Abstract
Post-translational modifications (PTM) are covalent modifications of proteins or peptides caused by proteolytic cleavage or the attachment of moieties to one or more amino acids. PTMs play essential roles in biological function and regulation and have been linked with several diseases. Modifications of protein acylation (Kac), a type of PTM, are known to induce epigenetic regulatory processes that promote various diseases. Thus, an increasing number of studies focusing on acylation modifications are being undertaken. Butyrylation (Kbu) is a new acylation process found in animals and plants. Kbu has been recently linked to the onset and progression of several diseases, such as cancer, cardiovascular diseases, diabetes, and vascular dementia. Moreover, the mode of action of certain drugs used in the treatment of lymphoma and colon cancer is based on the regulation of butyrylation levels, suggesting that butyrylation may play a therapeutic role in these diseases. In addition, butyrylation is also commonly involved in rice gene expression and thus plays an important role in the growth, development, and metabolism of rice. The tools and analytical methods that could be utilized for the prediction and detection of lysine butyrylation have also been investigated. This study reviews the potential role of histone Kbu, as well as the mechanisms underlying this process. It also summarizes various enzymes and analytical methods associated with Kbu, with the goal of providing new insights into the role of Kbu in gene regulation and diseases.
Keywords: Butyrylation, Gene regulation, Histone, Post-translational modification, Target treatment
Introduction
Post-translational modification (PTM) refers to the covalent modification of proteins or peptides via proteolytic cleavage or the addition of moieties to one or more amino acids. PTM changes the complexity of proteins or peptides in terms of activity state, subcellular localization, turnover, and interactions with other cellular molecules.1 PTM may reversibly regulate the conformation of proteins via peptides, thereby affecting protein function.2 PTMs are classified based on the type of chemical reactions involved, which may result in final products such as newly added chemical components, double bonds, and cross-linked products, as well as the hydrolysis and pyrolysis of proteins.3,4 A common PTM in proteins involves the addition of small or large chemical parts to a side chain.4 Hydroxylation, glycosylation, methylation, acylation, and phosphorylation remain the most common PTMs studied.5,6 PTMs play essential roles in biological function and regulation.7 For example, glycosylation impacts many biological processes, such as cell adhesion, proliferation, and differentiation,8 while methylation is a dynamic process that affects a variety of biological activities, including DNA damage repair, cell growth, metabolism, and signal transduction.9 Phosphorylation potently regulates innate inflammatory responses via the activation, cellular translocation, and interaction of innate receptors, adaptors, and downstream signaling molecules in response to infectious and harmful signals.10
Previous studies have indicated that PTMs are associated with several diseases. Crotonylation (Kcr) of lysine residues plays a role in psychiatric disorders, while the reduction of histone crotonylation is involved in regulating stress-induced depression. Crotonylation of lysine 27 on histone H3 (H3K27cr) is elevated in Alzheimer's disease, providing insights into the epigenetic role of crotonylation in Alzheimer's disease.11, 12, 13 Methylation of lysine residues in histones H3 and H4 promotes transcriptional activity. Histone methylation is associated with pancreatic cancer progression and related methyltransferase and demethylase inhibitors may be used to treat pancreatic cancer.14,15 Protein glycosylation is an essential PTM that occurs mainly in the Golgi apparatus and endoplasmic reticulum (ER).16 O-ligation-β-N-acetylglucosaminylation (O-GlcNAcylation) is a reversible dynamic modification that occurs primarily in the nucleus, cytoplasm, and mitochondria compared with conventional glycosylation. Studies have indicated that O-GlcNAcylation may potentially ameliorate cancer, diabetes, and neurodegenerative diseases.17, 18, 19, 20 Moreover, PTMs play a role not only in regulating human diseases but also in plants.21, 22, 23, 24 Different PTMs that are related to each other and involved in dynamic transcriptional reprogramming play a role in fruit ripening.25 PTMs alter the behavior of proteins, improving the function of proteomes. Lysine malonylation, a recently discovered PTM, plays an important role in cell metabolism.26 Malonyl modification is speculated to be regulated by lysine deacetylase and lysine acetyltransferase. Previous studies have shown that prokaryotic and eukaryotic histones displaying malonyl modifications may be involved in regulating gene transcription. In addition to histones, many other proteins found in mitochondria, chloroplasts, cytoplasm, and nuclei undergo malonyl modifications.27, 28, 29 Protein acylation (Kac), the post-translational and covalent attachment of acyl groups to proteins, is a widespread protein modification process. All acyl groups in cells, from acetate to modified long-chain fatty acids, can be activated and covalently bonded to different amino acid side chains to modulate protein activity.20 As acylation modifications are known to induce epigenetic regulatory processes that promote various diseases, an increasing number of studies have begun to focus on acylation modifications, leading to investigations not only into the association between each modification and disease, but also into methods for discovering new types and sites of protein PTMs, using analytical methods such as HPLC/MS/MS.
Recently, Zhao et al discovered butyrylation (Kbu), a newly reported acylation process found in animals and plants; Kbu is a PTM that involves both histone and non-histone proteins.30 Butyrylation refers to a biochemical interaction wherein the butyryl group covalently modifies the amino acid, lysine. Histone acetyltransferases (HATs) utilize acetyl-CoA as a coenzyme to catalyze the N-acetylation of lysine residues in histones H3 and H4. Moreover, short-chain fatty acids, such as butyrate, are involved in the development of human diseases, including vascular dementia, diabetes, psychiatric disorders, and male epigenome features, while several plant-based substances have also been potentially linked to Kbu of histones.31, 32, 33 Several regulatory enzymes have been identified as playing a crucial role in Kbu modification. Thus, Kbu may generally be considered a novel PTM that contributes to disease.
In this review, we provide a comprehensive summary of the recent findings on the role of Kbu in diseases. Furthermore, we discuss the association between plants and Kbu. Research techniques aimed at elucidating the Kbu process are constantly being refined and will likely serve as the foundation for future research. Moreover, several writers, erasers, and readers of Kbu that alter the regulatory role of Kbu in histones, have been identified. Drugs used to treat cancer are reportedly capable of altering the frequency of Kbu in histones. The current review may contribute to a better understanding of the role of Kbu in various diseases and the underlying mechanisms involved.
Enzymes in Kbu
Kbu writers
Enzymes that create histone marks are referred to as histone writers. Three major HAT families have been identified in metazoans: p300/CREB-binding protein (p300/CBP), MYST (Moz, Ybf2, Sas2, Tip60, HBO1, and hMOF), and GCN5-related N-acetyltransferase (GNAT).34 Chen et al found that p300 and CBP may be used as butyryl-modifying enzymes, causing histone H4 to undergo both propionylation and butyrylation. Since CBP and p300 are acetyltransferases, Chen et al30 incubated the core protein with butyryl-CoA and propionyl-CoA, and found that CBP and p300 could catalyze histone H4. To validate lysine, the in vitro modification reaction of acid residues was used to analyze CBP-catalyzed lysine modification residues in histone H4 using nano-HPLC/mass spectrometry (MS), and these residues could be propionylated and butyrylated. CBP and p300 were found to promote lysine Kbu in vitro.30 In addition, HBO1, a lysine acetyltransferase, reportedly promotes butyryl modification. HBO1 is the main source of histone H3 and H4 acetylation. Studies have shown that HBO1 exists in the form of two multipart complexes, consisting of ING4/5, hEaf6, and the scaffold proteins JADE1/2/3 or BRPF1/2/3.35,36 These two types of scaffolds enable HBO1 to acetylate the core histones H3 and H4 while showing different specificities in chromatin. Recent studies in bacteria that expressed and purified the complex BRPF2-HBO1, composed of the short N-terminal BRPF2 and the HBO1 MYST binding domain, and then used glucose and serum-starved HeLa cells to prepare the core histone substrate as the donor, demonstrated that the short N-terminal of HBO1 and BRFP2 are capable of binding to the HBO1 MYST domain and stimulating HAT activity. After subjecting a single acyl-CoA to an in vitro acylation assay, histone acylation was analyzed via western blotting using pantoyl-lysine-specific antibodies. The results showed that the BRPF2-HBO1 complex catalyzes Kbu, especially Kbu of histones H3 and H4. Moreover, further analysis of the experimental results indicated that the JADE1-HBO1 complex exhibited acylation abilities comparable to those of CBP in Kbu. Low HBO1 expression significantly decreased the Kbu of H3 and H4 in HeLa cells, but had no effect on the enzyme levels of other proteins. Furthermore, when HBO1 was expressed alone, the effect of Kbu on the scaffold protein JADE1 was insignificant. Similarly, BRPF2 expression alone had no effect on cell acylation. These results indicated that co-expression of both HBO1 and a scaffold protein was required to induce a significant increase in Kbu-facilitated PTM.37
The catalyzation of butyryl modification by p300/CBP is based on its acyl-binding pocket. Studies have demonstrated that GCN5, which is also a lysine acyltransferase, does not have an identical “back pocket,” like that of p300, that allows the displacement of the acyl chain (Table 1).38,39 Therefore, further research aimed at identifying more Kbu writers may be warranted.
