77.9%) vaccine efficacy at the end of the second epidemic season, in a randomized, double-blind, placebo-controlled multicenter phase III clinical trial among healthy children aged 6–13 weeks [11]. Besides, several inactivated and recombinant VP8 candidate vaccines have also been under developing [[40], [41], [42]] and should further verify the efficacy in human beings. In addition, the emergence of uncommonG8P[8] genotype will be another challenge to the broad and cross protection of vaccines.
Moreover, the shedding of vaccine strains was recorded. Vaccine virus shedding is a double-edged sword [43]. On the positive side, the occurrence of vaccine virus shedding not only indicates the effective replication of live RV in the intestine which is the basis for inducing effective immune response against the infection, but also realizes the potential herd immunity through the transmission of vaccine strains from vaccinated individuals to susceptible person. But the downside is the risk of vaccine derived infection in immunocompromised contacts. Here, we reported the first detection of RotaTeq vaccine strain from RVGE patients after it was imported to China. As reported, for live RV vaccines, vaccine-virus shedding occurs in approximately 5%–10% of the recipients after the first dose and very rarely thereafter [12,44,45]. With the rapid increasing lot-release of RotaTeq vaccine imported to China, the shedding of vaccine virus strains should be continuously monitored.
5. Conclusions
In summary, our study indicates that the third genotype shift of the predominant RV strains from G9P[8] toG8P[8] was occurred after 2021 among children younger than 5 years in the capital of China. It gives an early warning of RVA strains that could escape immune protection, which needs a continuous long-term monitoring of RVA genotype diversity around of China. Moreover, it also proposes a demand for the development of novel RV vaccines that are highly effective against newly emerged genotypes.
Author contribution statement
Yang Jiao: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Taoli Han; Xiao Qi; Yan Gao; Jianhong Zhao: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Yue Zhang; Beibei Li: Contributed reagents, materials, analysis tools or data.
Zheng Zhang: Analyzed and interpreted the data; Wrote the paper.
Jialiang Du; Lingli Sun: Conceived and designed the experiments; Wrote the paper.
Funding statement
This work was supported by the National Major Science & Technology Project (2019ZX09732002) and the National Key R&D Program of China (2022YFE0102400) sponsored by the Ministry of Science and Technology of the People’s Republic of China, as well as the Beijing Key Specialty Program for Major Epidemic Prevention and Control (2021), the Beijing Chaoyang District Science and Technology Project (CYSF2206), and the Special Scientific Research Project for Capital Health Development (2022-1G-4231).
Data availability statement
Data will be made available on request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e18236.
Contributor Information
Jialiang Du, Email: dujialiang@nifdc.org.cn.
Lingli Sun, Email: cycdpcsun@126.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Omatola C.A., Olaniran A.O. Genetic heterogeneity of group A rotaviruses: a review of the evolutionary dynamics and implication on vaccination. Expert Rev. Anti Infect. Ther. 2022;20:1587–1602. doi: 10.1080/14787210.2022.2139239. [DOI] [PubMed] [Google Scholar]
- 2.Omatola C.A., Olaniran A.O. Rotaviruses: from pathogenesis to disease control - a critical review. Viruses. 2022;14:875. doi: 10.3390/v14050875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cates J.E., Tate J.E., Parashar U. Rotavirus vaccines: progress and new developments. Expet Opin. Biol. Ther. 2022;22:423–432. doi: 10.1080/14712598.2021.1977279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yodmeeklin A., Khamrin P., Kumthip K., Malasao R., Ukarapol N., Ushijima H., Maneekarn N. Increasing predominance of G8P[8] species A rotaviruses in children admitted to hospital with acute gastroenteritis in Thailand, 2010-2013. Arch. Virol. 2018;163:2165–2178. doi: 10.1007/s00705-018-3848-0. [DOI] [PubMed] [Google Scholar]
