Abstract
The retinal pigment epithelium (RPE) and choroid are located behind the human retina and have multiple functions in the human visual system. Knowledge of the RPE and choroid cells and their gene expression profiles are fundamental for understanding retinal disease mechanisms and therapeutic strategies. Here, we sequenced the RNA of about 0.3 million single cells from human RPE and choroids across two regions and seven ages, revealing regional and age differences within the human RPE and choroid. Cell–cell interactions highlight the broad connectivity networks between the RPE and different choroid cell types. Moreover, the transcription factors and their target genes change during aging. The coding of somatic variations increases during aging in the human RPE and choroid at the single-cell level. Moreover, we identified ELN as a candidate for improving RPE degeneration and choroidal structure during aging. The mapping of the molecular architecture of the human RPE and choroid improves our understanding of the human vision support system and offers potential insights into the intervention targets for retinal diseases.
Keywords: Aging, Choroid, ELN, Human, RPE, scRNA-seq
Introduction
The retinal pigment epithelium (RPE) is a continuous monolayer of epithelial cells behind the retina. The RPE has multiple functions, such as absorption of light, storage and convention of vitamin A esters, and phagocytosis of rod and cone outer segments.1 The choroid is a heterogeneous connective tissue that supports both the RPE and the outer retina. It is located behind Bruch's membrane (with no cells) and RPE. The RPE, Bruch's membrane, and choroid are closely connected anatomically. The choroid contains a rich vascular system, which plays a critical role in providing oxygenated blood to the RPE and photoreceptor cells.2 The choroidal vascular bed provides about 65% of the blood to the whole retina3 and anatomically consists of a very dense superficial capillary system known as the choriocapillaris. Additionally, the choroid houses several cell types also found in other connective tissues, including fibroblasts, melanocytes, pericytes, and infiltrating immune cells.4 The RPE and the choroid work closely together both physiologically and pathologically.5 Dysfunction of both the RPE and the choroid has been widely implicated in age-related macular degeneration (AMD) pathogenesis.6,7
With aging, the human retina accumulates lipofuscin (an end product of phagosome breakdown). This accumulation debilitates RPE cells, and their lines of defense become weaker over time. It is believed that a reduced capability to absorb light and compensate for oxidative damage is an important factor contributing to the chain of events leading to the onset of AMD in the older population.8, 9, 10, 11 With age, the choroidal thickness decreases dramatically, accompanied by decreased choroidal blood flow.12 The age-related changes in RPE often lead to pathological tissue damage that warrants intervention.13 Although little is known about the mechanisms that trigger and accelerate this group of conditions, the amount of knowledge generated over the past few years in this field has been increasing.
The transcriptome of the human retina, RPE, and choroid has been reported using bulk tissue RNA-seq and single-cell RNA sequencing in a limited number of cells (scRNA-seq).6,14, 15, 16, 17, 18, 19, 20 However, the changes that take place across regions during development and aging are poorly understood. Here, we decode the molecular dynamic architecture of the human RPE and choroid by sequencing more than 0.3 million RPE and choroid cells at the single-cell level in seven samples of different ages. This work advances the current understanding of RPE and choroid regional differences, age correlations, cell–cell connectivity, transcriptional regulatory network, as well as somatic variations at the single-cell level and therefore provides clues for investigating human RPE and choroid development, anti-aging, and diseases.
Materials and methods
The main materials and methods are listed below. The detailed materials and methods are provided in Supplementary Methods.
Experimental procedures
Primary human RPE and choroid sample collection and scRNA sequencing
This study was approved by the Human Research Ethics Committee of the Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital (2019-327). Informed consent was obtained from all participants or their clients. The experiments conformed to the principles set out in the procedures and regulations for organ donation after the death of Chinese citizens (2019 edition). Donors with a documented history of ocular disease were excluded from the study. The RPE and choroid samples were dissociated and immediately processed for scRNA-seq using the 10× genomics platform.
For all RPE and choroid samples, scRNA-seq libraries were constructed using Chromium Single-Cell 3ʹ Reagent Kits v3 according to the manufacturer's protocol (CG000183 Rev C). Then, the PE 150 sequencing mode was adopted for about 30k read pairs/cell.
In situ RNA hybridization and quantification
Eyeballs were taken from the left eye of a donor aged 30 years (cause of death: accidental injury) and a donor aged 86 years (cause of death: lung cancer). In situ RNA hybridization was performed using the Advanced Cell Diagnostics RNAscope Multiplex Fluorescent Detection kit v2 (ACD, #323100) according to the manuals, following these steps: (i) prepare fresh frozen tissue sections, (ii) fix the sections, (iii) dehydrate the sections, (iv) create a hydrophobic barrier, (v) apply RNAscope® hydrogen peroxide, (vi) apply protease IV, and (vii) proceed immediately to the RNAscope® Assay.
For multiplex staining, the following probes were used: UGT2A1 (Hs-UGT2A1 814271-C1), DCN(Hs-DCN 589521-C1), F5 (Hs-F5 1000991-C2), and AOC2 (Hs-AOC2 485701-C3). Slides were counterstained with ProLong gold antifade reagent with DAPI (P36931, Life Technologies). Mounted slides were imaged on a ZEISS LSM900 fluorescent microscope (Carl Zeiss AG).
AAV vectors
For human ELN overexpression, pAV-CMV-P2A-GFP (NM_001081754.3) vector plasmid was constructed with AsisI/MluI enzyme digestion sites and packaged in AAV DJ. The final virus concentration was 2.2 × 1013 viral genomes/mL in a balanced salt solution (BSS) (Alcon, Fort Worth Texas) supplemented with 0.014% Tween-20. For ELN knockdown, four shRNA sequences were constructed in ADM-ELN-shRNA-mCMV-copGFP vector and transfected in HEK 293 cells for knockdown efficiency tests. Finally, sequence 5′GGAAACTGCCCTATGGCTATGTTCAAGAGACATAGCCATAGGGCAGTTTCCTTTTTT3′, which showed 75% knockdown efficiency, was used for packaging in adenovirus. For ELN knockdown control, ADM-U6-shRNA-mCMV-copGFP was used. The final virus concentration was 2.6 × 1011 viral genomes/mL in BSS (Alcon, Fort Worth Texas) supplemented with 0.014% Tween-20.
Cell culture and immunofluorescence
HSFCs (human embryonic scleral fibroblast cells; Wenzhou Medical University, China) and ARPE-19 (human retinal epithelial cells, national experimental cell resource sharing platform, China) were cultured in DMEM/F12 medium (hyclone, USA) containing 10% FBS and 1% streptomycin. HRECs (human retinal vascular endothelial cells, cell systems, USA, #acbri181) were cultured in an EBM-2 complete medium containing 5% FBS and 1% streptomycin. The cells were cultured in a moist 37 °C constant temperature incubator. Antibodies (ELN: elastin, Abcam, ab21610, 1:100), ghost pen cyclic peptide (Invitrogen, a12381, 1:40), and fluorescent secondary antibody (1:500) were added for laser confocal photography.
Mice
The C57BL/6J mice were raised in an environmentally controlled 12-h light–dark cycle in our laboratory with food and water available. The mice were divided into the old group (18 months old) and the young group (2 months old). One microliter of virus (virus titer: 2.2 × 1013 vg/mL) was injected into the vitreous with a microsyringe (syringe, Hamilton No. 7632-01; needle, Hamilton No. 7803-05, including fluorescent indicator), and the needle was removed after staying in the eye for 10 s. Three months later, the mice were anesthetized, and their eyeballs were used for experimental observation.
Mouse eyeball RPE wholemount
After the mice were sacrificed by anesthetization, the eyeballs were taken out and placed in a 1.5-mL EP tube, and PBS was added and soaked to remove hair and blood. The eyeballs were transferred to a 6-cm culture dish, PBS was added to float, and the connective tissue, optic nerve, and muscle outside the eyeball were removed under an anatomical microscope. The retina above the RPE was gently peeled off with tweezers, and the cut eyeballs were placed in 4% PFA and fixed at 4 °C for 24 h. Then, PFA was sucked from the tube, and the stationary solution was added (5% FBS, triton-x100, NaN3) and incubated at room temperature for 1 h. The blocking solution was then sucked off, ghost pen cyclic peptide was added (F-actin, 1:40), and the sample was left overnight at 4 °C. Finally, the eyeballs were placed on slides with a brush, and the slides were sealed.
Eyes immunofluorescence
Eyes were fixed in cold 4% paraformaldehyde (PFA) in PBS, placed into the embedding box filled with OCT at −80 °C, and frozen for 30 min. The eye tissue was then sectioned with a frozen slicer at a thickness of 12 μm. Next, the slides were baked at 37 °C for 30 min and washed with PBS three times for 5 min. A total of 50 μL of blocking solution was added (5% serum, 0.25% TritonX-100 + 0.06% sodium azide), followed by incubation for 2 h at room temperature. The same buffer was used to incubate tissues with the specified primary antibody (ELN: Elastin, Abcam, #ab21610, 1:100, F-Actin (Invitrogen, #A12381, 1:40) overnight at 4 °C. After washing in PBS, tissues were incubated with the appropriate secondary antibody for 1 h and rinsed in PBS before imaging.
Transmission electron microscopy (TEM)
Eyeballs were prefixed with a 3% glutaraldehyde, then postfixed in 1% osmium tetroxide, dehydrated in series acetone, infiltrated in Epox 812, and embedded. The semithin sections were stained with methylene blue, and ultrathin sections were cut with a diamond knife and stained with uranyl acetate and lead citrate. The sections were examined with a JEM-1400-FLASH transmission electron microscope.