Table 1.
Enzymes participating in butyrylation.
| Category | Gene | Cell | Function | Reference |
|---|---|---|---|---|
| p300/CBP | 293 cells | p53 can be butyrylated by p300/CBP in vitro | 30 | |
| Writer | HBO1-nBRF2/JADE1 | Hela and 293 T cells | Catalyze butyrylation | 35 |
| Sirt7 | Hela | Sirt7 removed acetyl and butyryl groups from H3K36/37 efficiently | 39 | |
| Eraser | HST2 | NA | Bind butyryl-lysine peptide and exhibit the slowest debutyrylase activity | 41 |
| Reader | BRD9, CECR2, TAF1, YEATS domain AF9 | NA | Associated with transcriptional regulation in Kbu | 43,45 |
Kbu erasers
Epigenetic marks induced by histone PTM and DNA covalent modification are not permanent. These marks may be deleted according to the expression status of the cell modification site. This is accomplished by a group of enzymes known as “erasers,” which act against writer activities. Erasers catalyze the removal of epigenetic marks, thereby reducing their effects on transcription and gene expression regulation.40
Zn2+-dependent histone deacetylases (HDACs) and nicotinamide adenine dinucleotide (NAD+)-dependent sirtuins constitute the two major families of lysine deacetylases. Soon after the discovery of Kbu, SIRT enzymes with debutyrylase activities were identified. Sirt7 is one of the most important debutyrylation enzymes.26 Location of the lysine residue influences the deacylase activity of sirtuins. Tanabe et al reported that sirt7 efficiently removed the butyryl groups of H3K36 and K37, compared with the butyryl groups of H3K18. H3K36 and K37 selectivity analysis of sirt7 indicated that sirt7 binds to DNA on nucleosomes via the C-terminal alkaline region and enhances the deacylation of H3K36/37.41,42 Little is known about the capacity of Zn2+-dependent HDACs to deacylate the entire repertoire of histone acylations, compared to that of sirtuins.
Previous studies have suggested that acetyl-lysine analog peptides, including butyl-lysine, may bind to the lysine deacetylase HST2. HST2 binds to butyl-lysine with greater affinity than to pancreatic lysine peptides, prompting us to model this binding in the active site of HST2. A significant spatial conflict exists between butyl-lysine and the residues in the binding pocket around HST2, requiring an angular adjustment that would allow a larger butyl group to be accommodated. Thus, tighter binding between butyl-lysine and HST2 suggests that butyl-lysine may act as a substrate of sirtuin, indicating the need for further investigation into the role of probable deacylation following Kbu modification in order to gain further insights into the functioning of this protein (Table 1).43
Because there have been few studies on Kbu deacylases, more research into Kbu modifications and diacylation appears to be warranted, to facilitate a better understanding of the regulatory role of Kbu modification in related proteins and diseases.
Kbu readers
Histone modifications carry key epigenetic information regarding gene regulation and cell fate decisions. These modifications are recognized by a large number of histone-bound “reader” modules.44 There are three major classes of histone Kac readers: bromodomain, YEATS (Yaf9, ENL, AF9, Taf14, and Sas5), and double plant homeodomain (PHD) finger proteins.26,45 The discovery of histone lysine acylation sparked an increased interest in domains that identify their specificity and functionality. Bromodomains are the archetypal histone acetylation readers. The bromodomain of BRD4 can bind to Kbu, but their binding affinities are significantly lower than those of Kac with the same residues according to a study.46
Recently identified as a novel Kac reader through the study of the human ALL1-fused gene from chromosome 9 protein (AF9), the YEATS domain is a conserved protein domain found in proteins associated with transcriptional regulation. The YEATS domain shows an enhanced binding affinity for Kbu compared with the bromine domain. Acylation isothermal titration calorimetry (ITC) was performed on H3K27 to elucidate the ability of YEATS2 to bind to different acylated histones. K27bu ranked second, behind K27cr, in the binding between the YEATS2 domain and H3, indicating that the YEATS domain of YEATS2 possessed certain Kbu-reading properties (Table 2).47, 48, 49
Table 2.
Functions and mechanisms of butyrylation in diseases.
| Disease | Cell | Function | Animal | Reference |
|---|---|---|---|---|
| Type 1 Diabetes | HEK 293T | Upregulate several genes associated with diabetes and so on | NA | 69 |
| Type 2 Diabetes | Primary hepatocytes | Increase H3K36me2 in DIO mice and change butyrylation in DIO | A high-fat diet-induced obese mice | 66 |
| Cardiac hypertrophy | Hap1 and neonatal cardiac myocytes | Reduced H3K9bu levels under an HFD and stress are detrimental to cardiovascular health | Transverse aortic constriction (TAC) in mice | 49 |
| Male epigenome features | Mouse spermatogenic cells | The interchangeable use of two closely related histone acylation marks at H4K5 | NA | 48 |
| Bladder cancer | The human epithelial bladder cancer cell line 5637 | The mechanism by which butyrylation is mediated is not well defined | NA | 70 |
Functions and potential mechanisms of Kbu in diseases
Several studies have demonstrated the role of Kbu in various physiological processes. The functions and mechanisms of butyrylation are summarized in Table 2.