- 5.Yuzhakov A., Yuzhakova K., Kulikova N., Kisteneva L., Cherepushkin S., Smetanina S., Bazarova M., Syroeshkin A., Grebennikova T. Prevalence and genetic diversity of group A rotavirus genotypes in moscow. Pathogens. 2021;10(674):2019–2020. doi: 10.3390/pathogens10060674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim K.G., Kee H.Y., Park H.J., Chung J.K., Kim T.S., Kim M.J. The long-term impact of rotavirus vaccines in Korea, 2008-2020; emergence of G8P[8] strain. Vaccines (Basel) 2021;9:406. doi: 10.3390/vaccines9040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okitsu S., Khamrin P., Hikita T., Thongprachum A., Pham N.T.K., Hoque S.A., Hayakawa S., Maneekarn N., Ushijima H. Changing distribution of rotavirus A genotypes circulating in Japanese children with acute gastroenteritis in outpatient clinic, 2014-2020. J. Infect. Public Health. 2022;15:816–825. doi: 10.1016/j.jiph.2022.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Kozawa K., Higashimoto Y., Kawamura Y., Miura H., Negishi T., Hattori F., Ihira M., Komoto S., Taniguchi K., Yoshikawa T. Rotavirus genotypes and clinical outcome of natural infection based on vaccination status in the post-vaccine era. Hum. Vaccines Immunother. 2022;18 doi: 10.1080/21645515.2022.2037983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohan K.V., Kulkarni S., Glass R.I., Zhisheng B., Atreya C.D. A human vaccine strain of lamb rotavirus (Chinese) NSP4 gene: complete nucleotide sequence and phylogenetic analyses. Virus Gene. 2003;26:185e92. doi: 10.1023/a:1023491514820. [DOI] [PubMed] [Google Scholar]
- 10.Soares-Weiser K., Bergman H., Henschke N., Pitan F., Cunliffe N. Vaccines for preventing rotavirus diarrhoea: vaccines in use. Cochrane Database Syst. Rev. 2019;2019:CD008521. doi: 10.1002/14651858.CD008521.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia S., Du J., Su J., Liu Y., Huang L., Yu Q., Xie Z., Gao J., Xu B., Gao X., Guo T., Liu Y., Zhou X., Yang H. Efficacy, immunogenicity and safety of a trivalent live human-lamb reassortant rotavirus vaccine (LLR3) in healthy Chinese infants: a randomized, double-blind, placebo-controlled trial. Vaccine. 2020;38:7393–7400. doi: 10.1016/j.vaccine.2020.04.038. [DOI] [PubMed] [Google Scholar]
- 12.Wu Z.W., Li Q.L., Zhou H.S., Duan K., Gao Z., Zhang X.J., Jiang Z.J., Hao Z.Y., Jin F., Bai X., Li Q., Xu G.L., Zhao Y.L., Yang X.M. Safety and immunogenicity of a novel oral hexavalent rotavirus vaccine: a phase I clinical trial. Hum. Vaccines Immunother. 2021;17:2311–2318. doi: 10.1080/21645515.2020.1861874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Z., Li Q., Liu Y., Lv H., Mo Z., Li F., Yu Q., Jin F., Chen W., Zhang Y., Huang T., Hu X., Xia W., Gao J., Zhou H., Bai X., Liu Y., Liang Z., Jiang Z., Chen Y.…Yang X. Efficacy, safety and immunogenicity of hexavalent rotavirus vaccine in Chinese infants. Virol. Sin. 2022;37:724–730. doi: 10.1016/j.virs.2022.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou X., Wang Y.H., Pang B.B., Chen N., Kobayashi N. Surveillance of human rotavirus in Wuhan, China (2011-2019): predominance of G9P[8] and emergence of G12. Pathogens. 2020;9:810. doi: 10.3390/pathogens9100810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian Y., Chughtai A.A., Gao Z., Yan H., Chen Y., Liu B., Huo D., Jia L., Wang Q., MacIntyre C.R. Prevalence and genotypes of group A rotavirus among outpatient children under five years old with diarrhea in Beijing, China, 2011-2016. BMC Infect. Dis. 2018;18:497. doi: 10.1186/s12879-018-3411-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S., Yin J., Yang J., Tian L., Li D., Zhang Q., Chen J., Xu W., Zhou X. Epidemiology and genetic diversity of group A rotavirus in acute diarrhea patients in pre-vaccination era in southwest China. J. Med. Virol. 2017;89:71–78. doi: 10.1002/jmv.24606. [DOI] [PubMed] [Google Scholar]
- 17.Phan T., Kobayashi M., Nagasawa K., Hatazawa R., Thi Kim Pham N., Miyashita H., Komoto S., Tajima T., Baba T., Okitsu S., Khamrin P., Maneekarn N., Kimura H., Kobayashi T., Hayakawa S., Ushijima H. Whole genome sequencing and evolutionary analysis of G8P[8] rotaviruses emerging in Japan. Virusdisease. 2022;33:215–218. doi: 10.1007/s13337-022-00765-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang Y., Cai Y. Epidemiology and genetic diversity of rotavirus in kunming, China, in 2015. Intervirology. 2018;61:9–13. doi: 10.1159/000489309. [DOI] [PubMed] [Google Scholar]