Scanning electron microscope (SEM)
The eye tissues were washed twice with water for 5 min and dehydrated with a gradient alcohol series (30%, 50%, 70%, 80%, 90%, 95%, and 100%), 10 min for each concentration. The samples were gently attached with conductive adhesive and sprayed with ion sputtering. Sections were examined with an ion sputtering device (E-1045, Japan) and an SEM (INSPECT, FEI, USA).
Computational methods
Data pre-processing
The Cell Ranger package (v3.1, 10× Genomics) was used to extract unique molecular identifiers, cell barcodes, and genomic reads from the raw sequence data of the 10× Chromium experiments. Reads were aligned against the human genome version GRCh38. Reads with the same cell barcode and unique molecular identifier were identified as unique transcripts. The Cell Ranger package v3.1 algorithm uses a cutoff based on the total UMI counts of each barcode to identify cells. Cells detected with more than 10,000 genes or fewer than 200 genes were removed. Cells with a mitochondrial gene ratio of greater than 10% or red blood cell marker gene ratios of greater than 10% were excluded. Doublets were detected by using the Scrublet package (version 2.0.2) using the scrub.scrub_doublets function. The batch effect was corrected using the FindIntegrationAnchors function for each sample and then running the IntegrateData function of Seurat v3.
Cell clustering and sub-clustering
The output of Cell Ranger aggr was loaded into R using Seurat v3 (Seurat_3.2.0) for modified integration analysis to remove batch effects and clustering. Cell types were assigned to each cluster using the canonical specific markers: RPE (RPE65), fibroblast (DCN), macrophage (AIF1), pericyte (ACTA2), Schwann (PLP1), melanocyte (MLANA), mast (KIT), endothelial (VWF), B cells (CD79A), and NK/Ts (CD2). The detailed pathways for each cell type are listed in Table S1.
Cell type annotation
The ten sets of cell markers were: RPE (RPE65), fibroblast (DCN), macrophage (AIF1), pericyte (ACTA2), Schwann (PLP1), melanocyte (MLANA), mast (KIT), endothelial (VWF, PECAM1), B cells (CD79A), and NK/Ts (CD2).
Regional transcriptomic comparative analysis
We performed DEG (differentially expressed gene) analysis on the RPE and each choroid cell type in the central region to the peripheral region. DEGs in the comparison group of samples were calculated using FindMarkers ((min.pct = 0.01, logfc. threshold = 0.1, test. use = “bimod”, assay = “RNA”); significant P-value ≤ 0.05 & abs (Log2_Fold_Change) ≥ 0.585). The average log (fold change) of each DEG (FDR < 0.05) was calculated as significant DEGs in that region (regional preferred genes) for pathway-enriched analysis using the metascape21 STRING: functional protein association networks.22
Transcriptomic comparative analysis across ages
To identify the dynamic changes in gene expression at different stages of development and aging, we used 21, 32, and 44 years as the adult group and the fourth day post-birth (P4th day), 7 years, 56 years, and 81 years for comparison with the adult group, respectively. We performed DEG analysis on each cell type in the comparison groups. The DEGs in each comparison group of samples (e.g., between the 7-year-old cells and the adult group) were calculated using FindMarkers ((min.pct = 0.01, logfc. threshold = 0.1, test. use = “bimod”, assay = “RNA”); significant P-value ≤ 0.05 & abs (Log2_Fold_Change) ≥ 0.585). The average log (fold change) of each DEG (FDR < 0.05) was calculated as significant DEGs for that age (age-preferred genes) for pathway-enriched analysis using the metascape21 STRING: functional protein association networks.22
Pairwise expression pattern of DEGs across ages
The pairwise distance matrix was calculated using R. This method calculates all pairwise distances between spatial points (in rows) and presents the distances as a matrix.
Correlation analysis of age with gene expression
The Pearson correlation coefficient was calculated between ages and gene expressions. The age of a person was calculated in days. The amount of gene expression for each cell type was calculated using the rank of its expression amount in the transcriptome.
Cell–cell communication analysis
The CellChat R package23 was used to infer global communications among RPE and choroid cell types, interactions among ligands, receptors, and their cofactors, which accurately represent known heteromeric molecular complexes.
Regulatory network analysis
The SCENIC R package can perform gene regulatory network reconstruction and cell-state identification from scRNA-seq data.24 We used this package to infer gene regulatory networks and cell types from scRNA-seq data for each age and combined ages. Briefly, the SCENIC pipeline consists of three steps. First, candidate regulatory modules were inferred from co-expression patterns between genes. Next, co-expression modules were refined by the elimination of indirect targets using TF motif information. Finally, the activity of these discovered regulons was measured in each cell and used for clustering.
Single-cell variant analysis (SNP and InDel calling)
A BAM file generated by CellRanger of each RPE and choroid single-cell, SNP/indel mutation analysis for the human genome GRCh38 version was carried out using Samtools (http://www.htslib.org/). SNP and InDel calling quality control were sequenced with a quality value > 10, base coverage depth > 20, snpgap set to 5, and indelgap set to 5. After the quality control, high-quality SNP and indel sets of single cells were obtained. Bcftools (https://samtools.github.io/bcftools/) was used to merge variants from multiple cells. Annovar25 (https://doc-openbio.readthedocs.io/projects/annovar/en/latest/) was used to annotate these variations sets.
Infer copy number variation from scRNA-Seq data
InferCNV (https://rdrr.io/bioc/infercnv/) was used to explore the scRNA-Seq data to identify evidence of large-scale chromosomal CNVs, such as gains or deletions of entire chromosomes or large segments of chromosomes. CNVs were obtained by exploring the expression intensity of genes across positions of the genome in comparison to the average cells. A heatmap was generated illustrating the relative expression intensities across each chromosome, and it became readily apparent which regions of the genome were over or less abundant compared to the average.
Results
Single-cell RNA-seq of 0.3 million human RPE and choroid cells
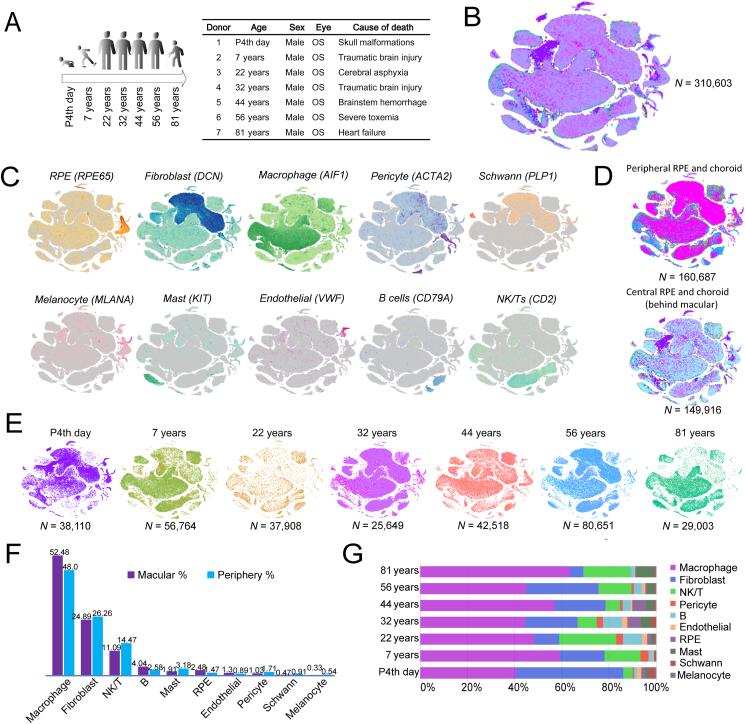
We generated the age-changed RPE and choroid transcriptome profiles of human beings at a single-cell resolution from seven separate donors of different ages with no ocular diseases. All seven donors were male Han Chinese, aged between P4th day and 81 years old (Fig. 1A). For each RPE and choroid, we dissected the RPE and choroid into the central choroid (behind macular region) and periphery choroid, separately. All the RPE and choroid samples were taken from the left eyes of male donors. A total of 310,603 qualified cells (Fig. 1B) were obtained for subsequent analyses (Fig. 1C). Then, ten main cell types were identified based on the expression of canonical specific markers: RPE (RPE65), fibroblast (DCN), macrophage (AIF1), pericyte (ACTA2), Schwann (PLP1), melanocyte (MLANA), mast (KIT), endothelial (VWF), B cells (CD79A), and NK/T (we combined natural killer cells and T lymphocytes as NK/T in this study) (CD2) (Fig. 1C). The numbers of sequenced cells and each cell proportion of the periphery or central RPE/choroid are presented in Figure 1D, F, respectively. RPE accounted for 1.47% in the periphery region and 2.48% in the central region, suggesting a higher percentage in the central region than in the periphery region. For the choroid, macrophages comprised about half of all cells (52.48% for macular choroid and 48.0% for periphery choroid) (Fig. 1F). The next-highest percentage cell type was fibroblasts, accounting for 24.89% of the macular choroid and 26.26% of the periphery choroid of all choroid cells. NK/T and B cells were the next largest populations. Minor cell populations, including masts, pericytes, Schwann, and melanocytes, had lower proportions in the macular choroid than that in the periphery choroid. A higher cell proportion of endothelial cells was distributed in the macular choroid than in the periphery choroid (1.30% vs. 0.89%) (Fig. 1F).
Figure 1.
Single-cell RNA sequencing (scRNA-seq) of 310,603 cells from the human RPE and choroid across two regions and seven ages. (A) scRNA-seq donors' information and workflow of the experimental scheme. (B) t-SNE visualization of 310,603 single cells from two human RPE and choroid regions colored by samples. (C) Annotation based on known marker genes of RPE and choroid cell types. (D) t-SNE visualization of cells from two human RPE and choroid regions (peripheral RPE and choroid, central RPE and choroid (behind the macular region)) colored by samples. (E) t-SNE visualization of cells in seven ages colored by samples. (F) The cell percentages of each of the main cell types behind the macular and peripheral RPE and choroid. (G) The cell percentages of each of the main cell types at each age. The most expressed genes of each subtype are shown in Table S1.