Role of Kbu in reproduction and development
Differential histone acylation plays a key role in cellular gene programming. Kbu is a ubiquitous modification in eukaryotic cells, and mouse spermatogenic cells are no exception. In mammals, highly specific gene expression programs are activated in meiotic and early post-meiotic cells during differentiation. Ten sites of histone Kbu were identified in mouse testis histones via mass spectrometry, while H4K5bu and H4K8bu were found in spermatogenic cells.50 During sperm cell differentiation, acetylation guides the binding of the bromodomain of the testis-specific bromodomain-containing motif protein (Brdt), whereas the butyryl group inhibits the binding of the bromodomain due to its structure being different from that of the acetyl group. Goudarzi et al investigated the acylation of histone H4 and lysines 5 and 8 at the gene promoter. Brdt stimulates the transcription of specific spermatogenic genes by recruiting P-TEFb complexes and directly binding to their transcription start sites (TSSs).50 During late spermatogenesis, the first bromodomain of Brdt is necessary to replace histones with non-histone sperm-specific transition proteins (TPs) and protamines (PRMs). Previous studies that explored the effect of histone exclusion acylation on sperm cell differentiation have described the key role of H4K5 and H4K8 acetylation in Brdt-driven activity. In elongating spermatids, Kbu of H4K5 and H4K8 is increased. In advanced sperm cells, acetylated histones are removed and degraded. Butyrylated H4 species abolish this wave of acetylation-dependent histone removal and eventually disappear in the condensed sperm cells. Kbu lasts longer than acetylation at H4K5 and H4K8 sites, where H4K5/K8 butyrylated nucleosomes are removed following H4K5/K8 acetylation of nucleosomes. The transcriptional activity of the corresponding genes determined the enrichment of the four H4 PTMs on TSSs.50
Role of Kbu in heart diseases
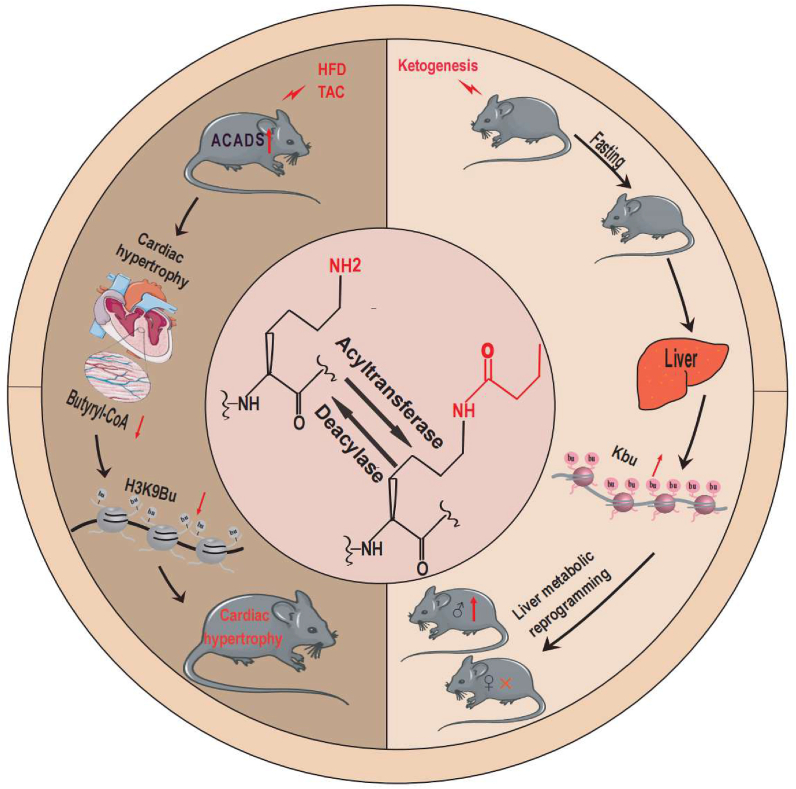
Yang et al found that Kbu of the promoter, H3K9, a histone modification that is negatively regulated by high fat and stress in an acyl-CoA dehydrogenase short-chain (ACADS)-dependent manner, moderates stress-regulated gene expression.51 ACADS reduces butyryl-CoA to crotonyl-CoA in the final stages of the β-oxidation spiral.52 Thus, the accumulation of butyryl-CoA during β-oxidation and Kbu of histones depends on the degree of influence exerted by the activity/abundance of ACADS. Histone acylation is influenced by β-oxidation enzymes, which are localized in both mitochondria and nuclei. H3K9Bu is enriched at transcription start sites and downregulated by dietary fat and stress. Gene set enrichment analysis results revealed that Kbu of H3K9 requires β-oxidation, where fatty acids synthesized from glucose-derived acetyl-CoA serve as a substitute in the absence of a fatty acid substrate. H3K9Bu was not only reduced by a high-fat diet but also by pressure overload-induced stress. With a fat-free diet, the heart exhibited much higher levels of H3K9Bu, which coincided with a reduction in the gene expression changes induced by stress. The responses of ACADS-deficient and wild-type mice to transverse aortic constriction (TAC)-induced changes in gene expression were compared. ACADS mice showed higher promoter-H3K9bu levels following TAC, similar to mice on fat-free diets, which was consistent with results of gene expression passivation. The findings suggested that higher promoter-H3K9bu levels inhibited stress-induced gene expression changes (Fig. 1), although the underlying mechanisms remained unclear.53 Although the association between changes in a high-fat diet and Kbu remains unclear, it leads to a better understanding of the role of diet and stress in the epigenetic modification of heart disease.
Figure 1.
Regulatory effects of Kbu under HFD and stress. Under HFD and the stress conditions of TAC, the protein level of ACADS will increase in cells, regulating butyryl-COA accumulation and butyrylation further. H3K9Bu will be significantly reduced, which will affect the normal systolic function of the mouse heart. Meanwhile, starvation of C57BL/6 mice and stimulation with ketogenesis resulted in an increase of Kbu in the liver cells of male mice, but there was no significant change in female mice.
Role of Kbu in vascular dementia
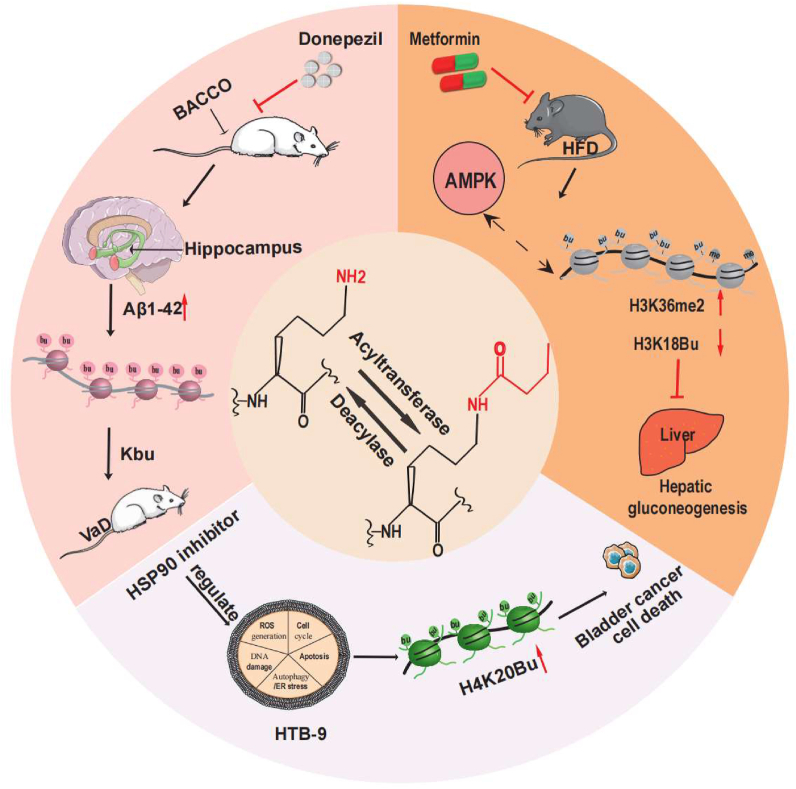
Vascular dementia (VaD) is a heterogeneous brain disorder caused by stroke or small vessel disease and is considered to be a universal form of dementia.53,54 Many clinical drugs have been used to treat vascular dementia55,56; however, studies have indicated that some of these drugs did not improve memory or executive ability.57 Some protein PTMs are associated with neurodegenerative diseases,58,59 and a deeper understanding of PTMs is essential for improving the targeted treatment of vascular dementia. Donepezil, a cholinesterase inhibitor, is an approved drug for Alzheimer's disease.59 Wang et al examined the effects of donepezil on vascular dementia and its underlying mechanisms in rats subjected to bilateral common carotid artery occlusion (BCCAO) via ligation.60 The Morris water maze (MWM) and food reward experiment demonstrated that donepezil reverses BCCAO-induced memory impairment and cognitive deficits in rats and attenuates Aβ1-42 levels. Further analyses showed that the levels of seven novel PTMs were significantly lower than those in the BCCAO group. In the donepezil treatment group, Western blot analysis of rat hippocampal lysates indicated significant changes in crotonylation, succinylation, propionylation, and butyrylation. Compared with that in the modification of other proteins, the Kbu level of the treatment group was significantly lower than that of the BACCO group. Thus, donepezil may attenuate Aβ1-42 levels in the rat hippocampus mainly by mediating changes in Kbu, thereby improving cognition and memory in rats (Fig. 2). This suggests that Kbu may play a potential regulatory role in neurodegenerative diseases such as vascular dementia, which may reveal novel targets for preventing VaD onset.
Figure 2.
Regulation of clinical drugs targeting Kbu. Donepezil can reduce the expression of Aβ1-42 in the hippocampus. Following donepezil treatment, the modified expression of Kbu in rat hippocampal histone was decreased, and the VaD of rats with BACCO was improved. In HFD-induced diabetic mice, H3K36me2 levels increased while H3K18bu levels decreased. After metformin treatment, their expression could be reversed. Moreover, liver gluconeogenesis could be inhibited to reduce blood glucose levels. In bladder cancer, the level of H4K20Bu in the histone of HTB-9 cells increased. HSP90 inhibitor treatment can promote apoptosis and necrosis of cancer cells by regulating oxidative stress, autophagy, and apoptosis. ACADS, Acyl-CoA dehydrogenase short chain; BCCAO, bilateral common carotid artery occlusion; CVDs, cardiovascular diseases; HFD, high-fat diet; Kbu, butyrylation; Kac, acetylation; Kcr, crotonylation; TAC, transverse aortic constriction; TSS, transcription start site; VaD, vascular dementia.