- 19.Wang S.J., Chen L.N., Wang S.M., Zhou H.L., Qiu C., Jiang B., Qiu T.Y., Chen S.L., von Seidlein L., Wang X.Y. Genetic characterization of two G8P[8] rotavirus strains isolated in Guangzhou, China, in 2020/21: evidence of genome reassortment. BMC Infect. Dis. 2022;22:579. doi: 10.1186/s12879-022-07542-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Y., Xie H., Wang D., Lu J. Nosocomial infection caused by a rare G8P[8] rotavirus subtype in a pediatric unit in Guangzhou, Southern China. Hum. Vaccines Immunother. 2021;17:3619–3622. doi: 10.1080/21645515.2021.1920771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen S., Ren S., Chen L., Xue J., Shao X., Zhang T., Zhao G. Rotavirus infection in children <5 Years of age in Suzhou, China, 2013-2019: disease burden, genotype distribution and seasonality. Pediatr. Infect. Dis. J. 2022;41:375–380. doi: 10.1097/INF.0000000000003463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiao Y., Qi X., Gao Y., Zhang S.Y., Zhang Y.S., Gu L., Sun L.L. Surveillance of enteric viral pathogens among infants with diarrhea in Chaoyang district of Beijing from 2011 to 2017. Chin. J. Viral Dis. 2018;8:275–281. doi: 10.16505/j.2095-0136.2018.0048. [DOI] [Google Scholar]
- 23.Mao T., Wang M., Wang J., Ma Y., Liu X., Wang M., Sun X., Li L., Li H., Zhang Q., Li D., Duan Z. Phylogenetic analysis of the viral proteins VP4/VP7 of circulating human rotavirus strains in China from 2016 to 2019 and comparison of their antigenic epitopes with those of vaccine strains. Front. Cell. Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.927490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X., Wang M., Li S., Li J., Xiao J., Li H., Zhang Q., Kong X., Wang H., Li D., Duan Z. Genomic and evolutionary characteristics of G9P[8], the dominant group a rotavirus in China (2016-2018) Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.997957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian Y., Gao Z., Li W., Liu B., Chen Y., Jia L., Yan H., Wang Q. Group A rotavirus prevalence and genotypes among adult outpatients with diarrhea in Beijing, China, 2011-2018. J. Med. Virol. 2021;93:6191–6199. doi: 10.1002/jmv.27100. [DOI] [PubMed] [Google Scholar]
- 26.Damanka S.A., Agbemabiese C.A., Dennis F.E., Lartey B.L., Adiku T.K., Enweronu-Laryea C.C., Armah G.E. Genetic analysis of Ghanaian G1P[8] and G9P[8] rotavirus A strains reveals the impact of P[8] VP4 gene polymorphism on P-genotyping. PLoS One. 2019;14 doi: 10.1371/journal.pone.0218790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., Wang H., Li D., Zhang Q., Liu N. Infection status and circulating strains of rotaviruses in Chinese children younger than 5-years old from 2011 to 2018: systematic review and meta-analysis. Hum. Vaccines Immunother. 2021;17:1811–1817. doi: 10.1080/21645515.2020.1849519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lestari F.B., Vongpunsawad S., Wanlapakorn N., Poovorawan Y. Rotavirus infection in children in Southeast Asia 2008-2018: disease burden, genotype distribution, seasonality, and vaccination. J. Biomed. Sci. 2020;27:66. doi: 10.1186/s12929-020-00649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawata K., Hoque S.A., Nishimura S., Yagyu F., Islam M.T., Sharmin L.S., Pham N., Onda-Shimizu Y., Quang T.D., Takanashi S., Okitsu S., Khamrin P., Maneekarn N., Hayakawa S., Ushijima H. Role of rotavirus vaccination on G9P[8] rotavirus strain during a seasonal outbreak in Japan. Hum. Vaccines Immunother. 2021;17:3613–3618. doi: 10.1080/21645515.2021.1925060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J., Lai S., Geng Q., Ye C., Zhang Z., Zheng Y., Wang L., Duan Z., Zhang J., Wu S., Parashar U., Yang W., Liao Q., Li Z. Prevalence of rotavirus and rapid changes in circulating rotavirus strains among children with acute diarrhea in China, 2009-2015. J. Infect. 2019;78:66–74. doi: 10.1016/j.jinf.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong S., Huang D., Wang Z., Zhang G., Zhang F., Sai L. Clinical and molecular epidemiological characterization of rotavirus infections in children under five years old in Shandong province, China. Arch. Virol. 2021;166:2479–2486. doi: 10.1007/s00705-021-05161-4. [DOI] [PubMed] [Google Scholar]