The cellular composition of RPE and choroid cells tends to change dynamically during their life span. The numbers of sequenced cells and the cell proportion at each age of the RPE/choroid are presented in Figure 1E, G, respectively. RPE cells decreased in the elderly ages (56 years, 0.80%; 81 years, 0.24%) compared to the average level (close to 2.0%). This is consistent with the previous findings that RPE cell density decreases with aging.1 The cellular composition of each choroid cell type also tends to vary with age. In general, macrophages accounted for the lowest proportion on the P4th day (38.8%) and the highest proportion at 81 years of age (63%). Similarly, masts and NK/T also showed the highest proportion at 81 years compared to other ages. These results suggest that the immune system is over-activated in the old choroid. On the contrary, the proportion of fibroblast cells in the human choroid was highest on the P4th day (46.5%) and lowest at the age of 81 years (5.4%) (Fig. 1G), indicating that the choroid structure degenerates in old age.
We next explored the biological implications of the 10 human RPE and choroid cell types using DEGs in each cell type for pathway enrichment analyses (Table S1). Each cell type has a specific physiological function related to signaling pathways. For example, the commonly expressed genes in the human RPE were enriched in pathways of energy metabolism (e.g., oxidative phosphorylation) (Table S1). Further, RPE cells are also involved in fatty acid degradation, steroid biosynthesis, etc., suggesting the multi-functional role of RPE tissue.8,26,27 In macrophages, its function was enriched in transport and catabolism (phagosome and lysosome) and immune response (antigen processing and presentation) (Table S1). Fibroblasts were involved in signal transduction (PI3K-Akt signaling, ECM-receptor interaction, and TGF-beta signaling).
The regional gene expression difference in the human RPE and choroid
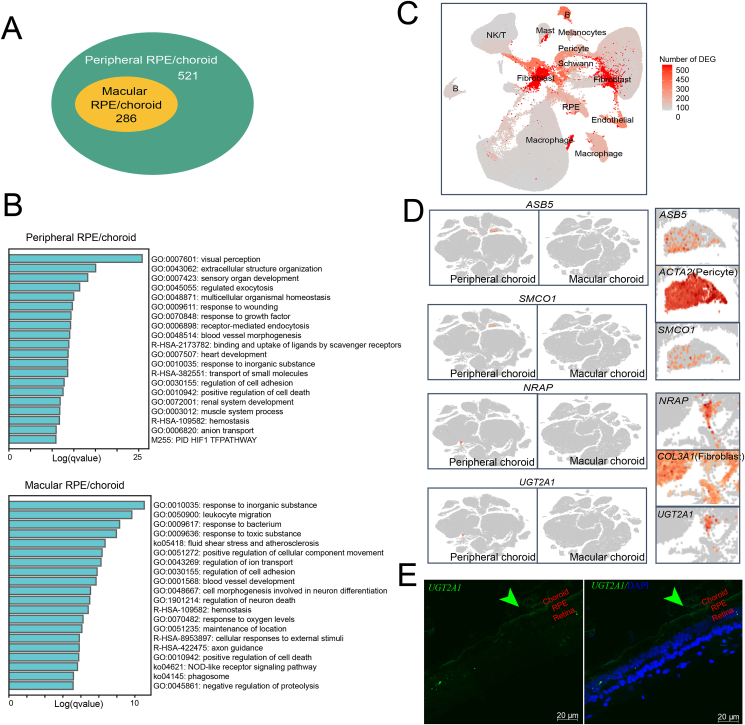
As shown in Figure 1D, the cell composition changes from behind the macular to peripheral regions of the RPE and choroid. To further investigate the functional preferences of these two regions, the preferred DEGs behind the macular and peripheral regions of the RPE and choroid were calculated (Fig. 2 and Table S2). The number of genes with preferred expression in the peripheral region was about twice that in the behind macular region (Fig. 2A). The preferred genes in the peripheral region were mainly related to response to visual perception and extracellular structure organization (Fig. 2B, the upper panel). The preferred genes expressed behind the macular RPE and choroid are mainly involved in the response to inorganic substances and leukocyte migration (Fig. 2B, the lower panel). This relatively higher enriched degree of these functions in the peripheral region than behind the macular region may indicate more active cellular events in the peripheral RPE and choroid in the peripheral region.
Figure 2.
Regional preferred genes of peripheral the RPE/choroid and central RPE/choroid. (A) The number of preferred genes of the peripheral RPE/choroid and central RPE/choroid (DEGs with higher expression in the corresponding region). (B) Top 20 functional annotations of the DEGs in each of the peripheral RPE/choroid and central RPE/choroid. (C) Cell type UMAP visualization of the number of DEGs of peripheral RPE/choroid and central RPE/choroid, colored by DEGs number. (D) t-SNE visualization of ASB5, SMCO1, NRAP, and UGT2A1, which are only expressed in the peripheral choroid. (E) RNAscope RNA in situ hybridization expression of UGT2A1 in the fibroblasts of the peripheral choroid. UGT2A1 is also rarely expressed in the outer segment of the photoreceptor layer.
This regional difference is associated with the cell type. Overall, the DEGs behind the macular and peripheral choroids were mainly concentrated in fibroblasts, partial macrophages, and masts (Fig. 2C). We also detected a few genes, which were only expressed in the peripheral choroid (Fig. 2D). For example, ASB5 and SMCO1 were specifically expressed in the pericytes of the peripheral choroid (Fig. 2D, the first two panels), while NRAP and UGT2A1 were specifically expressed in fibroblasts of the peripheral choroid (Fig. 2D, the second two panels). We validated the expression of UGT2A1 by RNA in situ hybridization, and the results suggested that UGT2A1 is expressed in fibroblasts in the peripheral choroid and the photoreceptor cell layer of the retina (Fig. 2E). These genes may contribute to the functional interval difference of human choroids.
Gene expression changes positively or negatively with age
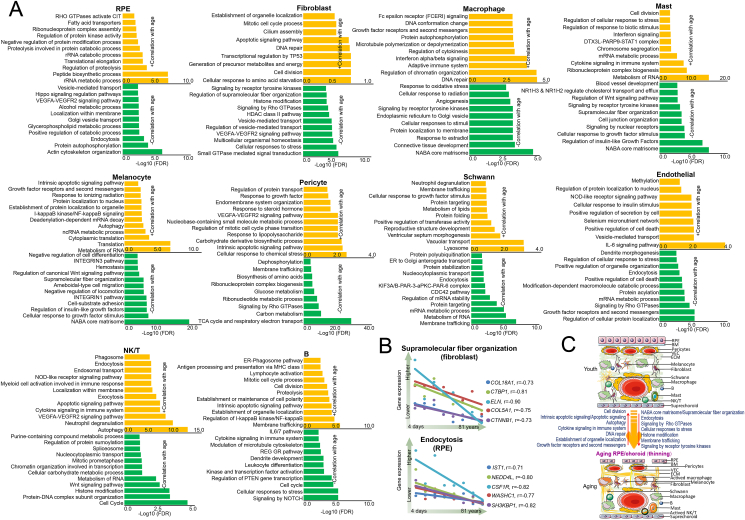
To investigate the genes and functions in the RPE and each of the choroid cell types, we calculated the correlation of gene expression in the RPE and choroid with age (Fig. 3A and Table S3). In general, there were similarities and differences in the enriched signal pathways and functions among cell types. The top negatively correlated genes (correlation = −0.65 to −0.99) were involved in the NABA core matrisome, endocytosis, signaling by Rho GTPases, supramolecular fiber organization, cellular responses to stress, and histone modification. For example, the NABA core matrisome, which is also related to the supramolecular fiber organization pathway, including ELN, COL1A1, etc. was negatively correlated with age for most cell types, such as fibroblasts, macrophages, RPE, HREC, mast, and menocypte cells (Fig. 3A, B-the first panel, C and Table S3). NABA core matrisome pathway genes encode the core extracellular matrix, including extracellular matrix (ECM), glycoproteins, collagens, and proteoglycans. Endocytosis, including the genes IST1 and WASHC1, were negatively correlated with age mainly in RPE, endothelial, and Swann cells (Fig. 3A, B-the second panel, C and Table S3). One hallmark of ECM proteins is their domain-based structure. These results suggested that the basic structural proteins of the choroid tissue degenerate with age.
Figure 3.
The top function/pathways of the negative or positive correlation of gene expression with age in each of the 10 cell types of the human RPE and choroid. (A) The most enriched function/pathways predicted by metascape (https://metascape.org) of the genes with correlation values between −0.6 and −1 (negative correlation) and 0.6–1 (positive correlation) for gene expression (using the rank of gene expression from high to low) and age (calculated as days). The abscissa is the −Log10(FDR) predicted by metascape; the ordinate indicates the function/pathways predicted by metascape. Detailed information regarding the enriched function/pathways of each cell type is provided in Table S4. (B) The genes associated with the supramolecular fiber organization function in fibroblasts (the first panel) and endocytosis in the RPE (the second panel). (C) Schematic diagram of representative changes in choroidal and RPE functions directly positively or negatively related to age (young and old age).