Role of Kbu in diabetes
Diabetes mellitus (DM) is prevalent worldwide. DM, compounded by its complications, not only affects the quality of life of patients but also contributes to a high mortality rate.61, 62, 63, 64, 65, 66 A combination of telmisartan and pancreatin ameliorates type 2 diabetes (T2D)-linked cardiomyopathy by reversing H3, H2A, and H2B histone modifications.67 This suggests the importance of decoding the role of epigenetics in diabetes. Metformin, which is widely used to treat T2D, is thought to inhibit gluconeogenesis via lysine acetyltransferase, which is considered to be its primary therapeutic mechanism.68 A comprehensive analysis of histone modifications in the liver tissues of diet-induced obese (DIO) mice and metformin-treated DIO (DIO-met) mice revealed an association between histone H3K36me2 levels and the development of prediabetes, and the antidiabetic activity of metformin (Fig. 2). Kbu levels were altered, and Kbu levels in the liver of mice in the DIO group were at least 1.5 times lower than those in the chow group. Kbu alterations were enriched in liver histones H3, H4, and H2B. There are three modification sites in histone H3 and H4, five in histone H2B, and one in histone H1. Therefore, butyrate may play an important role in the development of diabetes. In addition, the homeostasis of short-chain coenzyme-As, including malonyl-CoA and butyryl-CoA, also contributes to the development of diabetes.69,70 Therefore, changing short-chain coenzymes, which in turn affects Kbu levels, may present a new technique for improving insulin resistance and obesity. In addition, isobutyrylation is a novel PTM that increases histone isobutyrylation levels when treated with isobutyrate.71 The ability of isobutyrate treatment to mediate diabetes-related signaling in 293T cells was analyzed via RNA-seq. A recent study comparing Kbu and isobutyrylation suggests that these two modifications may play a regulatory role in diabetes.71
Role of Kbu in bladder cancer
Bladder cancer is the second-most common malignant tumor of the genitourinary system among Americans.72 PTMs regulate bladder cancer, and the mechanisms of phosphorylation, acetylation, glycosylation, and ubiquitination are well-defined.73 Overexpression of heat shock protein 90 (HSP90) in tumors is related to prognosis. Inhibition of HSP90, resulting in the loss of signal transduction, growth inhibition, anti-angiogenesis, and cell death, leads to the degradation of many oncogenic proteins. Several HSP90 inhibitors have demonstrated improved efficacy against bladder cancer in clinical oncology studies, and additional research indicates that these inhibitors may induce histone PTMs leading to global protein changes in tumor cells (Fig. 2). Kbu was differentially expressed in cancer cells treated with these inhibitors. Moreover, Kbu changes with lysine coenzyme A. H4K20 was identified as the site of histone Kbu modification, with the modified peptide sequence GAKRHRK(bu)VLRDNI. Kbu neutralizes the positive charge on the surface of lysine, thereby reducing histone-DNA contact, indicating that Kbu plays a regulatory role in bladder cancer. The changes in Kbu that are dependent on lysine coenzyme A may provide a new direction for the application of PTMs in the treatment of bladder cancer, although the precise mechanism underlying these changes remains unclear.72
Role of Kbu in starvation
Goudarzi et al74 found that after 48 h of fasting, Kbu levels in the hepatocytes of male C57BL/6 mice were significantly higher than those of female mice, possibly due to steroid hormones (Fig. 1). This suggested that higher histone Kbu levels may be an additional regulatory activity that ensures differential gene dosage expression in males and females. β-hydroxybutyrate (BHB), a fasting-induced metabolite, modifies histones and inhibits HDAC, exerting a regulatory effect on PTMs. BHB increased the concentration of MDA-MB231 cells in vitro but did not exert a significant effect on Kbu levels. Kbu of histones was significantly increased in hungry male mice, suggesting that butyryl-COA may be produced by ketones. Considered together, the differential expression of Kbu in hepatocytes suggests that histone Kbu contributes to liver metabolism.
Role of Kbu in plants
Rice (Oryza sativa), which is one of the most important food crops in the world, has recently been considered a monocot model plant in functional genomics research.75 Lu et al investigated Kbu and crotonylation of lysine in Arabidopsis, rice, corn, and tobacco, focusing primarily on Kbu-based research.76 The sites of Kbu modification were also identified as the N-terminal tails of rice histone H3 and H4 and the core region of H2B. Furthermore, Lu et al also investigated the association between histone acylation and replacement. Chip-seq analysis using anti-H3K9ac antibodies revealed more than 30,000 Kbu and Kcr peaks, indicating that Kbu is widespread in the rice genome. Rice was subjected to darkness for 48 h or starvation for 6 h, following which materials were collected for chip-seq analysis. ChIP-seq analysis revealed that the numbers of modified genes in the treatment and control groups were approximately similar. Under starvation and submergence, the Kbu levels of 279 and 277 genes increased, while the Kbu levels of 1260 and 304 genes decreased, indicating that gene expression of Kbu changes under external stress (Fig. S1). RNA-seq identified genes showing circadian rhythm expression in rice seedlings at four circadian time points. Compared with the functions of acetylation and crotonylation, Kbu is mainly involved in preparing genes for environmental stress-induced activation, as demonstrated by its ability to inhibit the expression of genes induced by external stress.
Previous reports have indicated that 23.87% of the Kbu modifications in histones were enriched in the promoter and intergenic regions of Dongjin rice, and the relationship between DNaseI hypersensitivity sites (DHSs) was studied. Over half the histone Kbu peaks in the rice genome overlapped with DHSs, indicating that histone Kbu may act as an active marker of the gene transcription that it promotes. In addition, considering acetylation modifications, Kbu combinations significantly increased gene expression compared with another acetylation modification group, indicating that Kbu was involved in transcriptional regulation and promoted transcription in cooperation with other modifications. GO analysis of specific Kbu-related genes indicated that genes are enriched in the processes of translation, transportation, and localization. Histone-modified genes in Dongjin rice are involved in transcription, binding processes, and stress responses. This indicates that Kbu is crucial for the growth, development, and metabolism of rice.69
In brief, Kbu closely resembles acetylation modification. The ratio of lysine acetylation to butyrylation is affected by environmental factors and functions in plants such as rice to regulate genes in response to starvation and flooding conditions. Dynamic regulation of metabolic signals may be a mechanism by which micro-epigenetics regulates the adaptation of plants to environmental changes.76
Experimental methods for identification and detection of butyrylation
Zhao et al contributed to much of the research on acylation modification. They developed high-energy specific antibodies against different types of lysine acylation, analyzed them using immunoprecipitation technology, and helped conduct in-depth research on the functions of these PTMs. Chemical probes may be used for covalent labeling of PTMs as a supplementary enrichment method for proteomics analyses using mass spectrometry. Existing endogenous PTM chemical proteomics analysis techniques can be roughly categorized into three types based on the chemical probes used: “reaction capture” of chemoselective probes, selection of markers for reactive amino acids in the proteome, and cross metabolism of PTM-like probes or direct labeling.77 In addition, PTM spectra may be used to perform in-depth chemical proteomics analyses of functional PTMs.
In yeast cells, researchers use the newly developed computer program PTMap to analyze mass spectrometry and protein sequence comparison. PTMap is the first method of sequence comparison algorithm that emphasizes the false positive identification of mismatched peaks. For the first time, they discovered multiple sites of propionyl and butyryl lysine in yeast histones H2B, H3, and H4.78 The presence of propionyl lysine and butyryl lysine in histones H3 and H4 was determined by Western blotting. Core histones were extracted after HDAC inhibitors were treated with yeast cells in vitro. Furthermore, the butyrylation and propionylation of yeast were confirmed by Western blot and LC/MS/MS analysis. In addition, HDAC inhibitors were used to extract core histone proteins from yeast cells via SDS-PAGE analysis, gel removal, gel digestion, and nano HPLC/MS–MS analysis. The PTMap algorithm is used for unrestricted sequence alignment of MS/MS data from yeast histones. Then, through the unique program of peak selection, automatic mass transfer adjustment, and accurate PTM positioning, the modification sites are identified by manual inspection. Subsequently, the sites of lysine butyrylation and propionylation of histones were further confirmed by MS/MS spectra of in vivo-derived peptides and synthetic counterparts. The butyrylation of lysine in histone H3K14 was confirmed by comparing the HPLC retention time of its corresponding acetylation difference. This study demonstrated that concentrated yeast histone contains multiple modification sites, indicating that butyrylation is a conserved histone marker in eukaryote evolution.