- 32.Dib F., Mayaud P., Chauvin P., Launay O. Online mis/disinformation and vaccine hesitancy in the era of COVID-19: why we need an eHealth literacy revolution. Hum. Vaccines Immunother. 2022;18:1–3. doi: 10.1080/21645515.2021.1874218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacDonald S.E., Paudel Y.R., Kiely M., Rafferty E., Sadarangani M., Robinson J.L., Driedger S.M., Svenson L.W., COVImm study team. Impact of the COVID-19 pandemic on vaccine coverage for early childhood vaccines in Alberta, Canada: a population-based retrospective cohort study. BMJ Open. 2022;12 doi: 10.1136/bmjopen-2021-055968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiao Y., Qi X., Han T.L., Gao Y., Zhang Y., Zhao J.H., Sun L.L. Tudy on the genetic characteristics of enteric viral pathogens of sporadic adult diarrhea in Chaoyang district, Beijing in 2019. Chin. J. Prev. Med. 2021;55(12):1404–1409. doi: 10.3760/cma.j.cn112150-20210224-00182. [DOI] [PubMed] [Google Scholar]
- 35.Wang L.P., Han J.Y., Zhou S.X., Yu L.J., Lu Q.B., Zhang X.A., Zhang H.Y., Ren X., Zhang C.H., Wang Y.F., Lin S.H., Xu Q., Jiang B.G., Lv C.L., Chen J.J., Li C.J., Li Z.J., Yang Y., Liu W., Fang L.Q.… Chinese centers for disease control and prevention (CDC) etiology of diarrhea surveillance study team. The changing pattern of enteric pathogen infections in China during the COVID-19 pandemic: a nation-wide observational study. Lancet Reg. Health West. Pac. 2021;16 doi: 10.1016/j.lanwpc.2021.100268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu C., He Q., Xu J., Xie H., Ding P., Hu W., Dong Z., Liu X., Wang M. Effectiveness of the Lanzhou lamb rotavirus vaccine against gastroenteritis among children. Vaccine. 2012;31:154–158. doi: 10.1016/j.vaccine.2012.10.078. [DOI] [PubMed] [Google Scholar]
- 37.Mo Z., Mo Y., Li M., Tao J., Yang X., Kong J., Wei D., Fu B., Liao X., Chu J., Qiu Y., Hille D.A., Nelson M., Kaplan S.S. Efficacy and safety of a pentavalent live human-bovine reassortant rotavirus vaccine (RV5) in healthy Chinese infants: a randomized, double-blind, placebo-controlled trial. Vaccine. 2017;35:5897–5904. doi: 10.1016/j.vaccine.2017.08.081. [DOI] [PubMed] [Google Scholar]
- 38.Vetter V., Gardner R.C., Debrus S., Benninghoff B., Pereira P. Established and new rotavirus vaccines: a comprehensive review for healthcare professionals. Hum. Vaccines Immunother. 2022;18 doi: 10.1080/21645515.2020.1870395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirkwood C.D., Ma L.F., Carey M.E., Steele A.D. The rotavirus vaccine development pipeline. Vaccine. 2019;37:7328–7335. doi: 10.1016/j.vaccine.2017.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y., Xue M., Yu L., Luo G., Yang H., Jia L., Zeng Y., Li T., Ge S., Xia N. Expression and characterization of a novel truncated rotavirus VP4 for the development of a recombinant rotavirus vaccine. Vaccine. 2018;36:2086–2092. doi: 10.1016/j.vaccine.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 41.Liu C., Huang P., Zhao D., Xia M., Zhong W., Jiang X., Tan M. Effects of rotavirus NSP4 protein on the immune response and protection of the SR69A-VP8* nanoparticle rotavirus vaccine. Vaccine. 2021;39:263–271. doi: 10.1016/j.vaccine.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y., Hu X., Chen R., Wu J., Lin X., Lu C., Yin N., Tang Y., Shi P., Song Z., Zhao Y., Sun M., Li H. Impact of maternal and pre-existing antibodies on immunogenicity of inactivated rotavirus vaccines. Vaccine. 2022;40:3843–3850. doi: 10.1016/j.vaccine.2022.05.036. [DOI] [PubMed] [Google Scholar]
- 43.Anderson E.J. Rotavirus vaccines: viral shedding and risk of transmission. Lancet Infect. Dis. 2008;8:642–649. doi: 10.1016/S1473-3099(08)70231-7. [DOI] [PubMed] [Google Scholar]
- 44.Kanchan V., Zaman K., Aziz A.B., Zaman S.F., Zaman F., Haque W., Khanam M., Karim M.M., Kale S., Ali S.K., et al. A randomized Phase I/II study to evaluate safety and reactogenicity of a heat-stable rotavirus vaccine in healthy adults followed by evaluation of the safety, reactogenicity, and immunogenicity in infants. Hum. Vaccines Immunother. 2020;16:693–702. doi: 10.1080/21645515.2019.1664239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vesikari T., Matson D.O., Dennehy P., Van Damme P., Santosham M., Rodriguez Z., Dallas M.J., Heyse J.F., Goveia M.G., Black S.B., et al. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N. Engl. J. Med. 2006;354:23–33. doi: 10.1056/NEJMoa052664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.