Conversely, other pathways were highly enriched in the top positively correlated groups in at least two kinds of cells, including cell division, the intrinsic apoptotic signaling pathway, the apoptotic signaling pathway, autophagy, cytokine signaling in the immune system, and DNA repair (Fig. 3A, C and Table S3). Apoptotic signaling pathway-related genes showed a positive correlation with age mainly in fibroblasts, melanocytes, and pericytes. The regulation of apoptosis is essential for maintaining the balance between cell survival and death signaling.28 The upregulation of the apoptosis pathway during aging in the RPE/choroid suggests that cell death stress increases with age. Cytokine signaling in immune system-related genes was positively correlated with age mainly in mast and NK/T cells.
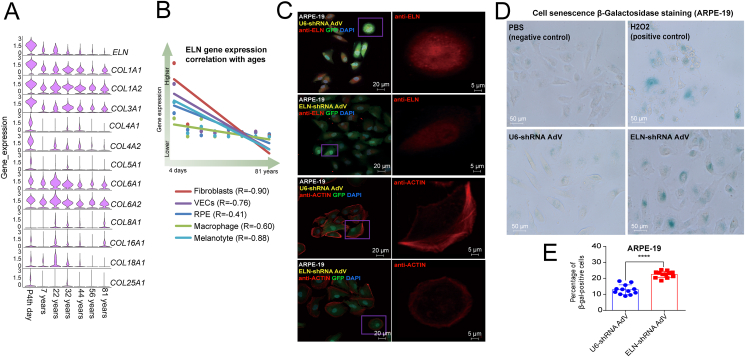
Gene expression profiles of supramolecular fiber organization
Prominent characteristics of cytoskeletal fibers in the aged RPE/choroid include size reduction, decreased elongation, and collapsed morphology.29 The ECM, and especially the connective tissue with its elastin, links tissues of the body together and plays an important role in force transmission and tissue structure maintenance, especially in the RPE/choroid. Our results suggested that the fibroblasts in the choroid degenerate with age. To further investigate the reduction of the ECM in the choroid, we focused on the supramolecular fiber organization. The structural integrity of the human RPE/choroid is largely dependent on the quality of the ECM, which is produced, organized, and maintained by fibroblasts and impacts the RPE and choroid. Therefore, we examined the expression patterns of individual genes that are thought to contribute to ECM supramolecular fiber organization, including elastin-coding gene ELN and collagen protein-coding genes (Fig. 4A). ELN, as mentioned above, is prominently reduced after 22 years of age. The expressions of most of the collagen coding genes, including COL1A1, COL1A2, COL3A1, COL4A1, COL4A2, COL5A1, COL6A1, and COL6A2, are reduced with aging. ELN expression is negatively correlated with age in several cell types (Fig. 4B). As ELN is a secretory protein and the main component of ECM tissue, to further investigate the effect of ELN reduction on the cytoskeleton, we knocked down the ELN gene using a lentivirus and carried out experiments in human RPE cells (ARPE-19 cell line; Fig. 4C). We observed that after ELN knockdown, the cytoskeleton of these cells was reduced to varying degrees. These results suggest that the decrease of ELN expression during aging may be one of the main reasons for RPE deformation in old age. The results of cell senescence β-Galactosidase staining in ARPE-19 (Fig. 4D, E) suggest that ELN knockdown promoted RPE cell senescence, indirectly proving that ELN overexpression may have anti-aging effects.
Figure 4.
Fiber changes with aging and the effect of ELN knockdown in vitro. (A) The violin plots of ELN and collagen coding genes' expression at each age. (B) The correlation between ELN expression and age in five cell types suggests a strong negative correlation with age. (C) The effect of ELN knockdown in vitro. Compared to the U6-shRNA control, the AdV driving ELN-shRNA knockdown reduced the density of elastin (staining by ELN antibody) and the cytoskeleton (staining by ACTIN antibody) in human RPE cells (ARPE-19). In the results of each cell, the figure on the right is an enlarged view of the figure in the box on the left. (D) Cell senescence β-Galactosidase staining of ARPE-19 cells treated with PBS (negative control), H2O2 (0.3 mM, positive control), U6-shRNA AdV, and ELN-shRNA AdV. (E) Statistical analysis of cell senescence β-Galactosidase staining of U6-shRNA AdV vs. ELN-shRNA AdV in Figure 4D.
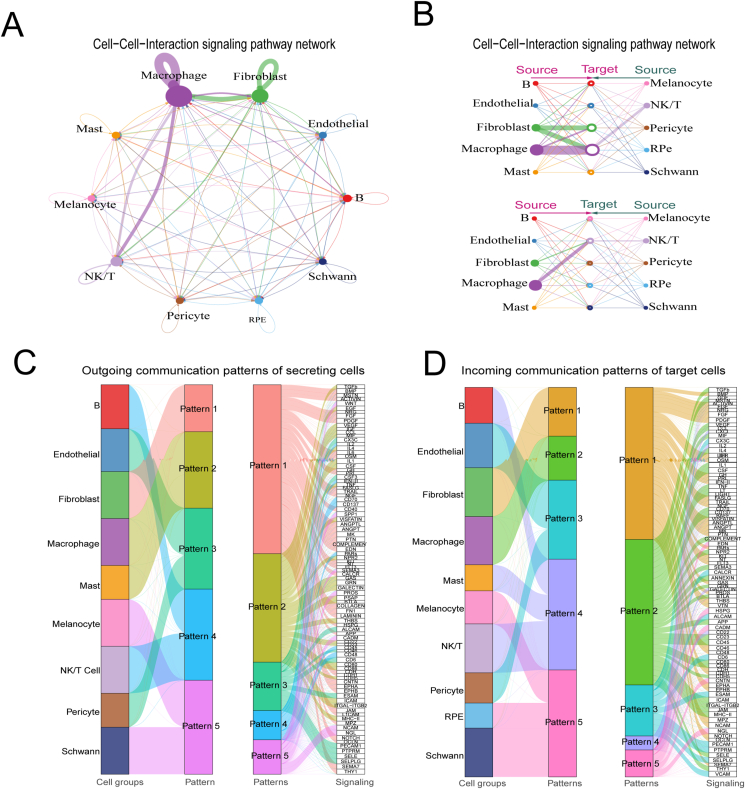
Cell–cell interaction in the human RPE and choroid
The RPE and choroid are responsible for photoelectric signal conduction from the retina to the brain. Currently, the molecular communication flow among RPE and choroid cell types is poorly understood. To investigate this issue, we analyzed the cell–cell communications using CellChat,23 which can identify key features of intercellular communications at the scRNA-seq level. In total, CellChat detected 412 significant ligand–receptor pairs among the 10 main RPE and choroid cell groups, which were further categorized into 102 signaling pathways, including VEGF and FGF (Fig. 5 and Table S4). A systematic network of the cell–cell communication of RPE and main choroid cell types is shown in Figure 5A and B. This analysis uncovered five patterns of outgoing signaling (Fig. 5C) and five patterns of incoming signaling (Fig. 5D). For example, the output revealed that the largest portion of outgoing fibroblast signaling was characterized by pattern #1, which represents multiple pathways, including but not limited to BMP and PDGF (Fig. 5C). Outgoing macrophage and mast signaling were characterized by pattern #2, which represents multiple pathways, including but not limited to TGFβ and VEGF (Fig. 5C). Outgoing endothelial cells and their pericytes signaling were characterized by pattern #3, which represents multiple pathways, including CX3C and CSF3 (Fig. 5C). Meanwhile, the communication patterns of target cells (Fig. 5D) showed that incoming macrophage signaling was dominated by pattern #2, while mast, B, and NK/T signaling was characterized by pattern #4, which represents multiple pathways, including but not limited to IL2 and CD22 (Fig. 5D). Incoming melanocyte, RPE, and Schwann signaling were characterized by pattern #5, which represents multiple pathways, including EDN and HSPG. These results show that (i) two distinct cell types can rely on largely overlapping signaling networks and that (ii) certain cell types, such as fibroblasts and macrophages, simultaneously activate multiple signaling patterns and pathways, while other cell types, such as melanocyte and Schwann cells, rely on fewer and more homogeneous communication patterns.
Figure 5.
Cell–cell interaction for 10 main cell types of the human RPE and choroid. (A) Circle visualization of human RPE and choroid cellular communication among the 10 main cell types, as predicted by CellChat. (B) Hierarchy visualization for human RPE and choroid cellular communication among the 10 main cell types, as predicted by CellChat. (C) The alluvial plot showing the outgoing communication patterns of secreting cells among the main human RPE and choroid cell types. (D) Incoming communication patterns of target cells in the human RPE and main choroid cell types.
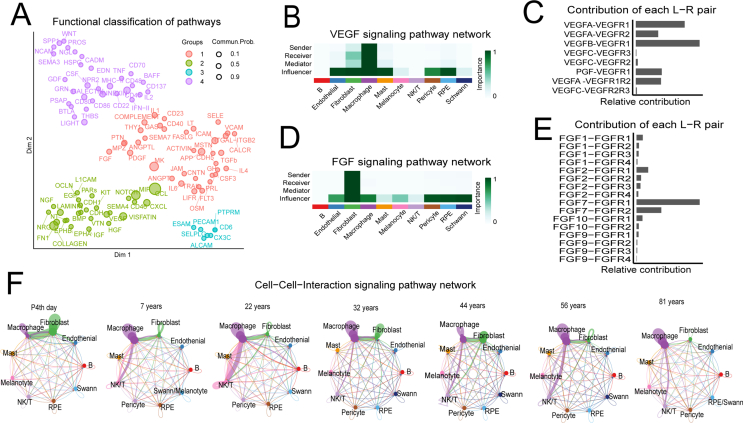
Further, we quantified the similarity between all significant signaling pathways and then grouped them based on their functional communication network similarity. This resulted in four groups of pathways (defined by colors) (Fig. S1A), which are mainly involved in immune responses. The grouping based on structural similarity revealed the general way in which sender and receiver cells utilize a given signaling pathway. Cell–cell interaction revealed 412 significant ligand–receptor pairs, which provides rich information regarding the molecular events in the human RPE and choroid (Table S4). For example, the VEGF and FGF networks in cell types, ligands, and receptors (L-R pair) are shown in Figure S1. We predicted that for VEGF signals, the sender, receiver, and mediator would mainly be macrophage cells; they would influence endothelial, fibroblast, macrophage, mast, pericyte, and RPE cells (Fig. S1B). VEGF signaling mainly occurs through the VEGFB and VEGFA pathways (Fig. S1C). We also predicted that fibroblasts send FGF signals, receive, and mediate them, influencing almost all cell types in the choroid except B and NK/T to varying degrees (Fig. S1D), mainly through the FGF1, FGF2, FGF7, FGF9, and FGF10 pathways (Fig. S1E). Cell-to-cell communication was similar at different ages, but there were slight differences (Fig. S1F). Collectively, we identified key features of the intercellular communications of the RPE and choroid and also predicted putative functions for poorly understood signaling pathways between them.