Kbu is essential for cellular regulation and metabolism. However, due to technical limitations, the identification of Kbu sites on a large scale is a challenging task. Huang et al proposed a method for calculating and identifying histone lysine Kbu based on information entropy, which is considered the first method developed for predicting Kbu.79 This is a four-part method. First, the histone sequence to be predicted is segmented, and the central amino acid is found. Second, proteins that pass butyrylation are regarded as positive samples and non-butyrylated negative samples. Third, the fragments that conform to the corresponding formula are transformed into numerical features according to the information entropy coding (IEE) and the k-space amino acid pair (CKSAAP). Finally, the random forest algorithm is assigned to the classifier according to the numerical characteristics, and the target protein is then introduced into the classifier for the final prediction. This calculation method revealed the potential and hidden characteristics of histone Kbu, indicating that histone Kbu modification binds to other molecules, thereby facilitating the development of systemic lupus erythematosus, viral carcinogenesis, and other pathways in cancer.
Identification of Kbu sites enables further exploration of their functions. Currently, analytical methods aimed at elucidating Kbu are relatively limited, as a result of which research on Kbu function remains in its infancy. Thus, more methods capable of predicting the effects of altered Kbu on diseases are needed in the future.60,68,74,80, 81, 82, 83
Clinic drugs
Currently, no drugs are available to treat butyrylation, but some clinical acetylation inhibitors have been reported to regulate butyrylation while exerting a therapeutic effect on diseases.
Suberoylanide hydroxamic acid (SAHA), an HDAC inhibitor clinically approved for the treatment of cutaneous T-cell lymphoma, demonstrates promising results in the treatment of neuroblastoma. A proteomic study by Xu et al showed that it could alter both acetylation and butyrylation levels, suggesting that in neuroblastoma, SAHA not only regulates acetylation but also exerts therapeutic effects, further suggesting that butyrylation is also involved in relevant biological pathways and provides insight into neuroblastoma.80 In addition, a novel HDAC inhibitor designed and prepared by replacing Val 1 with tyrosine, a novel largazole derivative, can modulate the level of histone acetylation in human colon cancer cells, thereby achieving therapeutic effects. HDAC inhibitors have also been shown to alter the abundance of butyrylation in histones. Nevertheless, specific regulatory mechanisms remain to be defined.81 Regardless, these drugs alter the abundance of histone butyrylation. In some cases, the role of butyrylation in disease can also be investigated.
Isobutyryl-COA deficiency (IBD) is a rare autosomal recessive metabolic disorder. Genetic metabolic disorders include those with elevated C4 acylcarnitine levels. IBD determined by neonatal screening in Korea is mainly associated with an ACAD8 mutation in PCR and sequencing analyses.82,83 Moreover, in cases from the UK, the level of C4 carnitine was increased, and the genetic metabolic disorder was diagnosed through a neonatal screening program. One patient had viral gastroenteritis and hypoglycemia and was diagnosed with IBD with dilated cardiomyopathy and anemia. However, after glucose polymer treatment, the blood glucose level was stable, and no illness was detected during the 10-year follow-up. Isobutyrylation depends on isobutyryl-CoA dehydrogenase, suggesting that isobutyrylation may regulate disease onset. Although there is no clinical drug targeting butyrylation, animal studies have demonstrated a significant role for butyrylation in disease models. Therefore, targeting butyrylation is a promising therapeutic approach that needs to be further studied.
Discussion
Histones are modified by an array of PTMs, including methylation, acetylation, ubiquitination, small ubiquitin-like modification, and ribosylation.84, 85, 86, 87 Histone coding, the combinatorial array of histone PTMs, determines the role of proteins in gene expression and chromatin dynamics. PTMs of histones have been studied using biochemical techniques and mass spectrometry.88,89
Studies investigating the association between Kbu and disease have revealed the involvement of Kbu in the regulation of cardiovascular diseases, nerves, male epigenetic characteristics, certain cancers, the transcription of gluconeogenic genes, multiple T2D phenotypes, protein expression, etc. Despite the fact that these studies have identified diseases associated with altered Kbu levels, no in-depth mechanisms have been elucidated, and no patients have been clinically diagnosed with altered Kbu levels, owing to a relative lack of diagnostic techniques. Therefore, analytical methods capable of diagnosing Kbu are required. Donepezil reduced Kbu levels in total histone of hippocampal lysates, which accounts for its efficacy in treating vascular dementia. Another study specifically analyzed the change in the Kbu of a certain histone and its mechanism and observed that targeted histone Kbu exerted a therapeutic effect on vascular dementia. Stress and a high-fat diet increased ACADS expression while decreasing butyryl-COA expression, lowering the Kbu of histone and causing myocardial damage. This provides an epigenetic direction for cardiovascular disease research. However, further research is required to determine whether the reduction of Kbu is specifically harmful to downstream proteins.
The degree of Kbu modification also affects changes in rice and other plants under conditions such as starvation and water inundation. Studies have demonstrated that Kbu is involved in the transcriptional regulation of rice, and gene expression analysis suggests that Kbu in conjunction with acetylation may regulate gene expression in rice. Gene analyses indicated that molecular functions associated with genes involved in Kbu may also be involved in the epigenetic inheritance of phosphorylation, thereby providing direction for future studies on plant diseases and functions.90, 91, 92
The dynamic equilibrium of acylation modification is mainly regulated by related acylases and deacetylases. Although certain related writers, readers, and erasers required for the lysine modification are known, knowledge regarding the molecules specifically recognized by the Kbu modification is still lacking. Currently, the majority of molecules recognized by Kbu modification are enzymes from the KAT family, such as p300, which plays a key regulatory role in Kbu modification. In patients with advanced T2D, the acetylation level of blood vessels is excessive, leading to the production of more ROS, which in turn causes vascular smooth muscle cell (VSMC) dysfunction. Garcinol, a lysine acetyltransferase inhibitor, has been recently shown to inhibit the p300/CBP association factor (PCAF), reducing ROS overproduction in VSMCs.93 The above-mentioned studies indicate that metformin affects Kbu levels in T2D patients. Thus, the regulation of Kbu by p300 may provide a direction for future research on the role of metformin in T2D-related Kbu as well as on potential targets for Kbu alterations that would enable more precise diabetes treatment. Deacetylation is the main activity of HDACS, including sirtuins, which are enzymes that regulate debutyrylation. Limited studies have been conducted on enzymes that modify Kbu or debutyrylation. Therefore, the role of the dynamic equilibrium between Kbu levels and debutyrylation levels in the regulation of related diseases may need further investigation. Additionally, specifically targeting butyrylation without affecting other types of acylations is difficult because a protein (writer, eraser, or reader) always regulates several types of acylations.
In addition to Kbu, the co-existence of two modifications also exerts a certain effect on the cellular processes. A dynamic and competitive balance between histone K4K5K8 acetylation and Kbu is a mark of highly active gene promoters. This is the first demonstration of the interchangeability of two closely related histone acylation marks at a specific site, indicating that in protein acylation modification, cellular processes are changed, not only by one modification but also by the competition between the two modifications, as both of these may affect cell transcription. Although some reports indicate that Kbu modification is associated with some diseases, the specific relationship between the regulation of Kbu modification and the occurrence and development of diseases remains unclear, and the relationship between Kbu modification and disease regulation needs further exploration.94,95
Conclusions
Kbu is a newly discovered histone modification with multiple modification sites in eukaryotes. Despite reports that Kbu is differentially expressed in a number of disease models, suggesting that it may play a regulatory role, the potential mechanism underlying Kbu in cellular processes remains unclear. Therefore, the focus should shift toward more detailed studies on the specific regulatory roles of Kbu as well as the mechanisms underlying Kbu in cellular processes and targeted substrates, which would provide new directions for the precise treatment of associated diseases.
Author contributions
Q.X. collected materials and wrote the manuscript. T.Y. and Y.Y. conceptualized the study. Q.X., T.J., and H.M. prepared the schematics. T.Y., Y.Y., X.L., J.W. H.Q., and H.L. helped with the final revision of the article. All authors reviewed and approved the final manuscript.
Conflict of interests
The authors have no conflict of interests to declare.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82270442 and 81870331) and the Qingdao Municipal Science and Technology Bureau Project (China) (No. 21-1-4-rkjk-12-nsh).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.10.025.