Transcriptional regulation in the human RPE and choroid
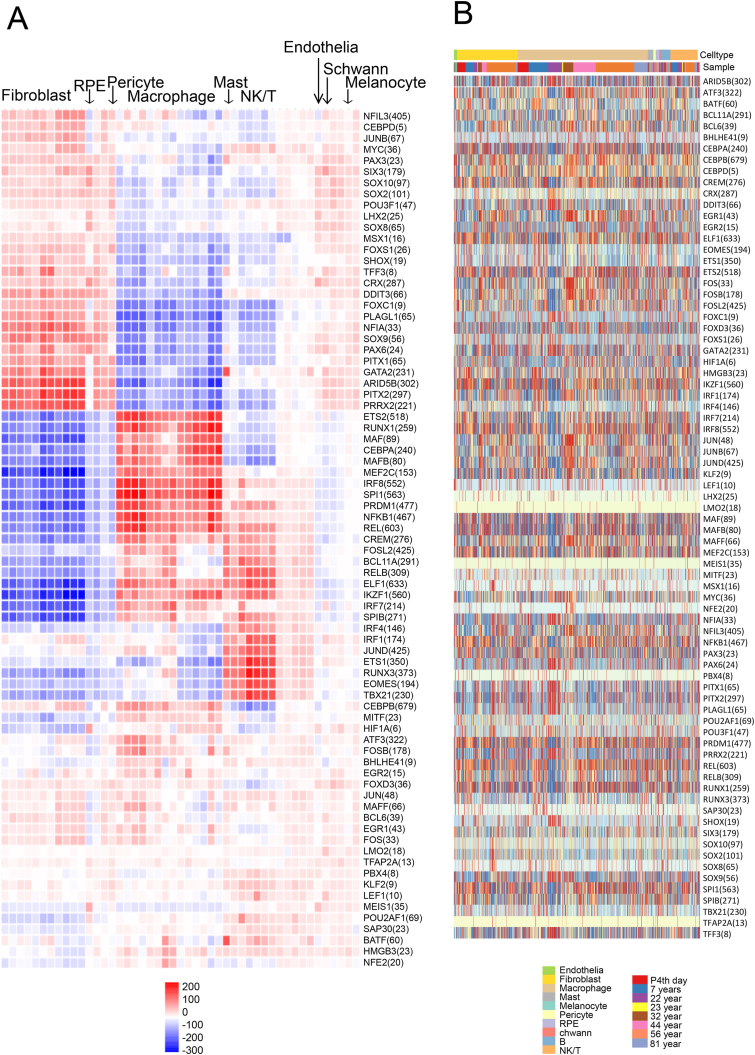
A regulon is a collection of genes or operons regulated by the same regulatory protein.30 SCENIC is a computational method for simultaneous gene regulatory network reconstruction and cell-state identification from single-cell RNA-seq data.24,31 Based on SCENIC regulatory network analysis, a total of 77 regulons (Fig. 6 and Table S5) were detected in combined RPE and choroid cells of different ages. The most enriched target genes (679) were detected in regulon CEBPB, followed by ELF1 (633), REL (603), SPI1 (563), and IKZF1 (560). Overall, fibroblasts and macrophages exhibited almost completely different regulation modes. Regulons NFIL3 (405), CEBPD (5), JUNB (67), MYC (36), etc. were highly expressed in fibroblasts but less expressed in macrophages. These genes may be important for modulating the growth and homeostasis of fibroblasts. Some of these 77 regulons showed cell-type specificity. For example, ETS2 (518) and RUNX1 (259) showed specific higher expression in macrophages compared to other cell types. Mast and NK/T cells showed similar expression patterns. MEIS1 (35) was highly expressed in RPE cells, while GATA2 (231) and BATF (60) were highly expressed in mast cells. VECs, Schwann, and melanocytes showed similar expression patterns (Fig. 6A). At different age stages, the expression patterns of each regulon were dynamic (Fig. 6B), suggesting that gene expression is regulated by age. Overall, the most variable regulons were associated with immunity and inflammation. For example, at 81 years of age, the IRF7 (214), IRF8 (552), NFKB1 (467), and REL (603) regulons were highly enriched in the choroid.
Figure 6.
Transcriptional regulation in the human RPE and choroid. (A) SCENIC master regulator results for the human RPE and choroid among 10 human RPE and choroid cell types. (B) The expression patterns of these genes across the 10 main cell types and different ages. The numbers in brackets indicate the number of target genes of each regulon.
Somatic nucleotide variants (SNVs) and copy number variants (CNVs) in single human retina cells
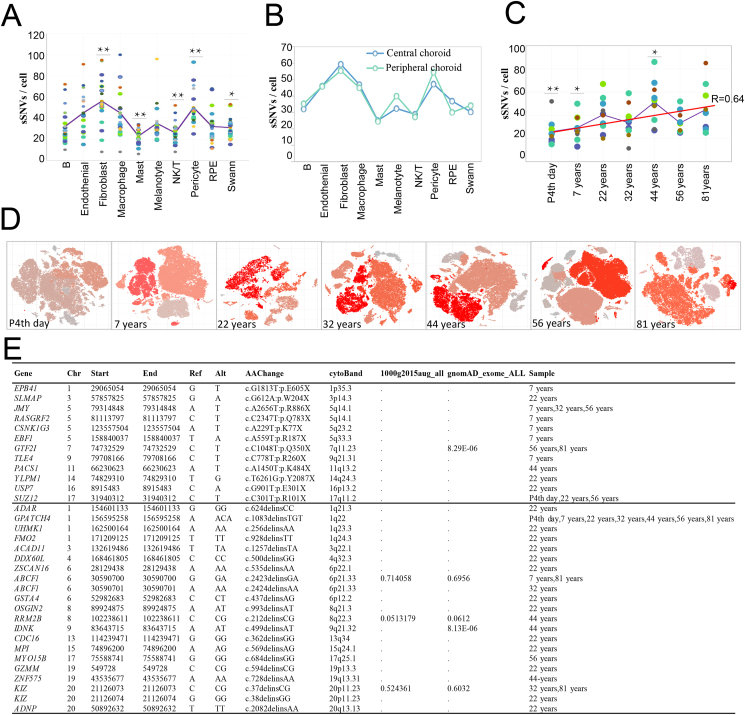
It has long been hypothesized that aging is associated with an accumulation of somatic mutations in somatic cells. A previous study found that SNVs increased approximately linearly with age in human brain neurons based on single-cell whole-genome sequencing.32 The identification of SNVs from scRNA-seq could also be used to find somatic variations at the single-cell level and with a large number of cells.33,34 Human bulk retina DNA is characterized by an accumulation of mitochondrial DNA deletions during aging,35 but it is not known whether somatic mutations or CNVs occur or accumulate with age in the human RPE and choroid. Here, we performed an analysis to delineate SNVs and CNVs in our scRNA-seq data on the human RPE and choroid to explore the variants in the targeted coding genomic regions (Fig. 7).
Figure 7.
Detection of sSNVs across human RPE and choroid regions and ages using scRNA-seq data. (A) sSNV counts are plotted for each of the 10 main cell types. More SNVs were detected in fibroblasts (P = 4.59 × 10−4, compared with the average). (B) sSNV counts are plotted in each of the two regions. They showed similar expression patterns. (C) sSNV counts are plotted at each age. The lowest number of somatic SNVs were found on the P4th day (P = 7.9 × 10−3). Overall, sSNVs increased approximately linearly with age in human RPE and choroid cells (R = 0.64). (D) t-SNE visualization of sSNVs in age colored by the degree of malignancy score. (E) Functional summary of the SNVs detected by age. Most SNVs were nonsynonymous and predicted to be benign.
We found that SNVs can be detected in numbers ranging from 11 to 68 (average 32 SNVs per cell) in a single human RPE cell. In the choroid, SNVs can be detected in numbers ranging from 6 to 100 (average 38 SNVs per cell) in a single choroid cell, suggesting a considerable amount of diversity among cells. More variants were detected in fibroblasts compared to the average cells (average 56 SNVs per cell, P = 4.59 × 10−4, compared to the whole choroid, - 38 SNVs per cell, the same below) and pericytes (50 SNVs per cell, P = 5.73 × 10−3) than other choroid cell types. On the contrary, mast (23 SNVs per cell, P = 1.11 × 10−8), NK/T (26 SNVs per cell, P = 7.29 × 10−5), and Schwann cells (31 SNVs per cell, P = 0.014) had fewer SNVs than other cell types (Fig. 7A).
Of the two RPE regions, more SNVs were detected in the central RPE (36 SNVs per cell) than in the peripheral RPE (29 SNVs per cell) (Fig. 7B), which might be due to the strong energy metabolism in the retina macular region resulting from a local high temperature-induced mutation in the RPE behind the macular region. In the choroid cell types, similar SNVs were detected between peripheral (37.4 SNVs per cell) and central choroid cells (38.9 SNVs per cell) (Fig. 7B). No statistical difference was detected between these two regions in the choroid. Overall, sSNVs increased approximately linearly with age in human RPE and choroid cells (R = 0.64) (Fig. 7C). The lowest number of somatic SNVs was detected on the P4th day (26 SNVs per cell, P = 7.9 × 10−3) (Fig. 7C). Further, a greater number of SNVs were detected at 44 years of age (51 SNVs per cell, P = 0.023) compared to the average (Fig. 7C). The TSNE plots of the SNVs of all the samples are presented in Figure 7D.
Most SNVs are expected to be functionally benign (Table S6). Indeed, very few SNVs are predicted to be pathogenic, suggesting that somatic random variants in the RPE and choroid are mostly harmless. The genes with pathogenic mutations are listed in Figure 7E (upper panel), including EPB41 (p.E605X), SLMAP (p.W204X), and JMY (p.R886X). All of these genes had stop-gain mutations in at least one sample. Other genes with frameshift substitution were also detected: ADAR (c.624delinsCC), GPATCH4 (c.1083delinsTGT, in almost all samples), and UHMK1 (c.256delinsAA) (Fig. 7E, lower panel). Among these genes, USP7, RRM2B, and JMY exhibited enriched transcriptional regulation by TP53 based on metascape analysis.21 EPB41, RASGRF2, and SLMAP were involved in the regulation of ion transport, while EPB41, USP7, and UHMK1 were involved in the regulation of cellular localization. The mechanisms that drive random RPE and choroidal SNVs and their functional impacts require further investigation. CNVs within the human RPE and choroidal from the P4th day to 81 years of age display similar cell types and age diversities to SNVs (Fig. S2). Across all cell types, the RPE showed the most loss of CNVs in chromosome six in individuals of all ages. Conversely, macrophages showed the greatest increase in CNVs in chromosome six in individuals of all ages. The impacts of these CNVs also need further investigation.
ELN AAV intravitreous injection rescues RPE and choroid degeneration during aging
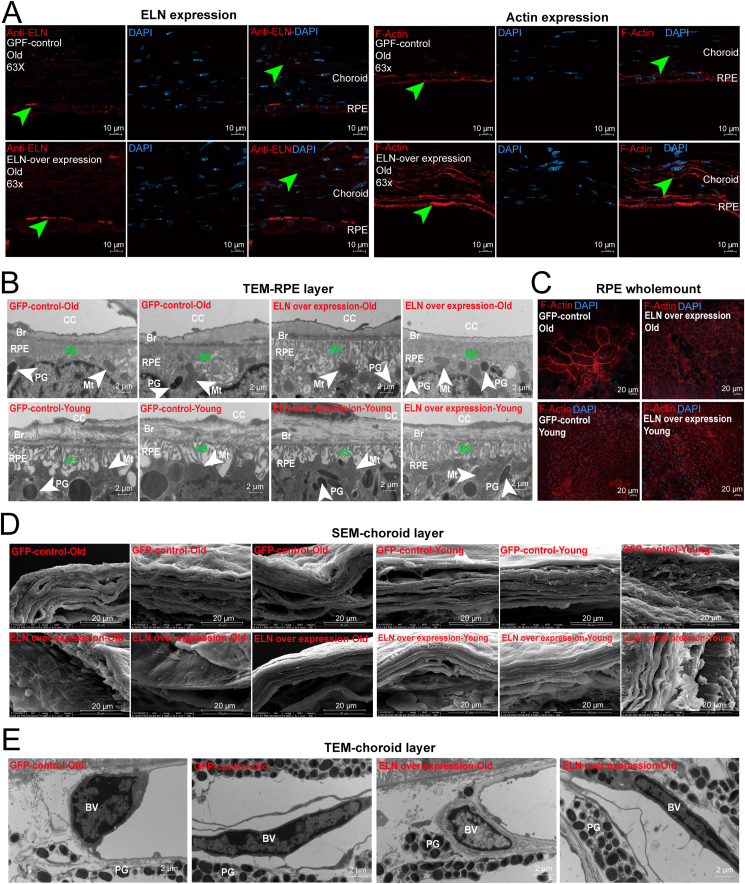
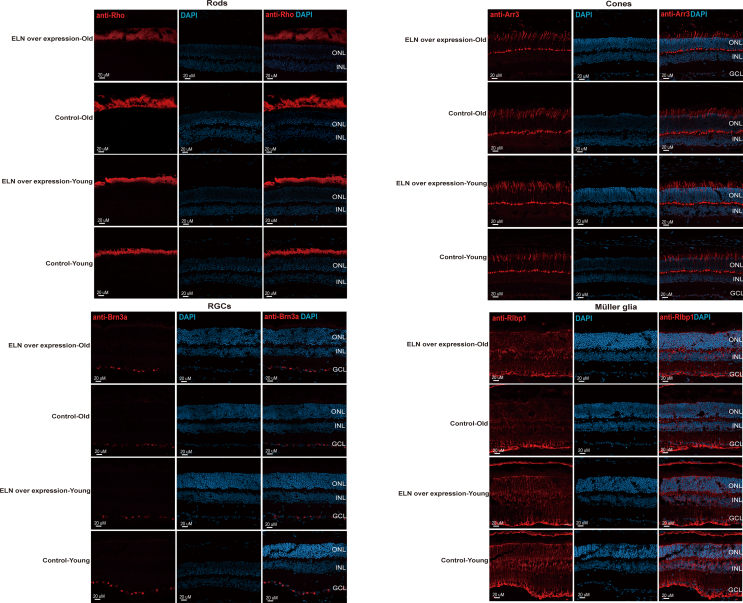
Previous studies have shown that the expression of secretory proteins can be detected in the choroid by vitreous injection of AAV using broad-spectrum promoters, such as SV40.36,37 To investigate whether enhanced expression of ELN is sufficient to protect structural elastic fibrin from aging-related degeneration, we performed intravitreal injection of AAV-mediated gene transfer of human ELN in both young (two-month-old C57BL/6 mice, young group) and old mice (18-month-old C57BL/6 mice, old group) and observed their organization structure three months after the AAV injection. We first checked their retina structure by Rho (labeled rods), Arr3 (labeled cones), Brn3a (labeled RGCs), and Rlbp1 (labeled Müller glia) immunofluorescence staining. No obvious abnormalities of these cell types were observed (Fig. S3). The results suggested that intravitreal injection of AAV-mediated gene transfer of human ELN may be safe for the mouse retina. We then examined the expression of ELN by antibody staining in the choroid and RPE and found the expression of ELN to be enhanced in RPE in the ELN overexpression group compared to the control group (Fig. 8A, the first panel of ELN, green arrowhead indicated). The expression of ELN in the choroidal layer was smoother compared to the normal group (Fig. 8A, the third panel of ELN, green arrowhead indicated). We further investigated the expression of actin, which is necessary for the formation of the cytoskeleton and always exists in the ECM, to investigate the tissue structures of the RPE and choroid. Actin expression in both the RPE and choroid layers was enhanced in the ELN overexpression mice compared to the control mice (Fig. 8A, actin panels, green arrowheads indicated). These results suggested that overexpression of human ELN by AAV-driving expression through vitreous injection in old mice could improve degenerating RPE and choroid structures in aging mice.
Figure 8.
AAV driving human ELN heterologous expression in vitreous can rescue the degenerated structure of the RPE and choroid in old mice. (A) Immunofluorescence staining of ELN and F-actin with GFP control AAV injection (the first line) or ELN overexpression AAV injection (the second line) in old mice (18 months) (RPE and choroid layers were observed with an oil lens, 63×). (B) TEM analysis of the RPE structure in old mice (18 months) and young mice (2 months) with control or ELN overexpression. Br, Bruch's membrane; PG, a pigment granule; Mt, mitochondrial; BSL, the basal surface of the RPE; CC, choroid. (C) Wholemount of RPE structure in old mice (18 months) and young mice (2 months) with control or ELN overexpression. (D) SEM analysis of the choroid structure in old mice (18 months) and young mice (2 months) with control or ELN overexpression. (E) TEM analysis of the choroid structure in old mice (18 months) with control or ELN overexpression. BV, blood vessel; PG, a pigment granule.
We further observed the effect of exogenous ELN expression on the fine structure of the RPE and choroidal layers using a transmission electron microscope (TEM). The basal labyrinth has a complex but ordered structural elements that break down with age and in choroideremia.38 In the RPE layer, we detected that the basal surface (BS) of the RPE layer was denser in the ELN overexpression group than in the control group in both the old (Fig. 8B, the first line) and young (Fig. 8B, the second line) groups. The wholemount RPE results also showed that ELN overexpression maintained the good RPE structure (Fig. 8C). These results suggested that ELN overexpression can cause the BS of RPE cells to reach a denser state. The choroidal layer was also examined using a scanning electron microscope (SEM), which showed that the fibrous structure of overexpressed ELN in old and young mice was smoother than in the controls, implying that ECM tissue has a better structure with enhanced ELN expression (Fig. 8D). TEM observation of the choroid layer revealed no obvious abnormalities of blood vessels (BVs) and pigment granules (PG) after ELN overexpression (Fig. 8E, the second line). Taken together, these results suggested that heterologous expression of human ELN into old mice RPE and choroids can maintain their structures in a tight and smooth state, which may delay the aging-related degeneration of the RPE and choroid, thus providing a new anti-aging target for the RPE and choroid.