Abbreviations
- ACADS
acyl-CoA dehydrogenase short chain
- AF9
ALL1-fused gene from chromosome 9 protein acylation
- BCCAO
bilateral common carotid artery occlusion
- BHB
β-hydroxybutyrate
- Brdt
bromodomain-containing motif protein
- CKSAAP
k-space amino acid pair
- DHS
DNaseI hypersensitivity sites
- DIO
diet-induced obesity
- DM
diabetes mellitus
- ER
endoplasmic reticulum
- GNAT
GCN5-related N-acetyltransferase
- HATs
histone acetyltransferases
- ICT
isothermal titration calorimetry
- IEE
information entropy coding
- Kac
protein acylation
- Kbu
butyrylation
- Kcr
crotonylation
- MS
mass spectrometry
- MWM
Morris water maze
- NAD+
nicotinamide adenine dinucleotide
- PCAF
p300/CBP association factor
- p300/CBP
p300/CREB-binding protein
- PHD
double plant homeodomain
- PRM
protamine
- PTM
post-translational modification
- T2D
type 2 diabetes
- TAC
transverse aortic constriction
- TPM
tags per kilobase of gene length per million mapped reads
- TPs
transition proteins
- TSSs
transcription start sites
- UDP-GlcNAc
uridine diphosphate-n-acetylglucosamine
- VaD
vascular dementia
- VSMC
vascular smooth muscle cell
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Gajjala P.R., Fliser D., Speer T., et al. Emerging role of post-translational modifications in chronic kidney disease and cardiovascular disease. Nephrol Dial Transplant. 2015;30(11):1814–1824. doi: 10.1093/ndt/gfv048. [DOI] [PubMed] [Google Scholar]
- 2.Liu J., Zhong L., Guo R. The role of posttranslational modification and mitochondrial quality control in cardiovascular diseases. Oxid Med Cell Longev. 2021;2021 doi: 10.1155/2021/6635836. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Bischoff R., Schlüter H. Amino acids: chemistry, functionality and selected non-enzymatic post-translational modifications. J Proteonomics. 2012;75(8):2275–2296. doi: 10.1016/j.jprot.2012.01.041. [DOI] [PubMed] [Google Scholar]
- 4.Müller M.M. Post-translational modifications of protein backbones: unique functions, mechanisms, and challenges. Biochemistry. 2018;57(2):177–185. doi: 10.1021/acs.biochem.7b00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Hondt C., Iyyathurai J., Vinken M., et al. Regulation of connexin- and pannexin-based channels by post-translational modifications. Biol Cell. 2013;105(9):373–398. doi: 10.1111/boc.201200096. [DOI] [PubMed] [Google Scholar]
- 6.Han K.K., Martinage A. Post-translational chemical modifications of proteins—III. Current developments in analytical procedures of identification and quantitation of post-translational chemically modified amino acid(s) and its derivatives. Int J Biochem. 1993;25(7):957–970. doi: 10.1016/0020-711x(93)90108-q. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y., Wang Y. Mapping histone modification-dependent protein interactions with chemical proteomics. Trends Biochem Sci. 2022;47(3):189–193. doi: 10.1016/j.tibs.2021.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Cai L., Hu F., Fu W., et al. Ginsenoside Rg2 ameliorates brain injury after intracerebral hemorrhage in a rat model of preeclampsia. Reprod Sci. 2021;28(12):3431–3439. doi: 10.1007/s43032-021-00692-2. [DOI] [PubMed] [Google Scholar]
- 9.Wu Z., Connolly J., Biggar K.K. Beyond histones - the expanding roles of protein lysine methylation. FEBS J. 2017;284(17):2732–2744. doi: 10.1111/febs.14056. [DOI] [PubMed] [Google Scholar]
- 10.Liu J., Qian C., Cao X. Post-translational modification control of innate immunity. Immunity. 2016;45(1):15–30. doi: 10.1016/j.immuni.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 11.Jiang G., Li C., Lu M., et al. Protein lysine crotonylation: past, present, perspective. Cell Death Dis. 2021;12(7):703. doi: 10.1038/s41419-021-03987-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y., Li M., Fan M., et al. Chromodomain Y-like protein-mediated histone crotonylation regulates stress-induced depressive behaviors. Biol Psychiatr. 2019;85(8):635–649. doi: 10.1016/j.biopsych.2018.11.025. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z., Zhao Y., Xu N., et al. NEAT1 regulates neuroglial cell mediating Aβ clearance via the epigenetic regulation of endocytosis-related genes expression. Cell Mol Life Sci. 2019;76(15):3005–3018. doi: 10.1007/s00018-019-03074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y., Yang M., Wang S., et al. An overview of epigenetic methylation in pancreatic cancer progression. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.854773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J.J., Zhu Y., Zhu Y., et al. Association of increased DNA methyltransferase expression with carcinogenesis and poor prognosis in pancreatic ductal adenocarcinoma. Clin Transl Oncol. 2012;14(2):116–124. doi: 10.1007/s12094-012-0770-x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang N., Jiang H., Zhang K., et al. OGT as potential novel target: structure, function and inhibitors. Chem Biol Interact. 2022;357 doi: 10.1016/j.cbi.2022.109886. [DOI] [PubMed] [Google Scholar]
- 17.Wang W., Okajima T., Takeuchi H. Significant roles of Notch O-glycosylation in cancer. Molecules. 2022;27(6):1783. doi: 10.3390/molecules27061783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jansson A.K., Chan L.X., Lubans D.R., et al. Effect of resistance training on HbA1c in adults with type 2 diabetes mellitus and the moderating effect of changes in muscular strength: a systematic review and meta-analysis. BMJ Open Diabetes Res Care. 2022;10(2) doi: 10.1136/bmjdrc-2021-002595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rebelo A.L., Chevalier M.T., Russo L., et al. Role and therapeutic implications of protein glycosylation in neuroinflammation. Trends Mol Med. 2022;28(4):270–289. doi: 10.1016/j.molmed.2022.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Kobeissy F., Kobaisi A., Peng W., et al. Glycomic and glycoproteomic techniques in neurodegenerative disorders and neurotrauma: towards personalized markers. Cells. 2022;11(3):581. doi: 10.3390/cells11030581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Contreras-de la Rosa P.A., Aragón-Rodríguez C., Ceja-López J.A., et al. Lysine crotonylation: a challenging new player in the epigenetic regulation of plants. J Proteonomics. 2022;255 doi: 10.1016/j.jprot.2022.104488. [DOI] [PubMed] [Google Scholar]
- 22.Boulias K., Greer E.L. Means, mechanisms and consequences of adenine methylation in DNA. Nat Rev Genet. 2022;23(7):411–428. doi: 10.1038/s41576-022-00456-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y., Song Y., Liu J., et al. Ubiquitination of receptorsomes, frontline of plant immunity. Int J Mol Sci. 2022;23(6):2937. doi: 10.3390/ijms23062937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi H., Yang J., Yu J., et al. Glucose-responsive nanogels efficiently maintain the stability and activity of therapeutic enzymes. Nanotechnol Rev. 2022;11(1):1511–1524. [Google Scholar]
- 25.Arc E., Galland M., Cueff G., et al. Reboot the system thanks to protein post-translational modifications and proteome diversity: how quiescent seeds restart their metabolism to prepare seedling establishment. Proteomics. 2011;11(9):1606–1618. doi: 10.1002/pmic.201000641. [DOI] [PubMed] [Google Scholar]
- 26.Sabari B.R., Zhang D., Allis C.D., et al. Metabolic regulation of gene expression through histone acylations. Nat Rev Mol Cell Biol. 2017;18(2):90–101. doi: 10.1038/nrm.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Y., Li Y., Yang K., et al. Antimicrobial peptide BCp12 inhibits Staphylococcus aureus growth by altering lysine malonylation levels in the arginine synthesis pathway. J Agric Food Chem. 2022;70(1):403–414. doi: 10.1021/acs.jafc.1c05894. [DOI] [PubMed] [Google Scholar]