Discussion
Due to the difficulty in obtaining human ocular tissues (e.g., RPE and choroid tissues), it has been challenging to determine the molecular mechanisms underlying the human vision system. Here, we sequenced the RNA of more than 0.3 million single cells from the human RPE and choroid across two regions and seven ages. Our results revealed the region-preferred or specific genes as well as the cellular compositions behind the macular region and the periphery RPE and choroid. These differences may underlie the supply disparity between the RPE and choroid to the retina. Our data revealed that while RPE and choroid structural cell components were decreased in the aged choroid (e.g., RPE and fibroblasts), the proportions of immune-related cells, such as macrophages and mast cells, were increased.
Previous studies have compared regional gene expression within the RPE and choroid at the bulk RNA level.39,40 However, due to the heterogeneity of different cell types, bulk-seq results might not describe the changes among cell types. Here we present the first single-cell RNA-Seq dataset for separately investigating the cellular composition and molecular mechanisms behind the macula and peripheral regions of the human RPE and choroid. We believe the datasets generated in this study are valuable for the research community and will be especially useful for the study of retinal inflammation and vascular diseases. Taken together, these results enhance our understanding of normal RPE and choroidal physiology.
RPE senescence is a hallmark of aging and degenerative retinal disease development.41 With aging, some RPE cells died, and the number of cells decreased. The elasticity of the human Bruch's membrane–choroid complex decreased linearly with age after the age of 21, with an approximate reduction of 1% per year.42 We also observed the expression of ELN deceased in the RPE with age (R = −0.41). The whole choroid becomes thinner during senescence. With aging, the numbers of fibroblasts, VECs, and pericytes continue to decrease. Given that fibroblasts contribute to the connective tissue stabilization of every organ's parenchyma, cellular senescence of fibroblasts may play a more general role in organ aging, organismal aging, and likely in many aging-related diseases.43 The proportion of macrophages, mast, VECs, NK/T, and B cells in choroids increased slightly with age. In macrophages, functions involved in DNA repair, the adaptive immune system, interferon–alpha/beta signaling, and the regulation of cytokinesis increased with age, suggesting that the expression of pro-inflammatory factors in macrophages is enhanced during aging. Activated macrophages, particularly of the M (IFNg and LPS) phenotype from nvAMD patients, were proangiogenic in vivo and in vitro.44 We found that pro-inflammatory factors increased in aged macrophage, mast, endothelial, and NK/T cells. By reanalyzing the correlation of gene expression with age using a previously published dataset of human bulk RNA sequencing of the choroid and RPE,18 we found that the change trends in many correlation pathways are consistent with our results (Fig. S4). For example, the expression of cytokine signaling in the immune system was positively correlated with age, and the expression of the NABA core matrisome was negatively correlated with age (Fig. S4). We further analyzed the intercellular communication networks generated from scRNA-seq data on the human RPE and choroid to investigate the major signaling inputs and outputs among cells. We also showed how those cells and signals coordinate in immune or other supply functions and pattern recognition approaches. Here we presented co-expression regulons of the human RPE and choroid during aging, assessing the activity of these discovered regulons in each of the 10 main human RPE and choroid cells.24,31 These regulons may provide valuable clues for reprogramming the human RPE to treat retinal diseases. Our analysis revealed the coding of random variation spectrums of the human RPE and choroid. Single-cell SNVs largely accumulated with age (R = 0.64). Fibroblasts had a higher frequency of accumulated variations compared to other cell types. Most random SNVs were benign.
Previous studies have only focused on a few transcription factors in developmental retinas,14, 15, 16,45,46 while the comprehensive regulatory network underlying human RPE and choroid aging has rarely been studied. The regulons revealed here will have broad implications for RPE and choroid reprogramming. For example, the use of regulon HDAC2 discovered in the P4th baby's RPE cells, combined with highly expressed TFs such as SOX9, which are specifically expressed in the RPE, may promote reprogramming of the aged RPE to cure dry AMD. Using the TFs NFIL3, CEBPD, MYC, PAX3, or SIX3 discovered in the fibroblasts may promote fibroblast reprogramming in the aged choroid, improving the constituent structure of the aged choroid, which tends to thin with age. The improved constituent structure may then strengthen the circulation system of the RPE and choroid, helping to avoid diseases such as AMD.
The natural aging process and various pathologies correlate with alterations in the composition, structural, and mechanical integrity of the connective tissue. Elastin (encoded by ELN) is an important protein of the extracellular matrix in higher vertebrates, which confers elasticity and resilience to various tissues and organs.47 Elastin is not only a structural protein, influencing the architecture and biomechanical properties of the ECM, but also plays a vital role in various physiological processes.47 Elastin accumulates damage during aging over the lifespan of an organism due to its low turnover and the effects of enzymatic degradation,48 oxidative damage,49 and the formation of advanced glycation end products.50 The aging of elastin involves fragmentation and thinning of elastin structures and may result in impaired elastic fiber function, reduced tissue elasticity, chronic inflammation, or eventual loss of function of tissues and organs, considerably increasing morbidity and mortality.51 In the human choroid, elastin is mainly secreted by fibroblasts, which constitute the principal component of the connective tissue of the choroid. The aging of elastin fibers involves enzymatic degradation, oxidative damage, glycation, calcification, aspartic acid racemization, binding of lipids and lipid peroxidation products, carbamylation, and mechanical fatigue. These processes can trigger impairment or loss of elastic protein function.52 The introduction of the human ELN gene to mice eyes significantly improved the microenvironment of the RPE and choroid in old mice. The rescue of ELN in aged RPE and choroid provides the physiological basis for protecting the RPE and choroid from aging degeneration.
This study also has some limitations. Because human choroidal samples were difficult to obtain and the number of samples used in this paper was limited, some quantitative analyses may not have had sufficient statistical analysis power. In addition, due to a large number of cells and information involved in this study, it was not possible to carry out detailed experimental verification of each observed result. It is hoped that in future research work, more samples and in-depth experimental verification will be carried out one by one to reveal the dynamic molecular changes of the human choroid with age.
In summary, this study provides a comprehensive single-cell transcriptomic atlas of the human RPE and choroid from two regions and seven ages, broadening our understanding of human RPE and choroid gene signatures. Importantly, it offers insights into the cell compositions and molecular mechanisms underlying the RPE and choroid region and aging in humans. Moreover, we identified ELN as a candidate for improvement in the degeneration of choroidal and RPE structures to combat aging. The single-cell molecular architecture from baby to aged RPE and choroid presented here can be a resource for determining the distinct gene-expression signatures and lays a foundation for RPE and choroid functional gene research. Moreover, the mechanistic insights arising from this study could establish new avenues for developing targeted interventions for anti-aging or ocular disease therapy.
Author contributions
Z.Y. and L.H. designed the experiments; L.H., C.Q., B.W., R.C., G.H., B.G., Z.L., H. Y., M. Y., Y.S., C. G., W.C., and Z.Y. recruited participants. L.H. performed the cell isolation experiments and analyzed the data; L.Y. and R.L. performed the ELN experiments. S.Z., S.L., and M.Y. performed the RNAscope experiments. L.H. and Z.Y. wrote the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81790643, 81970839, 82271105, and 82121003), the Sichuan Science and Technology Program (China) (No. 2021YFS0033, 2021YFS0404, 2021YFS0369, and 2020ZYD037), and the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (China) (No. 2019-I2M-5-032).
Data and material availability
All sequencing data are available on reasonable request. Please contact the corresponding author Zhenglin Yang for the raw data (yangzhenglin@cashq.sc.cn).
Conflict of interests
The authors have no competing interests to declare.