- 28.Xu M., Tian X., Ku T., et al. Global identification and systematic analysis of lysine malonylation in maize (Zea mays L.) Front Plant Sci. 2021;12 doi: 10.3389/fpls.2021.728338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peoples J.N., Ghazal N., Duong D.M., et al. Loss of the mitochondrial phosphate carrier SLC25A3 induces remodeling of the cardiac mitochondrial protein acylome. Am J Physiol Cell Physiol. 2021;321(3):C519–C534. doi: 10.1152/ajpcell.00156.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y., Sprung R., Tang Y., et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6(5):812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X.F., Chen X., Tang X. Short-chain fatty acid, acylation and cardiovascular diseases. Clin Sci (Lond) 2020;134(6):657–676. doi: 10.1042/CS20200128. [DOI] [PubMed] [Google Scholar]
- 32.Xu H., Wu M., Ma X., et al. Function and mechanism of novel histone posttranslational modifications in health and disease. BioMed Res Int. 2021;2021 doi: 10.1155/2021/6635225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X., Yang Y., Zhang B., et al. Lactate metabolism in human health and disease. Signal Transduct Targeted Ther. 2022;7(1):305. doi: 10.1038/s41392-022-01151-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao S., Zhang X., Li H. Beyond histone acetylation—writing and erasing histone acylations. Curr Opin Struct Biol. 2018;53:169–177. doi: 10.1016/j.sbi.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Han J., Lachance C., Ricketts M.D., et al. The scaffolding protein JADE1 physically links the acetyltransferase subunit HBO1 with its histone H3-H4 substrate. J Biol Chem. 2018;293(12):4498–4509. doi: 10.1074/jbc.RA117.000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao Y., Zhong C., Zhu J., et al. Structural and mechanistic insights into regulation of HBO1 histone acetyltransferase activity by BRPF2. Nucleic Acids Res. 2017;45(10):5707–5719. doi: 10.1093/nar/gkx142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao Y., Li W., Yang H., et al. HBO1 is a versatile histone acyltransferase critical for promoter histone acylations. Nucleic Acids Res. 2021;49(14):8037–8059. doi: 10.1093/nar/gkab607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du Y., Hou G., Zhang H., et al. SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function. Nucleic Acids Res. 2018;46(10):5195–5208. doi: 10.1093/nar/gky156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ringel A.E., Wolberger C. Structural basis for acyl-group discrimination by human Gcn5L2. Acta Crystallogr D Struct Biol. 2016;72(Pt 7):841–848. doi: 10.1107/S2059798316007907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biswas S., Rao C.M. Epigenetic tools (The Writers, the Readers and the Erasers) and their implications in cancer therapy. Eur J Pharmacol. 2018;837:8–24. doi: 10.1016/j.ejphar.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 41.Tanabe K., Liu J., Kato D., et al. LC-MS/MS-based quantitative study of the acyl group- and site-selectivity of human sirtuins to acylated nucleosomes. Sci Rep. 2018;8(1):2656. doi: 10.1038/s41598-018-21060-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang H.C., Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metabol. 2014;25(3):138–145. doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith B.C., Denu J.M. Acetyl-lysine analog peptides as mechanistic probes of protein deacetylases. J Biol Chem. 2007;282(51):37256–37265. doi: 10.1074/jbc.M707878200. [DOI] [PubMed] [Google Scholar]
- 44.Xiong X., Panchenko T., Yang S., et al. Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nat Chem Biol. 2016;12(12):1111–1118. doi: 10.1038/nchembio.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flynn E.M., Huang O.W., Poy F., et al. A subset of human bromodomains recognizes butyryllysine and crotonyllysine histone peptide modifications. Structure. 2015;23(10):1801–1814. doi: 10.1016/j.str.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 46.Vollmuth F., Geyer M. Interaction of propionylated and butyrylated histone H3 lysine marks with Brd4 bromodomains. Angew Chem Int Ed Engl. 2010;49(38):6768–6772. doi: 10.1002/anie.201002724. [DOI] [PubMed] [Google Scholar]
- 47.Li Y., Sabari B.R., Panchenko T., et al. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol Cell. 2016;62(2):181–193. doi: 10.1016/j.molcel.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao D., Guan H., Zhao S., et al. YEATS2 is a selective histone crotonylation reader. Cell Res. 2016;26(5):629–632. doi: 10.1038/cr.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andrews F.H., Shinsky S.A., Shanle E.K., et al. The Taf14 YEATS domain is a reader of histone crotonylation. Nat Chem Biol. 2016;12(6):396–398. doi: 10.1038/nchembio.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goudarzi A., Zhang D., Huang H., et al. Dynamic competing histone H4 K5K8 acetylation and butyrylation are hallmarks of highly active gene promoters. Mol Cell. 2016;62(2):169–180. doi: 10.1016/j.molcel.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Z., He M., Austin J., et al. Histone H3K9 butyrylation is regulated by dietary fat and stress via an Acyl-CoA dehydrogenase short chain-dependent mechanism. Mol Metabol. 2021;53 doi: 10.1016/j.molmet.2021.101249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abou-Abbass H., Abou-El-Hassan H., Bahmad H., et al. Glycosylation and other PTMs alterations in neurodegenerative diseases: current status and future role in neurotrauma. Electrophoresis. 2016;37(11):1549–1561. doi: 10.1002/elps.201500585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Brien J.T., Thomas A. Vascular dementia. Lancet. 2015;386(10004):1698–1706. doi: 10.1016/S0140-6736(15)00463-8. [DOI] [PubMed] [Google Scholar]
- 54.Bryant L., Li D., Cox S.G., et al. Histone H3.3 beyond cancer: Germline mutations in Histone 3 Family 3A and 3B cause a previously unidentified neurodegenerative disorder in 46 patients. Sci Adv. 2020;6(49) doi: 10.1126/sciadv.abc9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iadecola C. The pathobiology of vascular dementia. Neuron. 2013;80(4):844–866. doi: 10.1016/j.neuron.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gareri P., Cotroneo A.M., Orsitto G., et al. The CITIMEM study: a pilot study. Optimizing pharmacological treatment in dementia. Arch Gerontol Geriatr. 2020;89 doi: 10.1016/j.archger.2020.104073. [DOI] [PubMed] [Google Scholar]
- 57.Ghassab-Abdollahi N., Mobasseri K., Dehghani Ahmadabad A., et al. The effects of Huperzine A on dementia and mild cognitive impairment: an overview of systematic reviews. Phytother Res. 2021;35(9):4971–4987. doi: 10.1002/ptr.7126. [DOI] [PubMed] [Google Scholar]
- 58.Leijenaar J.F., Groeneveld G.J., Klaassen E.S., et al. Methylphenidate and galantamine in patients with vascular cognitive impairment-the proof-of-principle study STREAM-VCI. Alzheimer's Res Ther. 2020;12(1):10. doi: 10.1186/s13195-019-0567-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang H., Zong Y., Han Y., et al. Compared of efficacy and safety of high-dose donepezil vs standard-dose donepezil among elderly patients with Alzheimer's disease: a systematic review and meta-analysis. Expet Opin Drug Saf. 2022;21(3):407–415. doi: 10.1080/14740338.2022.2027905. [DOI] [PubMed] [Google Scholar]
- 60.Wang H., Lu J., Gao W.C., et al. Donepezil down-regulates propionylation, 2-hydroxyisobutyrylation, butyrylation, succinylation, and crotonylation in the brain of bilateral common carotid artery occlusion-induced vascular dementia rats. Clin Exp Pharmacol Physiol. 2020;47(10):1731–1739. doi: 10.1111/1440-1681.13352. [DOI] [PubMed] [Google Scholar]
- 61.Luís C., Soares R., Baylina P., et al. Underestimated prediabetic biomarkers: are we blind to their strategy? Front Endocrinol. 2022;13 doi: 10.3389/fendo.2022.805837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baradez C., Liska J., Brulle-Wohlhueter C., et al. Brief digital solutions in behavior change interventions for type 2 diabetes mellitus: a literature review. Diabetes Ther. 2022;13(4):635–649. doi: 10.1007/s13300-022-01244-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li X., Qi H., Cui W., et al. Recent advances in targeted delivery of non-coding RNA-based therapeutics for atherosclerosis. Mol Ther. 2022;30(10):3118–3132. doi: 10.1016/j.ymthe.2022.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li X., Yang Y., Wang Z., et al. Multistage-responsive nanocomplexes attenuate ulcerative colitis by improving the accumulation and distribution of oral nucleic acid drugs in the colon. ACS Appl Mater Interfaces. 2022;14(1):2058–2070. doi: 10.1021/acsami.1c21595. [DOI] [PubMed] [Google Scholar]