Acknowledgements
We gratefully acknowledge the people who donated their eye organs. We thank Genergy Bio for support with the scRNA sequencing and data analysis.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.11.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
References
- 1.Panda-Jonas S., Jonas J.B., Jakobczyk-Zmija M. Retinal pigment epithelial cell count, distribution, and correlations in normal human eyes. Am J Ophthalmol. 1996;121(2):181–189. doi: 10.1016/s0002-9394(14)70583-5. [DOI] [PubMed] [Google Scholar]
- 2.Lange C.A.K., Bainbridge J.W.B. Oxygen sensing in retinal health and disease. Ophthalmologica. 2012;227(3):115–131. doi: 10.1159/000331418. [DOI] [PubMed] [Google Scholar]
- 3.Nork T.M., Kim C.B.Y., Shanmuganayagam D., et al. Measurement of regional choroidal blood flow in rabbits and monkeys using fluorescent microspheres. Arch Ophthalmol. 2006;124(6):860–868. doi: 10.1001/archopht.124.6.860. [DOI] [PubMed] [Google Scholar]
- 4.Nickla D.L., Wallman J. The multifunctional choroid. Prog Retin Eye Res. 2010;29(2):144–168. doi: 10.1016/j.preteyeres.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lipecz A., Miller L., Kovacs I., et al. Microvascular contributions to age-related macular degeneration (AMD): from mechanisms of choriocapillaris aging to novel interventions. Geroscience. 2019;41(6):813–845. doi: 10.1007/s11357-019-00138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voigt A.P., Mulfaul K., Mullin N.K., et al. Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration. Proc Natl Acad Sci U S A. 2019;116(48):24100–24107. doi: 10.1073/pnas.1914143116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng Y., Qiao L., Du M., et al. Age-related macular degeneration: epidemiology, genetics, pathophysiology, diagnosis, and targeted therapy. Genes Dis. 2022;9(1):62–79. doi: 10.1016/j.gendis.2021.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanus J., Anderson C., Wang S. RPE necroptosis in response to oxidative stress and in AMD. Ageing Res Rev. 2015;24(Pt B):286–298. doi: 10.1016/j.arr.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datta S., Cano M., Ebrahimi K., et al. The impact of oxidative stress and inflammation on RPE degeneration in non-neovascular AMD. Prog Retin Eye Res. 2017;60:201–218. doi: 10.1016/j.preteyeres.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarrett S.G., Boulton M.E. Consequences of oxidative stress in age-related macular degeneration. Mol Aspect Med. 2012;33(4):399–417. doi: 10.1016/j.mam.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang L., Zhang H., Cheng C.Y., et al. A missense variant in FGD6 confers increased risk of polypoidal choroidal vasculopathy. Nat Genet. 2016;48(6):640–647. doi: 10.1038/ng.3546. [DOI] [PubMed] [Google Scholar]
- 12.Rozing M.P., Durhuus J.A., Krogh Nielsen M., et al. Age-related macular degeneration: a two-level model hypothesis. Prog Retin Eye Res. 2020;76 doi: 10.1016/j.preteyeres.2019.100825. [DOI] [PubMed] [Google Scholar]
- 13.Boulton M., Różanowska M., Wess T. Ageing of the retinal pigment epithelium: implications for transplantation. Graefes Arch Clin Exp Ophthalmol. 2004;242(1):76–84. doi: 10.1007/s00417-003-0812-8. [DOI] [PubMed] [Google Scholar]
- 14.Clark B.S., Stein-O’Brien G.L., Shiau F., et al. Single-cell RNA-seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron. 2019;102(6):1111–1126.e5. doi: 10.1016/j.neuron.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Y., Shiau F., Yi W., et al. Single-cell analysis of human retina identifies evolutionarily conserved and species-specific mechanisms controlling development. Dev Cell. 2020;53(4):473–491.e9. doi: 10.1016/j.devcel.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cowan C.S., Renner M., De Gennaro M., et al. Cell types of the human retina and its organoids at single-cell resolution. Cell. 2020;182(6):1623–1640.e34. doi: 10.1016/j.cell.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orozco L.D., Chen H.H., Cox C., et al. Integration of eQTL and a single-cell atlas in the human eye identifies causal genes for age-related macular degeneration. Cell Rep. 2020;30(4):1246–1259.e6. doi: 10.1016/j.celrep.2019.12.082. [DOI] [PubMed] [Google Scholar]
- 18.Voigt A.P., Whitmore S.S., Mulfaul K., et al. Bulk and single-cell gene expression analyses reveal aging human choriocapillaris has pro-inflammatory phenotype. Microvasc Res. 2020;131 doi: 10.1016/j.mvr.2020.104031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehmann G.L., Hanke-Gogokhia C., Hu Y., et al. Single-cell profiling reveals an endothelium-mediated immunomodulatory pathway in the eye choroid. J Exp Med. 2020;217(6) doi: 10.1084/jem.20190730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Y., Wang X., Hu B., et al. Dissecting the transcriptome landscape of the human fetal neural retina and retinal pigment epithelium by single-cell RNA-seq analysis. PLoS Biol. 2019;17(7) doi: 10.1371/journal.pbio.3000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y., Zhou B., Pache L., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D., Franceschini A., Kuhn M., et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database issue):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin S., Guerrero-Juarez C.F., Zhang L., et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aibar S., González-Blas C.B., Moerman T., et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85(3):845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 27.Fuhrmann S., Zou C., Levine E.M. Retinal pigment epithelium development, plasticity, and tissue homeostasis. Exp Eye Res. 2014;123:141–150. doi: 10.1016/j.exer.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plati J., Bucur O., Khosravi-Far R. Apoptotic cell signaling in cancer progression and therapy. Integr Biol. 2011;3(4):279–296. doi: 10.1039/c0ib00144a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramrattan R.S., van der Schaft T.L., Mooy C.M., et al. Morphometric analysis of Bruch's membrane, the choriocapillaris, and the choroid in aging. Invest Ophthalmol Vis Sci. 1994;35(6):2857–2864. [PubMed] [Google Scholar]
- 30.Novichkov P.S., Rodionov D.A., Stavrovskaya E.D., et al. RegPredict: an integrated system for regulon inference in prokaryotes by comparative genomics approach. Nucleic Acids Res. 2010;38(Web Server issue):W299–W307. doi: 10.1093/nar/gkq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van de Sande B., Flerin C., Davie K., et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc. 2020;15(7):2247–2276. doi: 10.1038/s41596-020-0336-2. [DOI] [PubMed] [Google Scholar]
- 32.Lodato M.A., Rodin R.E., Bohrson C.L., et al. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science. 2018;359(6375):555–559. doi: 10.1126/science.aao4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu F., Zhang Y., Zhang L., et al. Systematic comparative analysis of single-nucleotide variant detection methods from single-cell RNA sequencing data. Genome Biol. 2019;20(1):242. doi: 10.1186/s13059-019-1863-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tirosh I., Venteicher A.S., Hebert C., et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539(7628):309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barreau E., Brossas J.Y., Courtois Y., et al. Accumulation of mitochondrial DNA deletions in human retina during aging. Invest Ophthalmol Vis Sci. 1996;37(2):384–391. [PubMed] [Google Scholar]
- 36.Lee S.H.S., Kim H.J., Shin O.K., et al. Intravitreal injection of AAV expressing soluble VEGF receptor-1 variant induces anti-VEGF activity and suppresses choroidal neovascularization. Invest Ophthalmol Vis Sci. 2018;59(13):5398–5407. doi: 10.1167/iovs.18-24926. [DOI] [PubMed] [Google Scholar]
- 37.Li Z., Chen W., Zhang H., et al. The aquaporin-1 depletion downregulates the sclera biomechanical strength. Curr Eye Res. 2020;45(10):1240–1244. doi: 10.1080/02713683.2020.1730404. [DOI] [PubMed] [Google Scholar]
- 38.Hayes M.J., Burgoyne T., Wavre-Shapton S.T., et al. Remodeling of the basal labyrinth of retinal pigment epithelial cells with osmotic challenge, age, and disease. Invest Ophthalmol Vis Sci. 2019;60(7):2515–2524. doi: 10.1167/iovs.19-26784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitmore S.S., Wagner A.H., DeLuca A.P., et al. Transcriptomic analysis across nasal, temporal, and macular regions of human neural retina and RPE/choroid by RNA-Seq. Exp Eye Res. 2014;129:93–106. doi: 10.1016/j.exer.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian L., Kazmierkiewicz K.L., Bowman A.S., et al. Transcriptome of the human retina, retinal pigmented epithelium and choroid. Genomics. 2015;105(5–6):253–264. doi: 10.1016/j.ygeno.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jadeja R.N., Powell F.L., Jones M.A., et al. Loss of NAMPT in aging retinal pigment epithelium reduces NAD+ availability and promotes cellular senescence. Aging. 2018;10(6):1306–1323. doi: 10.18632/aging.101469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ugarte M., Hussain A.A., Marshall J. An experimental study of the elastic properties of the human Bruch's membrane-choroid complex: relevance to ageing. Br J Ophthalmol. 2006;90(5):621–626. doi: 10.1136/bjo.2005.086579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wlaschek M., Maity P., Makrantonaki E., et al. Connective tissue and fibroblast senescence in skin aging. J Invest Dermatol. 2021;141(4):985–992. doi: 10.1016/j.jid.2020.11.010. [DOI] [PubMed] [Google Scholar]
- 44.Hagbi-Levi S., Grunin M., Jaouni T., et al. Proangiogenic characteristics of activated macrophages from patients with age-related macular degeneration. Neurobiol Aging. 2017;51:71–82. doi: 10.1016/j.neurobiolaging.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 45.Peng Y.R., Shekhar K., Yan W., et al. Molecular classification and comparative taxonomics of foveal and peripheral cells in primate retina. Cell. 2019;176(5) doi: 10.1016/j.cell.2019.01.004. 1222–1237.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang L., Li R., Ye L., et al. Deep Sc-RNA sequencing decoding the molecular dynamic architecture of the human retina. Sci China Life Sci. 2023;66(3):496–515. doi: 10.1007/s11427-021-2163-1. [DOI] [PubMed] [Google Scholar]
- 47.Heinz A. Elastases and elastokines: elastin degradation and its significance in health and disease. Crit Rev Biochem Mol Biol. 2020;55(3):252–273. doi: 10.1080/10409238.2020.1768208. [DOI] [PubMed] [Google Scholar]
- 48.Antonicelli F., Bellon G., Debelle L., et al. Elastin-elastases and inflamm-aging. Curr Top Dev Biol. 2007;79:99–155. doi: 10.1016/S0070-2153(06)79005-6. [DOI] [PubMed] [Google Scholar]
- 49.Watanabe M., Sawai T., Nagura H., et al. Age-related alteration of cross-linking amino acids of elastin in human aorta. Tohoku J Exp Med. 1996;180(2):115–130. doi: 10.1620/tjem.180.115. [DOI] [PubMed] [Google Scholar]
- 50.Paul R.G., Bailey A.J. Glycation of collagen: the basis of its central role in the late complications of ageing and diabetes. Int J Biochem Cell Biol. 1996;28(12):1297–1310. doi: 10.1016/s1357-2725(96)00079-9. [DOI] [PubMed] [Google Scholar]
- 51.Green E.M., Mansfield J.C., Bell J.S., et al. The structure and micromechanics of elastic tissue. Interface Focus. 2014;4(2) doi: 10.1098/rsfs.2013.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heinz A. Elastic fibers during aging and disease. Ageing Res Rev. 2021;66 doi: 10.1016/j.arr.2021.101255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.