- 65.Yang Y., Li M., Liu Y., et al. The lncRNA punisher regulates apoptosis and mitochondrial homeostasis of vascular smooth muscle cells via targeting miR-664a-5p and OPA1. Oxid Med Cell Longev. 2022;2022 doi: 10.1155/2022/5477024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li M., Yang Y., Zong J., et al. MiR-564:a potential regulator of vascular smooth muscle cells and therapeutic target for aortic dissection. J Mol Cell Cardiol. 2022;170:100–114. doi: 10.1016/j.yjmcc.2022.06.003. [DOI] [PubMed] [Google Scholar]
- 67.Kadakol A., Malek V., Goru S.K., et al. Telmisartan and esculetin combination ameliorates type 2 diabetic cardiomyopathy by reversal of H3, H2A, and H2B histone modifications. Indian J Pharmacol. 2017;49(5):348–356. doi: 10.4103/ijp.IJP_710_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nie L., Shuai L., Zhu M., et al. The landscape of histone modifications in a high-fat diet-induced obese (DIO) mouse model. Mol Cell Proteomics. 2017;16(7):1324–1334. doi: 10.1074/mcp.M117.067553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nolan C.J., Madiraju M.S., Delghingaro-Augusto V., et al. Fatty acid signaling in the beta-cell and insulin secretion. Diabetes. 2006;55(Suppl 2):S16–S23. doi: 10.2337/db06-s003. [DOI] [PubMed] [Google Scholar]
- 70.Bandyopadhyay G.K., Yu J.G., Ofrecio J., et al. Increased malonyl-CoA levels in muscle from obese and type 2 diabetic subjects lead to decreased fatty acid oxidation and increased lipogenesis; thiazolidinedione treatment reverses these defects. Diabetes. 2006;55(8):2277–2285. doi: 10.2337/db06-0062. [DOI] [PubMed] [Google Scholar]
- 71.Zhu Z., Han Z., Halabelian L., et al. Identification of lysine isobutyrylation as a new histone modification mark. Nucleic Acids Res. 2021;49(1):177–189. doi: 10.1093/nar/gkaa1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Q.Q., Hao J.J., Zhang Z., et al. Proteomic analysis of proteome and histone post-translational modifications in heat shock protein 90 inhibition-mediated bladder cancer therapeutics. Sci Rep. 2017;7(1):201. doi: 10.1038/s41598-017-00143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oo H.Z., Seiler R., Black P.C., et al. Post-translational modifications in bladder cancer: expanding the tumor target repertoire. Urol Oncol Semin Orig Investig. 2020;38(12):858–866. doi: 10.1016/j.urolonc.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 74.Goudarzi A., Hosseinmardi N., Salami S., et al. Starvation promotes histone lysine butyrylation in the liver of male but not female mice. Gene. 2020;745 doi: 10.1016/j.gene.2020.144647. [DOI] [PubMed] [Google Scholar]
- 75.Shi J., Dong A., Shen W.H. Epigenetic regulation of rice flowering and reproduction. Front Plant Sci. 2015;5:803. doi: 10.3389/fpls.2014.00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu Y., Xu Q., Liu Y., et al. Dynamics and functional interplay of histone lysine butyrylation, crotonylation, and acetylation in rice under starvation and submergence. Genome Biol. 2018;19(1):144. doi: 10.1186/s13059-018-1533-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang F., Wang C. Profiling of post-translational modifications by chemical and computational proteomics. Chem Commun. 2020;56(88):13506–13519. doi: 10.1039/d0cc05447j. [DOI] [PubMed] [Google Scholar]
- 78.Zhang K., Chen Y., Zhang Z., et al. Identification and verification of lysine propionylation and butyrylation in yeast core histones using PTMap software. J Proteome Res. 2009;8(2):900–906. doi: 10.1021/pr8005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang G., Zheng Y., Wu Y.Q., et al. An information entropy-based approach for computationally identifying histone lysine butyrylation. Front Genet. 2020;10:1325. doi: 10.3389/fgene.2019.01325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu G., Wang J., Wu Z., et al. SAHA regulates histone acetylation, butyrylation, and protein expression in neuroblastoma. J Proteome Res. 2014;13(10):4211–4219. doi: 10.1021/pr500497e. [DOI] [PubMed] [Google Scholar]
- 81.Zhao Y., Fang X., Wang Y., et al. Comprehensive analysis for histone acetylation of human colon cancer cells treated with a novel HDAC inhibitor. Curr Pharmaceut Des. 2014;20(11):1866–1873. doi: 10.2174/13816128113199990531. [DOI] [PubMed] [Google Scholar]
- 82.Santra S., MacDonald A., Preece M.A., et al. Long-term outcome of isobutyryl-CoA dehydrogenase deficiency diagnosed following an episode of ketotic hypoglycaemia. Mol Genet Metab Rep. 2017;10:28–30. doi: 10.1016/j.ymgmr.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yun J.W., Jo K.I., Woo H.I., et al. A novel ACAD8 mutation in asymptomatic patients with isobutyryl-CoA dehydrogenase deficiency and a review of the ACAD8 mutation spectrum. Clin Genet. 2015;87(2):196–198. doi: 10.1111/cge.12350. [DOI] [PubMed] [Google Scholar]
- 84.Günes Günsel G., Conlon T.M., Jeridi A., et al. The arginine methyltransferase PRMT7 promotes extravasation of monocytes resulting in tissue injury in COPD. Nat Commun. 2022;13(1):1303. doi: 10.1038/s41467-022-28809-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Malebary S., Rahman S., Barukab O., et al. iAcety-SmRF: identification of acetylation protein by using statistical moments and random forest. Membranes. 2022;12(3):265. doi: 10.3390/membranes12030265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fiedler M., Ip W.H., Hofmann-Sieber H., et al. Protein-protein interactions facilitate E4orf6-dependent regulation of E1B-55K SUMOylation in HAdV-C5 infection. Viruses. 2022;14(3):463. doi: 10.3390/v14030463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang D., Kraus W.L. The expanding universe of PARP1-mediated molecular and therapeutic mechanisms. Mol Cell. 2022;82(12):2315–2334. doi: 10.1016/j.molcel.2022.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao Y., Zhang L., Ju C., et al. Quantitative multiplexed proteomics analysis reveals reshaping of the lysine 2-hydroxyisobutyrylome in Fusarium graminearum by tebuconazole. BMC Genom. 2022;23(1):145. doi: 10.1186/s12864-022-08372-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huo B., Liu Y., Li L., et al. A chemical method for genome- and proteome-wide enrichment and O-GlcNAcylation profiling of chromatin-associated proteins. Talanta. 2022;241 doi: 10.1016/j.talanta.2021.123167. [DOI] [PubMed] [Google Scholar]
- 90.Liu S., Liu G., Cheng P., et al. Genome-wide profiling of histone lysine butyrylation reveals its role in the positive regulation of gene transcription in rice. Rice. 2019;12(1):86. doi: 10.1186/s12284-019-0342-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.González-Grandío E., Álamos S., Zhang Y., et al. Chromatin changes in phytochrome interacting factor-regulated genes parallel their rapid transcriptional response to light. Front Plant Sci. 2022;13 doi: 10.3389/fpls.2022.803441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nounurai P., Afifah A., Kittisenachai S., et al. Phosphorylation of CAD1, PLDdelta, NDT1, RPM1 proteins induce resistance in tomatoes infected by Ralstonia solanacearum. Plants. 2022;11(6):726. doi: 10.3390/plants11060726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Carrillo-Sepulveda M.A., Maddie N., Johnson C.M., et al. Vascular hyperacetylation is associated with vascular smooth muscle dysfunction in a rat model of non-obese type 2 diabetes. Mol Med. 2022;28:30. doi: 10.1186/s10020-022-00441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zou L., Yang Y., Wang Z., et al. Lysine malonylation and its links to metabolism and diseases. Aging Dis. 2023;14(1):84–98. doi: 10.14336/AD.2022.0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xue Q., Yu T., Wang Z., et al. Protective effect and mechanism of ginsenoside Rg2 on atherosclerosis. J Ginseng Res. 2023;47(2):237–245. doi: 10.1016/j.jgr.2022.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.